Abstract
A prominent hurdle in developing small molecule probes against RNA is the relative scarcity of general screening methods. In this study, we demonstrate the application of a fluorescent peptide displacement assay to screen small molecule probes against four different RNA targets. The designed experimental protocol combined with statistical analysis provides a fast and convenient method to simultaneously evaluate small molecule libraries against different RNA targets and classify them based on affinity and selectivity patterns.
Introduction:
The discovery of non-protein-coding RNA and their participation in a variety of different biological processes has renewed interest in development of small molecules both for therapeutic targeting of RNA and to probe their cellular biology.1–3 Small molecule ligands for RNA provide several advantages over traditionally studied oligonucleotide probes, such as better cellular uptake and pharmacological availability.4–6 Although several advances are now being made towards the development of small molecules against RNA, several challenges remain.7 The relative paucity of three-dimensional structural characterization of RNA limits the application of structure-based methods on probe development.8–9 Moreover, the flexible structure of RNA and relatively low chemical diversity compared to protein targets complicates the development of highly selective small molecule ligands.10 Sustained efforts are thus critical in realizing the true therapeutic potential of RNA.
Screening commercial small molecule libraries against RNA targets is by far the most popular method to search for probes.4, 11 While several methodologies have been successfully utilized in screening small molecule libraries against RNA,12–16 fluorescence based assays are commonly employed due to several advantages such as sensitivity, speed, and the convenient availability of equipment. Conventional fluorescence assays, however, frequently involve incorporation of fluorescent probes in the RNA sequence, which can alter the native structure and often present synthetic challenges.17–19
One prominent screening method against nucleic acids is the fluorescent indicator displacement assay (FID) where the indicator displays different fluorescence properties in the presence and absence of the oligonucleotide and thus can be utilized to measure the binding properties of small molecules.20 Common fluorescent probes used in FID assays for nucleic acids include intercalating dyes such as ethidium bromide, Thiazole Orange (TO), and TO-PRO-1.21–25 These fluorescent probes show high nanomolar to micromolar binding affinities to RNA, which often necessitates the use of high quantities of RNA (>1 μM) in screening experiments.23, 26 Alternative probes with higher affinities can help reduce the amounts of RNA utilized in assays. The human immunodeficiency virus-1 (HIV-1) ribonucleoprotein Tat binds the Trans Activation Response element (TAR) RNA and facilitates viral transcription.27 A truncated peptide containing positively charged amino acid residues from the basic domain of Tat protein has been previously shown to bind small HIV-1-TAR RNA constructs,28 and displacement of the peptide has been utilized to screen small molecule libraries for TAR binding ligands.29–30 In addition to affinity, careful evaluation of the selectivity of RNA ligands is often overlooked. Studies that do evaluate selectivity often rely on using tRNA as a competitor as it is abundant in the cell and provides the opportunity for non-specific backbone and intercalation interactions. At the same time, few small molecule ligands have been reported that bind tRNA.31–35 Consequently, there remains a need for efficient assays that simultaneously screen small molecules against RNA targets and more stringently evaluate specificity.
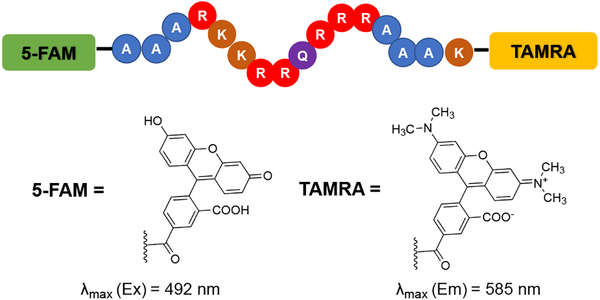
We chose to explore the suitability of the Tat peptide FID assay for this purpose as the high affinity allows for low RNA concentrations, and the peptide is known to bind other RNA secondary structures in vitro.36–37 Since its first introduction by Hamasaki and co-workers,38 the displacement assay involving a Tat peptide construct labeled with a Förster Resonance Enhancement Transfer (FRET) pair (Figure 1) has been extensively utilized for screening small molecules against HIV-1-TAR RNA.26, 39–45 This specific assay utilizes 5-carboxyfluorescein (FAM) at the N-terminus and 5-carboxytetramethylrhodamine (TAMRA) at the C-terminus of Tat, which can be commercially purchased. When the peptide is bound to RNA, the fluorophores are distant in space and FRET is facilitated, allowing for excitation of FAM and emission detection from TAMRA. When the small molecule probe displaces the peptide from the RNA, the two fluorophores are close in proximity and the emission of TAMRA is quenched. This quenching allows for quantification of the binding affinity of the small molecule towards TAR RNA. The Tat peptide displacement assay has also been utilized to evaluate the activity of tripeptide ligands against the analogous TAR RNA from the HIV-2 virus.46
Figure 1:
Sequence of FRET pair labeled Tat peptide used in displacement assays against HIV-1-TAR RNA. Structures of fluorophores 5-FAM, and TAMRA used at the N and C-termini shown.38 Amino acid abbreviations: A = Ala, R = Arg, K = Lys, Q = Gly.
We sought to extend the Tat peptide displacement assay to simultaneously screen small molecules ligands against multiple RNA targets of interest and thus assess both binding and selectivity in the same assay. Herein we describe the application of this assay for rapidly screening small molecule probes against four different RNA constructs. We begin by re-examining the binding of Tat to HIV-1-TAR and HIV-2-TAR and then test it against two other RNA structures, the bacterial ribosomal A-Site RNA and the IIB domain HIV-1-Rev response element RNA (RRE-IIB).47–50 We then describe our rapid small molecule screening protocol using the displacement of the Tat peptide from these RNA, which is subsequently validated by quantifying the binding of positive and negative hits against the 4 RNA targets. Observed activities across the four different RNA structures can be further utilized to classify small molecules based on their binding patterns and selectivity. Strong binding affinities of Tat towards RNA targets allows the use of significantly lower amounts of RNA than other assays, which provides a major economic advantage. Finally, this assay provides an easy method to assess the selectivity of small molecules against different RNA structures, which is critical to guide the future development of functional probes.
