Abstract
Much effort has been invested in standardizing medical terminology for representation of medical knowledge, storage in electronic medical records, retrieval, reuse for evidence-based decision making, and for efficient messaging between users. We only focus on those efforts related to the representation of clinical medical knowledge required for capturing diagnoses and findings from a wide range of general to specialty clinical perspectives (e.g., internists to pathologists). Standardized medical terminology and the usage of structured reporting have been shown to improve the usage of medical information in secondary activities, such as research, public health, and case studies. The impact of standardization and structured reporting is not limited to secondary activities; standardization has been shown to have a direct impact on patient healthcare.
Keywords: Medical terminology, ontology, SNOMED, standards, structured reporting
Data standards for terminology
In 2010, the Department of Health and Human Services (DHHS) assigned rules and regulations regarding medical recording through its electronic health record incentive program. These rules required all U.S. healthcare providers and institutions participating in Medicare and Medicaid programs to implement an electronic health record system by 2015.9 With increased adoption of electronic health records and the incentive to promote the “meaningful use” of health information technology by the DHHS, the volume of health information recorded in the organizations’ repositories will grow exponentially (Committee on Data Standards for Patient Safety, Key capabilities of an electronic health record system, 2003, https://goo.gl/NRYTvj).64 Therefore, healthcare in the United States will become an information-intensive industry. Moreover, a 2016 report by the Council for Affordable Quality Healthcare (a non-profit alliance of health plans and trade associations that intends to streamline the business of healthcare) showed that healthcare providers and commercial health plans could save $8.5 billion annually by transitioning from manual to electronic processes (https://goo.gl/cTvwdX). This transition from paper to electronic record systems provided an opportunity for these health records to be used in secondary activities, such as research, public health, and statistics. Reporting of clinical or pathologic findings in a systematized manner has been shown to improve the efficiency of these secondary uses.16 However, it is difficult to achieve improvement in secondary activities without corresponding standards for information storage and retrieval. It is also challenging to compare practice patterns and outcomes without a common language. The integration and maturation of clinical terminologies to standardize information recording will result in more accurate and detailed clinical information, which will facilitate more efficient and effective healthcare delivery and information recall.11,28
In 1994, the Board of Directors of the American Medical Informatics Association (AMIA) recommended specific approaches to standardization in the areas of the patient, provider, and site-of-care identifiers.6 Standards covered computerized healthcare message exchange, medical record content and structure, and medical codes and terminologies. The AMIA Board of Directors suggested coding systems for several subject domains, such as the World Health Organization (WHO) drug record codes for drugs, the International Classification of Diseases (ICD), and the Systematized Nomenclature of Medicine–Clinical Terms (SNOMED-CT) for diagnoses, Health Level 7 (HL7) for messaging, and others.
In healthcare message exchange, HL7 International is a standard-developing organization that provides framework and standards for the exchange, integration, sharing, and retrieval of electronic health information. The HL7 Reference Information Model (RIM) is an example of an object model created to provide conceptual standards that identify the life cycle that a message or groups of related messages will carry.71 HL7 was recommended by the AMIA Board of Directors to serve within-institution transmission of orders, clinical observations, and clinical data; admission, transfer, and discharge records; and charge and billing information. Moreover, in 2004, Consolidated Health Informatics (CHI) in the United States suggested the use of HL7 vocabulary standards for demographic information, units of measure, immunizations, and clinical encounters. CHI also suggested HL7’s Clinical Document Architecture standard for text-based reports (Digital Imaging and Communications in Medicine Committee, Standards adoption recommendation: multimedia information in patient records. 2011. Final Draft Report).26 Other standards were recommended by the CHI as well, such as the Laboratory Logical Observation Identifier Name Codes (LOINC) to standardize the electronic exchange of laboratory test orders and drug label section headers (Steindel S, et al. Introduction of a hierarchy to LOINC to facilitate public health reporting. Proc Am Med Inform Assoc; 2002).40
The ICD, which is maintained by WHO, has been one of the most common classification systems for epidemiology, health management, and clinical purposes. ICD is used by physicians, nurses, and other health providers to classify diseases and other health problems. ICD codes are currently being used by >50 countries to improve consistency in recording patients’ symptoms and diagnoses for the purposes of reimbursement and clinical research (WHO. International statistical classification of diseases and related health problems, 2010, https://goo.gl/vFzHzG).
