ABSTRACT
Brain organoids are self-assembled three-dimensional aggregates generated from pluripotent stem cells with cell types and cytoarchitectures that resemble the embryonic human brain. As such, they have emerged as novel model systems that can be used to investigate human brain development and disorders. Although brain organoids mimic many key features of early human brain development at molecular, cellular, structural and functional levels, some aspects of brain development, such as the formation of distinct cortical neuronal layers, gyrification, and the establishment of complex neuronal circuitry, are not fully recapitulated. Here, we summarize recent advances in the development of brain organoid methodologies and discuss their applications in disease modeling. In addition, we compare current organoid systems to the embryonic human brain, highlighting features that currently can and cannot be recapitulated, and discuss perspectives for advancing current brain organoid technologies to expand their applications.
KEY WORDS: Brain organoids, Neuroscience, Stem cell
Summary: In this Review, we discuss recent advances in the production of brain organoids, highlighting their potential applications as model systems for understanding disease states as well as normal brain development across species.
Introduction
The rapidly advancing field of stem cell biology continually provides new insights into basic biology and human disorders, and drives innovation for new treatments. Human pluripotent stem cells (hPSCs), including human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs), have emerged as invaluable tools for modeling human disorders, especially those with complex genetic origins that are challenging to model in animals (Takahashi et al., 2007; Takahashi and Yamanaka, 2006; Thomson et al., 1998). Under specific cues and favorable conditions, hPSCs have the potential to differentiate into any cell or tissue type found in the human body. They can also be used to generate organoids – organ-like three-dimensional (3D) tissue cultures containing multiple cell types that represent accessible systems for modeling organogenesis and developmental disorders.
Brain organoids are hPSC-derived organoids that self-assemble to form an organized architecture, composed of progenitor, neuronal and glial cell types, resembling the fetal human brain (Jo et al., 2016; Kadoshima et al., 2013; Lancaster et al., 2013; Mariani et al., 2015; Pasca et al., 2015; Qian et al., 2016). Unlike conventional two-dimensional (2D) cell cultures, brain organoids recapitulate the human brain not only at the cellular level, but also in terms of general tissue structure and developmental trajectory, therefore providing a unique opportunity to model human brain development and function, which are often inaccessible to direct experimentation. Combined with recent technological advances, such as patient-derived hiPSCs, genetic engineering, genomic editing, and high-throughput single cell transcriptomics and epigenetics, brain organoids have revolutionized our toolboxes to investigate development, diseases and evolution of the human brain.
In this Review, we first introduce recent advances in brain organoid technologies, and their applications as model systems and discovery tools. We then discuss the hurdles that current methodologies have yet to overcome, and offer feasible suggestions for future steps and objectives that would enable organoid technologies to progress further. Despite the tremendous promise of brain organoids, it is important to understand the limitations of organoid models. Therefore, this Review also aims to provide a balanced view between the advantages and disadvantages of brain organoids. Only when interpreted complementarily with other models can the results obtained using brain organoids shed light on our understanding of human biology.
Current brain organoid methodologies
Methodologies to induce neural differentiation from PSCs in 3D have been pursued since the early 1990s, but not all 3D neural culture systems should be considered brain organoids. Neurospheres, for instance, are 3D aggregations of several central nervous system (CNS) cell types derived from neural progenitor cells (NPCs) (Reynolds et al., 1992). Neurospheres contain various subtypes of neurons and glial cells, but distinctive cytoarchitectures are typically absent (Pamies et al., 2017). Neural cells can also be artificially assembled, using designed culture chambers and biomaterial scaffolds, into ‘organ-on-a-chip’ structures, which are advantageous for modeling cell-cell interactions in a highly controlled manner (Phan et al., 2017). Nevertheless, as far as this Review is concerned, brain organoids are distinct from neurosphere and microchip approaches most notably because they harness the self-organization capability of PSCs to mimic the cytoarchitecture and developmental trajectories found in vivo.
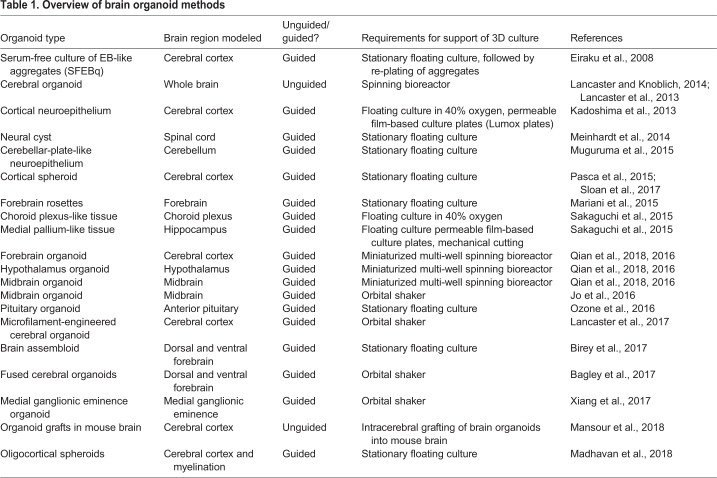
In general, two different types of methodologies can be used to generate brain organoids: unguided methods and guided methods (Fig. 1). Whereas unguided methods rely fully on spontaneous morphogenesis and intrinsic differentiation capacities within hPSC aggregates, guided organoid methods require supplementation of external patterning factors to induce hPSCs to differentiate towards desired lineages. The number and combination of external factors used in differentiation protocols varies, and the choice between unguided and guided approaches is often seen as a trade-off between diversity and consistency.
Fig. 1.
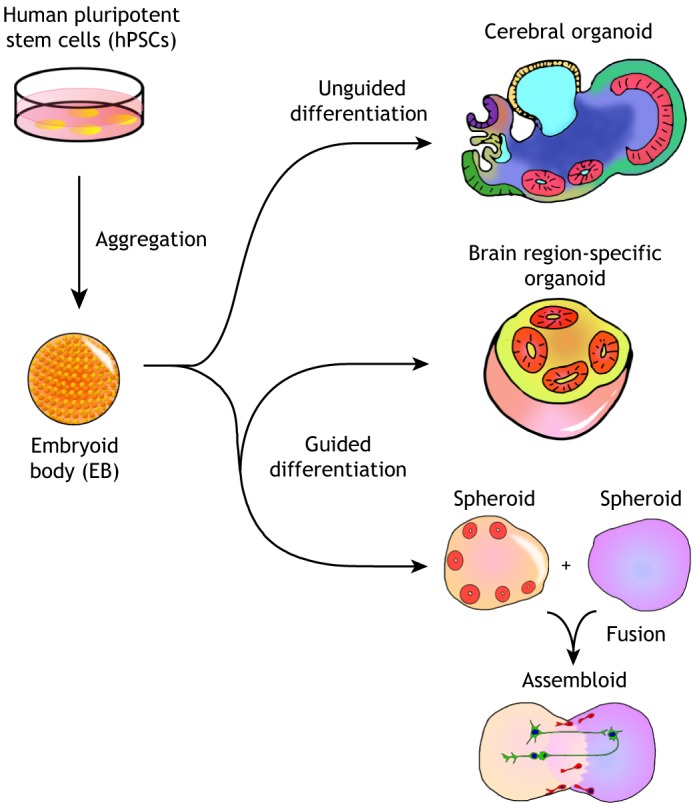
Unguided and guided approaches for making brain organoids. Unguided approaches (top) harness the intrinsic signaling and self-organization capacities of hPSCs to differentiate spontaneously into tissues mimicking the developing brain. The resulting cerebral organoids often contain heterogeneous tissues resembling various brain regions. By contrast, guided approaches (bottom) use small molecules and growth factors to generate spheroids that are specifically representative of one tissue type. Brain region-specific organoid methods involve the use of patterning factors at an early stage to specify progenitor fate; these factors are then removed in subsequent differentiation stages. Guided approaches can also be used to generate two or more spheroids/organoids representative of different brain region identities that can then be fused to form ‘assembloids’, which model interactions between different brain regions.
Unguided brain organoid methodologies
The unguided brain organoid, or the cerebral organoid, is inspired by methodologies developed for generating gastrointestinal organoids (Table 1) (Lancaster et al., 2013; Sato et al., 2011, 2009). In the protocol developed by the Knoblich group, embryoid bodies (EBs) derived from hPSC aggregates are embedded into an extracellular matrix (ECM), such as Matrigel, and subsequently cultured in spinning bioreactors to promote tissue expansion and neural differentiation (Lancaster and Knoblich, 2014). With minimal external interference, cerebral organoids produced by this approach give hPSCs the most freedom for self-organization, and sometimes give rise to very elongated neuroepithelial structures. They exhibit a variety of cell lineage identities ranging from forebrain, midbrain and hindbrain, to retina, choroid plexus and mesoderm (Camp et al., 2015; Lancaster et al., 2013). Large-scale single cell transcriptomic profiling has further revealed that cerebral organoids contain neural progenitors, excitatory neurons, inhibitory neurons, astrocytes and oligodendrocyte precursor cells (OPCs) found in the CNS, as well as photosensitive cells found in the retina (Quadrato et al., 2017). The stochastic nature of hPSC spontaneous differentiation, however, results in unpredictable proportions and a heterogeneous arrangement of each lineage and cell type across batches of differentiated organoids and across hPSC lines. Although this cell-type diversity in cerebral organoids offers a unique opportunity to model the interactions between different brain regions, the high variability and heterogeneity present significant challenges for systematic and quantitative studies.
Table 1.
Overview of brain organoid methods
Guided brain organoid methodologies
Guided methods for generating brain organoids were pioneered by the Sasai group, who developed a series of 3D differentiation protocols based on the serum-free culture of EB-like aggregates (Table 1) (Danjo et al., 2011; Eiraku et al., 2008; Kadoshima et al., 2013; Muguruma et al., 2015; Sakaguchi et al., 2015; Sasai, 2013). In these guided organoid, or ‘spheroid’ methods, small molecules and growth factors are used throughout the differentiation process to instruct hPSCs to form cells and tissues representative of certain brain regions, such as the cerebral cortex, hippocampus and midbrain (Jo et al., 2016; Mariani et al., 2015; Pasca et al., 2015; Sakaguchi et al., 2015; Yoon et al., 2019). These directed organoid cultures are sometimes capable of generating mixtures of cell types with relatively consistent proportions, exhibiting less variation across batches and cell lines (Sloan et al., 2017). However, directed organoids typically contain relatively small neuroepithelial structures and their cytoarchitecture is sometimes not well-defined, possibly owing to the interference of hPSC self-organization and cell-cell interactions by excessive use of external factors.
