Summary
Primary cilia are required for Smoothened to transduce vertebrate Hedgehog signals, but how Smoothened accumulates in cilia and is activated is incompletely understood. Here, we identify cilia-associated oxysterols that promote Smoothened accumulation in cilia and activate the Hedgehog pathway. Our data reveal that cilia-associated oxysterols bind to two distinct Smoothened domains to modulate Smoothened accumulation in cilia and tune the intensity of Hedgehog pathway activation. We find that the oxysterol synthase HSD11β2 participates in the production of Smoothened-activating oxysterols and promotes Hedgehog pathway activity. Inhibiting oxysterol biosynthesis impedes oncogenic Hedgehog pathway activation and attenuates the growth of Hedgehog pathway-associated medulloblastoma, suggesting that targeted inhibition of Smoothened activating oxysterol production may be therapeutically useful for patients with Hedgehog-associated cancers.
Introduction
Hedgehog proteins control developmental patterning and tissue homeostasis in evolutionarily diverse organisms (Briscoe and Therond, 2013). Misactivation of the Hedgehog (HH) pathway can lead to cancers, including medulloblastoma, the most common pediatric brain tumor, and basal cell carcinoma, the most common tumor in the United States. In vertebrates, HH signaling requires the primary cilium, an antenna-like projection on the surface of most cells. HH ligands relieve Patched1 (PTCH1) repression of Smoothened (SMO), allowing SMO to accumulate in cilia and activate GLI transcription factors (Briscoe and Therond, 2013). How SMO accumulates in cilia and is activated is incompletely understood.
Sterol lipids are required for vertebrate HH signaling, and both synthetic oxysterols and cholesterol can bind SMO to activate the downstream pathway (Byrne et al., 2016; Cooper et al., 2003; Corcoran and Scott, 2006; Dwyer et al., 2007; Huang et al., 2016, 2018; Luchetti et al., 2016; Myers et al., 2013, 2017; Nachtergaele et al., 2012, 2013; Nedelcu et al., 2013; Xiao et al., 2017). Synthetic oxysterols bind to the SMO N-terminal extracellular cysteine-rich domain (CRD), cause SMO to accumulate in cilia, activate the HH pathway, and stimulate the growth of cultured medulloblastoma cells (Corcoran and Scott, 2006; Dwyer et al., 2007; Myers et al., 2013; Nachtergaele et al., 2013; 2012; Nedelcu et al., 2013). Similarly, cholesterol binds to the SMO CRD and can induce HH signaling in neural progenitors, leading to the hypothesis that cholesterol is the endogenous ligand that activates SMO (Byrne et al., 2016; Huang et al., 2016, 2018; Luchetti et al., 2016). Phosphatidylinositol 4-phosphate is enriched in the ciliary membrane, revealing that the primary cilium can have a lipid composition distinct from that of other cellular membranes (Chavez et al., 2015; Garcia-Gonzalo et al., 2015). Therefore, we hypothesized that oxysterols that activate SMO may be present in primary cilia and may stimulate the HH pathway specifically in this subcellular context.
To define the oxysterol composition of cilia, we performed mass spectrometry of the membranes of isolated cilia. We identified endogenous cilia-associated oxysterols that bind SMO, cause SMO to accumulate in cilia, and activate the HH pathway. Moreover, we found that cilia-associated oxysterols activate the HH pathway through two separate domains of SMO. Either genetic or pharmacologic inhibition of HSD11β2, an oxysterol synthase, attenuates HH signal transduction and the growth of HH pathway-associated medulloblastoma. Thus, oxysterols found in primary cilia bind two distinct domains of SMO, cause SMO to accumulate in cilia, and activate the HH pathway to promote the growth of medulloblastoma.
Results
Identification of cilia-associated oxysterols
To identify ciliary oxysterols, we biochemically isolated cilia from sea urchin (Strongylocentrotus purpuratus) embryos, which require cilia to transduce HH signals (Figures 1A and 1B; Sigg et al., 2017; Warner et al., 2014). High-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) with normalization to protein content revealed that sea urchin embryo cilia are enriched in 7-keto-cholesterol (7k-C), 7β,27-dihydroxycholesterol (7β,27-DHC), 24-keto-cholesterol (24k-C), and 24,25-epoxycholesterol (24,25-EC) compared to either whole and de-ciliated embryos (Figures 1C, S1A, and S1B).
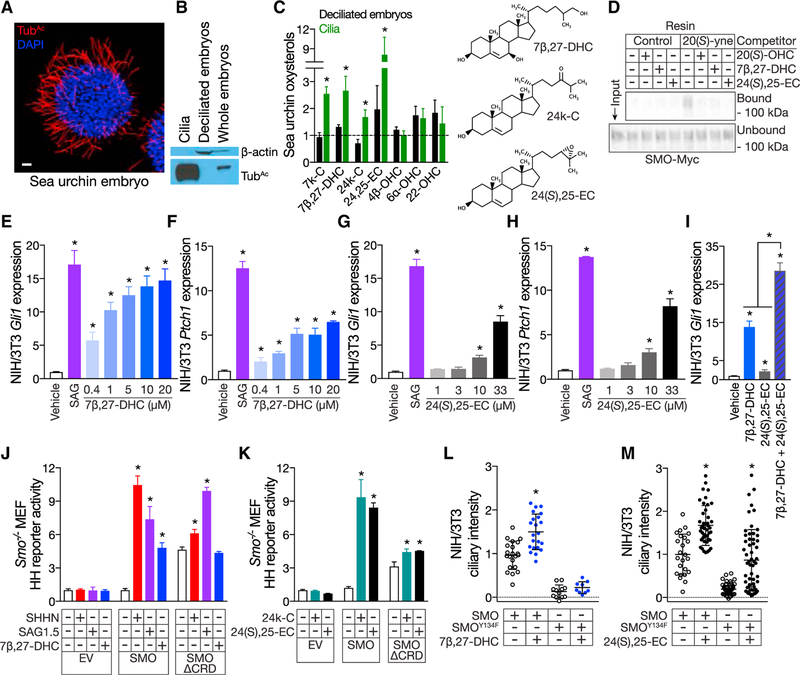
Figure 1. Cilia contain oxysterols that bind the CRD, activate the Hedgehog pathway and cause SMO to accumulate in cilia.
(A) Immunofluorescence of a sea urchin embryo stained for cilia (acetylated tubulin [TubAc], red) and nuclei (DAPI, blue). The scale bar represents 10μm.
(B) Immunoblot of lysates from sea urchin cilia, de-ciliated embryos, and whole sea urchin embryos. The ciliary fraction is enriched for TubAc and does not contain detectable components of the cytoplasm (β -actin).
(C) HPLC-MS/MS quantitation of oxysterols extracted from de-ciliated sea urchin embryos (black) and isolated cilia (green). Data are normalized to protein concentration and plotted relative to oxysterol levels in whole embryos (dashed line). 7k-C, 7β-DHC, 24k-C, and 24(S),25-EC are enriched in sea urchin embryo cilia.
(D) Anti-Myc immunoblot of detergent-solubilized membranes from HEK293S cells expressing SMO-Myc and incubated with 20(S)-yne affinity resin in the presence of 50 mM 20(S)-OHC, 7β-DHC, or 24(S),25-EC in ethanol. 20(S)-OHC, 7β-DHC, and 24(S),25-EC all interfere with the binding of SMO-Myc to 20(S)-yne affinity resin, indicating that they bind the CRD.
(E–H) qRT-PCR assessment of Gli1 (E and G) and Ptch1 (F and H) expression by ciliated NIH/3T3 cells treated with vehicle (ethanol), 100 nM SAG, 7β-DHC (E and F), or 24(S),25-EC (G and H). Data are normalized to vehicle control. 7β-DHC and 24(S),25-EC activate the HH pathway in a dose-dependent manner.
(I) qRT-PCR assessment of Gli1 expression by ciliated NIH/3T3 cells treated with vehicle (ethanol), 10 mM 7β-DHC, 10 mM 24(S),25-EC, or both. Data are normalized to vehicle control. 7β-DHC and 24(S),25-EC synergistically activate the HH pathway.
(J) Luciferase activity in ciliated Smo−/− MEFs co-transfected with Gli-luciferase reporter and empty vector (EV), SMO, or SMOΔCRD and treated with vehicle (ethanol), SHHN conditioned media, 10 nm SAG1.5, or ethanol complexed with 30 mM 7β-DHC. Data are normalized to activity in cells expressing SMO and treated with vehicle control. 7β-DHC requires the CRD to activate the HH pathway..
(K) Luciferase activity in ciliated Smo−/− MEFs co-transfected with Gli-luciferase reporter and EV, SMO, or SMOΔCRD and treated with vehicle (1 mM MβCD) or MβCD complexed with 30 μM cilia-associated oxysterols. Data are normalized to activity in cells expressing SMO and treated with vehicle control. 24k-C and 24(S),25-EC do not require the CRD to activate the HH pathway.
(L and M) Ciliary fluorescence intensity from NIH/3T3 cells stably expressing SMO-EGFP or SMOY134F-EGFP and treated with vehicle (ethanol) or 30 mM 7β,27-DHC (L) or 24(S),25-EC (M). Data are from 2 separate stable cell lines normalized to the average ciliary intensity of SMO-EGFP of cells treated with vehicle. Cilia-associated oxysterols induce SMO accumulation in cilia. Y134F substitution in the CRD blocks the effect of 7β,27-OHC, but not 24(S),25-EC, on ciliary accumulation, further suggesting that 24(S),25-EC can activate SMO independently of the CRD.
Histogram error bars show SEM. *p % 0.05; Student’s t test. See also Figures S1 and S2A.
Cilia-associated oxysterols bind to SMO, activate the Hedgehog pathway and cause SMO to accumulate in cilia
7k-C does not activate the HH pathway (Dwyer et al., 2007). As the other oxysterols that were enriched in sea urchin embryo cilia had not been previously investigated for roles in HH signaling, we examined whether they were able to bind to SMO. We incubated detergent-solubilized membranes from HEK293S cells expressing SMO with 20(S)-yne affinity resin and oxysterol competitors. Both 7β,27-DHC and 24(S),25-EC competed with 20(S)OHC for occupancy of the CRD, demonstrating that 7β,27-DHC and 24(S),25-EC bind to SMO (Figure 1D).
Treating ciliated NIH/3T3 cells with 7β,27-DHC and 24(S),25-EC showed that both can activate the HH pathway in a dose-dependent manner (Figures 1E–1H). Moreover, 7β-DHC and 24(S),25-EC synergized with one another to activate the HH pathway, suggesting that cilia-associated oxysterols may activate the HH pathway through multiple effectors or multiple domains within a single effector (Figure 1I).
Synthetic oxysterols and cholesterol require the SMO CRD to activate the HH pathway (Huang et al., 2016; Luchetti et al., 2016; Myers et al., 2013). To determine whether the SMO CRD is also necessary for cilia-associated oxysterols to activate the HH pathway, we co-transfected ciliated Smo−/− - mouse embryonic fibroblasts (MEFs) with a HH pathway luciferase reporter and SMO or SMO lacking the CRD (SMOΔCRD) and treated the cells with vehicle, 7β,27-DHC, 24k-C, or 24(S),25-EC. Similar to synthetic oxysterols and cholesterol, 7β,27-DHC was unable to activate the HH pathway through SMOΔCRD (Figure 1J). In contrast, 24k-C and 24(S),25-EC were able to activate the pathway through SMOΔCRD, albeit to a reduced extent, suggesting that these two oxysterols can function independently of the CRD through another domain of SMO (Figure 1K). Similar to 24k-C and 24(S),25-EC, Sonic Hedgehog (SHH) activated the HH pathway through SMOΔCRD to a reduced extent (Figure 1J). Together, these data indicate that 7β,27-DHC, 24k-C, and 24(S),25-EC are cilia-associated oxysterols capable of activating SMO.
To test whether any cilia-associated oxysterols promote the accumulation of SMO in cilia, we generated NIH/3T3 cell lines stably expressing EGFP fusions of wild-type SMO or SMO with the Y134F substitution (SMOY134F), which abolishes oxysterol interaction with the CRD (Nachtergaele et al., 2013). Quantification of ciliary immunofluorescence revealed that 7β,27-DHC, which requires the CRD to activate the HH pathway (Figure 1J), and 24(S),25-EC, which does not require the CRD (Figure 1K), both induced accumulation of SMO-EGFP in cilia (Figures 1L, 1M, and S2A). 7β,27-DHC did not cause SMOY134F-EGFP to accumulate in cilia (Figures 1L and S2A). In contrast, 24(S),25-EC caused SMOY134F-EGFP to accumulate in cilia, consistent with the ability of this oxysterol to activate the HH pathway independent of the SMO CRD (Figures 1M and S2A). Thus, 7β,27-DHC and 24(S),25-EC are oxysterols that are present in cilia and can promote the accumulation of SMO in cilia and activation of the HH pathway through distinct mechanisms.
