A goal of precision medicine1 is to stratify patients in order to improve diagnosis and medical treatment. Translational investigators are bringing to bear ever greater amounts of heterogeneous clinical data and scientific information to create classification strategies that enable the matching of intervention to underlying mechanisms of disease in subgroups of patients. Ontologies are systematic representations of knowledge that can be used to inte-grate and analyze large amounts of heterogeneous data, allowing precise classification of a patient. In this review, we describe ontologies and their use in computational reasoning to support precise classification of patients for diagnosis, care management, and translational research.
ABUNDANCE OF DATA
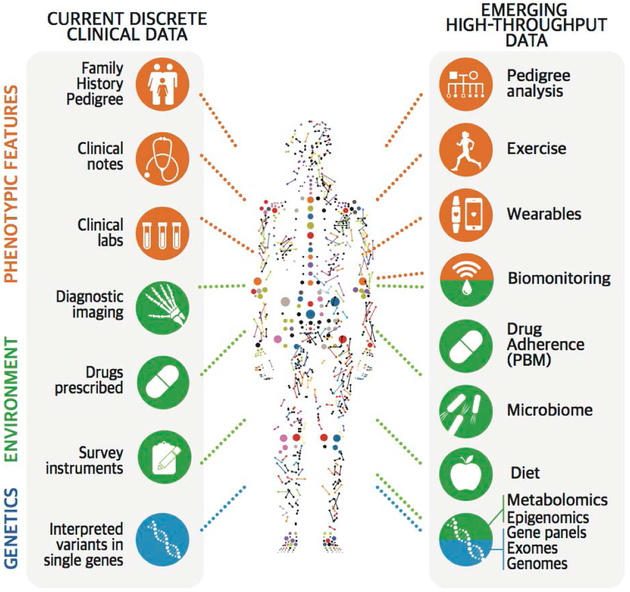
The widespread adoption of electronic health records (EHRs) affords an opportu-nity to collect objective and subjective observations related to demographic char-acteristics, findings, symptoms, diagnoses, test results, procedures, medications, nursing interventions, and so on. Very large amounts of high-throughput data, including those obtained through genomic, proteomic, and metabolomic analyses, are now being used in clinical analyses. Public data sets, such as those of the Cancer Genome Atlas and the 100,000 Genomes Project,2,3 provide a context (or baseline) for comparing clinical data, although such comparisons are seldom made. The volume and depth of data and the rate of its accrual are unprecedented in human history (Fig. 1).
Figure 1. Multimodal Clinical and High-Throughput Data, Captured in Diverse Ways.
The health trajectory of a person can be measured many times and in many ways, including by examining various aspects of genotypic, phenotypic, and environmental attributes. Clinical data (left side) currently include family history, notes, laboratory reports, imaging, clinical instrument outputs, drugs and drug doses, and interpreted variants in single genes. These features are now being complemented by emerging, high-throughput, dynamic data (right side) that have not yet been fully harnessed to a classification of disease. Sources of such data include wearable devices that track exercise, weight, heart rate, diet, geographic location, adherence to the administration of medications, and so forth, collected over a period of minutes to years. All collected information (except germline sequencing) is a reflection of a discrete point in time in a person’s health trajectory. PBM denotes pharmacy benefit manager.
Although EHRs document many types of data, they often impede analyses of patient-level, high-throughput or molecular data in combination with clinical data because the records are frequently incomplete, incorrect, of unknown provenance, or of insufficient level of detail. These problems are due in part to a design that is driven by billing concerns rather than a desire to document medically relevant biologic features of the patient.4 Data on behavioral phenotypes, environmental exposures, genome sequencing, and mobile health sensors are difficult to capture and are not systematically collected or integrated, especially since they are often “trapped” in PDF documents that can be difficult to parse into structured fields computationally.
In summary, phenotypic information about individual patients is often insufficiently detailed or inaccessible, thus obstructing the detection of similarities and the classification of patients into clinically useful groups. Such detection and clas-sification are both challenging and important in disorders with a spectrum of symptoms, signs, biomarkers, and genotypes that may not be present in all patients, but understanding how to use these data to stratify patients and to recognize similarities between distinct diseases is a major goal of precision medicine. For example, both BRAF-mutated melanoma and BRAF-mutated Langerhans-cell histiocytosis respond to the drug vemurafenib.5
MAKING SENSE OF DATA
Data without interpretation are facts without understanding. Methods of inference, such as statistical analyses or machine learning, require categorizing subjects according to covariates, features, or both. The challenge is to create useful classifications that combine a plethora of numerical or continuous variables, dichotomies, ordinal groups, and taxonomic categories. Classifications describe entities from domains of interest, such as diseases, phenotypes, medications, and exposures, by naming the entities in each domain and providing computational specifications of differing degrees of sophistication. Increasingly formal mechanisms exist for creating such names and specifying their relationships to one another, from simple terminologies to ontologies (Table 1).
Table 1.
