Abstract
Ran is a small signaling GTPase that is involved in nucleocytoplasmic transport. Two additional functions of animal Ran in the formation of spindle asters and the reassembly of the nuclear envelope in mitotic cells have been recently reported. In contrast to Ras or Rho, Ran is not associated with membranes. Instead, the spatial sequestering of its accessory proteins, the Ran GTPase-activating protein RanGAP and the nucleotide exchange factor RCC1, appears to define the local concentration of RanGTP vs. RanGDP involved in signaling. Mammalian RanGAP is bound to the nuclear pore by a mechanism involving the attachment of small ubiquitin-related modifier protein (SUMO) to its C terminus and the subsequent binding of the SUMOylated domain to the nucleoporin Nup358. Here we show that plant RanGAP utilizes a different mechanism for nuclear envelope association, involving a novel targeting domain that appears to be unique to plants. The N-terminal WPP domain is highly conserved among plant RanGAPs and the small, plant-specific nuclear envelope-associated protein MAF1, but not present in yeast or animal RanGAP. Confocal laser scanning microscopy of green fluorescent protein (GFP) fusion proteins showed that it is necessary for RanGAP targeting and sufficient to target the heterologous protein GFP to the plant nuclear rim. The highly conserved tryptophan and proline residues of the WPP motif are necessary for its function. The 110-aa WPP domain is the first nuclear-envelope targeting domain identified in plants. Its fundamental difference to its mammalian counterpart implies that different mechanisms have evolved in plants and animals to anchor RanGAP at the nuclear surface.
An emerging theme in signal transduction research is how the different pathways of signaling events are both separated and coordinated in a temporal and spatial manner in the living cell. It is becoming increasingly evident that discrete spatial positioning within the cell is a major aspect of this coordination. However, how this positioning is achieved for individual signaling molecules remains a fundamental question of molecular cell biology.
The small GTP-binding protein Ran is required for the trafficking of proteins and RNA in and out of the nucleus (1). It forms a complex with nuclear transport receptors and their cargoes and is involved in their directional passage through the nuclear pores. Like all small GTP-binding proteins, RanGTP has a very low intrinsic GTPase activity that requires stimulation by Ran GTPase activating protein (RanGAP) and its accessory factor RanBP1 (2). Replacement of GDP from RanGDP with GTP is accomplished by the Ran guanine nucleotide exchange factor RCC1, which closes the cycle. RanGTP and RanGDP have different roles in nuclear transport, and their respective abundance is regulated by a spatial separation of RanGAP and RanBP1 outside and RCC1 inside the nucleus (1). The nucleotide binding state of Ran thus serves as a marker for compartment identity. In contrast to other small signaling GTPases like Ras and Rho, Ran itself is not membrane bound. It is unique in that the sequestering of its accessory proteins provides the spatial information for its respective activities (3).
In animal cells, RanGAP is anchored to the outer basket of the nuclear pore by interaction with the nucleoporin Nup358, whereas RCC1 is sequestered in the nucleus through binding to chromatin (4). Vertebrate RanGAP contains a C-terminal Nup358-binding domain. Modification of this domain by the small ubiquitin-like protein SUMO causes a conformational change of RanGAP that allows Nup358 binding (5). The Nup358-binding domain is not present in Rna1p, the Saccharomyces cerevisiae and Schizosaccharomyces pombe RanGAP homolog (6), which appears to be localized in the cytoplasm (7, 8).
Besides the well-established role of Ran in animal and yeast nucleocytoplasmic transport, several exciting recent findings point toward a wider function of Ran in cellular signaling in animals (9, 10). RanGTP can induce microtubule self-organization and spindle assembly by releasing microtubule-assembly factors from inhibition by importin α and β (3, 11–13). At the end of mitosis, GTP hydrolysis by Ran and a high local concentration of RanGDP are required for the association of nuclear envelope material with the decondensing chromatin (14, 15).
Ran has been identified in plants and has been shown to complement the respective S. pombe mutant (16, 17). Plant Ran-binding proteins with high similarity to mammalian/yeast RanBP1 have been identified (18). No plant homolog for RCC1 is presently known, but several sequences for putative plant RanGAPs have been found. At present, nothing is known about the potential additional functions of Ran and its accessory proteins in plant spindle formation and nuclear envelope assembly.
We have shown previously that plant RanGAP sequences contain a unique N-terminal domain (WPP domain) not present in yeast or animal RanGAPs (19). The WPP domain shows strong similarity to the small nuclear envelope-associated protein MAF1, which also appears to be unique to plants (20). Here, we demonstrate that Arabidopsis RanGAP1 is localized at the nuclear envelope of plant interphase cells, similar to mammalian RanGAP and different from yeast Rna1p. We show that the WPP domain constitutes a targeting domain that is necessary and sufficient for anchoring AtRanGAP1 to the nuclear envelope and that the WPP motif is involved in its function. These findings indicate that the mechanism for subcellular anchoring of RanGAP differs fundamentally between plants and animals.
Materials and Methods
Sequence Comparison and Structural Modeling.
