Abstract
Purpose
Reduced nuclear expression of BRCA1 associated protein 1 (BAP-1) is associated with a high risk for metastasis in uveal melanoma. Manual assessment of the expression level may face issues with interobserver reproducibility. This could be improved with digital image analysis (DIA).
Methods
Thirty enucleated eyes with uveal melanoma from the Emory Eye Center (Atlanta, GA; years 2009–2017) were included and stained with BAP-1. Retrospective data on patient and tumor characteristics were retrieved. Patients were randomized to a training or validation cohort. Their tumor sections were digitally scanned and scored for percentage of BAP-1–positive cells with the QuPath Bioimage analysis software.
Results
Interobserver concordance was 75% (Cohen's κ 0.52) with manual BAP-1 scoring and 88% to 94% with DIA (Cohen's κ 0.75–0.88). Positive and negative predictive values for metastasis were 90% and 100% with DIA, 80% and 86% with manual scoring, and 78% and 88% with gene expression class 2. In binary logistic regression, manual and DIA of BAP-1 and gene expression class 2 were associated with metastasis, but none retained significance in multiple regression. Metastasis-free survival was significantly shorter with low BAP-1 expression as defined by DIA (log-rank P = 0.02), but not with manual scoring (log-rank P = 0.36) or with gene expression class 2 (log-rank P = 0.17).
Conclusions
DIA of BAP-1 is a competitive alternative to manual assessment as well as gene expression profiling in prognostication of enucleated specimens with uveal melanoma.
Translational Relevance
The emerging scope for automatization of qualified diagnostic tasks is applied to uveal melanoma.
Keywords: uveal melanoma, BAP-1, digital image analysis
Introduction
Uveal melanoma is the most common primary intraocular malignancy in adults.1 Two percent to 4% of patients have clinically detectable metastases at diagnosis.1 At a later stage however, up to 45% of patients will have metastases even if the eye containing the tumor has been removed.2 This is likely caused by micrometastases that have been seeded up to several years before primary tumor diagnosis.3 At the distant location, primarily the liver, these micrometastases then can remain dormant for several decades. Once they start growing into clinically detectable lesions, there is no effective treatment and median patient survival is only 4 to 12 months.4,5
Several methods for early prognostication have been proposed and partially implemented in clinical practice. In the absence of lymphatic spread, tumor thickness, diameter, and presence of distant metastasis are the bases for tumor staging.6,7 Loss of chromosome 3, as determined by fluorescence in situ hybridization or single-nucleotide polymorphism array, has a very high positive and negative predictive value for metastasis.8 Commercial gene expression tests, based on the expression of 12 classifier genes have been developed and show excellent prognostic use for identification of tumors with a high risk for metastasis.9 The latter tests, however, are not universally accessible.
Mutations in the SF3B1 or EIF1AX genes identify uveal melanomas with intermediate and low risk for metastasis.10 Inactivating mutations in the tumor suppressor BRCA associated protein-1 (BAP-1) gene located on chromosome 3p21.1 has been identified in >80% of metastasizing uveal melanomas and is strongly associated with gene expression classifications.11,12 Immunohistochemical staining of the BAP-1 protein in tumor tissue and assessment of its level of nuclear expression is a relatively inexpensive, quick, and readily available alternative to fluorescence in situ hybridizations, single-nucleotide polymorphism arrays and gene expression tests.13,14 The correlation between manual assessments of BAP-1 expression and metastasis has been confirmed in a cohort that included patients who had undergone brachytherapy before enucleation.13
As previously shown in other tumors, however, manual scoring of such immunohistochemical stains may face issues with interobserver reproducibility that can be reduced with digital image analysis (DIA).15,16 Therefore, we saw an opportunity to compare the concordance of manual and DIA-based scoring of BAP-1 expression in uveal melanoma patients from an American referral center with correlation to gene expression classification and metastasis-free survival.
Materials and Methods
Patients and Samples
The study adhered to the tenets of the Declaration of Helsinki. The protocol for collection of specimens and data from the Emory Eye Center (Atlanta, GA) was approved by the Emory institutional review board. To calculate the sample size, we examined our previous publication on manual assessments of BAP-1 expression, in which the 5-year cumulative metastasis-free survival probability of patients with tumors with high and low BAP-1 expression was approximately 0.8 and 0.3, respectively.13 To detect such a difference with a power of 0.80 (given a two-sided α of 0.05), a total sample size of 28 patients would be required.
