Abstract
Neurons are capable of degenerating their axons for the physiological clearance and refinement of unnecessary connections via the programmed degenerative pathways of apoptosis and axon pruning. While both pathways mediate axon degeneration they are however distinct. Whereas in apoptosis the entire neuron, both axons and cell body, degenerates, in the context of axon pruning only the targeted axon segments are selectively degenerated. Interestingly, the molecular pathways mediating axon degeneration in these two contexts have significant mechanistic overlap but also retain distinct differences. In this review, we describe the peripheral neuronal cell culture models used to study the molecular pathways of apoptosis and pruning. We outline what is known about the molecular mechanisms of apoptosis and axon pruning and focus on highlighting the similarities and differences of these two pathways.
Introduction to neuronal apoptosis and axon pruning
Neuronal apoptosis occurs extensively and plays a critical role during the development of the nervous system (Burek and Oppenheim, 1996). During nervous system development neurons are produced in excess to ensure productive connections are made to the appropriate targets. Neurons that are unable to successfully innervate their targets, or are no longer necessary, are selectively eliminated by the activation of the apoptotic pathway. This selective elimination of non-productive neurons ensures that the developing nervous system ends up with the appropriate number of neurons, and that these remaining neurons have formed complete and functional circuits. A prime example of developmental neuronal apoptosis occurs within the mammalian peripheral nervous system (PNS). Developing sympathetic neurons are acutely dependent on nerve growth factor (NGF) for survival, and, during the final stages of target innervation, target-derived NGF is the only source of survival signaling for these neurons. Thus, superfluous neurons that fail to innervate their targets die by apoptosis as a consequence of NGF deprivation (Oppenheim, 1991). Similarly, the central nervous system (CNS) also produces excess neurons during development that must be eliminated via apoptosis to ensure healthy brain development. However, the cues that regulate apoptosis in the CNS appear to be distinct from those in the PNS, as developmental apoptosis of cortical interneurons appears to be regulated by cell-autonomous triggers instead of target derived survival factors (Southwell et al., 2012). Despite requiring different cues, the removal of superfluous neurons in the CNS is an important event during development. Indeed, this importance is highlighted in mouse models that are genetically deleted for key apoptotic proteins (e.g. Apaf-1, Caspase-9, Caspase-3), which exhibit significant excess neurons and lethal neurodevelopmental abnormalities including exencephaly and cranial bone defects (Cecconi et al., 1998; Kuida et al., 1998; Kuida et al., 1996; Yoshida et al., 1998).
In contrast to apoptosis where the entire neuron (soma and axons) degenerates, there are many situations in which select axons degenerate while the rest of the neuron remains intact. This axon-specific degeneration, known as axon pruning in physiological contexts, selectively removes excessive, misguided, or unnecessary axon branches while maintaining the integrity of the cell body (Low and Cheng, 2006; Luo and O’Leary, 2005). Axon pruning is believed to be a key process mediating the successful development of optimally wired neuronal connections by allowing the selective elimination of undesired axonal branches or segments. Importantly, this axonal refinement can occur in both young and mature neurons and has been reported in a number of regions of the developing and adult nervous system, including the peripheral nervous system (Cusack et al., 2013; Singh et al., 2008), visual and motor cortices (O’Leary and Koester, 1993), hippocampus (Bagri et al., 2003), and the neuromuscular system (Bishop et al., 2004). Additionally, aberrant axon pruning is associated with neurodevelopmental disorders such as schizophrenia (Riccomagno and Kolodkin, 2015) and autism (Thomas et al., 2016). Axon degeneration also occurs after neuronal injury and in various neurodegenerative diseases including Alzheimer’s, Huntington’s, and Parkinson’s diseases, and Amyotrophic Lateral Sclerosis (Burke and O’Malley, 2013; Ferri et al., 2003; Fischer-Hayes et al., 2013; Li et al., 2001; Stokin et al., 2005; Vickers et al., 2009; Wang et al., 2012), although whether this resembles the physiological axon pruning pathway is not clear.
In this review, we will focus on the similarities and differences in the apoptosis and axon pruning pathways of axon degeneration in the mammalian nervous system. While not the focus of this current review, axon degeneration has also been well studied in the pathological context of axotomy (Coleman and Freeman, 2010). The mechanism of axotomy-induced axon degeneration, also known as Wallerian degeneration, appears to be distinct from the axon degeneration pathways of apoptosis and pruning and has been well covered by several recent reviews (Conforti et al., 2014; Geden and Deshmukh, 2016; Gerdts et al., 2016; Maor-Nof and Yaron, 2013; Neukomm and Freeman, 2014). Here, we will describe the primary models utilized to investigate the neuronal apoptosis and axon pruning pathways and highlight recent discoveries that have revealed similarities as well as unexpected differences in these two pathways.