Results and Discussion:
Fluorescently labeled Tat peptide binds different RNA structures:
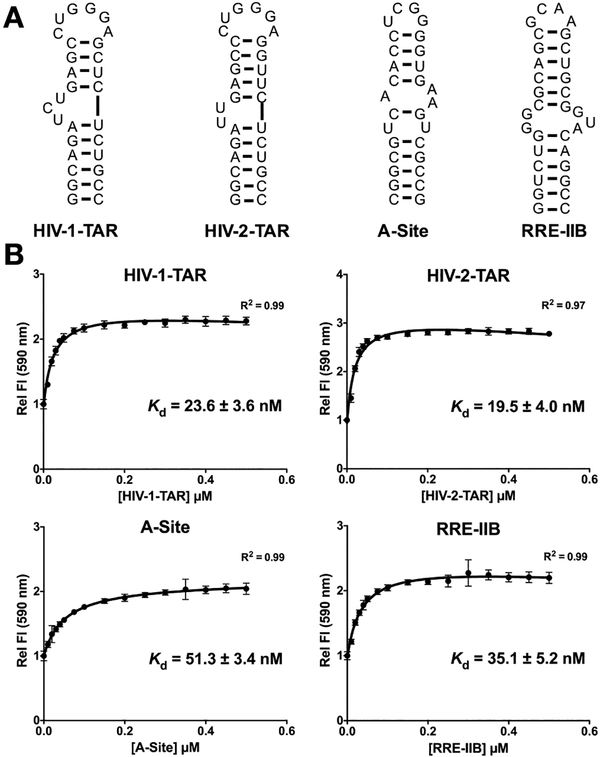
We began by exploring the direct binding of the Tat peptide (Figure 1) against the 4 RNA structures (Figure 2A). RNA constructs were selected based on disease-relevance and known small molecule binding partners and were designed to be of similar size (~30 n.t.). Selected RNA structures included: 1) HIV-1-TAR; 2) HIV-2-TAR RNA, which features a truncated bulge and a more dynamic upper stem containing a G-U base pair; 3) bacterial ribosomal A-Site RNA containing a 1X2 internal loop structure and a smaller 4 nucleotide apical loop, and 4) HIV-1-rev response element RNA fragment IIB (RRE-IIB), which features a larger 2X3 internal loop structure and a similar small apical loop.40 Figure 2B shows the binding curves observed for these 4 RNA against the Tat peptide. As expected, the Tat peptide showed strong binding interactions with HIV-1 and HIV-2-TAR RNA structures, which are native cellular binding partners (Kd = 23.6 ± 3.6 nM and 19.5 ± 3.0 nM, respectively). The affinity towards the other RNA structures studied, A-Site and RRE-IIB, was slightly diminished as compared to the two TAR structures (Kd = 51.3 ± 3.4 nM and 35.1 ± 5.2 nM, respectively) suggesting a binding preference for bulges. The low nanomolar Kd values observed for all four RNAs studied confirms that this basic and positively charged peptide is capable of interacting strongly with different RNA structures and can be utilized as a probe to screen small molecule libraries.
Figure 2:
A) Sequences and secondary structures of 4 RNAs used in assays.40 B) Binding curves for the 4 RNA targets against Tat peptide. Error bars shown are the standard errors of mean from three replicates. Conditions: 50 nM Tat, 0–500 nM RNA, Incubation 30 min, Ex = 485 nm, Em = 590 nm; Buffer: 50 mM Tris, 50 mM KCl, 0.01% Triton-X-100, pH = 7.4.44
The choice of appropriate buffer for displacement experiments is crucial as it can differentially influence RNA structure, small molecule binding affinities, and small molecule solubility.51 For example, ionic interactions between small molecules and RNA can be hindered or facilitated by high and low salt content buffers, respectively.39, 52 Previous studies on small molecule interactions with RNA using the Tat displacement assay have used low salt concentration buffers41–43 to increase the solubility of small molecule ligands and achieve stable photo-physical readouts in the bound state.53–54 However these less stringent conditions can lead to identification of non-specific interactions (false positives). On the other hand, high salt content can hinder the interaction between Tat and RNA and lead to false negatives. Finally, salt content is also known to impact the photophysical output of fluorophores.53 It is thus important to carefully optimize assay conditions such as buffer identity and salt concentration (see Table S1 of the Supporting Information).
Lowering the salt (KCl) content in Tris buffer improved the binding affinities of Tat for all 4 studied RNA while gradually increasing the amount of the bivalent ion magnesium in the buffer showed decreased binding affinities. These results are not surprising as the interaction between Tat and RNA is known to be ionic in nature,55 and magnesium is known to compact the tertiary structures of RNA and restrict available sites for interaction with ligands.56–57 Phosphate buffer, which is also commonly used in studying RNA binding to peptides and small molecules,54 showed a similar effect where low salt content improved the binding and increased magnesium content weakened it. Finally, PBS buffer, containing high concentrations of salts, showed significantly weakened binding of Tat to all 4 RNAs. An intermediary salt concentration of 50 mM KCl without additional magnesium or sodium was selected for our assays.
