ABSTRACT
Although the formation of RNA-protein bodies has been studied intensively, their mobility and how their number and size are regulated are still poorly understood. Here, we show significantly increased mobility of nuclear speckles after transcriptional inhibition, including long-range directed motion of one speckle towards another speckle, terminated by speckle fusion, over distances up to 4 µm and with velocities between 0.2 µm/min and 1.5 µm/min. Frequently, three or even four speckles follow very similar paths, with new speckles appearing along the path followed by a preceding speckle. Speckle movements and fusion events contribute to fewer, but larger, speckles after transcriptional inhibition. These speckle movements are not actin dependent, but occur within chromatin-depleted channels enriched with small granules containing the speckle marker protein SON. Similar long-range speckle movements and fusion events were observed after heat shock or heavy metal stress, and during late G2 and early prophase. Our observations suggest a mechanism for long-range, directional nuclear speckle movements, contributing to overall regulation of nuclear speckle number and size as well as overall nuclear organization.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Interchromatin granule clusters, Nuclear bodies, Nuclear speckles
Summary: Nuclear speckle fusions occur within chromatin-depleted channels enriched with speckle granules via repeated directional movements of nuclear speckles over several micrometers after transcriptional inhibition, other stresses and during G2.
INTRODUCTION
Intracellular compartmentalization in eukaryotic cells is thought to represent a key evolutionary advance by which complex biochemical processes can be optimized and regulated within highly crowded cellular environments (Banks and Fradin, 2005; Cremer and Cremer, 2001; Weidemann et al., 2003). Compartmentalization is achieved not only with membrane-bound organelles but also through membraneless ‘bodies’, including P-bodies and stress granules in the cytoplasm, and nucleoli, nuclear speckles, and Cajal and promyelocytic leukemia protein (PML) bodies in the nucleus. Most membraneless bodies are RNA-protein-rich complexes that function in transcription, RNA processing and/or protein modification (Hyman et al., 2014; Zhu and Brangwynne, 2015). Recent studies have led to the concept of RNA-protein bodies forming as the result of a phase transition between constituent RNA and proteins and surrounding cytoplasm or nucleoplasm, through a demixing driven by interactions between low-complexity sequences and multivalent molecules (Han et al., 2012; Kaiser et al., 2008; Kato et al., 2012; Schwartz et al., 2013). RNA-protein bodies have been proven to have general physical characteristics of liquid droplets (Hyman et al., 2014; Zhu and Brangwynne, 2015). For example, P-granules undergo rapid assembly/disassembly, and exhibit dripping, fusion and wetting by shearing force (Brangwynne et al., 2009), and nucleoli exhibit viscous fluid dynamics such as liquid bridge formation when they are ruptured (Brangwynne et al., 2011). Fluorescence recovery after photobleaching (FRAP) analysis of molecular dynamics showed that RNA-protein bodies continuously turn over many of their constituent molecules over a timescale from a few seconds to a few minutes (Aizer et al., 2008; Buchan and Parker, 2009; Dundr et al., 2004). These microscopy observations suggest that these bodies are in a liquid or gel state with a constant flux of constituent molecules.
Despite a number of studies examining liquid properties and related behaviors of other cellular bodies, both the physical properties and dynamics of nuclear speckle bodies have not been well studied. Defined originally as interchromatin granule clusters (IGCs) by electron microscopy (Spector, 1993; Thiry, 1995), nuclear speckles contain large clusters of ∼20-nm ribonucleoprotein (RNP) granules, and are enriched in RNA-processing factors and polyadenylated RNAs (Carter et al., 1991; Mintz et al., 1999; Saitoh et al., 2004). Nuclei typically contain 20–40 irregular-shaped nuclear speckles varying in size from ∼0.5 µm to several micrometers (Lamond and Spector, 2003).
Because ‘pure’ liquid droplet bodies theoretically are predicted to merge into progressively fewer and larger droplets, a major question regarding cell bodies in general is how their number and size are regulated in the cell. Interestingly, with transcriptional inhibition, nuclear speckles become rounder and larger with a reduction in their overall number (O'Keefe et al., 1994; Spector et al., 1991). Live-cell microscopy revealed that normally speckles were relatively stationary, but displayed extension and dissociating particles. However, no peripheral dynamics were observed in the absence of transcription (Misteli et al., 1997). These previous studies did not address directly how speckle morphology changes dynamically in terms of speckle numbers, size and shape, or how similar the physical properties of nuclear speckles were to the physical properties of other RNA-protein bodies described as showing liquid droplet-type behaviors. One previous study (Zhang et al., 2016) presented anecdotal evidence of a directional speckle movement based on limited visual inspection of live-cell movies; besides the lack of quantitative analysis of the observed speckle trajectories, the imaging conditions used in this study appeared to induce considerable cell stress, based on the considerable speckle rounding observed even at time 0 at the start of these movies.
Using the change in nuclear speckle morphology induced by transcriptional inhibition as a model system, we here undertook a quantitative analysis of the mobility and liquid-like behaviors of nuclear speckles under carefully controlled, low-light imaging conditions. Consistent with previous observations, we observed a general change in morphology after transcriptional inhibition, with speckles becoming bigger and rounder. However, we found that this was accompanied by a significant increase in speckle mobility within the nucleus.
This increase in speckle mobility was not random. Instead, we observed long-range, directional movement of multiple speckles over micrometer distances. Surprisingly, we observed repeated movement of multiple speckles over very similar trajectories, or ‘tracks’, followed by fusion to the same larger speckle. In many cases, this occurred through the nucleation of new speckles at sites previously occupied by a speckle prior to its movement towards and fusion to a larger speckle, or at sites along the path followed by a preceding speckle. These repeated fusion events contributed to the overall decrease in nuclear speckle number and increased mean speckle size as a function of time after transcriptional inhibition. Analysis of fusion events showed that speckles show a similar viscosity as measured previously for other RNA-protein bodies such as nucleoli and P-bodies (Brangwynne et al., 2009, 2011). Similar speckle dynamics were observed after treatment with other transcriptional inhibitors, after heat shock or heavy metal stress, or under normal growth conditions during late G2 and prophase.
Perturbing normal actin polymerization did not prevent speckle long-range motion, the directionality of this motion or speckle fusion events. Therefore, we excluded the possibility that actin filaments might provide ‘tracks’ for this speckle movement. Combining live-cell imaging with correlative super-resolution light microscopy revealed that speckles move within chromatin-depleted channels. Moreover, paths followed by more than one speckle as visualized by live-cell microscopy contained a concentration of small granules containing the speckle-marker SON within these DNA-depleted channels.
Our results demonstrating repeated nucleation, long-range directed motion and fusion of nuclear speckles in live cells reveal a likely trafficking mechanism of nuclear speckles contributing to the control of speckle number, size and nuclear localization. Overall, our results demonstrate regulated dynamics of nuclear speckles, with the intranuclear distribution of nuclear speckles significantly affected by translocation as well as dynamic nucleation and fusion of nuclear speckles, and possibly by changes in nuclear chromatin structure that may remove barriers to long-range speckle movements.
RESULTS
Inhibition of RNA polymerase II transcription changes speckle morphology and increases overall speckle mobility
We used SON protein as a speckle marker to visualize nuclear speckles with high contrast. The SON protein showed the highest concentration in nuclear speckles versus nucleoplasm of any speckle markers we tested, with less distributed diffusely or in foci outside of nuclear speckles than other RS-domain proteins such as SC35 (also known as SRSF2) or ASF/SF1 (Khanna et al., 2014; Saitoh et al., 2004). The SON protein contains an RS domain and six other tandem repeat regions unique to SON (Sharma et al., 2010), which may function as a scaffold for protein assembly (Sharma et al., 2010). The RS domain is found in a large number of mRNA-processing proteins, many enriched in nuclear speckles, as well as some proteins related to RNA polymerase 2 transcription (Boucher et al., 2001). Knockdown of SON results in a significant change in nuclear speckle morphology as visualized by immunostaining of SC35, another RS domain-containing speckle marker protein (Sharma et al., 2010).
Attempts to express EGFP-SON from a cDNA plasmid transgene resulted in variegated and unstable transgene expression. To obtain stable expression, we instead transfected Chinese hamster ovary (CHO) K1 cells with a bacterial artificial chromosome (BAC) transgene containing the full-length SON human genomic sequence engineered by BAC recombineering to express a EGFP-SON fusion protein (Khanna et al., 2014). Use of this ∼200 kb BAC containing large 5′ and 3′ human genomic sequences flanking the SON gene enabled isolation of stable clones expressing uniform levels of EGFP-SON.
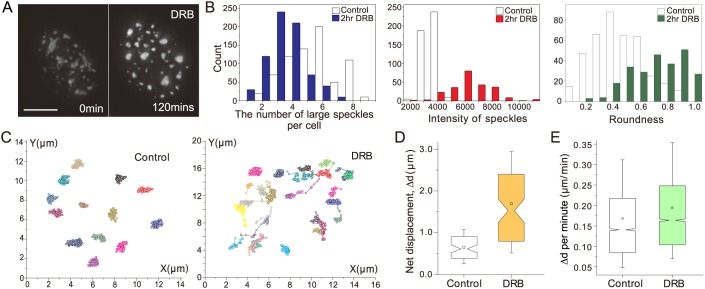
To study dynamics of speckles after transcriptional inhibition, we used 5,6-dichloro-1-β-ribofuranosyl benzimidazole (DRB). DRB blocks transcription via inhibition of kinases involved in transcription elongation (Bensaude, 2011; Yankulov et al., 1995). We confirmed in our CHO cell system previous work (O'Keefe et al., 1994; Spector et al., 1991) indicating that large speckles become brighter, rounder and fewer in number after DRB addition (Fig. 1A,B). These changes in speckle morphology after DRB treatment, visualized in a CHO cell clone expressing EGFP-SON, were comparable to the changes in speckle morphology after DRB treatment observed in wild-type CHO cells in which speckles were visualized by immunofluorescence (Fig. S1). The GFP-SON intensity within speckles became ∼2.5-fold higher on average 2 h after DRB addition. The roundness, defined as [4×area/π×(maximum axis length)2], increased ∼50% from 0.50 in control cells to 0.76 on average in DRB-treated cells. The roundness ranges from 1 for a perfect circle to 0 for a line. The transcriptional inhibitors α-amanitin and triptolide (TRL) and the heavy metal cadmium (Cd) had similar effects on speckle morphology as DRB (Fig. S2). However, we chose DRB for our imaging studies because it produced less rounding and change in shape of the nucleus in comparison to these other inhibitors. This reduced change in nuclear shape was critical for our tracking of speckle movement using alignment of nuclear images from different time points.