Compartmentalized chambers to investigate the molecular pathways of apoptosis versus axon pruning
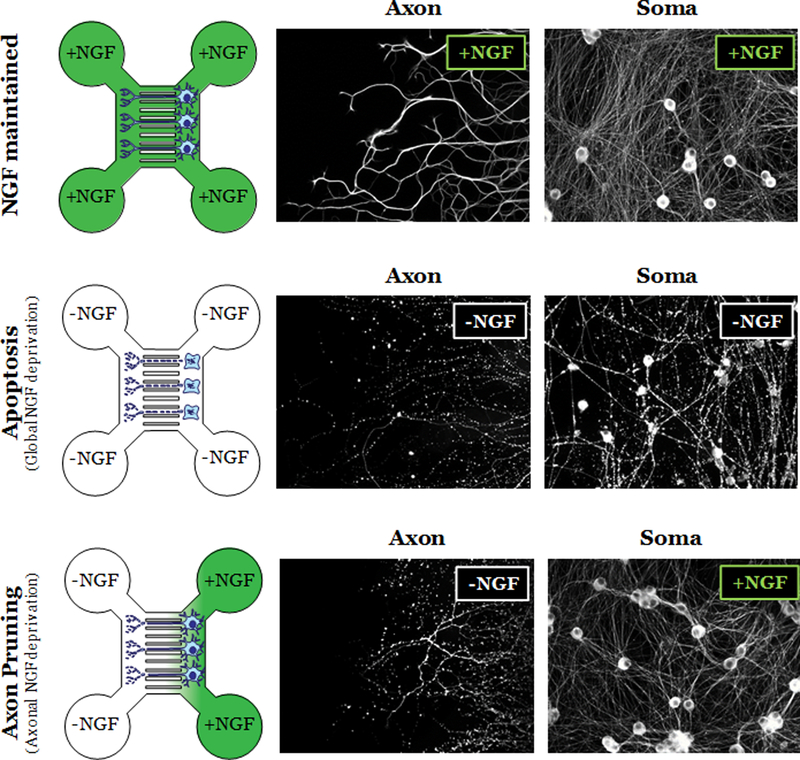
The model system most frequently used to study the molecular pathways of neuronal apoptosis and axon pruning are developing peripheral neurons (e.g. sympathetic neurons, Dorsal Root Ganglion neurons) in the context of NGF deprivation (Geden and Deshmukh, 2016; Kristiansen and Ham, 2014). Neurons globally deprived of NGF (from both soma and axons) activate the apoptotic pathway which results in the death and degeneration of the entire cell, both soma and axons. Indeed, NGF deprivation of neonatal mouse sympathetic neurons has been widely used as a model to study the mechanisms of neuronal apoptosis (Deshmukh and Johnson, 1997; Freeman et al., 2004; Greene et al., 2007; Ham et al., 2005; Jacobs et al., 2006; Kirkland and Franklin, 2003). In contrast, axonal deprivation of NGF (where distal axons are deprived but the proximal axons and soma are maintained in NGF) activates the axon pruning pathway instead, where only the deprived axon segments degenerate but the neurons survive. For in vitro study, this spatially controlled distal axon-only deprivation of NGF requires the neurons to be cultured in compartmentalized chambers (e.g. Campenot, microfluidic chambers) where the soma and proximal axons can be fluidically isolated and spatially separated from the distal axons (Campenot, 1977; Park et al., 2006; Taylor et al., 2005; Taylor and Jeon, 2011). For example, neurons from postnatal 0-day-old (P0) mouse sympathetic neurons can be plated in the soma compartment of the microfluidic device. These neurons are cultured in NGF containing media for the next 5 days, during which axons extend through the central microfluidic grooves and into the axon compartment. Axon pruning can then be induced by exposing the axon compartment to NGF-free media (along with the addition of anti-NGF antibodies), while maintaining the proximal axons and cell bodies in NGF-containing media in the soma compartment. This selective deprivation of NGF from only the axon compartment induces axon-specific degeneration of the distal axon segments (pruning) without affecting the survival of proximal axons and cell bodies in the soma compartment (Fig. 1)(Cusack et al., 2013). Apoptosis can also be induced in these chambers by depriving NGF globally from both the axon and soma compartments. While the use of microfluidic chambers is not necessary for studying apoptosis, the advantage of using the chambers is that the pathways of apoptosis and axon pruning can be examined in parallel in the same model system via global or axonal deprivation of NGF, respectively.
Fig. 1. Microfluidic chamber model for studying the pathways of apoptosis and axon pruning.
(Left) Illustration of microfluidic chambers in NGF maintained (top), Apoptosis (global NGF deprivation; middle), and Axon pruning (axonal NGF deprivation; bottom) conditions. (Right) Fluorescent images of neurons labeled with α-tubulin shows healthy and degenerated soma and axons in the indicated conditions.
It is important to note that while the mechanistic studies defining the axon pruning pathway have been done in the context of NGF deprivation, the fundamental axon pruning pathway is likely conserved and similarly engaged by other, non-NGF, triggers in vivo. However, NGF deprivation is currently the most common mechanism to activate mammalian axon pruning in vitro and provides a clear context in which to study the pathway. Importantly, mice deficient for key pruning pathway components, identified in the NGF model, were also found to have axon pruning defects in several other regions, including the CNS. For example, Casp3 and Casp6, which were identified to be required for axon pruning in the NGF pruning model in vitro are also important for mediating retinocollicular neuronal pruning in vivo (Cusack et al., 2013; Nikolaev et al., 2009; Simon et al., 2012).