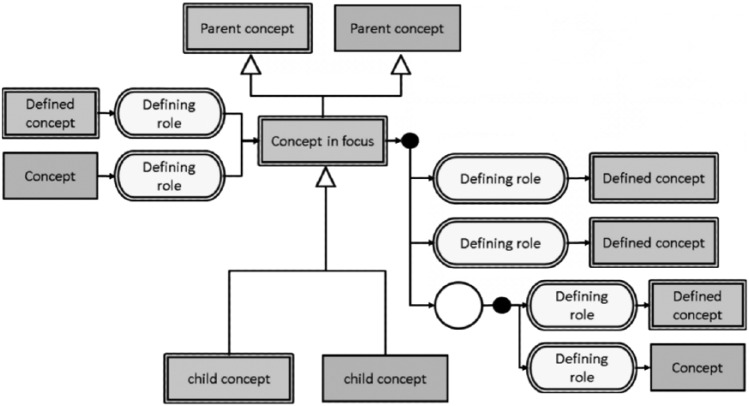
In 1965, the College of American Pathologists (CAP) published the Systematized Nomenclature of Pathology (SNOP) to describe morphology and anatomy. In 1975, led by the efforts of Dr. Roger Cote, CAP expanded SNOP to create the Systematized Nomenclature of Medicine (SNOMED). In 2000, CAP created a new logic-based version of SNOMED called SNOMED-RT. Over that the same time period, Dr. James Read developed the Read codes which evolved into Clinical Terms Version 3 (CTV-3) under the British National Health Service. In 2002, the CTV-3 and SNOMED-RT were combined to create SNOMED-CT, a joint development project of the British National Health Service (NHS) and CAP. SNOMED-CT is now the intellectual property of the International Health Terminology Standards Development Organization, which determines global standards for medical terms for clinical findings, procedures, body structures, organisms, qualifier values, and more (https://goo.gl/ocoSU3). With >344,000 unique medical concepts and 90,000 synonyms or alternative descriptions, SNOMED-CT is considered to be the most comprehensive clinical vocabulary that can be used to report and represent medical information. The components of SNOMED-CT define standard terms for health processes, indicate relationships between processes, and describe these processes through the use of synonyms. The terms for health processes are organized in multiple hierarchies with different degrees of granularity to provide flexibility in data recording and representation.24,51 SNOMED-CT’s current approach adopts a foundation based in descriptive logic (DL) that has many advantages, such as establishing formal semantics for SNOMED-CT’s assertions and suggesting a formal syntax. It also provides a basis for understanding expressiveness and computational complexity (Spackman, KA. Normal forms for description logic expressions of clinical concepts in SNOMED RT. Proc Am Med Inform Assoc Symp; 2001). Meanings of SNOMED-CT concepts are derived from their arrangement in the hierarchy and from axioms that connect them across hierarchies (Fig. 1). The connections across different hierarchies provide definitions, which classify concepts into sufficiently defined or non-sufficiently defined. A full explanation of the underlying DL used in SNOMED-CT is beyond the scope of this manuscript; suffice it to say that concepts are defined by their relationships to other concepts within the terminology (https://goo.gl/RKmzas).
Figure 1.
Diagrammatic representation of a SNOEMD-CT focus concept, its parent and children concepts, and relationships to other concepts used for its definition (https://goo.gl/6okNLC).
SNOMED-CT usage has grown with an increase in access facilitated by the National Library of Medicine’s U.S. public license free-access agreement through the Unified Medical Language System (UMLS).20 Moreover, in 2004, the CHI recommended the use of SNOMED-CT as the standard for diagnoses and problem lists, anatomy, and procedures.11 SNOMED-CT also provides an extension mechanism, in which organizations can create the concepts’ classes and descriptions that fulfill their needs and still fit into the SNOMED-CT framework. The logic-based framework of SNOMED-CT also increases its abilities by allowing for post-coordination (i.e., the creation of new concept classes by combining existing ones). The SNOMED-CT User and Starter Guides (https://goo.gl/ocoSU3) explain the concepts’ classes and relationships that developers can combine to post-coordinate clinical findings at the point of care (https://goo.gl/e2Jr4e). Post-coordination can be straightforward, and it can be complex; this potential complexity falls on the shoulders of coders and implementation experts who would have to code each single concept by more than one SNOMED-CT concept code.
A 40-y review of SNOMED showed no difference in SNOMED usage among various medical specialties.13 The study showed that SNOMED is broadening beyond pathology to be used to cover nursing, cancer, cardiology, primary care, gastroenterology, HIV, orthopedics, nephrology, and anesthesia. A previous review has shown that SNOMED-CT is reportedly used in >50 countries and its implementation has been steadily increasing.34 However, only a few publications showed SNOMED-CT utilization in operational settings. In data capture and implementation, SNOMED-CT has been very commonly used to represent terms from documents and forms (by concept mapping), as an interface terminology (where users browse through SNOMED-CT terms and descriptions), and as an automatic indexing system for clinical text (through the application of natural language processing). Previous studies facilitated mapping SNOMED-CT to >40 standard terminologies, with ICD (versions 9 and 10) being the most common one.22,34,47,65 SNOMED-CT medical domains of application reached >39, with problem list/diagnoses, nursing, drugs, and pathology being the most common ones.34 These studies showed that mapping SNOMED-CT to other terminologies identified gaps in concepts and synonyms that need to be incorporated within SNOMED-CT to improve its completeness.
As do physicians, veterinarians see the need for standards when recording animal patients’ information.5,15,30,66 A good example of standardizing recording of veterinary information is the use of SNOMED-CT as a standard nomenclature for clinical pathology reporting.72 The Veterinary Terminology Services Laboratory (VTSL) at Virginia Tech provides a browser to query SNOMED-CT content. VTSL maintains an extension of SNOMED-CT (http://vtsl.vetmed.vt.edu/) that integrates content that is considered to be “veterinary only” (is not applicable in human medicine). Because veterinary medical terminology is shared with human medicine, the VTSL browser presents an integrated view of the international release (core) of SNOMED-CT combined with the veterinary extension.