Types of Terminologies Used for Computational Analysis.*
| Term | Definition |
|---|---|
| Index | List of relevant terms pulled directly from a body of unstructured or semistructured text.An index is produced to improve the speed and relevance of search results. |
| Terminology | Set of preferred or official terms in a domain. A terminology may be a systematic nomenclature supported by a centralized body or as simple as the common usage that arises in a specific community of practice. |
| Thesaurus | Terminology that clusters synonyms and plesionyms (near synonyms) into categories. |
| Controlled vocabulary | Set of preferred terms created specifically for a domain or body of text. |
| Classification | Controlled vocabulary that is intended to comprehensively describe a topic or domain from a conceptual perspective and is not developed solely from a text corpus that it is meant to describe. |
| Statistical classification | Classification in which all concepts are mutually exclusive to avoid counting anything twice. This is typically achieved with the use of a monohierarchy, in which each concept has one and only one parent, such as the International Classification of Diseases (ICD).7 A statistical classification is exhaustive because it includes residual categories such as “unspecified” or “not elsewhere classified.” |
| Ontology | Controlled terminology invoking formal semantic relationships between and among concepts, manifested as a type of description logic, which is a subset of first-order predicate logic, chosen to accommodate computational tractability.8 A common example is OWL (Web Ontology Language; www.w3.org/OWL/). |
The information presented in the table is adapted from Cornet and Chute.6
Standards exist for the majority of types of data used for clinical medicine, including diagnoses, medications, adverse reactions, procedures, laboratory data, and imaging data, as well as signs, symptoms, and other phenotypic abnormalities (Table 2). However, these data-type standards are just the first step in making data computable and patients deeply classifiable. The larger challenge is to integrate formats and structures from different sources to make them compatible.
Table 2.
Standards, Terminologies, and Ontologies Widely Used in Clinical Medicine.*
| Type of Data | Ontology | Example of Term |
|---|---|---|
| Diagnoses | Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT) ICD Orphanet Rare Disease Ontology (ORDO) National Cancer Institute Thesaurus (NCIT) |
Triple-negative breast carcinoma (NCIT:C71732) |
| Phenotypic abnormalities | Human Phenotype Ontology (HPO) | Bronchopulmonary sequestration (HP:0010960) |
| Medications | RxNorm DrugBank ChEMBL |
Panobinostat (CHEMBL483254) |
| Adverse reactions | Ontology of Adverse Events (OAE) | Injection-site induration (OAE:0000323) |
| Procedures | Medical Dictionary for Regulatory Activities (MedDRA) | Cardiac aneurysm repair (MEDDRA/10007514) |
| Laboratory examinations | Logical Observation Identifiers Names and Codes (LOINC) | Creatinine in serum or plasma (LOINC:2160-0) |
| Imaging data | Digital Imaging and Communications in Medicine (DICOM) RadLex |
Periosteal cortical thinning (RID45761) |
SNOMED CT and some of the other ontologies cover multiple types of data (not shown). ChEMBL is a chemical database maintained by the European Molecular Biology Laboratory, and RadLex is a radiology-specific ontology produced by the Radiological Society of North America. Since 2015, RadLex and Logical Observation Identifiers Names and Codes (LOINC; maintained by the Regenstrief Institute) have been collaboratively producing a unified model for naming radiology procedures (https://loinc.org/collaboration/rsna/).
DATA STANDARDS FOR COMPARABLE AND CONSISTENT CLASSIFICATION
Data standards can ultimately be reduced to two components: structure and semantics. Conventionally, most of us think about structure as the arrangement of data, either on an EHR screen or as a database schema behind the scenes. Semantics, in turn, refers to concepts and the relationships between them. Software systems require assertions about term equivalence. For example, without equivalent terms, clinical laboratory data with codes local to a specific laboratory or hospital are difficult to compare with corresponding data elsewhere. Similarly, diagnoses captured in the Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT, a set of terms that describe clinical findings, symptoms, diagnoses, procedures, body structures, organisms and other causes of disease, substances, pharmaceuticals, devices, and specimens)9 do not always corre-spond with similarly named conditions in the International Classification of Diseases (ICD).7 In short, mapping clinical data across systems or to the basic science data needed for mechanistic classification is often compromised by differences in naming systems or structure.
Semantics and structure are not orthogonal but deeply intertwined. One may encounter a seemingly obvious term such as “myocardial infarction” in a patient’s medical record; however, the context must be taken into account. If the mention of “myocardial infarction” was nested within an EHR partition about the patient’s family history, the interpretation would be that someone in the patient’s family, not necessarily the patient, had a myocardial infarction. Correspondingly, the clinician-reported signs and symptoms (i.e., problem list) may contain a single phrase that combines context and diagnosis — in this case, “family history of myocardial infarction.” Although this example may seem trivial, the reality is that matching of information sources must not only accommodate different semantic foundations (terminologies and classifications) but also anticipate further modification of those semantics according to their local context. Sometimes the modification is extreme, such as in the case of negation (“no history of myocardial infarction”). Exploiting the promise of precision medicine will depend on our ability to align data across patients and systems with comparable and consistent formats and contextual meaning.
FROM TERMINOLOGY TO ONTOLOGY
Terminologies have a long history of use in information retrieval (i.e., the search for documents or database entries that match certain criteria). Some of the most important resources for information retrieval in the medical domain include Medical Subject Headings (MeSH) for indexing and searching PubMed; RxNorm,10 a terminology for generic and branded drugs; and the Unified Medical Language System (UMLS), which integrates more than 100 clinical terminologies and coding systems.11 These resources, which comprise standardized names and lists of synonyms and cross-references, provide the foundation for searching and indexing and are in common use in EHRs and public databases.
