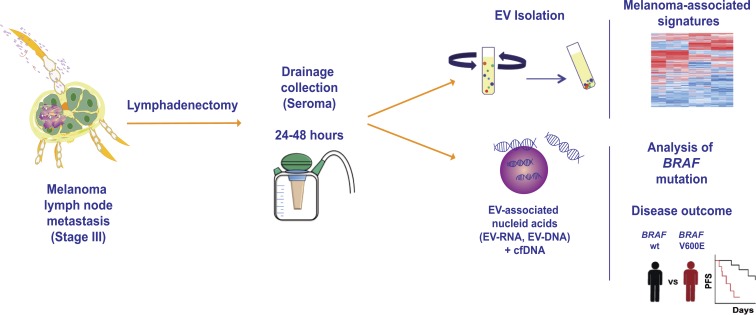
García-Silva et al. show for the first time that extracellular vesicles isolated from the exudative seroma obtained from the lymphatic drainage implanted in melanoma patients after lymphadenectomy can be interrogated for melanoma markers and BRAF mutations. Profiling the BRAFV600E mutation in this biofluid is a novel approach to predict disease relapse.
Abstract
Liquid biopsies from cancer patients have the potential to improve diagnosis and prognosis. The assessment of surrogate markers of tumor progression in circulating extracellular vesicles could be a powerful non-invasive approach in this setting. We have characterized extracellular vesicles purified from the lymphatic drainage also known as exudative seroma (ES) of stage III melanoma patients obtained after lymphadenectomy. Proteomic analysis showed that seroma-derived exosomes are enriched in proteins resembling melanoma progression. In addition, we found that the BRAFV600E mutation can be detected in ES-derived extracellular vesicles and its detection correlated with patients at risk of relapse.
Graphical Abstract
Introduction
LN metastases are associated with poor prognosis in cancer patients (Karaman and Detmar, 2014). The number of metastatic LNs is directly correlated with a decrease in overall survival, enhanced tumor aggressiveness, and the need for systemic therapies. In the clinical setting, exudative seroma (ES) obtained from the drainage placed at the anatomical location of the resected sentinel LN (sLN) has been used to profile disease markers in melanoma patients (Włodzimierz et al., 2004; Rutkowski et al., 2008). Tumor-derived extracellular vesicles (EVs), and more specifically exosomes, reach the sLNs, favoring metastatic spread (Hood et al., 2011; Pucci et al., 2016; Srinivasan et al., 2016). This is consistent with the role of tumor-secreted exosomes in the formation of premetastatic niches at distal sites (Hoshino et al., 2015; Peinado et al., 2017). However, EV profiling in human ES has not been reported.
In this work, we have characterized for the first time EVs from the ES obtained from the lymphatic drainage implanted after lymphadenectomy in melanoma patients. Proteomic profiling demonstrated that EVs carry melanoma-specific signatures. Importantly, BRAFV600E mutation is detected in ES-derived vesicles and related to a shorter disease-free survival.