GenBank and the Arabidopsis genome database were accessed through the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) and The Arabidopsis Information Resource (http://www.arabidopsis.org/), respectively. Sequence similarity searches were performed by using blast (21). Multiple sequence alignments were performed with the dnastar protein alignment protocol (DNASTAR, Madison, WI) by using the clustal algorithm. Parameters for clustal alignments were as follows. Pairwise alignment was: Ktuple, 1; gap Penalty, 3; windows, 5; and diagonals saved, 5. Multiple alignment was: gap penalty, 10; and gap length penalty, 10. The PAM250 weight table was used.
Structural fitting data were acquired by accessing Swiss-Model through the interfaced swiss pdb viewer (version 3.7b2) software (22, 23). The ≈10,000 protein structures deposited in the Protein Data Bank (PDB) were searched through Swiss-Model for a template to model AtRanGAP1 and OsRanGAP with the full-length amino acid sequences of the two proteins. The structure files 1YRGA and 1YRGB, representing the crystal structures of the two identical subunits of the SpRna1p crystal dimer, were identified with both sequences. Sequence identity of the regions identified for threading was 28.3% for AtRanGAP1 and 27.7% for OsRanGAP. 1YRGB was used for modeling. Preliminary threading was performed with magic fit in swiss pdb viewer, and the models were subsequently optimized on the Swiss-Model server (22, 23). The quality of the received models was confirmed with the what_check verification routine (24). Of the 48 parameters checked by the program, all parameters with impact on modeling by homology scored acceptable. Importantly, the Ramachandran Z score (measure for backbone structure) and the χ-1/χ-2 Z score (measure for side-chain conformation) were both in the acceptable range (−2.57 for Ramachandran Z score and −0.541 for χ-1/χ-2 Z score).
Reverse Transcription (RT)-PCR Cloning of AtRanGAP1.
The AtRanGAP1 cDNA was isolated by RT-PCR with the primers RanGAP1-F (5′-ATG GAT CAT TCA GCG AAA ACC-3′) and RanGAP1-R (5′-TCA TTC CTC CCC TTG CTT GAT-3′). RNA was prepared from Arabidopsis ecotype Columbia leaf tissue by using the RNeasy Plant Mini Kit (Qiagen, Chatsworth, CA). RT-PCR was performed by using the ProSTAR HF Single Tube RT-PCR System from Stratagene (La Jolla, CA) with 180 ng of total RNA, 100 ng of each primer, and 45°C annealing temperature. The resulting RT-PCR product of ≈1.6 kb length was cloned into the pCR II-TOPO vector, by using the TOPO TA Cloning Kit from Invitrogen, creating pCRII-TOPO-AtRanGAP1. The cDNA insert was sequenced.
Cloning of Green Fluorescent Protein (GFP) Fusion Constructs of AtRanGAP1 and AtRanGAP1 Deletion Clones.
To create AtRanGAP1-GFP, AtRanGAP1ΔC-GFP, and AtRanGAP1ΔN-GFP, the respective AtRanGAP1 fragments were amplified by PCR by using pCR II-TOPO-AtRanGAP1 as template and the primers RanGAP5.1 (5′-GCC ATG GAT CAT TCA GCG AAA ACC-3′) and RanGAP3.1 (5′-GGC CAT GGA TTC CTC CCC TTG CTT GAT-3′) for AtRanGAP1; RanGAP5.2 (5′-GCC ATG GAA GAA TCC GAG GTT GAG-3′) and RanGAP3.1 for AtRanGAP1ΔN; and RanGAP5.1 and RanGAP3.3 (5′-ACC CAT GGC CTC AAC CTC GGA TTC-3′) for AtRanGAP1ΔC. The resulting PCR fragments were cloned into pCR II-TOPO and sequenced for confirmation before cloning them into the single NcoI site of pRTL2-mGFPS65T (25) by using the internal NcoI sites of the PCR primers.
Site-Directed Mutagenesis.
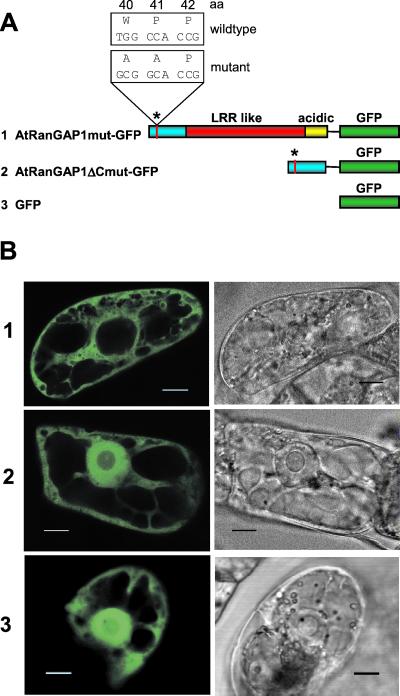
Point mutations were introduced by using the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene), the plasmids containing AtRanGAP1-GFP and AtRanGAPΔC-GFP as templates, and the mutagenic primers RanGAP-mu (5′-GTC AGT GAA GAT GGC GGC ACC GAG TAA GAG-3′) and RanGAP-mu-rev (5′-CTC TTA CTC GGT GCC GCC ATC TTC ACT GAC-3′) to replace the WPP motive with AAP (see Fig. 5A). The mutagenized plasmids were screened by using the destruction of a MscI site in the WPP motive, and the RanGAP inserts were sequenced for confirmation.
Figure 5.