Molecular Pathway of Neuronal Apoptosis
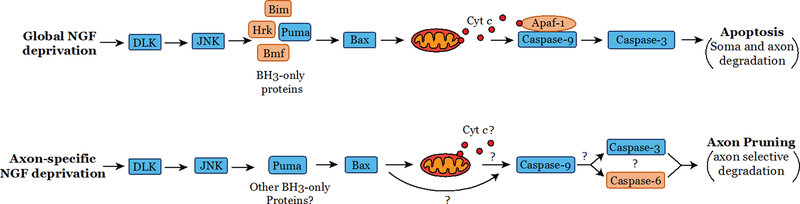
The key mediators of apoptosis are caspases, a family of cysteine proteases that cleave specific cellular substrates to cause rapid cell death (Denault and Salvesen, 2002)(Fig. 2). In sympathetic neurons undergoing apoptosis due to NGF deprivation, the mechanism by which caspases are activated has been well-studied and is known to occur via the mitochondrial (intrinsic) pathway (Fricker et al., 2018; Kristiansen and Ham, 2014). NGF deprivation results in decreased signaling from the survival promoting kinases PI3K (Phosphoinositide 3-kinase) and AKT (also known as Protein Kinase B), as well as activation of DLK (Dual leucine zipper-bearing kinase) and JNK (c-jun-N-terminal kinase) kinases. This results in the activation of transcription factors (e.g. c-jun, FOXO, Myb) that transcriptionally induce the death promoting BH3-only genes of the Bcl-2 family. Specifically, NGF deprivation is generally recognized to induce at least four BH3-only proteins including Bim (Gilley et al., 2003; Putcha et al., 2001), DP5/Hrk (Imaizumi et al., 1997), Puma (Besirli et al., 2005; Wyttenbach and Tolkovsky, 2006), and Bmf (Kole et al., 2011; Kristiansen et al., 2011), which together promote activation of the proapoptotic protein Bax. Of these, Bim and Puma are considered as “direct activators” since they can directly interact with Bax to induce a conformational change and promote Bax translocation to the mitochondria. DP5/Hrk and Bmf are sensitizers that indirectly enable Bax activation by binding to and inhibiting the anti-apoptotic proteins (Bcl-2, Bcl-XL, Bcl-w, Mcl-1)(Chipuk et al., 2010; Happo et al., 2012; Shamas-Din et al., 2011). Activated Bax then translocates to the mitochondria, causing the release of cytochrome c (cyt c) into the cytoplasm. Once in the cytosol, cyt c binds to Apaf-1 (Apoptotic protease activating factor 1) to promote Apaf-1 oligomerization and recruitment of procaspase-9 to form the apoptosome complex. Through close proximity and self-dimerization on the apoptosome, Caspase-9 (Casp9) becomes active and directly cleaves and activates Caspase-3 (Casp3). Active Casp3 then cleaves specific cellular substrates to induce rapid cell death (Kristiansen and Ham, 2014).
Fig. 2. The pathways of apoptosis and axon pruning induced by global or axonal NGF deprivation, respectively.
Top schematic shows the neuronal pathway of apoptosis activated with global NGF deprivation. Bottom schematic shows the neuronal pathway of axon pruning with axonal deprivation of NGF (from distal axons). The proteins shown in blue boxes are common to both apoptosis and axon pruning whereas the proteins shown in orange boxes highlights the proteins that are distinct between these two pathways.
While other details of the apoptotic pathway in neurons have also been identified (Fricker et al., 2018), we have focused here on the central components of the apoptotic pathway that have been shown to be important with inhibitor and mouse knock-out studies (Fig. 2). For example, inhibition or deletion of DLK, as well as inhibition of JNK, blocks neuronal apoptosis after NGF deprivation at an upstream point in the pathway (Ghosh et al., 2011; Watkins et al., 2013; Xu et al., 2001). Recently, the kinase inhibitor fortinib was also shown to strongly protect against neuronal apoptosis. While its exact mechanism of action for neuroprotection is still unclear (it appears to involve the inactivation a prodegenerative signal from the unligated NGF receptor TrkA), fortinib treatment blocks the induction of the BH3-only mRNA after NGF deprivation (Feinberg et al., 2017). Deletion of individual BH3-only genes (e.g. Bim, Puma, DP5/Hrk) has resulted in partial inhibition of neuronal apoptosis (Imaizumi et al., 2004; Kristiansen and Ham, 2014; Putcha et al., 2001; Wyttenbach and Tolkovsky, 2006), with additive effects seen with combined deletions (Ren et al., 2010), a finding that is consistent with the known redundant functions of these proteins. Interestingly, ectopic expression of miR-29, a microRNA that can target and inhibit multiple members of the BH3-only gene family, is effective in blocking neuronal apoptosis (Kole et al., 2011). Downstream of the BH3-only proteins, the apoptotic pathway converges on Bax as deletion of Bax alone is sufficient to inhibit apoptosis in neurons (Deckwerth et al., 1996). Likewise, deletion of Apaf-1, Casp9, or Casp3 inhibits neuronal apoptosis (Wright et al., 2007), demonstrating the essential function of these apoptosome pathway components in this context.