Ontologies differ from terminologies in that ontologies define relationships between concepts in a way that allows computational logical reasoning, enabling the drawing of conclusions from related assertions.12 For example, if an ontology classifies “virus” as an infectious agent and classifies “infectious meningitis” as a type of meningitis due to an infectious agent, then it would conclude that “viral meningitis” is a subclass of “infectious meningitis.” Aristotle developed conceptual taxonomies that are in some ways similar to modern bio-ontologies.13 More recently, scientists have used the word “ontology” to denote a computational representation describing specific domains of knowledge. An ontology consists of a set of concepts (terms) and their synonyms, as well as description-logic definitions that specify the formal relationships between the concepts (Fig. 2).
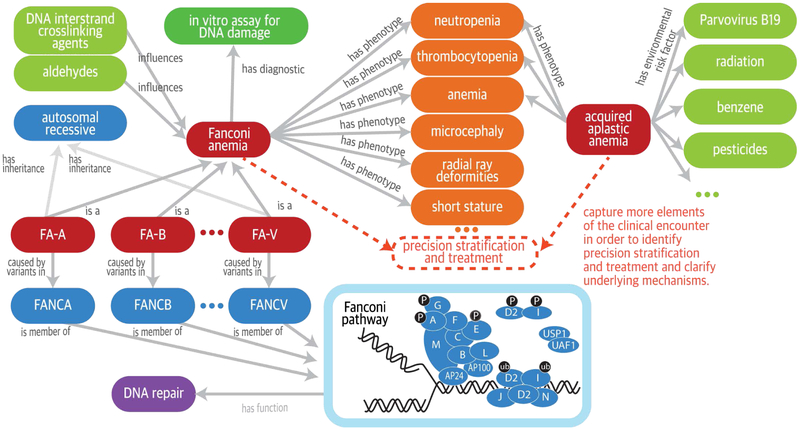
Figure 2. Ontology-Driven Representation of Fanconi Anemia and Acquired Aplastic Anemia.
Fanconi anemia and acquired aplastic anemia share several phenotypic features but have very different causal mechanisms. Computable relationships can be represented among diseases, phenotypic features, genes, and environmental exposures by interlinking terms (concepts) from sources including the Orphanet Rare Disease ontology (ORDO) (pink denotes diseases),14 the Human Phenotype Ontology (HPO)15 for phenotypic features (orange), the Chemical Entities of Biological Interest ontology for the chemical compounds (green denotes factors such as chemical exposures that can influence severity or trigger development of disease),16 and the Ontology for Biomedical Investigations for the comet assay (single-cell gel-electrophoresis assay) of DNA breakage (aqua),17 as well as the Gene Ontology (lavender denotes a biologic pathway)18 and the Reactome for molecular pathways (blue denotes disease genes and mode of inheritance).19 Ontologies can be used to support the integrative analysis of these data sources for precision stratification and treatment and to clarify underlying mechanisms, as suggested by the dashed lines. The labeled arrows between concepts represent description-logic definitions that specify the formal relationships between the concepts. FA-A denotes Fanconi anemia, complementation group A; FA-B, complementation group B; and so on. In the diagram of the Fanconi anemia pathway, the blue circles represent the proteins encoded by the Fanconi genes (e.g., A is the protein encoded by FANCA), and the grayish-blue ovals represent other interacting proteins; P denotes phosphorylation, and ub ubiquitination.
The use of description logics in an ontology can guarantee logical consistency, even with hundreds of thousands of concepts across multiple domains, enabling computational reasoning procedures to identify facts that are implied but not explicitly stated in the original data. Ontologies can thus help to leverage the latent knowledge within clinical big data by encoding the data with “computable” semantics, enabling machinelearning and other algorithms to address challenges in the analysis of multimodal, high-throughput data by integrating it with clinical meaning. Ontologies can be used in combination with natural language processing to disambiguate text concepts, such as those found in clinical notes, and improve knowledge extraction from EHRs and other sources.
Ontologies can also support integration of basic science data (e.g., data from animal models) and public knowledge (e.g., associations between genetic variants and diseases), enabling patient classification based on a corpus of data existing well beyond the EHR and permitting new clinical insights. The combination of massive data and affordable high-capacity computing provides an opportunity for unprecedented discovery of association and, increasingly, causal reasoning to gain diagnostic and therapeutic insight.
ONTOLOGIES FOR DISEASE CLASSIFICATION
The first modern medical classification that can be considered a true ontology of diseases (nosology) was developed by Carl Linnaeus (1707–1778), who divided diseases into 11 classes, 37 orders, and 325 species. Although Linnaeus’s classification contains some errors from a modern perspective, such as the notion that leprosy can be caught by eating herring worms,20 his classification laid the foundation for work that eventually led to the first edition of the ICD, in 1893.21 The ICD has advanced enormously since its creation as a cause-of-death inventory, but it continues to be intended as a standard for epidemiology, health management, and billing, not as a computational representation of the patient as a biologic subject. One of the biggest issues with the ICD is that historical editions are statistical classifications that are mutually exclusive (they do not double count things) and exhaustive (they provide a place to put everything). The ICDs have achieved exclusiveness through a monohierarchy (single parentage), with each code having one and only one parent. This precludes multiple counting but also creates arbitrary associations. For example, in the 10th edition of the ICD, malignant neoplasm of the thyroid gland (C73) is a child of malignant neoplasms but is not a child of disorders of the thyroid gland; it would be a child of both only if terms in ICD-10 could have multiple parents. Monohierarchies thus artificially constrain important axes of characterization and inquiry and impede meaningful analyses of disease and other phenotypes.
