ABSTRACT
Stomata are structures on the surfaces of most land plants that are required for gas exchange between plants and their environment. In Arabidopsis thaliana, stomata comprise two kidney bean-shaped epidermal guard cells that flank a central pore overlying a cavity in the mesophyll. These guard cells can adjust their shape to occlude or facilitate access to this pore, and in so doing regulate the release of water vapor and oxygen from the plant, in exchange for the intake of carbon dioxide from the atmosphere. Stomatal guard cells are the end product of a specialized lineage whose cell divisions and fate transitions ensure both the production and pattern of cells in aerial epidermal tissues. The stomatal lineage is dynamic and flexible, altering stomatal production in response to environmental change. As such, the stomatal lineage is an excellent system to study how flexible developmental transitions are regulated in plants. In this Cell Science at a Glance article and accompanying poster, we will summarize current knowledge of the divisions and fate decisions during stomatal development, discussing the role of transcriptional regulators, cell–cell signaling and polarity proteins. We will highlight recent work that links the core regulators to systemic or environmental information and provide an evolutionary perspective on stomata lineage regulators in plants.
KEY WORDS: Arabidopsis, Asymmetric cell division, SPEECHLESS, Cell–cell signaling, Stem cell, Stomata
Summary: This Cell Science at a Glance article describes stomatal development, focusing on the role of transcriptional regulators, cell–cell signaling and polarity proteins.
Introduction
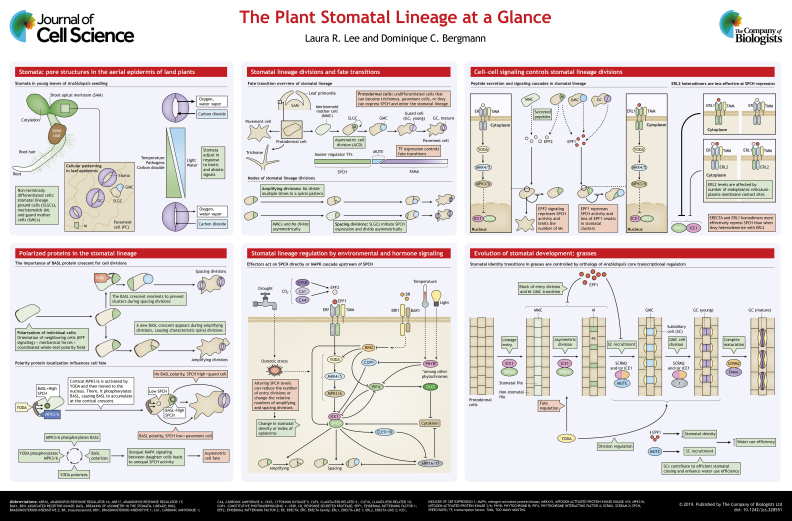
Stomata are pores in the plant surface that are flanked by two epidermal guard cells. They are found in the epidermal aerial portions of nearly all land plants and are critical regulators of gas and water vapor exchange between plants and their environments. Stomatal aperture is adjusted through a turgor-driven mechanism whereby ion channels in guard cells are activated to change osmotic potential – to open, guard cells import cations and the concomitant influx of water causes swelling (for review, see Jezek and Blatt, 2017). Stomatal aperture is responsive to many environmental stimuli including light, temperature, carbon dioxide levels and water availability (reviewed in Murata et al., 2015) (see poster). Stomatal development, too, is modulated by the environment, and whereas this phenomenon has been documented in many species and in the fossil record (McElwain and Steinthorsdottir, 2017), mechanistic details about stomatal development are best understood in the plant model Arabidopsis thaliana. Decades of work have thoroughly characterized the series of divisions and transitions in cellular identity that comprise the Arabidopsis stomatal lineage (see poster). Transitions between each intermediate stomatal lineage cell identity are mediated by closely-related master regulator basic helix-loop-helix (bHLH) class Ia transcription factors SPEECHLESS (SPCH), MUTE and FAMA and their heterodimerization partners, the class III bHLH transcription factors INDUCER OF CBF EXPRESSION 1 (ICE1, also known as SCRM) and/or SCREAM 2 (SCRM2) (Kanaoka et al., 2008; MacAlister et al., 2007; Ohashi-Ito et al., 2006; Pillitteri et al., 2007).
Here, we discuss the many factors that influence these core transcriptional regulators and describe recent advances in approaches to dissect these pathways (see also Box 1). We focus on the influence of cell–cell signaling, polarity and environmental and hormonal signals on stomatal divisions and cell fate in Arabidopsis, and highlight the evolution of stomata and their regulation throughout the plant kingdom.
Box 1. Emerging approaches to study the stomatal lineage.
The accessibility of the stomatal lineage has made it an attractive model for monitoring the emergence of cell identities and patterns in vivo. Time-lapse imaging of cell morphology, division behavior and cell identity (through transcriptional and translational reporter expression) in wild-type, transgenic and mutant plants are now being combined with quantitative data analysis and modeling to get a system-wide view of development (Bringmann and Bergmann, 2017; Mansfield et al., 2018; Robinson et al., 2011).