Molecular Pathway of Axon Pruning
The apoptosis and axon pruning pathways share many of the same proteins but with key differences that are highlighted here (Fig. 2). It is crucial to note here that while both apoptosis and axon pruning can be triggered in peripheral neurons with NGF deprivation, the context of NGF deprivation is essential to consider as it results in the engagement of distinct pathways. Whereas apoptosis is engaged in the context of global deprivation of entire neuron (both soma and axons), axon pruning is only engaged in the context of distal axon NGF deprivation (where only the distal axon segments are deprived of NGF and the proximal axons and soma are maintained). Thus, in vitro examination of axon pruning requires the use of the compartmentalized culture models we previously describe such that distal axons can be selectively deprived of NGF, while the proximal axons and cell bodies are maintained in NGF, so that the deprived axon segments are selectively pruned. As described below, this consideration is important in order to accurately define the specific molecular events that occur in pruning versus apoptosis (Geden and Deshmukh, 2016).
The upstream events in axon pruning occur similarly as in apoptosis, where axonal deprivation of NGF also activates the DLK and JNK signaling pathways that are both important for axon pruning (Ghosh et al., 2011; Simon et al., 2016). Likewise, c-jun is also activated during axon pruning, but whether it is essential for pruning is not yet known (Ghosh et al., 2011; Mok et al., 2009). Recently, the BH3-only protein Puma was identified as an important mediator for axon pruning where its deficiency conferred protection against axonal NGF deprivation (Maor-Nof et al., 2016; Simon et al., 2016). Whether any of the other BH3-only genes that are involved in apoptosis are also induced and important for axon pruning is not known. Interestingly, the kinase inhibitor foretinib, which blocks the induction of BH3-only genes during apoptosis in neurons, also inhibits axon pruning after axonal deprivation of NGF (Feinberg et al., 2017). Importantly, like apoptosis, the axon pruning pathway also converges on Bax as Bax-deficiency completely inhibits axon pruning (Cusack et al., 2013; Nikolaev et al., 2009; Schoenmann et al., 2010). Consistent with the known function of Bax in mediating mitochondrial permeabilization, cyt c is released from mitochondria in axons undergoing pruning (Cusack et al., 2013). However, whether cyt c is required for pruning is not known. This is particularly relevant as cyt c is known to activate Casp9 via Apaf-1 on the apoptosome (Wang, 2001). It is at this point that the pathway of axon pruning appears to show distinct differences from apoptosis, as our results have shown that Apaf-1 is not required for axon pruning (Cusack et al., 2013). While Apaf-1-deficient neurons are protected from undergoing apoptosis after global NGF deprivation (Simon et al., 2016; Wright et al., 2007), they still remain capable of undergoing axon pruning in response to local axonal NGF deprivation (Cusack et al., 2013). Interestingly, despite not requiring Apaf-1, axon pruning requires both Casp9 (Cusack et al., 2013) and Casp3 (Cusack et al., 2013; Simon et al., 2012). Additionally, axon pruning is dependent on Casp6 (Cusack et al., 2013; Nikolaev et al., 2009), a caspase which is not essential for apoptosis (Cusack et al., 2013). Importantly, Casp6 is also activated in degenerating axons in models of brain injury and disease (Akpan et al., 2011; Albrecht et al., 2009; Graham et al., 2011; Guo et al., 2004; LeBlanc et al., 1999; Uribe et al., 2012). However, exactly how caspases are activated via a novel, Apaf-1-independent mechanism during axon pruning, or even their precise order of activation, remains unknown (Fig. 2).
A striking aspect of axon pruning is that the degeneration is restricted to only the targeted axon segments that are deprived of NGF. At this point, we do not know the precise mechanism by which axon degeneration is spatially restricted during pruning. In the Drosophila model, localized degradation of the caspase inhibitory protein DIAP1 (Death-associated Inhibitor of Apoptosis-1) is known to enable localized caspase activation and degeneration during dendritic pruning (Kuo et al., 2006). A similar mechanism by which caspase activity is restricted to the targeted axons during pruning appears to be regulated via XIAP (X-linked Inhibitor of Apoptosis Protein). XIAP is known to strictly regulate caspase activation in neurons (Potts et al., 2003). Indeed, XIAP-deficient neurons not only exhibited enhanced axon degeneration than wild-type neurons but also aberrant caspase activation in the cell bodies during axon pruning (Cusack et al., 2013; Unsain et al., 2013). The importance of XIAP in regulating neuronal activity (presumably via regulation of axonal and/or synaptic pruning) is underscored by the observation that mice deleted for XIAP also exhibit reduced number of synapses and defects in memory and learning (Gibon et al., 2016; Martinez-Marmol et al., 2016). Together, these results suggest that differential modulation of XIAP function may enable either a permissive environment (e.g. low XIAP in axon regions targeted for pruning) or a repressive environment (e.g. high XIAP in soma and undeprived proximal axons) to spatially restrict caspase activity during axon pruning.