In review of the medical records, 56 enucleated eyes with histologically established uveal melanoma, gene expression classifications, and data on the presence or absence of metastasis were available. Twenty-five of these were excluded because their paraffin blocks were unavailable, and one was excluded because the tumor was fully necrotic. Our follow-up data were confirmed and further extended in telephone interviews with patients or relatives after verbal consent. Of the remaining 30 patients, none had undergone plaque brachytherapy before enucleation. When determining the relative size of the training and validation cohorts, we considered previous research indicating that the ratio should be inversely proportional to the square root of the number of free adjustable parameters.17 As we only evaluated one parameter (number of BAP-1–positive cells) and desired to include sufficient patients for survival analysis, we set the ratio to 0.8:1.
The patients randomized to the training cohort (n = 13) underwent enucleation from December 17, 2009 to June 22, 2017. The patients randomized to the validation cohort (n = 17) underwent enucleation from February 18, 2010 to November 22, 2017.
Immunohistochemistry
The paraffin blocks were cut into 4 μ sections, pretreated in ethylenediaminetetraacetic acid (EDTA) buffer at pH 9.0 for 20 minutes and incubated with mouse monoclonal antibodies against BAP-1 at dilution 1:40 (clone C-4; Santa Cruz Biotechnology, Dallas, TX) and a red chromogen, and finally counterstained with hematoxylin and rinsed with deionized water. The deparaffinization, pretreatment, primary staining, secondary staining, and counterstaining steps were run in a Bond III automated IHC/ISH stainer (Leica, Wetzlar, Germany). The dilutions had been gradually titrated until optimal staining was achieved, according to manual control.
Gene Expression Classification
Tumor tissue samples were obtained from freshly enucleated eyes by fine needle aspiration. The contents of the needle hub were transferred into one of two RNAse-free cryovials. Using the same needle, extraction buffer from the second cryovial was aspirated and expelled into the first. This then was placed in a specimen bag, immediately frozen to −80°C and shipped on dry ice for gene expression classification based on 12 discriminating and three control genes (Castle Biosciences, Inc., Friendswood, TX). All samples were processed during routine clinical testing for risk prognostication after obtaining patient consent.
Digital Image Analysis
After sectioning and staining, all glass slides were digitally scanned to the .ndpi file format at ×400, using identical digital scanners at both institutions (Nano Zoomer 2.0 HT; Hamamatsu Photonics K.K., Hamamatsu, Japan). The DIA software used was the QuPath Bioimage analysis v.0.1.2, which is an open-source software for digital pathology and whole slide image analysis developed at Northern Ireland Molecular Pathology Laboratory, Centre for Cancer Research and Cell Biology, Queen's University Belfast (Belfast, Northern Ireland, UK).18 The software was run on a standard off-the-shelf laptop computer (Apple, Inc., Cupertino, CA).
BAP-1 Scoring
Manual and DIA scoring of BAP-1 on the tumor slides was performed in a similar manner.
For manual scoring, the tissue sections were screened under low magnification (×40), and the three areas exhibiting the most intense BAP-1 staining were selected for grading. Nuclear immunoreactivity then was evaluated in approximately 100 cells in each of these areas (at ×200, corresponding to a field diameter of 1 mm) using a four-point scoring system described previously.13 Score 0 was given if <10% of the tumor cell nuclei were positive, score 1 if ≥10% to 33% were positive, score 2 if ≥33% to 66% were positive, and score 3 if ≥66% were positive. The tumor's BAP-1 expression was classified as low if the mean score in three fields was 0 or 1, and high if the mean score was 2 or 3. Two independent pathologists (GS, HG) performed the manual scoring blinded to patient outcome and gene expression class.
For DIA, one positive cell (red chromogen in nucleus) and one negative cell (hematoxylin, but no red chromogen in nucleus) was calibrated in each digitally scanned tissue section. All other parameters were left at default to limit time consumption and maintain ease of use. Tumors then were screened under low magnification (×40) and the three areas exhibiting the most intense BAP-1 staining selected for grading. Nuclear immunoreactivity was evaluated at ×200, in three circular 0.5 mm-diameter regions of interest (corresponding to a combined area of 0.59 mm2 per tumor) by automatic classification (positive cell detection). One pathologist (GS), one ocular oncology and pathology fellow (TRS), and one fourth year medical student without previous experience in pathology (SP) performed the DIA independently and blinded to patient outcome and gene expression class.