Guided organoid differentiation protocols can be carefully tailored to require the use of external patterning factors only at the early differentiation stage, thereby allowing hPSCs to be specified as progenitor cells exhibiting certain brain region identities with minimal heterogeneity. For these brain region-specific organoids, external factors are then removed or minimized after successful patterning during the initial stage of differentiation, and subsequent differentiation follows intrinsic programs similar to those operating in vivo after neural patterning. This approach has been used successfully to generate large ventricle-like structures with elaborate laminar organization and architecture (Lancaster et al., 2017; Qian et al., 2016). In addition to using chemical factors, synthetic biomaterials can be engineered to guide the formation of brain organoids physically. This is exemplified by the microfilament-engineered cerebral organoids method, in which elongated EBs form around scaffolds made from polymer microfilaments, resulting in more-consistent formation of enlarged ventricular structures and neuro-epithelium (Lancaster et al., 2017).
The use of spinning bioreactors can also provide enhanced nutrient and oxygen diffusion and sustained 3D suspension culture (Lancaster et al., 2013). However, commercial bioreactors are bulky and consume large volumes of culture medium, limiting the efficiency and throughput of organoid culture. To reduce the consumption of culture medium, multiple-well culture plates have been used together with orbital shakers placed in the incubator as an alternative to a spinning bioreactor (Lancaster and Knoblich, 2014). More recently, custom-designed miniaturized multi-well spinning bioreactors have been developed to both reduce the cost of maintaining organoid cultures and remove the need for bulky machines in the incubator; this approach allowed for protocol optimization with improved efficiency, and enabled the generation of brain region-specific organoids mimicking the dorsal forebrain, midbrain and hypothalamus (Qian et al., 2016). Notably, forebrain organoids generated via this approach consistently form cortical structures with distinct layers that resemble the ventricular zone (VZ), the inner and outer subventricular zone (iSVZ and oSVZ) and the cortical plate (CP) at molecular, cellular and structural levels. The recapitulation of primate/human-specific developmental features, such as an enlarged oSVZ, in forebrain organoids offers unique advantages for understanding human cortical development and developmental disorders.
Fused organoid technologies
Although cerebral organoid methods can produce tissues resembling various interacting brain regions, their proportion and spatial organization are highly heterogeneous and unpredictable. To improve modeling of inter-regional interactions, several groups concurrently developed new approaches, first differentiating hPSCs into different brain region-specific organoids separately, and then fusing them together to form organoids with multiple distinct region identities in a controlled manner (Bagley et al., 2017; Birey et al., 2017; Xiang et al., 2017). For example, fused dorsal and ventral forebrain organoids have been shown to form an ‘assembloid’ (Fig. 1) with two distinctive but interfacing domains (Birey et al., 2017). In these structures, interneurons generated from the ventral domain preferentially migrate towards the dorsal domain, resembling the tangential migration of interneurons from the subpallium to the cerebral cortex in vivo (Anderson et al., 1997). Moreover, electrophysiological characterization of forebrain assembloids revealed that migrating interneurons connect synaptically with local excitatory neurons to form microcircuits (Birey et al., 2017).
Overall, these recent advances in brain organoid methodologies have expanded our toolbox for modeling human development and disorders. The choice between guided and unguided methodologies will depend on the specific focus of investigation. For instance, unguided organoids are suitable for exploring cell-type diversity during whole-brain development, brain region-specific organoids better recapitulate brain cytoarchitecture with less heterogeneity, and assembloids allow for the investigation of interactions between specific brain regions with more consistent molecular and functional characterization.
Modeling cerebral cortex development using organoids: in vitro versus in vivo
Organoids mimicking the cerebral cortex have so far been better characterized and more frequently used than other brain organoids. Such cortical organoids have also attracted the most interest in the field because the cerebral cortex is the most unique and evolutionarily expanded region of the human brain, in comparison to that of other animals, and is often severely affected in many neurological disorders (Bystron et al., 2008; Hansen et al., 2010; Lui et al., 2011; Rakic, 2009). The presence of profound species differences in the cortex justifies the use of human cell-derived cortical organoids over animal models. However, like all in vitro models, these organoids are not identical replicas of their in vivo counterparts. Below, we use cortical organoids as an example to discuss the aspects of brain development that have been successfully reproduced in cortical organoids, and those aspects that are still lacking (Fig. 2).
Fig. 2.
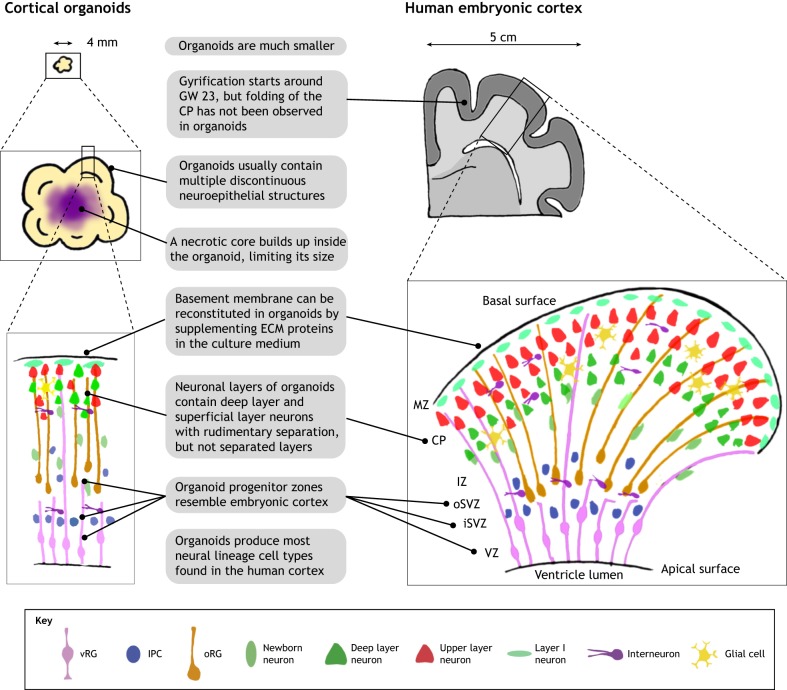
Structural comparison between cortical organoids and the human embryonic cortex. Cortical organoids resemble the cytoarchitecture of human developing cerebral cortex in early and mid-gestation with remarkable fidelity, despite their small size. A cortical organoid usually contains multiple short and independent neuroepithelial structures. Within each structure, well-defined layers resembling the VZ, iSVZ, oSVZ, CP and MZ can be observed. Major neural lineage cell types in the embryonic cortex can also be detected in cortical organoids, but vascular and immune cells are absent. Late-gestational features of corticogenesis, such as the formation of cortical folding and the six separated cortical layers are not observed in cortical organoids generated by currently available approaches. CP, cortical plate; IPC, intermediate progenitor cells; iSVZ, inner subventricular zone; IZ, intermediate zone; MZ, marginal zone; oRG, outer radial glia; oSVZ, outer subventricular zone; vRG, ventricular radial glia; VZ, ventricular zone.
Size and general architecture
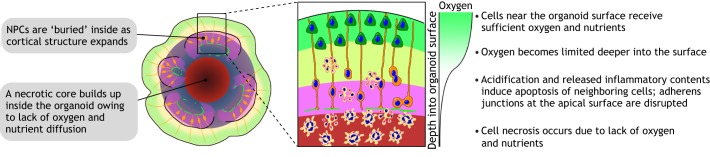
First of all, cortical organoids are much smaller in size compared with human cerebral cortex (Fig. 2). Whereas cortical organoids can at most expand to ∼4 mm in diameter, the human neocortex is about 15 cm in diameter, with the thickness of gray matter alone being 2-4 mm (Lancaster et al., 2017; Qian et al., 2016; Rakic, 2009). Moreover, owing to a lack of vascularization and circulation, a necrotic core inevitably builds up in the organoid's interior and the actual viable thickness is further limited (Fig. 3). The dramatically different sizes and numbers of cells in cortical organoids suggest that it is almost impossible for current organoid technologies to represent fully the complexity of a bona fide cortex.
Fig. 3.
The diffusion limit depletes progenitors and prohibits organoid expansion. Brain organoids grown as a sphere in 3D suspension cultures can expand to 3-4 mm in diameter, but only those cells within a limited distance from the surface can receive sufficient oxygen and nutrients via diffusion. The build-up of a necrotic core is therefore common in brain organoids. As the outer layers expand, the more metabolically demanding progenitors in the interior will eventually deplete, resulting in a loss of proliferation and structural disorganization over long-term cultures.
Surprisingly, however, cortical organoids do recapitulate the organization of neural progenitor zones with considerable accuracy, especially when analyzed in vertical columns from the apical (inner) surface to the basal (outer) surface (Fig. 2). Indeed, cortical organoids contain a large VZ formed by SOX2+ ventricular radial glia cells (vRGs) organized into polarized radial structures with a lumen and apical surfaces expressing adherens junction proteins (Kadoshima et al., 2013; Lancaster et al., 2017; Qian et al., 2016). Immediately adjacent to the VZ, a thin and dense layer of TBR2 (EOMES)+ intermediate progenitor cells (IPCs) can be found, resembling the iSVZ (Lui et al., 2011). A well-defined oSVZ, populated by IPCs, immature neurons and unipolar NPCs expressing human outer radial glia (oRG)-specific markers, such as HOPX and PTPRZ1, can also be observed in cortical organoids (Bershteyn et al., 2017; Lancaster et al., 2017; Li et al., 2017a,b; Qian et al., 2016; Watanabe et al., 2017). Notably, in forebrain organoids developed by Qian et al., the oSVZ expands disproportionally as organoids age, and by 80 days of differentiation, the oSVZ becomes several times larger than the VZ, similar to human corticogenesis where the oSVZ is the dominant source of neurogenesis in the second and third trimester (Hansen et al., 2010; Qian et al., 2016). Moreover, the basal processes of RGs in cortical organoids are radially aligned, forming migrational tracks for newborn neurons to reach the CP, as occurs in vivo (Qian et al., 2018). Live imaging of fluorescently labeled vRGs and oRGs in cortical organoids has revealed their mitotic behavior and dynamics, again suggesting that they resemble RGs in vivo (Bershteyn et al., 2017; Subramanian et al., 2017).