While the details of synaptic remodeling are beyond the scope of this review, it is interesting to note that synaptic pruning also appears to be regulated by many of these same pathway components as pruning and apoptosis. For example, both Bax and Casp3 have been shown to be important for synaptic pruning and in the regulation of long-term depression (Jiao and Li, 2011; Li et al., 2010; Li and Sheng, 2012). As previously mentioned, XIAP appears to regulate synaptic remodeling in hippocampal neurons, presumably by mediating the activation of Casp3 in synapses (Gibon et al., 2016). These findings indicate that the pathways for refinement and elimination of synapses have similarity to the elimination of axon segments via pruning. However, whether the pathways of synaptic and axon pruning are identical is still unclear.
Thus, increasing evidence indicates that although the neuronal pathways of apoptosis and axon pruning utilize many of the same components to promote axon degeneration, the two pathways are distinct. Interestingly, both pathways engage very similar signaling upstream events to trigger Bax activation. However, downstream of Bax activation the pathways diverge, with apoptosis requiring Apaf-1 but not Casp6, and axon pruning requiring Casp6 but not Apaf-1. Many of the interesting details about pruning and axon degeneration pathways remain to be discovered. Some questions that remain unclear in the axon pruning pathway include: What is the essential role for Casp6? How are caspases activated independently of Apaf-1? and, Why is Bax activation required while formation of the apoptosome is not required? While the primary discussion in this review is focused on the mechanisms of programmed axon degeneration, it is important to note that activation of these pathways that result in aberrant pruning could also lead to the underlying pathology of neurodevelopmental disorders and neurodegenerative disease. Research into these mechanisms will undoubtedly reveal new insights and remain an exciting area of investigation.
Acknowledgement
This work was supported by grant from National Institutes of Health (GM118331) to MD.
References
- Akpan N, Serrano-Saiz E, Zacharia BE, Otten ML, Ducruet AF, Snipas SJ, Liu W, Velloza J, Cohen G, Sosunov SA, Frey WH, Salvesen GS, Connolly ES, Troy CM, 2011. Intranasal Delivery of Caspase-9 Inhibitor Reduces Caspase-6-Dependent Axon/Neuron Loss and Improves Neurological Function after Stroke. J. Neurosci 31, 8894–8904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht S, Bogdanovic N, Ghetti B, Winblad B, LeBlanc AC, 2009. Caspase-6 activation in familial alzheimer disease brains carrying amyloid precursor protein or presenilin i or presenilin II mutations. J. Neuropathol. Exp. Neurol 68, 1282–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagri A, Cheng H-J, Yaron A, Pleasure SJ, Tessier-Lavigne M, 2003. Stereotyped Pruning of Long Hippocampal Axon Branches Triggered by Retraction Inducers of the Semaphorin Family. Cell 113, 285–299. [DOI] [PubMed] [Google Scholar]
- Besirli CG, Wagner EF, Johnson EM Jr., 2005. The limited role of NH2-terminal c-Jun phosphorylation in neuronal apoptosis: identification of the nuclear pore complex as a potential target of the JNK pathway. J. Cell Biol 170, 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DL, Misgeld T, Walsh MK, Gan W-B, Lichtman JW, 2004. Axon Branch Removal at Developing Synapses by Axosome Shedding. Neuron 44, 651–661. [DOI] [PubMed] [Google Scholar]
- Burek MJ, Oppenheim RW, 1996. Programmed cell death in the developing nervous system. Brain Pathol. 6, 427–446. [DOI] [PubMed] [Google Scholar]
- Burke RE, O’Malley K, 2013. Axon degeneration in Parkinson’s disease. Exp. Neurol 246, 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campenot RB, 1977. Local control of neurite development by nerve growth factor. Proc. Natl. Acad. Sci. USA 74, 4516–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi F, Alvarez BG, Meyer BI, Roth KA, Gruss P, 1998. Apaf1 (CED-4 homolog) regulates programmed cell death in mammalian development. Cell 94, 727–737. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR, 2010. The BCL-2 Family Reunion. Mol. Cell 37, 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman MP, Freeman MR, 2010. Wallerian Degeneration, WldS, and Nmnat. Annu. Rev. Neurosci 33, 245–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforti L, Gilley J, Coleman MP, 2014. Wallerian degeneration: an emerging axon death pathway linking injury and disease. Nat. Rev. Neurosci 15, 394–409. [DOI] [PubMed] [Google Scholar]
- Cusack CL, Swahari V, Hampton Henley W, Michael Ramsey J, Deshmukh M, 2013. Distinct pathways mediate axon degeneration during apoptosis and axon-specific pruning. Nat. Commun 4, 1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckwerth TL, Elliott JL, Knudson CM, Johnson EM Jr., Snider WD, Korsmeyer SJ, 1996. Bax is required for neuronal death after trophic factor deprivation and during development. Neuron 17, 401–411. [DOI] [PubMed] [Google Scholar]