Cilia-associated oxysterols activate the Hedgehog pathway through separate SMO domains
Although the SMO CRD is critical for 7β,27-DHC to activate SMO, the dispensability of the CRD for SMO activation by 24k-C and 24(S),25-EC raises the possibility that other domains of SMO may interact with oxysterols. Apart from the CRD, vertebrate SMO contains a site within the transmembrane domains of the heptahelical bundle (HHB) that binds small molecules, such as cyclopamine (CYA) (Chen et al., 2002a, 2002b). In addition, certain B-ring oxysterols can inhibit SMO at a site distinct from either the CRD or the HHB (Chen et al., 2002a; Sever et al., 2016), raising the possibility that there is another site within SMO that interacts with 24k-C and 24(S),25-EC.
To probe whether a cilia-associated oxysterol can bind to a domain of SMO other than the CRD, we synthesized 24k-CBODIPY (Figure S2B) and assessed its ability to interact with recombinant SMO or SMOΔCRD by fluorescence polarization anisotropy. Either SMO or SMOΔCRD increased the polarization of CYA-BODIPY fluorescence, consistent with the ability of CYA to interact with either the CRD or the HHB (Figure 2A; Chen et al., 2002a; Huang et al., 2016). Consistently, CYA-BODIPY demonstrated positively cooperative binding to SMO (Hill coefficient 1.6), but not to SMOΔCRD (Hill coefficient 0.8), and the micromolar Kd of CYA-BODIPY binding to SMO was four-fold lower than to SMOΔCRD (Byrne et al., 2016; Chen et al., 2002a). Like CYA-BODIPY fluorescence polarization, 24k-C-BODIPY fluorescence polarization increased in the presence of SMO (Figure 2B). 24k-C-BODIPY fluorescence polarization also increased in the presence of SMOΔCRD with a Kd equivalent to that for full-length SMO, indicating that 24k-CBODIPY can bind to a domain of SMO other than the CRD.
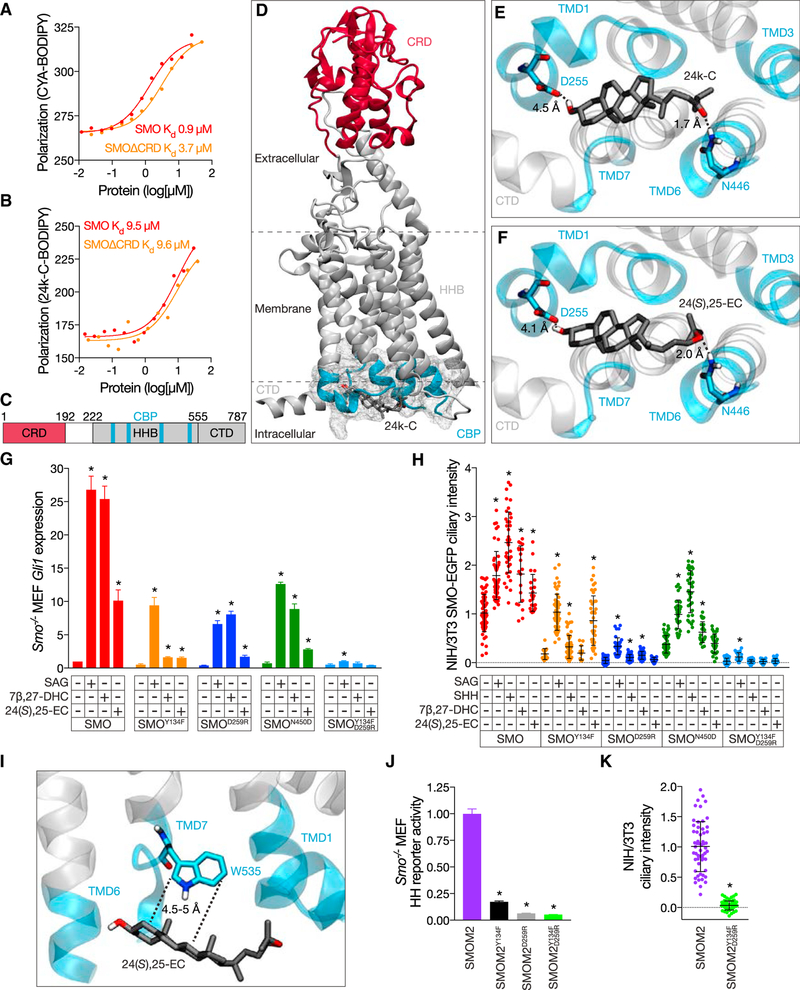
Figure 2. Cilia-associated oxysterols and the SMOM2 substitution function through the CBP.
(A and B) Florescence polarization anisotropy of human SMO and SMOΔCRD in response to 20 nM CYA-BODIPY (A) or 50 nM 24k-C-BODIPY (B). Data are shown in a.u. CYA-BODIPY binding to SMO is positively cooperative (Hill coefficient 1.6) with higher affinity than to SMOΔCRD (Hill coefficient 0.8). 24k-C-BODIPY binds to SMO and SMOΔCRD (Hill coefficients 1) with equivalent affinities.
(C) Schematic of SMO with relative positions of CRD (red) and a predicted oxysterol binding pocket at the membrane-proximal cytoplasmic surface (CBP, aqua) within the HHB (gray). The C-terminal domain (CTD) is also depicted in gray. Residue numbers demarcating domains are indicated.
(D–F) Docking of 24k-C and 24(S),25-EC against human SMO (PDB: 5L7D) predicts oxysterol binding to the CBP (gray mesh encompasses residues in aqua). The CBP is composed of intracellular loops and portions of transmembrane domains (TMDs) 1 (T251-I266), 3 (W339-L346), 6 (N446-I454), and 7 (W535-T553) and is distant from the CRD (red). Residues D255 and N446 (corresponding to D259 and N450 in mouse SMO) are predicted to form hydrogen bonds (dashed lines) with the carbon 3 hydroxyl and iso-octyl tail oxygens, respectively, of 24k-C (E) and 24(S),25-EC (F). Hydrogen bond lengths are shown in angstroms (Å).
(G) qRT-PCR assessment of Gli1 expression in ciliated Smo−/− MEFs expressing wild-type mouse SMO, SMOY134F, SMOD259R, SMON450D, or SMOY134F, D259R and treated with vehicle (ethanol), 100 nM SAG, or 30 μM 7β-DHC or 24(S),25-EC. Data are normalized to Gli1 expression in wild-type SMO-expressing cells treated with vehicle. Y134F substitution in the CRD attenuates the ability of 7β-DHC and 24(S),25-EC to induce Gli1, whereas D259R and N450D substitutions in the CBP attenuate the effect of 24(S),25-EC. Combined substitutions in the CRD and CBP block the effects of cilia-associated oxysterols and attenuate the effect of SAG.
(H) Ciliary fluorescence intensity of NIH/3T3 cells stably expressing mouse SMO-, SMOY134F-, SMOD259R-, SMON450D -, or SMOY134F, D259R-EGFP treated with vehicle (ethanol), 100 nM SAG, 1 μg/mL SHH, or 30 μM 7β-DHC or 24(S),25-EC. Data are from 2 separate cell lines normalized to the average intensity in cells expressing wild-type SMO-EGFP and treated with vehicle. Y134F substitution in the CRD specifically blocks the ability of 7β-DHC to induce SMO accumulation in cilia, whereas the D259R and N450D substitutions in the CBP block the effect 24(S),25-EC. Combined substitutions in the CRD and CBP block the effect of cilia-associated oxysterols and SHH and substantially attenuate the effect of SAG.
(I) Docking of 24(S),25-EC against human SMO predicts van der Waals interactions between cilia-associated oxysterols and W535 of the CBP, the residue mutated in SMOM2. Interaction lengths are shown in Å.
(J) Luciferase activity of ciliated Smo−/− MEFs co-transfected with Gli-luciferase reporter and oncogenic constitutively active form of SMO, SMOM2 (SMOW535L), SMOM2Y134F, SMOM2D259R, or SMOM2Y134F, D259R. Data are normalized to SMOM2 activity. Y134F substitution in the CRD, D259R substitution in the CBP, and combined substitutions in the CRD and CBP inhibit the activity of SMOM2.
(K) Ciliary fluorescence intensity of NIH/3T3 cells stably expressing mouse SMOM2- or SMOM2Y134F, D259R-EGFP. Data are from 2 separate cell lines normalized to the average ciliary intensity of cells expressing SMOM2-EGFP. Combined substitutions in the CRD and CBP block SMOM2 accumulation in cilia.
Histogram error bars show SEM. *p % 0.05; Student’s t test. See also Figures S2B–S2E.
To identify possible oxysterol binding sites outside of the CRD, we computationally docked the oxysterols 24k-C and 24(S),25-EC in human SMO with 4Å relaxation of side chains to mimic induced fit. We identified a possible binding pocket comprised of cytoplasmic-facing portions of transmembrane domain 1 (TMD1) (T251-I266), TMD3 (W339-L346), TMD6 (N446-I454), and TMD7 (W535-T553; Figures 2C and 2D). We named this site the cytoplasmic binding pocket (CBP). Within the CBP, modeling suggested that residues D255 and N446 in human SMO (corresponding to D259 and N450 in mouse SMO) could hydrogen bond with the carbon 3 hydroxyl and iso-octyl tail oxygens of 24k-C and 24(S),25-EC, respectively (Figures 2E and 2F). Therefore, we hypothesized that 24k-C and 24(S),25-EC activate SMO by binding to the CBP.
To test whether the CBP participates in oxysterol-mediated activation of SMO, we generated mutant forms of SMO with substitutions at either of the two residues predicted to hydrogen bond with oxysterols (D259R or N450D; Figure S2C). We transfected ciliated Smo−/− MEFs with wild-type SMO, SMOY134F, SMOD259R, or SMON450D; stimulated the HH pathway by adding SAG, SHH, 7β,27-DHC, or 24(S),25-EC; and assessed HH pathway activity. SAG activated wild-type and mutant SMO, although mutants showed decreased activity (Figures 2G and S2D). 7β,27-DHC activated SMOD259R and SMON450D equivalently to SAG but activated SMOY134F less than SAG, providing additional evidence that 7β,27-DHC functions through the CRD (Figure 2G). In contrast, 24(S),25-EC activation of SMOY134F, SMOD259R, and SMON450D was less than wild-type SMO, suggesting that activation by 24(S),25-EC involves both the CRD and the CBP (Figure 2G). Further supporting the idea that the CRD and CBP have partly overlapping functions in promoting SMO activity, neither 7β,27-DHC nor 24(S),25-EC could activate SMO containing substitutions in both the CRD and CBP (SMOY134F, D259R; Figure 2G). These data suggest that cilia-associated oxysterols activate SMO through both the CRD and CBP.
To determine whether the CBP regulates accumulation of SMO in cilia, we generated additional NIH/3T3 lines stably expressing EGFP fusions of SMOD259R, SMON450D, and SMOY134F, D259R (Figure S2C). Quantitative immunofluorescence showed that SHH and SAG caused SMOY134F-EGFP, SMOD259R-EGFP, and SMON450D-EGFP to accumulate in cilia, although to a lesser extent than wild-type SMO-EGFP (Figure 2H). 7β,27-DHC caused SMOD259R-EGFP and SMON450D-EGFP to accumulate in cilia, but not SMOY134F-EGFP, again consistent with 7β,27DHC requiring the CRD to activate the HH pathway (Figure 2H). In contrast, 24(S),25-EC failed to induce ciliary accumulation of either SMOD259R-EGFP or SMON450D-EGFP but did cause SMOY134F-EGFP to accumulate in cilia, further suggesting that 24(S),25-EC activity is dependent on the CBP (Figure 2H). Combining substitutions that inhibit the function of both the CRD and the CBP (SMOY134F, D259R) abolished the ability of either 7β,27DHC or 24(S),25-EC to promote ciliary accumulation (Figure 2H). Together, these results suggest that 7β,27-DHC acts through the CRD, 24(S),25-EC primarily acts through the CBP, and the CRD and CBP have overlapping functions in promoting the ciliary accumulation of SMO in cilia. In further support of that hypothesis, ciliary accumulation of SMOD259R-EGFP, SMON450D-EGFP, and SMOY134F-EGFP by SHH and SAG were diminished, and SHH failed to cause SMOY134F, D259R-EGFP to accumulate in cilia (Figure 2H).