Simultaneous screening of small molecule libraries against 4 RNA targets:
The relatively tight binding affinities of the Tat peptide to all studied RNA supported the utility of this system for rapid screening of small molecule libraries. Using these observed Kd values, we first calculated the fraction of Tat bound to each RNA (Table S2) to choose RNA concentrations where approximately half of the Tat was complexed.25 The Z’ Scores of the 4 systems were then determined to evaluate the quality of the screening assay (See Table S3).58 Acceptable Z’-score values were found for HIV-1-TAR (Z’ = 0.54), HIV-2-TAR (Z’ = 0.65) and RRE-IIB (Z’ = 0. 48). For A-Site RNA, the Z’ score was distinctly lower (Z’ = 0.35), which can be attributed to the lower fluorescence intensity of the bound A-Site:Tat complex. The result implies that screening against A-Site RNA under these conditions may lead to higher false positives / negatives. While a lower salt content buffer improved the Z’ Score for A-Site RNA assay to 0.44, presumably due to tighter Tat binding, we chose to maintain constant salt conditions across the screen. Three libraries with 10 molecules each were chosen for initial screening experiments: 1) aminoglycosides,59 which are frequently found as hits in screening experiments against RNA; 2) other known RNA binding molecules from the literature;60–61 and 3) the in-house developed amiloride derivative library shown to be active against HIV-1-TAR44 (See Figure S1 for structures of small molecule ligands studied). All small molecules were tested against the 4 RNA structures at concentrations of 10 μM and 50 μM to identify both strong and weak RNA binders as measured by the % displacement of Tat from RNA. Percent Fluorescence Indicator Displacement (%FID) was calculated using Equation 2 (see supporting information). Small molecules showing >25% FID were designated as hits in keeping with the literature.22, 24
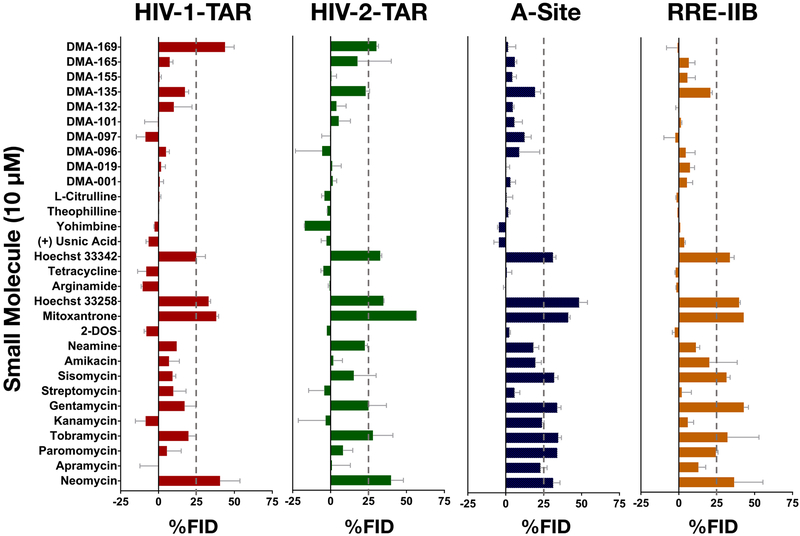
Figure 3 shows the results of screening experiments at 10 μM concentrations. This stringent screening condition is first chosen to clearly identify the stronger RNA binders and assess selectivity. First, a distinction can be made between the observed hits for the two bulge-stem-loop structures in HIV-1 and HIV-2-TAR RNA, and the two internal-loop containing A-Site and RRE-IIB RNA structures. Aminoglycoside analogs showed more preference for targeting the latter RNA structures as compared to the former with only one aminoglycoside (neomycin) being identified as hit against all 4 RNA structures. No other aminoglycoside led to >25% FID from HIV-1-TAR RNA while gentamycin and tobramycin were identified as hits for HIV-2-TAR RNA. This difference may be attributed to the more flexible RNA secondary structure of HIV-2-TAR resulting from the non-canonical G-U base pair in the upper stem region of this RNA. Overall these results were in line with previous reports of strong binding of aminoglycosides to internal loop structures, particularly A-Site RNA.62 The known RNA binding small molecules mitoxantrone, Hoechst 33258, and Hoechst 33342 appeared to bind to all 4 RNA with similar % FID values; however, control experiments in the absence of RNA suggested that the fluorescence of these three small molecules is likely to interfere with the Tat peptide fluorescence at the concentrations used here, and these dyes were thus excluded from further analysis (See page S18 of the supporting information). None of the other studied molecules in this class show appreciable displacement under these stringent screening conditions. From the in-house synthesized amiloride derivative library, DMA-169 was identified as a hit for both HIV-1 and HIV-2-TAR RNAs while showing no appreciable displacement of Tat from A-Site and RRE-IIB RNAs. This result further confirms the observation that this molecule is a selective binder to the pyrimidine rich bulge residues of TAR.44 As can be expected, more compounds are identified as hits under the non-stringent 50 μM screening condition (See Figure S2). Finally, the negative %FID values seen in select cases can be attributed to the enhancement in fluorescence emission of the fluorophore TAMRA as a result of interactions of small molecules with the peptide or enhanced affinity of Tat to RNA in the presence of the small molecule.25
Figure 3:
Results of screening of 30 small molecules against HIV-1-TAR (Panel 1: Red), HIV-2-TAR (Panel 2: Green), A-Site (Panel 3: Blue), RRE-IIB (Panel 4: Orange) at 10 μM small molecule concentration. Conditions: 50 nM Tat-peptide; 40 nM HIV-1-TAR, 40 nM HIV-2-TAR, 120 nM A-Site, 75 nM RRE-IIB; 0, 10, 50 μM Small molecule; λ(Ex): 485 nm, λ(Em): 590 nm; Buffer: 50 mM Tris, 50 mM KCl, 5% DMSO, 0.01% Triton-X-100, pH = 7.4. %FID calculated using Equation 2 shown in the SI. See tables S4 and S5 and Figure S2 for other results of the Tat peptide displacement based screening experiments.