Results and discussion
ES collected after lymphadenectomy is enriched in exosomes
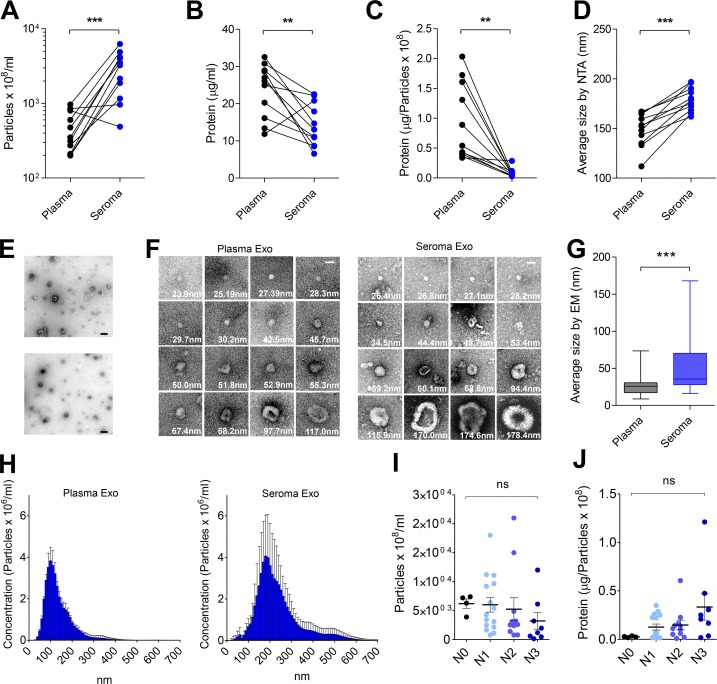
We characterized the number of circulating exosomes in matched frozen ES and plasma samples from melanoma patients using nanoparticle tracking analysis (NTA). ES contains a significantly increased number of particles than plasma (n = 11 patients; Fig. 1 A and Table S1). Additionally, we profiled frozen plasma (n = 20) or frozen ES (n = 40; Table S2) from independent cohorts of melanoma patients, observing that ES is highly enriched in exosomes compared with plasma (Fig. S1 A). Protein content and protein content per particle were reduced in ES-derived exosomes, suggesting that the increased number of particles observed in ES is not related to protein contamination (Fig. 1, B and C). The size of particles in ES was also significantly increased (median size: 148 nm; mean size: 179 nm) compared with plasma (median size: 126 nm; mean size: 152 nm; Fig. 1 D and Fig. S1 B). We performed electron microscopy (EM) after standard or gradient ultracentrifugation (Fig. 1, E–G; and Fig. S1 C), demonstrating the integrity of exosomes and the increased size. Moreover, ES-derived exosomes display a more heterogeneous size distribution than plasma-circulating exosomes (Fig. 1 H). Notably, Broggi et al. in this issue reported comparable results. Analysis of particle number and protein stratified by LN involvement demonstrated that particle concentration was not significantly affected across the different substages (Fig. 1 I). However, the amount of proteins per particle tended to increase through tumor progression (Fig. 1 J), as described for plasma-circulating vesicles (Peinado et al., 2012).
Figure 1.
Characterization of ES-derived EVs. (A) Quantification of the number of particles per milliliter in samples. (B) Quantification of total protein cargo in samples. (C) Ratio of protein per particle in samples. (D) Size (diameter, mean) of particles measured by NTA in samples. (E) Representative EM images of exosomes purified by ultracentrifugation (upper image) and by density gradient centrifugation (lower image). Bars, 500 nm. (F) Representative EM images of individual purified exosomes. Bars, 50 nm. (G) Size (diameter, mean) quantification determined by EM. (H) Size distribution of particles determined by NTA. (I and J) Analysis of the concentration of particles (I) and estimation of the ratio of exosomal protein stratified by LN involvement substage (J). N0, no metastatic LNs; N1, 1 metastatic LN; N2, 2–3 metastatic LNs; N3, ≥4 more metastatic LNs (N0, n = 4; N1, n = 15; N2, n = 11; N3, n = 8). All data are shown as mean ± SEM. ***, P < 0.001; **, P < 0.01; ns, not significant as determined by paired Student’s t test (A–D), unpaired Student’s t test (G), or ANOVA (I and J). Exo, exosomes.