Site-directed mutagenesis of the WPP motif. (A) Constructs in pRTL2-mGFPS65T used for transient transformation of BY-2 cells. The wild-type sequence of the WPP motif (see Fig. 1B) and the introduced base pair and amino acid changes are indicated above the bar diagram of AtRanGAP1mut-GFP. The asterisk indicates the position of the identical mutation in AtRanGAP1ΔCmut-GFP. Color code of the AtRanGAP1 domains is as in Fig. 1. (B) GFP fluorescence of BY-2 cells transiently transformed with the constructs shown in A. (Left) GFP fluorescence. (Right) Transmitted light images. (Bar equals 10 μm.)
Ballistic Transient Transformation of Tobacco BY-2 Cells.
Tobacco BY-2 cells were cultured in Murashige and Skoog medium (MS salts; Life Technologies, Gaithersburg, MD) supplemented with 0.3% (wt/vol) sucrose, 1.87 mM KH2PO4, 100 mg/liter myo-inositol, 1 mg/liter thiamine, and 0.2 mg/liter 2,4-D, with a final pH of 5.0 adjusted with KOH. Cells were maintained by shaking at 200 rpm in constant light at 24°C and subcultured weekly by 1:50 dilution. Transient transformation of BY-2 cells was performed essentially as described for NT-1 cells (20). For DNA staining, cells were incubated with SYTO 82 orange fluorescent nucleic acid stain (Molecular Probes) at a final concentration of 500 nM for 45 min at room temperature before microscopic imaging.
Confocal Laser Scanning Microscopy.
Digitized confocal images were acquired at 1024 × 1024 pixel resolution with a Nikon Plan Fluor ×40/0.75 air objective (1 pixel = 0.3 μm) or Nikon Plan Fluor ×100/1.30 oil objective (1 pixel = 0.12 μm) on a PCM 2000/Nikon Eclipse E600 confocal laser scanning microscope (Nikon Bioscience Confocal Systems, Melville, NY). For the detection of GFP in the green channel, the 488-nm excitation line of an Argon laser was used in combination with a 515/30-nm bandpass emission filter (EM515/30HQ). To detect SYTO 82 orange in the red channel, a 543-nm He-Ne laser was used for excitation in combination with a 565-nm long pass emission filter (E565LP). The simplepci software (Compix Imaging Systems, Cranberry Township, PA) was used for image capture. Photomultiplier tube black levels were kept at a default of 350 for fluorescence images and 600 for transmitted light images, and gain settings for the green and red channel were adjusted between 1,200 and 2,400, according to the intensity of the fluorescence signal. Images were captured in 1 × exposure mode with 4 × rolling average processing during capture for single layer images, and 2 × rolling average processing for stacks of images. Further image processing and assembly were done by using Adobe photoshop and Adobe imageready software (Adobe Systems, Mountain View, CA).
Results
Plant RanGAPs Have a Unique Domain Structure.
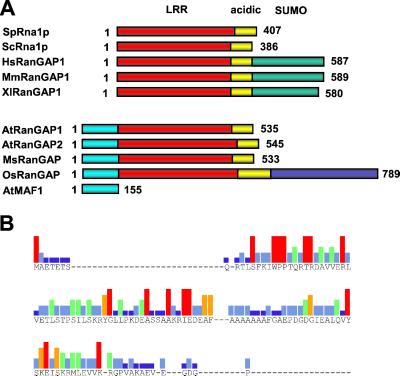
Sequences for four putative plant RanGAPs have been deposited in GenBank (Medicago sativa RanGAP (MsRanGAP), AF215731; Oryza sativa RanGAP (OsRanGAP), AAD27557; Arabidopsis RanGAP1 (AtRanGAP1), AF214559; and Arabidopsis RanGAP2 (AtRanGAP2), AF214560). Fig. 1A shows the domain structure of plant RanGAPs as derived from sequence alignments and structural fitting (see below) in comparison with yeast and vertebrate RanGAPs. The S. cerevisiae and S. pombe RanGAP orthologs ScRna1p and SpRna1p consist of a leucine-rich repeat domain, followed by an acidic domain, both involved in RanGAP function (6, 26). Vertebrate RanGAPs contain the additional C-terminal Nup358-binding domain not present in their yeast counterparts (6).
Figure 1.
Plant RanGAPs contain a unique N-terminal domain. (A) Schematic comparison of the domain structure of yeast, vertebrate, and plant RanGAPs. LRR, red; acidic domain, yellow; SUMO-attachment domain (SUMO), green; MAF1-like WPP domain, cyan; and unique C-terminal domain of OsRanGAP, blue. The S. pombe (SpRna1p), S. cerevisiae (ScRna1p), Homo sapiens (HsRanGAP1), Mus musculus (MsRanGAP1), and Xenopus laevis (XlRanGAP1) RanGAPs are grouped. The plant sequences are derived from A. thaliana (AtRanGAP1 and AtRanGAP2), M. sativa (MsRanGAP), and rice (OsRanGAP). Arabidopsis MAF1 (AtMAF1) is included for size comparison. The number of total residues for each protein is indicated. (B) Graphic depiction of the consensus sequence of the WPP domain derived from an alignment of the N-terminal 120 aa of AtRanGAP1, AtRanGAP2, MsRanGAP, OsRanGAP, and full-length MAF1 from Arabidopsis (AtMAF1), tomato (LeMAF1), soybean (GmMAF1), maize (ZmMAF1), and Canna edulis (CeMAF1; ref. 19). Amino acid residues of the consensus are indicated in one letter code. Bar colors represent consensus strength: red, nine of nine; orange, eight of nine; green, six or seven of nine; light blue, four or five of nine; dark blue, two or three of nine. Dashes, no consensus, including gaps in the alignment.
