Abstract
Fungal infections can lead to severe clinical outcomes such as multiple organ failure and septic shock. Rapid detection of fungal infections allows clinicians to treat patients in a timely manner and improves clinical outcomes. Conventional detection methods include blood culture followed by plate culture and polymerase chain reaction. These methods are time-consuming and require expensive equipment, hence, they are not suitable for point-of-care and clinical settings. There is an unmet need to develop a rapid and inexpensive detection method for fungal infections such as candidemia. We developed an innovative immuno-based microfluidic device that can rapidly detect and capture Candida albicans from phosphate-buffered saline (PBS) and human whole blood. Our microchip technology showed an efficient capture of C. albicans in PBS with an efficiency of 61–78% at various concentrations ranging from 10 to 105 colony-forming units per milliliter (cfu/mL). The presented microfluidic technology will be useful to screen for various pathogens at the point-of-care and clinical settings.
1. Introduction
Candida albicans (C. albicans) are human commensals of genitourinary and gastrointestinal tracts, and skin.1 However, C. albicans is an opportunistic fungal pathogen that can cause invasive fungal infections.2,3 Mortality associated with C. albicans infection is greater than 50% making Candida a leading cause of healthcare-associated bloodstream infections in the United States.3 Neutrophils are an essential element of the innate immune system. Low count of neutrophils in blood (neutropenia) and its lengthy and repeated episodes provide favorable conditions for Candida invasion, especially in immunocompromised patients.4 Because of the lack of rapid diagnosis, these bloodstream infections require prolonged hospital stays that significantly increase treatment and hospitalization costs.3,5,6
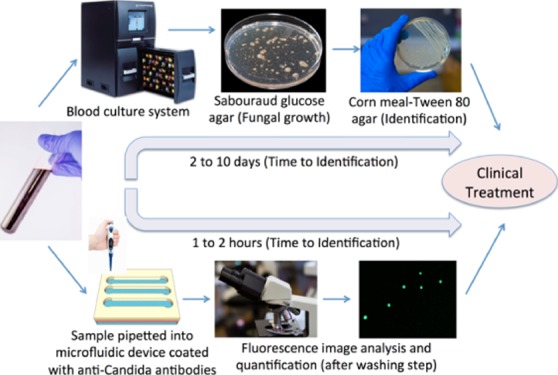
Rapid detection of Candida is urgently needed, especially in the cases of preterm neonates and immunocompromised patients; candidemia can lead to multiple organ failure and septic shock.7−9 Conventionally, in hospital settings, about 10 mL of blood from the patients is cultured in blood culture incubators using specific media to culture either aerobic or anaerobic organism (Figure 1).5 If the blood culture is positive, then pathogen identification can be determined by follow-up pathogen cultures including Sabouraud glucose agar and cornmeal agar.5 Chromogenic medium-based culture can further improve Candida differentiation as the medium contains chromogenic substrates that react with enzymes produced by different pathogens and produce colonies of varying colors and morphologies.10 Additionally, carbohydrate assimilation and fermentation reactions can be used for Candida identification, however, the test takes up to 72 h incubation. Overall, the conventional laboratory methods for Candida detection are highly time-consuming, which delays patient treatment and can take 2–10 days.
Figure 1.
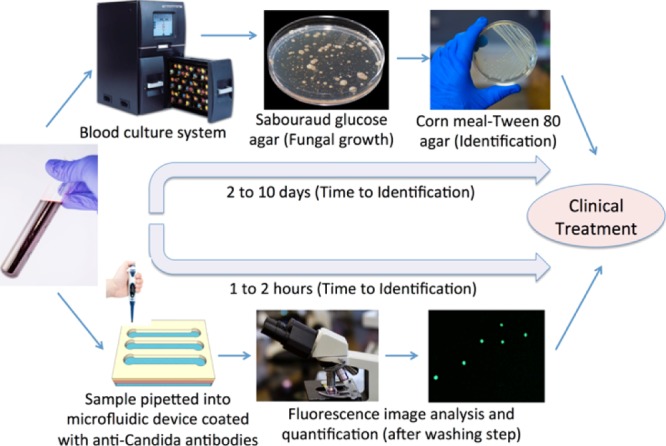
Comparison of the conventional culture method and the microchip-based C. albicans detection. In conventional procedure for Candida detection in clinical settings, the following protocol is followed; (1) blood sample is collected from patients. (2) Blood samples are incubated in an automated blood culture system. (3) Pathogens are grown on a Sabouraud glucose agar plate. (4) Sample is subcultured into a Corn meal-Tween 80 agar plate for morphological identification. In the point-of-care testing approach, the following protocol is developed; (1) blood sample collection [spiked with green fluorescent protein (GFP)-expressing Candida as a model microorganism]. (2) Blood sample is analyzed in microchannels functionalized with anti-Candida antibodies. Candida cells were specifically captured by antibodies on the microchannel surface. Unbound Candida cells are washed away with PBS in the washing step. (3) GFP-Candida is imaged and quantified under a fluorescence microscope. Some images are taken from freerangestock.com and pixabay.com.
It is important to rapidly diagnose fungal infections with high accuracy to initiate timely treatment. The nucleic acid real-time polymerase chain reaction (PCR), nucleic acid sequence-based amplification (NASBA), and loop-mediated isothermal amplification (LAMP) methods take around 3–6 h for detection of C.albicans. Microfluidic devices have emerged as a potential candidate for rapid disease diagnostics in the current era.11−19 Although, microfluidic real-time PCR provides a rapid Candida detection,20 it relies on thermal cycles and the effective monitoring and control of various temperatures during experimentation are quite challenging. The fluidic manipulations and utilization of magnetic beads also increase the complexity of the devices. Table 1 shows comparison of different Candida detection technologies.