The cortical organoids developed by Kadoshima et al. demonstrated a cell-sparse region filled by neuronal processes between the SVZ and the CP, resembling the intermediate zone (Kadoshima et al., 2013). In early stages of cortical organoid development, the neuronal layer made from TBR1+ and CTIP2 (BCL11B)+ early-born neurons and reelin+ Cajal–Retzius cells is highly reminiscent of the preplate (PP) seen in first trimester corticogenesis (Lancaster et al., 2013; Qian et al., 2016). As this neuronal layer expands in size and becomes more condensed, it exhibits a compact morphology and starts to resemble the CP of the fetal cortex, with the help of ECM proteins supplemented in culture medium to reconstitute the basement membrane on the organoid surface (Kadoshima et al., 2013; Lancaster et al., 2017; Qian et al., 2018). Moreover, the temporal order of neurogenesis in cortical organoids follows the ‘inside-out’ rules that have been demonstrated in vivo: neurons expressing deep layer markers are born early, whereas upper layer neurons, expressing markers such as SATB2 and CUX1, are found later. However, extensive mixing and colocalization of upper and deep layer neurons are often observed, indicating that the specification of cortical lamination is incomplete in these organoids (Lancaster et al., 2017; Qian et al., 2016). Interestingly, such co-expression and intermingling of upper and deep layer fate are similarly observed in the CP of mid-gestation human embryonic cortex (Ip et al., 2011; Nowakowski et al., 2017; Ozair et al., 2018; Zhong et al., 2018). However, at the perinatal period, the separation of upper and deep cortical layers becomes evident, suggesting that the establishment of distinct cortical layers takes place near the end of the second trimester (Saito et al., 2011; Zeng et al., 2012). Furthermore, unlike in the perinatal neocortex, the cortical layers in organoids cannot be discriminated based on their neuron distribution and density, partially owing to them exhibiting drastically fewer layers of CP neurons (Molyneaux et al., 2007). Organoid neurons also lack layer-specific dendritic morphology and axonal projection patterns (Zhong et al., 2018). It is also difficult to divide the neuronal layer of cortical organoids unambiguously into distinctive subplate (SP) and CP regions, likely owing to the fact that the SP-like region is interspersed between the CP and the SVZ (Ozair et al., 2018).
Neuronal function and activity
Excitatory and inhibitory neurons in cortical organoids are functional, but unsurprisingly less mature, compared to adult neurons (Birey et al., 2017; Pasca et al., 2015; Qian et al., 2016). Because cortical organoids are a model for the early-mid stages of embryonic development, during which time newborn neurons are continuously produced, electrophysiological characterization often reveals highly variable functionality and maturation parameters when cells are randomly chosen. The neurons in more mature organoids, for example, fire trains of action potentials upon current injection and show robust spontaneous excitatory/inhibitory postsynaptic currents (Pasca et al., 2015; Qian et al., 2016), and extracellular recording using a multi-electrode probe has revealed spontaneous burst-like firing activities in these neurons (Quadrato et al., 2017). Sparse labeling sometimes reveals neurons with pyramidal-like morphology and dendritic spines (Lancaster et al., 2017; Qian et al., 2016). Synaptogenesis also appears to be abundant when examined using immunostaining or electron microscopy (Birey et al., 2017; Pasca et al., 2015; Quadrato et al., 2017; Sloan et al., 2017). However, neuron subtype-specific firing patterns and plasticity are yet to be observed. Nonetheless, despite their large variability, the basic electrophysiological properties of neurons, such as resting membrane potential, resistance, capacitance, amplitude and frequency of spontaneous post-synaptic currents, show statistically significant trends of maturation over time as organoids age (Qian et al., 2016). This is in line with a recent study, using patch-clamping on organotypic slices of human fetal cortex, which reported that most neurons at the end of the second trimester are immature, and only early-born SP and deep layer neurons are able to fire action potentials and exhibit spontaneous synaptic transmission (Zhong et al., 2018). Thus, neurons generated in organoids might reflect the development and maturation of neuronal functionalities during embryonic stages in vivo.
Cell-type diversity
In addition to generating various types of RGs and neurons, organoids recapitulate the cellular diversity of the cerebral cortex by producing interneurons, astrocytes and OPCs (Birey et al., 2017; Qian et al., 2016; Quadrato et al., 2017; Sloan et al., 2017). Similar to rodents, the majority of human cortical interneurons originate from the ganglionic eminence of the ventral forebrain and tangentially migrate to the cerebral cortex (Hansen et al., 2013; Ma et al., 2013), but several studies have provided evidence for dorsal origins of some cortical interneurons, which is an observation unique to primates (Jakovcevski et al., 2011; Letinic et al., 2002; Petanjek et al., 2009; Radonjic et al., 2014; Yu and Zecevic, 2011). Although NKX2.1-expressing ventral progenitors are absent in dorsally patterned forebrain organoids at early stages, a small population of interneurons is detected later, suggesting that dorsal forebrain progenitors are also potentially capable of producing interneurons in vitro (Qian et al., 2016). In addition, a recent study described a protocol to specifically enrich OPCs and oligodendrocytes to generate oligo-cortical spheroids with myelination (Madhavan et al., 2018). Together, these findings highlight that cortical organoids appear to generate all cell types from the neuroectoderm lineage. By contrast, cortical organoids lack cells found in the brain with non-neural origins, including vascular cells and immune cells. Endothelial cells (ECs) can be cultured separately and added to organoids during the EB formation stage, and can self-organize to form network-like structures (Nzou et al., 2018; Pham et al., 2018). Such primitive endothelial networks, however, are incapable of functionally facilitating nutrient delivery into organoids and alleviating the build-up of a necrotic core, although they do instead provide a potential platform for modeling the neurovascular niche and blood brain barrier (Nzou et al., 2018; Pham et al., 2018). Human iPSC-derived microglia can also be co-cultured with brain organoids; they adhere to the organoid surface and migrate to the interior, suggesting chemotaxis of microglia towards CNS cues (Abud et al., 2017). Remarkably, when brain organoids are mechanically injured, the microglia cluster near the injury site and adopt a round morphology, in contrast to their normal ramified morphology, resembling the activated microglia morphology observed upon CNS injury in vivo (Abud et al., 2017). It remains a challenge, however, to develop protocols to generate non-neural cell types within brain organoids intrinsically without introducing further heterogeneity, and artificially combining separately cultured cell types is thus likely to be the most promising approach.
Gene expression
Transcriptomic profiling of entire organoids through bulk or single cell RNA sequencing (scRNA-seq) has revealed encouraging similarities, at the gene expression level, between cortical organoids and human fetal brain tissues (Camp et al., 2015; Jeong and Tiwari, 2018; Quadrato et al., 2017; Sloan et al., 2017). Similarly, brain organoids appear to share some epigenetic signatures with in vivo cortical development (Luo et al., 2016). These studies have revealed that the developmental trajectory of the organoid transcriptome and epigenome dynamically parallels the trajectory of human cortical development: for instance, after 100 days in culture, the expression signature of forebrain organoids resembles that of the gestational week (GW) 22 prefrontal cortex (Qian et al., 2016). However, growing forebrain organoids beyond 100 days does not necessarily mimic the third trimester or neonatal stages, because expansion of the organoid is stalled owing to depletion of proliferative NPCs (caused by the aforementioned diffusion limitation and necrosis observed in organoids). We should be cautious, however, when making conclusions based solely on gene expression analysis, as bulk gene expression patterns alone are not enough to specify function.
Mimicking arealization
Our discussion so far has regarded the cerebral cortex as one entity, but the cerebral cortex is specified, in a process termed arealization, into various anatomically and functionally distinct areas along the anterior-posterior (A-P) and medial-lateral (M-L) axes (Chambers and Fishell, 2006; O'Leary et al., 2007; Sur and Rubenstein, 2005). Studies in rodents have revealed that morphogens and signaling molecules, such as WNT and BMP, expressed in early patterning centers are responsible for establishing areal identities, but the spatiotemporal mechanisms regulating arealization of the human neocortex remain unclear (Caronia-Brown et al., 2014; De Clercq et al., 2018; Ozair et al., 2018; Zembrzycki et al., 2015). Another challenge in modeling cortical arealization in organoids is the lack of distinguishable features of cortical areas during early- and mid-gestational stages. Indeed, cortical progenitors fated to give rise to different areas express very similar markers, and the morphological differences between cortical areas are not distinct until postnatal stages (Elsen et al., 2013; Rakic et al., 2009). Recent studies on human fetal cortical tissues have identified differences in the expression and segregation patterns of upper and deep layer neuron markers in the second trimester along the A-P axis, and demonstrated the involvement of WNT signals in controlling areal identities post-mitotically (Nowakowski et al., 2017; Ozair et al., 2018). Based on this knowledge, cortical organoids resembling a specific cortical area or areas along the A-P axis could theoretically be engineered by tailoring differentiation protocols further.
Cortical folding
Finally, cortical folding remains an unachieved ‘holy grail’ for cortical organoids. The dramatic increase in cortical neurogenesis from the oSVZ has been considered the source of cortical surface area expansion and gyrification in gyrencephalic mammals, but oSVZ-containing cortical organoids have not shown folding of the pial surface and CP layers underneath (Borrell, 2018; Lewitus et al., 2013; Zilles et al., 2013). This may be because current cortical organoids do not reach the developmental stage at which gyrification occurs; indeed, it is known that the demarcation of primary gyri does not begin until around GW 23 in humans, with secondary gyri and sulci emerging later in the third trimester (Lewitus et al., 2013). Attempts have been made to induce neuroepithelial ‘crinkling’ or ‘pseudo-folding’ in early organoid differentiation, either by induction of NPC overgrowth at the cost of neurogenesis via knockout of PTEN, or by mechanical spatial confinement in microchips (Chenn and Walsh, 2002; Karzbrun et al., 2018; Li et al., 2017b). However, this induced neuroepithelial crinkling does not lead to the formation of gyrus- and sulcus-like structures in the CP (Chenn and Walsh, 2002). A variety of factors, including uneven neurogenesis from the neuroepithelium, cell-cell adhesion, and transformation of migration scaffolds and mechanical forces, have been found to influence the complex but highly controlled process of gyrification during late human corticogenesis (Bae et al., 2014; Bayly et al., 2014; Del Toro et al., 2017; Florio et al., 2015; Nowakowski et al., 2016; Stahl et al., 2013; Zilles et al., 2013). A better understanding of these mechanisms may shed light on how we can engineer cortical organoids with early features of gyrification. However, it is unlikely that current organoid structures can fully recapitulate the folding of the human neocortex, as statistical analyses have revealed that the degree of folding across mammalian species scales with the surface area and the thickness of the CP, and organoids – at least those generated using currently available approaches – may just be too small to achieve this (Lewitus et al., 2016; Mota and Herculano-Houzel, 2015).