Plant RanGAPs do not contain this C-terminal domain, but contain the N-terminal MAF1-like WPP domain instead (ref. 20 and Fig. 1B). All plant sequences have a domain with similarity to the leucine-rich repeat (LRR) domain, followed by an acidic domain (Fig. 1A and Fig. 2A). The rice sequence contains an additional C-terminal domain with no homology to the C-terminal domains of vertebrate RanGAPs (Fig. 1A). However, as this sequence is derived from conceptual translation, the presence of this protein domain awaits confirmation.
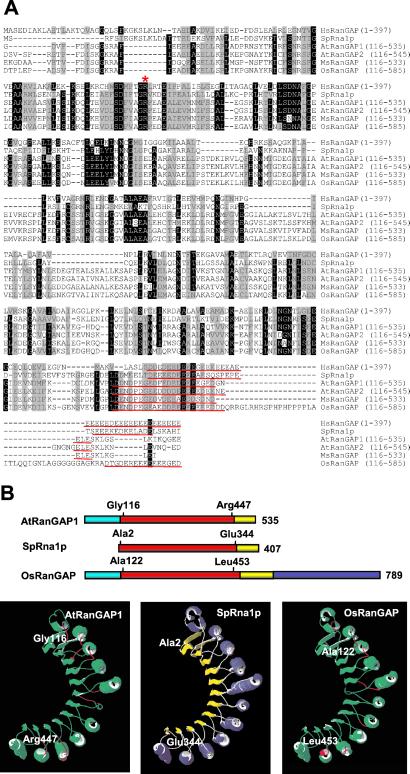
Figure 2.
Plant RanGAPs share a core domain with the animal and yeast proteins. (A) Amino acid-sequence alignment of human, yeast, and plant RanGAPs after trimming the kingdom-specific domains. Amino acid identities and similarities in at least five sequences are highlighted in black and gray, respectively. The arginine residue necessary for RanGAP activity is marked by a red asterisk; the acidic domains are underlined in red. (B) Molecular modeling of AtRanGAP1 and OsRanGAP onto the crystal structure of SpRna1p. Bar diagrams show the domains in the same color code as in Fig. 1A. The first and last amino acids represented in the structure are indicated above the bars. (B) Ribbon representation of the crystal structure of SpRna1p (Center), the predicted structure of amino acid 116 to 447 of AtRanGAP1 (Left), and of amino acid 122 to 453 of OsRanGAP (Right). In SpRna1p, α helices are labeled in blue and β sheets in yellow. In the two predicted structures, successful fit onto the SpRna1p structure is indicated in green, nonfitting areas in red.
Sequence similarity between vertebrate, yeast, and plant RanGAPs is low (≈30% identity between human RanGAP (HsRanGAP) and ScRna1p, ≈20% between plant RanGAPs and either HsRanGAP or ScRna1p). To prevent false alignments due to limited sequence similarity, we deleted the unique N-terminal and C-terminal domains and aligned the “core” LRR-like and acidic domains only (indicated in red and yellow in Fig. 1A). Fig. 2A shows that the plant sequences contain a number of evolutionary conserved residues shared with the mammalian and yeast sequences, including the arginine residue (asterisk in Fig. 2A) necessary in human and yeast RanGAP for binding of Ran and activation of its GTPase activity (6, 26). All plant sequences contain an acidic domain comparable to those in yeast and mammalian RanGAP (underlined in Fig. 2A).
Plant RanGAP Core Domains Can Be Modeled onto the Crystal Structure of SpRna1p.
One dicot (AtRanGAP1) and the monocot (OsRanGAP) sequence were tested for structural fit to crystal structures available in the Swiss-Model three-dimensional database (22, 23). swiss pdb viewer interfaced with Swiss-Model was used to search for a template, and 1YRG (the crystal structure of SpRna1p) was identified by the program. The structure of SpRna1p has been resolved at 2.66 Å and contains 11 leucine-rich repeats, which adopt a crescent-like structure of alternating α helices and β sheets (6), but does not include the acidic domain (amino acid 2 to amino acid 344; Fig. 2B Center).
The results of fitting the two plant sequences onto the 1YRG structure by Swiss-Model are shown in Fig. 2B. Successful fit was observed for amino acid 116 to amino acid 447 of AtRanGAP1 and amino acid 122 to amino acid 453 of OsRanGAP (Fig. 2B, top). The predicted structures of the AtRanGAP1 and OsRanGAP sequences threaded onto the SpRna1p structure are depicted in the Left and Right panels of Fig. 2B, respectively. Except for small areas indicated in red, the majority of the molecules are predicted to adopt the structure of SpRna1p (indicated in green). The successful fit begins immediately after the end of the WPP domain in both plant RanGAPs and ends right before the acidic domain. Together with the conservation of the crucial arginine residue and the acidic domain, as well as the complementation of the yeast rna1 mutant PSY714 by MsRanGAP and AtRanGAP1 (Ferenc Nagy, personal communication), these data indicate that the putative plant RanGAPs are true orthologs of their yeast and animal counterparts.