Exosomes derived from ES and plasma display different proteomic profiles
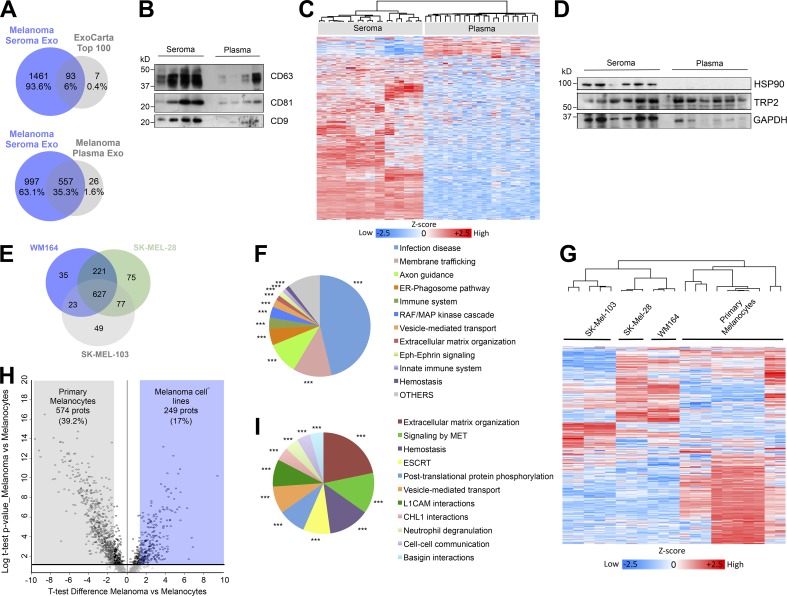
We compared the protein cargo in ES- and plasma-derived vesicles from matched samples (n = 11 patients; Table S1) by liquid chromatography–tandem mass spectrometry (LC-MS/MS). Exosomal origin of ES preparations was verified by the identification of 93 out of 100 most-frequent proteins found in exosomes from the ExoCarta database (Simpson et al., 2012; Fig. 2 A, upper Venn diagram). In plasma samples, we identified 75 proteins from the top 100 proteins specified in the ExoCarta database (Fig. S1 D). Proteins detected in ES-derived exosomes outnumbered proteins in plasma-derived exosomes (Fig. 2 A, lower Venn diagram; Fig. S1 E; and Table S3). Exosomal markers CD63, CD81, and CD9 were analyzed in both fluids by Western blot (Fig. 2 B). We performed unsupervised hierarchical cluster analysis and observed that samples clustered by fluid, reasserting the different characteristics of plasma and ES (Fig. 2 C). We found 745 proteins significantly more abundant in ES- than in plasma-derived exosomes, including HSP90B, Annexin A1, S100 A4, NRAS, and Lactoferrin (Fig. S1 F). Functional analysis showed a significant enrichment in several pathways in ES-derived exosomes related to antigen presentation, endoplasmic reticulum (ER)–phagosome pathway, G2/M transition, and IL-12 family signaling (Fig. S1 G). We analyzed HSP90 and TRP-2, previously described as enriched in plasma-circulating exosomes from melanoma patients and correlated with progression (Peinado et al., 2012). While TRP-2 was found in both fluids, HSP90 was only detected in ES-derived exosomes (Fig. 2 D).
Figure 2.
Proteomic analysis of ES- and melanoma-derived exosomes. (A) Venn diagrams showing the overlap between proteins identified in ES-derived exosomes and the top 100 proteins in the ExoCarta database (upper diagram) and proteins from ES- and plasma-derived exosomes (lower diagram). (B) Western blot of exosomal markers in paired samples of ES- and plasma-derived exosomes. (C) Unsupervised hierarchical cluster analysis of the proteomic profiles of ES- and plasma-derived exosomes of the same cohort of patients. (D) Western blot of melanoma markers HSP90 and TRP-2 in paired samples of ES- and plasma-derived exosomes. (E) Venn diagram showing the identified proteins between the indicated human melanoma cell lines. (F) ClueGO analysis of the Reactome pathways enriched in melanoma cell line–derived exosomes. (G) Unsupervised hierarchical clustering of the proteomic data of primary melanocyte- and melanoma cell line–derived exosomes. (H) Volcano plot of proteins differentially expressed between exosomes secreted by primary melanocytes and melanoma cell lines. Black circles represent proteins differentially expressed with P < 0.05. (I) ClueGO analysis of the Reactome pathways associated to the proteins upregulated in melanoma cells-derived exosomes versus primary melanocyte-derived exosomes. *** indicates group-corrected P < 0.001 as determined by Bonferroni stepdown test applied to pathway networks (F and I). ESCRT, endosomal sorting complex required for transport; Exo, exosomes; prots, proteins.