SMOW535L, also called SMOM2, is a constitutively active, oncogenic form of SMO associated with HH pathway-associated medulloblastoma and basal cell carcinoma (Lam et al., 1999; Xie et al., 1998). How the W535L substitution activates SMO is unknown. We discovered that W535 is located within the CBP, and modeling suggested that W535 may form van der Waals interactions with oxysterols (Figure 2I). To determine whether residues involved in oxysterol binding are necessary for SMOM2 activity, we expressed SMOM2, SMOM2Y134F, SMOM2D259R, or SMOM2Y134F, D259R in ciliated Smo−/− MEFs. Y134F and D259R substitutions reduced SMOM2 activity, and combined Y134F and D259R substitutions blocked SMOM2 activity, suggesting that the CRD and CBP are both important for oncogenic HH signaling by SMOM2 (Figures 2J and S2D).
To determine whether the CRD and CBP are important for ciliary accumulation of SMOM2, we generated stable NIH/3T3 lines expressing fusions of EGFP with SMOM2 or SMOM2 with combined Y134F and D259R substitutions. Consistent with the functional data, quantitative immunofluorescence showed that SMOM2Y134F, D259R-EGFP failed to accumulate in cilia, indicating that the CRD and CBP are critical for mediating the constitutive ciliary localization of SMOM2 (Figures 2K and S2E). Thus, as with wild-type SMO, the CRD and CBP promote the ability of SMOM2 to activate the HH pathway.
The oxysterol synthase HSD11β2 participates in SMO-mediated activation of the Hedgehog pathway
We hypothesized that enzymes involved in cilia-associated oxysterol biosynthesis would be enriched in domains of active HH signaling. To begin to test that hypothesis, we performed RNA sequencing of the mouse Math1-Cre SmoM2c/WT model of HH-pathway-associated medulloblastoma, in which constitutively active SMOM2 is specifically expressed in the cerebellar external granule layer (EGL). Among the diverse oxysterol synthases expressed in medulloblastoma, hydroxysteroid 11-β dehydrogenase 2 (Hsd11β2) displayed the highest differential expression and was 864± 82-fold higher in Math1-Cre SmoM2c/WT medulloblastomas than in control cerebella (Figures 3A, S3A, and S3B). Similar to mouse medulloblastoma, HSD11β2 was also enriched in human HH pathway-associated medulloblastoma (Figures 3B, S3C, and S3D).
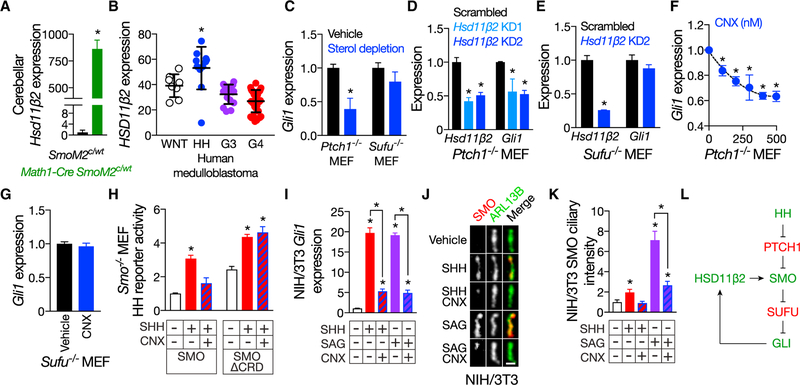
Figure 3. HSD11β2 promotes Hedgehog pathway activation and cause SMO to accumulate in cilia in a manner that requires the CRD.
(A) RNA sequencing of medulloblastomas from P35 Math1-Cre SmoM2c/WT mice compared to control cerebella of P35 SmoM2c/WT littermates indicates that HSD11β2 expression is 864 ± 82-fold higher in HH pathway-associated medulloblastoma.
(B) Re-analysis of human transcriptome data SPR008292 indicates that HSD11β2 expression is highest in HH pathway-associated medulloblastoma. Data are shown in a.u.
(C) qRT-PCR assessment of Gli1 expression in ciliated Ptch1−/− and Sufu−/− MEFs treated with vehicle (water) or 1% MβCD and 20 μM pravastatin to deplete sterols. Data are normalized to vehicle treatment. Sterol depletion inhibits HH pathway activation caused by loss of PTCH1, but not loss of SUFU, consistent with a critical role for sterols in SMO activation.
(D) qRT-PCR assessment of HSD11β2 and Gli1 expression in ciliated Ptch1−/− MEFs transduced with 1 of 2 different HSD11β2 shRNAs. Data are normalized to expression in scrambled shRNA control-transduced cells. HSD11β2 knockdown (KD) inhibits HH pathway activation downstream of PTCH1.
(E) qRT-PCR assessment of HSD11β2 and Gli1 expression in ciliated Sufu−/− MEFs transduced with HSD11β2 shRNAs. Data are normalized to expression in scrambled shRNA control-transduced cells. HSD11β2 KD does not inhibit HH pathway activation caused by loss of SUFU, consistent with HSD11β2 acting at the level of SMO.
(F) qRT-PCR assessment of Gli1 expression in ciliated Ptch1−/− MEFs treated with vehicle (water) or CNX. Data are normalized to expression in vehicle-treated cells. Pharmacologic inhibition of HSD11β2 inhibits HH signaling downstream of PTCH1 in a dose-dependent manner.
(G) qRT-PCR assessment of Gli1 expression in ciliated Sufu−/− MEFs treated with vehicle (water) or 400 nM CNX. Data are normalized to expression in vehicle- treated cells. Pharmacologic inhibition of HSD11β2 does not inhibit HH pathway activation caused by loss of SUFU.
(H) Luciferase activity of ciliated Smo−/− MEFs co-transfected with Gli-luciferase reporter and SMO or SMOΔCRD and treated with vehicle (water) or 1 μg/mL SHH, with or without 400 nM CNX. Data are normalized to luciferase activity of SMO-expressing cells treated with vehicle. The CRD is required for CNX to block HH pathway stimulation by SHH.
(I) qRT-PCR assessment of Gli1 expression in ciliated NIH/3T3 cells treated with vehicle (water), 1 μg/mL SHH, or 100 nM SAG, with or without 400 nM CNX. Data are normalized to expression in vehicle-treated cells. Pharmacologic inhibition of HSD11β2 blocks HH pathway stimulation by SHH or SAG.
(J) Immunofluorescence of endogenous SMO (red) localization to cilia (ARL13B, green) in NIH/3T3 cells treated with vehicle (water), 1 μg/mL SHH, or 100 nM SAG, with or without 400 nM CNX. The scale bar represents 1 μm.
(K) Quantitation of SMO ciliary immunofluorescence normalized to intensity in vehicle-treated cells. Pharmacologic inhibition of HSD11β2 blocks SMO accumulation in cilia by SHH or SAG.
(L) Model of the HSD11β2-mediated HH pathway activation. Pathway activators are shown in green. Pathway inhibitors are shown in red. Histogram error bars show SEM. *p % 0.05; Student’s t test. See also Figure S3.
During development, HSD11β2 is expressed in domains of active HH signaling, including the EGL (Heine and Rowitch, 2009; Naray-Fejes-Toth and Fejes-Toth, 2007). Given the developmental expression pattern of HSD11β2, and the finding that HSD11β2 is dramatically upregulated in HH pathway-associated medulloblastoma, we hypothesized that HSD11β2 could participate in the production of SMO-activating oxysterols.
PTCH1 represses the HH pathway upstream of SMO, whereas SUFU represses the HH pathway by binding to GLI transcription factors downstream of SMO (Briscoe and Therond, 2013). Sterol depletion blocked HH pathway activity in ciliated Ptch1−/− MEFs but had no effect in ciliated Sufu−/− MEFs, consistent with a role for sterols downstream of PTCH1 and upstream of SUFU (Figure 3C). To test the hypothesis that HSD11β2 potentiates HH pathway induction, we depleted HSD11β2 using short hairpin RNAs (shRNAs) in ciliated Ptch1−/− and Sufu−/− MEFs (Figure S3E). Like sterol depletion, depleting HSD11β2 attenuated HH signaling in Ptch1−/− MEFs, but not Sufu−/− MEFs, suggesting that HSD11β2 modulates HH signaling downstream of PTCH1 and upstream of SUFU (Figures 3D and 3E).
HSD11β2 is inhibited by a compound in licorice (Farese et al., 1991; Monder et al., 1989). We hypothesized that carbenoxolone (CNX), a derivative of the HSD11β2 inhibitor in licorice, would block HH signaling by inhibiting oxysterol production. In support of that hypothesis, we found that CNX reduced HH pathway activity in a dose-dependent manner in Ptch1−/− MEFs, but not Sufu−/− MEFs (Figures 3F and 3G).
To determine whether HSD11β2 regulates SMO, we expressed SMO or SMOΔCRD in ciliated Smo−/− MEFs, stimulated with SHH with or without HSD11β2 inhibition, and measured pathway activity. CNX inhibition of HSD11β2 reduced HH signaling through SMO but had no effect on signaling through SMOΔCRD, suggesting that HSD11β2 promotes the production of oxysterols that act through the CRD (Figure 3H). Competition assays with BODIPY-CYA demonstrated that CNX does not directly antagonize SMO at the CYA-binding pocket (Figures S3G and S3H).
Given that cilia-associated oxysterols can bind the CRD and induce SMO accumulation in cilia, we hypothesized that HSD11β2 would also regulate SMO accumulation in cilia. Indeed, in NIH/3T3 cells stimulated with either SHH or SAG, pharmacologic inhibition of HSD11β2 reduced the ciliary accumulation of SMO (Figures 3J, 3K, and S3F). Together, these data indicate that HH signaling induces expression of HSD11β2, which promotes SMO activity through production of oxysterols that act at the CRD (Figure 3L).
HSD11β2 and CYP27A1 participate in the production of SMO-activating oxysterols
Overexpression of HSD11β2 in ciliated NIH/3T3 cells conferred resistance to CNX but did not activate the HH pathway (Figures S3I and S3J). One interpretation of these data is that HSD11β2 generates a precursor molecule that requires downstream processing by a rate-limiting enzyme to activate SMO. 7k-C is present in sea urchin embryo cilia (Figure 1C), and we hypothesized that HSD11β2 converts 7β-OHC to 7k-C as a precursor for biosynthesis of 7-keto,27-hydroxycholesterol (7k,27OHC) and 7β,27-DHC, both of which bind to SMO and promote SMO activity (Myers et al., 2013) (Figure 4A). To test whether HSD11β2 can generate 7k-C, we cultured HEK293T cells overexpressing HSD11β2 with 7β-OHC or the related oxysterol 7a-OHC and used HPLC-MS/MS to assess 7k-C production. HSD11β2 increased 7k-C production in 7β-OHC-treated cells, but not in 7a-OHC-treated cells (Figure 4B). 7k-C production in HEK293T cells overexpressing HSD11β2 was inhibited by CNX (Figure 4B), suggesting that HSD11β2 converts 7β-OHC to 7k-C.
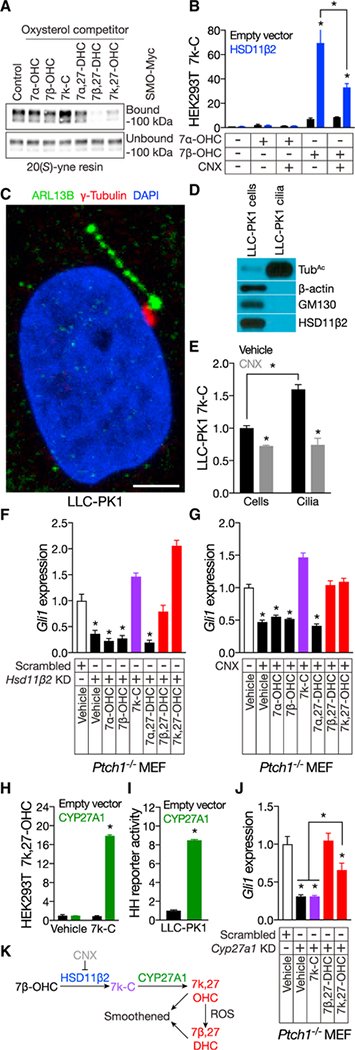
Figure 4. HSD11β2 and CYP27A1 participate in the biosynthesis of SMO-activating oxysterols.
(A) Anti-Myc immunoblot of detergent-solubilized membranes from HEK293S cells expressing Myc-tagged SMO and incubated with 20(S)-yne affinity resin in the presence of 50 mM of the indicated oxysterols. 7β-DHC and 7k,27-OHC, but not 7k-C, compete for occupancy of 20(S)-yne affinity resin, indicating that 7β-DHC and 7k,27-OHC bind the CRD.