Tat displacement assay for quantitative measurement of activity of small molecules:
The activity of a set of 10 small molecules featuring both hits and non-hits from the screening assay was then studied by performing small molecule titrations with Tat peptide displacement assay. Table 1 shows the observed competitive displacement at 50% fluorescence (CD50) for these 10 molecules against the 4 RNAs studied.
Table 1:
Tat peptide displacement assays of a representative set of small molecules with HIV-1-TAR, HIV-2-TAR, A-Site, RRE-IIB.
| Entry | Small Molecule | CD50 (μM)a,b | |||
|---|---|---|---|---|---|
| HIV-1-TAR | HIV-2-TAR | A-Site | RRE-IIB | ||
| 1 | Neomycin | 0.48 ± 0.03 | 0.20 ± 0.02 | 0.49 ± 0.05 | 0.10 ± 0.02 |
| 2 | Paromomycin | 5.6 ± 0.6 | 3.3 ± 0.3 | 0.71 ± 0.08 | 0.58 ± 0.14 |
| 3 | Tobramycin | 2.8 ± 0.4 | 1.4 ± 0.2 | 0.64 ± 0.12 | 0.25 ± 0.08 |
| 4 | Gentamycin | 4.2 ± 0.3 | 1.8 ± 0.14 | 0.42 ± 0.06 | 0.75 ± 0.24 |
| 5 | Sisomycin | 3.5 ± 1.9 | 2.0 ± 0.3 | 0.49 ± 0.11 | 0.82 ± 0.11 |
| 6 | Streptomycin | 148 ± 122 | 206 ± 90 | 15.3 ± 3.9 | 66.4 ± 30 |
| 7 | Amikacin | 30.3 ± 4.4 | 50.0 ± 6.2 | 4.5 ± 1.04 | 6.2 ± 1.7 |
| 8 | Argininamide | >300 | >300 | >300 | >300 |
| 9 | DMA-135 | 15.3 ± 1.7 | 12.9 ± 1.7 | 60.2 ± 13.8 | 42.8 ± 8.7 |
| 10 | DMA-169 | 3.9 ± 0.7 | 9.3 ± 1.3 | 18.7 ± 5.8 | 29.0 ± 7.1 |
Conditions: 50 nM Tat, 40 nM HIV-1-TAR, 40 nM HIV-2-TAR, 0–300 μM small molecule. Buffer: 50 mM Tris, 50 mM KCl, 0.01% Triton-X-100, 5% DMSO, pH = 7.4.
CD50: competitive dosage required for displacement of 50% of Tat peptide from preformed TAR:Tat complex, as measured by fluorescence of FAM- and TAMRA-labeled Tat peptide at 590 nm, the emission λmax of TAMRA.
Errors are standard errors of mean for three independent replicates.
The aminoglycosides neomycin, paromomycin, tobramycin, gentamicin, and sisomicin (Entries 1–5) showed nanomolar to low-micromolar CD50 values for all 4 RNAs studied. While the observed activity of neomycin was comparable against all 4 RNA structures (Entry 1), the CD50 values of the latter four aminoglycosides were slightly lower for A-Site and RRE-IIB than the two TAR RNAs. On the other hand, the aminoglycoside streptomycin was a significantly weaker RNA binder (Entry 6) with high micromolar CD50 values observed for all four RNA. The selectivity towards the two internal-loop containing RNA was more pronounced in case of amikacin (Entry 7), where the CD50 values for the two TAR RNA were over ~10 times weaker than those for A-Site and RRE-IIB. The relative RNA binding affinity observed for aminoglycosides generally correlated with previous literature.61, 63–65 Similarly, the slight preference for internal loop structures has been previously reported.24, 59 The known TAR binding amino-acid derivative argininamide did not show appreciable displacement of Tat peptide from any of the four RNAs. This could be indicative of weaker binding (Kd >1 mM),66–67 or of a binding event that does not lead to displacement of the indicator Tat peptide.22, 25 Interestingly, the amiloride derivative DMA-135 showed similar CD50 values for all 4 RNAs in contrast to the selectivity previous observed using 100 fold excess of tRNA.44 Finally, pronounced selectivity towards the bulge containing TAR RNA was observed for DMA-169, which was consistent with the observed bulge selectivity of this compound in previous studies.44 These results indicate that only the small molecules with CD50 ≤ 1 μM are identified as hits at the stringent screening condition of 10 μM small molecule concentration, while molecules with CD50 values >1μM are identified as hits at the 50 μM screening conditions (See Figure S2 of the supporting information). Screening at two different small molecule concentrations thus identifies hits and their approximate affinities for each RNA. Finally, the comparison of CD50 values can be used to quantify their RNA motif selectivity.