In parallel, advances in DNA sequencing technology have enabled unprecedented high-throughput genomic profiling in the stomatal lineage. Datasets generated using these techniques include profiles of plants enriched or depleted in the stomatal lineage (Bergmann et al., 2004; Pillitteri et al., 2011), genome-wide maps of SPCH binding sites (Lau et al., 2014), identification of MUTE targets by RNA-seq following MUTE induction (Han et al., 2018), and cell type-specific transcriptional profiles (Adrian et al., 2015). From these large-scale data sets, new genes and new regulatory modules have emerged. For instance, following up on these data (Adrian et al., 2015) led to identification of the stomatal lineage-specific CYCLIN D7;1 (CYCD7;1), which is part of a system that ensures GMCs divide once – and only once – to produce a pair of GCs (Weimer et al., 2018; and further elaborated in Han et al., 2018). A role for ICE1 in anther dehydration regulation was revealed by over-representation of GC-expressed genes among ICE1-regulated genes in anthers (Wei et al., 2018). GC cell walls are unique among Arabidopsis cells with regards to the composition and modification of cell wall matrix polymers; the stomatal lineage transcriptional map was noted as a valuable resource for identification of GC-specific cell wall-modifying enzymes (Rui et al., 2018). Regulatory systems feeding back into the core stomatal bHLHs were also revealed. For example, the POLAR family of potential BR signaling scaffolds emerged first from stomatal lineage profiles (Pillitteri et al., 2011; Houbaert et al. 2018) and genome-wide maps of SPCH targets (Lau et al., 2014) inspired research that established roles for cytokinin signaling in tuning stomatal lineage divisions (Vatén et al., 2018) and new connections between temperature and regulation of stomatal development (Lau et al., 2018).
Divisions and fate transitions of the stomatal lineage
The stomatal lineage initiates in leaf primordia. Here, many young epidermal cells express SPCH RNA, and a subset of these cells enter the stomatal lineage upon stable expression of SPCH protein (MacAlister et al., 2007). These stomatal lineage initial cells are likely chosen stochastically in a patterning mechanism that is dependent on feedback interactions among SPCH and ICE1 and/or SCRM2 (Horst et al., 2015). The remaining young epidermal cells differentiate into pavement cells or trichomes. High SPCH levels enable cells to undergo an asymmetric cell division (ACD), producing daughter cells of unequal cell size and fate. The larger daughter cell is the stomatal lineage ground cell (SLGC) (Shpak et al., 2005), whereas the smaller cell is a meristemoid (Nadeau and Sack, 2002). The latter can continue to express SPCH, giving it the capacity to divide asymmetrically again several more times in a process of amplifying divisions (Robinson et al., 2011) (see poster). SLGCs either differentiate into pavement cells or reinitiate SPCH expression and undergo spacing divisions by dividing asymmetrically again. These ACDs produce a loosely patterned tissue with dispersed adult stem cells that drive growth throughout the epidermis. When a meristemoid ceases ACDs, it will stop expressing SPCH protein, start expressing MUTE, and become a round guard mother cell (GMC) (Pillitteri et al., 2007). GMCs then initiate FAMA expression, cease MUTE expression, and symmetrically divide exactly once to produce a pair of guard cells (GCs), which remodel a central pore and establish kidney-shaped morphology (Han et al., 2018; Ohashi-Ito et al., 2006).
Stomata lineage regulators: cell–cell signaling
Stomatal patterning is critical for efficient regulation of gas exchange (Dow et al., 2014). Stomata are always separated from one another by one non-GC, a phenomenon known as the one-cell-spacing rule (Geisler et al., 2000). The placement and number of stomata in the epidermis are partly determined by the number of entry, amplifying and spacing divisions that occur; cell–cell signaling is largely responsible for tuning this process (see poster). Cell–cell communication among stomatal lineage cells is mediated by secreted peptides in the EPIDERMAL PATTERNING FACTOR (EPF) family. Receptors for these peptides include the membrane-bound receptor-like kinases (RLKs) in the ERECTA family (ERf), along with their epidermal-specific co-regulator and heterodimerization partner TOO MANY MOUTHS (TMM) (Shpak et al., 2004, 2005) and general co-receptors of the SOMATIC EMBRYOGENESIS RECEPTOR KINASE (SERK) family (Meng et al., 2015). EPF2, secreted by meristemoid mother cells (MMCs) and meristemoids, is detected by ERECTA–TMM heterodimeric complexes in protodermal cells (Hunt and Gray, 2009; Lee et al., 2012). EPF1 is detected by ERECTA-LIKE 1 (ERL1)–TMM heterodimeric complexes in SLGCs (Lee et al., 2012). Recent work indicates that EPF1 is also perceived by GMCs, which is critical for timing stomatal differentiation (Qi et al., 2017). Signaling is further modulated by structural features such as plasma membrane–endoplasmic reticulum contact sites, which regulate ERL2 levels in order to fine-tune EPF perception, possibly by changing the relative amounts of ERECTA family homodimers and heterodimers (Ho et al., 2016). TMM also binds another secreted peptide, STOMAGEN (also known as EPFL9), which is a positive regulator of stomatal development and is expressed in the inner tissue mesophyll, as opposed to stomata-bearing epidermal layers (Sugano et al., 2010).
Several intracellular signaling cascades affect stomatal development, primarily through the phosphorylation and subsequent downregulation of SPCH. Genetic data indicate that a mitogen-activated protein kinase (MAPK) cascade –comprising the MAPK-kinase kinase YODA (YDA), mitogen-activated protein kinase kinases 4 and 5 (MKK4, MKK5), and mitogen-activated protein kinases 3 and 6 (MPK3, MPK6) – acts downstream of the EPF–ERECTA signaling (Gudesblat et al., 2012; Lampard et al., 2008; Wang et al., 2007). Simultaneously, kinases of the shaggy-like kinase family such as BRASSINOSTEROID-INSENSITIVE 2 (BIN2, also known as ASK7), which are connected to brassinosteroid (BR) signaling, also target and downregulate SPCH protein (Gudesblat et al., 2012).
How developmental specificity is mediated by broadly expressed factors such as YODA remains an open question. Domain swaps among MAPK family members engineered to signal only in specific stages of the stomatal lineage identified unique portions of MKK5 and MKK7 that enabled them to affect divergent developmental decisions, possibly by mediating interaction with different scaffold proteins (Wengier et al., 2018). Components of the MAPK cascade depend on scaffold proteins to obtain asymmetric inheritance during ACDs, and their differential presence in daughters of an ACD could be responsible for mediating cell fate asymmetry (Houbaert et al., 2018; Zhang et al., 2015). This links MAPK signaling to polarity proteins, which we discuss in the next section. The MAPK cascade, along with secreted peptides and membrane-bound receptors described here, are critical for regulating stomatal patterning.