SNOMED-CT applications
Searching medical records using traditional information retrieval methodologies (such as keyword filtering or text-mining) is a challenge. There is a need for a more robust data retrieval methodology, such as semantic searches, that would be enabled by using terminologies such as SNOMED-CT. A previous study has shown that SNOMED-CT’s concept-based approach can effectively deal with the 2 types of queries: keyword mismatch and specialization versus generalization (granularity).33 In another study, SNOMED-CT searching terms were used to recall incidents from a database related to neuromuscular blockade in anesthesia4; the authors concluded that a keyword search is only as good as the terms selected, but the use of SNOMED-CT reveals more incidents.
Another retrospective study examined the feasibility of using electronic laboratory and admission-discharge-transfer data to retrieve human cases with Clostridium difficile infection.4 Data recalled from 44 hospitals showed that 40 hospitals sent test results recorded using LOINC codes, and 5 hospitals sent SNOMED-CT codes. Retrieval of cases used 25 LOINC and 20 SNOMED-CT codes. The authors emphasized the need for widespread adoption of standard vocabularies to facilitate public health use of electronic data. Other studies used SNOMED-CT to retrieve terms and classify cases from information systems for chronic diseases, infectious disease symptoms, and cancer morphologies, such as breast and prostate malignancies.36,39,48,61,63,73 Although SNOMED-CT has been shown to be very comprehensive and useful for many domains, its usage as a knowledge base ontology has been a challenge.57 SNOMED-CT has shown the ability to facilitate semantic interoperability (i.e., the ability of 2 parties to exchange data with unambiguous and shared meaning) aspects, such as classifying terms by meaning, placing terms with similar meanings into one hierarchy, and enriching them with synonyms or descriptions.25 Logic-based concept definitions were then introduced and consolidated in SNOMED-CT (Spackman, A. Normal forms for description logic expressions of clinical concepts in SNOMED RT. Proc Am Med Inform Assoc Symp; 2001). As a result, the current SNOMED-CT constitutes a blend of diverging and even contradicting architectural principles. Other challenges related to using SNOMED-CT as a knowledge base ontology have been reported.57 In histopathology, a 2014 study investigated the use of SNOMED-CT to represent histologic diagnoses.8 Authors of that study investigated the ability to represent diagnostic tissue morphologies and tissue architectures described within a pathologist’s microscopic examination report for 24 breast biopsy cases. Results found that 75% of the 95 diagnostic statements could be represented by one valid SNOMED-CT pre-coordinated expression and 73 SNOMED-CT post-coordinated expressions. The authors concluded that development of the SNOMED-CT model and content to cover microscopic examination of histologic tissues is required in order for SNOMED-CT to be effective in surgical pathology knowledge capture.
In veterinary medicine, reference and terminology standards are core components of the informatics infrastructure. In 2002, The American Veterinary Medical Association endorsed the use of SNOMED, HL7, and LOINC as the official informatics standards in veterinary medicine (https://goo.gl/FAPRgj). This subset of SNOMED-CT contains >5,600 clinical signs and diagnostic terms commonly used in companion animal practice in North America. The subset was drawn from both the SNOMED-CT core and Veterinary Extension. With the availability of different standard terminologies for veterinary medicine, several studies tested the implementation, evaluation, and application of these standards. One study highlighted some of the needs and challenges surrounding different areas of veterinary informatics.56 The authors suggested that the application of LOINC, HL7, and SNOMED-CT for laboratory tests and messaging would promote the use of evidence-based practice in veterinary medicine. SNOMED-CT contains veterinary concepts in addition to a great number of anatomic structures for nonhumans.
A previous study examined SNOMED-CT coverage of veterinary clinical pathology concepts taken from textbooks and industry pathology reports.72 Authors of the study found that SNOMED-CT representation of veterinary clinical pathology content was limited (45–48% good matching). However, the missing and problem concepts were related to a relatively small area of terminology. The authors concluded that revising and enriching some of the SNOMED-CT contents will optimize it for veterinary clinical pathology applications.
Another study examined organizing microbiome data from mammalian hosts using biomedical informatics methodologies and the Foundational Model of Anatomy (FMA) and SNOMED-CT (Bodenreider O, Zhang S. Comparing the representation of anatomy in the FMA and SNOMED CT. Proc Am Med Inform Assoc Symp; 2006:46).54 The study concluded that researchers can use the current biomedical infrastructure in organizing microbiome data from animals, therefore, bringing a new source of knowledge to facilitate testing comparative biomedical hypotheses pertaining to human health.
Another study tested the applications of utilizing standardized technologies, such as HL7 and SNOMED-CT, in veterinary hospital information systems.69 A survey showed an improvement (89% agreement) in the management of hospital data and the retrieval of useful clinical information when following standards.