Table 1. Comparison of Developed Technology with Existing Methods of Candida Detection.
| method | sensitivity | assay time | limit of detection | description |
|---|---|---|---|---|
| blood culture-based detection | 42–60%21,22 | 1–7 days | 1 cfu/mL23 | it is time consuming (takes several days) and labor-intensive method that may delay treatment options. Blood culture-based methods require large volumes of blood |
| PCR | 85–90%24 | 4–6 h | ≤10 cfu/mL23 | PCR is very sensitive method and provides low limit of detection. This method requires breaking of fungal cell wall. The difficulty in breaking Candida cell wall makes the DNA extraction step quite difficult. Furthermore, PCR is highly temperature dependent process, thus requires strict temperature control and thermal cycling, which makes PCR unsuitable for POC settings |
| NASBA | higher than 90% | ∼2–3 h | 1–10 cfu/mL25,26 | it is an isothermal amplification process that eliminates the need of thermocycler.25 It reduces the Candida detection time as compared to PCR and other culture-based methods. NASBA has quicker nucleic acid extraction as compared to PCR based methods.27 As this method requires nucleic acid extraction and purification, hence has limited application in POC settings |
| LAMP | similar to NASBA and PCR method28 | ∼2–3 h | similar to NASBA and PCR29 | LAMP provides a cost-effective and rapid isothermal amplification method for the detection of C. albicans.30 Similarly to NASBA, it requires nucleic acid extraction and purification step, hence has limited application in POC settings |
| T2Candida | 91.6%31 | 3–5 h | 1–3 cfu/mL | it can rapidly detect the C. albicans from blood samples. The detection time is 3–5 h. However the system is costly |
| Developed method | not tested with clinical samples | ∼2 h | 10–105 cfu/mL | the developed assay can be used as a cost-effective and rapid Candida detection method at POC as well as clinical settings. It requires only 50 μL sample volume and up to 2 h maximum for detection of Candida. It can also work with larger volumes with extended assay times. |
To overcome the limitations of conventional culture-based Candida detection methods, we developed an inexpensive microfluidic device functionalized with antibodies to C. albicans. This technology allows for rapid detection, capture, and isolation of C. albicans in PBS and blood in an efficient manner. Low sample consumption because of the higher surface to volume ratio of blood within the microchannels supports rapid capturing of targeted pathogens. Our device overcomes the limitation of long incubation times. We demonstrated that C. albicans can be detected within 2 h at a minimum of 10 cfu/mL.
2. Materials and Methods
2.1. Microfluidic Device Fabrication
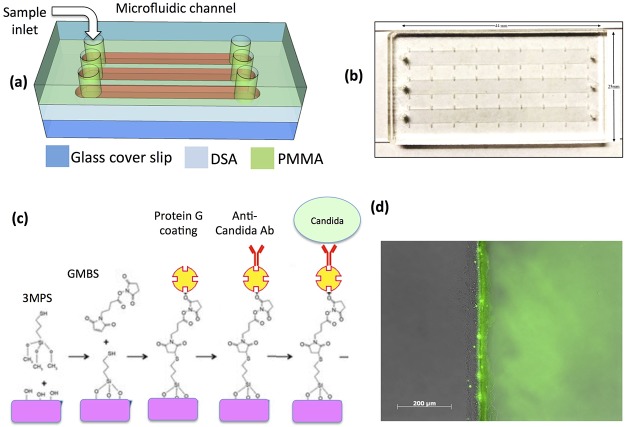
The microfluidic device was fabricated by using plastics layers and polymer adhesives as previously reported (Figure 2a,b).17 The design for the device was created in AutoCAD 2015 and uploaded to the UCP Software for cutting the device using a laser cutter. Poly(methyl methacrylate) (PMMA) (McMaster-Carr, Atlanta, GA and ePlastics, San Diego, CA 1.5 mm thick) and the double-sided adhesive (DSA) (3 M, St. Paul, MN, 76 μm thick) were cut using a VLS 2.30 laser cutter (VersaLaser, Scottsdale, AZ). In each microfluidic device, three parallel channels (dimensions: 44 mm × 5 mm × 76 μm) were cut in DSA. One side of the DSA film was attached to glass cover slide [70% ethanol in distilled (DI) water and dried by nitrogen gas], whereas other side was attached to a PMMA. Three inlet and three outlet holes (0.65 mm diameter) were also cut in PMMA before assembly. The complete assembled device is shown in (Figure 2b).
Figure 2.
(a) Schematic of the microfluidic chip consisting of PMMA, DSA, and glass cover. (b) Actual image of the assembled microchip containing three microfluidic channels, inlets, and outlets. (c) Protein G-based surface chemistry was used to immobilize anti-Candida antibodies on the surface. (d) Fluorescent image showing clear green signal (right side of microchannel) that indicates successful immobilization of fluorescein isothiocyanate (FITC)-conjugated protein.
2.2. Microfluidic Channel Surface Functionalization with Antibodies
Protein G-based surface chemistry was used to immobilize antibodies (Figure 2c). Protein G is an immunoglobulin-binding protein that binds to the fragment crystallization region of antibodies with high efficiency. For surface functionalization, glass cover slide was cleaned with 70% ethanol in DI water and dried by nitrogen gas. Glass cover slide was then treated with oxygen plasma (100 W, 1% oxygen) for 2 min in a PX-250 chamber (March instruments, Concord, MA) to form the hydroxyl (OH) surface functional groups followed by a 30 min incubation with silanization solution [4% (v/v) 3-MPS ((3-mercaptopropyl)trimethoxysilane, CN: 175617)) in ethanol] in a Petri dish at room temperature for covalent binding. After incubation, the cover slide was washed with ethanol and was allowed to dry for 3–4 min at room temperature. The microfluidic device was assembled by sandwiching DSA between PMMA and cover slide. Channels were washed 3 times with PBS. GMBS (N-g-maleimidobutyryloxy succinimide ester) solution (4% (w/v) GMBS dissolved in 10% DMSO in PBS) was pipetted into microfluidic channels. Devices were incubated for 30 min at room temperature. From now onward, channels were washed 3 times with PBS after each incubation step. Then Protein G (1 mg/mL in PBS, Thermo Fisher Scientific) was pipetted into microfluidic channels followed by 2 h incubation at 4 °C. For capturing C. albicans, we tested two different anti-Candida antibodies; one was monoclonal (Abcam, ab82704) and the second was polyclonal (Thermo Fisher Scientific, Catalog: PA1-27158). Then 30 μL (5 μg/mL solution) of each anti-C. albicans antibody was pipetted into each microchannel followed by a 1 h incubation at room temperature. Microchannels were washed with PBS 3 times. Then 2% (w/v) bovine serum albumin in PBS was injected into microchannels followed by 30 min incubation at room temperature and subsequent washing with PBS was performed. The devices were ready for Candida capture experiments.