Fig. 1.
Inhibition of RNA polymerase II transcription by DRB changes speckle morphology and increases speckle mobility. (A) Changes in speckle morphology, visualized using GFP-SON, before and after DRB addition. Scale bar: 5 µm. (B) Measurement of speckle number (>1 μm in diameter) (left), intensity (middle) and roundness (right), before and after 2 h DRB treatment. Large speckles reduce in number (blue), and become brighter (∼2.5-fold, red) and rounder (1.5-fold increase in roundness, green) after DRB addition. (C) Speckle trajectories before and after DRB addition. Speckles, each assigned a different color, were tracked over a 1-h period in control (left) and DRB-treated (right) cells. (D) Net displacement (Δd) of speckles measured in control (white) and DRB-treated (yellow) cells. Speckle displacements increase after DRB addition (P=1.007×10–12; paired Student's t-test). Boxplots: boxes, mean (square inside box), median (notch of box), 25 (bottom) and 75 (top) percentiles; ends of error bars, 10 (bottom) and 90 (top) percentiles. n=75 speckles from ten control cells, n=80 speckles from ten DRB-treated cells from a single experiment. (E) Displacement (Δd) per minute increases from before (white) to after (green) DRB addition (P=2.2×10–16; paired Wilcoxon signed rank test). Boxplots as in D. n=4487 steps from 49 speckles (control), n=4375 steps from 50 speckles (DRB) from a single experiment.
Live-cell imaging revealed an obvious increase in nuclear speckle mobility during DRB treatment (Fig. 1C–E). To facilitate analysis, at each 1-min time point, we used 2D projections of 3D z-stacks. We used a cross-correlation approach to best correct for nuclear rotations and translations between adjacent time points. In comparison to control cells, in which nuclear speckles showed restricted movements over 1 h (Fig. 1C, left), in DRB-treated cells, a significant fraction of nuclear speckles showed longer-range net displacements (Fig. 1C, right). Overlapping trajectories corresponded to examples in which nuclear speckles came together and merged (Fig. 1C). DRB treatment increased the mean net displacement (Δd) of nuclear speckles over 1.5 h from 0.64 μm to 1.69 μm (P=1.007×10–12, paired Student's t-test; Fig. 1D), with the increase in the mean distance change per 1-min time point from 0.17 μm to 0.19 μm (P=2.2×10–16, paired Wilcoxon signed rank test; Fig. 1E).
DRB treatment induces long-range, directional speckle movements
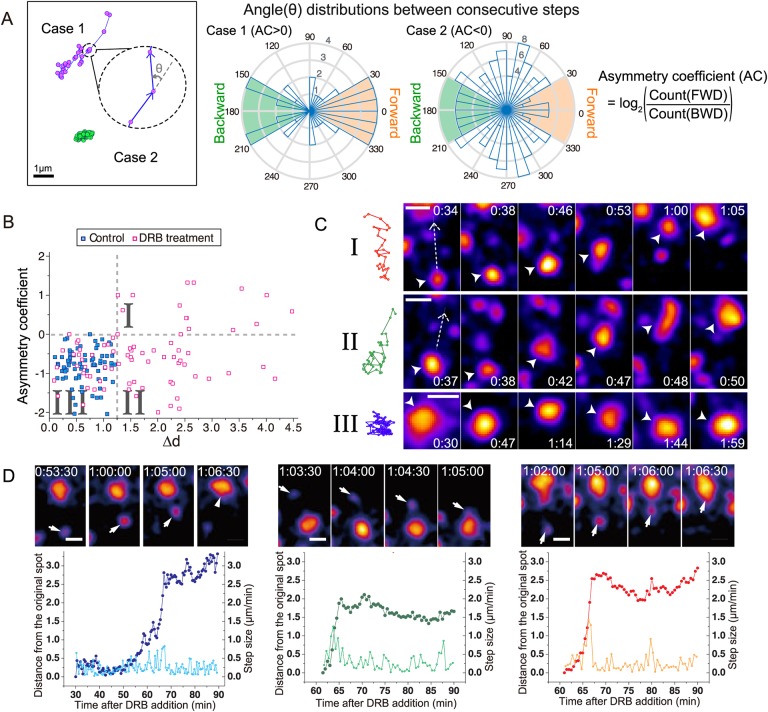
We characterized the movement of individual nuclear speckles after DRB addition by measuring the asymmetry coefficient (AC) together with the net displacement, Δd. We observed speckles for 1.5 h beginning 30 min after DRB addition using a 1-min time interval, tracking only the speckles that could be observed in at least 20 consecutive time points. To calculate the AC, we first measured the angle between the directions of speckle displacements for two consecutive time points (Fig. 2A). The AC was then defined as the logarithm to the base 2 of the ratio between the frequencies of backward (180±30°) versus forward (±30°) movements over the entire speckle trajectory (Izeddin et al., 2014). Random diffusion produces AC values near 0, because forward motion and backward motion are equally likely. AC values higher than 0 indicate a bias towards forward movements, whereas AC values below 0 suggest constrained diffusion.
Fig. 2.
DRB treatment induces long-range, directional speckle movements. (A) Analysis of angular distributions for two speckle trajectory examples. Left: two speckle trajectories, Case 1 (magenta) and Case 2 (green). The angle (θ) between every two consecutive steps is calculated (cartoon blowup). Middle: histogram distributions of angles for Case 1 (left) compared to Case 2 (right) trajectories. The radius of the shaded sectors (blue) shows the number of adjacent steps with a given angle. Backward (BWD) steps are defined by angles in green; forward (FWD) steps are defined by angles in orange. Right: asymmetry coefficients (AC) for Case 1 and Case 2 are negative and positive, respectively, as AC is defined as the log2 ratio of FWD over BWD steps. (B) Scatter plot of asymmetry coefficient (AC, y-axis) versus net displacement (Δd, x-axis) for speckle trajectories from control (blue squares) versus DRB-treated (pink squares) cells. Three trajectory types I–III are defined by their placement within three quadrants (I–III) defined by dotted lines (AC=0; Δd=1.25). Control cells fall into negative AC and low Δd quadrant (I). n=75 speckles from 10 control cells; n=80 speckles from 10 DRB-treated cells. (C) Speckle trajectories (left) and corresponding time-lapse images (right) from each of the three types I–III in B. Time (h:min) is after DRB addition. Scale bars: 1 µm. Dashed line arrows mark the direction of movement; arrowheads mark the moving speckle. Top: Type I: forward motion-dominant, long-range directional motion. Middle: Type II: long-range directional motion, but fewer forward steps. Bottom: Type III: confined speckle motion. See Movie 1. (D) Three examples of long-range directed speckle motion. Top: time-lapse images. Time (h:min:s) is after DRB addition. Scale bars: 1 µm. Arrowheads mark the speckle that moves. Bottom: distance (µm, vivid colors, left y-axis), measured from the starting spot position, and velocities (µm/min, light colors, right y-axis) as a function of time after DRB addition.
To compare speckle mobility in control versus DRB-treated cells, we created a scatter plot summarizing AC values versus net distance displacement for speckles (Fig. 2B). In cases in which a speckle moved and then merged with a second, stationary speckle, if the merged speckle remained stationary for 5 min or longer, the trajectory was terminated with this fusion event. In a small fraction of cases (∼5%), a speckle moved, fused with a second speckle, and then the fused speckle continued to move; these examples were counted as a single long trajectory of the first speckle.
This scatter plot reveals three motion regimes (Fig. 2B,C): Type I trajectories show large net displacements (≥1.25 µm) with positive AC values as a result of subtrajectories that contain a large number of discrete forward steps. Type II trajectories show large net displacements but negative AC values. In contrast, Type III trajectories (Fig. 2B,C; Movie 1) show small net displacements with negative AC values, consistent with a confined motion of speckles.
In control cells, essentially all speckles showed locally confined, Type III trajectories. No speckles (0/75) traveled further than 1.25 µm, and AC values were all below 0, with a total approximate AC range of −2.0–0.0. In contrast, in DRB-treated cells, speckle mobility and AC values increased significantly; 63% of speckle trajectories (Type I+Type II, 50/80) exhibited net distance displacements, Δd, greater than 1.25 µm, with 14 of 50 of these trajectories showing AC values greater than 0 (Type I, Fig. 2B). Several of these speckles moved as far as 3–4 µm, nearly three times as far as the largest displacement seen in control cells.
Plotting the distance moved as a function of time revealed that the subtrajectories containing periods of long-range movement were predominately unidirectional, with larger steps than stationary movements shown before or after the long-range movement. (Fig. 2D). The imprecision of the spatial alignment of nuclei between time points, due to changes in nuclear shape, prevents us from determining to what degree small reverse speckle movements represent true reverse speckle movements versus alignment errors. Projecting over time the 2D spatial projections from multiple time points revealed the linear path of these subtrajectories containing long-range directional movements (Fig. S3A,B).
Repeated long-range, directional speckle movements all terminate with speckle fusion
Strikingly, all the examples of long-range speckle movements shown in Fig. 2 and Fig. S3 terminate with a fusion between the smaller, mobile speckle and a larger, relatively immobile speckle. These are not exceptional examples, but rather the general rule observed for nearly all long-range movements contained within a single observation period.
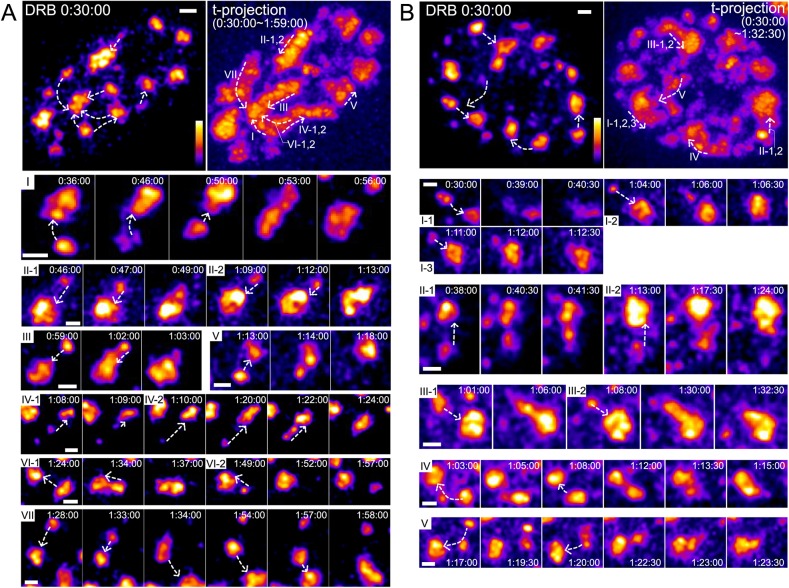
Fig. 3 shows all long-range speckle movements in two representative nuclei as examples. Ten long-range speckle movements in nucleus 1 (Fig. 3A; Movie 2) end with fusion to seven larger speckles; in three cases, two different small speckles move long-distance, and merge with the same larger speckle. Nine long-range speckle movements in nucleus 2 (Fig. 3B; Movie 3) end with fusion to five larger speckles; three smaller speckles merge with the same larger speckle in one case and two smaller speckles merge with the same larger speckle in two other cases. Thus, 19/19 long-range speckle trajectories terminate with fusion to a larger, mostly immobile nuclear speckle.