Stomatal lineage regulators: polarity
Mechanistic connections between signaling components and stomatal ‘polarity proteins’ in coordinating ACDs come from studies of BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE (BASL) (Dong et al., 2009) and POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION (POLAR) (Pillitteri et al., 2011) proteins. In the stomatal lineage, ACDs are coordinated by these polarity proteins, which localize to specific subdomains of the cell membrane in stomatal lineage cells prior to ACDs. This is critical because errors in stomatal lineage ACDs result in stomata contacting each other that, as a consequence, do not open or close efficiently (Dow et al., 2014). The localization of the accumulated proteins at specific cortical subdomains, referred to as the polarity domain or crescent, is predictive and instructive of the orientation of an ACD (Dong et al., 2009) (see poster). BASL and POLAR have partially overlapping polarity domains (Houbaert et al., 2018), and BASL activity is required for POLAR to be polarized (Pillitteri et al., 2011). When stomatal lineage cells divide asymmetrically, the polarity crescent is always found in the SLGC (Dong et al., 2009). BASL polarization is tightly regulated by phosphorylation on several residues (Zhang et al., 2016a). Changing phosphorylation status alters the relative amount of polarized cortical to nuclear BASL with varying ability to rescue the basl phenotype (Zhang et al., 2015, 2016a,b). Evidence supports a mechanism wherein BASL is phosphorylated by MPK3 and/or MPK6 and serves as a scaffold to hold MPK3/6 and YODA at the polarity crescent (Zhang et al., 2015), resulting in differential signaling capacity and, ultimately, differential SPCH protein levels in the two daughter cells (Zhang et al., 2016b). Interestingly, POLAR appears to regulate stomatal lineage ACDs by altering subcellular localization of BIN2 (Houbaert et al., 2018). This change can relieve the repressive effects of BIN2 on SPCH (discussed below) by sequestering BIN2 to the polarity crescent and freeing SPCH to drive ACD (Houbaert et al., 2018).
Observations of polarity protein dynamics in live tissues have also revealed tissue-level growth coordination in Arabidopsis leaves and cotyledons. Fluorescently tagged BASL that is expressed under the control of a ubiquitous promoter successfully polarizes in all cells and reveals a coordinated whole-leaf polarity field (Mansfield et al., 2018). However, the degree to which individual cells polarize relative to this overall field varies; in meristemoids and SLGCs, it appears that orientation relative to neighbor cells, as dictated by EPF signaling, and potentially mechanical forces, dominates over global alignments (Bringmann and Bergmann, 2017).
Auxin transport has long been recognized as a critical cue for coordinating growth in Arabidopsis (Sabatini et al., 1999). It is tempting to think that auxin and the polarized localization of its transporters might have a role in stomatal lineage polarity, but current data do not completely support this. Auxin transporter PIN-FORMED 1 (PIN1) is polarized in very young leaves (Kuchen et al., 2012) and BASL, when ectopically expressed at this early stage, is polarized to the opposite side of the cell (Mansfield et al., 2018). However, during the leaf stages where the stomatal lineage produces the largest number of asymmetric divisions, PIN1 is no longer expressed, and PIN3, which can be seen throughout the epidermis, is not polarized (Le et al., 2014; Robinson et al., 2011). Auxin signaling does affect stomatal fates (Le et al., 2014), but it is not clear whether this is through polar transport.
Overall, current research indicates that polarity proteins critically regulate ACDs and do so in collaboration with BR and MAPK signaling. In the next section, we will explore how hormonal and environmental signals regulate stomatal lineage ACDs.
Regulators: hormonal and environmental signaling
Plant hormones also mediate environmental responses. Stomata are critical gatekeepers between plants and their environments. As such, stomatal development integrates a tremendous number of hormonal and environmental signals to optimize the number and placement of stomata for the ambient environment of a plant (see poster). Much of this regulation converges on stomatal lineage initiation by regulating SPCH expression or SPCH protein levels. Drought and the ‘drought hormone’ abscisic acid (ABA) regulate stomatal closing (reviewed in Cutler et al., 2010). ABA mediates signaling of drought conditions in Arabidopsis and can inhibit progression of stomatal development (Tanaka et al., 2013). Interestingly, recent work has demonstrated that osmotic stress, which mimics drought, activates the YODA–MKK4/5–MPK3/6 signaling cascade, thereby downregulating SPCH activity and limiting the number of stomatal lineage ACDs (Kumari et al., 2014). Therefore, drought also regulates stomata behaviorally and developmentally.
Increasing levels of carbon dioxide (CO2) induce expression of the extracellular protease CO2 RESPONSE SECRETED PROTEASE (CRSP), which represses stomatal development by activating extracellular EPF2 peptide through cleavage of its pro-peptide form (Engineer et al., 2014). Furthermore, CARBONIC ANHYDRASE 1 and 4 (CA1, also known as BCA1; CA4, also known as BCA4), which have been shown to regulate stomatal aperture in response to CO2 levels (Hu et al., 2010), are also required together to induce EPF2 expression when atmospheric CO2 is high (Engineer et al., 2014). This indicates dual roles for CO2 in stomatal physiology and development.
Temperature and light affect stomatal development through phytochrome B (PHYB), whereas light also signals to stomatal development through other phytochrome and cryptochrome receptors to positively regulate stomatal development (Kang et al., 2009). The E3 ubiquitin-protein ligase COP1 degrades ICE1 in the absence of light, which inhibits stomatal development (Lee et al., 2017). This response may be auxin-dependent, because auxin-insensitive mutant seedlings that are grown in darkness fail to repress stomatal development (Balcerowicz et al., 2014). Mechanistically, activated PHYB represses PHYTOCHROME INTERACTING FACTOR 4 (PIF4), which downregulates SPCH levels. PIF4 itself is a SPCH transcriptional target, creating a negative feedback loop (Lau et al., 2018) and further linking light and temperature signals to BR signaling. PIF4, along with SPCH and YODA, is a target of BIN2 repression (Kim et al., 2012). These targets are de-repressed upon BR signaling through repression of BIN2 via the BRI1 SUPPRESSOR 1 (BSU1) family of phosphatases, in turn. Thus, light and temperature signal to stomatal development through many of the same components. Moving forward, it will be interesting to see how stomatal development responds to apparently conflicting signals, such as low light and high temperature.