Applications of brain organoids
Disease modeling
Brain organoids derived from hPSCs, especially patient-derived iPSCs, have been extensively explored for the potential to model neurodevelopmental brain disorders (Fig. 4) (Chen et al., 2018). They have been particularly successful for recapitulating disease-related phenotypes of conditions in which structural malformations are apparent at early embryonic stages. The mechanisms of such disorders are frequently attributed to disrupted progenitor cell regulation, including premature differentiation, reduced proliferation, and cell cycle disruption, all of which can be analyzed reliably in brain organoids.
Fig. 4.
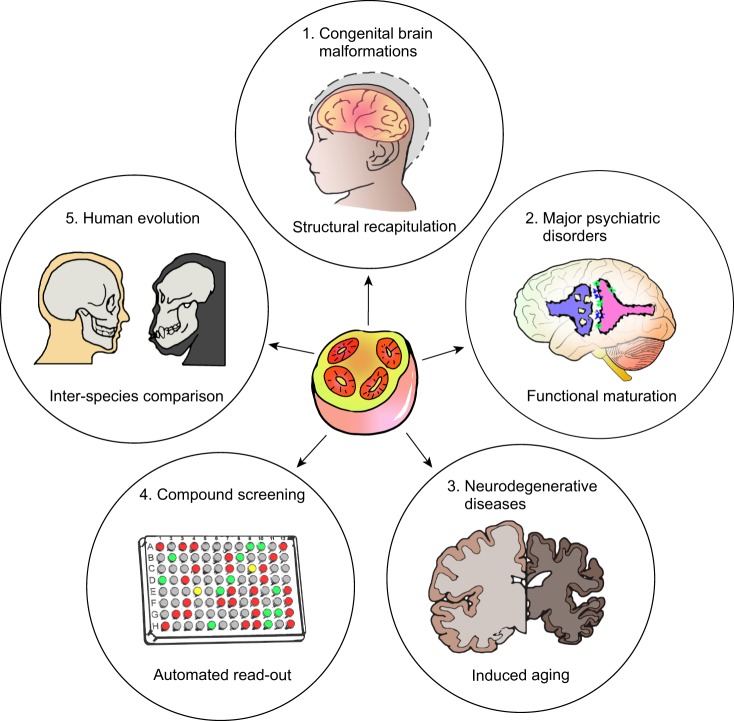
Applications of brain organoids. (1) Brain organoids have proven to be particularly informative for modeling congenital brain malformations caused by genetic deficits or infectious disease, because the organoid cytoarchitecture provide a direct read-out for disease-relevant phenotypes. (2) Primitive microcircuits are detected in brain organoids (Birey et al., 2017), but further promoting functional maturation will be key for modeling psychiatric disorders such as autism and schizophrenia. (3) It remains challenging to model age-dependent neurodegenerative diseases with current brain organoids, because they mimic mostly embryonic brain development. A method of artificially inducing aging in vitro could potentially allow organoids to represent disease-relevant phenotypes. (4) The ability to generate brain organoids in large quantities with high consistency raises the possibility of using organoids for compound screening and subsequent validation. The development of automated high-throughput platforms could expedite such advances. (5) Organoids offer the unique opportunity to understand the basis of human brain formation and evolution in comparison to other species. For instance, brain samples from great apes are largely inaccessible, but organoids generated from great ape iPSCs can be compared with human cell-derived organoids to discover uniquely human features.
Organoids generated from iPSCs derived from microcephaly patients with mutations in mitosis-associated genes show dysregulation of the cell division plane, resulting in early depletion of NPCs and smaller organoids (Gabriel et al., 2016; Lancaster et al., 2013; Li et al., 2017a). Deletion of the tumor-suppressor gene PTEN in hPSCs results in over-proliferation and delayed neurogenesis of NPCs in organoids, resulting in abnormally large organoids reminiscent of macrocephalic brains (Li et al., 2017b). In addition to genetic conditions, neurotrophic pathogens affecting brain development can be modeled using brain organoids. For example, exposure of brain organoids to Zika virus results in preferential infection of NPCs, which suppresses proliferation and causes an increase in cell death, ultimately leading to drastically reduced organoid size (Cugola et al., 2016; Dang et al., 2016; Garcez et al., 2016; Qian et al., 2017, 2016). Moreover, such infected cortical organoids display a series of characteristics identified in congenital Zika syndrome, including thinning of the neuronal layer, disruption of apical surface adherens junctions and dilation of the ventricular lumen, offering direct evidence for the causal relationship between embryonic Zika exposure and neurological disorders (Ming et al., 2016; Nguyen et al., 2016; Qian et al., 2017, 2016; Rasmussen et al., 2016; Wen et al., 2017; Zhang et al., 2016). Subsequently, brain organoids have been used as platforms for validating results of compound screening in search of anti-viral drug candidates (Watanabe et al., 2017; Xu et al., 2016; Zhou et al., 2017).
When combined with genetic manipulations using viral vectors or electroporation, organoids also provide an accessible model with which to specifically probe the function of one protein or pathway, and to investigate the underlying molecular mechanisms of diseases (Ye et al., 2017; Yoon et al., 2017). However, because current organoids cannot form cortical folds, studies to model diseases such as Miller–Dieker syndrome, a severe form of lissencephaly, are more difficult. Nonetheless, although recent attempts to model this disease using patient-derived organoids do not reveal lissencephalic features, they do instead show abnormalities in radial glia division dynamics that may be linked to disrupted gyrification (Bershteyn et al., 2017; Iefremova et al., 2017).
Comprehensive modeling of neurodevelopmental disorders that do not involve dramatic structural malformations is more challenging. Nevertheless, studies using brain organoids have been able to provide glimpses into the cellular and molecular mechanisms involved in disease development. For example, forebrain organoids derived from cells of individuals with autism spectrum disorder (ASD) show an imbalance of excitatory neuron and inhibitory neuron proportions, which is associated with overexpression of the forebrain transcription factor FOXG1 (Choi et al., 2017; Mariani et al., 2015). Defects in interneuron migration are observed in dorsal-ventral forebrain ‘assembloids’ generated from iPSCs derived from individuals with Timothy syndrome that harbor a mutation in the L-type calcium channel gene (Birey et al., 2017). Moreover, hypothalamic organoids containing nuclei-like clusters of neuropeptidergic neuron subtypes are promising models for studying metabolic disorders and obesity (Qian et al., 2016; Rajamani et al., 2018).
Brain organoids have also attracted great interest as neurodegenerative disease models, but attempts so far have had limited success (Arber et al., 2017; Gonzalez et al., 2018). Because most neurodegenerative diseases are late onset and age related, brain organoids mimicking embryonic brain development may not robustly reproduce disease-relevant endophenotypes. Although not strictly classified as organoids, human neural cultures in ECM gel or neurospheres derived from individuals with Alzheimer's disease (AD) have been shown to recapitulate AD-like pathologies, including amyloid aggregation, hyperphosphorylated tau protein, and endosome abnormalities (Choi et al., 2014; Jorfi et al., 2018; Raja et al., 2016). Midbrain organoids containing tyrosine hydroxylase (TH)-positive dopaminergic neurons, when combined with pharmacological treatment to induce neurodegeneration, may potentially serve as a relevant model for Parkinson's disease and a cellular source for cell-replacement therapies (Monzel et al., 2017; Qian et al., 2016; Xiao et al., 2016).
In summary, although brain organoids clearly provide a unique platform for understanding complicated neurological diseases, it is important to keep in mind that the current organoids are very simplistic and immature, and the knowledge obtained from them may be biased owing to their in vitro nature. Further development of technologies, such as accelerating functional maturation and incorporating other cell and tissue types, will push the field towards more comprehensive and faithful models.
Understanding human evolution
The human brain, especially the neocortex, has evolved to be disproportionally larger compared with that of other species (Rakic, 2009; Sousa et al., 2017). A better understanding of this species-dependent difference will help us to understand better the mechanisms that make humans unique, and may aid the translation of findings made in animal models into therapeutic strategies (Fig. 3). Brain organoids open up new possibilities for comparative studies of the brain across species. Moreover, brain samples from human's close relatives, great apes, are almost impossible to obtain, and iPSC-derived organoids may be the only feasible method to gain access to great ape brain development. Indeed, a comparison between human, chimpanzee and macaque iPSC-derived organoids has revealed differences in the proliferative dynamics of NPCs that lead to different neurogenesis outputs, which may explain the neocortical size differences among primates (Otani et al., 2016). Consistent with this finding, scRNA-seq studies detected upregulation of genes involved in proliferation of radial glia in human organoids compared with chimpanzee organoids (Kronenberg et al., 2018; Mora-Bermudez et al., 2016). One limitation of this approach is that the differentiation protocol followed for both human and non-human primate organoids has been developed and optimized exclusively for hPSCs, and the observed differences in non-human organoids might simply be a result of suboptimal differentiation conditions, rather than species-dependent distinctions. Moreover, as human pregnancy is longer than that in many species, and neurodevelopment continues until adulthood, the difference in the temporal control of brain development is difficult to gauge when performing inter-species comparison. Establishing hallmarks for developmental stages that are shared across primate species could provide a more solid basis for such comparisons.
With the help of genome-editing technologies, it is now possible to investigate the function of human-specific genes or genetic variants in organoid models (Ran et al., 2013). Two recent studies identified high expression of human-specific paralogs of the NOTCH2 receptor, NOTCH2NL genes in human radial glia, suggesting important functions for these genes in neurogenesis in humans (Fiddes et al., 2018; Suzuki et al., 2018). Indeed, deletion of NOTCH2NL results in premature neurogenesis and microcephaly-like deficits in hESC-derived organoids; on the other hand, ectopic expression of NOTCH2NL in mouse ESCs gives rise to organoids with delayed neural differentiation (Fiddes et al., 2018; Suzuki et al., 2018). Future studies could further exploit genome engineering to knock-in human genes into iPSCs of other species and generate ‘humanized’ organoids, in order to understand the evolutionary significance of human-specific genetic variants.
Conclusions and perspectives
Tremendous breakthroughs have been made in brain organoid technologies in the past few years. Although these brain organoids faithfully recapitulate a number of key features of the human brain, they are not perfect replica, and overcoming current limitations to engineer ‘better’ organoids will greatly expand our ability to investigate human brain development and disorders. The definition of ‘better’ organoids may vary based on specific applications, but the benchmark is to make organoids that more faithfully recapitulate features of the human brain. Therefore, continued systematic characterization of human fetal and postnatal brain tissue is fundamental. Despite the difficulty in collecting human brain samples with consistent quality, analyses of these samples have continued to expand our knowledge of normal and diseased human brains (Calvet et al., 2016; Hansen et al., 2010; Moore et al., 2017; Nowakowski et al., 2016; Ozair et al., 2018; Pollen et al., 2015; Stoner et al., 2014). Remarkably, large-scale single cell transcriptome profiling has resulted in unprecedented resolution, revealing the extent of cellular diversity and molecular identities of the embryonic human brain (Nowakowski et al., 2017; Zhong et al., 2018). Pioneered by The Allen Institute for Brain Science, the eventual establishment of a comprehensive human brain atlas containing immunohistology, in situ hybridization and transcriptomics data will provide an invaluable resource for organoid engineers (Ding et al., 2017).