Fig. 3.
Long-range speckle movements are directed towards other speckles and terminated by speckle fusion. (A) Overview of speckle motions in one nucleus (cell clone E8). All images show maximum-intensity 2D projections of optical sections, using pseudocolor intensity scale. Time (h:min:s) is after DRB addition. Scale bars: 1 µm. Dashed line arrows show the direction of speckle movements. Top left: first time-lapse image taken 30 min after DRB addition. Top right: maximum-intensity projection over all time-lapse 2D images. Roman numerals indicate different regions showing speckle movements, ordered by the time of speckle movements. In some regions, multiple speckles appear and then move. Each of these speckles is tagged by a number following the roman numeral (i.e. II-1 and II-2 show the path of two different speckles moving in region II). Bottom: time-lapse images of each speckle movement marked by roman numeral and speckle number. See Movie 2. (B) Same as A for the second nucleus (cell clone D6). See Movie 3.
Again, projecting over time, the 3D spatial projections from multiple time points revealed a largely curvilinear path for these long-range speckle trajectories, within the resolution of the nuclear alignment between time points (Fig. 3A,B, top right).
Even more strikingly, in many cases, we saw repeated long-range speckle movements in which different nuclear speckles followed a very similar path to merge with the same single, larger speckle. In the two nuclei shown in Fig. 3, 6/12 larger speckles are the fusion targets for two or three mobile, smaller speckles. In each of these six examples, the second or even third smaller nuclear speckle moves along essentially the same linear path, within the approximate resolution of our nuclear alignment, to fuse with the larger speckle. Typically, after one speckle began to move on a long-range trajectory, a new speckle would nucleate and enlarge, either at the site of the first speckle prior to its movement (Fig. 4A,B; Movie 4) or along the trajectory followed by the first speckle (Fig. 4C,D; Movie 4, Fig. S3C). The second speckle would later begin to move along a similar trajectory to the first and then merge with the same speckle with which the first speckle moved.
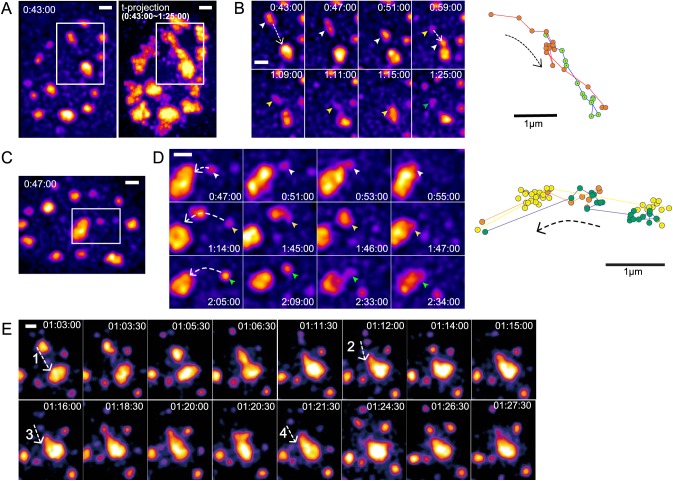
Fig. 4.
Multiple speckles undergo long-range, directional movements along similar paths: repeated cycles of speckle nucleation, motion and fusion to the same target speckle. 2D images represent 2D maximum-intensity projections of z-stacks. GFP-SON intensities are pseudocolored to increase dynamic range. Time (h:min:s) represents the time after DRB addition. Scale bars: 1 µm. (A) Left: image of first cell nucleus 43 min after DRB addition, with the boxed region containing the repeated long-range speckle motions shown in B. Maximum-intensity projection of multiple 2D projected images over time from 43 min to 1 h 25 min. An elongated high intensity ‘trail’ is generated by the time projection of repeated speckle movements. (B) Left: time-lapse images of speckle movements within the boxed region in A. Dashed line arrows show the direction of speckle movements. The first speckle (white arrowheads) moves and merges with the target speckle. The second speckle (yellow arrowheads) appears along the path followed by the first speckle and moves to same target speckle. The green arrowhead marks the appearance of the third speckle along the same path. Right: trajectories of the first (red) and second (green) moving speckles, showing their close overlap. (C) Image of the second cell nucleus 47 min after DRB addition, with the boxed region containing the repeated speckle motions shown in D. (D) Left: time-lapse images of speckle movements within the boxed region in C. Dashed line arrows show the direction of speckle movements. White, yellow and green arrowheads point to three different speckles that move over similar paths. Right: trajectories of these three speckles showing their close overlap. Second (yellow) and third (green) speckles nucleate at similar location but at different times. (E) An additional example of four repeated speckle movements along similar paths. See Movie 4 for the images shown in B, D and E.
We observed a number of cases in which three or even four speckles would appear in the same or similar location, observing long-range movements of two, three or even four of these speckles along a similar trajectory before these speckles terminated their motion by fusion with the same larger speckle (Fig. 4E; Movie 4). Furthermore, we observed examples in which one speckle would merge with another, and then this merged speckle would move to fuse with a third speckle (Figs 3A,VII,B,V and 4D, 1:14:00–1:46:00).
These observations of long-range movements occurring through largely unidirectional steps along curvilinear trajectories argue strongly for directional speckle movements along or within a nuclear path or channel. This is strongly supported further by these observations of repeated speckle movements along the same path or channel. Finally, as a further suggestion for a mechanism leading to directed movements along a possible path, we observed that speckles undergoing long-range movements would sometimes elongate in the direction of motion (Fig. 4B, 00:47:00, 1:09:00, E, 1:06:30) or the recipient speckle would form a long protrusion along the path of the original smaller speckle trajectory (Figs 3A,I, 00:53:00, III, 1:02:00, B,III, 1:06:00, 1:30:00 and 4D, 2:33:00, E, 1:18:30).
Viscoelastic behaviors of speckles and estimation of speckle viscosity
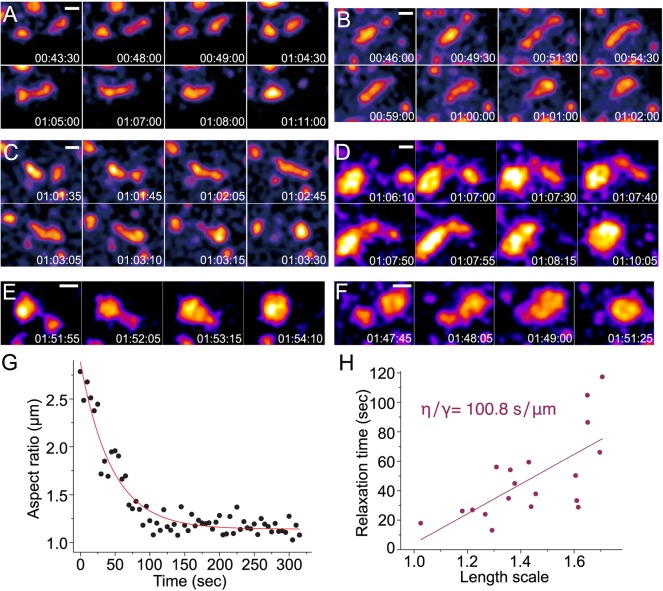
During long-ranged speckle movements, we observed distinctive viscoelastic behaviors such as ‘inchworm’ or reptation motion, suggesting that speckles are liquid-like bodies. For example, a speckle would elongate in the direction of another speckle and then retract at the elongated end nearest the target speckle to form a rounded speckle now closer to the target speckle; this was then followed by a second round of elongation towards the target speckle, followed by fusion and then rounding into a single, larger speckle (Fig. 5A). In other cases, one speckle would significantly elongate towards another target speckle until a long, linear speckle was formed that would fuse with the target speckle; this would be followed by retraction of the elongated speckle into the target speckle to form a single, rounded and larger speckle (Fig. 5B–D).
Fig. 5.
Viscoelastic behaviors of nuclear speckles. GFP-SON images represent maximum-intensity 2D projections of 3D image stacks. Time (h:min:s) is after DRB treatment. Scale bars: 1 µm. (A) ‘Inchworm’-like motion: a nuclear speckle repeat cycle of elongation and translocation until it fuses with another speckle located ∼3 µm away from the starting position of the first speckle. (B–D) Reptation motion: a speckle elongates towards and then fuses with another speckle prior to speckle rounding. (E,F) Two examples of the most commonly observed speckle fusion. (G) Plot of the aspect ratio versus time for two speckles fusing with each other. The aspect ratio is defined as the ratio of the long (llong) to short (lshort) axis lengths of the ellipse approximating the morphology of the fusing speckles. Fitting this plot to a single exponential decay curve yields an exponential decay constant corresponding to the relaxation time of the speckle fusion. In this example, the relaxation time was calculated as 44.9 s. (H) Plot of relaxation time versus length scale for multiple speckle fusion events (N=19). The slope of this linear fit of relaxation time versus length scale corresponds to the inverse capillary velocity, equal to the ratio of viscosity to surface tension (η/γ). The measured inverse capillary velocity from this plot=100.8 s/µm.