(B) HPLC-MS/MS measurement of 7k-C in HEK293T cells transfected with empty vector or expressing HSD11β2 and incubated with vehicle (ethanol) or 10 mM 7a-OHC or 7β-OHC, with or without 400 nM CNX. Data are normalized to deuterated 7k-C internal standards and vehicle-treated cells transfected by CNX.
(C) Immunofluorescence of a primary cilium (ARL13B, green), basal body (γ-tubulin, red), and nucleus (DAPI, blue) of a LLC-PK1 cell. The scale bar represents 5 μm.
(D) Immunoblot of lysates from LLC-PK1 cells and isolated cilia. The ciliary fraction is enriched for the ciliary component acetylated tubulin (TubAc) and does not contain detectable components of the cytoplasm (β-actin) or Golgi (GM130) or HSD11β2.
(E) HPLC-MS/MS measurement of 7k-C in LLC-PK1 cells and isolated cilia treated with vehicle (ethanol) or 400 nM CNX. Data are normalized to protein concentration in each sample relative to 7k-C of cells treated with vehicle. LLC-PK1 cilia are enriched in 7k-C, and pharmacologic inhibition of HSD11β2 reduces cellular and ciliary 7k-C.
(F) qRT-PCR assessment of Gli1 expression in ciliated Ptch1−/− MEFs transduced with scrambled control or HSD11β2 shRNAs and treated with 1 mM MβCD vehicle or 30 μM of the indicated oxysterols. Data are normalized to expression in cells transduced with scrambled shRNA and treated with vehicle. 7k-C, 7β-DHC, and 7k,27-OHC restore HH signaling to HSD11β2-depleted cells.
(G) qRT-PCR assessment of Gli1 expression in ciliated Ptch1−/− MEFs treated with 1 mM MβCD vehicle, 400 nM CNX, and 30 μM of the indicated oxysterols. Data are normalized to Gli1 expression in vehicle-treated cells. 7k-C, 7b,27-DHC, and 7k,27-OHC restore HH signaling after pharmacologic inhibition of HSD11β2.
(H) HPLC-MS/MS measurement of 7k,27-OHC in HEK293S cells transfected with empty vector or expressing CYP27A1 and incubated with vehicle (ethanol) or 10 μM 7k-C. Data are normalized to deuterated 7k,27-OHC internal standards and vehicle-treated cells transfected with empty vector. CYP27A1 converts 7k-C to 7k,27-OHC.
(I) Luciferase activity of ciliated LLC-PK1 cells co-transfected with Gli-luciferase reporter and empty vector or CYP27A1. Data are normalized to activity in empty vector transfected cells. CYP27A1 expression in LLC-PK1 cells is sufficient to activate HH pathway activity.
(J) qRT-PCR assessment of Gli1 expression in ciliated Ptch1−/− MEFs transduced with scrambled control or Cyp27a1 shRNAs and treated with 1 mM MβCD vehicle or 30 μM of the indicated oxysterols. Data are normalized to expression in cells transduced with scrambled shRNA and treated with vehicle. 7β-DHC and 7k,27-OHC, but not 7k-C, restore HH pathway activity in Cyp27a1-depleted cells.
(K) Model of the biosynthesis of SMO-activating oxysterols.
Histogram error bars show SEM. *p % 0.05; Student’s t test. See also Figures S1 and S3.
To assess whether HSD11β2 participates in the production of cilia-associated oxysterols, we biochemically isolated cilia from pig (Sus scrofa) LLC-PK1 renal cells (Figures 4C and 4D), which express Hsd11β2 (Figure S3K). HPLC-MS/MS analysis revealed that LLC-PK1 cilia, like sea urchin cilia, were enriched in 7k-C (Figures 4E, S1C, and S1D). Pharmacologic inhibition of HSD11β2 reduced 7k-C in LLC-PK1 cells and cilia (Figure 4E), further indicating that HSD11β2 generates 7k-C.
To test the hypothesis that HSD11β2 converts 7β-OHC to 7k-C as a precursor for the biosynthesis of SMO-activating oxysterols, we cultured ciliated Ptch1−/− MEFs in oxysterols and Hsd11β2 shRNAs or CNX. Following genetic or pharmacologic inhibition of HSD11β2, the addition of 7k-C, 7β,27-DHC, and 7k,27-OHC restored HH pathway activity (Figures 4F, 4G, and S3L). Thus, 7k-C, 7β,27-DHC, and 7k,27-OHC function downstream of HSD11β2 to activate the HH pathway.
Sterol 27-hydroxylase (CYP27A1) converts 7k-C into 7k,27OHC (Figure 4H; Heo et al., 2011), and we hypothesized that CYP27A1 acts downstream of HSD11β2 to generate SMO-activating oxysterols. LLC-PK1 cells do not transduce HH signals and do not express Cyp27a1 (Figure S3K). However, expression of CYP27A1 in LLC-PK1 cells was sufficient to activate the HH transcriptional program, suggesting that absence of CYP27A1 limits the production of a SMO-activating oxysterol in LLC-PK1 cells (Figure 4I). Like HSD11β2, shRNA-mediated depletion of CYP27A1 inhibited HH pathway activity in Ptch1−/− MEFs (ures 4J and S3M). 7β-DHC and 7k,27-OHC, but not 7k-C, restored HH signaling following depletion of CYP27A1 (ure 4J). Thus, CYP27A1 functions downstream of 7k-C in the pathway of SMO-activating oxysterol biosynthesis. A model that accounts for these data is that (1) HSD11β2 oxidizes 7β-OHC to 7k-C, (2) CYP27A1 oxidizes 7k-C to 7k,27-OHC, and (3) reactive oxygen species reduce 7k,27-OHC to 7β,27-DHC (Figure 4K). Both 7k,27-OHC and 7β,27-DHC bind to the CRD, cause SMO to accumulate in cilia, and induce HH pathway activity.
Oxysterol biosynthesis potentiates cerebellar and oncogenic Hedgehog signaling
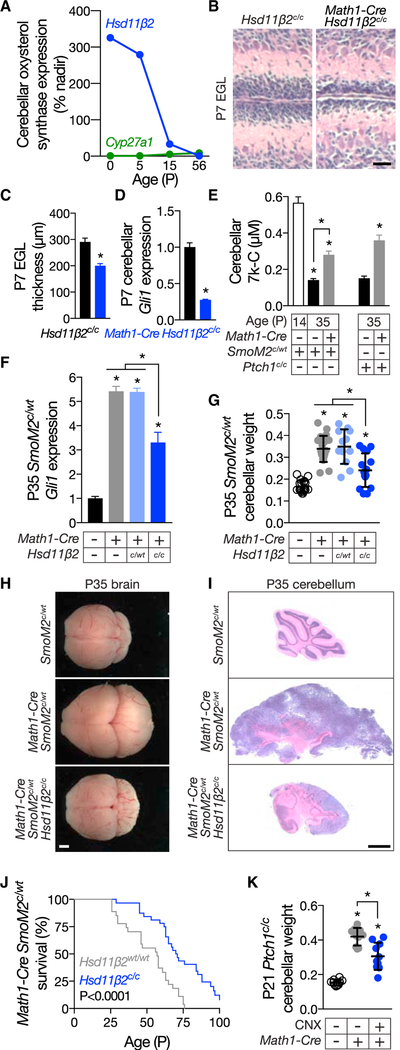
Hsd11β2 is strongly expressed in the neonatal cerebellum, raising the possibility that it participates in cerebellar development (Figure 5A; Pal et al., 2011). To test that hypothesis, we conditionally deleted Hsd11β2 (Hsd11β2c/c; Jiang et al., 2013) in the EGL using Math1-Cre and found that Math1-Cre Hsd11β2c/c mice are subviable (p = 0.01; chi-square test). HH signaling promotes EGL proliferation (Corrales et al., 2006), and in mice surviving to P7, deletion of Hsd11β2 decreased EGL thickness and reduced cerebellar HH pathway activity (Figures 5B–5D).
Figure 5. HSD11β2 promotes developmental and oncogenic Hedgehog signaling.
(A) Re-analysis of mouse transcriptome data GSE23525 indicates that cerebellar Hsd11β2 expression decreases from birth to postnatal day 56. In contrast, cerebellar Cyp27a1 expression remains largely stable. Data are shown as percent of nadir.
(B) H&E-stained micrographs of P7 control Hsd11β2c/c and Math1-Cre Hsd11β2c/c cerebella. The scale bar represents 30 μm.
(C) Quantitation of EGL thickness in Hsd11β2c/c and Math1-Cre Hsd11β2c/c cerebella. Homozygous genetic deletion of Hsd11β2 restricts EGL growth.
(D) qRT-PCR assessment of Gli1 expression in P7 control Hsd11β2c/c and Math1-Cre Hsd11β2c/c cerebella. Data are normalized to expression in control cerebella. Homozygous deletion of Hsd11β2 attenuates HH pathway activity during cerebellar development.
(E) HPLC-MS/MS measurement of 7k-C in control SmoM2c/WT and Ptch1c/c cerebella during development (P14) and/or adulthood (P35) as compared to Math1-Cre SmoM2c/WT and Math1-Cre Ptch1c/c medulloblastomas. 7k-C levels are normalized to sample weights. 7k-C levels are elevated at P14 cerebella and in HH-pathway-associated medulloblastoma.
(F) qRT-PCR assessment of Gli1 expression in P35 control SmoM2c/WT cerebella and Math1-Cre SmoM2c/WT, Math1-Cre SmoM2c/WT Hsd11β2c/WT, and Math1-Cre SmoM2c/WT Hsd11β2c/c medulloblastomas. Data are normalized to expression in control cerebella. Homozygous deletion of Hsd11β2 attenuates HH pathway activity in medulloblastoma.
(G) Weight of P35 control SmoM2c/WT cerebella and Math1-Cre SmoM2c/WT, Math1-Cre SmoM2c/WT Hsd11β2c/WT, and Math1-Cre SmoM2c/WT Hsd11β2c/c medulloblastomas normalized to total brain weight. Homozygous deletion of Hsd11β2 blocks the growth of HH-pathway-associated medulloblastoma caused by activation of SMO.
(H) Gross images of P35 control SmoM2c/WT, Math1-Cre SmoM2c/WT, and Math1-Cre SmoM2c/WT Hsd11β2c/c brains. Homozygous deletion of Hsd11β2 attenuates the growth of HH-pathway-associated medulloblastoma caused by activation of SMO. The scale bar represents 5 mm. (I) Sagittal H&E-stained sections of P35 control SmoM2c/WT cerebella and Math1-Cre SmoM2c/WT and Math1-Cre SmoM2c/WT HSD11β2c/c medulloblastoma. Homozygous deletion of HSD11β2 reduces the number of small round blue tumor cells and partially restores cerebellar architecture in HH-pathway-associated medulloblastoma. The scale bar represents 2 mm. (J) Kaplan-Meier curves of 27 Math1-Cre SmoM2c/WT and 32 Math1-Cre SmoM2c/WT HSD11β2c/c mice. Homozygous deletion of HSD11β2 prolongs the survival of mice with HH-pathway-associated medulloblastoma by 25% (56 days versus 70 days; p < 0.0001; log rank test). (K) Weight of P21 Ptch1c/c control cerebella and Math1-Cre Ptch1c/c medulloblastomas treated with vehicle (water) or 100 mg/g CNX by intraperitoneal injection for 2 weeks, normalized to total brain weight. Pharmacologic inhibition of HSD11β2 attenuates the growth of HH-pathway-associated medulloblastoma caused by loss of Ptch1. Histogram error bars show SEM. *p % 0.05; Student’s t test. See also Figures S4 and S5.
Using HPLC-MS/MS and SmoM2c/WT cerebella, we found that 7k-C was elevated at P14, when the cerebellum is still developing (Figures 5E and S4). At P35, cerebellar 7k-C decreased, paralleling decreased Hsd11β2 expression (Figures 5A and 5E). Constitutive HH pathway activation in the EGL of Math1-Cre SmoM2c/WT or Math1-Cre Ptch1c/c mouse models of HH pathway-associated medulloblastoma increased 7k-C in the P35 cerebella (Figures 5E and S4), consistent with HH pathway stimulation of HSD11β2 expression in cerebellar granule cell precursors (Heine and Rowitch, 2009).
HSD11β2 converts cortisol to cortisone in the kidney, raising the possibility that HSD11β2 promotes granule cell proliferation by metabolizing glucocorticoids (Heine and Rowitch, 2009). However, deletion of HSD11β2 in the cerebellum did not affect glucocorticoid target gene expression (Figures S5A and S5B), suggesting that HSD11β2 may not be critical for restraining physiological levels of glucocorticoids in the cerebellum. In further contrast to glucocorticoids, pharmacologic inhibition of HSD11β2 in vivo did not increase medulloblastoma apoptosis (Figures S5C and S5D).