Statistical analysis of FID data:
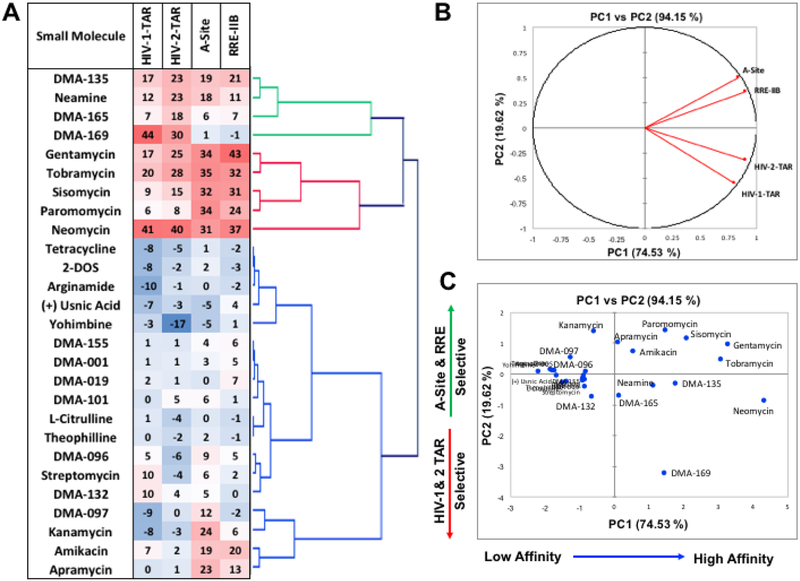
Finally, to assess activity and selectivity patterns within the screening data we performed statistical analysis on the %FID values obtained for the library of 27 small molecules against each RNA at 10 μM concentration. Two separate analyses were performed on the data: agglomerative hierarchical clustering analysis (AHC), which clusters data based on similarities within the observations, and principal component analysis (PCA), which is a multivariate statistical analysis method for reducing the dimensionality of large datasets.
As seen in Figure 4A, AHC identifies 3 distinct clusters within the data. Cluster 1 includes DMA-135, neamine, and DMA-165, which show moderate %FID numbers and DMA-169, which shows selectivity towards the two TAR-RNA structures studied. Cluster 2 includes five aminoglycosides that show higher %FID values towards all studied RNA and are more selective towards the internal loop containing A-Site and RRE-IIB RNAs. Cluster 3 includes all other small molecules that show minimal %FID scores, and two aminoglycosides amikacin and apramycin that show moderate %FID values and selectivity towards A-Site and RRE-IIB. AHC can thus classify these molecules based on both their displacement activity and RNA selectivity. Figure 4B shows the loading plots for the PCA data along the PC1 and PC2 axes, which clearly separates the data into two clusters between the internal loop containing A-Site and RRE-IIB RNA and the bulge-stem-loop containing HIV-1 and HIV-2-TAR RNAs, with strongest contributions in the positive PC1 direction. Figure 4C shows the scatter plots of the PCA data along the PC1 and PC2 axes where the data can be seen to separate along the PC1 axis based on the affinity towards RNA targets, and along the PC2 axis based on selectivity. Combined PCA with the screening data at both 10 μM and 50 μM small molecule concentration shows similar classification based on affinity and selectivity along the PC1 and PC2 axes respectively (See page S22 of the supporting information). These simple statistical analyses demonstrate the potential of this method to not only screen small molecule ligands for different RNA targets but also to provide information-rich data sets that can lend insights into selectivity trends of the tested small molecules.
Figure 4:
A) Heat map and agglomerative hierarchical clustering (AHC) data with the displacement screening (%FID) data obtained at 10 μM small molecule concentration. B) Loading plots from the principal component analysis (PCA) data along PC1 and PC2 C) Scatter plot of the data along the PC1 and PC2 axes.
Conclusions and future directions:
In this study, we demonstrated the displacement of a basic, positively charged fluorescently labeled Tat peptide as a general method for screening small molecules against RNA targets. Four similarly sized RNA targets possessing varied secondary structure motifs were chosen and were shown to bind to the Tat peptide with low nanomolar affinities, which allowed the utilization of low amounts of RNA as compared to related FID assays. The effect of varying buffer salt concentrations on the binding of Tat to these 4 RNA was then examined. Based on the observed variability in affinity with respect to salt content, a buffer with intermediate salt concentration and no magnesium was chosen to minimize the false positive and negative results. Fluorescence change upon displacement was utilized in establishing a screening protocol for small molecules against the aforementioned RNA structures. A library of 30 small molecules was tested that included aminoglycosides, known RNA binding small molecules, and in-house developed amiloride derivatives. The screening assay was successful in identifying hits against all four RNA structures as well as indiscriminate and differential binding of individual small molecules and confirmed the importance of using multiple RNA targets to evaluate specificity. As with any fluorescent displacement assay, limitations include the potential formation of ternary complexes that do not displace the indicator and fluorescence background interference. Importantly, binding results correlated with known literature results where available. Finally, AHC and PCA statistical analysis were used to assess patterns within the data and clarify the relationship between small molecule structures and RNA binding affinity and selectivity. We believe that the low concentrations of small molecule and RNA needed, the ease of set up, and the rapid assessment of small molecule:RNA binding patterns render this assay a powerful screen for evaluating small molecule:RNA interactions, whether to identify leads or to analyze trends guiding selectivity.
Supplementary Material
Acknowledgements:
We would like to thank Prof. Hashim Al-Hashimi for providing initial samples of the fluorescently labeled Tat peptide. We also thank Laura Ganser for helpful advice on assay design and implementation. The authors acknowledge financial support for this work from Duke University, the US National Institute of Health Grant/Award Number: U54GM10329, and the National Institute of General Medical Sciences (NIGMS) Maximizing Investigator’s Research Award (MIRA) Grant/Award Number: R35GM124785. Finally, we would like to thank all members of the Hargrove lab, particularly Sarah Wicks, for stimulating discussions on the project and for providing helpful feedback on the manuscript.
Footnotes
Conflicts of Interest: There are no conflicts of interest to declare.