Furthermore, examination of speckle fusion events (Fig. 5E,F) supports the concept of nuclear speckles as liquid-like nuclear bodies that have a similar viscosity to that measured previously for other liquid-like RNP bodies such as nucleoli and P-granules (Brangwynne et al., 2009, 2011; Hubstenberger et al., 2013). We estimated the viscosity of speckles by analyzing the time required after speckle fusion for relaxation to a circular shape in cells treated with DRB (Fig. 5G). As expected, relaxation times (τ) were linearly proportional to the characteristic length scale (L), with a slope, which should approximate the inverse capillary velocity, of 101 s/µm (Fig. 5H). This value is similar to previously measured inverse capillary velocities of 2 s/µm for germline P-granules of Caenorhabditis elegans (Brangwynne et al., 2009), 46.1 s/µm for nucleoli of Xenopus oocytes (Brangwynne et al., 2011) and 125 s/µm for large RNP assemblies (grPB) in C. elegans oocytes (Hubstenberger et al., 2013). Assuming a speckle RNP granule size of ∼20 nm, we obtained an estimated viscosity (η) of speckles of ∼1×103 Pa·s, close to the measured viscosities of nucleoli (∼2×103 Pa·s) and grPBs (5×103 Pa·s).
Long-range speckle motion does not require polymerized actin
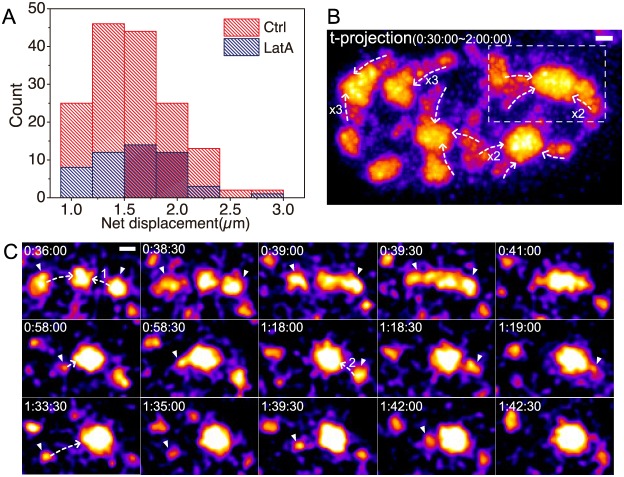
Previous examples of long-range chromosomal movements within interphase nuclei suggested a direct or indirect role of actin, and, in certain cases, nuclear myosin 1C, in these movements (Chuang et al., 2006; Dundr et al., 2007). To determine whether actin was involved in the long-range motion of speckles, we inhibited actin polymerization using latrunculin A (latA), and then measured the frequency and length distributions of long-range speckle movements.
LatA addition resulted in a decrease in long-range movements (Fig. 6A; Table S1). After latA addition, long-range speckle movements greater than 1 µm were observed an average of 2.7 times per nucleus. This compared to an average of 9.8 long-range speckle movements per nucleus in control cells. Although the frequency of long-range nuclear speckle movements was significantly reduced, the distance distribution of these long-range speckle movements did not significantly change with latA addition relative to that observed in control cells (Fig. 6A). Similar types of long-distance speckle trajectories, also ending in fusion with a target speckle, were observed after latA treatment (Fig. 6B,C; Movie 5).
Fig. 6.
Long-range speckle motion does not require polymerized actin. (A) Histogram of speckle net displacements after DRB addition with or without latA. LatA treatment decreases the frequency of long-range speckle motion, but not the length distribution of their net displacements. (B) Maximum-intensity projection over time (t-projection) of GFP-SON nuclear 2D projections from 30 min to 2 h after DRB and latA treatment. This t-projection shows curvilinear paths of long-range speckle motions that terminate at another speckle. Dashed line arrows indicate the direction of speckle motions. Numbers show the number of speckles that move along the direction of the arrow over time (no number for one speckle). Time (h:min:s) represents the time after DRB addition. Scale bar: 1 µm. See Movie 5. (C) Time-lapse images from the boxed region in B. Arrowheads point to moving speckles, and dashed line arrows indicate the direction of speckle movements. The numbers on the dashed lines distinguish different speckles that move on the same path. Time (h:min:s) represents the time after DRB addition. Scale bar: 1 µm.
To further test the possible involvement of actin in speckle motion, we transiently transfected cells with plasmid constructs of actin fused to mRFP and a nuclear localization signal (NLS) to concentrate this actin fusion protein in the nucleus (Posern et al., 2004, 2002). Different constructs were used for expression of the wild-type β-actin sequence, a nonpolymerizable actin mutant (Posern et al., 2002) (G13R) or an actin mutant that favors polymerized actin F-actin (Posern et al., 2004) (S14C).
Long-range nuclear speckle movements were observed in cells transfected with all three mRFP-NLS-actin mutants or mRFP-NLS-wild-type actin as well as nontransfected control cells (Table S1). The average number of long-range movements nuclear speckle movements ranged from ∼12 to 14 per nucleus. Mean distances of these speckle movements were also similar (1.28–1.48 µm). The velocities of speckle movement during the periods of long-range movement were also similar (∼0.5–1.5 µm/min, data not shown).
Thus, we conclude that the observed long-range, directional movements of nuclear speckles after DRB treatment are not actin dependent.
Nuclear speckles move in DAPI-depleted regions that are enriched with SON granules
To explore a possible structural basis for the repeated directed movement of small nuclear speckles along the same apparent path towards a larger nuclear speckle, we used super-resolution light microscopy. We complemented 3D structured illumination microscopy (SIM) and stimulated emission depletion (STED) microscopy on fixed cells with correlative live-cell imaging, in which we used conventional wide-field microscopy on live cells followed by fixation and then SIM and STED imaging on the same cells.
Inspection of nuclear speckle movement in live-cell movies suggested the formation of transient, faint connections of GFP-SON between nearby speckles (Movies 6 and 7). Using STED microscopy, we could visualize individual, distinct GFP-SON granules that are in the size range of STED resolution of ∼60 nm and are distributed nonrandomly in the nucleoplasm outside of nuclear speckles (Fig. 7). We used a combination of 3D SIM and STED to visualize the relative distribution of chromatin and SON. After 3D SIM of 4′,6-diamidino-2-phenylindole (DAPI)-stained, fixed nuclei, STED imaging of the same cells on a different STED microscope was performed. These DAPI SIM and anti-GFP-SON STED images were then aligned using a cross-correlation procedure.
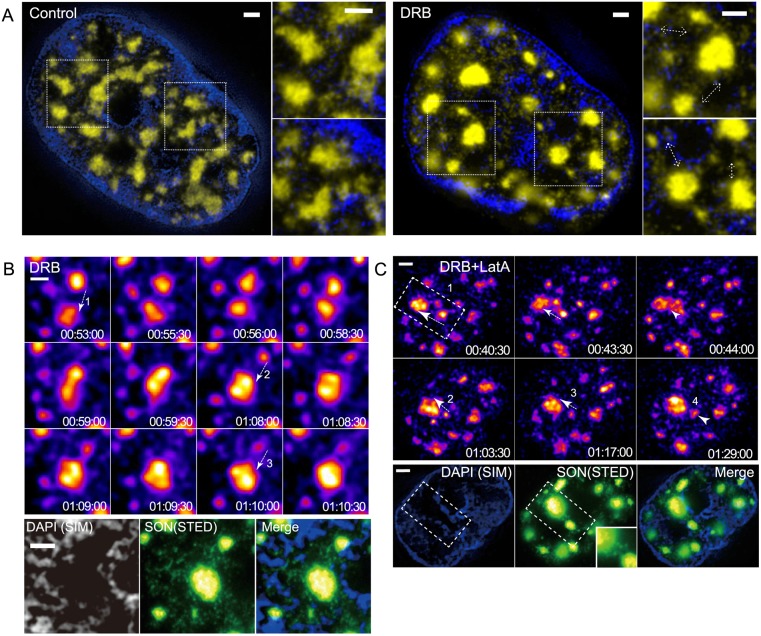
Fig. 7.
Repeated nuclear speckle motions occur within DAPI-depleted regions enriched in SON granules. Scale bars: 1 μm. (A) Correlative SIM (DAPI, blue) and STED (SON, yellow) images of single optical sections, showing heterogeneous distribution of nuclear speckles and SON granules in DAPI-poor regions before (left) and after (right) DRB treatment. Regions of interest are marked with dotted line boxes and shown enlarged in panels to the right of nuclear images. Dashed line arrows are drawn adjacent to local accumulations of SON granules running between two nearby nuclear speckles. (B) Correlation between the path of repeated speckle motions, DAPI-depleted region and local concentration of SON granules in a DRB-treated cell. Top: time-lapse images of repeated speckle motions. Each speckle motion is marked by a dashed line arrow and number. Time (h:min:s) is after DRB addition. See Movie 6. Bottom: correlative super-resolution images after fixation in the region of repeated speckle movements (see top) occurred in the DAPI-depleted region (left), enriched in SON granules (middle). The merged image is shown on the right. (C) Same as B but for DRB+latA-treated cell. See Movie 7.
In these combined DAPI-SON images, we observed enrichment of SON granules in DAPI-depleted spaces connecting nearby nuclear speckles, with most speckles appearing to be connected by higher local concentrations of these SON granules (Fig. 7A). These connections persisted even after DRB treatment, even becoming clearer due to the reduced number of granules overall in the nucleoplasm (Fig. 7A, right). The spatial distribution of these granules suggests they are related to the faint, transient GFP-SON connections between nuclear speckles visualized in our live-cell imaging.
DAPI preferentially binds AT-rich regions in DNA. To rule out the possibility that the apparent DNA-depleted spaces connecting nuclear speckles were occupied by GC-rich DNA, we used 3D SIM to compare DAPI staining with 7-aminoactinomycin D (7-AAD) staining, which preferentially binds GC-rich regions in DNA, before and after 1 h DRB treatment. Although the relative ratio of staining differed for different chromatin regions, as expected, the correlation between both staining distributions was high (Pearson correlation coefficient ∼0.7–0.9) (Fig. S4). Indeed, after 1 h DRB treatment, most DAPI-poor regions (with just one to two exceptions per nucleus), were also depleted of 7-AAD staining structures (Fig. S4C). We therefore conclude that most of these DAPI-poor regions between nuclear speckles in fact represent spaces depleted of chromatin.
We next used correlative live-cell and super-resolution imaging to determine the relationship between the path followed by repeated nuclear speckle trajectories and these DAPI-depleted, SON granule-enriched spaces connecting neighboring nuclear speckles. After DRB treatment, we followed live-cell imaging with immediate fixation of cells using paraformaldehyde and staining. Inspection of the live-cell movies identified cells containing examples of repeated speckle movements along a similar path, terminated by fusion with the same larger nuclear speckle. These cells were then identified and imaged by 3D SIM and STED.