- Denault JB, Salvesen GS, 2002. Caspases: keys in the ignition of cell death. Chem. Rev 102, 4489–4500. [DOI] [PubMed] [Google Scholar]
- Deshmukh M, Johnson EM Jr., 1997. Programmed cell death in neurons: focus on the pathway of nerve growth factor deprivation-induced death of sympathetic neurons. Mol. Pharm 51, 897–906. [DOI] [PubMed] [Google Scholar]
- Feinberg K, Kolaj A, Wu C, Grinshtein N, Krieger JR, Moran MF, Rubin LL, Miller FD, Kaplan DR, 2017. A neuroprotective agent that inactivates prodegenerative TrkA and preserves mitochondria. J Cell Biol 216, 3655–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri A, Sanes JR, Coleman MP, Cunningham JM, Kato AC, 2003. Inhibiting axon degeneration and synapse loss attenuates apoptosis and disease progression in a mouse model of motoneuron disease. Curr. Biol 13, 669–673. [DOI] [PubMed] [Google Scholar]
- Fischer-Hayes LR, Brotherton T, Glass JD, 2013. Axonal degeneration in the peripheral nervous system: Implications for the pathogenesis of amyotrophic lateral sclerosis. Exp. Neurol 246, 6–13. [DOI] [PubMed] [Google Scholar]
- Freeman RS, Burch RL, Crowder RJ, Lomb DJ, Schoell MC, Straub JA, Xie L, 2004. NGF deprivation-induced gene expression: after ten years, where do we stand? Prog. Brain Res 146, 111–126. [DOI] [PubMed] [Google Scholar]
- Fricker M, Tolkovsky AM, Borutaite V, Coleman M, Brown GC, 2018. Neuronal Cell Death. Physiol. Rev 98, 813–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geden MJ, Deshmukh M, 2016. Axon degeneration: context defines distinct pathways. Curr. Opin. Neurobiol 39, 108–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdts J, Summers Daniel W., Milbrandt J, DiAntonio A, 2016. Axon Self-Destruction: New Links among SARM1, MAPKs, and NAD+ Metabolism. Neuron 89, 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh AS, Wang B, Pozniak CD, Chen M, Watts RJ, Lewcock JW, 2011. DLK induces developmental neuronal degeneration via selective regulation of proapoptotic JNK activity. J. Cell Biol 194, 751–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon J, Unsain N, Gamache K, Thomas RA, De Leon A, Johnstone A, Nader K, Seguela P, Barker PA, 2016. The X-linked inhibitor of apoptosis regulates long-term depression and learning rate. FASEB J 30, 3083–3090. [DOI] [PubMed] [Google Scholar]
- Gilley J, Coffer PJ, Ham J, 2003. FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J. Cell Biol 162, 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RK, Ehrnhoefer DE, Hayden MR, 2011. Caspase-6 and neurodegeneration. Trends Neurosci. 34, 646–656. [DOI] [PubMed] [Google Scholar]
- Greene LA, Liu DX, Troy CM, Biswas SC, 2007. Cell cycle molecules define a pathway required for neuron death in development and disease. Biochim. Biophys. Acta 1772, 392–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Albrecht S, Bourdeau M, Petzke T, Bergeron C, LeBlanc AC, 2004. Active caspase-6 and caspase-6-cleaved tau in neuropil threads, neuritic plaques, and neurofibrillary tangles of Alzheimer’s disease. Am. J. Pathol 165, 523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham J, Towers E, Gilley J, Terzano S, Randall R, 2005. BH3-only proteins: key regulators of neuronal apoptosis. Cell Death Differ. 12, 1015–1020. [DOI] [PubMed] [Google Scholar]
- Happo L, Strasser A, Cory S, 2012. BH3-only proteins in apoptosis at a glance. J. Cell Sci 125, 1081–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi K, Benito A, Kiryu-Seo S, Gonzalez V, Inohara N, Lieberman AP, Kiyama H, Nunez G, 2004. Critical role for DP5/Harakiri, a Bcl-2 homology domain 3-only Bcl-2 family member, in axotomy-induced neuronal cell death. J. Neurosci 24, 3721–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi K, Tsuda M, Imai Y, Wanaka A, Takagi T, Tohyama M, 1997. Molecular cloning of a novel polypeptide, DP5, induced during programmed neuronal death. J. Biol. Chem 272, 18842–18848. [DOI] [PubMed] [Google Scholar]
- Jacobs WB, Kaplan DR, Miller FD, 2006. The p53 family in nervous system development and disease. J. Neurochem 97, 1571–1584. [DOI] [PubMed] [Google Scholar]
- Jiao S, Li Z, 2011. Nonapoptotic Function of BAD and BAX in Long-Term Depression of Synaptic Transmission. Neuron 70, 758–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland RA, Franklin JL, 2003. Bax, reactive oxygen, and cytochrome c release in neuronal apoptosis. Antioxid. Redox Signal 5, 589–596. [DOI] [PubMed] [Google Scholar]