References:
- 1.Connelly Colleen M.; Moon Michelle H.; Schneekloth John S. Jr., The Emerging Role of RNA as a Therapeutic Target for Small Molecules. Cell Chem. Biol 2016, 23 (9), 1077–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rizvi NF; Smith GF, RNA as a small molecule druggable target. Bioorg. Med. Chem. Lett 2017, 27 (23), 5083–5088. [DOI] [PubMed] [Google Scholar]
- 3.Donlic A; Hargrove AE, Targeting RNA in mammalian systems with small molecules. Wiley Interdiscip. Rev.: RNA 2018, 9 (4), e1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas JR; Hergenrother PJ, Targeting RNA with small molecules. Chem. Rev 2008, 108 (4), 1171–1224. [DOI] [PubMed] [Google Scholar]
- 5.Hermann T, Small molecules targeting viral RNA. Wiley Interdiscip. Rev.: RNA 2016, 7 (6), 726–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kole R; Krainer AR; Altman S, RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discovery 2012, 11, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morgan BS; Forte JE; Hargrove AE, Insights into the development of chemical probes for RNA. Nucleic Acids Res. 2018, 46 (16), 8025–8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Velagapudi SP; Gallo SM; Disney MD, Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat. Chem. Biol 2014, 10 (4), 291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warner KD; Hajdin CE; Weeks KM, Principles for targeting RNA with drug-like small molecules. Nat. Rev. Drug Discovery 2018, 17, 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgan BS; Hargrove AE, Chapter 7: Synthetic receptors for oligonucleotides and nucleic acids In Synthetic Receptors for Biomolecules: Design Principles and Applications, The Royal Society of Chemistry: 2015; pp 253–325. [Google Scholar]
- 11.Guan L; Disney MD, Recent advances in developing small molecules targeting RNA. ACS Chem. Biol 2012, 7 (1), 73–86. [DOI] [PubMed] [Google Scholar]
- 12.Rizvi NF; Howe JA; Nahvi A; Klein DJ; Fischmann TO; Kim H-Y; McCoy MA; Walker SS; Hruza A; Richards MP; Chamberlin C; Saradjian P; Butko MT; Mercado G; Burchard J; Strickland C; Dandliker PJ; Smith GF; Nickbarg EB, Discovery of Selective RNA-Binding Small Molecules by Affinity-Selection Mass Spectrometry. ACS Chem. Biol 2018, 13 (3), 820–831. [DOI] [PubMed] [Google Scholar]
- 13.Seth PP; Miyaji A; Jefferson EA; Sannes-Lowery KA; Osgood SA; Propp SS; Ranken R; Massire C; Sampath R; Ecker DJ; Swayze EE; Griffey RH, SAR by MS: Discovery of a New Class of RNA-Binding Small Molecules for the Hepatitis C Virus: Internal Ribosome Entry Site IIA Subdomain. J. Med. Chem 2005, 48 (23), 7099–7102. [DOI] [PubMed] [Google Scholar]
- 14.Garavís M; López-Méndez B; Somoza A; Oyarzabal J; Dalvit C; Villasante A; Campos-Olivas R; González C, Discovery of Selective Ligands for Telomeric RNA G-quadruplexes (TERRA) through 19F-NMR Based Fragment Screening. ACS Chem. Biol 2014, 9 (7), 1559–1566. [DOI] [PubMed] [Google Scholar]
- 15.Sztuba-Solinska J; Shenoy SR; Gareiss P; Krumpe LRH; Le Grice SFJ; O’Keefe BR; Schneekloth JS, Identification of biologically active, HIV TAR RNA-binding small molecules using small molecule microarrays. J. Am. Chem. Soc 2014, 136 (23), 8402–8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Velagapudi SP; Pushechnikov A; Labuda LP; French JM; Disney MD, Probing a 2-aminobenzimidazole library for binding to RNA internal loops via two-dimensional combinatorial screening. ACS Chem. Biol 2012, 7 (11), 1902–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin PZ; Pyle AM, Site-Specific Labeling of RNA with Fluorophores and Other Structural Probes. Methods 1999, 18 (1), 60–70. [DOI] [PubMed] [Google Scholar]
- 18.Paredes E; Evans M; Das SR, RNA labeling, conjugation and ligation. Methods 2011, 54 (2), 251–259. [DOI] [PubMed] [Google Scholar]
- 19.Schmitz AG; Zelger-Paulus S; Gasser G; Sigel RKO, Strategy for Internal Labeling of Large RNAs with Minimal Perturbation by Using Fluorescent PNA. ChemBioChem 2015, 16 (9), 1302–1306. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen BT; Anslyn EV, Indicator–displacement assays. Coord. Chem. Rev 2006, 250 (23), 3118–3127. [Google Scholar]
- 21.Boger DL; Fink BE; Brunette SR; Tse WC; Hedrick MP, A Simple, High-Resolution Method for Establishing DNA Binding Affinity and Sequence Selectivity. J. Am. Chem. Soc 2001, 123 (25), 5878–5891. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J; Umemoto S; Nakatani K, Fluorescent Indicator Displacement Assay for Ligand-RNA Interactions. J. Am. Chem. Soc 2010, 132 (11), 3660–3661. [DOI] [PubMed] [Google Scholar]