2.3. C. albicans Strain and Growth
To validate the surface chemistry and isolation experiments, a genetically modified C. albicans, SC5314, expressing GFP was used.32C. albicans was grown to the log phase in yeast extract–peptone–dextrose medium overnight at 30 °C in a shaker incubator at 250 rpm.33 Yeast was harvested, washed, and resuspended in PBS and blood for use. The initial Candida count was determined first by haemocytometer. Then, C. albicans were thoroughly mixed with PBS/blood to obtain a homogeneous solution. After Candida capture experiments, counting was performed manually. The counting approach was optimized (Figure S1). Capture efficiency was calculated by dividing the number of Candida cells captured by number of Candida cell spiked. Only 50 μL of the Candida-spiked sample was used for these experiments. Candida counts were normalized to the sample volume used.
2.4. Sample Preparation for Microfluidic Experiments
GFP-expressing C. albicans (GFP-Candida) was spiked into 1× PBS and whole blood with the final concentrations ranging from 10 to 5 × 105 cfu/mL for analysis on the chip. Discarded deidentified whole blood (purchased from Research Blood Components, LLC, Cambridge, MA) from healthy individuals was used in this study. For lysed blood experiments, we did the following: (1) GPF-Candida cells were spiked into whole blood at 5 × 105 cfu/mL and mixed thoroughly by inverting to enable homogenous distribution. (2) The spiked blood sample was mixed with ACK (ammonium–chloride–potassium) lysis buffer at 1:10 ratio (v/v) (Thermo Fisher Scientific, A1049201) and incubated for 3 min at room temperature. Candida cells remained intact in ACK lysis buffer. This is primarily because of the chitin found in their cell wall.34 (3) Centrifugation at 3000 rpm for 3 min was performed; Candida cell remained intact and made a pellet. (4) Supernatant was aspirated, leaving approximately 50 μL to avoid disturbing the pellet. Then, 5 mL of PBS was added followed by 3000 rpm centrifugation (3 min) and the supernatant was aspirated again leaving approximately 50 μL of the sample including pellet. (5) The pellet was disturbed with a pipette and mixed gently. The complete blood lysis and Candida enrichment process take about 10 min.
2.5. Operation of Microfluidic Experiments, Candida Capture, and Quantification
To optimize the capture efficiency, 50 μL of the GFP-Candida sample was pipetted into the functionalized microchannels, and then incubated at ambient temperature for 15 min. Following the incubation, microchannels were washed with PBS at a flow rate of 5 or 10 μL/min using a syringe pump (Harvard Apparatus, Holliston, MA) for 60 min. After washing, captured GFP-Candida was imaged using an inverted fluorescence microscope (Zeiss Observer optical microscope) through a GFP fluorescence filter (excitation wavelength 470 nm). For comparison, bright-field images were also taken (Figure 5c). All images were taken (Figure 5c and 5d) with 10× objective except Figure 5e (100× objective). The number of GFP-Candida detected using a GFP filter was counted manually.
Figure 5.
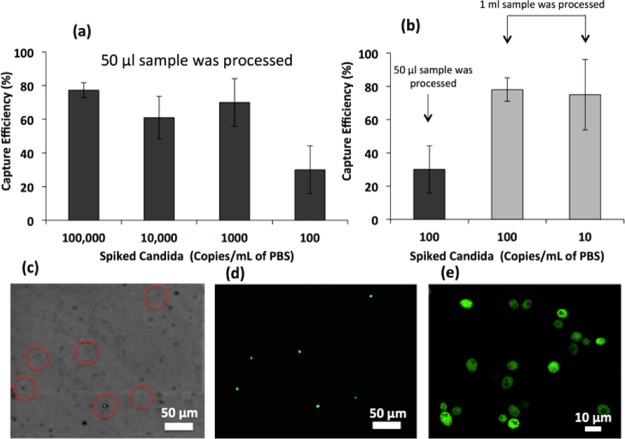
(a) Capture efficiency of C. albicans cells inside microfluidic channels functionalized with polyclonal antibodies at various concentrations ranging from 100 to 100 000 cfu/mL. We processed 50 μL of spiked PBS. (b) Capture efficiency of Candida was increased when 1 mL of the spiked PBS sample was processed by injecting 50 μL sample multiple times in the same channel followed by incubation after each injection. (c) Image of the captured GFP-Candida inside microchannel at 10× magnification under bright field. (d) Image of the captured GFP-Candida inside microchannel at 10× magnification under a fluorescence microscope. (e) Image of the captured GFP-Candida at 100× magnification under a fluorescence microscope.
2.6. Statistical Analysis
Statistical analysis was performed using one-way analysis of variance (ANOVA). Each experiment was repeated at least three times. A p-value of less than 0.05 was considered statistically significant.