We next profiled the protein cargo of exosomes from human melanoma cell lines (WM164, SK-MEL-28, and SK-MEL-103). 1,107 proteins were identified, 627 proteins were common to all of the cell lines (Fig. 2 E and Table S4), and 96 of the top 100 proteins defined by ExoCarta were among them (Fig. S1 H). The protein classes included guanosine triphosphate (GTP)–binding and chaperones, among others (Fig. S1 I). Functional analysis revealed that pathways related to membrane trafficking, RAF/MAPK signaling, ER–phagosome pathway, and immune system, among others, were enriched in melanoma cell line–derived exosomes (Fig. 2 F).
Unsupervised hierarchical clustering of melanoma model– and primary melanocyte–secreted exosomes showed the segregation between exosomes from tumor and non-tumor cells (Fig. 2 G). Principal component analysis (PCA) confirmed robust differences in the proteomic patterns of melanoma- and melanocyte-derived exosomes (Fig. S1 J). 249 proteins were significantly up-regulated in melanoma-derived exosomes (Fig. 2 H and Table S5). Functional analysis showed a significant enrichment in networks of pathways related to extracellular matrix organization and MET signaling, among others (Fig. 2 I). Interestingly, we found that 605 proteins present in melanoma exosomes were also detected in ES-derived EVs, while only 256 were identified in plasma-derived exosomes (Fig. S1 K).
Melanoma signaling pathways in ES-derived exosomes
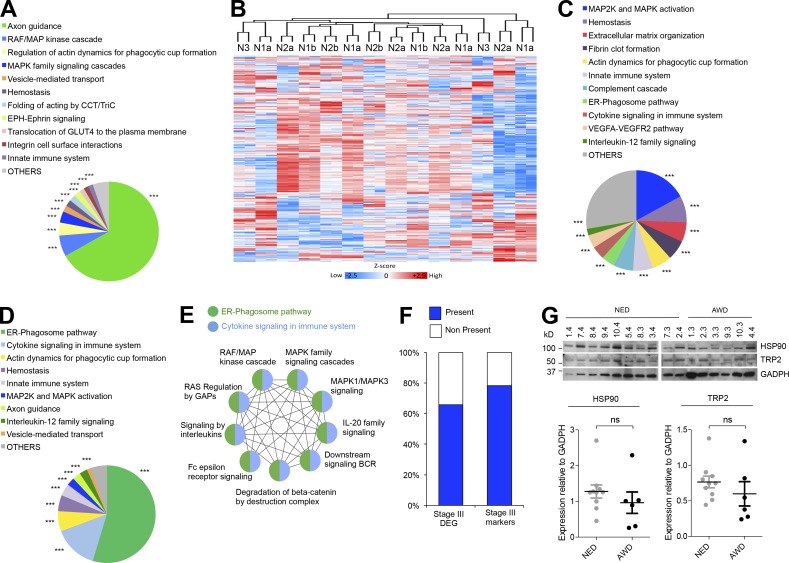
We next profiled by LC-MS/MS exosomes purified from ES of 14 melanoma patients with different regional LN involvement (Table S6). We found a total of 1,041 proteins (Table S7); 702 of these proteins have not been identified previously in lymph proteome-related datasets (Hansen et al., 2015; Fig. S2 A and Table S8). There was a significant enrichment in GDP-binding related proteins, endopeptidases, and enzyme inhibitors in ES-derived EVs (Fig. S2 B). The analysis showed an enrichment in pathways involved in immune response, protein activation, and proteolysis (Fig. S2 C). Interestingly, axon guidance and activation of RAF/MAPK cascades were among the pathway networks significantly associated with the proteins shared between ES-derived exosomes and melanoma cell–derived exosomes (Fig. 3 A). We observed that unsupervised hierarchical clustering or PCA did not stratify samples according to their LN involvement (Fig. 3 B and Fig. S2 D). The intragroup coefficient of variation ranged from 0.5043 to 0.6021 for all groups analyzed here (Fig. S2 E), suggesting that the protein profiles did not vary considerably across substages or that interpatient heterogeneity precluded the identification of distinct patterns.