Tumor areas with intense inflammation, heavy pigmentation, bleeding, necrosis, or poor fixation were avoided for manual scoring and DIA.
Statistical Methods
In addition to the cutoffs for manual BAP-1 classification provided by the previous report,13 we calibrated cutoffs for BAP-1 high versus low for manual and DIA scoring after adjustments by points on receiver operating characteristic (ROC) curves. The calibration was performed in the training cohort based on melanoma-related death and subsequently applied to the validation cohort, blinded to patient outcome. For measurement of interobserver concordance, the percentage of identically classified cases and Cohen's κ statistics computed.19 For correlation with outcome, manual, DIA and gene expression classifications were evaluated with Cox proportional hazards analysis adjusted for tumor thickness as a marker for tumor size,20 binary logistic and multiple logistic regressions for association with metastasis, and with Kaplan-Meier metastasis-free survival. Follow-up was defined as the time in months from enucleation to metastasis, or in the absence of metastasis, to the last occasion patient was seen or in contact alive. Event-free follow-up was defined as the time in months from enucleation to the last occasion patients without metastases were seen or in contact alive. Differences with a P < 0.05 were considered significant, all P values being two-sided. All statistical analyses were performed using IBM SPSS statistics version 25 (Armonk, NY).
Results
Descriptive Statistics
Mean age at enucleation of patients in the training and validation cohorts was 66 and 60 years, respectively (P = 0.26). Nine females and seven males had been randomized to the training cohort and eight females and 12 males to the validation cohort (P = 0.50). Two of the 20 patients in the validation cohort had undergone plaque brachytherapy before enucleation. Mean follow-up was 34 months for the training cohort and 42 months for the validation cohort (P = 0.48). Mean event-free follow-up was 27 and 18 months, respectively (P = 0.61; overall mean 23 months; Supplementary Fig. S1).
There were no significant differences in tumor location, histologic cell type, mean tumor thickness, diameter, AJCC T-category,6 gene expression class or manual BAP-1 classification between the training and validation cohorts (Table 1).
Table 1.
Characteristics of Patients and Tumors Included in This Study
|
Training Cohort |
Validation Cohort |
P |
|
| n | 13 | 17 | |
| Mean age at diagnosis, years (min—max) | 65 (43–88) | 60 (24–92) | 0.32a |
| Sex, n (%) | 0.15b | ||
| Female | 8 (62) | 6 (35) | |
| Male | 5 (38) | 11 (65) | |
| Tumor location, n (%) | 0.52c | ||
| Choroid | 12 (92) | 16 (94) | |
| Ciliary body | 1 (8) | 1 (6) | |
| Iris | 0 (0) | 0 (0) | |
| Cell type, n (%) | 0.59c | ||
| Spindle | 0 (0) | 0 (0) | |
| Epitheloid | 2 (15) | 2 (12) | |
| Mixed | 11 (85) | 15 (88) | |
| Mean tumor thickness, mm (range) | 8.7 (1.1–15.9) | 9.0 (0.8–17.4) | 0.81a |
| Mean tumor diameter, mm (range) | 16.3 (4.8–22.5) | 16.7 (1.4–21.5) | 0.65a |
| Previous brachytherapy, n (%) | 1.0b | ||
| No | 13 | 17 | |
| Yes | 0 | 0 | |
| AJCC T-category, n (%) | 0.07c | ||
| 1 | 1 (8) | 1 (5) | |
| 2 | 0 (0) | 5 (29) | |
| 3 | 6 (46) | 9 (53) | |
| 4 | 6 (46) | 2 (12) | |
| Gene expression class, n (%) | 0.43c | ||
| 1a | 4 (31) | 3 (18) | |
| 1b | 2 (15) | 6 (35) | |
| 2 | 7 (54) | 8 (47) | |
| Manual BAP1 classification, n (%) | 0.74b | ||
| High | 6 (38) | 9 (45) | |
| Low | 10 (63) | 11 (55) | |
| Follow-up monthsd, mean (SD, range)d | 22 (16, 0–50) | 21 (20, 1–70) | 0.56a |
| Event-free follow-up monthse, mean (SD, range) | 27 (19, 0–50) | 18 (21, 1–56) | 0.61a |
Statistical tests were used for comparisons of continuous variables, categorical variables in two-by-two tables and for categorical variables with >2 expected values, respectively. Tumor location signifies the assessed primary locale.