3. Results
We developed a microfluidic device having 3 microchannels functionalized with protein G-based surface chemistry (Figure 2). The microfluidic channels provide a high surface to volume ratio that would allow efficient Candida capture on the surface of the channel. 3MPS–GMBS (3-mercaptopropyl trimethoxysilane-N-g-maleimidobutyryloxy succinimide ester)-based surface chemistry was used to immobilize protein on the microchannels of the device as previously reported.17 The reaction between the amine group and GMBS allow protein immobilization. To determine protein conjugation to the surface of microchannels, we incubated FITC-conjugated protein inside microchannel after the GMBS step. The channels were washed with PBS after 2 h of incubation at 4 °C. We visualized fluorescence using the fluorescent microscope, which showed that protein was successfully immobilized inside microchannels (Figure 2d).
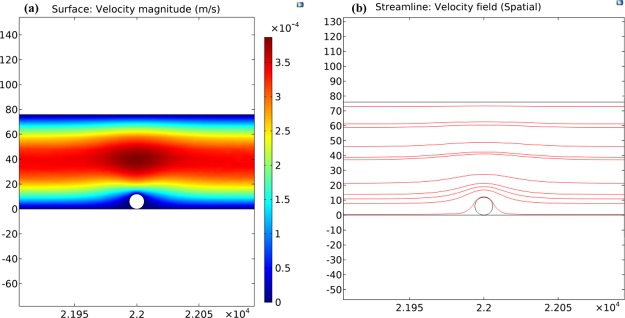
COMSOL simulations were performed to determine the effects of shear stress on the C. albicans captured inside the microfluidic device. A single Candida cell was modeled as a spherical-shaped object (radius 6 μm). The microfluidic device with exact dimensions was considered and a laminar flow was assumed. The no slip boundary condition was applied to the walls of the microfluidic channel. Two flow rates (5 and 10 μL/min) were assumed. A boundary condition with pressure = 0 was set for the outlet. The Naiver–Stokes equation was used to simulate the motion of fluid past the captured Candida. Various simulations were carried out to calculate the velocity and pressure profiles. The velocity magnitude and streamlines are shown in Figure 3a,b, respectively. The shear stress was measured for the two flow rates (5 and 10 μL/min) and drag force was also calculated. The value of drag force was 20.47 pN for 5 μL/min. When the flow rate was increased to 10 μL/min, drag force became 40.96 pN. Higher flow rates resulted in increased drag force on the captured Candida. This increase in drag force reduces the capture efficiency of Candida as also observed in experiments (Figure 4).
Figure 3.
COMSOL simulation results for 5 μL/min. (a) Simulated flow velocity representation inside the microfluidic device. (b) Streamlines for the velocity field.
Figure 4.
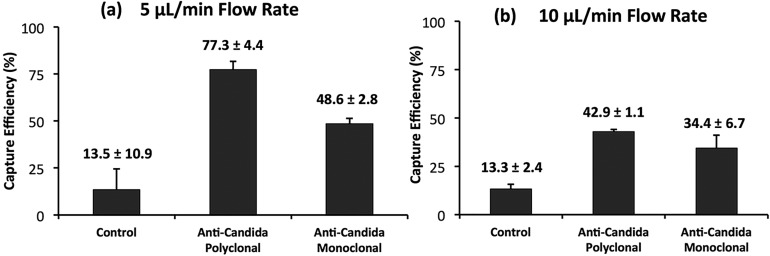
Comparison of the capture efficiency of C. albicans by two different anti-Candida antibodies; polyclonal and monoclonal. The Candida sample was incubated inside microchannels to allow binding with a functionalized surface for 15 min at room temperature. (a) Capture efficiency was significantly higher when the washing step was performed at (a) 5 μL/min as compared to (b) 10 μL/min.
We evaluated two different antibodies (monoclonal and polyclonal) for their efficiency to capture C. albicans from spiked samples inside microfluidic channels. The capture efficiency via polyclonal anti-Candida antibodies (77.4 ± 4.4%) was observed to be significantly higher (p-value < 0.05) than monoclonal anti-Candida antibodies (48.6 ± 2.8%) (Figure 4a). For all further experiments, we used polyclonal antibody because of its higher capture efficiency compared to monoclonal.
We observed that the 5 μL/min flow rate (during washing) gave significantly higher capture efficiencies compared to 10 μL/min (Figure 4). The lower efficiency observed at higher flow rates may be related to the correspondingly higher shear stress within the microchannels as also shown in simulation graphs (Figure 3). For all further experiments, we used a flow rate of 5 μL/min.
To determine microchip’s limit of detection for Candida capture, we spiked GFP-Candida into PBS at various clinically relevant concentrations ranging from 10 to 105 cfu/mL (Figure 5).35−37
In the first set of experiments, we tested only 50 μL of the spiked PBS sample. We observed similar capture efficiencies of 77.4 ± 4.4, 61 ± 12.7, and 70 ± 13.2% for 105, 104, and 103 cfu/mL samples respectively, however, capture efficiency was reduced to 30 ± 14.2% for the 102 cfu/mL sample. One possible reason for lower capture efficiency in the case of the 102 cfu/mL sample may have originated from losing Candida during sampling from 1 mL to 50 μL. To investigate this hypothesis, we tested whole 1 mL of spiked samples and observed higher capture efficiencies of 78 ± 13.2 and 75 ± 21.1% for 102 and 10 cfu/mL samples (Figure 5b). From these results, we observed that increasing the sample volume resulted in increase in capture efficiencies at lower concentration samples (10 and 102 cfu/mL).
To further investigate the effect of the sample matrix and the presence of other cells on the capture efficiency, we spiked 105 cfu/mL GFP-Candida into whole human blood and processed the sample using the microfluidic device. We observed the capture efficiency of 40.5 ± 4.7% from blood samples (Figure 6), which was significantly lower than when spiked PBS was used 77.4 ± 4.4%. This decrease in capture efficiency from the blood sample may be due to the presence of millions of blood cells that hindered Candida–antibody interactions. To overcome the effect of blood cells, we lysed the spiked blood sample and isolated the pellet as described in the Materials and Methods section. The pellet containing Candida cells was mixed and processed through the microfluidic device. We observed significantly higher capture efficiency of 74.6 ± 6.8% compared to the spiked whole blood sample.