Finally, adopting a consistent coding system will enable tracking animal health data on a national or international scale, which would facilitate public health use of human and animal records and allow for the usage of records in secondary activities.51,60 Such systems should be interoperable and should receive widespread adoption to be beneficial. Recommendations for perfect standard terminologies to improve interoperability and receive widespread adoption have been studied, including terminology content, concept orientation, polyhierarchy structure, and granularity.12
Structured reporting
Electronic health data comprises various data types from structured information, such as patient blood test results or diagnoses, to unstructured (free-text) data such as admission notes, description of patient physical examination, and histologic descriptions of tissue specimens. This variation in data types and the use of a free format presents a challenge for data integration and reuse. When it comes to processing unstructured information, such as free-text documents (which have been shown to account for 80% of the data in business and health record systems), the task consumes more time and effort (https://goo.gl/Hdo8sq). Previous studies showed techniques to learn from free-text and used them widely to identify special patterns and features that can be of interest. One study showed a way of converting scanned images of pathology reports into a free-text report so researchers can analyze information and classify reports computationally into cancer or non-cancer categories.73 However, applying text-mining techniques ignores some relationships between information stored in the text. A more efficient computational approach (such as natural language processing) requires human guidance for information to be extracted and interpreted properly from free text.
Although unstructured (free-text) reports can be more fluid and explicit of findings, they are not easily converted into a structured, computable data format.7 Recording standards will help ensure consistent reporting of patients’ information (such as physicians’ observations, diagnoses, and treatment) to improve data integration and uses. Standards have to be followed when recording patients’ information in order to automate computer-based technology tools that can assist in a variety of hospital processes and secondary uses, such as reminders, procedures, and decision-making activities. Such computer-based systems have been shown to help minimize errors in medical practice, control costs, and ensure correct billing of claims.41
Clinicians tend to value flexibility and efficiency in reporting information, whereas informaticists (users of information in secondary activities) value the information structure. Structured reports can require more time than unstructured ones at the point of care; however, structured reporting saves time and effort when it comes to secondary activities. More importantly, supporting automation of secondary activities would have a good impact at the point of care (the primary activity). One study showed that information could be found 4 times faster if recorded using a structured format than using an unstructured one.19 In information coverage, a previous assessment across the 2 formats showed that 10% of the studied features could not be found in unstructured clinical records.41 Another study suggested that structured, well-organized data can influence care decisions by offering a complete picture of the patient state to the decision maker.67 The well-organized display of information provided to physicians was shown in another study to reduce physician-ordered tests by 15%, therefore reducing the cost and invasiveness of diagnostic tests on patients.68
Although structured reports have been shown to have some advantages over unstructured ones, structured reports may lack the expressiveness of the narrative format. This lack of expressiveness can make it challenging to gain physician’s acceptance of the structured format. However, a partial capture of information in a structured manner has been more readily accepted. For example, a 1988 study showed that the best the institution could do was to capture some value or items from physicians in a structured format with the rest being coded as free-text.41 However, the right (computationally sufficient and medically representative of a particular scenario) amount of information to be structured presents a challenge. The right balance between report expressiveness and report structure has been shown to be an optimization problem.23,28,38 In order to achieve the optimal balance, a study conducted in 2008 suggested the use of a structured narrative design, in which unstructured text and coded data are fused into a single model.29 A structured narrative report is marked up with standardized codes used within a narrative structure to enable efficient retrieval of information while keeping the report in its original expressive shape. Natural language processing (NLP) and reliance on post-hoc text processing can then be used to identify fine structure. However, NLP can’t ensure the full understanding achieved using the structured format and its optimal accuracy. In other words, analyzing and understanding yes/no-based attributes is straightforward for machines, but it is more challenging for machines to draw a similar conclusion if the information was reported using natural language.
Previous studies have discussed the effects of structured versus unstructured (free-text) data on the retrieval and analysis of records.3,18 Studies concluded that a structured report, or the integration of unstructured information in a structured format, has to be adopted in order to provide actionable data. Other studies examined the use of structured reporting formats in reporting a variety of laboratory information, such as clinical notes, history and physical examination, magnetic resonance imaging, fracture risk assessment, endoscopy, radiology, and cytology.1,23,29,42,46,53,55
Studies concluded that structured reports are associated with a lower missing data rate than unstructured reports, contain significantly more relevant information, and produce superior reports, probably as a result of their reminder effect. Studies have shown that structured reporting has the capacity to provide a technically reliable basis for good reporting, and that free-form reports yielded reports with an unsatisfactory quality. One study showed that automated structured information reports can produce assessments that agree with experts on most routine exams, and highlighted that experts may diverge from the guidelines when not using the structured reports. Although some authors see that using structured reports is a must, others proposed a structured narrative format to facilitate capturing information coded in a narrative way and therefore integrate unstructured information in a structured framework. Studies highlighted factors to consider when choosing a format, including fitness of workflow, information coverage, time efficiency, ease of use, narrative expressiveness, machine readability, document structure, and cost.
In the field of pathology, the pathology report is the formal communication link between pathologist and clinician. An unstructured format has been commonly used for reporting pathologic findings. Standardizing the terminology used within pathology reports has been evaluated in many studies. In one study, researchers surveyed 96 veterinary clinical pathologists.10 The study showed that 68 unique terms were used to express probability or likelihood of cytologic diagnoses, among them were: “possible,” “suggestive of,” “consistent with,” “most likely,” “probable,” and more. By including a numerical interpretation of the terms, the study drew a good probability diagram by translating each term into numerical expression.