- 23.Asare-Okai PN; Chow CS, A modified fluorescent intercalator displacement assay for RNA ligand discovery. Anal. Biochem 2011, 408 (2), 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tran T; Disney MD, Identifying the preferred RNA motifs and chemotypes that interact by probing millions of combinations. Nat. Commun 2012, 3, 1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.del Villar-Guerra R; Gray RD; Trent JO; Chaires JB, A rapid fluorescent indicator displacement assay and principal component/cluster data analysis for determination of ligand–nucleic acid structural selectivity. Nucleic Acids Res. 2018, 46 (7), e41–e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar S; Kellish P; Robinson WE; Wang D; Appella DH; Arya DP, Click Dimers To Target HIV TAR RNA Conformation. Biochemistry 2012, 51 (11), 2331–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J; Tamilarasu N; Hwang S; Garber ME; Huq I; Jones KA; Rana TM, HIV-1 TAR RNA Enhances the Interaction between Tat and Cyclin T1. J. Biol. Chem 2000, 275 (44), 34314–34319. [DOI] [PubMed] [Google Scholar]
- 28.Weeks K; Ampe C; Schultz S; Steitz T; Crothers D, Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science 1990, 249 (4974), 1281–1285. [DOI] [PubMed] [Google Scholar]
- 29.Mei H-Y; Mack DP; Galan AA; Halim NS; Heldsinger A; Loo JA; Moreland DW; Sannes-Lowery KA; Sharmeen L; Truong HN; Czarnik AW, Discovery of selective, small-molecule inhibitors of RNA complexes—1. The tat protein/TAR RNA complexes required for HIV-1 transcription. Biorg. Med. Chem 1997, 5 (6), 1173–1184. [DOI] [PubMed] [Google Scholar]
- 30.Gelus N; Bailly C; Hamy F; Klimkait T; Wilson WD; Boykin DW, Inhibition of HIV-1 Tat-TAR interaction by diphenylfuran derivatives: effects of the terminal basic side chains. Biorg. Med. Chem 1999, 7 (6), 1089–1096. [DOI] [PubMed] [Google Scholar]
- 31.Das A; Bhadra K; Suresh Kumar G, Targeting RNA by Small Molecules: Comparative Structural and Thermodynamic Aspects of Aristololactam-β-D-glucoside and Daunomycin Binding to tRNAphe. PLOS ONE 2011, 6 (8), e23186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun T; Zhang Y, Pentamidine binds to tRNA through non-specific hydrophobic interactions and inhibits aminoacylation and translation. Nucleic Acids Res. 2008, 36 (5), 1654–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walter F; Pütz J; Giegé R; Westhof E, Binding of tobramycin leads to conformational changes in yeast tRNAAsp and inhibition of aminoacylation. The EMBO Journal 2002, 21 (4), 760–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirillov S; Vitali LA; Goldstein BP; Monti F; Semenkov Y; Makhno V; Ripa S; Pon CL; Gualerzi CO, Purpuromycin: an antibiotic inhibiting tRNA aminoacylation. RNA 1997, 3 (8), 905–913. [PMC free article] [PubMed] [Google Scholar]
- 35.Mikkelsen NE; Johansson K; Virtanen A; Kirsebom LA, Aminoglycoside binding displaces a divalent metal ion in a tRNA–neomycin B complex. Nat. Struct. Biol 2001, 8, 510. [DOI] [PubMed] [Google Scholar]
- 36.Weeks KM; Crothers DM, RNA recognition by Tat-derived peptides: Interaction in the major groove? Cell 1991, 66 (3), 577–588. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y; Varani G, Protein families and RNA recognition. The FEBS Journal 2005, 272 (9), 2088–2097. [DOI] [PubMed] [Google Scholar]
- 38.Matsumoto C; Hamasaki K; Mihara H; Ueno A, A high-throughput screening utilizing intramolecular fluorescence resonance energy transfer for the discovery of the molecules that bind HIV-1-TAR RNA specifically. Bioorg. Med. Chem. Lett 2000, 10 (16), 1857–1861. [DOI] [PubMed] [Google Scholar]
- 39.Riguet E; Désiré J; Boden O; Ludwig V; Göbel M; Bailly C; Décout J-L, Neamine dimers targeting the HIV-1 TAR RNA. Bioorg. Med. Chem. Lett 2005, 15 (21), 4651–4655. [DOI] [PubMed] [Google Scholar]
- 40.Stelzer AC; Frank AT; Kratz JD; Swanson MD; Gonzalez-Hernandez MJ; Lee J; Andricioaei I; Markovitz DM; Al-Hashimi HM, Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nat. Chem. Biol 2011, 7 (8), 553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ranjan N; Kumar S; Watkins D; Wang D; Appella DH; Arya DP, Recognition of HIV-TAR RNA using neomycin–benzimidazole conjugates. Bioorg. Med. Chem. Lett 2013, 23 (20), 5689–5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeiger M; Stark S; Kalden E; Ackermann B; Ferner J; Scheffer U; Shoja-Bazargani F; Erdel V; Schwalbe H; Göbel MW, Fragment based search for small molecule inhibitors of HIV-1 Tat-TAR. Bioorg. Med. Chem. Lett 2014, 24 (24), 5576–5580. [DOI] [PubMed] [Google Scholar]
- 43.Abulwerdi FA; Shortridge MD; Sztuba-Solinska J; Wilson R; Le Grice SFJ; Varani G; Schneekloth JS, Development of Small Molecules with a Noncanonical Binding Mode to HIV-1 Trans Activation Response (TAR) RNA. J. Med. Chem 2016, 59 (24), 11148–11160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patwardhan NN; Ganser LR; Kapral GJ; Eubanks CS; Lee J; Sathyamoorthy B; Al-Hashimi HM; Hargrove AE, Amiloride as a new RNA-binding scaffold with activity against HIV-1 TAR. MedChemComm 2017, 8 (5), 1022–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ganser LR; Lee J; Rangadurai A; Merriman DK; Kelly ML; Kansal AD; Sathyamoorthy B; Al-Hashimi HM, High-performance virtual screening by targeting a high-resolution RNA dynamic ensemble. Nat. Struct. Mol. Biol 2018, 25 (5), 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ludwig V; Krebs A; Stoll M; Dietrich U; Ferner J; Schwalbe H; Scheffer U; Dürner G; Göbel MW, Tripeptides from Synthetic Amino Acids Block the Tat–TAR Association and Slow Down HIV Spread in Cell Cultures. ChemBioChem 2007, 8 (15), 1850–1856. [DOI] [PubMed] [Google Scholar]