Mann-Whitney U test.
Fisher's exact test.
Pearson χ2 test.
Follow-up terminated at time of metastasis, or in the absence of metastasis, at last occasion patient was seen or in contact alive.
Follow-up time for patients without metastasis.
Training Cohort
With DIA in the training cohort (Fig. 1), the mean proportion of BAP-1–positive cells was 43.9% (standard deviation [SD], 33.0; range, 3.2%–98.7%). The mean number of cells automatically analyzed was 1820 (SD, 1323; range, 432–4815) in each of the circular 0.5 mm-diameter regions of interest and 5461 (SD, 3969; range, 1269–14,445) in the three regions of interest per tumor combined.
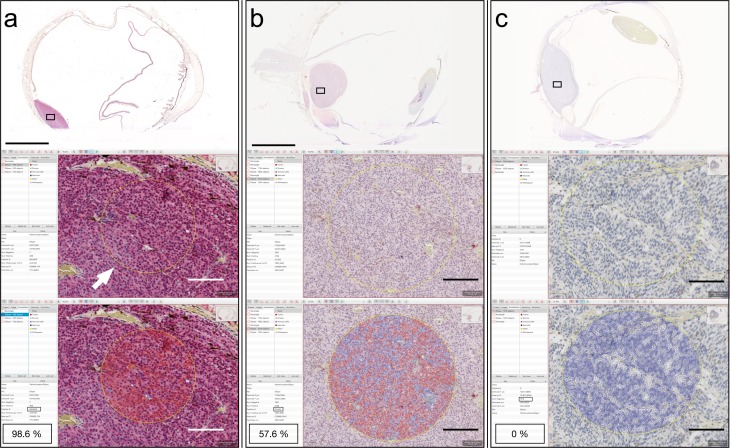
Figure 1.
Example of digital image analysis in three tumors with different levels of BAP-1 expression. (a) In a tumor with intensive stain on overview, a circular region of interest has been defined in ×20 (arrow). In this circular area, the software then automatically evaluates the number of cells with nuclear stain (cell borders and nuclei marked red by the software). As indicated in the annotation window 98.6% of the cells were classified as positive. (b) In a tumor with intermediate stain on overview, both cells with (cell borders and nuclei marked red by the software) and without detected stain (cell borders and nuclei marked blue by the software) are present. As indicated in the annotation window, 57.6% of the cells were classified as positive. (c) In a tumor with no stain on overview, only cells without detected stain (cell borders and nuclei marked blue by the software) are found. As indicated in the annotation window, 0% of the cells were classified as positive. Scale bars: top = 5 mm, lower = 100 μm.
The mean proportion of BAP-1–positive cells was 17.2% (SD, 16.5; range, 3.9%–35.7%) in patients who suffered metastasis before the end of follow-up and 52.8% (SD 32.8; range, 3.2–98.7%; Mann-Whitney U test, P = 0.15) in patients who did not have metastasis.
The mean proportion of BAP-1–positive cells was 71.3% (SD, 20.8; range, 35.7–98.7%) in tumors of gene expression class 1a or 1b and 12.5% (SD, 9.0; range, 3.2–22.3%, Mann-Whitney U test, P = 0.004) in tumors of gene expression class 2.