These correlative images revealed that the path or channel along which nuclear speckles had repeatedly moved was relatively DAPI depleted but surrounded by DAPI-stained chromatin (Fig. 7B). Moreover, a relatively high concentration of SON granules was present within these paths or channels relative to surrounding nuclear regions. Similar correlative results were observed in cells treated with latA prior to addition of DRB (Fig. 7C).
Our observations of nuclear speckles moving within DAPI-depleted regions suggested that chromatin might act as a barrier to speckle motion. We next used live-cell imaging to examine the temporal correlation between changes in the DNA distribution surrounding nuclear speckles and speckle movement and fusion events (Fig. 8). We used the cell-permeable, far-red SiR-Hoechst dye that binds DNA in the minor groove like DAPI. Chromatin shows condensation with time after DRB treatment, increasing the size of DNA-depleted intranuclear spaces (Fig. 8A). Fusion between nearby large speckles indeed is temporally correlated with the disappearance of the DNA staining observed in preceding time points within the space separating these same speckles (Fig. 8B–E; Movies 8 and 9).
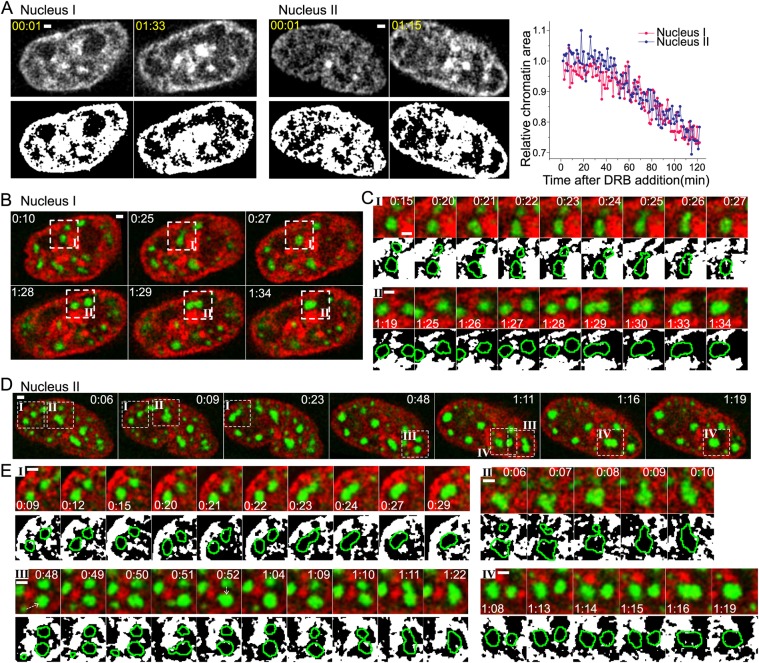
Fig. 8.
Speckle motions are spatially limited by chromatin structure. All images show single optical sections. Time (h:min) is after DRB addition. Scale bars: 1 μm. (A) Chromatin compaction over time after DRB addition. Left, middle: grayscale (top) and binary (bottom) images of chromatin (SiR-Hoechst) in nucleus I (left) and nucleus II (middle) at the indicated time points. Change in relative chromatin area (y) over time after DRB addition (x). (B) Time-lapse images of nucleus I showing speckle (GFP-SON, green) motions and fusion following loss of chromatin (SiR-Hoechst, red) between the speckles that fuse. See Movie 8. Regions of interest (ROI) I and II marked by dashed line boxes are shown enlarged in C. (C) Top row for ROI I and II: time-lapse images showing that speckle-merging events occurred after depletion of chromatin in the space between speckles. Bottom row for ROI I and II: binary images of thresholded SiR-Hoechst staining (white) relative to speckle boundaries (green lines). (D) Same as B for nucleus II. See Movie 9. Nucleus II contains four ROI (I–IV). (E) Same as C for nucleus II.
Similar long-range and repeated speckle movements after heat shock or heavy metal addition, or during transition from late G2 to prophase
To place our observations within a more physiological context, we examined speckle dynamics during heat shock. After heat shock, although most genes are transcriptionally downregulated 5-fold or more (Mahat et al., 2016), heat-shock genes are induced up to 200-fold (Lis et al., 1981; Mahat et al., 2016). Previously, we demonstrated that heat-shock transcriptional induction of Hsp70 plasmid transgene arrays was greatly enhanced after nuclear speckle contact (Khanna et al., 2014). Similar to DRB treatment, after heat shock we observed multiple examples of long-range motions of smaller nuclear speckles terminating with fusion with larger nuclear speckles. Fig. S5 shows a striking example of repeated nucleation of new speckles adjacent to the Hsp70 transgene array, followed by long-range motion of these speckles towards a larger, distant nuclear speckle. Later, speckles nucleate along this speckle trajectory and then merge to form a continuous, ∼4-µm-long connection between the transgene array and distant speckle. Fig. S6 shows a long-range speckle motion terminating after association with the Hsp70 BAC transgene array, which then shows an abrupt increase in its transcription.
Similar speckle dynamics were observed using other transcriptional inhibitors (α-amanitin, TRL; data not shown) and in response to other stress conditions, specifically heavy metal stress induced by Cd (Fig. S7).
As previously described, live-cell imaging revealed that speckle movements and fusion events after DRB-induced transcriptional inhibition correlated with increased chromatin condensation and the appearance of larger interchromatin channels, suggesting that decondensed interphase chromatin might inhibit speckle movements. Chromatin condensation also occurs under normal physiological conditions immediately prior to mitosis during the G2 to early prophase transition, and entry into mitosis is also accompanied by significant transcriptional repression (Gottesfeld and Forbes, 1997; Parsons and Spencer, 1997). If long-range speckle movements are facilitated by the appearance of larger interchromatin channels, then we would predict similar long-range speckle movements during this late G2 to prophase stage of normal cell growth.
Indeed, live-cell movies during the transition from G2 to prophase showed a similar transition to fewer larger and rounder nuclear speckles concomitant with increased speckle long-range movements and similar observations of small speckles moving towards and merging with larger speckles as seen after transcriptional inhibition, heat shock or Cd treatment (Fig. S8). Live-cell movies also showed that nuclear speckles are excluded from regions of chromatin compaction during prophase, while nuclear speckle fusions occurred within long chromatin-free channels between compact chromatin structures. Further experiments would be needed to dissect to what degree these similar speckle dynamics correlate with the increased chromosome condensation, transcriptional inhibition and/or other events accompanying entry into mitosis. Regardless of mechanism, though, these observations demonstrate that this phenomenology of small speckles moving long distances and merging with larger speckles is observed not only after exposure to transcriptional inhibitors or in response to stress, but also occurs normally at certain points in the cell cycle.
DISCUSSION
Here, we showed an increased mobility of nuclear speckles after transcriptional inhibition by DRB, including prominent long-range movements of smaller speckles over distances of 1–4 µm with velocities of 0.2–1.5 µm/min that typically terminated through fusion with a larger speckle. These long-range movements appeared directional, following roughly linear trajectories pointing to and ending with merging in pre-existing stationary speckles. Speckles frequently elongated in the direction of movement during these movements, suggesting the possibility of an active transport mechanism. Directionality was also inferred based on our observations of repeated cycles of nucleation of new speckles and their movement along similar trajectories followed by previous moving speckles. These long-range, speckle movements occurred independent of polymerized nuclear actin as their length distribution was unchanged after latA treatment. Correlation of live-cell imaging with super-resolution microscopy on fixed samples revealed that speckles moved within channels relatively depleted of DNA but enriched in SON protein-containing granules.
As observed previously, upon transcriptional inhibition, speckles became rounder and larger. This increase in speckle size was imagined to occur as a result of RNA processing components being released from previous sites of transcription, diffusing into speckles and then accumulating within the speckle through binding to other components (Jimenez-Garcia and Spector, 1993; Misteli et al., 1997). Dephosphorylation of SR proteins was proposed to lead to their accumulation in speckles, whereas phosphorylation of SR proteins was proposed to lead to their release from speckles, diffusion into the nucleoplasm and binding to local transcription sites (Colwill et al., 1996; Misteli et al., 1998). Consistent with this model, modeling the mobility of speckle proteins as measured by FRAP led to the conclusion that differential nuclear distribution of splicing factors was controlled by changes in their binding to either nucleoplasmic or speckle sites (Kruhlak et al., 2000; Phair and Misteli, 2000; Rino et al., 2007).
Not explained by these studies was how the number of speckles decreases with transcriptional inhibition. Also, these previous studies proposed that the increased size of speckles after transcriptional inhibition is entirely due to diffusion of speckle components from the nucleoplasm followed by binding to sites within speckles.
Our results instead suggest that a significant cause of both the increase in individual speckle size and brightness and the reduction in speckle number is through the directed, long-range movements of speckles towards other speckles followed by their fusion. Further work will be needed to determine the relative contributions of diffusion and binding of speckle components versus speckle fusion events to the observed increase in speckle size observed after transcriptional inhibition. Further work will also be needed to determine whether post-translational modifications of speckle proteins are connected to directional motion and/or fusion of speckles.
One previous study, using transformed MCF-10A human breast epithelial cells, described the long-range movement of smaller nuclear speckles and subsequent fusion with larger speckles (Zhang et al., 2016). However, these movements were not characterized other than by visual inspection of several movies and by display of net vector velocities averaged over an unspecified number of time points for several speckles; specifically, no analysis or display of individual trajectories was shown. Curiously, in this previous study, long-range speckle movements were reported to occur under normal physiological growth conditions. This directly contrasted with our observations of similar long-range speckle movements in CHO cells only after transcriptional inhibition, heat-shock or heavy metal stress, or during the transition from late G2 into prophase. We believe that this discrepancy in results was due to cell stress induced by the transient transfection and/or live-cell imaging conditions used in Zhang et al. (2016). Indeed, in this previous study, nuclear speckles appeared unusually round, large and bright (see supplementary movies 1–3 in Zhang et al., 2016), similar to the nuclear speckle appearance we observed only after extended exposure to transcriptional inhibitors. Using this same MCF-10A cell line, we observed ‘normal’, nonround speckles in fixed cells after immunostaining against SON, similar in appearance to the nuclear speckles we observe routinely in multiple cell lines (CHO, mouse NIH 3T3, and human WI-38, Tig3, HFF, HCT116, K562, H1 embryonic stem cells and Hap1).