In support of the possibility that oxysterol synthases producing SMO-activating ligands participate in pathogenic signaling, homozygous genetic deletion of Hsd11β2 from the EGL of the Math1-Cre SmoM2c/WT mice reduced HH pathway activity in medulloblastoma (Figure 5F). Homozygous genetic deletion of Hsd11β2 also reduced tumor weight and the prevalence of small round blue cells characteristic of medulloblastoma, as well as partially restored cerebellar architecture (Figures 5G–5I). These changes were associated with prolongation in survival of Math1-Cre SmoM2c/WT Hsd11β2c/c mice relative to Math1-Cre SmoM2c/WT animals (70 days versus 56 days; p < 0.0001; log rank test; Figure 5J).
We hypothesized that, like genetic deletion of Hsd11β2, CNX-mediated inhibition of HSD11β2 might inhibit the growth of HH pathway-associated cancer. Treatment of cultured basal cell carcinoma (ASZ) and medulloblastoma (Med1) cells with CNX reduced HH pathway activity (Figures S5E and S5F). Mass spectrometry confirmed that CNX crosses the blood brain barrier in vivo (Figure S5G; Heine and Rowitch, 2009). Therefore, we treated Math1-Cre Ptch1c/c mice with CNX. Similar to homozygous genetic deletion of Hsd11β2 in Math1-Cre SmoM2c/WT mice, pharmacological inhibition of HSD11β2 reduced tumor weight and the prevalence of small round blue cells in Math1-Cre Ptch1c/c medulloblastomas (Figures 5K, S5H, and S5I).
Discussion
We have identified cilia-associated oxysterols that promote ciliary accumulation of SMO and activate the HH pathway through two separate domains of SMO. The CRD, an N-terminal extracellular domain, was previously identified as a site of action for synthetic oxysterols and cholesterol (Myers et al., 2013; Nachtergaele et al., 2012, 2013; Nedelcu et al., 2013). The second site, the CBP, is at the membrane-cytoplasmic interface, and like the CRD, substitutions within the CBP abrogate SMO activity. HSD11β2, an oxysterol synthase expressed in regions of active HH signaling, participates in the production of SMO-activating oxysterols that activate the CRD. Consistently, we find that either genetic deletion of HSD11β2 or pharmacologic inhibition with a compound from licorice diminishes HH signal transduction and the growth of HH pathway-associated cancer. Recent structures of SMO with cholesterol or 20(S)-OHC in the CRD reveal interactions between the 3β-hydroxyl group of sterols and a SMO CRD aspartate (D99 in mouse SMO; Byrne et al., 2016; Huang et al., 2016, 2018; Luchetti et al., 2016). SMO D99 is essential for the ability of the CRD to bind sterols, indicating that CRD binding of the 3b-hydroxyl group of sterols is important for SMO activation (Huang et al., 2016; Nedelcu et al., 2013). The 3β-hydroxyl group is also present in cilia-associated oxysterols, raising the possibility that cilia-associated oxysterols bind the CRD in a similar conformation. In the CBP, modeling and mutagenesis similarly suggest that interaction between the oxysterol 3β-hydroxyl group and SMO D259 are important for SMO activation.
Interestingly, W535, commonly mutated to L to oncogenic activate SMO, is located within the CBP (Xie et al., 1998). How the W535L substitution constitutively activates SMOM2 is unknown. As the activity of SMOM2 depends on other CBP residues, we speculate that W535L induces a conformational change that mimics oxysterol binding. In support of the possibility that SMOM2 does not depend on oxysterol binding to the CBP, SMOM2 activity is independent of a cholesterol biosynthetic enzyme required for wild-type SMO activity (Blassberg et al., 2016).
For an oxysterol to be a SMO agonist, it must be present in HH-responsive cells, it must bind and activate SMO, and it must be required for pathway activity. We have shown that 7β-DHC, 24k-C, and 24,25-EC are produced by sea urchin and mammalian cells and bind and activate SMO. Moreover, the contribution of HSD11β2 to oncogenic HH signaling suggests that the oxysterols produced by enzymes are required for high-level pathway activity, providing evidence that oxysterols are relevant endogenous SMO agonists.
We propose that the SMO CRD and CBP are activated through binding oxysterols. As SMO-activating oxysterols are enriched in the ciliary membrane, perhaps SMO only encounters its cognate oxysterols once it has accessed this specialized compartment. Alternatively, SMO may encounter oxysterols elsewhere where they induce a conformation that stimulates SMO accumulation in cilia. In either model, the involvement of oxysterols activating SMO through two sites is reminiscent of the “two-person concept,” which requires two operators to unlock separate locks to launch a missile, a system designed to increase the fidelity of critical decisions (Woodward, 2013).
The ability of CNX to suppress medulloblastoma growth suggests that inhibiting oxysterol biosynthesis may be a useful approach to targeting HH pathway-associated cancers. Given the selective enrichment of HSD11β2 among molecular subgroups of medulloblastoma, we anticipate that oxysterol biosynthesis inhibition may only be useful for HH pathway-associated tumors. Indeed, the expression of HSD11β2 is highly cell-type specific (Heine and Rowitch, 2009; Naray-Fejes-Toth and Fejes-Toth, 2007). Therefore, inhibiting oxysterol synthases may be a strategy to inhibit the HH pathway in a tissue-specific manner, thereby lessening the toxicity of HH pathway inhibition that has limited the clinical utility of SMO antagonists in pediatric cancer patients (Lucas and Wright, 2016).
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jeremy F. Reiter (jeremy.reiter@ucsf.edu).
METHOD DETAILS
Cell Culture
Med1 cells, and Ptch1−/−, Smo−/− and Sufu−/− MEFs, were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Thermo Fisher Scientific, Waltham, MA) supplemented with 10% fetal bovine serum (FBS) and glutamine. NIH/3T3, Flp-IN-3T3 and SHH Light II cells were cultured similarly except in 10% fetal calf serum. ASZ cells were cultured in 154-CF media (Thermo Fisher Scientific) supplemented with glutamine, pen/strep and 4% FBS chelexed to remove calcium (6.7 parts chelexed FBS plus 1 part non-chelexed FBS). LLC-PK1 cells were cultured in Media 199 (Thermo Fisher Scientific) supplemented with 3% FBS, bicarbonate and glutamine.
To promote ciliogenesis, cell lines other than LLC-PK1 cells were cultured in Opti-MEM (Thermo Fisher Scientific). SHH, SAG, CYA and vismodegib were added for 24 hr; glucocorticoids, oxysterols, and sterols were added for 30–36 hr; and CNX was added for 48 hr.
Chemistry
Reagents and solvents were purchased from Sigma-Aldrich and used as received unless otherwise indicated. A synthetic mechanism is diagramed in Figure S2B. Compound 1 was prepared as previously described (Windsor et al., 2013). Flash column chromatography was carried out using a Biotage Isolera Four system and SiliaSep silica gel cartridges from Silicycle. 1H NMR spectra were recorded on a Varian INOVA-400 400MHz spectrometer. Chemical shifts are reported in d units (ppm) relative to residual solvent peak. Coupling constants (J) are reported in hertz (Hz). Characterization data are reported as follows: chemical shift, multiplicity (s = singlet, d = doublet, t = triplet, m = multiplet), coupling constants, number of protons and mass to charge ratio. LCMS analyses were performed on a Waters Micromass ZQ/Waters 2795 Separation Module/Waters 2996 Photodiode Array Detector/Waters 2424 Evaporative Light Scattering Detector system.
To synthesize Compound 2 (Figure S2B), a 1 M solution of tetrabutyl ammonium fluoride in tetrahydrofuran (0.126 ml, 0.1 mmol) was added to a solution of (6R)-6-[(2R,5S,15R)-5-[(tert-butyldimethylsilyl)oxy]-2,15-dimethyltetracyclo[8.7.0.02,7.011,15]heptadec-7en-14-yl]hept-1-yn-3-ol (1)(0.045 g, 0.1 mmol) in tetrahydrofuran (1 mL) cooled to 0°C. After stirring at room temperature for 18 h, the reaction mixture was diluted with ethyl acetate and washed with water and brine. The organic layers were dried over magnesium sulfate, concentrated under reduced pressure and purified by flash column chromatography (0%–50% ethyl acetate/hexanes) to obtain 31 mg (90%) of Compound 2 as a white solid.
1H NMR (400 MHz, CDCl3) mixture of isomers d 5.37–5.39 (m, 1H), 4.35 (t, J = 6.5 Hz, 1H), 3.54–3.56 (m, 1H), 2.49 (t, J = 2.3 Hz, 1H), 2.25–2.31 (m, 2H), 2.00–2.04 (m, 2H), 1.77–1.90 (m, 5H), 1.42–1.65 (m, 12H), 1.10–1.30 (m, 9H), 1.03 (s, 3H), 0.95–0.99 (m, 4H), 0.71 (s, 3H); 13C NMR (100 MHz, d6-DMSO) d 140.77, 121.70, 85.03, 72.94, 72.79, 71.82, 62.88, 62.73, 56.76, 55.81, 50.11, 42.35, 42.31, 39.79, 37.26, 36.51, 35.38, 35.36, 34.29, 34.22, 31.91, 31.68, 31.13, 31.01, 28.15, 24.28, 21.09, 19.42, 18.72, 18.67, 11.89; LCMS m/z 407.06 (MNa+).
To synthesize 24k-C-yne (Figure S2B), manganese (IV) oxide, activated (0.13 g, 1.5 mmol), was added to a solution of Compound 2 in tetrahydrofuran (2mL) and stirred at room temperature for 48 h. The reaction mixture was filtered through celite, the filtrate concentrated under reduced pressure and purified by flash column chromatography (0%–35% ethyl acetate/hexanes) to obtain 9 mg (31%) of 24k-C-yne as colorless oil.
1H NMR (400 MHz, CDCl3) d 5.37 (d, J = 5.1 Hz, 1H), 3.52–3.58 (m, 1H), 3.23 (s, 1H), 2.50–2.67 (m, 2H), 2.22–2.34 (m, 2H), 1.98–2.03 (m, 2H), 1.85–1.88 (m, 4H), 1.47–1.66 (m, 9H), 1.28–1.34 (m, 2H), 0.95–1.21 (m, 10H), 0.70 (s, 3H); 13C NMR (100 MHz, CDCl3) d 188.01, 140.76, 121.67, 78.28, 77.24, 71.81, 56.73, 55.74, 50.07, 42.53, 42.41, 42.30, 39.74, 37.26, 36.50, 35.20, 31.90, 31.66, 29.79, 28.11, 24.26, 21.08, 19.41, 18.42, 11.89; LCMS m/z 405.10 (MNa+).
To synthesize 24k-C-BODIPY (Figure S2B), a solution of 0.3 M aqueous solution of copper(II) sulfate pentahydrate (4 mL, 0.0013 mmol) and 1M aqueous solution of sodium ascorbate (26 mL, 0.026) was added to a solution of BODIPY-FL-azide (5 mg, 0.013 mmol) and 24k-C-yne in N,N’-dimethylformamide/water (1 mL, 1:1). After stirring at 80oC for 18 h, the reaction mixture was diluted with dicholoromethane (5 mL). The organic layers were washed with brine, dried over magnesium sulfate, concentrated under reduced pressure and purified by flash column chromatography (0%–50% ethyl acetate/hexanes followed by 5% methanol/ dichloromethane) to obtain 6 mg (61%) of compound 4 as bright orange solid.
1H NMR (400 MHz, CDCl3) d 7.11 (s, 1), 6.90 (d, J = 3.9 Hz, 1H), 6.33 (d, J = 3.9 Hz, 1H), 6.17 (s, 1H), 5.38 (d, J = 5.1 Hz, 1H), 4.25 (t, J = 6.9 Hz, 2H), 3.25–3.32 (m, 4H), 3.04–3.15 (m, 2H), 2.73 (t, J = 7.2 Hz, 2H), 2.26–2.31 (m, 4H), 2.02–2.05 (m, 4H), 1.86–1.91 (m, 4H), 1.46–1.54 (m, 16H), 0.98–1.28 (m, 12H), 0.71 (s, 3H); LCMS m/z 757.37 (MH+), 759.39 (MH-).