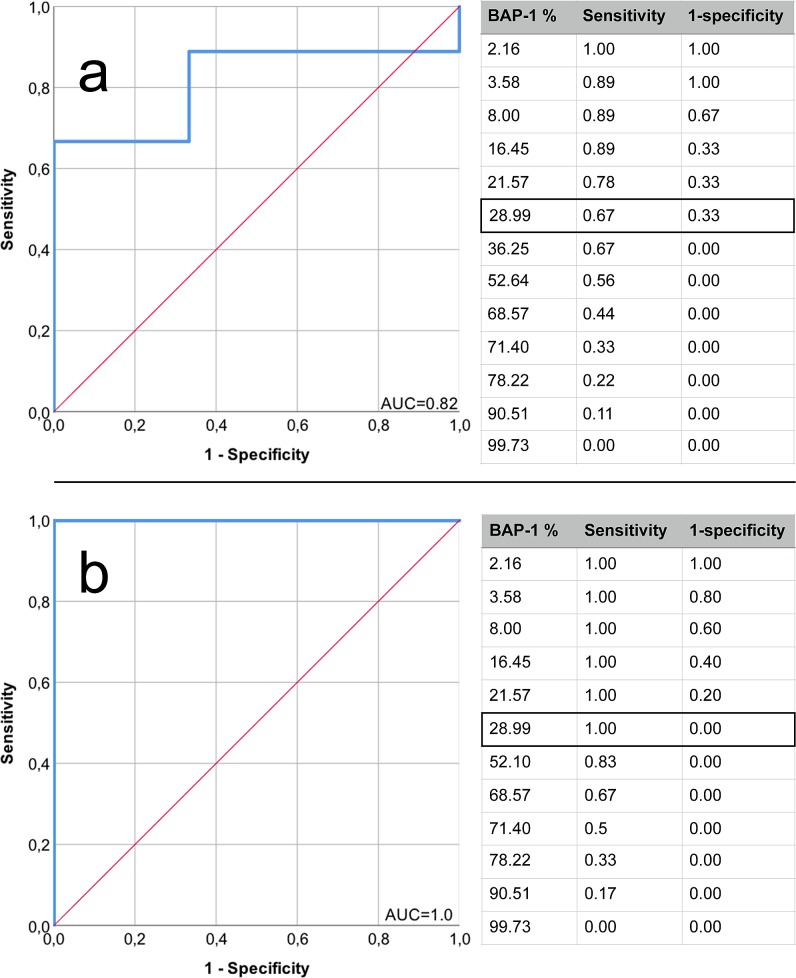
In analysis of ROCs of the BAP-1 scores, maximum sensitivity and specificity for metastasis status and gene expression class 2 were given equal importance. A cutoff of 29% BAP-1–positive cells was deemed optimal for discrimination of patients with and without metastases, and tumors of gene expression class 2 from classes 1a and 1b (Fig. 2). This yielded a sensitivity and specificity of 67% for metastasis (area under the curve [AUC] = 0.82), and a sensitivity and specificity of 100% for gene expression class 2 (AUC = 1.00). For practical use in comparisons of interobserver concordance and in the validation cohort, the cutoff was rounded to <30%.
Figure 2.
ROC curves, DIA of the proportion of BAP-1–positive cells versus metastasis status and gene expression class in the training cohort. (a) Separation of patients with from those without metastases. AUC = 0.82. Sensitivity 67% and specificity 67% with a cutoff of 29% BAP-1–positive cells. (b) Separation of tumors with gene expression class 2 from tumors with class 1a or 1b. AUC = 1.0. Sensitivity 100% and specificity 100% with a cutoff of 29% BAP-1–positive cells.
Interobserver Concordance
When comparing the two pathologists' manual scoring of BAP-1, 12 of 16 tumors (75%) had an identical classification (seven classified as low, five as high) while four tumors had dissimilar classification, yielding a Cohen's κ statistic indicating moderate agreement (κ = 0.52).
With DIA, the first and second observer (pathologist and ocular oncology and pathology fellow) obtained an identical classification for 15 of 16 tumors (94%) with only one tumor having a dissimilar classification, yielding a Cohen's κ statistic indicating almost perfect agreement (κ = 0.88). Between the first and third observers (pathologist and medical student), an identical classification was obtained for 14 tumors (88%), yielding a Cohen's κ statistic indicating substantial agreement (κ = 0.75).
Validation Cohort
The mean time required for calibration of one positive and one negative cell in each tumor was 40 seconds (SD, 12; range, 20–60) and for definition of three circular regions of interest and automated scoring it was 32 seconds (SD, 15; range, 15–60) adding up to a mean time consumption for DIA of 72 seconds per tumor (SD, 21; range, 40–120). The mean time required for manual assessment of a total of 300 cells in three high power fields was 264 seconds (SD, 61; range, 212–348).