During the dynamic movement of speckles, we confirmed liquid-state properties of nuclear speckles. Speckles exhibited nucleation, deformation in the direction of translocation and fusion (Marzahn et al., 2016). Strikingly, we found that the motion of speckles after transcriptional inhibition always ended with fusion with another speckle. After fusion, speckles regained their circular shape but were bigger and showed an increased intensity, suggesting the mixing of constituent molecules as seen in liquid-phase cellular bodies (Brangwynne, 2013). Analysis of the time interval between speckle fusion and rounding of the merged speckles allowed us to estimate the viscosity of nuclear speckles, which we found was similar to previously measured viscosities for Xenopus oocyte nucleoli and grPBs of C. elegans oocytes. Not considered in our analysis was the possibility that adjacent chromatin interacting with nuclear speckles might slow the rounding of the nuclear speckle after fusion events. Therefore, our estimates of nuclear speckle viscosity might be best considered an upper bound on the actual speckle viscosity, where the actual viscosity nuclear speckles might be somewhat lower depending on how significant these chromatin interactions might be on the actual relaxation time of the speckles after fusion. Overall, though, these measurements indicate that nuclear speckles possess viscoelastic properties and behaviors similar to those observed in other RNP bodies.
Significantly, although the frequency of long-range speckle movements decreased after perturbation of actin polymerization, the length distributions of these speckle movements did not change. Therefore, the long-range, directed movement of speckles after transcriptional inhibition is not dependent on polymerized actin. This rules out a more conventional actin-myosin-based mechanism for speckle motility, despite reports of nuclear myosin I isoforms in nuclear speckles and enrichment of these isoforms in speckles after transcription inhibition (Ihnatovych et al., 2012).
Fusion between nearby speckles to form larger speckles could be explained as driven by collisions between speckles mediated through random Brownian motion. However, our observation of directed motion of speckles over distances up to several micrometers in length, in many cases followed by a new speckle moving along a similar path, requires an explanation other than simple random diffusion.
One possible clue pointing to a potential mechanism underlying speckle long-range motion might be our observation of SON-containing granules concentrated in DAPI-poor channels connecting nearby speckles. Correlative live-cell/fixed-cell imaging revealed that these channels roughly colocalized to the path of actual speckle movements observed in live cells shortly before fixation. We can envision more than one scenario that might give rise to these localized SON granule accumulations on the path of speckle movements.
A flow of SON granules between neighboring speckles, through some unknown mechanism(s), might establish this concentration of granules. Numerical modeling of a simple binary fluid mixture predicts a net flux of individual components through diffusion from small to larger droplets, due to the greater stability of the larger droplets, because of their lower interface free energy. Moreover, this net flux generates a composition correlation between two neighboring droplets of different sizes, leading to an intradroplet gradient of interfacial tension and producing a hydrodynamic flow of small droplets towards larger droplets followed by droplet fusion (Shimizu and Tanaka, 2015). Interestingly, this modeled behavior of droplet movements in simple binary fluid mixtures mirrors our observation of small speckles moving towards larger speckles. Additionally, movement of one speckle along a trajectory might leave SON granules along its path – analogous to tiny droplets of water left behind by movement of a large water droplet along a surface. These pre-positioned SON-granule speckles might work as ‘steps’ biasing movements of other speckles at later times. Finally, we cannot rule out an active, but unknown, transport mechanism driving speckle movements. Additional experiments including future live-cell imaging at higher temporal and spatial resolution will be needed to explore these ideas.
Our correlative imaging using SIM, STED and live-cell imaging also allowed us to determine the relative spatial arrangement between chromatins and speckles, but, furthermore, we found that the movement of speckles indeed occurred within the interchromatin space, with chromatin seen surrounding the speckles and the apparent channels through which the speckles moved, suggesting that chromatin may act as a spatial barrier to speckle movement and confine these movements to linear trajectories. Indeed, under normal growth conditions, we observed speckle mobility consistent with confined motion. The observed directional motion after transcriptional inhibition is consistent with a release of constraints to speckle motion as a result of the chromatin condensation observed in live-cell movies after transcriptional inhibition. Thus, we suspect that chromatin in nuclei plays a similar constraining role in restricting translocation and fusion of nuclear speckles as actin networks have been shown to play in constraining movement and coalescence of RNP droplets and nucleoli in oocytes (Brangwynne et al., 2011; Feric and Brangwynne, 2013).
In conclusion, in this study, the most striking aspects of the long-range speckle movement we observed were the repeated cycles of speckle nucleation, translocation of the newly nucleated speckles along a similar trajectory as that followed by preceding speckles, and then fusion with the same target speckle. Together, these observations point to a previously unsuspected cellular trafficking system for movement and nuclear positioning of nuclear speckles. Although we observed these events during periods of increased nuclear speckle reorganization induced by transcriptional inhibition or stress responses, similar dynamics were observed under normal growth conditions during the entry into mitosis. We anticipate that these nonrandom speckle dynamics may provide rapid and effective transport of RNA processing and/or transcription factors to specific nuclear sites, as well as recycling of speckle components to nuclear speckles and regulation of speckle number and size. Moreover, recent identification of genomic regions that associate at near 100% frequencies with nuclear speckles (Chen et al., 2018) suggest that the nuclear positioning of speckles may also drive nuclear genome organization. The phenomenology of nuclear speckle movements described here should allow future development of assays that can be used to identify the molecular components responsible for this nuclear speckle cellular trafficking system.
MATERIALS AND METHODS
Cell culture and establishment of cell line
CHO-K1 cells were grown in F12 medium (Cell Media Facility, University of Illinois at Urbana-Champaign) supplemented with 10% fetal bovine serum (Sigma-Aldrich, F2442) at 37°C in 5% CO2. To generate a stable cell line expressing EGFP fused to the SON protein, we used a BAC EGFP-SON transgene (EGFP-SON-Zeo BAC) in which a starting BAC containing a human SON genomic insert (RP11-165J2, Invitrogen) was retrofitted using BAC recombineering to add a Zeocin selectable marker and the EGFP sequence in frame with the SON NH2 terminus (Khanna et al., 2014). We purified the EGFP-SON-Zeo BAC using a Large-Construct Kit (Qiagen). The purified construct (5 μg) was linearized with BsiWI (NEB), followed by incubation at 65°C for 20 min to inactivate BsiWI. CHO-K1 cells were transfected with the linearized EGFP-SON-Zeo BAC using Lipofectamine 2000 (Invitrogen) 1 day after passaging while at ∼60% confluency, following the manufacturer's suggested protocol. After 36 h, transfected cells were trypsinized (Trypsin-EDTA 0.25%, Gibco) and transferred to a larger flask to which selection medium (200 μg/ml Zeocin, Thermo Fisher Scientific) was added. After 10 days of selection, cells were subcloned by serial dilution into 96-well plates. Individual subclones were screened by microscopy using a Deltavision wide-field microscope (GE Healthcare) to select clones that showed uniform and stable EGFP-SON expression.
Drug treatments
To add drugs without adding new serum, we removed half the medium from the cell culture dishes, added chemicals to a 2× concentration using this medium and then returned this medium back to the same dishes. The final DRB working concentration was 50 μg/ml [Sigma-Aldrich, 50 mg/ml stock solution in dimethyl sulfoxide (DMSO)]. Other drugs were used at the following working (and stock) concentrations: 50 μg/ml α-amanitin [A2263, Sigma-Aldrich; 1 mg/ml in deionized (DI) water], 0.1 μg/ml TRL (T3652, Sigma-Aldrich; 1 mg/ml in DMSO), 200 μM Cd solution (20920, Sigma-Aldrich; 100 mM in DI water). For live-cell imaging, chemicals were prepared in the same way and returned to the live-cell dish on the microscope.
For latA treatment (L5163, Sigma-Aldrich), cells were seeded on poly-L-lysine-coated coverslips or glass-bottom dishes (MatTek). Coverslips or glass-bottom dishes (MatTek) were covered with 0.01% poly-L-lysine solution (P4707, Sigma-Aldrich) for 10 min, rinsed with sterilized DI water and dried in the tissue culture hood. Cells were grown for 2 days on these coated surfaces, treated with 1 μM latA for 30 min and then treated with DRB as described above.
Sample preparation for fixed-cell imaging
For quantitative analysis of speckle morphology, cells were seeded on coverslips (Fisher) and fixed 48 h later at ∼90–100% confluency using freshly prepared 3.6% paraformaldehyde (PFA; P6148-500G, Sigma-Aldrich) in PBS for 20 min at room temperature. After washing 3× for 5 min each in PBS, the fixed cells were mounted in a Mowiol-DABCO anti-fade medium (Harlow and Lane, 1988).
For STED and SIM imaging, immunofluorescence and DAPI staining were performed after fixation and 3×5 min washes in PBS. For immunofluorescence staining, cells were incubated in blocking buffer (0.5% Triton X-100, Thermo Fisher Scientific) and 0.5% normal goat serum (Sigma-Aldrich) in PBS for 30 min. Cells were then incubated with monoclonal anti-SON primary antibody (1:300 in PBS; HPA023535, Sigma-Aldrich) overnight at 4°C. After 3×5 min washes in PBS, cells were incubated with Atto647N-conjugated goat anti-rabbit IgG (1:300 in PBS; 611-156-122S, Rockland) for 2 h at room temperature and then washed 3× for 5 min each in PBS. The cells were postfixed with 3.6% PFA for 15 min, and washed 3×5 min in PBS. Cells were counterstained with 1 µg/ml DAPI (Sigma-Aldrich) in PBST (0.1% Triton X-100 in PBS) for 0.5–1 h at room temperature, and then washed 5× for 5 min each in PBS. For 7-AAD staining, after fixation with 3.6% PFA for 15 min and permeabilization with PBS with 0.5% Triton X-100, cells were counterstained with 40 µM 7-AAD (Thermo Fisher Scientific) in PBS for 4 h at room temperature, and then washed 3× for 5 min each in PBST and 2× for 5 min each in PBS. We equilibrated the cell samples with Vectashield antifade mounting medium (H-1000, Vector Laboratories) for 10 min, aspirated the medium and added fresh mounting medium. This was repeated three to five times to completely infiltrate the cells with the mounting medium to avoid refractive index changes over the cells. Coverslips were mounted on slides and sealed with nail polish.
Microscopy of fixed samples and analysis of cell morphology
We used a Deltavision wide-field microscope (GE Healthcare), equipped with a Xenon lamp, 60×/1.4 NA oil immersion objective (Olympus) and CoolSNAP HQ CCD camera (Roper Scientific). Then, 1024×1024 pixel 2D images were acquired as 3D z-stacks and processed using the ‘Enhanced’ version of the iterative, nonlinear deconvolution algorithm provided by the Softworx software (Agard et al., 1989) (GE Healthcare), and 3D deconvolved image stacks were projected into 2D using a maximum intensity algorithm.