PIF4 might also connect light or temperature signaling to cytokinin signaling through induction of the cytokinin-degrading enzyme CYTOKININ OXIDASE 5 (CKX5) (Nomoto et al., 2012). Cytokinin, a well-known promoter of cell divisions and vascular patterning, was recently shown to regulate the balance between stomatal lineage amplifying and spacing divisions, tuning the cellular composition of the epidermis (Vatén et al., 2018). Cell type-specific profiling of gene expression within the stomatal lineage and SPCH target profiling revealed stomatal lineage-specific repressive type A ARABIDOPSIS RESPONSE REGULATORs (ARRs), ARR16 and ARR17. These ARRs, together with CLAVATA3/ESR-RELATED 9 (CLE9) and CLE10, regulate cytokinin levels (Vatén et al., 2018). Active cytokinin signaling increases SPCH levels to promote spacing divisions; increased SPCH then positively regulates ARR16 and ARR17 to suppress cytokinin levels. Furthermore, increased SPCH levels promote CLE9 and CLE10 expression, which negatively regulates ARR16 and ARR17. Thus, SPCH and cytokinin signaling constitutes a complex feedback loop (see poster).
Taken together, a change in stomatal density or pattern can be attributed to experimental manipulation of nearly all the classic plant hormones, mainly by modulating SPCH protein and RNA levels. A future challenge will be to decipher how SPCH can integrate so many different inputs.
Evolution of stomatal development
Much of our knowledge regarding stomatal development comes from the model dicot Arabidopsis. Yet, stomata are found in nearly all land plants, with reasonably consistent morphology: stomata in non-vascular plants and in dicots have a characteristic kidney bean shape, whereas monocot stomata are more typically dumbbell shaped. Evidence from the fossil record indicates stomata morphologically similar to those in dicots were found as far back as 410 million years ago (Edwards et al., 1992). Recent work has taken advantage of this deep evolutionary conservation to expand our knowledge into plants that are less experimentally tractable, but more important as food crops such as rice, corn and wheat. Furthermore, as stomatal structure has been largely preserved through evolution (Chater et al., 2016,b11, 2017; Rudall et al., 2013), increased understanding of stomatal development across species can reveal essential evolutionary mechanisms for stomatal development. Many key stomatal lineage regulators are conserved (see poster and Box 2). Orthologs of the core stomatal lineage regulators control grass stomatal development as well, with some variation in functionality.
Box 2. Evolution of stomatal development: deep ancestry and divergence.
Mosses and angiosperms last shared a common ancestor over 400 million years ago (Morris et al., 2018). Stomatal development in the moss Physcomitrella patens requires PpSMF1 (SPEECHLESS, MUTE and FAMA-like) and PpSCREAM1, two orthologs of genes that were first characterized in Arabidopsis, SPCH and MUTE (Chater et al., 2016). Patterning of stomata in Physcomitrella involves orthologs of TMM, EPF1 and ERECTA (Caine et al., 2016). Thus, the core stomatal lineage regulators have been preserved across vast evolutionary distances, indicating control of stomatal development is deeply fundamental in the plant kingdom. This dovetails with previous observations that bHLH transcription factor cascades regulate development across the kingdoms of life; closely related bHLH transcription factors also regulate muscle development in animals, suggesting the foundations of this strategy were already in place when plants and animals diverged 1600 million years ago (Matos and Bergmann, 2014).
Monocot and dicot stomata have distinct morphology and patterning, but both employ orthologs of the same set of core transcriptional regulators – SPCH, MUTE, FAMA, ICE1 and SCRM2 (reviewed in Hepworth et al., 2018). What then accounts for the divergent epidermal patterning in these species? The polarity proteins BASL and POLAR are critical patterning regulators (Dong et al., 2009; Houbaert et al., 2018; Pillitteri et al., 2011). Orthologs of these proteins have not been reported in non-dicots, and protein searches with BLAST (Altschul et al., 1990) do not reveal any obvious non-dicot candidates. In parallel, repeated asymmetric amplifying divisions are common in Arabidopsis and other rosid eudicots, but not in grasses or earlier-derived angiosperms (reviewed in Rudall et al., 2013). So, although stomatal lineage identity regulators are conserved, BASL and POLAR do not appear to be. No cross-species functional tests have yet been done, but it is intriguing to speculate that the polarity proteins may have evolved as part of a patterning mechanism that exists only in dicots.
Stomatal development has been described in several cereal crops and in the diploid biofuel model purple false brome (Brachypodium distachyon). Grass leaves differ from Arabidopsis in that their stomatal lineage cells are typically restricted to specific cell files, and they have a strict base-to-tip growth orientation, with earlier phases of stomatal development occurring closer to the base of the leaf (for details of grass leaf development see Hepworth et al., 2018). In Brachypodium, stomatal cell files are specified by BdSPCH1 and BdSPCH2, which precede and drive asymmetric entry divisions with putative binding partner BdICE1 (Raissig et al., 2016). There are no repeated rounds of ACDs, so the smaller cells produced in this phase act as GMCs and the larger cells become intervening pavement cells. BdYODA mutants fail to establish this fate asymmetry, producing clusters of GMCs that eventually produce incorrectly spaced GCs (Abrash et al., 2018). As in Arabidopsis, Brachypodium GMCs express BdMUTE (Raissig et al., 2017). Mature grass stomata are four-celled structures, where the GC pair is flanked on each side by a subsidiary cell (SC) (see poster). BdMUTE further serves to recruit SCs, which contribute to improved environmental responsiveness of stomata in Brachypodium (Raissig et al., 2017). In barley and rice, both the entry ACD and SC recruitment are repressed by overexpression of the EPF peptides HvEPF1 and OsEPF2, respectively, suggesting parallel roles in repressing stomatal progression between grasses and dicots. Importantly, for agriculture, these manipulations can improve water use efficiency (Caine et al., 2019; Hughes et al., 2017; Lu et al., 2019).