With DIA in the validation cohort and a cutoff for mean proportion of BAP-1–positive cells of <30%, 10 tumors were classified as low and seven as high. The mean number of cells analyzed was 3389 (SD, 1310; range, 532–5685) in each high power field and 8713 (SD 3969; range, 1269–14445) in the three combined high power fields of each tumor.
When manual and digital scores of the same cases were compared, identical classification was obtained in 15 of 17 cases (88%), yielding a Cohen's κ statistic indicating substantial agreement (κ = 0.76).
Sensitivity and specificity for gene expression class 2 was 100% and 78% respectively, and the positive and negative predictive values for metastasis were 90% and 100%, respectively. This can be compared to manual BAP-1 classification, which had a sensitivity and specificity for gene expression class 2 of 100% and 80%, respectively, and a positive and negative predictive value for metastasis of 80% and 86%, respectively. Gene expression class 2 had a positive and negative predictive value for metastasis of 78% and 88%, respectively.
Regression Analysis
To evaluate the prognostic value of manual and DIA of BAP-1 expression and gene expression class within our data set, we performed univariate Cox proportional hazards analysis with metastasis-free survival as the end point, adjusted for tumor thickness. We then excluded time-dependence from the model and performed binary logistic regressions with metastasis as the dependent variable. Manual and DIA scoring of BAP-1 and gene expression class 2 were individually associated with metastasis. In a multiple logistic regression, none of these methods was a significant independent predictor (Table 2).
Table 2.
Regression Analyses
|
Regression Coefficient, β (SE) |
Wald Statistic |
P |
Hazard Coefficient, Exp(b) (95% CI) |
|
| Univariate Cox proportional hazards | ||||
| DIA BAP-1 IHC low | 3.8 (3.1) | 1.5 | 0.22 | 45.0 (0.1 > 1000) |
| Manual BAP-1 IHC low | 1.0 (1.1) | 0.8 | 0.38 | 2.6 (0.3–21.4) |
| Gene expression class 2 | 1.0 (0.8) | 1.5 | 0.22 | 2.8 (0.6–13.8) |
| Binary logistic regression | ||||
| Patient age | 0 (0.1) | 0 | 0.83 | 1.0 (0.8–1.2) |
| Male sex | 0.1 (0.3) | 0 | 0.90 | 1.1 (0.5–2.0) |
| Tumor thickness | 0 (0.1) | 0.1 | 0.82 | 1.0 (0.9–1.2) |
| LBD | 0 (0) | 0 | 0.86 | 1.0 (1.0–1.1) |
| Epithelioid cell type | −21.6 (>1000) | 0 | 0.99 | 0 (0) |
| DIA BAP-1 IHC low | 2.2 (1.1) | 4.3 | 0.04 | 9.0 (1.1–71.0) |
| Manual BAP-1 IHC low | 3.2 (1.3) | 5.6 | 0.02 | 24 (1.7–330.8) |
| Gene expression class 2 | 3.2 (1.3) | 5.7 | 0.02 | 24.5 (1.8–336.2) |
| Multiple logistic regression | ||||
| DIA BAP-1 IHC low | 40.5 (35662) | 0.0 | 0.99 | 0 (0-) |
| Manual BAP-1 IHC low | −20.3 (25216) | 0.0 | 0.99 | 0 (0-) |
| Gene expression class 2 | −18.3 (25216) | 0.0 | 0.99 | 0 (0-) |
IHC, immunohistochemistry; LBD, largest basal diameter; CI, confidence interval.
The top portion of Table 2 shows the Cox proportional hazards analysis of the association between metastasis-free survival and DIA classification of BAP-1 expression, manual classification of BAP-1 expression, and gene expression class 2. Tumor thickness was included as a covariate. No method was individually associated with shortened metastasis-free survival. The middle portion of Table 2 shows binary logistic regressions with metastasis as a dependent variable. Manual and DIA BAP-1-scoring and gene expression class 2 were individually associated with metastasis. Tumor thickness is indicated in 2 mm intervals and age in 10-year intervals. The bottom portion shows multiple logistic regression, none of the methods was a significant independent predictor for metastasis.