Analysis of speckle morphology used custom MATLAB (MathWorks) codes for image segmentation in several sequential steps. Multiple intensity thresholds and image segmentation steps allowed us to segment speckles of varying sizes and intensity levels. We first automatically chose an initial intensity threshold using MATLAB’s ‘graythresh’ function (graythresh×1.5–1.8) based on Otsu's method (Otsu, 1979). This allowed us to produce a binary image from which we measured area, perimeter and roundness [4×area/π ×(maximum axis length)2] of the larger, brighter speckles. To calculate intensities of these speckles, we used the segmented areas as a mask, which was applied to a new 2D projected image generated by an intensity projection from the original 3D raw image stacks. The normalized speckle intensity was calculated by summing the intensities of all pixels within a speckle, subtracting the background intensity over an equivalent area and then normalizing by the speckle area {[sum of pixel values in speckle–(mean value of nuclear background×size of the speckle)]/size of speckle}. Once these large speckles were segmented and analyzed, we removed them from the deconvolved projected image by replacing their pixel intensities equal to the cellular background level. We then repeated the image thresholding, setting a new intensity threshold using the ‘graythresh’ function (graythresh×0.8–1.8), and repeating the same process as described above to measure a new set of speckles and then remove them from the image. If necessary, this process was repeated one more time to select the remaining small speckles. To prevent the situation in which part of a speckle that was not segmented in a previous cycle was counted as a new speckle, at each step, we recorded the coordinates of segmented speckle centers and discarded any new speckles in a subsequent cycle that were within 0.3 μm of a previous speckle center. Through three such segmentation cycles, we were able to identify and measure the morphology of most speckles present in the nucleus. However, very small speckles less than 0.4 μm in diameter were not considered in our measurements of changes in speckle morphology before and after transcriptional inhibition.
Live-cell imaging and tracking
For live-cell imaging, we used a V3 OMX (GE Healthcare) microscope, equipped with a 100×/1.4 NA oil immersion objective (Olympus), two Evolve EMCCDs (Photometrics), a live-cell incubator chamber for CO2 perfusion, and two temperature controllers for the incubator and the objective lens heaters. Temperatures of the live-cell chamber and lens were maintained at 37°C. Cells were seeded in glass-bottom dishes (MatTek) to reach ∼90–100% confluency 48 h later. We acquired 3D stacks (z-spacing 300 nm) in the conventional wide-field imaging mode at given time intervals, followed for each time point by 3D deconvolution and 2D maximum intensity projection using the Softworx software. ImageJ (https://imagej.nih.gov/ij/) was used first to smooth these projections with a Gaussian filter (σ=2). Then, a rigid body registration (ImageJ plugin ‘StackReg’) was applied to correct for any x–y nuclear rotation and/or translational displacement between sequential time points. A region including a full speckle motion was cropped from the image manually using ImageJ, and the single speckle was tracked by custom MATLAB code. The cropped image was transformed into a binary image by the intensity threshold selected by MATLAB's ‘graythresh’ function. The center of mass of the speckle was then determined over time by MATLAB's ‘regionprops’ function and used for speckle tracking.
Viscosity analysis
To estimate nuclear speckle viscosity, we acquired live-cell movies at 5-s time intervals over 90 min. We identified nuclear speckles undergoing fusion and measured their long (llong) and short (lshort) axes to calculate, as a function of time, their aspect ratio (AR), defined as AR=llong/lshort. A relaxation time (τ) for the speckle fusion was estimated by fitting the measured AR to the exponential decay curve defined by AR=P+(AR0–P)·e−t/τ, where P is the plateau of the exponential curve and AR0 is the AR at time=0.
The length scale was calculated as L=[(llong−lshort)×lshort]0.5 at time=0, and plotted against τ. From a linear fit of τ versus L by least-squares estimation, we obtained the slope that corresponds to the inverse capillary velocity (η∕γ), where η is the viscosity and γ the surface tension (Eggers, 1997). We could estimate γ≈kbΤ/d2 (Aarts et al., 2004), where kb is the Boltzmann constant, Τ is the temperature and d is molecular length scale [∼10 nm used for RNPs in previous studies (Brangwynne et al., 2011; Hubstenberger et al., 2013)]. We used ∼20 nm for the characteristic RNP granules contained within IGCs (Spector, 1993; Thiry, 1995). From the estimated γ and the inverse capillary velocity (η∕γ), we estimated the viscosity of nuclear speckles.
Correlative live-cell imaging and super-resolution imaging
To combine live-cell imaging with STED and SIM, cells were seeded in glass-bottom dishes engraved with a 50-μm grid pattern, letters and numbers (81148, Ibidi), reaching ∼60–70% confluency 2 days later. Prior to live-cell imaging, we recorded locations of target cells using the alphanumeric characters to find the same target cells later when we used different microscopes. After live-cell imaging, cells were immediately fixed by adding 2× PFA solution in PBS to the cell medium. The final PFA concentration was 4%. Immunofluorescence and DAPI staining followed by mounting in Vectashield anti-fade medium were performed as described above.
For STED images, we used a custom-made STED microscope (Han and Ha, 2015), and acquired 3D z-stacks through the entire SON immunofluorescent-stained nucleus using 300 nm z-spacing.
We acquired 3D SIM images on the V3 OMX microscope (see above) using 0.125 µm z-spacing and sequential excitation at 488 nm and 405 nm on each image plane. Each z-stack image contained 15 images at five different phases per angle and three different angles per slice. We collected a full 3D SIM image of GFP-SON and DAPI staining and reconstructed 3D SIM image stacks with Softworx. The chromatic aberration offset between the GFP and DAPI wavelengths was measured with the alignment slide provided by GE Healthcare and used to correct the 3D SIM images using the OMX Image Registration function in Softworx.
To align the STED SON and SIM DAPI images, we selected a single STED optical z-section showing speckles of interest and then matched it to the most similar optical z-section from the complete stack of the 3D SIM GFP-SON image. Because 2D STED imaging was performed using a 300 nm z-interval (z resolution≈600 nm), we used the 2D projection of three adjacent SIM z-sections, consisting of the most similar SIM optical section plus the SIM optical sections immediately above and below this optical section. Because the ‘StackReg’ registration tool in ImageJ required a combined stack of images, we matched x–y sizes of SIM (1024×1024 pixels, pixel size=40 nm after reconstruction) and STED (varying, pixel size=20–40 nm) images by adding pixels of background intensity or by taking a certain size of subregion from a large image. We then generated a RGB SIM image from DAPI (blue) and SON after saturating SON intensity to be used as a reference for alignment with the STED image. We finally combined the RGB SIM image and STED SON image as z-stacks, and ran ‘StegReg’ with rigid body mode. In this way, a STED SON image could be easily aligned on the strong SON signal of the SIM image, where a SIM DAPI image and SIM SON image were grouped during the alignment process. From the aligned z-stacks, we took the SIM DAPI image and STED SON image.
Live-cell imaging and analysis of chromatin compaction after DRB treatment
To visualize chromatin structure in live cells, we incubated cells growing in glass-bottom dishes with 100 nM SiR-Hoechst (SC007, Spirochrome) and 10 µM verapamil (V4629, Sigma-Aldrich) for 1 h before imaging. We started 3D live-cell imaging immediately after adding DRB, acquiring simultaneously both GFP-SON (excitation at 488 nm) and SiR-Hoechst (excitation at 642 nm) images for 2 h at 1-min time intervals. Deconvolution was performed by Softworx as described above. We converted these images into binary images by segmentation using the ImageJ plugin ‘Threshold’ based on the Otsu thresholding method (Otsu, 1979). From these segmented binary chromatin images, we computed the chromatin area as a function of time using MATLAB's ‘regionprops’ function.
Supplementary Material
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.K., A.S.B.; Methodology: J.K., A.S.B.; Software: J.K.; Validation: J.K.; Formal analysis: J.K.; Investigation: J.K., A.S.B.; Resources: J.K., K.Y.H, N.K., T.H., A.S.B.; Writing: J.K., A.S.B.; Visualization - live cell imaging and SIM: J.K.; Visualization - STED: J.K., K.Y.H.; Supervision: T.H., A.S.B.; Project administration: A.S.B.; Funding acquisition: T.H., A.S.B.