Figure 6.
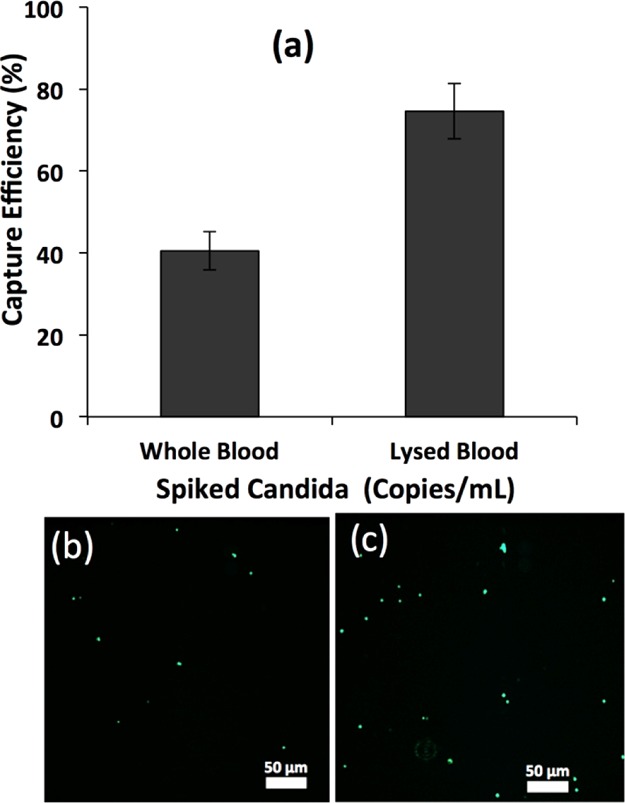
(a) Capture efficiency of GFP-C. albicans cells inside microfluidic channels functionalized with polyclonal antibodies from 16 μL of whole and lysed blood. Blood was lysed after spiking GFP-Candida. (b) Image of the captured GFP-Candida from whole blood inside the microchannel at 10× magnification under a fluorescence microscope. (c) Image of the captured GFP-Candida from lysed blood inside microchannel at 10× magnification under a fluorescence microscope. GFP-Candida was spiked into blood at the concentration of 105 cfu/mL.
To show that the developed microfluidic device can be used to capture unstained Candida from samples, unstained Candida spiked in buffer (not expressing GFP) was utilized. Precapture and postcapture staining were performed with FITC conjugated anti-Candida antibody (ab21164). The results are shown in the Figure S2. C. albicans not producing GFP were initially captured using the polyclonal antibody. Then staining was performed with FITC-conjugated anti-Candida antibody. The capture efficiency was recorded 68.8 ± 6.8%. It was observed that precapture staining resulted in lower capture efficiency, that is, 43.2 ± 4.5%. One possible reason can be the blocking of some capture sites of the Candida strains. The C. albicans captured by monoclonal antibody in the microfluidic channel were also stained with the secondary antibody. The postcapture staining resulted in a capture efficiency of 29.6 ± 5.7%. The precapture staining resulted in slightly decreased capture efficiency 22.4 ± 4.5%.
4. Discussion
Using the developed microfluidic device, we were able to efficiently isolate and quantify C. albicans from spiked PBS and whole blood sample. The whole capture experiment takes about 1.5 h (in the case of PBS and whole blood) and less than 2 h (in the case of blood lysis protocol). Following blood lysis protocol developed herein, we were able to capture 74.6 ± 6.8% of Candida from blood samples in significantly lesser time (2 h) as compared to conventional blood culture followed by plate culture methods (more than a week) (Figure 1). There are other existing methods that can detect Candida at lesser time than conventional culture methods including PCR,38 magnetic resonance (T2Candida by T2 Biosystems),3,39 and isothermal amplification methods such as LAMP,40,41 NASBA,42 and rolling circle amplification.43 PCR and isothermal amplification-based detection provide high specificity and sensitivity; however, these approaches are multistep, require sample purification, and nucleic acid extraction prior to the detection step, a time-consuming process. More importantly, during nucleic acid amplification Candida cells are lysed, hence viability of Candida cannot be tested and drug resistance and susceptibility cannot be analyzed. The T2Candida system utilizes magnetic resonance to detect aggregation of magnetic particles in the presence of the target.39 T2Candida also relies on amplification of genetic information where nucleic acids are first isolated and purified from Candida cells using beads; however, T2Candida is a sample-to-answer system where all the processing steps are automated. This system provides high sensitivity and specificity and detection can be performed in 3–5 h.3 However, similar to other nucleic acid-based detection systems, Candida cells are lysed in T2Candida and drug resistance and susceptibility testing cannot be performed, which are becoming very important for personalized therapy. The developed microfluidic device can address some of these limitations by allowing whole Candida capture directly from the lysed blood sample as Candida cells are not lysed during the isolation step, hence culture and drug resistance and susceptibility testing may be possible.
Although blood is a complex biological matrix; we have successfully captured and detected spiked Candida using the microfluidic approach. The presented method can also be applied to other matrices such as saliva and urine. In the case of urinary tract infection, high concentration of Candida cells (104 to 105 cfu/mL)37 may be present in urine that can be rapidly isolated and quantified using the developed microfluidic devices. In this study, GFP-expressing C. albicans was used to facilitate the detection and quantification under the fluorescent microscope. The extension from a GFP-expressing Candida strain to wild-type strains can be simply achieved using an ELISA or peptide nucleic acid-based fluorescence in situ hybridization (PNA-FISH)-based detection method, as previously reported.44−46 The characterized microchip can be integrated with rapid detection methods such as lensless imaging17,47 and smartphone-based imaging48−50 to enable point-of-care testing. The presented microfluidic approach can be adapted to detect other microorganisms that cause sepsis such as Gram-negative and Gram-positive bacteria.51 Also, the cost to fabricate one functionalized chip is significantly lower than other assays (PCR, T2Candida); current material cost (excluding other related costs such as labor and equipment cost) includes 10¢ of glass, 1¢ of PMMA, and 80¢ of antibodies per device. The antibody cost could be lowered with large-scale production and ordering.