In other studies, different formats used in reporting pathology information have been shown to have an effect on the clarity of the message transferred. A 1991 study assessed the completeness of 1,565 upper endoscopy structured reports in comparison to 360 free-text reports23; structured reports had a lower missing data percentage (18%) compared to free-text reports (45%) and contained 60% more relevant information. Another study of radiology reports concluded that free-text reports contained 13–75% of forensically relevant findings compared to 100% coverage by structured reports.58
Since 1986, CAP has been working to establish guidelines for the assessment and recording of pathology examinations.2 In 1992, one study by CAP examined 15,940 pathology reports for colorectal carcinoma from 532 laboratories70; standardized or checklist structure of pathology reports was the only factor significantly associated with increased likelihood of providing complete or adequate pathology information. In the same year, a study was published declaring the adequacy needed within structured surgical pathology reports.31 Following the release of the 1992 CAP standards, and using them as reference, a study conducted in 1993 proposed standardized surgical pathology forms for reporting major tumor types. The study included recommendations for reporting histologic grade, patterns of growth, local invasion, vascular invasion, and others for many body sites.52 A study published in 1994 described the use of standardized, synoptic surgical pathology reports to improve the accuracy with which critical information can be obtained consistently and easily regardless of the institution of origin.35 The authors of the study reviewed variables found to be relevant to the clinical management of patients, proposed a standardized reporting format, and showed a simple query for extracting information about tumors from a database for patients with right partial mastectomies, and the standardized format has been researched by many groups.31,54,69
Another study in 1996 by CAP reviewed >8,300 lung carcinoma cases from 464 institutions.21 The study assessed the adequacy of lung carcinoma surgical pathology reports in covering gross and microscopic findings, and examined the presence or absence of 23 descriptors. A standardized or a checklist format was used in 21% of the cases and was associated with increased likelihood of recording 9 of the 23 descriptors. The authors proposed a standardized reporting format for resected bronchogenic carcinomas and recommended the use of a standardized report or a checklist report. This work has been cited in >42 other manuscripts with respect to contributions made to improving communication and patient health.
In 1999, to assist pathologists in making diagnoses, CAP published a checklist reporting format to be used when recording specimen information for patients with carcinoma of renal tubular origin.17 In the same year, a study published guidelines for a more precise morphologic evaluation and reporting of celiac disease diagnoses.49 Authors developed guidelines for the histologic evaluation of biopsy specimens from patients with celiac disease as well as a structured report composed of a checklist of findings to be recorded. Researchers also developed other reporting standards for cancer pathology; among them is a report that was released by the Center for Disease Control and Prevention (CDC) on reporting of pathology protocols for colon and rectal cancers (Department of Health and Human Services. Report on the reporting pathology protocols for colon and rectum cancers project, 2005, https://goo.gl/JDE8Nv). Although the structured data format seemed to be the appropriate reporting structure, a 2000 study highlighted a communication gap between pathologists and surgeons as a result of unfamiliarity with the structured representation; they emphasized the need for a more complete, clear, and standardized format.50
In 2008, CAP established the Diagnostic Intelligence and Health Information Technology Committee with a mission “to advance the implementation of the CAP Cancer Checklists using health information technology.” In 2009, the number of available checklists was expanded to 55. Extensible markup language versions of the checklists became available to improve compatibility of checklists with a variety of platforms and to integrate checklist records into information systems.2 In the same year, another study showed that implementing some of the CAP standards resulted in improvements in rates of synoptic reporting and completeness of cancer pathology reporting.62 The study showed a significant increase in completeness rates when structured reports were used to report cancer of the prostate, colon\rectum, lung, and breast.
A 2010 CAP study evaluated the frequency with which surgical pathology cancer reports contain all of the scientifically validated elements by the American College of Surgery Commission on Cancer.27 Authors evaluated 2,125 cancer reports generated by 86 institutions. The study found that institutions in which practitioners routinely used checklist reports reported all required elements at a higher rate (88%) than those that did not use checklists (34%). The same study also showed that the institutions that had a system in place to track errors, which can only be achieved by computable reports, reported all required elements at a higher rate (88%) compared with those that did not have such a system (68%). A study conducted in 2014 found expressions of uncertainty in 35% of 1,500 human surgical pathology reports.37 In the same study, authors surveyed 76 clinicians seeking a percentage of certainty to be given to expressions of uncertainty reported within diagnoses and was found to be a significant source of miscommunication.37 Results of these 2 studies suggest that the use of checklists and defined reporting features with avoidance of certainty percentages may help reduce errors of omission and miscommunication among clinicians.
Although a “diagnosis” is the major product of pathology reports, a previous study found that the pathologist’s product is not simply the correct diagnosis; there is pertinent and credible information that is useful in addressing patient care needs in other areas of the report, and structured reports can help ensure its capture.14 A 2016 study of microscopic assessments of rectal tumor specimens examined the effect of structured reporting formats on the data recorded.32 The authors found that a structured reporting dictation template improves data collection and may reduce the subsequent administrative burden when evaluating rectal specimens.