The small size of current organoids remains the fundamental limiting factor that prohibits us from using organoids to fully recapitulate late stages of human brain development. The viable thickness of organoids is restricted by the physical distance over which oxygen and nutrients can diffuse from the surface, which is typically less than 400 µm (Rambani et al., 2009). Because NPCs with high metabolic demands are often located in the most interior part of the cortical structures, they are the first to succumb to the diffusion limit, and neurogenesis cannot be sustained in long-term organoid cultures (Fig. 3). The formation of a CP with six distinct layers and cortical folding is therefore still out of reach. Methods to create a more permissive environment to alleviate this condition include the use of spinning bioreactors or orbital shakers to enhance diffusion, and to provide a higher oxygen concentration in the incubator (Kadoshima et al., 2013; Lancaster et al., 2017; Qian et al., 2016). Current organoid preparations also lack vascular cells and, although endothelial cells can be exogenously incorporated into brain organoids, this alone is insufficient to improve long-term culture as the primitive endothelial network that forms is not functional (Pham et al., 2018). Future work to incorporate biomaterials and microfluidic systems could therefore be used to engineer vascular-like networks with perfusion to supply the organoid interior with adequate oxygen and nutrients. The transplantation of organoids into animals, allowing the host vasculature to grow into the organoid graft, has also proven to be a promising approach to promote organoid viability and maturation (Mansour et al., 2018).
The fact that brain organoids dynamically mimic the temporal progression of human brain development is both an advantage and a disadvantage for researchers. On the one hand, brain organoids of different ages recapitulate their corresponding in vivo counterparts, offering researchers a versatile platform to probe different developmental stages. On the other hand, from a practical point of view, brain organoids take a long time to grow and mature, raising the cost and hindering the efficiency of experiments. Methods for speeding up the maturation process thus need to be explored. For instance, the use of NOTCH inhibitors is very effective in accelerating neuronal differentiation in vitro and could be applied to brain organoids. But this poses a dilemma because such a manipulation could interfere with the intrinsic program of neural differentiation, making the resulting organoids no longer representative of their in vivo counterparts (Borghese et al., 2010). Furthermore, to faithfully model age-dependent neurodegenerative diseases requires inducing aging in organoids by pharmacological or genetic methods (Studer et al., 2015). One such strategy for inducing aging-related features in human iPSC-derived organoids, used to model PD, involves the expression of progerin, a truncated form of lamin A associated with premature aging (Miller et al., 2013). Telomere shortening induced via downregulation of telomerase has also been shown to result in age-related and disease-relevant phenotypes in human iPSC-derived neurons (Vera et al., 2016). However, whether these ‘induced’ aging events accurately reflect the aging process that occurs naturally in vivo remains to be determined.
The introduction of fused organoids or ‘assembloid’ systems provides a path to a modular design approach to investigate inter-brain-region and inter-organ crosstalk. Assembly of cortical organoids and organoids with subcortical identities, such as the thalamus, may offer insights into the development of corticofugal projections of deep layer cortical neurons (Pasca, 2018). More complex assembloid systems composed of three or more brain regions are feasible, and the ultimate goal is the assembly of a whole-brain organoid for comprehensive modeling of brain development and function. The combination of organoids from different tissue types could also capture the interface between the nervous system and other organs. Such is the case for the recently reported hPSC-derived intestinal organoids containing neural crest cells, which self-organize to resemble the enteric nervous system and produce rhythmic waves to regulate contraction of the organoids (Workman et al., 2017). Moreover, oncogene manipulation using CRISPR/Cas9 can be applied to initiate tumorigenesis in human brain organoids as an innovative approach to model brain tumors (Bian et al., 2018; Ogawa et al., 2018). Alternatively, brain tumor organoids can be generated from primary glioblastoma specimens that are dissected into smaller pieces and cultured in a 3D environment (Hubert et al., 2016). The subsequent fusion of these tumor organoids with normal brain organoids may create a scalable in vitro model for cancer metastasis, providing the means to screen for therapies that specifically block cancer cells' invasion into surrounding tissue.
Lastly, it should be noted that organoid differentiation protocols that rely on the self-organization of hPSCs, and stochasticity in their spontaneous differentiation, lead to inherently variable outcomes. Therefore, unlike organs that arise from the precisely controlled process of in vivo organogenesis, no two organoids are identical. To improve quality control, variables in the system should be eliminated wherever possible (Jabaudon and Lancaster, 2018). Feeder-dependent hPSC cultures are more technique dependent, and properties of each hPSC line are sometimes inconsistent. A shift to using feeder-free hPSC culture is likely to significantly improve reproducibility across laboratories and cell lines (Lancaster et al., 2017; Nakagawa et al., 2014; Yoon et al., 2019). The use of variable ingredients, such as animal-derived ECM (Matrigel), in culture protocols should also be minimized, and recombinant growth factors should be replaced by small molecules whenever applicable (Cruz-Acuña et al., 2017; Gjorevski et al., 2016). Furthermore, newly developed organoid generation methods need to be quantitatively characterized to show consistent results with multiple hPSC lines and independent batches before they are ready for publication. As mentioned above, the development of a miniaturized multi-well spinning bioreactor has enabled efficient optimization of organoid protocols and scalable organoid production (Qian et al., 2016), but current methods for organoid characterization are labor intensive and prevent scaling-up of organoid experiments. Moving forward, the development of systems with automated read-outs for high-throughput analyses will be instrumental for transformation of organoid models into high-throughput platforms suitable for compound screening and drug discovery.
Together, the rapid advances in brain organoid technologies have opened up new avenues for gaining a better understanding of human brain development, function, evolution and disorders. The brain organoid field has made exciting steps to empower researchers with new tools to address questions that are difficult to address in other model systems, but there remains a long way to go towards obtaining a more faithful in vitro representation of the developing human brain. It is important to keep in mind that no model is perfect. Only through synergy across different model systems can we truly gain knowledge that will light the path to overcoming neurological diseases.
Acknowledgements
We would like to thank members of the Ming and Song laboratories for comments and suggestions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
The research in the authors’ laboratories was supported by grants from the National Institutes of Health (R37NS047344, P01NS097206, U19MH106434 to H.S. and R01MH105128, R35NS097370, U19AI131130 to G.M.), The Simons Foundation (575050 to H.S.) and The Dr Miriam and Sheldon G. Adelson Medical Research Foundation (to G.M.). Deposited in PMC for release after 12 months.
References
- Abud E. M., Ramirez R. N., Martinez E. S., Healy L. M., Nguyen C. H. H., Newman S. A., Yeromin A. V., Scarfone V. M., Marsh S. E., Fimbres C. et al. (2017). iPSC-Derived human microglia-like cells to study neurological diseases. Neuron 94, 278-293.e279. 10.1016/j.neuron.2017.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S. A., Eisenstat D. D., Shi L. and Rubenstein J. L. (1997). Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science 278, 474-476. 10.1126/science.278.5337.474 [DOI] [PubMed] [Google Scholar]
- Arber C., Lovejoy C. and Wray S. (2017). Stem cell models of Alzheimer's disease: progress and challenges. Alzheimers Res. Ther. 9, 42 10.1186/s13195-017-0268-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae B.-I., Tietjen I., Atabay K. D., Evrony G. D., Johnson M. B., Asare E., Wang P. P., Murayama A. Y., Im K., Lisgo S. N. et al. (2014). Evolutionarily dynamic alternative splicing of GPR56 regulates regional cerebral cortical patterning. Science 343, 764-768. 10.1126/science.1244392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagley J. A., Reumann D., Bian S., Levi-Strauss J. and Knoblich J. A. (2017). Fused cerebral organoids model interactions between brain regions. Nat. Methods 14, 743-751. 10.1038/nmeth.4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly P. V., Taber L. A. and Kroenke C. D. (2014). Mechanical forces in cerebral cortical folding: a review of measurements and models. J. Mech. Behav. Biomed. Mater 29, 568-581. 10.1016/j.jmbbm.2013.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershteyn M., Nowakowski T. J., Pollen A. A., Di Lullo E., Nene A., Wynshaw-Boris A. and Kriegstein A. R. (2017). Human iPSC-derived cerebral organoids model cellular features of lissencephaly and reveal prolonged mitosis of outer radial glia. Cell Stem Cell 20, 435-449. 10.1016/j.stem.2016.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian S., Repic M., Guo Z. M., Kavirayani A., Burkard T., Bagley J. A., Krauditsch C. and Knoblich J. A. (2018). Genetically engineered cerebral organoids model brain tumor formation. Nat. Methods 15, 631 10.1038/s41592-018-0070-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birey F., Andersen J., Makinson C. D., Islam S., Wei W., Huber N., Fan H. C., Metzler K. R. C., Panagiotakos G., Thom N. et al. (2017). Assembly of functionally integrated human forebrain spheroids. Nature 15, 631-639. 10.1038/nature22330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghese L., Dolezalova D., Opitz T., Haupt S., Leinhaas A., Steinfarz B., Koch P., Edenhofer F., Hampl A. and Brustle O. (2010). Inhibition of notch signaling in human embryonic stem cell-derived neural stem cells delays G1/S phase transition and accelerates neuronal differentiation in vitro and in vivo. Stem Cells 28, 955-964. 10.1002/stem.408 [DOI] [PubMed] [Google Scholar]