AtRanGAP1 Is Located at the Plant Nuclear Envelope.
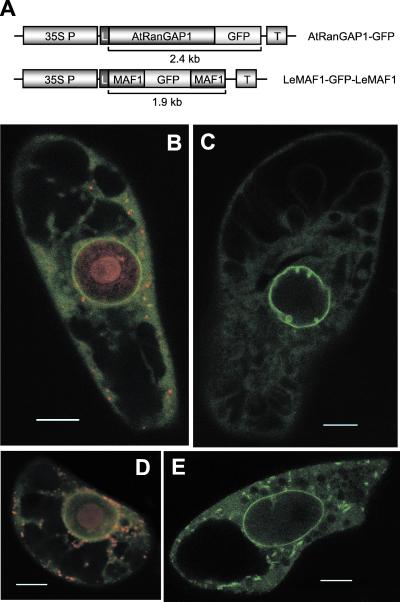
We have investigated whether in plants RanGAP is located at the nuclear envelope like mammalian RanGAP, or localized in the cytoplasm like yeast Rna1p. We have therefore compared the localization of GFP fusions of AtRanGAP1 and tomato MAF1 (LeMAF1), the latter having previously been shown to be associated with the plant nuclear envelope (20). The GFP-fusion constructs outlined in Fig. 3A were transiently transformed into tobacco BY-2 cells. Fig. 3B shows that AtRanGAP1-GFP localizes to the nuclear rim, with a weaker, uniform staining in the cytoplasm and none in the nucleus (counterstained with SYTO 82 orange). In some cells expressing AtRanGAP1-GFP, prominent invaginations at the nuclear rim were observed (Fig. 3C), which might correspond to the extended grooves and invaginations of the nuclear envelope recently reported for plant cells (27). The AtRanGAP1 localization pattern was found to be very similar to the localization of LeMAF1 (Fig. 3 D and E) except for cytoplasmic speckles of LeMAF1-GFP-LeMAF1 fluorescence, which were rarely observed with AtRanGAP1-GFP (compare Fig. 3 C and E), and some fluorescence of LeMAF1-GFP-LeMAF1 inside the nucleus. Together, these data show that AtRanGAP1 has a subcellular localization very similar to both mammalian RanGAP (28) and MAF1.
Figure 3.
AtRanGAP1 is located at the nuclear envelope. (A) Expression cassettes for transient transformation of AtRanGAP1-GFP and LeMAF1-GFP-LeMAF1 fusions. The LeMAF1-GFP-LeMAF1 sandwich expression cassette, designed to prevent passive diffusion into the nucleus, has been described previously (20). 35S P, cauliflower mosaic virus 35S promoter; L, tobacco etch virus translational leader; GFP, mGFPS65T from pRTL2-mGFPS65T (25); T, 35S terminator. (B–E) GFP fluorescence of BY-2 cells transiently transformed with AtRanGAP1-GFP (B and C) and LeMAF1-GFP-LeMAF1 (D and E). In B and D, SYTO 82 orange was used to counterstain for DNA, labeling the nucleus, as well as mitochondria and plastids in the cytoplasm. (In B–E, bars equal 10 μm.) See Movie 1, which is published as supporting information on the PNAS web site, www.pnas.org. Sequence 1 shows a scan through a cell expressing AtRanGAP1-GFP, sequence 2 a scan through a cell expressing LeMAF1-GFP-LeMAF1.
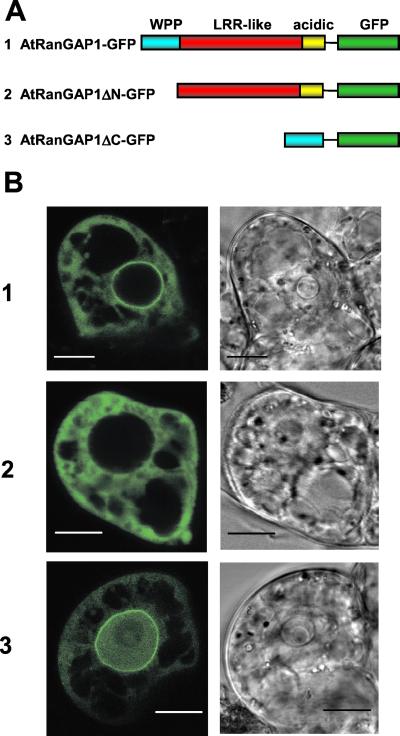
The WPP Domain Is Necessary and Sufficient for Nuclear Envelope Targeting of AtRanGAP1.
To test the hypothesis that the WPP domain shared between plant RanGAP and MAF1 is involved in nuclear-envelope targeting, deletion constructs of AtRanGAP1 were fused to GFP (Fig. 4A), and their localization in BY-2 cells was investigated (Fig. 4B). In contrast to AtRanGAP1-GFP, AtRanGAP1ΔN-GFP, which had the N-terminal WPP domain deleted, predominantly accumulated in the cytoplasm without a concentration at the nuclear rim. AtRanGAP1ΔC-GFP, which contained only the WPP domain fused to GFP, was concentrated at the nuclear rim like AtRanGAP1-GFP. The observed presence of AtRanGAP1ΔC-GFP inside the nucleus is likely due to passive diffusion of this 37-kDa fusion protein into the nucleus (exclusion size ≈40 kDa; ref. 29).