In veterinary pathology reports, the unstructured format has been the traditional format used in recording histopathologic findings. In 2005, the World Small Animal Veterinary Association (WSAVA) GI International Standardization Group undertook the responsibility of standardizing the histologic evaluation of the gastrointestinal tract of dogs and cats (Washabau RJ. 2005 report from: WSAVA Gastrointestinal Standardization Group, 2005, https://goo.gl/MsXQPq). In 2008, the group proposed standards for the assessment of microscopic findings from GI biopsy samples.15 The standards were found to minimize variation among pathology reports of microscopic findings in the GI tract. The group established criteria for quantifying and ranking key endoscopic, histologic observations from the gastric body, antrum, duodenum, and colon. These criteria provided specific numerical intervals to categorize the severity (mild, moderate, and marked) of different observations. In 2010, the same group published endoscopic, biopsy, and histopathologic guidelines for the evaluation and partial reporting of GI inflammation in companion animals.66 However, the WSAVA has not proposed standardization for recording the histologic diagnosis, and there has been no formal assessment of the effect of the WSAVA structured format on information capture.
It is our hope that the reader has gained some appreciation for the wide variety of organizations working to improve clarity and understanding of content in medical documents and electronic healthcare records. These ongoing efforts continue to evolve with the increased use of electronic healthcare data but fundamentally still depend upon use of controlled terminologies and establishment of reporting stands by domain experts. In this review, we have given multiple examples of how these basic informatics tools have been used to help ensure reporting of required elements, to increase data interoperability across different systems, and to standardize healthcare practices among clinicians and pathologists in hopes of improving patient care in veterinary and human medicine.35,43–45,52,59,64
Footnotes
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
References
- 1. Allin S, et al. Evaluation of automated fracture risk assessment based on the Canadian association of radiologists and osteoporosis Canada assessment tool. J Clin Densitom 2016;19:332–339. [DOI] [PubMed] [Google Scholar]
- 2. Amin MB. The 2009 version of the cancer protocols of the College of American Pathologists. Arch Pathol Lab Med 2010;134:326–330. [DOI] [PubMed] [Google Scholar]
- 3. Baars H, et al. Management support with structured and unstructured data—an integrated business intelligence framework. Inf Syst Manage 2008;25:132–148. [Google Scholar]
- 4. Benoit SR, et al. Automated surveillance of clostridium difficile infections using biosense. Infect Control Hosp Epidemiol 2011;32:26–33. [DOI] [PubMed] [Google Scholar]
- 5. Berendt M, et al. International veterinary epilepsy task force consensus report on epilepsy definition, classification and terminology in companion animals. BMC Vet Res 2015;11:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Board of Directors of the American Medical Informatics Association. Standards for medical identifiers, codes, and messages needed to create an efficient computer-stored medical record. J Am Med Inform Assoc 1994;1:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Branavan SRK, et al. Learning document-level semantic properties from free-text annotations. J Artificial Intell Res 2009;34:569–603. [Google Scholar]
- 8. Campbell WS, et al. Semantic analysis of SNOMED CT for a post-coordinated database of histopathology findings. J Am Med Inform Assoc 2014;21:885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Centers for Medicare & Medicaid Services. Medicare and Medicaid programs; electronic health record incentive program. Final rule. Fed Regist 2010;75:44313–44588. [PubMed] [Google Scholar]
- 10. Christopher MM, et al. Cytologic diagnosis: expression of probability by clinical pathologists. Vet Clin Pathol 2004;33:84–95. [DOI] [PubMed] [Google Scholar]
- 11. Chute CG, et al. A framework for comprehensive health terminology systems in the United States: Development guidelines, criteria for selection, and public policy implications. ANSI Healthcare Informatics Standards Board Vocabulary Working Group and the Computer-Based Patient Records Institute Working Group on Codes and Structures. J Am Med Inform Assoc 1998;5:503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med 1998;37:394–403. [PMC free article] [PubMed] [Google Scholar]
- 13. Cornet R, et al. Forty years of SNOMED: a literature review. BMC Med Inform Decis Mak 2008;8(Suppl 1):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cowan DF. Quality assurance in anatomic pathology. An information system approach. Arch Pathol Lab Med 1990;114:129–134. [PubMed] [Google Scholar]
- 15. Day MJ, et al. Histopathological standards for the diagnosis of gastrointestinal inflammation in endoscopic biopsy samples from the dog and cat: a report from the World Small Animal Veterinary Association Gastrointestinal Standardization Group. J Comp Pathol 2008;138(Suppl 1):S1–S43. [DOI] [PubMed] [Google Scholar]
- 16. Elkin PL, et al. Secondary use of clinical data. Stud Health Technol Inform 2010;155:14–29. [PubMed] [Google Scholar]