- 47.Ratmeyer L; Zapp ML; Green MR; Vinayak R; Kumar A; Boykin DW; Wilson WD, Inhibition of HIV-1 Rev-RRE Interaction by Diphenylfuran Derivatives. Biochemistry 1996, 35 (42), 13689–13696. [DOI] [PubMed] [Google Scholar]
- 48.Zapp ML; Young DW; Kumar A; Singh R; Boykin DW; Wilson WD; Green MR, Modulation of the rev-RRE interaction by aromatic heterocyclic compounds. Biorg. Med. Chem 1997, 5 (6), 1149–1155. [DOI] [PubMed] [Google Scholar]
- 49.Carter AP; Clemons WM; Brodersen DE; Morgan-Warren RJ; Wimberly BT; Ramakrishnan V, Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 2000, 407, 340. [DOI] [PubMed] [Google Scholar]
- 50.Fernandes J; Jayaraman B; Frankel A, The HIV-1 Rev response element. RNA Biology 2012, 9 (1), 6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lipfert J; Doniach S; Das R; Herschlag D, Understanding Nucleic Acid–Ion Interactions. Annu. Rev. Biochem 2014, 83 (1), 813–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen H; Meisburger SP; Pabit SA; Sutton JL; Webb WW; Pollack L, Ionic strength-dependent persistence lengths of single-stranded RNA and DNA. Proc. Natl. Acad. Sci. U. S. A 2012, 109 (3), 799–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu W-T; Wu J-H; Li ES-Y; Selamat ES, Emission Characteristics of Fluorescent Labels with Respect to Temperature Changes and Subsequent Effects on DNA Microchip Studies. Appl. Environ. Microbiol 2005, 71 (10), 6453–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eubanks CS; Hargrove AE, Sensing the impact of environment on small molecule differentiation of RNA sequences. Chem. Commun 2017, 53 (100), 13363–13366. [DOI] [PubMed] [Google Scholar]
- 55.Doetsch M; Fürtig B; Gstrein T; Stampfl S; Schroeder R, The RNA annealing mechanism of the HIV-1 Tat peptide: conversion of the RNA into an annealing-competent conformation. Nucleic Acids Res. 2011, 39 (10), 4405–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laing LG; Gluick TC; Draper DE, Stabilization of RNA Structure by Mg Ions: Specific and Non-specific Effects. J. Mol. Biol 1994, 237 (5), 577–587. [DOI] [PubMed] [Google Scholar]
- 57.Olejniczak M; Gdaniec Z; Fischer A; Grabarkiewicz T; Bielecki Ł; Adamiak RW, The bulge region of HIV‐1 TAR RNA binds metal ions in solution. Nucleic Acids Res. 2002, 30 (19), 4241–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang J-H; Chung TDY; Oldenburg KR, A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screening 1999, 4 (2), 67–73. [DOI] [PubMed] [Google Scholar]
- 59.Disney MD; Labuda LP; Paul DJ; Poplawski SG; Pushechnikov A; Tran T; Velagapudi SP; Wu M; Childs-Disney JL, Two-Dimensional Combinatorial Screening Identifies Specific Aminoglycoside-RNA Internal Loop Partners. J. Am. Chem. Soc 2008, 130 (33), 11185–11194. [DOI] [PubMed] [Google Scholar]
- 60.Disney MD; Winkelsas AM; Velagapudi SP; Southern M; Fallahi M; Childs-Disney JL, Inforna 2.0: A platform for the sequence-based design of small molecules targeting structured RNAs. ACS Chem. Biol 2016, 11 (6), 1720–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chittapragada M; Roberts S; Ham YW, Aminoglycosides: Molecular Insights on the Recognition of RNA and Aminoglycoside Mimics. Perspect. Med. Chem 2009, 3, 21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wong C-H; Hendrix M; Scott Priestley E; Greenberg WA, Specificity of aminoglycoside antibiotics for the A-site of the decoding region of ribosomal RNA. Chem. Biol. (Oxford, U. K.) 1998, 5 (7), 397–406. [DOI] [PubMed] [Google Scholar]
- 63.Cho J; Rando RR, Specificity in the Binding of Aminoglycosides to HIV-RRE RNA. Biochemistry 1999, 38 (26), 8548–8554. [DOI] [PubMed] [Google Scholar]
- 64.Hyun Ryu D; Rando RR, Aminoglycoside binding to human and bacterial A-Site rRNA decoding region constructs. Biorg. Med. Chem 2001, 9 (10), 2601–2608. [DOI] [PubMed] [Google Scholar]
- 65.Kulik M; Goral Anna M.; Jasiński M; Dominiak Paulina M.; Trylska J, Electrostatic Interactions in Aminoglycoside-RNA Complexes. Biophys. J 2015, 108 (3), 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tao J; Frankel AD, Specific binding of arginine to TAR RNA. Proc. Natl. Acad. Sci. U. S. A 1992, 89 (7), 2723–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pitt SW; Majumdar A; Serganov A; Patel DJ; Al-Hashimi HM, Argininamide Binding Arrests Global Motions in HIV-1 TAR RNA: Comparison with Mg2+-induced Conformational Stabilization. J. Mol. Biol 2004, 338 (1), 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.