Figure 4.
The WPP domain is necessary and sufficient for nuclear-envelope targeting of AtRanGAP1. (A) Fusion constructs in pRTL2-mGFPS65T. Color code of AtRanGAP1 domains is as in Fig. 1. (B) GFP fluorescence of BY-2 cells transiently transformed with AtRanGAP1-GFP (1), AtRanGAP1ΔN-GFP (2), and AtRanGAP1ΔC-GFP (3). (Left) GFP fluorescence. (Right) Transmitted light images of cells. (Bars equal 10 μm.)
Approximately 80% of the cells transformed with either AtRanGAP1-GFP (82 cells total) or AtRanGAP1ΔC-GFP (112 cells total) showed bright fluorescence at the nuclear rim. In the remaining ≈20% of GFP-expressing cells, no clear nuclear-rim fluorescence was detected. In contrast, 87% cells transformed with AtRanGAP1ΔN-GFP (80 cells total) showed diffuse cytoplasmic fluorescence. The remaining 13% showed some enhanced fluorescence at the nuclear rim. It is not known whether this result was due to the developmental state of the cells or to their morphology, because some cells contained only a very narrow rim of cytoplasm between the nucleus and surrounding vacuoles, which could be mistaken for nuclear envelope-staining. Together, these data indicate that the WPP domain is both necessary and sufficient for targeting AtRanGAP1 to the plant nuclear envelope.
Identification of Amino Acid Residues Required for Nuclear Envelope Targeting of AtRanGAP1.
To determine whether the highly conserved WPP motive in the WPP domain is involved in nuclear envelope targeting, W40 and P41 were replaced with two alanines in the AtRanGAP1-GFP and AtRanGAP1ΔC-GFP constructs (Fig. 5A). Transient transformation of these constructs into BY-2 cells revealed that the mutation completely disrupted nuclear envelope targeting of both fusion proteins (Fig. 5B). The mutations did not alter the behavior of the two proteins with respect to nuclear import, and the distribution of AtRanGAP1ΔCmut-GFP is indistinguishable from free GFP (Fig. 5B3). These data indicate that the conserved tryptophan and proline residues are essential for nuclear envelope targeting of plant RanGAP, likely by being involved in creating a binding surface for an acceptor protein of RanGAP at the nuclear rim.
Discussion
Based on the data presented here, eukaryotic RanGAPs can be divided into at least three classes, based on the presence or absence of specific nuclear envelope-targeting domains. Yeast RanGAPs represent the most simple form, consisting of an LRR domain followed by an acidic domain. Both domains are necessary for binding of Ran and activation of the Ran GTPase activity (6, 26). Vertebrate and plant RanGAPs share this core domain with the yeast proteins, including critical amino acid residues.
Vertebrate and plant RanGAPs contain additional, kingdom-specific domains necessary for association with the nuclear envelope in the respective organism. Their fundamentally different structures imply that different mechanisms to anchor RanGAP to the outer surface of the nuclear envelope have evolved in plants and animals. In vertebrates, SUMOylation of the C-terminal domain leads to a conformational change that allows the protein to bind to Nup358 (5, 28). In plants, the N-terminal WPP domain is necessary and sufficient to target RanGAP to the nuclear rim, and the WP amino acid pair is critical for this function. The WPP domain has no sequence similarity to the vertebrate nuclear envelope-targeting domain. It does not contain a consensus motif for SUMOylation ((I/V/L)KXE; ref. 30), suggesting that nuclear envelope targeting in plants does not involve SUMOylation of RanGAP. However, because no plant substrate for SUMO has yet been identified, it cannot be fully excluded that a different sequence motif for SUMOylation exists in plants (31).
Interestingly, no non-RanGAP protein sequences in GenBank show significant similarity with the C-terminal domain of vertebrate RanGAPs, whereas in plants MAF1 has strong similarity to the WPP domain of RanGAP and colocalizes with RanGAP at the nuclear rim. The function of MAF1 is presently unknown, but it is tempting to speculate that it could be involved in regulating RanGAP-attachment to an acceptor protein, a function that could be provided in vertebrates by regulated SUMOylation of RanGAP.
The acceptor protein of mammalian RanGAP, Nup358, is a complex, multifunctional nucleoporin that does not appear to have an ortholog in yeast (32, 33). It consists of four RanBP1-like Ran-binding sites, zinc fingers that bind DNA (34), a leucine-rich region, a cyclophilin A-homologous domain, and an inverted peptide repeat, and binds both mammalian Ran and RanGAP (5, 28, 33–35). The RanGAP-binding surface has been mapped to a 300 aa region including the inverted peptide repeats (28).
At present, no nucleoporins have been identified from plants. A surprisingly low degree of similarity exists between the known 32 yeast and 25 mammalian nuclear pore complex proteins. Only six proteins have a convincing ortholog in the other organism (32). We have searched the Arabidopsis full genome database with the sequence of human Nup358 (GenBank accession no. A57545). The only proteins with similarity are cyclophilins and related cis-trans isomerases that align with the C-terminal 200 aa of Nup358 and the already described family of RanBP1-like proteins that share the Ran-binding domains (18). No protein with the combination of domains indicative of Nup358 exists in the annotated Arabidopsis genome. This finding is consistent with the notion that plant RanGAP utilizes a different interaction partner for anchoring to the nuclear rim.