- 17. Farrow G, et al. Protocol for the examination of specimens from patients with carcinomas of renal tubular origin, exclusive of Wilms tumor and tumors of urothelial origin: a basis for checklists. Cancer Committee, College of American Pathologists. Arch Pathol Lab Med 1999;123:23–27. [DOI] [PubMed] [Google Scholar]
- 18. Fonseca F, et al. Semantic granularity in ontology-driven geographic information systems. Ann Math Artif Intell 2002;36:121–151. [Google Scholar]
- 19. Fries JF. Alternatives in medical record formats. Med Care 1974;12:871–881. [DOI] [PubMed] [Google Scholar]
- 20. Fung KW, et al. Integrating SNOMED CT into the UMLS: an exploration of different views of synonymy and quality of editing. J Am Med Inform Assoc 2005;12:486–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gephardt GN, et al. Lung carcinoma surgical pathology report adequacy: a College of American Pathologists Q-probes study of over 8300 cases from 464 institutions. Arch Pathol Lab Med 1996;120:922–927. [PubMed] [Google Scholar]
- 22. Giannangelo K, et al. Mapping SNOMED CT to ICD-10. Stud Health Technol Inform 2012;180:83–87. [PubMed] [Google Scholar]
- 23. Gouveia-Oliveira A, et al. Longitudinal comparative study on the influence of computers on reporting of clinical data. Endoscopy 1991;23:334–337. [DOI] [PubMed] [Google Scholar]
- 24. Gu H, et al. Quality assurance of UMLS semantic type assignments using SNOMED CT hierarchies. Methods Inf Med 2016;55:158–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heiler S. Semantic interoperability. ACM Computing Surveys (CSUR) 1995;27:271–273. [Google Scholar]
- 26. Henry WL, et al. Report of the American Society of Echocardiography Committee on Nomenclature and Standards in two-dimensional echocardiography. Circulation 1980;62:212–217. [DOI] [PubMed] [Google Scholar]
- 27. Idowu MO, et al. Adequacy of surgical pathology reporting of cancer: a College of American Pathologists Q-probes study of 86 institutions. Arch Pathol Lab Med 2010;134:969–974. [DOI] [PubMed] [Google Scholar]
- 28. Jensen PB, et al. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet 2012;13:395–405. [DOI] [PubMed] [Google Scholar]
- 29. Johnson SB, et al. An electronic health record based on structured narrative. J Am Med Inform Assoc 2008;15:54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kelton DF, et al. Recommendations for recording and calculating the incidence of selected clinical diseases of dairy cattle. J Dairy Sci 1998;81:2502–2509. [DOI] [PubMed] [Google Scholar]
- 31. Kempson RL. The time is now. Checklists for surgical pathology reports. Arch Pathol Lab Med 1992;116:1107–1108. [PubMed] [Google Scholar]
- 32. King S, et al. Structured pathology reporting improves the macroscopic assessment of rectal tumour resection specimens. Pathology 2016;48:349–352. [DOI] [PubMed] [Google Scholar]
- 33. Koopman B, et al. Towards semantic search and inference in electronic medical records: an approach using concept-based information retrieval. Australas Med J 2012;5:482–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee D, et al. Literature review of SNOMED CT use. J Am Med Inform Assoc 2014;21:e11–e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Leslie KO, et al. Standardization of the surgical pathology report: formats, templates, and synoptic reports. Semin Diagn Pathol 1994;11:253–257. [PubMed] [Google Scholar]
- 36. Liaw ST, et al. Health reform: is routinely collected electronic information fit for purpose? Emerg Med Australas 2012;24:57–63. [DOI] [PubMed] [Google Scholar]
- 37. Lindley SW, et al. Communicating diagnostic uncertainty in surgical pathology reports: disparities between sender and receiver. Pathol Res Pract 2014;210:628–633. [DOI] [PubMed] [Google Scholar]
- 38. Mann R, et al. Standards in medical record keeping. Clin Med (Lond) 2003;3:329–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Matheny ME, et al. Detection of infectious symptoms from VA emergency department and primary care clinical documentation. Int J Med Inform 2012;81:143–156. [DOI] [PubMed] [Google Scholar]
- 40. McDonald CJ, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin Chem 2003;49:624–633. [DOI] [PubMed] [Google Scholar]
- 41. McDonald CJ, et al. Computer-stored medical records. Their future role in medical practice. J Am Med Assoc 1988;259:3433–3440. [PubMed] [Google Scholar]
- 42. McKinley M, et al. Observations on the application of the Papanicolaou Society of Cytopathology standardised terminology and nomenclature for pancreaticobiliary cytology. Pathology 2016;48:353–356. [DOI] [PubMed] [Google Scholar]
- 43. Miller PR. Inpatient diagnostic assessments: 2. Interrater reliability and outcomes of structured vs. unstructured interviews. Psychiatry Res 2001;105:265–271. [DOI] [PubMed] [Google Scholar]
- 44. Miller PR. Inpatient diagnostic assessments: 3. Causes and effects of diagnostic imprecision. Psychiatry Res 2002;111:191–197. [DOI] [PubMed] [Google Scholar]
- 45. Miller PR, et al. Inpatient diagnostic assessments: 1. Accuracy of structured vs. unstructured interviews. Psychiatry Res 2001;105:255–264. [DOI] [PubMed] [Google Scholar]
- 46. Montoliu-Fornas G, et al. Magnetic resonance imaging structured reporting in infertility. Fertil Steril 2016;105:1421–1431. [DOI] [PubMed] [Google Scholar]