- Kole AJ, Swahari V, Hammond SM, Deshmukh M, 2011. miR-29b is activated during neuronal maturation and targets BH3-only genes to restrict apoptosis. Genes Dev. 25, 125–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen M, Ham J, 2014. Programmed cell death during neuronal development: the sympathetic neuron model. Cell Death Differ. 21, 1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen M, Menghi F, Hughes R, Hubank M, Ham J, 2011. Global analysis of gene expression in NGF-deprived sympathetic neurons identifies molecular pathways associated with cell death. BMC Genomics 12, 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuida K, Haydar TF, Kuan CY, Gu Y, Taya C, Karasuyama H, Su MS, Rakic P, Flavell RA, 1998. Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell 94, 325–337. [DOI] [PubMed] [Google Scholar]
- Kuida K, Zheng TS, Na S, Kuan C, Yang D, Karasuyama H, Rakic P, Flavell RA, 1996. Decreased apoptosis in the brain and premature lethality in CPP32-deficient mice. Nature 384, 368–372. [DOI] [PubMed] [Google Scholar]
- Kuo CT, Zhu S, Younger S, Jan LY, Jan YN, 2006. Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron 51, 283–290. [DOI] [PubMed] [Google Scholar]
- LeBlanc A, Liu H, Goodyer C, Bergeron C, Hammond J, 1999. Caspase-6 Role in Apoptosis of Human Neurons, Amyloidogenesis, and Alzheimer’s Disease. J. Biol. Chem 274, 23426–23436. [DOI] [PubMed] [Google Scholar]
- Li H, Li SH, Yu ZX, Shelbourne P, Li XJ, 2001. Huntingtin aggregate-associated axonal degeneration is an early pathological event in Huntington’s disease mice. J. Neurosci 21, 8473–8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Jo J, Jia JM, Lo SC, Whitcomb DJ, Jiao S, Cho K, Sheng M, 2010. Caspase-3 activation via mitochondria is required for long-term depression and AMPA receptor internalization. Cell 141, 859–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Sheng M, 2012. Caspases in synaptic plasticity. Mol. Brain 5, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low LK, Cheng H-J, 2006. Axon pruning: an essential step underlying the developmental plasticity of neuronal connections. Phil. Trans. Royal Soc. B: Biol. Sci 361, 1531–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, O’Leary DD, 2005. Axon retraction and degeneration in development and disease. Annu. Rev. Neurosci 28, 127–156. [DOI] [PubMed] [Google Scholar]
- Maor-Nof M, Romi E, Shalom Hadas S., Ulisse V, Raanan C, Nof A, Leshkowitz D, Lang R, Yaron A, 2016. Axonal Degeneration Is Regulated by a Transcriptional Program that Coordinates Expression of Pro- and Anti-degenerative Factors. Neuron 92, 991–1006. [DOI] [PubMed] [Google Scholar]
- Maor-Nof M, Yaron A, 2013. Neurite pruning and neuronal cell death: spatial regulation of shared destruction programs. Curr. Opin. Neurobiol 23, 990–996. [DOI] [PubMed] [Google Scholar]
- Martinez-Marmol R, Barneda-Zahonero B, Soto D, Andres RM, Coccia E, Gasull X, Planells-Ferrer L, Moubarak RS, Soriano E, Comella JX, 2016. FAIM-L regulation of XIAP degradation modulates Synaptic Long-Term Depression and Axon Degeneration. Sci Rep 6, 35775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok SA, Lund K, Campenot RB, 2009. A retrograde apoptotic signal originating in NGF-deprived distal axons of rat sympathetic neurons in compartmented cultures. Cell Res. 19, 546–560. [DOI] [PubMed] [Google Scholar]
- Neukomm LJ, Freeman MR, 2014. Diverse cellular and molecular modes of axon degeneration. Trends Cell Biol. 24, 515–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev A, McLaughlin T, O’Leary DD, Tessier-Lavigne M, 2009. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature 457, 981–989. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- O’Leary DDM, Koester SE, 1993. Development of projection neuron types, axon pathways, and patterned connections of the mammalian cortex. Neuron 10, 991–1006. [DOI] [PubMed] [Google Scholar]
- Oppenheim RW, 1991. Cell death during development of the nervous system. Annu. Rev. Neurosci 14, 453–501. [DOI] [PubMed] [Google Scholar]
- Park JW, Vahidi B, Taylor AM, Rhee SW, Jeon NL, 2006. Microfluidic culture platform for neuroscience research. Nat. Protoc 1, 2128–2136. [DOI] [PubMed] [Google Scholar]
- Potts PR, Singh S, Knezek M, Thompson CB, Deshmukh M, 2003. Critical function of endogenous XIAP in regulating caspase activation during sympathetic neuronal apoptosis. J. Cell Biol 163, 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putcha GV, Moulder KL, Golden JP, Bouillet P, Adams JA, Strasser A, Johnson EM, 2001. Induction of BIM, a proapoptotic BH3-only BCL-2 family member, is critical for neuronal apoptosis. Neuron 29, 615–628. [DOI] [PubMed] [Google Scholar]