However, unlike in Arabidopsis, where ICE1 and SCRM2 broadly and redundantly regulate stomatal development, loss of BdICE1 alone prevents stomatal lineage initiation and BdSCRM2 functionality is restricted to stomatal lineage termination (Raissig et al., 2016). BdSCRM2 is expressed throughout the Brachypodium stomatal lineage, but is only required for the differentiation of mature stomata (Raissig et al., 2016). In rice OsFAMA mutants, GMCs can divide once symmetrically, but fail to establish GC morphology (Liu et al., 2009). This is unlike Arabidopsis, where GMCs in FAMA mutants divide uncontrollably while failing to achieve GC morphology (Ohashi-Ito et al., 2006). The mutant phenotypes of BdSCRM2 and OsFAMA – four-celled stomatal complexes where GCs never attain their characteristic dumbbell shape – suggest that these proteins may function cooperatively in monocot stomatal lineage terminal differentiation.
These studies reveal the deep conservation of stomata, underlining the physiological importance of stomata in land plants. The regulatory mechanisms we describe here – cell–cell signaling, polarity, and environmental and hormonal signals – all modulate the fundamental and highly conserved regulators of the series of divisions and identity changes known as the stomatal lineage.
Conclusions and future perspectives
Studying stomatal development reveals regulatory strategies by which plants optimize developmental trajectories. This might provide broad insight into plant development strategies, since many of these mechanisms are alternatively purposed in other tissues in plants. For instance, cytokinin signaling regulates the cellular composition in the Arabidopsis epidermis (Vatén et al., 2018). This hormone is also critical for phyllotactic patterning (Giulini et al., 2004) and in the regulation of vascular cell identity and pattern (Bishopp et al., 2011). BR signaling impacts stomatal development at several levels (Houbaert et al., 2018; Kim et al., 2012) and regulates multiple aspects of root growth (reviewed in Wei and Li, 2016). This makes the stomatal lineage a valuable model system for studying general developmental strategies in plants, but it also poses a problem. Classical approaches define gene function by mutant phenotype. Many of the early-identified stomatal lineage regulators were specific to the stomatal lineage and were identified in mutant screens for stomatal lineage defects (Dong et al., 2009; MacAlister et al., 2007; Ohashi-Ito et al., 2006; Pillitteri et al., 2007). However, mutant phenotypes of broadly expressed stomatal lineage regulators will also have non-stomatal lineage defects, which can lead to confounding pleiotropic effects. We discussed recent advancements that circumvented this hurdle by using genome-wide profiling of stomatal lineage cells (see Box 1) to identify stomatal lineage-specific functions for broadly employed developmental regulators, which can potentially be validated by tissue-specific gene editing (Decaestecker et al., 2018 preprint). Moving forward, novel discoveries of stomatal lineage regulators will likely be aided by the production of cell type-specific proteomes as well as finer-scale genome-wide datasets.
Acknowledgements
We would like to thank members of the Bergmann Lab for their insightful comments. Yan Gong provided helpful commentary on the manuscript. Dirk Spencer and Andrea Mair gave valuable feedback on poster panel design.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
L.R.L. was funded by National Institutes of Health training grant NIH5T32GM007276 to Stanford University; D.C.B. is an investigator of the Howard Hughes Medical Institute. Deposited in PMC for release after 12 months.
Cell science at a glance
A high-resolution version of the poster and individual poster panels are available for downloading at http://jcs.biologists.org/lookup/doi/10.1242/jcs.228551.supplemental.
References
- Abrash E., Anleu Gil M. X., Matos J. L. and Bergmann D. C. (2018). Conservation and divergence of YODA MAPKKK function in regulation of grass epidermal patterning. Development 145, dev165860 10.1242/dev.165860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrian J., Chang J., Ballenger C. E., Bargmann B. O. R., Alassimone J., Davies K. A., Lau O. S., Matos J. L., Hachez C., Lanctot A. et al. (2015). Transcriptome dynamics of the stomatal lineage: birth, amplification, and termination of a self- renewing population. Dev. Cell 33, 107-118. 10.1016/j.devcel.2015.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. and Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403-410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Balcerowicz M., Ranjan A., Rupprecht L., Fiene G. and Hoecker U. (2014). Auxin represses stomatal development in dark-grown seedlings via Aux/IAA proteins. Development 141, 3165-3176. 10.1242/dev.109181 [DOI] [PubMed] [Google Scholar]
- Bergmann D. C., Lukowitz W. and Somerville C. R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304, 1494-1497. 10.1126/science.1096014 [DOI] [PubMed] [Google Scholar]
- Bishopp A., Help H., El-Showk S., Weijers D., Scheres B., Friml J., Benková E., Mähönen A. P. and Helariutta Y. (2011). A mutually inhibitory interaction between Auxin and Cytokinin specifies vascular pattern in roots. Curr. Biol. 21, 917-926. 10.1016/j.cub.2011.04.017 [DOI] [PubMed] [Google Scholar]
- Bringmann M. and Bergmann D. C. (2017). Tissue-wide mechanical forces influence the polarity of stomatal stem cells in Arabidopsis. Curr. Biol. 27, 877-883. 10.1016/j.cub.2017.01.059 [DOI] [PubMed] [Google Scholar]