Cilia Isolation
Gastrula stage Strongylocentrotus purpuratus embryos were concentrated by centrifugation at 170–200 × g for 4–10 min at 4oC and washed 3–4 times with artificial seawater. To isolate cilia, embryos were gently resuspended in 0.5M NaCl in artificial seawater approximately 10 times the volume of the embryo pellet. The samples were immediately centrifuged at 400 × g for 5 min at 4oC to pellet de-ciliated embryos. The supernatant was transferred to a fresh tube and centrifuged at 400 × g for another 5 min at 4oC to pellet any remaining embryos. The supernatant was then centrifuged at 10,000 × g for 20 min at 4oC and the cilia pellet was collected.
LLC-PK1 cells were cultured for 3 weeks after confluence for ciliary elongation, and at least three 15-cm diameter plates were pooled for each treatment condition. Cells were washed in PBS, and cilia were isolated by shear force on a rotary shaker at 37oC for 4 min at 360 rotations per minute. Cell debris were removed by centrifugation at 4oC for 10 min at 1,000 × g, then cilia were isolated from the supernatant by ultracentrifugation at 4oC for 30 min at 40,000 × g (Mitchell, 2013).
Cloning
The Flp-IN-3T3 system was used to stably integrate SMO constructs in pgLAP5 (Thermo Fisher Scientific). pcDNA3.1 (Addgene, Cambridge, MA) was used for all transient transfections. Site-directed mutagenesis with the QuikChange XL kit (Agilent Technologies, Santa Clara, CA) was used to introduce substitutions into constructs.
Competition Assays
20(S)-yne affinity resin was prepared and competition experiments were performed as described previously (Myers et al., 2013). For BODIPY-CYA competition assays, cells were washed in PBS, fixed for 8 min in 4% paraformaldehyde, blocked for 10 min in Opti-MEM without phenol red (Thermo Fisher Scientific) supplemented with 0.5% FBS, and incubated for 1 hr in blocking buffer supplemented with small molecules at room temperature. Coverslips were mounted with Gelvatol mounting media following 2 additional washes in PBS.
Expression Analysis
RNA sequencing was performed from 3 female Math1-Cre SmoM2c/WT and 3 SmoM2c/WT female P35 littermates. In brief, RNA was isolated using TRIzol (Thermo Fisher Scientific) followed by RNeasy Mini Kit cleanup (QIAGEN, Valencia, CA). Library preparation was performed using the TruSeq RNA Library Prep Kit v2 (Illumina, San Diego, CA) and sequenced on an Illumina HiSeq 2500 to at least 30 million unique reads per sample. Input sequences were analyzed in FASTQ format, with trimming of known adapters and low-quality regions using Fastq-mcf.; sequence quality control using FastQC and RseQC; alignment to the UCSC mm10 mouse genome assembly with Bowtie 2.2.4; gene assignment using featureCounts; and Ensembl gene annotation in GTF format (Langmead and Salzberg, 2012; Liao et al., 2014; Wang et al., 2012). EdgeR was used to calculate differential expression p values, and p.adjust was used to calculate the false discovery rate for each p value using the Benjamini-Hochberg method (Robinson et al., 2010; Robinson and Oshlack, 2010; Robinson and Smyth, 2008, 2007).
Fluorescence Polarization Assays
Fluorescence polarization of BODIPY FL conjugated ligands was measured on a Biotek H4 plate reader with 384 well low-volume microplates. All experiments were equilibrated for 10 to 15 min at room temperature before reading. Samples were excited from the top using a 485/20 nm filter set for excitation, and a 528/20 nm filter set for the emission. Serial dilutions for binding experiments were done in protein storage buffer (10 mM HEPES pH 7.5, 150 mM NaCl, 10% glycerol, 0.03% n-dodecyl-b-D-maltoside [DDM], 0.006% cholesteryl hemisuccinate [CHS]).
Histology and Microscopy
Gross light micrographs were obtained from fresh mouse brains suspended in PBS. For histology, mouse cerebella were fixed overnight at 4oC in 4% paraformaldehyde, washed in PBS, and embedded in paraffin. Sagittal sections were taken through the midline, and stained with hematoxylin and eosin (H&E) or by terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) using the In Situ Cell Death Detection Kit with Fluorescein (Roche, Basel, CH). Gross and microscopic samples were imaged using an inverted light microscope (Zeiss, Oberkochen, DE). Fluorescent microscopy was performed on an SP5 confocal microscope (Leica, Wetzlar, DE). Image processing was completed using ImageJ (Schneider et al., 2012). For fluorescence quantifications, regions of interest were selected and quantified with normalization to background fluorescence.
Immunoblotting
Immunoblots were performed as described previously (Raleigh et al., 2018). In brief, samples were boiled in 2x Laemmli reducing buffer for 5 min, separated on 4%–15% gradient TGX precast gels (Bio-Rad), transferred onto nitrocellulose (Whatman, Pittsburgh, PA) and subjected to immunoblot.
Immunofluorescence
Cells on glass coverslips were washed in PBS and fixed in 4% paraformaldehyde for 8 min. Following incubation in blocking buffer for 30 min at room temperature (2.5% BSA, 0.1% Triton X-100 and 0.03% NaN3), cells were incubated with primary antibodies in blocking buffer overnight at 4oC. The next day, cells were washed 3 times in PBS and incubated with secondary antibodies and DNA dyes in blocking buffer at room temperature for 1 hr. Following 3 final washes in PBS, coverslips were mounted with Gelvatol mounting media.
Luciferase Assays
SHH Light II cells stably express a GLI-responsive Firefly luciferase reporter and pRL-TK (Promega, Madison, WI), which constitutively expresses Renilla luciferase. For other cell lines, pRL-TK and reporter constructs were transiently expressed using Lipofectamine LTX with Plus Reagent (Thermo Fisher Scientific). HH reporter activity was quantified from pGL3-Luciferase (Promega) containing 8 consecutive GLI binding sites. Luciferase assays with 1% control renilla construct (5 ng) and 99% reporter luciferase construct (495 ng) were performed 72 hr after transfection using the Dual Luciferase Reporter Assay System (Promega) and a GloMax 96 Microplate Luminometer with Dual Injectors (Promega). Firefly luciferase reporter activities were calculated relative to internal renilla luciferase controls.
Mass Spectrometry
For mass spectrometry of sea urchin samples, a modified fractionation method was used to enhance detection of dihydroxysterols (McDonald et al., 2012). In brief, we used a two-step elution process from the aminopropyl SPE column. Column conditioning, sample loading, and column rinsing with hexane were all performed as previously described. To elute our compounds of interest, we first used 2×3mL of 30% diethyl ether in hexane to elute the sterol fraction, followed by 3mL of chloroform/methanol 23:1 to elute the oxysterol fraction. Separating the cholesterol from the oxysterols allowed us to load more of the oxysterol fraction onto the HPLC column, enhancing our detection and quantitation of trace-level oxysterols (see Figure S1 for representative chromatograms).
For mass spectrometry of LLC-PK1 and cerebellum samples, oxysterol quantification was performed relative to a d7–7-ketocholesterol (d7–7kC) internal standard that was preapared as previously reported (Xu et al., 2011). The protein content of LLC-PK1 cells and cilia was determined using the BioRad-DC protein Assay Kit (BioRad, Hercules, CA). The average protein weight of the LLC-PK1 cells and cilia were 5.29 ± 0.62 mg/mL and 0.30 ± 0.09 mg/mL, respectively. Prior to lipid extraction, d7–7k-C internal standard was added to each sample (150 ng for cells; 25 ng for cilia). Lipid extraction was performed using the Folch method, in which 1 mL of 0.9% NaCl and 4 mL of Folch solution (2:1 chloroform/methanol) was added to each sample. The resulting mixture was vortexed briefly and centrifuged at 1000 rpm and 10oC for 5 min. The organic layer was recovered and dried under vacuum. The dried extracts were reconstituted in methylene chloride (300 mL for cells; 200 mL for cilia). For UHPLC-APCI-MS/MS analysis, 100 mL of LLC-PK1 cell and cilia lipid extracts was transferred into glass LC vials, dried under a stream of Argon, and reconstituted in 90% methanol with 0.1% formic acid (200 mL for LLC-PK1 cells; 50 mL for LLC-PK1 cilia). The cerebellums of P14 and P35 mice were homogenized in 5 mL of 4:1 Folch/0.9% NaCl with a blade homogenizer in the presence of d7–7k-C internal standard (2 mg for P14; 1 mg for P35). Following centrifugation, the organic layer was recovered and dried under vacuum. The dried extracts were reconstituted in 1 mL of methylene chloride. For UHPLC-APCI-MS/MS analysis, 50 mL of lipid extract was transferred into glass LC vials, dried under a stream of Argon and reconstituted in 50 mL of 90% methanol with 0.1% formic acid. Analysis of 7-kC and other oxysterols was performed by UHPLC-MS/MS on a triple quadrupole mass spectrometer (Sciex 6500) equipped with atmospheric pressure chemical ionization (APCI), as described previously (Fliesler et al., 2018). Briefly, oxysterols were separated by reversed phase chromatography on a C18 column (1.7 mm, 2.1 3 100 mm, Phenomenex Kinetex) using an isocratic gradient of 90% methanol with 0.1% formic acid at a flow of 0.4 mL/min. The APCI parameters were as follows: nebulizer current, 3 mA; temperature, 350oC; curtain gas, 20 psi; ion source gas, 55 psi. Selective reaction monitoring (SRM) was used to monitor the dehydration of the oxysterol [M+H]+ ion to generate [M+H-H2O]+ ions (d7–7k-C, m/z 408.3 / 390.3; 7k-C, m/z 401.3 / 383.3; 7OH-C, m/z 385.3 / 367.3; 7k,27-OHC, m/z 417.3 / 399.3; 24or 25-OHC, m/z 385.3 / 367.3; 24k-C and 24,25-EC, m/z 383.3 / 365.3), as described previously (Fliesler et al., 2018; Xu et al., 2011, 2013) (see Figure S1 for representative chromatograms). The MS conditions for SRM analysis were as follows: declustering potential, 80 V; entrance potential, 10 V; collision energy, 25 V; collision cell exit potential, 20 V. Data analysis was performed with Analyst (v. 1.6.2) Quantitation Wizard. Quantitation of each oxysterol was performed against the d7–7k-C internal standard using a relative response factor (RRF) determined from an equal mixture of each oxysterol and d7–7k-C. The resulting 7k-C concentration was normalized to either protein content (for LLC-PK1 cells and cilia) or tissue weight (for cerebellums). Sterols, such as cholesterol and demosterol, were analyzed using a similar LC-MS/MS method as described previously (Fliesler et al., 2018).
Targeted mass spectrometry of oxysterols in HEK293 cells was performed as described previously using deuterated 7k-C and 7k,27-OHC standards (Myers et al., 2013). Targeted mass spectrometry of CNX in mouse brain homogenate used LC-MS/MS in positive electrospray ionization mode. Test samples and calibration standards in each matrix were processed by protein precipitation with two volumes of acetonitrile containing 50 ng/mL dextromethorphan internal standard (IS). The precipitated samples were vortexed, centrifuged at 6,100 × g for 30 min, diluted with two volumes 0.2% formic acid in water, and subsequently analyzed by LC-MS/MS. The analyte/IS peak area ratios versus the nominal analyte concentrations of the calibration samples in each matrix were used to fit a calibration curve by power regression. The analyte concentrations for the calibration standards and unknown samples were calculated using the established calibration equation for each matrix.
Mice
B6.Cg-Tg(Atoh1-cre)1Bfri/J (Math1-Cre), GT(ROSA)26Sortm1(Smo/EYFP)Amc/J (SmoM2c) and Ptch1tm1Mps/J (Ptch1c) alleles were obtained from The Jackson Laboratory (Bar Harbor, ME). The HSD11β2tm1.1Mzz allele was obtained as a generous gift from Dr. Ming-Zhi Zhang. Animals were monitored for survival until death or protocol-defined neurologic endpoints including hydrocephalus or ataxia, and tumor weight was normalized to brain weight. CNX was injected intraperitoneally in water vehicle at 100 mg/g for 14 consecutive days from P7 to P21.
Molecular Modeling
The Small Molecule Drug Discovery Suite 2016–1 was used for the computational modeling (Small-Molecule Drug Discovery Suite 2016–1: Schrodinger Suite 2016–1 Virtua Screening Workflow protocol; LigPrep, version 3.7; Epik, version 3.5, Glide version 7.0; Prime version 4.3; Schrodinger, LLC, New York, NY, 2016.). The protein from PDB 5L7D was prepared in a ready-to-dock-format with the Protein Preparation Wizard workflow, missing side chains were added, bond orders were assigned, hydrogens were added and disulfide bonds were created. The orientation of hydroxyl groups and proper protonation state were assigned at pH 7, using the PROPKA version implemented in Epik version 3.5 (Olsson et al., 2011).