- Ren D, Tu HC, Kim H, Wang GX, Bean GR, Takeuchi O, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH, 2010. BID, BIM, and PUMA are essential for activation of the BAX- and BAK-dependent cell death program. Science 330, 1390–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccomagno MM, Kolodkin AL, 2015. Sculpting neural circuits by axon and dendrite pruning. Annu. Rev. Cell Dev. Biol 31, 779–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenmann Z, Assa-Kunik E, Tiomny S, Minis A, Haklai-Topper L, Arama E, Yaron A, 2010. Axonal degeneration is regulated by the apoptotic machinery or a NAD+-sensitive pathway in insects and mammals. J. Neurosci 30, 6375–6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamas-Din A, Brahmbhatt H, Leber B, Andrews DW, 2011. BH3-only proteins: Orchestrators of apoptosis. Biochim. Biophys. Acta 1813, 508–520. [DOI] [PubMed] [Google Scholar]
- Simon David J., Pitts J, Hertz Nicholas T., Yang J, Yamagishi Y, Olsen O, Tešić Mark M, Molina H, Tessier-Lavigne M, 2016. Axon degeneration gated by retrograde activation of somatic pro-apoptotic signaling. Cell 164, 1031–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DJ, Weimer RM, McLaughlin T, Kallop D, Stanger K, Yang J, O’Leary DD, Hannoush RN, Tessier-Lavigne M, 2012. A caspase cascade regulating developmental axon degeneration. J. Neurosci 32, 17540–17553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK, Park KJ, Hong EJ, Kramer BM, Greenberg ME, Kaplan DR, Miller FD, 2008. Developmental axon pruning mediated by BDNF-p75NTR-dependent axon degeneration. Nat. Neurosci 11, 649–658. [DOI] [PubMed] [Google Scholar]
- Southwell DG, Paredes MF, Galvao RP, Jones DL, Froemke RC, Sebe JY, Alfaro-Cervello C, Tang Y, Garcia-Verdugo JM, Rubenstein JL, Baraban SC, Alvarez-Buylla A, 2012. Intrinsically determined cell death of developing cortical interneurons. Nature 491, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokin GB, Lillo C, Falzone TL, Brusch RG, Rockenstein E, Mount SL, Raman R, Davies P, Masliah E, Williams DS, Goldstein LS, 2005. Axonopathy and transport deficits early in the pathogenesis of Alzheimer’s disease. Science 307, 1282–1288. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, Jeon NL, 2005. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat. Methods 2, 599–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Jeon NL, 2011. Microfluidic and compartmentalized platforms for neurobiological research. Crit. Rev. Biomed. Eng 39, 185–200. [DOI] [PubMed] [Google Scholar]
- Thomas MSC, Davis R, Karmiloff‐Smith A, Knowland VCP, Charman T, 2016. The over‐pruning hypothesis of autism. Dev. Sci 19, 284–305. [DOI] [PubMed] [Google Scholar]
- Unsain N, Higgins Julia M., Parker Kristen N., Johnstone Aaron D., Barker Philip A., 2013. XIAP regulates caspase activity in degenerating axons. Cell Rep. 4, 751–763. [DOI] [PubMed] [Google Scholar]
- Uribe V, Wong BKY, Graham RK, Cusack CL, Skotte NH, Pouladi MA, Xie Y, Feinberg K, Ou Y, Ouyang Y, Deng Y, Franciosi S, Bissada N, Spreeuw A, Zhang W, Ehrnhoefer DE, Vaid K, Miller FD, Deshmukh M, Howland D, Hayden MR, 2012. Rescue from excitotoxicity and axonal degeneration accompanied by age-dependent behavioral and neuroanatomical alterations in caspase-6 deficient mice. Human Mol. Genet 21, 1954–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers JC, King AE, Woodhouse A, Kirkcaldie MT, Staal JA, McCormack GH, Blizzard CA, Musgrove REJ, Mitew S, Liu Y, Chuckowree JA, Bibari O, Dickson TC, 2009. Axonopathy and cytoskeletal disruption in degenerative diseases of the central nervous system. Brain Res. Bull 80, 217–223. [DOI] [PubMed] [Google Scholar]
- Wang JT, Medress ZA, Barres BA, 2012. Axon degeneration: molecular mechanisms of a self-destruction pathway. J. Cell Biol 196, 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, 2001. The expanding role of mitochondria in apoptosis. Genes Dev. 15, 2922–2933. [PubMed] [Google Scholar]
- Watkins TA, Wang B, Huntwork-Rodriguez S, Yang J, Jiang Z, Eastham-Anderson J, Modrusan Z, Kaminker JS, Tessier-Lavigne M, Lewcock JW, 2013. DLK initiates a transcriptional program that couples apoptotic and regenerative responses to axonal injury. Proc. Natl. Acad. Sci 110, 4039–4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KM, Vaughn AE, Deshmukh M, 2007. Apoptosome dependent caspase-3 activation pathway is non-redundant and necessary for apoptosis in sympathetic neurons. Cell Death Differ. 14, 625–633. [DOI] [PubMed] [Google Scholar]
- Wyttenbach A, Tolkovsky AM, 2006. The BH3-only protein Puma is both necessary and sufficient for neuronal apoptosis induced by DNA damage in sympathetic neurons. J. Neurochem 96, 1213–1226. [DOI] [PubMed] [Google Scholar]
- Xu Z, Maroney AC, Dobrzanski P, Kukekov NV, Greene LA, 2001. The MLK family mediates c-Jun N-terminal kinase activation in neuronal apoptosis. Mol. Cell. Biol 21, 4713–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Kong YY, Yoshida R, Elia AJ, Hakem A, Hakem R, Penninger JM, Mak TW, 1998. Apaf1 is required for mitochondrial pathways of apoptosis and brain development. Cell 94, 739–750. [DOI] [PubMed] [Google Scholar]