- 47. Nadkarni PM, et al. Migrating existing clinical content from ICD-9 to SNOMED. J Am Med Inform Assoc 2010;17:602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nguyen A, et al. Classification of pathology reports for cancer registry notifications. Stud Health Technol Inform 2012;178:150–156. [PubMed] [Google Scholar]
- 49. Oberhuber G, et al. The histopathology of coeliac disease: time for a standardized report scheme for pathologists. Eur J Gastroenterol Hepatol 1999;11:1185–1194. [DOI] [PubMed] [Google Scholar]
- 50. Powsner SM, et al. Clinicians are from Mars and pathologists are from Venus: clinician interpretation of pathology reports. Arch Pathol Lab Med 2000;124:1040–1046. [DOI] [PubMed] [Google Scholar]
- 51. Richesson RL, et al. Use of SNOMED CT to represent clinical research data: a semantic characterization of data items on case report forms in vasculitis research. J Am Med Inform Assoc 2006;13:536–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rosai J. Standardized reporting of surgical pathology diagnoses for the major tumor types. A proposal. The Department of Pathology, Memorial Sloan-Kettering Cancer Center. Am J Clin Pathol 1993;100:240–255. [DOI] [PubMed] [Google Scholar]
- 53. Rosenbloom ST, et al. Data from clinical notes: a perspective on the tension between structure and flexible documentation. J Am Med Inform Assoc 2011;18:181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rosse C, et al. A reference ontology for biomedical informatics: the foundational model of anatomy. J Biomed Inform 2003;36:478–500. [DOI] [PubMed] [Google Scholar]
- 55. Ryu KH, et al. Cervical lymph node imaging reporting and data system for ultrasound of cervical lymphadenopathy: a pilot study. Am J Roentgenol 2016;206:1286–1291. [DOI] [PubMed] [Google Scholar]
- 56. Santamaria SL, et al. Uses of informatics to solve real world problems in veterinary medicine. J Vet Med Educ 2011;38:103–109. [DOI] [PubMed] [Google Scholar]
- 57. Schulz S, et al. SNOMED reaching its adolescence: ontologists’ and logicians’ health check. Int J Med Inform 2009;78(Suppl 1):S86–S94. [DOI] [PubMed] [Google Scholar]
- 58. Schweitzer W, et al. Virtopsy approach: structured reporting versus free reporting for PMCT findings. J Forensic Radiol Imag 2014;2:28–33. [Google Scholar]
- 59. Scolyer RA, et al. Pathology of melanocytic lesions: new, controversial, and clinically important issues. J Surg Oncol 2004;86:200–211. [DOI] [PubMed] [Google Scholar]
- 60. Smith-Akin KA, et al. Toward a veterinary informatics research agenda: an analysis of the PubMed-indexed literature. Int J Med Inform 2007;76:306–312. [DOI] [PubMed] [Google Scholar]
- 61. Spasic I, et al. Text mining of cancer-related information: review of current status and future directions. Int J Med Inform 2014;83:605–623. [DOI] [PubMed] [Google Scholar]
- 62. Srigley JR, et al. Standardized synoptic cancer pathology reporting: a population-based approach. J Surg Oncol 2009;99:517–524. [DOI] [PubMed] [Google Scholar]
- 63. Strauss JA, et al. Identifying primary and recurrent cancers using a SAS-based natural language processing algorithm. J Am Med Inform Assoc 2013;20:349–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Van der Meijden MJ, et al. Development and implementation of an EPR: how to encourage the user. Int J Med Inform 2001;64:173–185. [DOI] [PubMed] [Google Scholar]
- 65. Wade G, et al. Experiences mapping a legacy interface terminology to SNOMED CT.BMC Med Inform Decis Mak 2008;8(Suppl 1):S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Washabau R, et al. Endoscopic, biopsy, and histopathologic guidelines for the evaluation of gastrointestinal inflammation in companion animals. J Vet Intern Med 2010;24:10–26. [DOI] [PubMed] [Google Scholar]
- 67. Whiting-O’Keefe QE, et al. A computerized summary medical record system can provide more information than the standard medical record. J Am Med Assoc 1985;254:1185–1192. [PubMed] [Google Scholar]
- 68. Wilson GA, et al. The effect of immediate access to a computerized medical record on physician test ordering: a controlled clinical trial in the emergency room. Am J Public Health 1982;72:698–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zaninelli M, et al. The O3-Vet project: integration of a standard nomenclature of clinical terms in a veterinary electronic medical record for veterinary hospitals. Comput Methods Programs Biomed 2012;108:760–772. [DOI] [PubMed] [Google Scholar]
- 70. Zarbo RJ. Interinstitutional assessment of colorectal carcinoma surgical pathology report adequacy. A College of American Pathologists Q-probes study of practice patterns from 532 laboratories and 15,940 reports. Arch Pathol Lab Med 1992;116:1113–1119. [PubMed] [Google Scholar]
- 71. Zhang YF, et al. Integrating HL7 RIM and ontology for unified knowledge and data representation in clinical decision support systems. Comput Methods Programs Biomed 2016;123:94–108. [DOI] [PubMed] [Google Scholar]
- 72. Zimmerman KL, et al. SNOMED representation of explanatory knowledge in veterinary clinical pathology. Vet Clin Pathol 2005;34:7–16. [DOI] [PubMed] [Google Scholar]
- 73. Zuccon G, et al. The impact of OCR accuracy on automated cancer classification of pathology reports. Stud Health Technol Inform 2012;178:250–256. [PubMed] [Google Scholar]