Survival
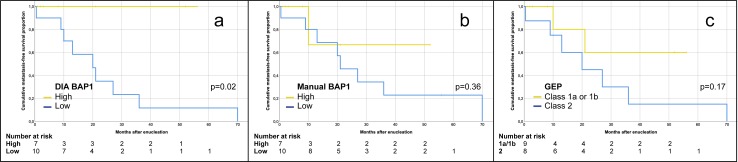
In Kaplan-Meier analysis, patients had significantly shorter metastasis-free survival if their tumors had low BAP-1 expression as defined by DIA (log-rank P = 0.02), but not with manual scoring (log-rank P = 0.36) or with gene expression class 2 versus 1a or 1b (log-rank P = 0.17, Fig. 3).
Figure 3.
Kaplan-Meier curves, cumulative metastasis-free survival. (a) Patients with tumors with high (yellow) versus low (blue) BAP-1 expression, as defined by DIA using a mean of <30% stained tumor cells over three high power fields as cutoff (log-rank P = 0.02). (b) Patients with tumors with high (yellow) versus low (blue) BAP-1 expression, as defined by manual scoring using a mean of <33% stained tumor cells over three high power fields as cutoff (log-rank P = 0.36). (c) Patients with tumors with gene expression class 1a or 1b (yellow) versus class 2 (blue; log-rank P = 0.17). GEP, gene expression profiling.
Discussion
In this study, DIA of BAP-1 expression outperformed the manual method in interobserver concordance and time required for classification. Further, it outperformed the manual method and gene expression classifications in positive and negative predictive values for metastasis, hazard for metastasis, and separation of patients with long versus short metastasis-free survival. Clinically, this is valuable for patients who wish to be informed about their prognosis, and possibly in the development of future adjuvant treatments for patients with high risk of metastasis.
One could perhaps argue that the addition of DIA to evaluation of uveal melanoma specimens is the addition of another burden on pathologists, who often already are skilled and experienced in manual assessments. Training and substantial investments in digital scanning and storage capacity are required before effective use of the technology can be expected. However, after these investments, DIA comes with the cost of immunohistochemistry only, which is universally available and substantially cheaper than gene expression tests. Further, we have shown here that even an untrained individual can perform DIA with reliable results. In doing so, more than fifteen times as many cells are assessed in less than a third of the time required for manual scoring. Therefore, the potential extra burden can be reduced to the preanalytical delay of digital scanning. This will not necessarily affect pathologists, who can be allocated to more qualified tasks.
As the stained tumor tissue is visible throughout its handling in the DIA software, one might fairly argue that our improved results over manual scoring are actually a matter of a more meticulous examination of the BAP-1 expression, rather than an intrinsic superiority of the method itself. Another natural objection to these results is the small number of patients included, and the short follow-up for a substantial proportion of these. This limits any far-reaching conclusions from this study.
Last, we only investigated the feasibility of DIA on tumors in eyes that have been enucleated, typically because of their size. A large proportion of patients with uveal melanoma have smaller tumors with lower metastatic risk, and undergo primary plaque brachytherapy or proton beam radiotherapy. For these patients, tissue for analysis typically is obtained with fine needle aspirations. The prognostic use of immunohistochemistry and DIA on such specimens remains unclear.
In summary, DIA of BAP-1 immunohistochemistry was a competitive, if not superior, alternative to both manual scoring and gene expression class. Accordingly, we strongly encourage further studies to confirm these results in larger, prospective populations.
Supplementary Material
Acknowledgments
Supported by St. Erik Eye Hospital (GS), the St. Erik Research Foundation (S:t Eriks Ögonforskningsstiftelse), the Swedish Ophthalmological Society, Cronqvists stiftelse (Cronqvist Foundation), the Swedish Eye Foundation (Ögonfonden) and Karolinska Institutet (Karolinska Institutets Stiftelsemedel för Ögonforskning).