Why would a protein with presumably similar function in nucleocytoplasmic transport have such different modes of localization in different organisms? In contrast to mammalian and plant RanGAP, which are both clearly associated with the nuclear rim, Rna1p in both S. cerevisiae and S. pombe is predominantly cytoplasmic (7, 36, 37). It therefore appears that yeast RanGAP does not have a specific mechanism for nuclear envelope anchoring, and indeed a purely or predominantly cytoplasmic location might suffice for the suggested function in establishing a RanGTP/RanGDP gradient across the interphase nuclear envelope. In contrast to yeast, both higher plants and animals undergo open mitosis. The newly discovered functions of animal Ran in spindle formation and nuclear vesicle fusion imply a specific role for both RanGTP and RanGDP in mitotic cells (10). Plant and animal cells, but not yeast, require therefore the means to spatially separate the Ran accessory proteins RanGAP and RCC1 in the absence of an intact nuclear envelope. Attachment of RCC1 to the chromosomes and of RanGAP to structures derived from the dissociated nuclear envelope might provide such means.
It has been noted previously that the protein composition of the nuclear envelope differs surprisingly between organisms from different kingdoms. An example is the complete lack of homologs of lamins and lamina-associated proteins in both yeast and plants (19, 38). The vast differences in nuclear pore components between yeast and mammalian cells were a similarly unexpected finding (32). If the process of open mitosis had evolved independently after the separation of plants and animals one billion years ago, it could explain why many players at the nuclear envelope have no counterpart in the other kingdoms. In plant evolution, higher plant-like open mitosis appears first in the Charophycea (stoneworts), multicellular freshwater algae and the predicted ancestors of land plants, whereas the euglenophytae (Euglena) have closed mitosis and the chlorophycae (Volvox, Chlamydomonas) show all three forms of closed, fenestral, and open mitosis (39). This late occurrence of open mitosis in plants argues indeed strongly for a separate evolution of the mechanism in the plant and animal kingdoms.
A subset of the mammalian nuclear pore complex proteins, including Nup358, is among the first proteins to reassemble on the decondensing chromosomes in early telophase (40–43). Whereas none of them have been shown to be functionally required for nuclear assembly, it is conceivable that especially the DNA-binding Nups may play a role in the association of nuclear envelope components with the decondensing chromatin. A recruitment during evolution of different proteins into roles in nuclear envelope and nuclear pore dynamics during open mitosis in plants and animals might explain why RanGAP utilizes different anchor surfaces for nuclear-envelope attachment. The finding that RanGTP hydrolysis is required for the fusion of nuclear vesicles points to a role for the RanGAP-anchoring factor—and RanGAP—at an early stage of nuclear envelope reformation. The fundamental differences in nuclear envelope targeting of RanGAP in plants and animals presented here lead us to expect different players in the spatial organization of Ran signaling in the plant and animal kingdoms.
Supplementary Material
Acknowledgments
We thank Dr. Biao Ding for generous user time of his confocal microscope and advice in its use and Dr. David Somers for critical reading of the manuscript. This work was supported by grants from the National Science Foundation (MCB-0079577) and the U.S. Department of Agriculture (Plant Growth and Development no. 2001-01901).
Abbreviations
- GFP
green fluorescent protein
- RanGAP
Ran GTPase activating protein
- LRR
leucine-rich repeat
- SUMO
small ubiquitin-related modifier protein
- RT
reverse transcription
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Görlich D, Kutay U. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 2.Takai Y, Sasaki T, Matozaki T. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 3.Kahana J A, Cleveland D W. J Cell Biol. 1999;146:1205–1210. doi: 10.1083/jcb.146.6.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohtsubo M, Okazaki H, Nishimoto T. J Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matunis M J, Wu J, Blobel G. J Cell Biol. 1998;140:499–509. doi: 10.1083/jcb.140.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hillig R C, Renault L, Vetter I R, Drell T T, Wittinghofer A, Becker J. Mol Cell. 1999;3:781–791. doi: 10.1016/s1097-2765(01)80010-1. [DOI] [PubMed] [Google Scholar]
- 7.Hopper A K, Traglia H M, Dunst R W. J Cell Biol. 1990;111:309–321. doi: 10.1083/jcb.111.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melchior F, Weber K, Gerke V. Mol Biol Cell. 1993;4:569–581. doi: 10.1091/mbc.4.6.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sazer S, Dasso M. J Cell Sci. 2000;113:1111–1118. doi: 10.1242/jcs.113.7.1111. [DOI] [PubMed] [Google Scholar]