- Caine R. S., Chater C. C., Kamisugi Y., Cuming A. C., Beerling D. J., Gray J. E. and Fleming A. J. (2016). An ancestral stomatal patterning module revealed in the non-vascular land plant Physcomitrella patens. Development 143, 3306-3314. 10.1242/dev.135038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caine R. S., Yin X., Sloan J., Harrison E. L., Mohammed U., Fulton T., Biswal A. K., Dionora J., Chater C. C., Coe R. A. et al. (2019). Rice with reduced stomatal density conserves water and has improved drought tolerance under future climate conditions. New Phytol. 221, 371-384. 10.1111/nph.15344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater C. C., Caine R. S., Tomek M., Wallace S., Kamisugi Y., Cuming A. C., Lang D., MacAlister C. A., Casson S., Bergmann D. C. et al. (2016). Origin and function of stomata in the moss Physcomitrella patens. Nat. Plants 2, 16179 10.1038/nplants.2016.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater C. C. C., Caine R. S., Fleming A. J. and Gray J. E. (2017). Origins and evolution of stomatal development. Plant Physiol. 174, 624-638. 10.1104/pp.17.00183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S. R., Rodriguez P. L., Finkelstein R. R. and Abrams S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651-679. 10.1146/annurev-arplant-042809-112122 [DOI] [PubMed] [Google Scholar]
- Decaestecker W., Buono R. A., Pfeiffer M. L., Vangheluwe N., Jourquin J., Karimi M., Van Isterdael G., Beeckman T., Nowack M. K. et al. (2018). CRISPR-TSKO facilitates efficient cell type-, tissue-, or organ-specific mutagenesis in Arabidopsis. bioRxiv 474981 10.1101/474981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., MacAlister C. A. and Bergmann D. C. (2009). BASL controls asymmetric cell division in arabidopsis. Cell 137, 1320-1330. 10.1016/j.cell.2009.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow G. J., Berry J. A. and Bergmann D. C. (2014). The physiological importance of developmental mechanisms that enforce proper stomatal spacing in Arabidopsis thaliana. New Phytol. 201, 1205-1217. 10.1111/nph.12586 [DOI] [PubMed] [Google Scholar]
- Edwards D., Davies K. L. and Axe L. (1992). A vascular conducting strand in the early land plant Cooksonia. Nature 357, 683-685. 10.1038/357683a0 [DOI] [Google Scholar]
- Engineer C. B., Ghassemian M., Anderson J. C., Peck S. C., Hu H. and Schroeder J. I. (2014). Carbonic anhydrases, EPF2 and a novel protease mediate CO2 control of stomatal development. Nature 513, 246-250. 10.1038/nature13452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M., Nadeau J. and Sack F. D. (2000). Oriented Asymmetric Divisions That Generate the Stomatal Spacing Pattern in Arabidopsis Are Disrupted by the too many mouths Mutation. Plant Cell Online 12, 2075-2086. 10.1105/tpc.12.11.2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulini A., Wang J. and Jackson D. (2004). Control of phyllotaxy by the cytokinin-inducible response regulator homologue ABPHYL1. Nature 430, 1031-1034. 10.1038/nature02778 [DOI] [PubMed] [Google Scholar]
- Gudesblat G. E., Schneider-Pizoń J., Betti C., Mayerhofer J., Vanhoutte I., van Dongen W., Boeren S., Zhiponova M., de Vries S., Jonak C. et al. (2012). SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat. Cell Biol. 14, 548-554. 10.1038/ncb2471 [DOI] [PubMed] [Google Scholar]
- Han S.-K., Qi X., Sugihara K., Dang J. H., Endo T. A., Miller K. L., Kim E.-D., Miura T. and Torii K. U. (2018). MUTE directly orchestrates cell-state switch and the single symmetric division to create stomata. Dev. Cell 45, 303-315.e5. 10.1016/j.devcel.2018.04.010 [DOI] [PubMed] [Google Scholar]
- Hepworth C., Caine R. S., Harrison E. L., Sloan J. and Gray J. E. (2018). Stomatal development: focusing on the grasses. Curr. Opin. Plant Biol. 41, 1-7. 10.1016/j.pbi.2017.07.009 [DOI] [PubMed] [Google Scholar]
- Ho C.-M. K., Paciorek T., Abrash E. and Bergmann D. C. (2016). Modulators of stomatal lineage signal transduction alter membrane contact sites and reveal specialization among ERECTA kinases. Dev. Cell 38, 345-357. 10.1016/j.devcel.2016.07.016 [DOI] [PubMed] [Google Scholar]
- Horst R. J., Fujita H., Lee J. S., Rychel A. L., Garrick J. M., Kawaguchi M., Peterson K. M. and Torii K. U. (2015). Molecular framework of a regulatory circuit initiating two-dimensional spatial patterning of stomatal lineage. PLoS Genet. 11, e1005374 10.1371/journal.pgen.1005374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaert A., Zhang C., Tiwari M., Wang K., de Marcos Serrano A., Savatin D. V., Urs M. J., Zhiponova M. K., Gudesblat G. E., Vanhoutte I. et al. (2018). POLAR-guided signalling complex assembly and localization drive asymmetric cell division. Nature 563, 574-578. 10.1038/s41586-018-0714-x [DOI] [PubMed] [Google Scholar]
- Hu H., Boisson-Dernier A., Israelsson-Nordström M., Böhmer M., Xue S., Ries A., Godoski J., Kuhn J. M. and Schroeder J. I. (2010). Carbonic anhydrases are upstream regulators of CO2-controlled stomatal movements in guard cells. Nat. Cell Biol. 12, 87-93. 10.1038/ncb2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J., Hepworth C., Dutton C., Dunn J. A., Hunt L., Stephens J., Waugh R., Cameron D. D. and Gray J. E. (2017). Reducing stomatal density in barley improves drought tolerance without impacting on yield. Plant Physiol. 174, 776-787. 10.1104/pp.16.01844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L. and Gray J. E. (2009). The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 19, 864-869. 10.1016/j.cub.2009.03.069 [DOI] [PubMed] [Google Scholar]