Figure 3.
ES-derived exosomes from melanoma patients are enriched in tumor features. (A) Reactome pathways associated with the proteins shared between ES-derived exosomes and melanoma cell line–derived exosomes. (B) Unsupervised hierarchical clustering of ES-derived exosomes from stage III melanoma patients with different LN involvement. (C) Reactome pathways significantly enriched in ES-derived exosomes from N3 compared with N1a patients. (D) Pie chart representing the same analysis as in C but only considering the proteins upregulated in N3 ES-derived exosomes also present in melanoma cell line–derived exosomes. (E) Analysis by ClueGO and representation by Cytoscape of the enriched RAS/RAF/MAPK-related pathways related to ER-phagosome and cytokine networks. (F) Percentage of proteins differentially expressed in stage III patients with bad prognosis (stage III DEG; Mactier et al., 2014) or potential prognostic markers (stage III markers) present in ES-derived exosomes. (G) Representative Western blot of HSP90 and TRP-2 in ES-derived exosomes of melanoma subjects. GAPDH was used as loading control. Quantification is shown below. Data are shown as mean ± SEM. ns, not significant as determined by unpaired Student’s t test (G). In pie charts, *** indicates group-corrected P < 0.001 as determined by Bonferroni stepdown test applied to pathway networks (B–D). AWD, alive with disease; NED, no evidence of disease; BCR, B cell receptor signaling; GAPs, GTPase activating proteins; DEG, differentially expressed genes.
We ruled out the presence of contaminants such as blood-related proteins in ES-derived EVs. Deconvolution of proteomic data using the GlioVis tool (Bowman et al., 2017) revealed blood cell signatures (Fig. S2 F), but their contribution was <2% in our samples (Fig. S2 G). The contribution of other contaminants, such as apolipoproteins, was also ∼1.27% (Fig. S2 G). We found that 38% of the proteins in exosomes derived from human lymphatic endothelial cells (Brown et al., 2018) were also present in our ES samples as well as the 43% of associated inflammatory markers (Fig. S2 H). We found 17 groups of pathways significantly enriched in N3 versus N1a patients, including MAPK2/MAPK signaling, hemostasis, platelet activation, and complement cascade pathway, among others (Fig. 3 C and Table S9). Several signaling networks were still significantly enriched after restricting the analysis to the up-regulated proteins in N3 ES-derived exosomes common to the melanoma cell line–derived exosome dataset (Fig. 3 D). Importantly, several RAS/RAF/MAPK-related pathways were enriched in ES-derived exosomes from N3 melanoma patients compared with N1a patients (Fig. 3 E).
Comparison of the ES-derived exosome dataset with a list of proteins differentially expressed in LNs from the American Joint Committee on Cancer (AJCC) melanoma stage III patients with good and poor prognosis (Mactier et al., 2014) showed that 65% of these proteins were identified in ES-derived exosomes (Fig. 3 F and Table S10). Furthermore, ∼80% of the 21 prognostic markers for stage III melanoma selected in this study were also present in ES-derived EVs (Fig. 3 F and Table S11). Interestingly, HSP90 was one of the common markers between the two datasets. However, no differences in HSP90 protein levels were observed among patients with no evidence of disease or patients alive with disease, and similar results were obtained for TRP-2 (Fig. 3 G and Fig. S2 I). Our data suggest that, although the proteomic cargo in ES-derived exosomes cannot discriminate between patients with different LN involvement or different outcome, signaling pathways associated with melanoma tumor cells and their secreted EVs were overrepresented in ES-derived EVs from N3 compared with N1a patients. Remarkably, Broggi et al. (2019) have reported a similar enrichment in melanoma-associated proteins and pathways.