5. Conclusions
To overcome the limitations of culture-based detection methods for fungal infections, we have developed a microfluidic immunoassay to capture C. albicans (a Yeast) from PBS and blood samples with reliable capture efficiency. We observed that polyclonal antibody captured a significantly high number of Candida cells as compared to monoclonal antibody. The washing flow rate can also affect final capture efficiency, and we observed that a flow rate of 5 μL/min provides higher capture efficiency as compared to 10 μL/min. To enable efficient detection of Candida from blood samples, the lysis step was used that significantly improved the capture efficiency from whole blood samples. The presented technology allows the capture and isolation of whole Candida cells, hence enabling potentially drug resistance and susceptibility testing. The microfluidic platform can be potentially adapted to detect various other microorganisms and pathogens rapidly at the point-of-care settings.
Acknowledgments
We acknowledge Carly Boltin for her help in proofreading the manuscript and providing helpful comments. We also acknowledge Saad Shaukat (Massachusetts Institute of Technology) for his help in microfluidic experiments. We acknowledge research support from NIH R15AI127214, Institute for Sensing and Embedded Networking Systems Engineering (I-SENSE) Research Initiative Award, FAU Faculty Mentoring Award, Humanity in Science Award, and a start-up research support from College of Engineering and Computer Science, Florida Atlantic University, Boca Raton, FL. Dr. Demirci would like to acknowledge CIMIT U54 1U54EB015408-01.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.9b00499.
Candida quantification process and Candida capture efficiency (PDF)
The authors declare the following competing financial interest(s): U.D. is a founder of, and has an equity interest in (i) DxNow Inc., a company that is developing microfluidic and imaging technologies, (ii) Koek Biotech, a company that is developing microfluidic IVF technologies for clinical solutions, and (iii) LEVITAS Inc., a company that develops biotechnology tools for genomic analysis in cancer. U.D.' s interests were viewed and managed in accordance with the conflict of interest policies. All other authors declare no conflict of interest.
Supplementary Material
References
- Murray P. R.; Rosenthal K. S.; Pfaller M. A., Medical Microbiology, 7th ed.; Elsevier Saunders: Philadelphia, PA, 2013; pp 621–630; 677–683. [Google Scholar]
- Zilberberg M. D.; Shorr A. F.; Kollef M. H. Secular Trends in Candidemia-Related Hospitalization in the United States, 2000-2005. Infection Control and Hospital Epidemiology 2008, 29, 978–980. 10.1086/591033. [DOI] [PubMed] [Google Scholar]
- Safavieh M.; Coarsey C.; Esiobu N.; Memic A.; Vyas J. M.; Shafiee H.; Asghar W. Advances in Candida detection platforms for clinical and point-of-care applications. Crit. Rev. Biotechnol. 2017, 37, 441–458. 10.3109/07388551.2016.1167667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas P. G. Opportunistic fungi: a view to the future. Am. J. Med. Sci. 2010, 340, 253–257. 10.1097/maj.0b013e3181e99c88. [DOI] [PubMed] [Google Scholar]
- Hall K. K.; Lyman J. A. Updated review of blood culture contamination. Clin. Microbiol. Rev. 2006, 19, 788–802. 10.1128/cmr.00062-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan J.; Meltzer M. I.; Plikaytis B. D.; Sofair A. N.; Huie-White S.; Wilcox S.; Harrison L. H.; Seaberg E. C.; Hajjeh R. A.; Teutsch S. M. Excess mortality, hospital stay, and cost due to candidemia: a case-control study using data from population-based candidemia surveillance. Infect. Control 2005, 26, 540–547. 10.1086/502581. [DOI] [PubMed] [Google Scholar]
- Jarvis W. R. Epidemiology of nosocomial fungal infections, with emphasis on Candida species. Clin. Infect. Dis. 1995, 20, 1526–1530. 10.1093/clinids/20.6.1526. [DOI] [PubMed] [Google Scholar]
- Jarvis W. R.; Martone W. J. Predominant pathogens in hospital infections. J. Antimicrob. Chemother. 1992, 29, 19. 10.1093/jac/29.suppl_a.19. [DOI] [PubMed] [Google Scholar]
- Leleu G.; Aegerter P.; Guidet B. Systemic candidiasis in intensive care units: a multicenter, matched-cohort study. J. Crit. Care 2002, 17, 168–175. 10.1053/jcrc.2002.35815. [DOI] [PubMed] [Google Scholar]
- Adam H. J.; Richardson S. E.; Roscoe M.; Boroumandi S.; Gris M.; Yau Y. C. W. An implementation strategy for the use of chromogenic media in the rapid, presumptive identification of Candida species. Open Mycol. J. 2010, 4, 33–38. 10.2174/1874437001004010033. [DOI] [Google Scholar]
- Chin C. D.; Laksanasopin T.; Cheung Y. K.; Steinmiller D.; Linder V.; Parsa H.; Wang J.; Moore H.; Rouse R.; Umviligihozo G.; Karita E.; Mwambarangwe L.; Braunstein S. L.; van de Wijgert J.; Sahabo R.; Justman J. E.; El-Sadr W.; Sia S. K. -based diagnostics of infectious diseases in the developing world. Nat. Med. 2011, 17, 1015. 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- Pandey C. M.; Augustine S.; Kumar S.; Kumar S.; Nara S.; Srivastava S.; Malhotra B. D. Microfluidics Based Point-of-Care Diagnostics. Biotechnol. J. 2018, 13, 1700047. 10.1002/biot.201700047. [DOI] [PubMed] [Google Scholar]