Rational disease classification dates back to Hippocrates, though it remains an active field of study today.6 Conventionally, phenotype has denoted observable characteristics of a person, often attributable to genotype. Increasingly, the term is being co-opted by translational researchers, working at the boundaries of “omics” data and clinical records, to define a cohort of patients with the same “diagnosis” on the basis of similar clinical and “omics” features.22,23
The Electronic Medical Records and Genomics (eMERGE) study, funded by the National Human Genome Research Institute, showed the usefulness of computable phenotyping algorithms24,25 across medical centers with different EHR systems.26 Such pragmatic and reproducible methods use standard coding systems for phenotypic abnormalities, diseases, laboratory values, medications, adverse effects, and natural language processing of free text. However, most algorithms are presented as a set of English instructions, rules, and filters, making their translation to computation and integration with multimodal, high-throughput data an exercise for each user. A limitation is that the ICD codes used in these algorithms are captured for billing purposes and not for differential diagnosis. Statistical and machine-learning approaches to cohort definitions based on mining of structured data and free-text descriptions represent another promising method of semiautomated phenotyping for large-scale case–control studies.27
SNOMED CT is a compositional system, meaning that it can represent complex concepts by combining discrete facts and observations. For instance, acute perforated appendicitis can be represented by combining the concepts for “acute inflammation,” “perforation,” and “appendix structure.” SNOMED CT uses description logic to recognize logically equivalent ways of expressing “acute perforated appendicitis,” thereby providing a consistent and computable framework.28,29 In this manner, SNOMED CT can be used in different ways within different systems and contexts and by different users and still result in the same conceptual and computational meaning, thus offering an advantage over simpler terminologies. Currently, however, less than a quarter of the content of SNOMED CT is logically defined; the remainder is primitive and not amenable to this method.
The increasing use of wearable health devices30 and biomonitoring, as well as advances in medical digital imaging, portends growth in the volume of clinical data. Ontologies can help to organize and analyze vast quantities of data that are too large for an individual physician to manage. Patient-reported information has provided robust findings in some areas, such as genomewide association studies.31 Therefore, formally encoding lay synonyms of medical terminology within an ontology32 may improve our ability to classify patients in meaningful disease groupings by integrating patient-reported information with standard medical terminology.
RARE DISEASES AND THE HUMAN PHENOTYPE ONTOLOGY
Ontologies have made a substantial contribution to translational research and the genetic diagnosis of rare disease. The sequencing of exomes and genomes has enabled the discovery of hundreds of novel disease-associated genes, and the diagnostic yield (percentage of patients who receive a molecular diagnosis) in many large-scale exome- or genome-sequencing studies is now at least 35% for some disease groups.33-37 In some cases, the diagnosis results in a change in clinical management, as well as family counseling.38-40
Variant-driven analysis aims to identify a disease-causing variant among the roughly 25,000 to 100,000 variants in a typical exome or approximately 4.5 million variants in a typical genome. Realizing the full clinical value of these data requires additional information about diseases and phenotypic abnormalities. Ontologies provide a computational-analysis framework that contextualizes the molecular data within an evaluation of the phenotypic presentation. The Human Phenotype Ontology (HPO) enables a deep phenotyping approach wherein computable phenotypic profiles of human diseases and individual patients allow the linking of terms that are close to one another in the hierarchy and weighted according to the specificity of individual phenotypic abnormalities15,41 (Fig. 3). There are more than 13,500 terms in a subtype hierarchy, which also encodes the specificity of each term for dif-ferential diagnosis as a function of the frequency of the term across all diseases in the HPO data-base. For example, the Marfan syndrome is characterized by relatively specific HPO terms such as ectopia lentis, which is found in 40 mendelian diseases in the HPO database, as well as less specific terms such as scoliosis, which is a characteristic of 424 mendelian diseases.
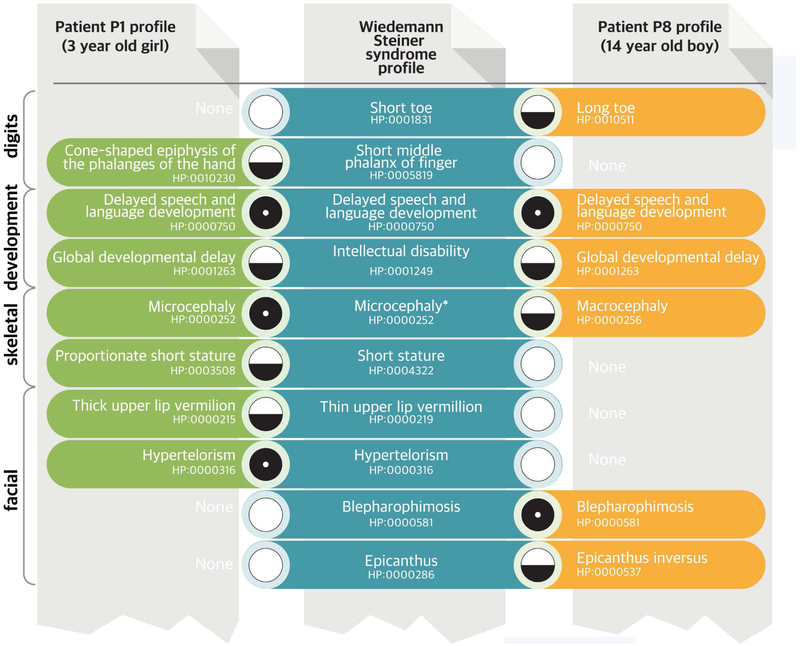
Figure 3. “Fuzzy” Matching of Phenotypic Profiles.