Each small molecule was drawn with the Maestro graphical user interface version 10.5, and protonation states and tautomers at pH 7.0 ± 2 were predicted by Epik. Three dimensional conformers were assigned using LigPrep version 3.7 together with the OPLS force field version 3 (Harder et al., 2016). Binding sites in the protein were identified using the SiteMap tool with default parameters resulting in four sites. Virtual screening was carried out at each site.
For docking, search grids were erected around each of the 4 putative binding pockets with a box size of 20×20×20 Å 3. Small molecule poses were scored using the Glide XP scoring function, and strain energy was included in the energy value. Aromatic groups were forced to remain planar, and a maximum of 10 poses were identified for subsequent post-docking minimization. Five lowest energy poses were saved. The best binding poses identified from docking were all located in the cytoplasmic binding pocket (CBP) composed of residues T251-I266, W339-L346, N446-I454 and W535-T553. These CBP poses were subsequently rescored with the MM/GBSA protocol (Graves et al., 2008) using the VSGB solvent model and an implicit representation of the membrane.
We allowed protein flexibility up to 4A from the ligand to mimic induced-fit effects. The membrane embedded residues used to define the boundaries of the implicit membrane were determined with the Orientations of Proteins in Membranes (OPM) database (Lomize et al., 2006).
Quantitative Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR)
cDNA was synthesized from tissue and cell culture samples using the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). qRT-PCR primers are described in Table S1. qRT-PCR was performed with SYBR Green Real-Time PCR Master Mix (Thermo Fisher Scientific) and Applied Biosystems (Foster City, CA) and Life Technologies (Grand Island, NY) real-time PCR systems using the DDCt method relative to GAPDH or β-ACTIN expression.
Reagents
Antibodies and fluorescent molecules were obtained from the following suppliers: TubAc, Sigma-Aldrich (T7451, St. Louis, MO); Alexa-conjugated secondary antibodies, Life Technologies; ARL13B, Abcam (ab136648, Cambridge, UK); BODIPY-CYA, BioVision (Milpitas, CA); N3-BODIPY FL, Tocris; b-actin, Abcam (ab8227); DAPI, Thermo Fisher Scientific; GFP, Abcam (ab290); GM130, BD Biosciences (610822, San Jose, CA); Hoechst 3342, Life Technologies; HRP-conjugated secondary antibodies, Jackson ImmunoResearch Laboratories (West Grove, PA) and Cell Signaling (Grand Island, NY); HSD11β2, Abcam (ab37800 and ab115696). Small molecules were obtained from the following suppliers: 20(S)-yne Tocris Biosciences (Bristol, UK); CNX, Torcris Biosciences; CYA, Tocris Biosciences; vismodegib, Genentech (South San Francisco, CA); SAG, Merck Millipore (Billercia, MA); SAG1.5, Cellagen Technology (San Diego, CA); SHH, R&D Systems (Minneapolis, MN).
Sterols were obtained from Avanti Polar Lipids (Alabaster, AL). Mission short hairpin RNAs (shRNAs), which are described in Table S1, were obtained from Sigma-Aldrich. SHH N terminus (SHHN) was produced from HEK293-SHHN stable cells as described previously and diluted 10-fold for cell treatments (Myers et al., 2013).
Statistics
All experiments were performed with at least 3 biologic replicates. Histograms show mean ± standard error of the mean. Scatterplots show median ± 95% confidence intervals. Overall survival was estimated using the Kaplan-Meyer method and compared by Logrank tests. Student’s unpaired t test or Chi-square test was used, as indicated, to compare groups. In all cases, statistical significance, as denoted by (*), was defined as p<0.05.
Study Approval
Animal experiments were conducted in a Laboratory Animal Resource Center as per Institutional Animal Care and Use Committee approved protocol AN098101. Human tumor samples were obtained and handled as per the Institutional Committee on Human Research approved protocol 10–03204.
Supplementary Material
Figure S1. The expanded lipidomic composition of sea urchin and mammalian cilia.
HPLC-MS/MS quantification and chromatograms of diverse sterols from ciliated sea urchin embryos (A, B) and LLC-PK1 cells (C, D).
(*) = P ≤ 0.05, Student’s t test.
See also Figure 1 and Figure 4.
Figure S2. Cilia-associated oxysterols cause SMO to accumulate in cilia.
Ciliary (ARL13B, red) and nuclear (DAPI, blue) immunofluorescence of NIH/3T3 cells stably expressing mouse SMO-EGFP or SMOY134F-EGFP treated with vehicle (ethanol) or 30 μM 7β,27-OHC or 24(S),25-EC 30 μM. Cilia-associated oxysterols cause SMO to accumulate in cilia. Y134F substitution in the CRD blocks the effect of 7β,27-OHC but not 24(S),25-EC on ciliary accumulation. Scale bars: main, 10μm; insert, 1μm.
See also Figure 1.
Figure S3. Cilia-associated oxysterols cause SMO to accumulate in cilia and activate the Hedgehog pathway through the CBP.
(A) Cilia-associated oxysterol probe synthesis. Copper(II) sulfate pentahydrate, CuSO4 5H2O; dichloromethane, DCM; 4-dimethylaminopyridine, DMAP; N,N’-dimethylformamide, DMF; ethynylmagnesium bromide, HCCMgBr; lithium aluminum hydride, LAH; manganese dioxide, MnO2; pyridinium chlorochromate, PCC; tert-butyldimethylsilyl chloride, TBS-Cl; tetrahydropyran, THF; and tetra-n-butylammonium, TBAF.
(B) Ciliary (ARL13B, red) and nuclear (DAPI, blue) immunofluorescence of NIH/3T3 cells stably expressing mouse SMO-, SMOD259R-, SMON450D-, or SMOY134F,D259R-EGFP. Substitutions of the CBP, and combined substitution in the CRD and CBP, block SMO accumulation in cilia. Scale bars: main, 10μm; insert, 1μm.
(C) Luciferase activity of ciliated Smo−/− MEFs co-transfected with Gli-luciferase reporter and SMO SMOM2 with the indicated substitutions and treated with vehicle (DMSO), 1μg/ml SHH, or 100nM SAG. Data are normalized to luciferase activity of SMO-expressing cells treated with vehicle. Substitution of the CRD and/or CBP attenuates the activity of SHH and SAG.
(D) Ciliary (ARL13B, red) and nuclear (DAPI, blue) immunofluorescence of NIH/3T3 cells stably expressing mouse SMOM2-EGFP or SMOY134F, D259R-EGFP. Combined substitutions of the CRD and CBP block SMOM2 accumulation in cilia. Scale bars: main, 10μm; insert, 1μm.
(*) = P ≤ 0.05, Student’s t test.
See also Figure 2.
Figure S4. Sterol and oxysterol synthases are expressed in Hedgehog pathway-associated medulloblastoma.
(A) RNA sequencing of medulloblastomas from P35 Math1-Cre SmoM2c/wt mice compared to control cerebella of P35 SmoM2c/wt littermates shows diverse changes in sterol and oxysterol synthase gene expression in Hedgehog-associated medulloblastoma.
(B) qRT-PCR assessment of Hsd11β2 expression in P35 control SmoM2c/wt cerebella and Math1-Cre SmoM2c/wt medulloblastomas. Data are normalized to expression in control cerebella. Hsd11β2 expression is 585±45-fold higher in Hedgehog-associated medulloblastoma.
(C) Immunoblot of equal quantities of protein lysate from human Hedgehog (n=2), Group 3 (n=3) and Group 4 (n=3) medulloblastomas shows that HSD11β2 expression is specific to Hedgehog-associated medulloblastoma.
(D) Re-analysis of human transcriptome data SPR008292 (G. Robinson et al., 2012) indicates sterol and oxysterol synthases are expressed in all molecular subgroups of human medulloblastoma (G. Robinson et al., 2012). Data are shown in arbitrary units.
(E) Immunoblot of lysates from Ptch1−/− MEFs transduced with scrambled or anti-Hsd11β2 shRNAs demonstrate of HSD11β2 without altering β-actin expression.
(*) = P ≤ 0.05, Student’s t test.
See also Figure 3.
Figure S5. Pharmacologic inhibition of HSD11β2 blocks accumulation of SMO in cilia.
Immunofluorescence of endogenous SMO (red) localization to cilia (ARL13B, green) in NIH/3T3 cells treated with vehicle (water), 1μg/ml SHH or 100nM SAG, with or without 400nM CNX. Pharmacologic inhibition of HSD11β2 blocks accumulation of SMO in cilia by SHH or SAG. Scale bar, 1μm.
See also Figure 3.
Table S1. qRT-PCR primers and shRNA sequences.
Acknowledgements
We thank Dr. Ming-Zhi Zhang for providing the Hsd11β2c mouse allele, Dr. Ljiljana Milenkovic for providing Med1 cells, Dr. Rune Toftgard for providing Sufu−/− MEFs, Kaitlyn Eckert for developing the modified oxysterol extraction method used for mass spectrometry analyses done at the University of Texas Southwestern Medical Center, and Dr. Lindsay Raleigh for providing natural sea water for sea urchin embryo culture. This work was supported by grants from the NIH (HL007731 and CA212279–01), the UCSF Physician Scientist Scholar Program, the American Society of Clinical Oncology, the Rally Foundation for Childhood Cancer Research, and the American Brain Tumor Association to D.R.R.; the NIH program Project Grant to Molecular Genetics (HL20948) to J.G.M.; Cancer Research UK (C20724 and A14414) and the European Research Council (647278) to C.S.; the NIH (AR065409 and HD092659) to S.Y.W. and L.X., respectively; the NIH (R01GM102498), the Ludwig Cancer Institute, and the Howard Hughes Medical Institute to P.A.B.; and the NIH (AR054396 and GM095941), the Burroughs Wellcome Fund, and the Packard Foundation to J.F.R.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- Blassberg R, Macrae JI, Briscoe J, and Jacob J (2016). Reduced cholesterol levels impair Smoothened activation in Smith-Lemli-Opitz syndrome. Hum. Mol. Genet 25, 693–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, and Therond PP (2013). The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol 14, 416–429. [DOI] [PubMed] [Google Scholar]
- Byrne EFX, Sircar R, Miller PS, Hedger G, Luchetti G, Nachtergaele S, Tully MD, Mydock-McGrane L, Covey DF, Rambo RP, et al. (2016). Structural basis of Smoothened regulation by its extracellular domains. Nature 535, 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez M, Ena S, Van Sande J, de Kerchove d’Exaerde A, Schurmans, tent regulates trafficking and Sonic Hedgehog signaling output. Dev. Cell 34, 338–350. [DOI] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Cooper MK, and Beachy PA (2002a). Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 16, 2743–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Young KE, Maiti T, and Beachy PA (2002b). Small molecule modulation of Smoothened activity. Proc. Natl. Acad. Sci. USA 99, 14071–14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MK, Wassif CA, Krakowiak PA, Taipale J, Gong R, Kelley RI, Porter FD, and Beachy PA (2003). A defective response to Hedgehog signaling in disorders of cholesterol biosynthesis. Nat. Genet 33, 508–513. [DOI] [PubMed] [Google Scholar]
- Corcoran RB, and Scott MP (2006). Oxysterols stimulate Sonic hedgehog signal transduction and proliferation of medulloblastoma cells. Proc. Natl. Acad. Sci. USA 103, 8408–8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales JD, Blaess S, Mahoney EM, and Joyner AL (2006). The level of sonic hedgehog signaling regulates the complexity of cerebellar foliation. Development 133, 1811–1821. [DOI] [PubMed] [Google Scholar]
- Dwyer JR, Sever N, Carlson M, Nelson SF, Beachy PA, and Parhami F (2007). Oxysterols are novel activators of the hedgehog signaling pathway in pluripotent mesenchymal cells. J. Biol. Chem 282, 8959–8968. [DOI] [PubMed] [Google Scholar]
- Farese RV Jr., Biglieri EG, Shackleton CH, Irony I, and Gomez-Fontes R (1991). Licorice-induced hypermineralocorticoidism. N. Engl. J. Med 325, 1223–1227. [DOI] [PubMed] [Google Scholar]