- Herrada C. A.; Kabir M. A.; Altamirano R.; Asghar W. Advances in Diagnostic Methods for Zika Virus Infection. J. Med. Dev. 2018, 12, 040802. 10.1115/1.4041086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.; Asghar W.; Kim Y.; Iqbal S. M. Cell elasticity-based microfluidic label-free isolation of metastatic tumor cells. Br. J. Med. Med. Res. 2014, 4, 2129–2140. 10.9734/bjmmr/2014/7392. [DOI] [Google Scholar]
- Yu S.; Rubin M.; Geevarughese S.; Pino J. S.; Rodriguez H. F.; Asghar W. Emerging technologies for home-based semen analysis. Andrology 2018, 6, 10–19. 10.1111/andr.12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coarsey C. T.; Esiobu N.; Narayanan R.; Pavlovic M.; Shafiee H.; Asghar W. Strategies in Ebola virus disease (EVD) diagnostics at the point of care. Crit. Rev. Microbiol. 2017, 43, 779–798. 10.1080/1040841x.2017.1313814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghar W.; Yuksekkaya M.; Shafiee H.; Zhang M.; Ozen M. O.; Inci F.; Kocakulak M.; Demirci U. Engineering long shelf life multi-layer biologically active surfaces on microfluidic devices for point of care applications. Sci. Rep. 2016, 6, 21163. 10.1038/srep21163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyas A.; Asghar W.; Ahmed S.; Lotan Y.; Hsieh J.-T.; Kim Y.-t.; Iqbal S. M. Electrophysiological analysis of biopsy samples using elasticity as an inherent cell marker for cancer detection. Anal. Methods 2014, 6, 7166–7174. 10.1039/c4ay00781f. [DOI] [Google Scholar]
- Asghar W.; Ramachandran P. P.; Adewumi A.; Noor M. R.; Iqbal S. M. Rapid nanomanufacturing of metallic break junctions using focused ion beam scratching and electromigration. J. Manuf. Sci. Eng. 2010, 132, 030911. 10.1115/1.4001664. [DOI] [Google Scholar]
- Schell W. A.; Benton J. L.; Smith P. B.; Poore M.; Rouse J. L.; Boles D. J.; Johnson M. D.; Alexander B. D.; Pamula V. K.; Eckhardt A. E.; Pollack M. G.; Benjamin D. K.; Perfect J. R.; Mitchell T. G. of a digital microfluidic real-time PCR platform to detect DNA of Candida albicans in blood. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 2237–2245. 10.1007/s10096-012-1561-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posch W.; Heimdörfer D.; Wilflingseder D.; Lass-Flörl C. Invasive candidiasis: future directions in non-culture based diagnosis. Expert Rev. Anti-Infect. Ther. 2017, 15, 829–838. 10.1080/14787210.2017.1370373. [DOI] [PubMed] [Google Scholar]
- Clancy C. J.; Nguyen M. H. Finding the “missing 50%” of invasive candidiasis: how nonculture diagnostics will improve understanding of disease spectrum and transform patient care. Clin. Infect. Dis. 2013, 56, 1284–1292. 10.1093/cid/cit006. [DOI] [PubMed] [Google Scholar]
- Pfeiffer C. D.; Samsa G. P.; Schell W. A.; Reller L. B.; Perfect J. R.; Alexander B. D. Quantitation of Candida colony forming units in initial positive blood cultures. J. Clin. Microbiol. 2011, 49, 2879–2883. 10.1128/JCM.00609-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy C.; Nguyen M. H. Non-culture diagnostics for invasive candidiasis: Promise and unintended consequences. J. Fungi 2018, 4, 27. 10.3390/jof4010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst A.; Leverstein-Van Hall M. A.; Verhoef J.; Fluit A. C. Detection of Candida spp. in blood cultures using nucleic acid sequence-based amplification (NASBA). Diagn. Microbiol. Infect. Dis. 2001, 39, 155–160. 10.1016/s0732-8893(01)00211-5. [DOI] [PubMed] [Google Scholar]
- Loeffler J.; Hebart H.; Cox P.; Flues N.; Schumacher U.; Einsele H. Nucleic Acid Sequence-Based Amplification of Aspergillus RNA in Blood Samples. J. Clin. Microbiol. 2001, 39, 1626–1629. 10.1128/jcm.39.4.1626-1629.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P. L.; Shetty A.; Barnes R. A. Detection of seven Candida species using the Light-Cycler system. J. Med. Microbiol. 2003, 52, 229–238. 10.1099/jmm.0.05049-0. [DOI] [PubMed] [Google Scholar]
- Goodarzi M.; Shahhosseiny M. H.; Bayat M.; Hashemi S. J.; Ghahri M., Comparison between molecular methods (PCR vs LAMP) to detect Candida albicans in bronchoalveolar lavage samples of suspected tuberculosis patients. Microbiol. Res. 2018,8 (). 10.4081/mr.2017.7306 [DOI] [Google Scholar]
- Zhou J.; Liao Y.; Li H.; Lu X.; Han X.; Tian Y.; Chen S.; Yang R. Development of a loop-mediated isothermal amplification assay for rapid detection of Trichosporon asahii in experimental and clinical samples. BioMed Res. Int. 2015, 2015, 1–9. 10.1155/2015/732573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Xu H. Development and evaluation of loop-mediated isothermal amplification (LAMP) for the rapid diagnosis of Candida parapsilosis. Afr. J. Microbiol. Res. 2014, 8, 3850–3855. 10.1128/JCM.02739-13. [DOI] [Google Scholar]
- Pfaller M. A.; Wolk D. M.; Lowery T. J. T2MR and T2Candida: novel technology for the rapid diagnosis of candidemia and invasive candidiasis. Future Microbiol. 2016, 11, 103–117. 10.2217/fmb.15.111. [DOI] [PubMed] [Google Scholar]