- 10.Dasso M. Cell. 2001;104:321–324. doi: 10.1016/s0092-8674(01)00218-5. [DOI] [PubMed] [Google Scholar]
- 11.Wiese C, Wilde A, Moore M S, Adam S A, Merdes A, Zheng Y. Science. 2001;291:653–656. doi: 10.1126/science.1057661. [DOI] [PubMed] [Google Scholar]
- 12.Gruss O J, Carazo-Salas R E, Schatz C A, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj I W. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 13.Nachury M V, Maresca T J, Salmon W C, Waterman-Storer C M, Heald R, Weis K. Cell. 2001;104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- 14.Hetzer M, Bilbao-Cortes D, Walther T C, Gruss O J, Mattaj I W. Mol Cell. 2000;5:1013–1024. doi: 10.1016/s1097-2765(00)80266-x. [DOI] [PubMed] [Google Scholar]
- 15.Zhang C, Clarke P R. Science. 2000;288:1429–1432. doi: 10.1126/science.288.5470.1429. [DOI] [PubMed] [Google Scholar]
- 16.Ach R A, Gruissem W. Proc Natl Acad Sci USA. 1994;91:5863–5867. doi: 10.1073/pnas.91.13.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merkle T, Haizel T, Matsumoto T, Harter K, Dallmann G, Nagy F. Plant J. 1994;6:555–565. doi: 10.1046/j.1365-313x.1994.6040555.x. [DOI] [PubMed] [Google Scholar]
- 18.Haizel T, Merkle T, Pay A, Fejes E, Nagy F. Plant J. 1997;11:93–103. doi: 10.1046/j.1365-313x.1997.11010093.x. [DOI] [PubMed] [Google Scholar]
- 19.Meier I. Plant Physiol. 2000;124:1507–1510. doi: 10.1104/pp.124.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gindullis F, Peffer N J, Meier I. Plant Cell. 1999;11:1755–1768. doi: 10.1105/tpc.11.9.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guex N, Peitsch M C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 23.Guex N, Diemand A, Peitsch M C. Trends Biochem Sci. 1999;24:364–367. doi: 10.1016/s0968-0004(99)01427-9. [DOI] [PubMed] [Google Scholar]
- 24.Hooft R W, Vriend G, Sander C, Abola E E. Nature (London) 1996;381:272. doi: 10.1038/381272a0. (lett.). [DOI] [PubMed] [Google Scholar]
- 25.von Arnim A G, Deng X W, Stacey M G. Gene. 1998;221:35–43. doi: 10.1016/s0378-1119(98)00433-8. [DOI] [PubMed] [Google Scholar]
- 26.Haberland J, Becker J, Gerke V. J Biol Chem. 1997;272:24717–24726. doi: 10.1074/jbc.272.39.24717. [DOI] [PubMed] [Google Scholar]
- 27.Collings D A, Carter C N, Rink J C, Scott A C, Wyatt S E, Allen N S. Plant Cell. 2000;12:2425–2440. doi: 10.1105/tpc.12.12.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matunis M J, Coutavas E, Blobel G. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grebenok R J, Pierson E, Lambert G M, Gong F C, Afonso C L, Haldeman-Cahill R, Carrington J C, Galbraith D W. Plant J. 1997;11:573–586. doi: 10.1046/j.1365-313x.1997.11030573.x. [DOI] [PubMed] [Google Scholar]
- 30.Poukka H, Karvonen U, Janne O A, Palvimo J J. Proc Natl Acad Sci USA. 2000;97:14145–14150. doi: 10.1073/pnas.97.26.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vierstra R D, Callis J. Plant Mol Biol. 1999;41:435–442. doi: 10.1023/a:1006323317890. [DOI] [PubMed] [Google Scholar]
- 32.Vasu S K, Forbes D J. Curr Opin Cell Biol. 2001;13:363–375. doi: 10.1016/s0955-0674(00)00221-0. [DOI] [PubMed] [Google Scholar]
- 33.Wu J, Matunis M J, Kraemer D, Blobel G, Coutavas E. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- 34.Yokoyama N, Hayashi N, Seki T, Pante N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, et al. Nature (London) 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]
- 35.Yaseen N R, Blobel G. J Biol Chem. 1999;274:26493–26502. doi: 10.1074/jbc.274.37.26493. [DOI] [PubMed] [Google Scholar]
- 36.Traglia H M, O'Connor J P, Tung K S, Dallabrida S, Shen W C, Hopper A K. Proc Natl Acad Sci USA. 1996;93:7667–7672. doi: 10.1073/pnas.93.15.7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melchior F, Weber K, Gerke V. Mol Biol Cell. 1993;4:569–581. doi: 10.1091/mbc.4.6.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen M, Lee K K, Wilson K L, Gruenbaum Y. Trends Biochem Sci. 2001;26:41–47. doi: 10.1016/s0968-0004(00)01727-8. [DOI] [PubMed] [Google Scholar]
- 39.Kubai D F. Int Rev Cytol. 1975;43:167–227. doi: 10.1016/s0074-7696(08)60069-8. [DOI] [PubMed] [Google Scholar]
- 40.Haraguchi T, Koujin T, Hayakawa T, Kaneda T, Tsutsumi C, Imamoto N, Akazawa C, Sukegawa J, Yoneda Y, Hiraoka Y. J Cell Sci. 2000;113:779–794. doi: 10.1242/jcs.113.5.779. [DOI] [PubMed] [Google Scholar]
- 41.Bodoor K, Shaikh S, Enarson P, Chowdhury S, Salina D, Raharjo W H, Burke B. Biochem Cell Biol. 1999;77:321–329. [PubMed] [Google Scholar]
- 42.Bodoor K, Shaikh S, Salina D, Raharjo W H, Bastos R, Lohka M, Burke B. J Cell Sci. 1999;112:2253–2264. doi: 10.1242/jcs.112.13.2253. [DOI] [PubMed] [Google Scholar]
- 43.Wiese C, Goldberg M W, Allen T D, Wilson K L. J Cell Sci. 1997;110:1489–1502. doi: 10.1242/jcs.110.13.1489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.