Shown in this example are portions of the HPO profiles (green and brown) of two patients for whom clinical dysmorphologic analysis did not help establish the diagnosis, even though they were seen in the same clinic within weeks of each other. Clinical exome sequencing showed a mutation in KMT2A in both patients, which in combination with the phenotype comparisons led to a diagnosis of the Wiedemann–Steiner syndrome.42 Each set of HPO terms is compared with all other phenotypic profiles in the HPO database to find the best nonexact (“fuzzy”) match.41 Each patient has a distinct phenotypic profile that only partially matches the computational model of the Wiedemann–Steiner syndrome derived from the literature (blue). Patient 1 had microcephaly, whereas Patient 2 was found to have macrocephaly; microcephaly had been observed in only one previously described patient.43 Some of the matches are relatively specific for this syndrome, such as blepharophimosis, and contribute more to the matching score than do features that are common to many diseases, such as intellectual disability. The final matching score can be calculated from the matching score for each query term and represents the proximity of the query term to its best match in the computational disease definition. A perfect match between a phenotypic feature of a patient and a feature of the disease is symbolized here by a black circle and would be assigned a high match score. A nonexact match (a fuzzy match, in which the patient has a feature that is similar to a feature of the disease but is not an exact match) is symbolized by a circle that is half black and half white and would be assigned a lower matching score. A lack of a match (a patient who does not have a feature that characterizes a disease) is symbolized by a white circle and can be penalized by some computational similarity algorithms. Existing algorithms exploit information in the ontology and annotations in many ways; commonly, they take into account the specificity of the term, usually calculated as the information content (not shown).
The HPO differs from other clinical terminologies and ontologies in that it provides a substantially more detailed representation of clinical phenotypes44 and it is designed for computational analysis by linking to computational disease definitions and to ontologies of gene function, anatomy, biochemistry, and other biologic attributes.15 For instance, “neutropenia” is logically defined with the use of terms from three ontologies covering the domains of cell types, gross anatomy, and attributes: “neutrophil,” “blood,” and “decreased count.” With the use of this multiattribute classification, a matching procedure can be applied to every disease in the database to find the closest match, or it can assist phenotypeclustering efforts by finding patients with similar phenotypic manifestations. Because this method represents the patient as a biologic subject, the Monarch Initiative has extended the approach to human–mouse phenotype comparisons on the basis of a cross-species ontology of anatomy 45,46 and the Mammalian Phenotype Ontology.47-49
HPO-based phenotypic profile matching used in combination with genome sequencing has allowed prioritization of candidate genes with predicted pathogenic variants.50-54 This approach helped establish diagnoses in 28% of children who might have otherwise remained on “diagnostic odysseys.”42 Use of the HPO and data from animal models yielded about 10 to 20% more molecular diagnoses than those obtained by manual curation of sequence data from patients in the Undiagnosed Diseases Program.55 Similarly, a machine-learning approach was applied to a cohort of 2045 persons with bleeding and platelet disorders to identify novel “fuzzy” phenotypic profiles associated with rare pathogenic variants, enabling the identification of disease-associated genes.56-58
THE FUTURE OF ONTOLOGIES IN MEDICINE
The ability to leverage extremely large amounts of data in order to answer the question, “Have I seen a case like that?” and to identify effective, safe treatments is a long-cherished aspiration.59 But what does it mean to be the patient in “a case like that”? Which axes of patients’ characteristics bear scrutiny: demographic data, signs, symptoms, family history, diagnoses, anthropometrics, test results, radiographic studies, or “omics” measures? How much of this information is already in my patient’s record? How much is in the records of putative patients similar to mine? How big is the universe of corresponding data that I can examine, in my practice, in my hospital, in my group, in my state, in my country? This is the logical extension of the “learning health system,”60 taken to the level of nearly homogeneous groups of patients. The National Research Council convened a forum on precision medicine,1 which proposed a “new taxonomy” for biology and medicine that would be structured to recognize and avail the multiple axes of basic science and clinical characteristics as a matrix defining disease endotypes (Fig. 4). The annotation of data with ontologic tags must also become an automated, background task; this is already a reality for some data sources61,62 and is tantalizingly close for others.