- Wheeler R. T.; Kombe D.; Agarwala S. D.; Fink G. R. Dynamic, Morphotype-Specific Candida albicans β-Glucan Exposure during Infection and Drug Treatment. PLoS Pathog. 2008, 4, e1000227 10.1371/journal.ppat.1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasperkovitz P. V.; Khan N. S.; Tam J. M.; Mansour M. K.; Davids P. J.; Vyas J. M. Toll-like receptor 9 modulates macrophage antifungal effector function during innate recognition of Candida albicans and Saccharomyces cerevisiae. Infect. Immun. 2011, 79, 4858–4867. 10.1128/iai.05626-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J. L.; Miller G. R. Quality control slide for potassium hydroxide and cellufluor fungal preparations. J. Clin. Microbiol. 1989, 27, 1411–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffler J.; Henke N.; Hebart H.; Schmidt D.; Hagmeyer L.; Schumacher U.; Einsele H. Quantification of fungal DNA by using fluorescence resonance energy transfer and the light cycler system. J. Clin. Microbiol. 2000, 38, 586–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir A.; Forrest G.; Clarkson J.; Wheals A. Detection of Candida albicans DNA from blood samples using a novel electrochemical assay. J. Med. Microbiol. 2011, 60, 467–471. 10.1099/jmm.0.026229-0. [DOI] [PubMed] [Google Scholar]
- Kauffman C. A. Candiduria. Clin. Infect. Dis. 2005, 41, S371–S376. 10.1086/430918. [DOI] [PubMed] [Google Scholar]
- Kan V. L. Polymerase chain reaction for the diagnosis of candidemia. J. Infect. Dis. 1993, 168, 779–783. 10.1093/infdis/168.3.779. [DOI] [PubMed] [Google Scholar]
- Hamula C. L.; Hughes K.; Fisher B. T.; Zaoutis T. E.; Singh I. R.; Velegraki A. T2Candida provides rapid and accurate species identification in pediatric cases of candidemia. Am. J. Clin. Pathol. 2016, 145, 858–861. 10.1093/ajcp/aqw063. [DOI] [PubMed] [Google Scholar]
- Inacio J.; Flores O.; Spencer-Martins I. Efficient identification of clinically relevant Candida yeast species by use of an assay combining panfungal loop-mediated isothermal DNA amplification with hybridization to species-specific oligonucleotide probes. J. Clin. Microbiol. 2008, 46, 713–720. 10.1128/jcm.00514-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safavieh M.; Kanakasabapathy M. K.; Tarlan F.; Ahmed M. U.; Zourob M.; Asghar W.; Shafiee H. Emerging loop-mediated isothermal amplification-based microchip and microdevice technologies for nucleic acid detection. ACS Biomater. Sci. Eng. 2016, 2, 278–294. 10.1021/acsbiomaterials.5b00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widjojoatmodjo M. N.; Borst A.; Schukkink R. A. F.; Box A. T. A.; Tacken N. M. M.; Van Gemen B.; Verhoef J.; Top B.; Fluit A. C. Nucleic acid sequence-based amplification (NASBA) detection of medically important Candida species. J. Microbiol. Methods 1999, 38, 81–90. 10.1016/s0167-7012(99)00079-2. [DOI] [PubMed] [Google Scholar]
- Zhou X.; Kong F.; Sorrell T. C.; Wang H.; Duan Y.; Chen S. C. A. Practical method for detection and identification of Candida, Aspergillus, and Scedosporium spp. by use of rolling-circle amplification. J. Clin. Microbiol. 2008, 46, 2423–2427. 10.1128/jcm.00420-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekhotiaeva N.; Awasthi S. K.; Nielsen P. E.; Good L. Inhibition of Staphylococcus aureus gene expression and growth using antisense peptide nucleic acids. Mol. Ther. 2004, 10, 652–659. 10.1016/j.ymthe.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Bisha B.; Kim H. J.; Brehm-Stecher B. F. Improved DNA-FISH for cytometric detection ofCandidaspp. J. Appl. Microbiol. 2011, 110, 881–892. 10.1111/j.1365-2672.2011.04936.x. [DOI] [PubMed] [Google Scholar]
- Trnovsky J.; Merz W.; Della-Latta P.; Wu F.; Arendrup M. C.; Stender H. Rapid and accurate identification of Candida albicans isolates by use of PNA FISHFlow. J. Clin. Microbiol. 2008, 46, 1537–1540. 10.1128/jcm.00030-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennel R.; Asghar W. Image sensor road map and solid-state imaging devices. NanoWorld J. 2017, 1, 10–14. [Google Scholar]
- Coleman B.; Coarsey C.; Kabir M. A.; Asghar W. Point-of-care colorimetric analysis through smartphone video. Sens. Actuators, B 2019, 282, 225–231. 10.1016/j.snb.2018.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coarsey C.; Coleman B.; Kabir M. A.; Sher M.; Asghar W. Development of a flow-free magnetic actuation platform for an automated microfluidic ELISA. RSC Adv. 2019, 9, 8159–8168. 10.1039/c8ra07607c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman B.; Coarsey C.; Asghar W., Cell phone based colorimetric analysis for point-of-care settings. Analyst 2019, 144–1947. 10.1039/c8an02521e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalanabis M.; Al-Muayad H.; Kulinski M. D.; Altman D.; Klapperich C. M. Cell lysis and DNA extraction of gram-positive and gram-negative bacteria from whole blood in a disposable microfluidic chip. Lab Chip 2009, 9, 2811–2817. 10.1039/b905065p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.