Detection of BRAF mutation in ES-derived EVs
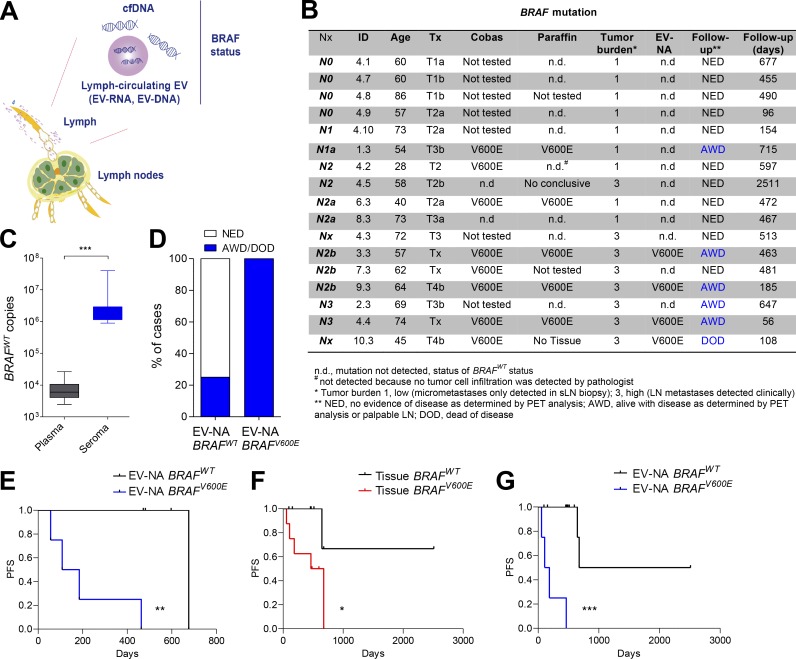
Finally, we assessed whether DNA in seroma-derived EVs could be used to detect surrogate markers of disease progression. Mutations in the V600 codon of BRAF are found in 35–50% of human melanomas (Luke et al., 2017). Previous studies have demonstrated a significant association between mutant BRAF levels in plasma cell–free DNA (cfDNA) and melanoma outcome in patients (Santiago-Walker et al., 2016; Calapre et al., 2017). EV-derived DNA has been analyzed in lung cancer, glioblastoma, and pancreatic cancer (Allenson et al., 2017; Figueroa et al., 2017; Yang et al., 2017; Castellanos-Rizaldos et al., 2018). Since mutations in coding regions can be found in cfDNA and exosomal RNA/DNA (Thakur et al., 2014; Möhrmann et al., 2018), we combined all these nucleic acid fractions, analyzing EV-associated nucleic acids (EV-NAs) for BRAF mutation in ES (Fig. 4 A).
Figure 4.
Detection of BRAF mutation in ES EV-NAs. (A) Scheme of the nucleic acid test used. (B) Table summarizing the clinical status and follow-up information of patients included in the BRAF study performed in tissue and EV-NAs. (C) Number of BRAFWT copies per milliliter detected in samples. (D) Percentage of patients with disease progression according to BRAF status in ES-derived EVs. (E) Kaplan–Meier curve for PFS only in diagnosed BRAFV600E patients according to the BRAF status in EV-NAs; n = 8. (F) Kaplan–Meier plot for PFS of patients according to the BRAF status in solid biopsy; n = 16. (G) Kaplan–Meier plot for PFS of patients according to BRAF status in ES EV-NAs independently of BRAF status in tumor or LNs; n = 17. Data are shown as mean ± SEM. ***, P < 0.001 by unpaired Student’s t test (C). Mantel-Cox log-rank test was applied to the survival curves: **, P = 0.0067 (E); *, P = 0.0418 (F); and ***, P < 0.0001 (G). AWD, alive with disease; DOD, dead of disease; NED, no evidence of disease.
We analyzed BRAF mutation in ES EV-NAs obtained from 17 melanoma patients with different levels of LN involvement (from N0 to N3) after lymphadenectomy (Table S12). Of the 17 patients, 10 were tested also for BRAF mutation in tumor tissue by PCR at diagnosis (Fig. 4 B). We verified BRAF mutation in paraffin-embedded LN sections and confirmed the status of all patients (Fig. S3 A). Next, we analyzed the BRAF mutation in EV-NAs performing an allele-specific quantitative PCR with a limit of detection of 0.002% mutant allele frequency and a limit of quantification of 10 copies, 0.01% mutant allele frequency. We found ∼700 times more BRAFWT copies present in ES- than in plasma-derived EVs (Fig. 4 C). Notably, the analysis of BRAF mutation in the cfDNA fraction of ES failed to detect mutant BRAF even in the sample 4.4 that presented the largest number of BRAFV600E copies per milliliter (Fig. S3 B).