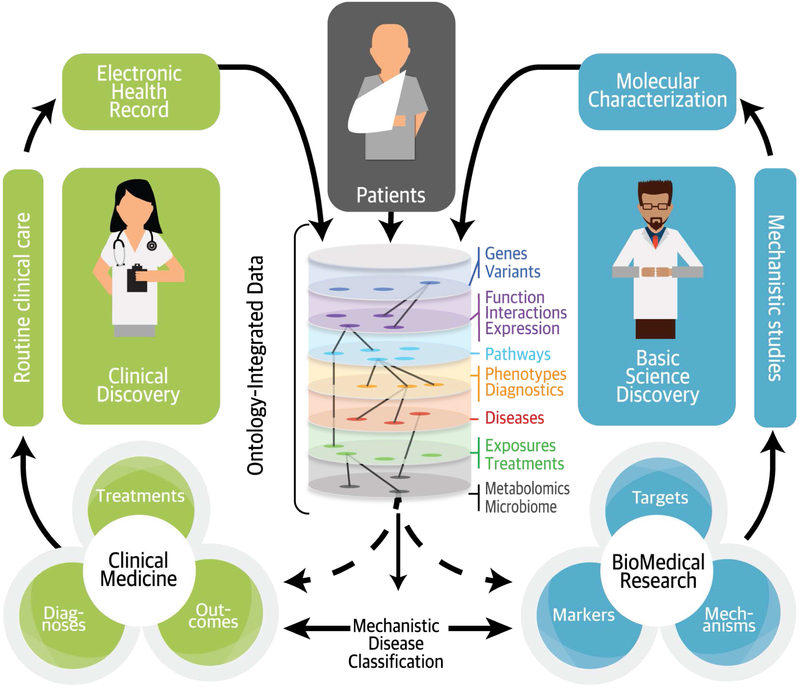
Figure 4. Ontology-Based Mechanistic Classification of Disease.
Well-structured clinical data can be readily integrated with discovery research data by using ontologies, which make clinical and basic science observations “computable” in a way that reflects present knowledge and allows new inferences. Integrating the two streams of data enables a mechanistic classification of disease across many data types, making a more refined and dynamic classification of patients possible.1
What are the barriers blocking progress toward the goal of individualized medicine? The first and most challenging is a set of privacy laws, enacted without consideration for the data-rich and data-dependent world in which we now find ourselves. Patient privacy and confidentiality are necessary to maintain the cooperation of our patients and their trust in us. Nevertheless, a new ethical framework may be in order, balancing the needs of society and future patients with legitimate expectations of privacy63 and the wishes of those who want to share their medical data for the betterment of society.64
The second barrier is the cost and effort of getting data into and out of EHRs. Manual input of structured data by clinicians is not scalable and is not a good use of clinicians’ time. Emerging efforts on standard application interfaces with EHRs from devices and data sources65 could help, as could patient-collected and patient-entered information.32,66,67 Systematically harvesting signs, symptoms, severity, and other clinical details from dictated notes or even from audio capture of the patient encounter is becoming increasingly practical.68 The third barrier is a lack of comparability and consistency among data and knowledge resources (e.g., public databases and clinical references, EHR systems and implementations, and clinical laboratories), which translates to a lack of interoperability. Harmonization can best be achieved through consistent invest-ment and community participation in computa-tional resources for translational research.69,70
Acknowledgments
We thank Julie McMurry, Rose Relevo, Mark Lawler, Maureen E. Hoatlin, Bill Hersh, Hannah Blau, Sebastian Köhler, John Mattison, and Mark Wanner for perceptive contributions.
Contributor Information
Melissa A. Haendel, Oregon Clinical and Transla-tional Research Institute, Oregon Health and Science University, Portland, and the Linus Pauling Institute and the Center for Genome Research and Biocomputing, Ore-gon State University, Corvallis
Christopher G. Chute, Johns Hopkins University Schools of Medi-cine, Public Health, and Nursing, Balti-more
Peter N. Robinson, Jackson Laboratory for Genomic Medicine and the Institute for Systems Genomics, University of Connect-icut — both in Farmington
REFERENCES
- 1.National Research Council, Committee on a Framework for Developing a New Taxonomy of Disease. Toward precision medicine: building a knowledge network for biomedical research and a new taxonomy of disease. Washington, DC: National Academies Press, 2011. [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marx V The DNA of a nation. Nature 2015;524:503–5. [DOI] [PubMed] [Google Scholar]
- 4.Goroll AH. Emerging from EHR purgatory — moving from process to outcomes. N Engl J Med 2017;376:2004–6. [DOI] [PubMed] [Google Scholar]
- 5.Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N Engl J Med 2015;373:726–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornet R, Chute CG. Health concept and knowledge management: twenty-five years of evolution. Yearb Med Inform 2016;Suppl 1:S32–S41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nadkarni PM, Darer JA. Migrating existing clinical content from ICD-9 to SNOMED. J Am Med Inform Assoc 2010;17:602–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baader F, Calvanese D, McGuinness DL, Nardi D, Patel-Schneider PF, eds. The description logic handbook: theory, implementation, and applications. New York: Cambridge University Press, 2003. (https://dl.acm.org/citation.cfm?id=885746). [Google Scholar]
- 9.National Library of Medicine. SNOMED CT. 2016. (https://www.nlm.nih.gov/healthit/snomedct). [DOI] [PubMed]
- 10.Nelson SJ, Zeng K, Kilbourne J, Powell T, Moore R. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc 2011;18:441–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bodenreider O The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004;32(Database issue):D267–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chute CG. Clinical classification and terminology: some history and current observations. J Am Med Inform Assoc 2000;7:298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson PN, Bauer S. Introduction to biol-ontologies. Boca Raton, FL: CRC Press, 2011. [Google Scholar]
- 14.Rath A, Olry A, Dhombres F, Brandt MMC, Urbero B, Ayme S. Representation of rare diseases in health information systems: the Orphanet approach to serve a wide range of end users. Hum Mutat 2012;33:803–8. [DOI] [PubMed] [Google Scholar]