Funding
This work was supported by the National Institutes of Health [R01 GM058460 to A.S.B.; GM112659 to T.H.] and the National Science Foundation [PHY-1430124 to T.H.]. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.226563.supplemental
References
- Aarts D. G. A. L., Schmidt M. and Lekkerkerker H. N. W. (2004). Direct visual observation of thermal capillary waves. Science 304, 847-850. 10.1126/science.1097116 [DOI] [PubMed] [Google Scholar]
- Agard D. A., Hiraoka Y., Shaw P. and Sedat J. W. (1989). Fluorescence microscopy in 3 dimensions. Methods Cell Biol. 30, 353-377. 10.1016/S0091-679X(08)60986-3 [DOI] [PubMed] [Google Scholar]
- Aizer A., Brody Y., Ler L. W., Sonenberg N., Singer R. H. and Shav-Tal Y. (2008). The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol. Biol. Cell 19, 4154-4166. 10.1091/mbc.e08-05-0513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks D. S. and Fradin C. (2005). Anomalous diffusion of proteins due to molecular crowding. Biophys. J. 89, 2960-2971. 10.1529/biophysj.104.051078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaude O. (2011). Inhibiting eukaryotic transcription: which compound to choose? How to evaluate its activity? Transcription 2, 103-108. 10.4161/trns.2.3.16172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher L., Ouzounis C. A., Enright A. J. and Blencowe B. J. (2001). A genome-wide survey of RS domain proteins. RNA 7, 1693-1701. [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C. P. (2013). Phase transitions and size scaling of membrane-less organelles. J. Cell Biol. 203, 875-881. 10.1083/jcb.201308087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C. P., Eckmann C. R., Courson D. S., Rybarska A., Hoege C., Gharakhani J., Julicher F. and Hyman A. A. (2009). Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729-1732. 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- Brangwynne C. P., Mitchison T. J. and Hyman A. A. (2011). Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. USA 108, 4334-4339. 10.1073/pnas.1017150108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J. R. and Parker R. (2009). Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932-941. 10.1016/j.molcel.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter K. C., Taneja K. L. and Lawrence J. B. (1991). Discrete nuclear domains of poly(A) RNA and their relationship to the functional-organization of the nucleus. J. Cell Biol. 115, 1191-1202. 10.1083/jcb.115.5.1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhang Y., Wang Y., Zhang L., Brinkman E. K., Adam S. A., Goldman R., van Steensel B., Ma J. and Belmont A. S. (2018). Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J. Cell Biol. 217, 4025-4048. 10.1083/jcb.201807108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C.-H., Carpenter A. E., Fuchsova B., Johnson T., de Lanerolle P. and Belmont A. S. (2006). Long-range directional movement of an interphase chromosome site. Curr. Biol. 16, 825-831. 10.1016/j.cub.2006.03.059 [DOI] [PubMed] [Google Scholar]
- Colwill K., Pawson T., Andrews B., Prasad J., Manley J. L., Bell J. C. and Duncan P. I. (1996). The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 15, 265-275. 10.1002/j.1460-2075.1996.tb00357.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T. and Cremer C. (2001). Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2, 292-301. 10.1038/35066075 [DOI] [PubMed] [Google Scholar]
- Dundr M., Hebert M. D., Karpova T. S., Stanek D., Xu H. Z., Shpargel K. B., Meier U. T., Neugebauer K. M., Matera A. G. and Misteli T. (2004). In vivo kinetics of Cajal body components. J. Cell Biol. 164, 831-842. 10.1083/jcb.200311121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Ospina J. K., Sung M.-H., John S., Upender M., Ried T., Hager G. L. and Matera A. G. (2007). Actin-dependent intranuclear repositioning of an active gene locus in vivo. J. Cell Biol. 179, 1095-1103. 10.1083/jcb.200710058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers J. (1997). Nonlinear dynamics and breakup of free-surface flows. Rev. Mod. Phys. 69, 865-929. 10.1103/RevModPhys.69.865 [DOI] [Google Scholar]
- Feric M. and Brangwynne C. P. (2013). A nuclear F-actin scaffold stabilizes ribonucleoprotein droplets against gravity in large cells. Nat. Cell Biol. 15, 1253-1259. 10.1038/ncb2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld J. M. and Forbes D. J. (1997). Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 22, 197-202. 10.1016/S0968-0004(97)01045-1 [DOI] [PubMed] [Google Scholar]
- Han K. Y. and Ha T. (2015). Dual-color three-dimensional STED microscopy with a single high-repetition-rate laser. Opt. Lett. 40, 2653-2656. 10.1364/OL.40.002653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T. N. W., Kato M., Xie S. H., Wu L. C., Mirzaei H., Pei J. M., Chen M., Xie Y., Allen J., Xiao G. H. et al. (2012). Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768-779. 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane D. (1988). Antibodies. A Laboratory Manual. New York: Cold Spring Harbor Laboratory. [Google Scholar]
- Hubstenberger A., Noble S. L., Cameron C. and Evans T. C. (2013). Translation repressors, an RNA helicase, and developmental cues control RNP phase transitions during early development. Dev. Cell 27, 161-173. 10.1016/j.devcel.2013.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman A. A., Weber C. A. and Jüelicher F. (2014). Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39-58. 10.1146/annurev-cellbio-100913-013325 [DOI] [PubMed] [Google Scholar]
- Ihnatovych I., Migocka-Patrzalek M., Dukh M. and Hofmann W. A. (2012). Identification and characterization of a novel myosin Ic isoform that localizes to the nucleus. Cytoskeleton 69, 555-565. 10.1002/cm.21040 [DOI] [PubMed] [Google Scholar]
- Izeddin I., Recamier V., Bosanac L., Cisse I. I., Boudarene L., Dugast-Darzacq C., Proux F., Benichou O., Voituriez R., Bensaude O. et al. (2014). Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. Elife 3, e02230 10.7554/eLife.02230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Garcia L. F. and Spector D. L. (1993). In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell 73, 47-59. 10.1016/0092-8674(93)90159-N [DOI] [PubMed] [Google Scholar]
- Kaiser T. E., Intine R. V. and Dundr M. (2008). De novo formation of a subnuclear body. Science 322, 1713-1717. 10.1126/science.1165216 [DOI] [PubMed] [Google Scholar]
- Kato M., Han T. W., Xie S., Shi K., Du X., Wu L. C., Mirzaei H., Goldsmith E. J., Longgood J., Pei J. et al. (2012). Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753-767. 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna N., Hu Y. and Belmont A. S. (2014). Hsp70 transgene directed motion to nuclear speckles facilitates heat shock activation. Curr. Biol. 24, 1138-1144. 10.1016/j.cub.2014.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak M. J., Lever M. A., Fischle W., Verdin E., Bazett-Jones D. P. and Hendzel M. J. (2000). Reduced mobility of the alternate splicing factor (ASF) through the nucleoplasm and steady state speckle compartments. J. Cell Biol. 150, 41-51. 10.1083/jcb.150.1.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A. I. and Spector D. L. (2003). Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4, 605-612. 10.1038/nrm1172 [DOI] [PubMed] [Google Scholar]
- Lis J. T., Neckameyer W., Dubensky R. and Costlow N. (1981). Cloning and characterization of nine heat-shock-induced mRNAs of Drosophila melanogaster. Gene 15, 67-80. 10.1016/0378-1119(81)90105-0 [DOI] [PubMed] [Google Scholar]
- Mahat D. B., Salamanca H. H., Duarte F. M., Danko C. G. and Lis J. T. (2016). Mammalian heat shock response and mechanisms underlying its genome-wide transcriptional regulation. Mol. Cell 62, 63-78. 10.1016/j.molcel.2016.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzahn M. R., Marada S., Lee J., Nourse A., Kenrick S., Zhao H. Y., Ben-Nissan G., Kolaitis R. M., Peters J. L., Pounds S. et al. (2016). Higher-order oligomerization promotes localization of SPOP to liquid nuclear speckles. EMBO J. 35, 1254-1275. 10.15252/embj.201593169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz P. J., Patterson S. D., Neuwald A. F., Spahr C. S. and Spector D. L. (1999). Purification and biochemical characterization of interchromatin granule clusters. EMBO J. 18, 4308-4320. 10.1093/emboj/18.15.4308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T., Cáceres J. F. and Spector D. L. (1997). The dynamics of a pre-mRNA splicing factor in living cells. Nature 387, 523-527. 10.1038/387523a0 [DOI] [PubMed] [Google Scholar]
- Misteli T., Cáceres J. F., Clement J. Q., Krainer A. R., Wilkinson M. F. and Spector D. L. (1998). Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J. Cell Biol. 143, 297-307. 10.1083/jcb.143.2.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe R. T., Mayeda A., Sadowski C. L., Krainer A. R. and Spector D. L. (1994). Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J. Cell Biol. 124, 249-260. 10.1083/jcb.124.3.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu N. (1979). Threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 9, 62-66. 10.1109/TSMC.1979.4310076 [DOI] [Google Scholar]
- Parsons G. G. and Spencer C. A. (1997). Mitotic repression of RNA polymerase II transcription is accompanied by release of transcription elongation complexes. Mol. Cell. Biol. 17, 5791-5802. 10.1128/MCB.17.10.5791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair R. D. and Misteli T. (2000). High mobility of proteins in the mammalian cell nucleus. Nature 404, 604-609. 10.1038/35007077 [DOI] [PubMed] [Google Scholar]
- Posern G., Miralles F., Guettler S. and Treisman R. (2004). Mutant actins that stabilise F-actin use distinct mechanisms to activate the SRF coactivator MAL. EMBO J. 23, 3973-3983. 10.1038/sj.emboj.7600404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posern G., Sotiropoulos A. and Treisman R. (2002). Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor. Mol. Biol. Cell 13, 2001-2015. 10.1091/mbc.02-05-0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rino J., Carvalho T., Braga J., Desterro J. M. P., Lührmann R. and Carmo-Fonseca M. (2007). A stochastic view of spliceosome assembly and recycling in the nucleus. PLoS Comput. Biol. 3, 2019-2031. 10.1371/journal.pcbi.0030201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh N., Spahr C. S., Patterson S. D., Bubulya P., Neuwald A. F. and Spector D. L. (2004). Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell 15, 3876-3890. 10.1091/mbc.e04-03-0253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J. C., Wang X., Podell E. R. and Cech T. R. (2013). RNA seeds higher-order assembly of FUS protein. Cell Rep. 5, 918-925. 10.1016/j.celrep.2013.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Takata H., Shibahara K.-I., Bubulya A. and Bubulya P. A. (2010). Son is essential for nuclear speckle organization and cell cycle progression. Mol. Biol. Cell 21, 650-663. 10.1091/mbc.e09-02-0126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu R. and Tanaka H. (2015). A novel coarsening mechanism of droplets in immiscible fluid mixtures. Nat. Commun. 6, 7407 10.1038/ncomms8407 [DOI] [PubMed] [Google Scholar]
- Spector D. L. (1993). Macromolecular domains within the cell-nucleus. Annu. Rev. Cell Biol. 9, 265-315. 10.1146/annurev.cb.09.110193.001405 [DOI] [PubMed] [Google Scholar]
- Spector D. L., Fu X. D. and Maniatis T. (1991). Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 10, 3467-3481. 10.1002/j.1460-2075.1991.tb04911.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry M. (1995). The interchromatin granules. Histol. Histopathol. 10, 1035-1045. [PubMed] [Google Scholar]
- Weidemann T., Wachsmuth M., Knoch T. A., Müller G., Waldeck W. and Langowski J. (2003). Counting nucleosomes in living cells with a combination of fluorescence correlation spectroscopy and confocal imaging. J. Mol. Biol. 334, 229-240. 10.1016/j.jmb.2003.08.063 [DOI] [PubMed] [Google Scholar]
- Yankulov K., Yamashita K., Roy R., Egly J.-M. and Bentley D. L. (1995). The transcriptional elongation inhibitor 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole inhibits transcription factor IIH-associated protein kinase. J. Biol. Chem. 270, 23922-23925. 10.1074/jbc.270.41.23922 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Kota K. P., Alam S. G., Nickerson J. A., Dickinson R. B. and Lele T. P. (2016). Coordinated dynamics of RNA splicing speckles in the nucleus. J. Cell. Physiol. 231, 1269-1275. 10.1002/jcp.25224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L. and Brangwynne C. P. (2015). Nuclear bodies: the emerging biophysics of nucleoplasmic phases. Curr. Opin. Cell Biol. 34, 23-30. 10.1016/j.ceb.2015.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.