- Jezek M. and Blatt M. R. (2017). The membrane transport system of the guard cell and its integration for stomatal dynamics. Plant Physiol. 174, 487-519. 10.1104/pp.16.01949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka M. M., Pillitteri L. J., Fujii H., Yoshida Y., Bogenschutz N. L., Takabayashi J., Zhu J.-K. and Torii K. U. (2008). SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20, 1775-1785. 10.1105/tpc.108.060848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C.-Y., Lian H.-L., Wang F.-F., Huang J.-R. and Yang H.-Q. (2009). Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21, 2624-2641. 10.1105/tpc.109.069765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.-W., Michniewicz M., Bergmann D. C. and Wang Z.-Y. (2012). Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482, 419-422. 10.1038/nature10794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchen E. E., Fox S., de Reuille P. B., Kennaway R., Bensmihen S., Avondo J., Calder G. M., Southam P., Robinson S., Bangham A. et al. (2012). Generation of leaf shape through early patterns of growth and tissue polarity. Science 335, 1092-1096. 10.1126/science.1214678 [DOI] [PubMed] [Google Scholar]
- Kumari A., Jewaria P. K., Bergmann D. C. and Kakimoto T. (2014). Arabidopsis reduces growth under osmotic stress by decreasing SPEECHLESS protein. Plant Cell Physiol. 55, 2037-2046. 10.1093/pcp/pcu159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard G. R., MacAlister C. A. and Bergmann D. C. (2008). Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322, 1113-1116. 10.1126/science.1162263 [DOI] [PubMed] [Google Scholar]
- Lau O. S., Davies K. A., Chang J., Adrian J., Rowe M. H., Ballenger C. E. and Bergmann D. C. (2014). Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science 345, 1605-1609. 10.1126/science.1256888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O. S., Song Z., Zhou Z., Davies K. A., Chang J., Yang X., Wang S., Lucyshyn D., Tay I. H. Z., Wigge P. A. et al. (2018). Direct control of SPEECHLESS by PIF4 in the high-temperature response of stomatal development. Curr. Biol. 28, 1273-1280.e3. 10.1016/j.cub.2018.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le J., Liu X.-G., Yang K.-Z., Chen X.-L., Zou J.-J., Wang H.-Z., Wang M., Vanneste S., Morita M., Tasaka M. et al. (2014). Auxin transport and activity regulate stomatal patterning and development. Nat. Commun. 5, 3090 10.1038/ncomms4090 [DOI] [PubMed] [Google Scholar]
- Lee J. S., Kuroha T., Hnilova M., Khatayevich D., Kanaoka M. M., McAbee J. M., Sarikaya M., Tamerler C. and Torii K. U. (2012). Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev. 26, 126-136. 10.1101/gad.179895.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H., Jung J.-H. and Park C.-M. (2017). Light inhibits COP1-mediated degradation of ICE transcription factors to induce stomatal development in arabidopsis. Plant Cell 29, 2817-2830. 10.1105/tpc.17.00371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Ohashi-Ito K. and Bergmann D. C. (2009). Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development 136, 2265-2276. 10.1242/dev.032938 [DOI] [PubMed] [Google Scholar]
- Lu J., He J., Zhou X., Zhong J., Li J. and Liang Y.-K. (2019). Homologous genes of epidermal patterning factor regulate stomatal development in rice. J. Plant Physiol. 234–235, 18-27. 10.1016/j.jplph.2019.01.010 [DOI] [PubMed] [Google Scholar]
- MacAlister C. A., Ohashi-Ito K. and Bergmann D. C. (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445, 537-540. 10.1038/nature05491 [DOI] [PubMed] [Google Scholar]
- Mansfield C., Newman J. L., Olsson T. S. G., Hartley M., Chan J. and Coen E. (2018). Ectopic BASL reveals tissue cell polarity throughout leaf development in Arabidopsis thaliana. Curr. Biol. 28, 2638-2646.e4. 10.1016/j.cub.2018.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J. and Bergmann D. (2014). Convergence of stem cell behaviors and genetic regulation between animals and plants: insights from the Arabidopsis thaliana stomatal lineage. F1000Prime Rep. 6, doi:10.12703/P6-53 10.12703/P6-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwain J. C. and Steinthorsdottir M. (2017). Paleoecology, ploidy, paleoatmospheric composition, and developmental biology: a review of the multiple uses of fossil stomata. Plant Physiol. 174, 650-664. 10.1104/pp.17.00204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Chen X., Mang H., Liu C., Yu X., Gao X., Torii K. U., He P. and Shan L. (2015). Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr. Biol. 25, 2361-2372. 10.1016/j.cub.2015.07.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. L., Puttick M. N., Clark J. W., Edwards D., Kenrick P., Pressel S., Wellman C. H., Yang Z., Schneider H. and Donoghue P. C. J. (2018). The timescale of early land plant evolution. Proc. Natl. Acad. Sci. USA 115, E2274-E2283. 10.1073/pnas.1719588115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y., Mori I. C. and Munemasa S. (2015). Diverse stomatal signaling and the signal integration mechanism. Annu. Rev. Plant Biol. 66, 369-392. 10.1146/annurev-arplant-043014-114707 [DOI] [PubMed] [Google Scholar]
- Nadeau J. A. and Sack F. D. (2002). Stomatal development in Arabidopsis. Arab. B. 1, e0066 10.1199/tab.0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomoto Y., Kubozono S., Yamashino T., Nakamichi N. and Mizuno T. (2012). Circadian clock- and PIF4-controlled plant growth: a coincidence mechanism directly integrates a hormone signaling network into the photoperiodic control of plant architectures in Arabidopsis thaliana. Plant Cell Physiol. 53, 1950-1964. 10.1093/pcp/pcs137 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K., Bergmann D. C. and Bergmann D. C. (2006). Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18, 2493-2505. 10.1105/tpc.106.046136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L. J., Sloan D. B., Bogenschutz N. L. and Torii K. U. (2007). Termination of asymmetric cell division and differentiation of stomata. Nature 445, 501-505. 10.1038/nature05467 [DOI] [PubMed] [Google Scholar]