- 15.Köhler S, Vasilevsky NA, Engelstad M, et al. The Human Phenotype Ontology in 2017. Nucleic Acids Res 2017;45(D1):D865–D876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hastings J, de Matos P, Dekker A, et al. The ChEBI reference database and ontology for biologically relevant chemistry: enhancements for 2013. Nucleic Acids Res 2013;41(Database issue):D456–D463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bandrowski A, Brinkman R, Brochhausen M, et al. The Ontology for Biomedical Investigations. PLoS One 2016;11(4):e0154556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res 2015;43(Database issue):D1049–D1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabregat A, Sidiropoulos K, Garapati P, et al. The Reactome pathway Knowledgebase. Nucleic Acids Res 2016;44(D1):D481–D487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pulteney R, Maton WG, Troilius C, von Linné C. A general view of the writings of Linnaeus. London: J. Mawman, 1805. (https://www.worldcat.org/title/general-view-of-the-writings-of-linnaeus/oclc/718424031&referer=brief_results). [Google Scholar]
- 21.Knibbs GH. The International classification of disease and causes of death and its revision. Med J Aust 1929;1:2–12. [Google Scholar]
- 22.Rea S, Pathak J, Savova G, et al. Building a robust, scalable and standards-driven infrastructure for secondary use of EHR data: the SHARPn project. J Biomed Inform 2012;45:763–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pathak J, Bailey KR, Beebe CE, et al. Normalization and standardization of electronic health records for high-throughput phenotyping: the SHARPn consortium. J Am Med Inform Assoc 2013;20(e2):e341–e348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conway M, Berg RL, Carrell D, et al. Analyzing the heterogeneity and complexity of Electronic Health Record oriented phenotyping algorithms. AMIA Annu Symp Proc 2011;2011:274–83. [PMC free article] [PubMed] [Google Scholar]
- 25.Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc 2013;20(e1):e147–e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pathak J, Wang J, Kashyap S, et al. Mapping clinical phenotype data elements to standardized metadata repositories and controlled terminologies: the eMERGE Network experience. J Am Med Inform Assoc 2011;18:376–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smoller JW. The use of electronic health records for psychiatric phenotyping and genomics. Am J Med Genet B Neuropsychiatr Genet 2017. May 30 (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans DA, Cimino JJ, Hersh WR, Huff SM, Bell DS. Toward a medical-concept representation language. J Am Med Inform Assoc 1994;1:207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campbell KE, Cohn SP, Chute CG, Rennels G, Shortliffe EH. Gálapagos: computer-based support for evolution of a convergent medical terminology. Proc AMIA Annu Fall Symp 1996:269–73. [PMC free article] [PubMed] [Google Scholar]
- 30.Pourzanjani A, Quisel T, Foschini L. Adherent use of digital health trackers is associated with weight loss. PLoS One 2016;11(4):e0152504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eriksson N, Macpherson JM, Tung JY, et al. Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet 2010;6(6):e1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vasilevsky NA, Foster ED, Engelstad ME, et al. Plain-language medical vocabulary for precision diagnosis. Nat Genet 2018;50:474–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rauch A, Wieczorek D, Graf E, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 2012;380:1674–82. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med 2013;369:1502–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 2014;312:1870–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu X, Petrovski S, Xie P, et al. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet Med 2015;17:774–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dragojlovic N, Elliott AM, Adam S, et al. The cost and diagnostic yield of exome sequencing for children with suspected genetic disorders: a benchmarking study. Genet Med 2018. January 4 (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 38.Willig LK, Petrikin JE, Smith LD, et al. Whole-genome sequencing for identification of Mendelian disorders in critically ill infants: a retrospective analysis of diagnostic and clinical findings. Lancet Respir Med 2015;3:377–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meng L, Pammi M, Saronwala A, et al. Use of exome sequencing for infants in intensive care units: ascertainment of severe single-gene disorders and effect on medical management. JAMA Pediatr 2017;171(12):e173438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan TY, Dillon OJ, Stark Z, et al. Diagnostic impact and cost-effectiveness of whole-exome sequencing for ambulant children with suspected monogenic conditions. JAMA Pediatr 2017;171:855–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Köhler S, Schulz MH, Krawitz P, et al. Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am J Hum Genet 2009;85:457–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zemojtel T, Köhler S, Mackenroth L, et al. Effective diagnosis of genetic disease by computational phenotype analysis of the disease-associated genome. Sci Transl Med 2014;6:252ra123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koç A, Karaer K, Ergün MA, Cinaz P, Perçin EF. A new case of hairy elbows syndrome (hypertrichosis cubiti). Genet Couns 2007;18:325–30. [PubMed] [Google Scholar]
- 44.Winnenburg R, Bodenreider O. Coverage of phenotypes in standard terminologies. In: Proceedings of the Joint Bio-Ontologies and BioLINK ISMB’2014 SIG session “Phenotype Day.” Bethesda, MD: National Library of Medicine, 2014:41–4. [Google Scholar]