- Borrell V. (2018). How cells fold the cerebral cortex. J. Neurosci. 38, 776-783. 10.1523/JNEUROSCI.1106-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystron I., Blakemore C. and Rakic P. (2008). Development of the human cerebral cortex: boulder committee revisited. Nat. Rev. Neurosci. 9, 110-122. 10.1038/nrn2252 [DOI] [PubMed] [Google Scholar]
- Calvet G., Aguiar R. S., Melo A. S. O., Sampaio S. A., de Filippis I., Fabri A., Araujo E. S., de Sequeira P. C., de Mendonca M. C., de Oliveira L. et al. (2016). Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. Lancet Infect. Dis. 16, 653-660. 10.1016/S1473-3099(16)00095-5 [DOI] [PubMed] [Google Scholar]
- Camp J. G., Badsha F., Florio M., Kanton S., Gerber T., Wilsch-Brauninger M., Lewitus E., Sykes A., Hevers W., Lancaster M. et al. (2015). Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. USA 112, 15672-15677. 10.1073/pnas.1520760112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caronia-Brown G., Yoshida M., Gulden F., Assimacopoulos S. and Grove E. A. (2014). The cortical hem regulates the size and patterning of neocortex. Development 141, 2855-2865. 10.1242/dev.106914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers D. and Fishell G. (2006). Functional genomics of early cortex patterning. Genome Biol. 7, 202 10.1186/gb-2006-7-1-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. I., Song H. and Ming G. L. (2018). Applications of human brain organoids to clinical problems. Dev. Dyn. 248, 53-64. 10.1002/dvdy.24662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A. and Walsh C. A. (2002). Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science 297, 365-369. 10.1126/science.1074192 [DOI] [PubMed] [Google Scholar]
- Choi S. H., Kim Y. H., Hebisch M., Sliwinski C., Lee S., D'Avanzo C., Chen H., Hooli B., Asselin C., Muffat J. et al. (2014). A three-dimensional human neural cell culture model of Alzheimer's disease. Nature 515, 274-278. 10.1038/nature13800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H., Song J., Park G. and Kim J. (2017). Modeling of autism using organoid technology. Mol. Neurobiol. 54, 7789-7795. 10.1007/s12035-016-0274-8 [DOI] [PubMed] [Google Scholar]
- Cruz-Acuña R., Quirós M., Farkas A. E., Dedhia P. H., Huang S., Siuda D., García-Hernández V., Miller A. J., Spence J. R., Nusrat A. et al. (2017). Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat. Cell Biol. 19, 1326-1335. 10.1038/ncb3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cugola F. R., Fernandes I. R., Russo F. B., Freitas B. C., Dias J. L., Guimarães K. P., Benazzato C., Almeida N., Pignatari G. C., Romero S. et al. (2016). The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534, 267-271. 10.1038/nature18296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang J., Tiwari S. K., Lichinchi G., Qin Y., Patil V. S., Eroshkin A. M. and Rana T. M. (2016). Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell 19, 258-265. 10.1016/j.stem.2016.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danjo T., Eiraku M., Muguruma K., Watanabe K., Kawada M., Yanagawa Y., Rubenstein J. L. and Sasai Y. (2011). Subregional specification of embryonic stem cell-derived ventral telencephalic tissues by timed and combinatory treatment with extrinsic signals. J. Neurosci. 31, 1919-1933. 10.1523/JNEUROSCI.5128-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq S., Keruzore M., Desmaris E., Pollart C., Assimacopoulos S., Preillon J., Ascenzo S., Matson C. K., Lee M., Nan X. et al. (2018). DMRT5 together with DMRT3 directly controls hippocampus development and neocortical area map formation. Cereb. Cortex 28, 493-509. 10.1093/cercor/bhw384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Toro D., Ruff T., Cederfjall E., Villalba A., Seyit-Bremer G., Borrell V. and Klein R. (2017). Regulation of cerebral cortex folding by controlling neuronal migration via FLRT adhesion molecules. Cell 169, 621-635.e616. 10.1016/j.cell.2017.04.012 [DOI] [PubMed] [Google Scholar]
- Ding S.-L., Royall J. J., Sunkin S. M., Ng L., Facer B. A. C., Lesnar P., Guillozet-Bongaarts A., McMurray B., Szafer A., Dolbeare T. A. et al. (2017). Comprehensive cellular-resolution atlas of the adult human brain. J. Comp. Neurol. 525, 407 10.1002/cne.24130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M., Watanabe K., Matsuo-Takasaki M., Kawada M., Yonemura S., Matsumura M., Wataya T., Nishiyama A., Muguruma K. and Sasai Y. (2008). Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3, 519-532. 10.1016/j.stem.2008.09.002 [DOI] [PubMed] [Google Scholar]
- Elsen G. E., Hodge R. D., Bedogni F., Daza R. A. M., Nelson B. R., Shiba N., Reiner S. L. and Hevner R. F. (2013). The protomap is propagated to cortical plate neurons through an Eomes-dependent intermediate map. Proc. Natl. Acad. Sci. USA 110, 4081-4086. 10.1073/pnas.1209076110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiddes I. T., Lodewijk G. A., Mooring M., Bosworth C. M., Ewing A. D., Mantalas G. L., Novak A. M., van den Bout A., Bishara A., Rosenkrantz J. L. et al. (2018). Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell 173, 1356-1369.e1322. 10.1016/j.cell.2018.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florio M., Albert M., Taverna E., Namba T., Brandl H., Lewitus E., Haffner C., Sykes A., Wong F. K., Peters J. et al. (2015). Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 347, 1465-1470. 10.1126/science.aaa1975 [DOI] [PubMed] [Google Scholar]
- Gabriel E., Wason A., Ramani A., Gooi L. M., Keller P., Pozniakovsky A., Poser I., Noack F., Telugu N. S., Calegari F. et al. (2016). CPAP promotes timely cilium disassembly to maintain neural progenitor pool. EMBO J. 35, 803-819. 10.15252/embj.201593679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcez P. P., Loiola E. C., Madeiro da Costa R., Higa L. M., Trindade P., Delvecchio R., Nascimento J. M., Brindeiro R., Tanuri A. and Rehen S. K. (2016). Zika virus impairs growth in human neurospheres and brain organoids. Science 352, 816-818. 10.1126/science.aaf6116 [DOI] [PubMed] [Google Scholar]
- Gjorevski N., Sachs N., Manfrin A., Giger S., Bragina M. E., Ordóñez-Morán P., Clevers H. and Lutolf M. P. (2016). Designer matrices for intestinal stem cell and organoid culture. Nature 539, 560-564. 10.1038/nature20168 [DOI] [PubMed] [Google Scholar]
- Gonzalez C., Armijo E., Bravo-Alegria J., Becerra-Calixto A., Mays C. E. and Soto C. (2018). Modeling amyloid beta and tau pathology in human cerebral organoids. Mol. Psychiatry 23, 2363-2374. 10.1038/s41380-018-0229-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D. V., Lui J. H., Parker P. R. L. and Kriegstein A. R. (2010). Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554-561. 10.1038/nature08845 [DOI] [PubMed] [Google Scholar]
- Hansen D. V., Lui J. H., Flandin P., Yoshikawa K., Rubenstein J. L., Alvarez-Buylla A. and Kriegstein A. R. (2013). Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat. Neurosci. 16, 1576-1587. 10.1038/nn.3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert C. G., Rivera M., Spangler L. C., Wu Q. L., Mack S. C., Prager B. C., Couce M., McLendon R. E., Sloan A. E. and Rich J. N. (2016). A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 76, 2465-2477. 10.1158/0008-5472.CAN-15-2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iefremova V., Manikakis G., Krefft O., Jabali A., Weynans K., Wilkens R., Marsoner F., Brandl B., Muller F. J., Koch P. et al. (2017). An Organoid-based model of cortical development identifies non-cell-autonomous defects in Wnt signaling contributing to miller-dieker syndrome. Cell Rep. 19, 50-59. 10.1016/j.celrep.2017.03.047 [DOI] [PubMed] [Google Scholar]
- Ip B. K., Bayatti N., Howard N. J., Lindsay S. and Clowry G. J. (2011). The corticofugal neuron-associated genes ROBO1, SRGAP1, and CTIP2 exhibit an anterior to posterior gradient of expression in early fetal human neocortex development. Cereb. Cortex 21, 1395-1407. 10.1093/cercor/bhq219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabaudon D. and Lancaster M. (2018). Exploring landscapes of brain morphogenesis with organoids. Development 145, dev172049 10.1242/dev.172049 [DOI] [PubMed] [Google Scholar]
- Jakovcevski I., Mayer N. and Zecevic N. (2011). Multiple origins of human neocortical interneurons are supported by distinct expression of transcription factors. Cereb. Cortex 21, 1771-1782. 10.1093/cercor/bhq245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H. and Tiwari V. K. (2018). Exploring the complexity of cortical development using single-cell transcriptomics. Front. Neurosci. 12, 31 10.3389/fnins.2018.00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo J., Xiao Y., Sun A. X., Cukuroglu E., Tran H.-D., Göke J., Tan Z. Y., Saw T. Y., Tan C.-P., Lokman H. et al. (2016). Midbrain-like organoids from human pluripotent stem cells contain functional dopaminergic and neuromelanin-producing neurons. Cell Stem Cell 19, 248-257. 10.1016/j.stem.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorfi M., D'Avanzo C., Tanzi R. E., Kim D. Y. and Irimia D. (2018). Human neurospheroid arrays for in vitro studies of Alzheimer's disease. Sci. Rep. 8, 2450 10.1038/s41598-018-20436-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoshima T., Sakaguchi H., Nakano T., Soen M., Ando S., Eiraku M. and Sasai Y. (2013). Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. USA 110, 20284-20289. 10.1073/pnas.1315710110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzbrun E., Kshirsagar A., Cohen S. R., Hanna J. H. and Reiner O. (2018). Human brain organoids on a chip reveal the physics of folding. Nat. Phys. 14, 515-522. 10.1038/s41567-018-0046-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg Z. N., Fiddes I. T., Gordon D., Murali S., Cantsilieris S., Meyerson O. S., Underwood J. G., Nelson B. J., Chaisson M. J. P., Dougherty M. L. et al. (2018). High-resolution comparative analysis of great ape genomes. Science 360, eaar6343 10.1126/science.aar6343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M. A. and Knoblich J. A. (2014). Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 9, 2329-2340. 10.1038/nprot.2014.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M. A., Renner M., Martin C.-A., Wenzel D., Bicknell L. S., Hurles M. E., Homfray T., Penninger J. M., Jackson A. P. and Knoblich J. A. (2013). Cerebral organoids model human brain development and microcephaly. Nature 501, 373-379. 10.1038/nature12517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M. A., Corsini N. S., Wolfinger S., Gustafson E. H., Phillips A. W., Burkard T. R., Otani T., Livesey F. J. and Knoblich J. A. (2017). Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 35, 659-666. 10.1038/nbt.3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letinic K., Zoncu R. and Rakic P. (2002). Origin of GABAergic neurons in the human neocortex. Nature 417, 645-649. 10.1038/nature00779 [DOI] [PubMed] [Google Scholar]