Disclosure: G. Stålhammar, None; T.R.O. See, None; S. Phillips, None; S. Seregard, None; H.E. Grossniklaus, None
References
- 1.Singh N, Bergman L, Seregard S, Singh AD. Epidemiologic Aspects. In: Damato B, Singh AD, editors. Clinical Ophthalmic Oncology: Uveal Tumors. Berlin, Heidelberg: Springer;; 2014. pp. 75–87. [Google Scholar]
- 2.Kujala E, Mäkitie T, Kivelä T. Very long-term prognosis of patients with malignant uveal melanoma. Invest Ophthalmol Vis Sci. 2003;44:4651–4659. doi: 10.1167/iovs.03-0538. [DOI] [PubMed] [Google Scholar]
- 3.Singh AD. Uveal melanoma: implications of tumor doubling time. Ophthalmology. 2001;108:829–830. doi: 10.1016/s0161-6420(00)00607-2. [DOI] [PubMed] [Google Scholar]
- 4.Carvajal RD, Schwartz GK, Tezel T, Marr B, Francis JH, Nathan PD. Metastatic disease from uveal melanoma: treatment options and future prospects. Br J Ophthalmol. 2017;101:38–44. doi: 10.1136/bjophthalmol-2016-309034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Augsburger JJ, Corrêa ZM, Shaikh AH. Effectiveness of treatments for metastatic uveal melanoma. Am J Ophthalmol. 2009;148:119–127. doi: 10.1016/j.ajo.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 6.Kivelä T, Simpson ER, Grossniklaus HE, et al. AJCC Cancer Staging Manual. Chicago: Springer;; 2017. Uveal melanoma; pp. 805–817. [Google Scholar]
- 7.Arnljots TS, Al-Sharbaty Z, Lardner E, All-Eriksson C, Seregard S, Stålhammar G. Tumour thickness, diameter, area or volume? The prognostic significance of conventional versus digital image analysis-based size estimation methods in uveal melanoma. Acta Ophthalmol. 2018;96:510–518. doi: 10.1111/aos.13668. [DOI] [PubMed] [Google Scholar]
- 8.Bornfeld N, Prescher G, Becher R, Hirche H, Jöckel KH, Horsthemke B. Prognostic implications of monosomy 3 in uveal melanoma. Lancet. 1996;347:1222–1225. doi: 10.1016/s0140-6736(96)90736-9. [DOI] [PubMed] [Google Scholar]
- 9.Onken MD, Worley LA, Char DH, et al. Collaborative Ocular Oncology Group report number 1: prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology. 2012;119:1596–1603. doi: 10.1016/j.ophtha.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yavuzyigitoglu S, Koopmans AE, Verdijk RM, et al. Uveal melanomas with SF3B1 mutations: a distinct subclass associated with late-onset metastases. Ophthalmology. 2016;123:1118–1128. doi: 10.1016/j.ophtha.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 11.Decatur CL, Ong E, Garg N, et al. Driver mutations in uveal melanoma: associations with gene expression profile and patient outcomes. JAMA Ophthalmol. 2016;134:728–733. doi: 10.1001/jamaophthalmol.2016.0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szalai E, Wells JR, Ward L, Grossniklaus HE. Uveal melanoma nuclear BRCA1-associated protein-1 immunoreactivity is an indicator of metastasis. Ophthalmology. 2018;125:203–209. doi: 10.1016/j.ophtha.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koopmans AE, Verdijk RM, Brouwer RW, et al. Clinical significance of immunohistochemistry for detection of BAP1 mutations in uveal melanoma. Mod Path. 2014;27 doi: 10.1038/modpathol.2014.43. [DOI] [PubMed] [Google Scholar]
- 15.Stålhammar G, Fuentes Martinez N, Lippert M, et al. Digital image analysis outperforms manual biomarker assessment in breast cancer. Mod Path. 2016;29:318. doi: 10.1038/modpathol.2016.34. [DOI] [PubMed] [Google Scholar]
- 16.Stålhammar G, Robertson S, Wedlund L, et al. Digital image analysis of Ki67 in hot spots is superior to both manual Ki67 and mitotic counts in breast cancer. Histopathology. 2018;72:974–989. doi: 10.1111/his.13452. [DOI] [PubMed] [Google Scholar]
- 17.Guyon I. A Scaling Law for the Validation-Set Training-Set Size Ratio. Berkeley, California: AT&T Bell Laboratories;; 1997. pp. 1–11. [Google Scholar]
- 18.Bankhead P, Loughrey M, Fernández J, et al. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7:16878–16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen J. A coefficient of agreement for nominal scales. Ed Psych Meas. 1960;20:37–46. [Google Scholar]
- 20.Shields CL, Furuta M, Thangappan A, et al. Metastasis of uveal melanoma millimeter-by-millimeter in 8033 consecutive eyes. Arch Ophthalmol. 2009;127:989–998. doi: 10.1001/archophthalmol.2009.208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.