- 45.Haendel MA, Balhoff JP, Bastian FB, et al. Unification of multi-species vertebrate anatomy ontologies for comparative biology in Uberon. J Biomed Semantics 2014;5:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Washington NL, Haendel MA, Mungall CJ, Ashburner M, Westerfield M, Lewis SE. Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol 2009;7(11):e1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith CL, Eppig JT. The Mammalian Phenotype Ontology as a unifying standard for experimental and high-throughput phenotyping data. Mamm Genome 2012;23:653–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mungall CJ, McMurry JA, Köhler S, et al. The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res 2016;45(D1):D718–D722. abstract (http://nar.oxfordjournals.org/content/early/2016/11/29/nar.gkw1128). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meehan TF, Conte N, West DB, et al. Disease model discovery from 3,328 gene knockouts by the International Mouse Phenotyping Consortium. Nat Genet 2017;49:1231–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson PN, Köhler S, Oellrich A, et al. Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome Res 2014;24:340–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smedley D, Jacobsen JO, Jäger M, et al. Next-generation diagnostics and disease-gene discovery with the Exomiser. Nat Protoc 2015;10:2004–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smedley D, Schubach M, Jacobsen JOB, et al. A whole-genome analysis framework for effective identification of pathogenic regulatory variants in Mendelian disease. Am J Hum Genet 2016;99:595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sifrim A, Popovic D, Tranchevent LC, et al. eXtasy: variant prioritization by genomic data fusion. Nat Methods 2013;10:1083–4. [DOI] [PubMed] [Google Scholar]
- 54.Singleton MV, Guthery SL, Voelkerding KV, et al. Phevor combines multiple biomedical ontologies for accurate identification of disease-causing alleles in single individuals and small nuclear families. Am J Hum Genet 2014;94:599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gall T, Valkanas E, Bello C, et al. Defining disease, diagnosis, and translational medicine within a homeostatic perturbation paradigm: the National Institutes of Health Undiagnosed Diseases Program experience. Front Med (Lausanne) 2017;4:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greene D, Richardson S, Turro E. Phenotype similarity regression for identifying the genetic determinants of rare diseases. Am J Hum Genet 2016;98:490–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stritt S, Nurden P, Turro E, et al. A gain-of-function variant in DIAPH1 causes dominant macrothrombocytopenia and hearing loss. Blood 2016;127:2903–14. [DOI] [PubMed] [Google Scholar]
- 58.Turro E, Greene D, Wijgaerts A, et al. A dominant gain-of-function mutation in universal tyrosine kinase SRC causes thrombocytopenia, myelofibrosis, bleeding, and bone pathologies. Sci Transl Med 2016;8:328ra30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chute CG. Clinical data retrieval and analysis: I’ve seen a case like that before. Ann N Y Acad Sci 1992;670:133–40. [DOI] [PubMed] [Google Scholar]
- 60.Lu CY, Williams MS, Ginsburg GS, Toh S, Brown JS, Khoury MJ. A proposed approach to accelerate evidence generation for genomic-based technologies in the context of a learning health system. Genet Med 2018;20:390–6. [DOI] [PubMed] [Google Scholar]
- 61.Hassanpour S, Langlotz CP. Information extraction from multi-institutional radiology reports. Artif Intell Med 2016;66:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burger G, Abu-Hanna A, de Keizer N, Cornet R. Natural language processing in pathology: a scoping review. J Clin Pathol 2016. July 22 (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 63.Faden RR, Kass NE, Goodman SN, Pronovost P, Tunis S, Beauchamp TL. An ethics framework for a learning health care system: a departure from traditional research ethics and clinical ethics. Hastings Cent Rep 2013;Spec No:S16–S27. [DOI] [PubMed] [Google Scholar]
- 64.Goodman D, Johnson CO, Bowen D, Smith M, Wenzel L, Edwards K. De-identified genomic data sharing: the research participant perspective. J Community Genet 2017;8:173–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Index. FHIR, version 3.0.1 (https://www.hl7.org/fhir/).
- 66.Quint JK, Donaldson GC, Hurst JR, Goldring JJ, Seemungal TR, Wedzicha JA. Predictive accuracy of patient-reported exacerbation frequency in COPD. Eur Respir J 2011;37:501–7. [DOI] [PubMed] [Google Scholar]
- 67.Beach WR. Patient Reported Outcomes Measurement Information System (PROMIS) may be our promise for the future. Arthroscopy 2017;33:1775–6. [DOI] [PubMed] [Google Scholar]
- 68.Chung AE, Basch EM. Incorporating the patient’s voice into electronic health records through patient-reported outcomes as the “review of systems.” J Am Med Inform Assoc 2015;22:914–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lloyd KC, Robinson PN, MacRae CA. Animal-based studies will be essential for precision medicine. Sci Transl Med 2016;8(352):352ed12. [DOI] [PubMed] [Google Scholar]
- 70.McMurry JA, Köhler S, Washington NL, et al. Navigating the phenotype frontier: the Monarch Initiative. Genetics 2016;203:1491–5. [DOI] [PMC free article] [PubMed] [Google Scholar]