- Lewitus E., Kelava I. and Huttner W. B. (2013). Conical expansion of the outer subventricular zone and the role of neocortical folding in evolution and development. Front. Hum. Neurosci. 7, 424 10.3389/fnhum.2013.00424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewitus E., Kelava I., Kalinka A. T., Tomancak P. and Huttner W. B. (2016). Comment on “Cortical folding scales universally with surface area and thickness, not number of neurons”. Science 351, 825 10.1126/science.aad2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Sun L., Fang A., Li P., Wu Q. and Wang X. (2017a). Recapitulating cortical development with organoid culture in vitro and modeling abnormal spindle-like (ASPM related primary) microcephaly disease. Protein Cell 8, 823-833. 10.1007/s13238-017-0479-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Muffat J., Omer A., Bosch I., Lancaster M. A., Sur M., Gehrke L., Knoblich J. A. and Jaenisch R. (2017b). Induction of expansion and folding in human cerebral organoids. Cell Stem Cell 20, 385-396.e383. 10.1016/j.stem.2016.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui J. H., Hansen D. V. and Kriegstein A. R. (2011). Development and evolution of the human neocortex. Cell 146, 18-36. 10.1016/j.cell.2011.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Lancaster M. A., Castanon R., Nery J. R., Knoblich J. A. and Ecker J. R. (2016). Cerebral organoids recapitulate epigenomic signatures of the human fetal brain. Cell Rep. 17, 3369-3384. 10.1016/j.celrep.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T., Wang C., Wang L., Zhou X., Tian M., Zhang Q., Zhang Y., Li J., Liu Z., Cai Y. et al. (2013). Subcortical origins of human and monkey neocortical interneurons. Nat. Neurosci. 16, 1588-1597. 10.1038/nn.3536 [DOI] [PubMed] [Google Scholar]
- Madhavan M., Nevin Z. S., Shick H. E., Garrison E., Clarkson-Paredes C., Karl M., Clayton B. L. L., Factor D. C., Allan K. C., Barbar L. et al. (2018). Induction of myelinating oligodendrocytes in human cortical spheroids. Nat. Methods 15, 700-706. 10.1038/s41592-018-0081-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour A. A. F., Gonçalves J. T., Bloyd C. W., Li H., Fernandes S., Quang D., Johnston S., Parylak S. L., Jin X. and Gage F. H. (2018). An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 36, 432-441. 10.1038/nbt.4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J., Coppola G., Zhang P., Abyzov A., Provini L., Tomasini L., Amenduni M., Szekely A., Palejev D., Wilson M. et al. (2015). FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell 162, 375-390. 10.1016/j.cell.2015.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt A., Eberle D., Tazaki A., Ranga A., Niesche M., Wilsch-Bräuninger M., Stec A., Schackert G., Lutolf M. and Tanaka E. M. (2014). 3D reconstitution of the patterned neural tube from embryonic stem cells. Stem Cell Rep. 3, 987-999. 10.1016/j.stemcr.2014.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. D., Ganat Y. M., Kishinevsky S., Bowman R. L., Liu B., Tu E. Y., Mandal P. K., Vera E., Shim J.-W., Kriks S. et al. (2013). Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell 13, 691-705. 10.1016/j.stem.2013.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming G.-L., Tang H. and Song H. (2016). Advances in Zika virus research: stem cell models, challenges, and opportunities. Cell Stem Cell 19, 690-702. 10.1016/j.stem.2016.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux B. J., Arlotta P., Menezes J. R. L. and Macklis J. D. (2007). Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 8, 427-437. 10.1038/nrn2151 [DOI] [PubMed] [Google Scholar]
- Monzel A. S., Smits L. M., Hemmer K., Hachi S., Moreno E. L., van Wuellen T., Jarazo J., Walter J., Brüggemann I., Boussaad I. et al. (2017). Derivation of human midbrain-specific organoids from neuroepithelial stem cells. Stem Cell Rep. 8, 1144-1154. 10.1016/j.stemcr.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. A., Staples J. E., Dobyns W. B., Pessoa A., Ventura C. V., Fonseca E. B., Ribeiro E. M., Ventura L. O., Neto N. N., Arena J. F. et al. (2017). Characterizing the pattern of anomalies in congenital Zika syndrome for pediatric clinicians. JAMA Pediatr 171, 288-295. 10.1001/jamapediatrics.2016.3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Bermudez F., Badsha F., Kanton S., Camp J. G., Vernot B., Kohler K., Voigt B., Okita K., Maricic T., He Z. et al. (2016). Differences and similarities between human and chimpanzee neural progenitors during cerebral cortex development. Elife 5, e18683 10.7554/eLife.18683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota B. and Herculano-Houzel S. (2015). Brain structure. Cortical folding scales universally with surface area and thickness, not number of neurons . Science 349, 74-77. 10.1126/science.aaa9101 [DOI] [PubMed] [Google Scholar]
- Muguruma K., Nishiyama A., Kawakami H., Hashimoto K. and Sasai Y. (2015). Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 10, 537-550. 10.1016/j.celrep.2014.12.051 [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Taniguchi Y., Senda S., Takizawa N., Ichisaka T., Asano K., Morizane A., Doi D., Takahashi J., Nishizawa M. et al. (2014). A novel efficient feeder-free culture system for the derivation of human induced pluripotent stem cells. Sci. Rep. 4, 3594 10.1038/srep03594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H. N., Qian X., Song H. and Ming G.-L. (2016). Neural stem cells attacked by Zika virus. Cell Res. 26, 753-754. 10.1038/cr.2016.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski T. J., Pollen A. A., Sandoval-Espinosa C. and Kriegstein A. R. (2016). Transformation of the radial glia scaffold demarcates two stages of human cerebral cortex development. Neuron 91, 1219-1227. 10.1016/j.neuron.2016.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski T. J., Bhaduri A., Pollen A. A., Alvarado B., Mostajo-Radji M. A., Di Lullo E., Haeussler M., Sandoval-Espinosa C., Liu S. J., Velmeshev D. et al. (2017). Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318-1323. 10.1126/science.aap8809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nzou G., Wicks R. T., Wicks E. E., Seale S. A., Sane C. H., Chen A., Murphy S. V., Jackson J. D. and Atala A. J. (2018). Human cortex spheroid with a functional blood brain barrier for high-throughput neurotoxicity screening and disease modeling. Sci. Rep. 8, 7413 10.1038/s41598-018-25603-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa J., Pao G. M., Shokhirev M. N. and Verma I. M. (2018). Glioblastoma model using human cerebral organoids. Cell Rep. 23, 1220-1229. 10.1016/j.celrep.2018.03.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary D. D., Chou S. J. and Sahara S. (2007). Area patterning of the mammalian cortex. Neuron 56, 252-269. 10.1016/j.neuron.2007.10.010 [DOI] [PubMed] [Google Scholar]
- Otani T., Marchetto M. C., Gage F. H., Simons B. D. and Livesey F. J. (2016). 2D and 3D stem cell models of primate cortical development identify species-specific differences in progenitor behavior contributing to brain size. Cell Stem Cell 18, 467-480. 10.1016/j.stem.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozair M. Z., Kirst C., van den Berg B. L., Ruzo A., Rito T. and Brivanlou A. H. (2018). hPSC Modeling reveals that fate selection of cortical deep projection neurons occurs in the subplate. Cell Stem Cell 23, 60-73.e66. 10.1016/j.stem.2018.05.024 [DOI] [PubMed] [Google Scholar]
- Ozone C., Suga H., Eiraku M., Kadoshima T., Yonemura S., Takata N., Oiso Y., Tsuji T. and Sasai Y. (2016). Functional anterior pituitary generated in self-organizing culture of human embryonic stem cells. Nat. Commun. 7, 10351 10.1038/ncomms10351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamies D., Barreras P., Block K., Makri G., Kumar A., Wiersma D., Smirnova L., Zang C., Bressler J., Christian K. M. et al. (2017). A human brain microphysiological system derived from induced pluripotent stem cells to study neurological diseases and toxicity. ALTEX 34, 362-376. 10.14573/altex.1609122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca S. P. (2018). The rise of three-dimensional human brain cultures. Nature 553, 437-445. 10.1038/nature25032 [DOI] [PubMed] [Google Scholar]
- Pasca A. M., Sloan S. A., Clarke L. E., Tian Y., Makinson C. D., Huber N., Kim C. H., Park J.-Y., O'Rourke N. A., Nguyen K. D. et al. (2015). Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 12, 671-678. 10.1038/nmeth.3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petanjek Z., Berger B. and Esclapez M. (2009). Origins of cortical GABAergic neurons in the cynomolgus monkey. Cereb. Cortex 19, 249-262. 10.1093/cercor/bhn078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham M. T., Pollock K. M., Rose M. D., Cary W. A., Stewart H. R., Zhou P., Nolta J. A. and Waldau B. (2018). Generation of human vascularized brain organoids. Neuroreport 29, 588-593. 10.1097/WNR.0000000000001014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan D. T. T., Bender R. H. F., Andrejecsk J. W., Sobrino A., Hachey S. J., George S. C. and Hughes C. C. W. (2017). Blood-brain barrier-on-a-chip: Microphysiological systems that capture the complexity of the blood-central nervous system interface. Exp. Biol. Med. (Maywood) 242, 1669-1678. 10.1177/1535370217694100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen A. A., Nowakowski T. J., Chen J., Retallack H., Sandoval-Espinosa C., Nicholas C. R., Shuga J., Liu S. J., Oldham M. C., Diaz A. et al. (2015). Molecular identity of human outer radial glia during cortical development. Cell 163, 55-67. 10.1016/j.cell.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Nguyen H. N., Song M. M., Hadiono C., Ogden S. C., Hammack C., Yao B., Hamersky G. R., Jacob F., Zhong C. et al. (2016). Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165, 1238-1254. 10.1016/j.cell.2016.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Nguyen H. N., Jacob F., Song H. and Ming G. L. (2017). Using brain organoids to understand Zika virus-induced microcephaly. Development 144, 952-957. 10.1242/dev.140707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Jacob F., Song M. M., Nguyen H. N., Song H. and Ming G. L. (2018). Generation of human brain region-specific organoids using a miniaturized spinning bioreactor. Nat. Protoc. 13, 565-580. 10.1038/nprot.2017.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrato G., Nguyen T., Macosko E. Z., Sherwood J. L., Min Yang S., Berger D. R., Maria N., Scholvin J., Goldman M., Kinney J. P. et al. (2017). Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545, 48-53. 