- Fliesler SJ, Peachey NS, Herron J, Hines KM, Weinstock NI, Ramachandra Rao S, and Xu L (2018). Prevention of retinal degeneration in a rat model of Smith-Lemli-Opitz syndrome. Sci. Rep 8, 1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, Phua SC, Roberson EC, Garcia G 3rd, Abedin M, Schurmans S, Inoue T, and Reiter JF (2015). Phosphoinositides regulate ciliary protein trafficking to modulate Hedgehog signaling. Dev. Cell 34, 400–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves AP, Shivakumar DM, Boyce SE, Jacobson MP, Case DA, and tions and experimental testing. J. Mol. Biol 377, 914–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder E, Damm W, Maple J, Wu C, Reboul M, Xiang JY, Wang L, Lupyan D, Dahlgren MK, Knight JL, et al. (2016). OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput 12, 281–296. [DOI] [PubMed] [Google Scholar]
- Heine VM, and Rowitch DH (2009). Hedgehog signaling has a protective effect in glucocorticoid-induced mouse neonatal brain injury through an 11betaHSD2-dependent mechanism. J. Clin. Invest 119, 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine VM, Griveau A, Chapin C, Ballard PL, Chen JK, and Rowitch DH (2011). A small-molecule smoothened agonist prevents glucocorticoid- induced neonatal cerebellar injury. Sci. Transl. Med 3, 105ra104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo GY, Bederman I, Mast N, Liao W-L, Turko IV, and Pikuleva IA nant CYP27A1 and retinal pigment epithelial cells. J. Lipid Res 52, 1117–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Nedelcu D, Watanabe M, Jao C, Kim Y, Liu J, and Salic A (2016). Cellular cholesterol directly activates Smoothened in Hedgehog signaling. Cell 166, 1176–1187.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Zheng S, Wierbowski BM, Kim Y, Nedelcu D, Aravena L, Liu J, Kruse AC, and Salic A (2018). Structural basis of Smoothened activation in Hedgehog signaling. Cell 174, 312–324.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Yang S, Yin H, Fan X, Wang S, Yao B, Pozzi A, Chen X, Harris RC, and Zhang M-Z (2013). Epithelial-specific deletion of 11b-HSD2 hinders Apcmin/+ mouse tumorigenesis. Mol. Cancer Res 11, 1040–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam CW, Xie J, To KF, Ng HK, Lee KC, Yuen NW, Lim PL, Chan LY, Tong SF, and McCormick F (1999). A frequent activated smoothened mutation in sporadic basal cell carcinomas. Oncogene 18, 833–836. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lomize MA, Lomize AL, Pogozheva ID, and Mosberg HI (2006). OPM: orientations of proteins in membranes database. Bioinformatics 22, 623–625. [DOI] [PubMed] [Google Scholar]
- Lucas JT Jr., and Wright KD (2016). Vismodegib and physeal closure in a pediatric patient. Pediatr. Blood Cancer 63, 2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchetti G, Sircar R, Kong JH, Nachtergaele S, Sagner A, Byrne EF, Covey DF, Siebold C, and Rohatgi R (2016). Cholesterol activates the G-protein coupled receptor Smoothened to promote Hedgehog signaling. eLife 5, 1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JG, Smith DD, Stiles AR, and Russell DW (2012). A compre- hensive method for extraction and quantitative analysis of sterols and secos- teroids from human plasma. J. Lipid Res 53, 1399–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell KA (2013). Isolation of primary cilia by shear force. Curr. Protoc. Cell Biol Chapter 3. Unit 3.42.1–3.42.9. [DOI] [PubMed] [Google Scholar]
- Monder C, Stewart PM, Lakshmi V, Valentino R, Burt D, and Edwards CR (1989). Licorice inhibits corticosteroid 11 beta-dehydrogenase of rat kidney and liver: in vivo and in vitro studies. Endocrinology 125, 1046–1053. [DOI] [PubMed] [Google Scholar]
- Myers BR, Sever N, Chong YC, Kim J, Belani JD, Rychnovsky S, tiple lipid binding sites on the smoothened effector of signal response. Dev. Cell 26, 346–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers BR, Neahring L, Zhang Y, Roberts KJ, and Beachy PA (2017). lation by membrane cholesterol and extracellular sodium. Proc. Natl. Acad. Sci. USA 114, E11141–E11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtergaele S, Mydock LK, Krishnan K, Rammohan J, Schlesinger PH, Covey DF, and Rohatgi R (2012). Oxysterols are allosteric activators of the oncoprotein Smoothened. Nat. Chem. Biol 8, 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtergaele S, Whalen DM, Mydock LK, Zhao Z, Malinauskas T, Krishnan K, Ingham PW, Covey DF, Siebold C, and Rohatgi R (2013). Structure and function of the Smoothened extracellular domain in vertebrate Hedgehog signaling. eLife 2, e01340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naray-Fejes-Toth A, and Fejes-Toth G (2007). Novel mouse strain with Cre recombinase in 11beta-hydroxysteroid dehydrogenase-2-expressing cells. Am. J. Physiol. Renal Physiol 292, F486–F494. [DOI] [PubMed] [Google Scholar]
- Nedelcu D, Liu J, Xu Y, Jao C, and Salic A (2013). Oxysterol binding to the extracellular domain of Smoothened in Hedgehog signaling. Nat. Chem. Biol 9, 557–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson MHM, Søndergaard CR, Rostkowski M, and Jensen JH (2011). PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput 7, 525–537. [DOI] [PubMed] [Google Scholar]
- Pal S, Gupta R, Kim H, Wickramasinghe P, Baubet V, Showe LC, Dahmane N, and Davuluri RV (2011). Alternative transcription exceeds alternative splicing in generating the transcriptome diversity of cerebellar development. Genome Res. 21, 1260–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh DR, Choksi PK, Krup AL, Mayer W, Santos N, and Reiter JF (2018). Hedgehog signaling drives medulloblastoma growth via CDK6. J. Clin. Invest 128, 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, and Oshlack A (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, and Smyth GK (2007). Moderated statistical tests for as- sessing differences in tag abundance. Bioinformatics 23, 2881–2887. [DOI] [PubMed] [Google Scholar]
- Robinson MD, and Smyth GK (2008). Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9, 321–332. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever N, Mann RK, Xu L, Snell WJ, Hernandez-Lara CI, Porter NA, and Beachy PA (2016). Endogenous B-ring oxysterols inhibit the Hedgehog component Smoothened in a manner distinct from cyclopamine or side-chain oxysterols. Proc. Natl. Acad. Sci. USA 113, 5904–5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigg MA, Menchen T, Lee C, Johnson J, Jungnickel MK, Choksi SP, Garcia G 3rd, Busengdal H, Dougherty GW, Pennekamp P, et al. (2017). Evolutionary proteomics uncovers ancient associations of cilia with signaling pathways. Dev. Cell 43, 744–762.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang S, and Li W (2012). RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185. [DOI] [PubMed] [Google Scholar]
- Warner JF, McCarthy AM, Morris RL, and McClay DR (2014). Hedgehog signaling requires motile cilia in the sea urchin. Mol. Biol. Evol 31, 18–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windsor K, Genaro-Mattos TC, Kim H-YH, Liu W, Tallman KA, tion with alkynyl surrogates: application to Smith-Lemli-Opitz syndrome. J. Lipid Res 54, 2842–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward MH (2013). Nuclear Surety Tamper Control and Detection Programs. Air Force Instruction 91–104, September 10, 2010. https://fas.org/irp/doddir/usaf/afi91-104.pdf. [Google Scholar]
- Xiao X, Tang J-J, Peng C, Wang Y, Fu L, Qiu Z-P, Xiong Y, Yang L-F, Cui H-W, He X-L, et al. (2017). Cholesterol modification of Smoothened is required for Hedgehog signaling. Mol. Cell 66, 154–162.e10. [DOI] [PubMed] [Google Scholar]
- Xie J, Murone M, Luoh SM, Ryan A, Gu Q, Zhang C, Bonifas JM, tations in sporadic basal-cell carcinoma. Nature 391, 90–92. [DOI] [PubMed] [Google Scholar]
- Xu L, Korade Z, Rosado DA Jr., Liu W, Lamberson CR, and Porter NA (2011). An oxysterol biomarker for 7-dehydrocholesterol oxidation in cell/mouse models for Smith-Lemli-Opitz syndrome. J. Lipid Res 52, 1222–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Korade Z, Rosado DA Jr., Mirnics K, and Porter NA (2013). cholesterol in cells. J. Lipid Res 54, 1135–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The expanded lipidomic composition of sea urchin and mammalian cilia.
HPLC-MS/MS quantification and chromatograms of diverse sterols from ciliated sea urchin embryos (A, B) and LLC-PK1 cells (C, D).
(*) = P ≤ 0.05, Student’s t test.
See also Figure 1 and Figure 4.
Figure S2. Cilia-associated oxysterols cause SMO to accumulate in cilia.
Ciliary (ARL13B, red) and nuclear (DAPI, blue) immunofluorescence of NIH/3T3 cells stably expressing mouse SMO-EGFP or SMOY134F-EGFP treated with vehicle (ethanol) or 30 μM 7β,27-OHC or 24(S),25-EC 30 μM. Cilia-associated oxysterols cause SMO to accumulate in cilia. Y134F substitution in the CRD blocks the effect of 7β,27-OHC but not 24(S),25-EC on ciliary accumulation. Scale bars: main, 10μm; insert, 1μm.
See also Figure 1.
Figure S3. Cilia-associated oxysterols cause SMO to accumulate in cilia and activate the Hedgehog pathway through the CBP.
(A) Cilia-associated oxysterol probe synthesis. Copper(II) sulfate pentahydrate, CuSO4 5H2O; dichloromethane, DCM; 4-dimethylaminopyridine, DMAP; N,N’-dimethylformamide, DMF; ethynylmagnesium bromide, HCCMgBr; lithium aluminum hydride, LAH; manganese dioxide, MnO2; pyridinium chlorochromate, PCC; tert-butyldimethylsilyl chloride, TBS-Cl; tetrahydropyran, THF; and tetra-n-butylammonium, TBAF.
(B) Ciliary (ARL13B, red) and nuclear (DAPI, blue) immunofluorescence of NIH/3T3 cells stably expressing mouse SMO-, SMOD259R-, SMON450D-, or SMOY134F,D259R-EGFP. Substitutions of the CBP, and combined substitution in the CRD and CBP, block SMO accumulation in cilia. Scale bars: main, 10μm; insert, 1μm.
(C) Luciferase activity of ciliated Smo−/− MEFs co-transfected with Gli-luciferase reporter and SMO SMOM2 with the indicated substitutions and treated with vehicle (DMSO), 1μg/ml SHH, or 100nM SAG. Data are normalized to luciferase activity of SMO-expressing cells treated with vehicle. Substitution of the CRD and/or CBP attenuates the activity of SHH and SAG.
(D) Ciliary (ARL13B, red) and nuclear (DAPI, blue) immunofluorescence of NIH/3T3 cells stably expressing mouse SMOM2-EGFP or SMOY134F, D259R-EGFP. Combined substitutions of the CRD and CBP block SMOM2 accumulation in cilia. Scale bars: main, 10μm; insert, 1μm.
(*) = P ≤ 0.05, Student’s t test.
See also Figure 2.
Figure S4. Sterol and oxysterol synthases are expressed in Hedgehog pathway-associated medulloblastoma.
(A) RNA sequencing of medulloblastomas from P35 Math1-Cre SmoM2c/wt mice compared to control cerebella of P35 SmoM2c/wt littermates shows diverse changes in sterol and oxysterol synthase gene expression in Hedgehog-associated medulloblastoma.
(B) qRT-PCR assessment of Hsd11β2 expression in P35 control SmoM2c/wt cerebella and Math1-Cre SmoM2c/wt medulloblastomas. Data are normalized to expression in control cerebella. Hsd11β2 expression is 585±45-fold higher in Hedgehog-associated medulloblastoma.
(C) Immunoblot of equal quantities of protein lysate from human Hedgehog (n=2), Group 3 (n=3) and Group 4 (n=3) medulloblastomas shows that HSD11β2 expression is specific to Hedgehog-associated medulloblastoma.
(D) Re-analysis of human transcriptome data SPR008292 (G. Robinson et al., 2012) indicates sterol and oxysterol synthases are expressed in all molecular subgroups of human medulloblastoma (G. Robinson et al., 2012). Data are shown in arbitrary units.
(E) Immunoblot of lysates from Ptch1−/− MEFs transduced with scrambled or anti-Hsd11β2 shRNAs demonstrate of HSD11β2 without altering β-actin expression.
(*) = P ≤ 0.05, Student’s t test.
See also Figure 3.
Figure S5. Pharmacologic inhibition of HSD11β2 blocks accumulation of SMO in cilia.
Immunofluorescence of endogenous SMO (red) localization to cilia (ARL13B, green) in NIH/3T3 cells treated with vehicle (water), 1μg/ml SHH or 100nM SAG, with or without 400nM CNX. Pharmacologic inhibition of HSD11β2 blocks accumulation of SMO in cilia by SHH or SAG. Scale bar, 1μm.
See also Figure 3.
Table S1. qRT-PCR primers and shRNA sequences.