- Pillitteri L. J., Peterson K. M., Horst R. J. and Torii K. U. (2011). Molecular profiling of stomatal meristemoids reveals new component of asymmetric cell division and commonalities among stem cell populations in arabidopsis. Plant Cell 23, 3260-3275. 10.1105/tpc.111.088583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Han S.-K., Dang J. H., Garrick J. M., Ito M., Hofstetter A. K. and Torii K. U. (2017). Autocrine regulation of stomatal differentiation potential by EPF1 and ERECTA-LIKE1 ligand-receptor signaling. Elife 6, e24102 10.7554/eLife.24102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig M. T., Abrash E., Bettadapur A., Vogel J. P. and Bergmann D. C. (2016). Grasses use an alternatively wired bHLH transcription factor network to establish stomatal identity. Proc. Natl. Acad. Sci. USA 113, 8326-8331. 10.1073/pnas.1606728113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig M. T., Matos J. L., Anleu Gil M. X., Kornfeld A., Bettadapur A., Abrash E., Allison H. R., Badgley G., Vogel J. P., Berry J. A. et al. (2017). Mobile MUTE specifies subsidiary cells to build physiologically improved grass stomata. Science 355, 1215-1218. 10.1126/science.aal3254 [DOI] [PubMed] [Google Scholar]
- Robinson S., Barbier de Reuille P., Chan J., Bergmann D., Prusinkiewicz P. and Coen E. (2011). Generation of spatial patterns through cell polarity switching. Science 333, 1436-1440. 10.1126/science.1202185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudall P. J., Hilton J. and Bateman R. M. (2013). Several developmental and morphogenetic factors govern the evolution of stomatal patterning in land plants. New Phytol. 200, 598-614. 10.1111/nph.12406 [DOI] [PubMed] [Google Scholar]
- Rui Y., Chen Y., Kandemir B., Yi H., Wang J. Z., Puri V. M. and Anderson C. T. (2018). Balancing strength and flexibility: how the synthesis, organization, and modification of guard cell walls govern stomatal development and dynamics. Front. Plant Sci. 9, 1202 10.3389/fpls.2018.01202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P. et al. (1999). An auxin-dependent distal organizer of pattern and polarity in the arabidopsis root. Cell 99, 463-472. 10.1016/S0092-8674(00)81535-4 [DOI] [PubMed] [Google Scholar]
- Shpak E. D., Berthiaume C. T., Hill E. J. and Torii K. U. (2004). Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131, 1491-1501. 10.1242/dev.01028 [DOI] [PubMed] [Google Scholar]
- Shpak E. D., McAbee J. M., Pillitteri L. J. and Torii K. U. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309, 290-293. 10.1126/science.1109710 [DOI] [PubMed] [Google Scholar]
- Sugano S. S., Shimada T., Imai Y., Okawa K., Tamai A., Mori M. and Hara-Nishimura I. (2010). Stomagen positively regulates stomatal density in Arabidopsis. Nature 463, 241-244. 10.1038/nature08682 [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Nose T., Jikumaru Y. and Kamiya Y. (2013). ABA inhibits entry into stomatal-lineage development in Arabidopsis leaves. Plant J. 74, 448-457. 10.1111/tpj.12136 [DOI] [PubMed] [Google Scholar]
- Vatén A., Soyars C. L., Tarr P. T., Nimchuk Z. L. and Bergmann D. C. (2018). Modulation of asymmetric division diversity through Cytokinin and SPEECHLESS regulatory interactions in the stomatal lineage. Dev. Cell 47, 53-66.e5. 10.1016/j.devcel.2018.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ngwenyama N., Liu Y., Walker J. C. and Zhang S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19, 63-73. 10.1105/tpc.106.048298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z. and Li J. (2016). Brassinosteroids regulate root growth, development, and symbiosis. Mol. Plant 9, 86-100. 10.1016/j.molp.2015.12.003 [DOI] [PubMed] [Google Scholar]
- Wei D., Liu M., Chen H., Zheng Y., Liu Y., Wang X., Yang S., Zhou M. and Lin J. (2018). Inducer of CBF expression 1 is a male fertility regulator impacting anther dehydration in Arabidopsis. PLoS Genet. 14, e1007695 10.1371/journal.pgen.1007695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer A. K., Matos J. L., Sharma N., Patell F., Murray J. A. H., Dewitte W. and Bergmann D. C. (2018). Lineage- and stage-specific expressed CYCD7;1 coordinates the single symmetric division that creates stomatal guard cells. Development 145, dev160671 10.1242/dev.160671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengier D. L., Lampard G. R. and Bergmann D. C. (2018). Dissection of MAPK signaling specificity through protein engineering in a developmental context. BMC Plant Biol. 18, 60 10.1186/s12870-018-1274-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang P., Shao W., Zhu J.-K. and Dong J. (2015). The BASL polarity protein controls a MAPK signaling feedback loop in asymmetric cell division. Dev. Cell 33, 136-149. 10.1016/j.devcel.2015.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Bergmann D. C. and Dong J. (2016a). Fine-scale dissection of the subdomains of polarity protein BASL in stomatal asymmetric cell division. J. Exp. Bot. 67, 5093-5103. 10.1093/jxb/erw274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Guo X. and Dong J. (2016b). Phosphorylation of the polarity protein BASL differentiates asymmetric cell fate through MAPKs and SPCH. Curr. Biol. 26, 2957-2965. 10.1016/j.cub.2016.08.066 [DOI] [PMC free article] [PubMed] [Google Scholar]