10.1038/nature22047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radonjic N. V., Ayoub A. E., Memi F., Yu X., Maroof A., Jakovcevski I., Anderson S. A., Rakic P. and Zecevic N. (2014). Diversity of cortical interneurons in primates: the role of the dorsal proliferative niche. Cell Rep. 9, 2139-2151. 10.1016/j.celrep.2014.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja W. K., Mungenast A. E., Lin Y. T., Ko T., Abdurrob F., Seo J. and Tsai L. H. (2016). Self-organizing 3D human neural tissue derived from induced pluripotent stem cells recapitulate Alzheimer's disease phenotypes. PLoS ONE 11, e0161969 10.1371/journal.pone.0161969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamani U., Gross A. R., Hjelm B. E., Sequeira A., Vawter M. P., Tang J., Gangalapudi V., Wang Y., Andres A. M., Gottlieb R. A. et al. (2018). Super-obese patient-derived iPSC hypothalamic neurons exhibit obesogenic signatures and hormone responses. Cell Stem Cell 22, 698-712.e699. 10.1016/j.stem.2018.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. (2009). Evolution of the neocortex: a perspective from developmental biology. Nat. Rev. Neurosci. 10, 724-735. 10.1038/nrn2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P., Ayoub A. E., Breunig J. J. and Dominguez M. H. (2009). Decision by division: making cortical maps. Trends Neurosci. 32, 291-301. 10.1016/j.tins.2009.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambani K., Vukasinovic J., Glezer A. and Potter S. M. (2009). Culturing thick brain slices: an interstitial 3D microperfusion system for enhanced viability. J. Neurosci. Methods 180, 243-254. 10.1016/j.jneumeth.2009.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A. and Zhang F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281-2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S. A., Jamieson D. J., Honein M. A. and Petersen L. R. (2016). Zika virus and birth defects--reviewing the evidence for causality. N. Engl. J. Med. 374, 1981-1987. 10.1056/NEJMsr1604338 [DOI] [PubMed] [Google Scholar]
- Reynolds B. A., Tetzlaff W. and Weiss S. (1992). A multipotent EGF-responsive striatal embryonic progenitor cell produces neurons and astrocytes. J. Neurosci. 12, 4565-4574. 10.1523/JNEUROSCI.12-11-04565.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Hanai S., Takashima S., Nakagawa E., Okazaki S., Inoue T., Miyata R., Hoshino K., Akashi T., Sasaki M. et al. (2011). Neocortical layer formation of human developing brains and lissencephalies: consideration of layer-specific marker expression. Cereb. Cortex 21, 588-596. 10.1093/cercor/bhq125 [DOI] [PubMed] [Google Scholar]
- Sakaguchi H., Kadoshima T., Soen M., Narii N., Ishida Y., Ohgushi M., Takahashi J., Eiraku M. and Sasai Y. (2015). Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat. Commun. 6, 8896 10.1038/ncomms9896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai Y. (2013). Next-generation regenerative medicine: organogenesis from stem cells in 3D culture. Cell Stem Cell 12, 520-530. 10.1016/j.stem.2013.04.009 [DOI] [PubMed] [Google Scholar]
- Sato T., Vries R. G., Snippert H. J., van de Wetering M., Barker N., Stange D. E., van Es J. H., Abo A., Kujala P., Peters P. J. et al. (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262-265. 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- Sato T., Stange D. E., Ferrante M., Vries R. G. J., Van Es J. H., van den Brink S., van Houdt W. J., Pronk A., van Gorp J., Siersema P. D. et al. (2011). Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 141, 1762-1772. 10.1053/j.gastro.2011.07.050 [DOI] [PubMed] [Google Scholar]
- Sloan S. A., Darmanis S., Huber N., Khan T. A., Birey F., Caneda C., Reimer R., Quake S. R., Barres B. A. and Pasca S. P. (2017). Human astrocyte maturation captured in 3D cerebral cortical spheroids derived from pluripotent stem cells. Neuron 95, 779-790.e776. 10.1016/j.neuron.2017.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa A. M. M., Meyer K. A., Santpere G., Gulden F. O. and Sestan N. (2017). Evolution of the human nervous system function, structure, and development. Cell 170, 226-247. 10.1016/j.cell.2017.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl R., Walcher T., De Juan Romero C., Pilz G. A., Cappello S., Irmler M., Sanz-Aquela J. M., Beckers J., Blum R., Borrell V. et al. (2013). Trnp1 regulates expansion and folding of the mammalian cerebral cortex by control of radial glial fate. Cell 153, 535-549. 10.1016/j.cell.2013.03.027 [DOI] [PubMed] [Google Scholar]
- Stoner R., Chow M. L., Boyle M. P., Sunkin S. M., Mouton P. R., Roy S., Wynshaw-Boris A., Colamarino S. A., Lein E. S. and Courchesne E. (2014). Patches of disorganization in the neocortex of children with autism. N. Engl. J. Med. 370, 1209-1219. 10.1056/NEJMoa1307491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer L., Vera E. and Cornacchia D. (2015). Programming and reprogramming cellular age in the era of induced pluripotency. Cell Stem Cell 16, 591-600. 10.1016/j.stem.2015.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L., Bershteyn M., Paredes M. F. and Kriegstein A. R. (2017). Dynamic behaviour of human neuroepithelial cells in the developing forebrain. Nat. Commun. 8, 14167 10.1038/ncomms14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur M. and Rubenstein J. L. (2005). Patterning and plasticity of the cerebral cortex. Science 310, 805-810. 10.1126/science.1112070 [DOI] [PubMed] [Google Scholar]
- Suzuki I. K., Gacquer D., Van Heurck R., Kumar D., Wojno M., Bilheu A., Herpoel A., Lambert N., Cheron J., Polleux F. et al. (2018). Human-specific NOTCH2NL genes expand cortical neurogenesis through Delta/Notch regulation. Cell 173, 1370-1384.e1316. 10.1016/j.cell.2018.03.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K. and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663-676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K. and Yamanaka S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861-872. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S. and Jones J. M. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282, 1145-1147. 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- Vera E., Bosco N. and Studer L. (2016). Generating late-onset human iPSC-based disease models by inducing neuronal age-related phenotypes through telomerase manipulation. Cell Rep. 17, 1184-1192. 10.1016/j.celrep.2016.09.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Buth J. E., Vishlaghi N., de la Torre-Ubieta L., Taxidis J., Khakh B. S., Coppola G., Pearson C. A., Yamauchi K., Gong D. et al. (2017). Self-Organized cerebral organoids with human-specific features predict effective drugs to combat Zika virus infection. Cell Rep. 21, 517-532. 10.1016/j.celrep.2017.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z., Song H. and Ming G.-L. (2017). How does Zika virus cause microcephaly? Genes Dev. 31, 849-861. 10.1101/gad.298216.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman M. J., Mahe M. M., Trisno S., Poling H. M., Watson C. L., Sundaram N., Chang C. F., Schiesser J., Aubert P., Stanley E. G. et al. (2017). Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat. Med. 23, 49-59. 10.1038/nm.4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Tanaka Y., Patterson B., Kang Y. J., Govindaiah G., Roselaar N., Cakir B., Kim K. Y., Lombroso A. P., Hwang S. M. et al. (2017). Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell 21, 383-398.e387. 10.1016/j.stem.2017.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B., Ng H. H., Takahashi R. and Tan E. K. (2016). Induced pluripotent stem cells in Parkinson's disease: scientific and clinical challenges. J. Neurol. Neurosurg. Psychiatry 87, 697-702. 10.1136/jnnp-2015-312036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Lee E. M., Wen Z., Cheng Y., Huang W.-K., Qian X., Tcw J., Kouznetsova J., Ogden S. C., Hammack C. et al. (2016). Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat. Med. 22, 1101-1107. 10.1038/nm.4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F., Kang E., Yu C., Qian X., Jacob F., Yu C., Mao M., Poon R. Y. C., Kim J., Song H. et al. (2017). DISC1 Regulates neurogenesis via modulating kinetochore attachment of Ndel1/Nde1 during Mitosis. Neuron 96, 1041-1054.e1045. 10.1016/j.neuron.2017.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K. J., Song G., Qian X., Pan J., Xu D., Rho H. S., Kim N. S., Habela C., Zheng L., Jacob F. et al. (2017). Zika-virus-encoded NS2A disrupts mammalian cortical neurogenesis by degrading adherens junction proteins. Cell Stem Cell 21, 349-358.e346. 10.1016/j.stem.2017.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S.-J., Elahi L. S., Pasca A. M., Marton R. M., Gordon A., Revah O., Miura Y., Walczak E. M., Holdgate G. M., Fan H. C. et al. (2019). Reliability of human cortical organoid generation. Nat. Methods 16, 75-78. 10.1038/s41592-018-0255-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. and Zecevic N. (2011). Dorsal radial glial cells have the potential to generate cortical interneurons in human but not in mouse brain. J. Neurosci. 31, 2413-2420. 10.1523/JNEUROSCI.5249-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zembrzycki A., Perez-Garcia C. G., Wang C.-F., Chou S.-J. and O'Leary D. D. M. (2015). Postmitotic regulation of sensory area patterning in the mammalian neocortex by Lhx2. Proc. Natl. Acad. Sci. USA 112, 6736-6741. 10.1073/pnas.1424440112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H., Shen E. H., Hohmann J. G., Oh S. W., Bernard A., Royall J. J., Glattfelder K. J., Sunkin S. M., Morris J. A., Guillozet-Bongaarts A. L. et al. (2012). Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell 149, 483-496. 10.1016/j.cell.2012.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Hammack C., Ogden S. C., Cheng Y., Lee E. M., Wen Z., Qian X., Nguyen H. N., Li Y., Yao B. et al. (2016). Molecular signatures associated with ZIKV exposure in human cortical neural progenitors. Nucleic Acids Res. 44, 8610-8620. 10.1093/nar/gkw765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Zhang S., Fan X., Wu Q., Yan L., Dong J., Zhang H., Li L., Sun L., Pan N. et al. (2018). A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524-528. 10.1038/nature25980 [DOI] [PubMed] [Google Scholar]
- Zhou T., Tan L., Cederquist G. Y., Fan Y., Hartley B. J., Mukherjee S., Tomishima M., Brennand K. J., Zhang Q., Schwartz R. E. et al. (2017). High-content screening in hPSC-neural progenitors identifies drug candidates that inhibit Zika virus infection in fetal-like organoids and adult brain. Cell Stem Cell 21, 274-283.e275. 10.1016/j.stem.2017.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilles K., Palomero-Gallagher N. and Amunts K. (2013). Development of cortical folding during evolution and ontogeny. Trends Neurosci. 36, 275-284. 10.1016/j.tins.2013.01.006 [DOI] [PubMed] [Google Scholar]