All patients found to be BRAF negative in tumor samples were also BRAF negative in ES-EVs, indicating that specificity was 100% (Fig. 4 B). In four out of the eight cases diagnosed as BRAFV600E in tumor or LN tissue, the mutation was also detected in ES-derived EVs (Fig. 4 B). Interestingly, all these patients relapsed with distal metastases between 56 to 463 d after lymphadenectomy (Fig. 4, B and D). Only two out of all cases in which BRAF mutation was not detected in EV-NAs progressed after lymphadenectomy but in a time over 647 d (Fig. 4 B). Remarkably, in patients diagnosed as BRAFV600E by tissue biopsy, the EV-NA test for BRAF mutation showed that patients positive for this test had a median survival of 146.5 d versus 715 d for patients negative for BRAF mutation Fig. 4 E; log-rank P = 0.0067). Although tumor burden in those BRAFV600E patients predicted a similar PFS, it did not reach statistical significance (Fig. S3 D). The median survival of patients was 715 d for low tumor burden (tumor burden 1, micrometastasis) versus 185 d for high tumor burden (tumor burden 3, macrometastasis), with log-rank P = 0.0532 (Fig. S3 D). Although the presence of the BRAF mutation in tissue biopsy indicated a faster relapse (569.5 d) compared with noncarriers (Fig. 4 F; log-rank P = 0.0418), our analysis of BRAFV600E mutation in seroma EV-NAs stratified patients independently of BRAF status in tumor and/or LNs. The analysis showed a significantly shorter PFS for EV-NA BRAFV600E patients (146.5 d), whereas median PFS for EV-NA BRAFWT patients was 1,594 d with a log-rank P < 0.0001 (Fig. 4 G and Fig. S3 E). Thus, detection of the BRAFV600E mutation in ES-derived EV nucleic acids could serve as a minimal residual disease/prognostic indicator, with added value over the current tissue biopsies being an almost real-time predictor of risk right after lymphadenectomy.
In summary, we demonstrate for the first time that ES-derived EVs may represent a useful surrogate marker of melanoma progression and are suitable to detect melanoma-specific mutations. The ES obtained through lymphatic drainage is a biofluid enriched in EVs compared with plasma. We also found a remarkable overlap between proteins and pathways in ES-derived exosomes and melanoma cell line–secreted exosomes. Furthermore, detection of BRAF mutation in ES vesicles obtained through lymphatic drainage may be a novel parameter to identify patients at risk of relapse probably due to the presence of residual disease. These patients could subsequently benefit from specific adjuvant therapies right after surgery.
Materials and methods
Patient and sample collection
The study included several independent cohorts of melanoma patients (n = 40 for ES collection and n = 20 for plasma collection) and a cohort of matched plasma/ES samples obtained after lymphadenectomy from 11 patients. The samples were provided by Dr. P. Rutkowski (Maria Sklodowska-Curie Institute Oncology Center, Warsaw, Poland) and Dr. P.L. Ortiz-Romero and Dr. J.L. Rodriguez-Peralto (Hospital 12 de Octubre, Madrid, Spain). Patients were considered eligible if they were diagnosed with cutaneous melanoma with regional LN involvement (stage III) according to the AJCC determined by histological analysis and if they underwent radical LN dissection. Complete LN dissection was performed due to clinical nodal metastases or positive sLN biopsy together with the exclusion of distant metastases by imaging (computed tomography, or magnetic resonance, and/or positron emission tomography–computed tomography). Lymphadenectomy was performed within 4–8 wk after sLN biopsy or after cytological/pathological diagnosis of clinical (palpable) metastases. All patients were followed carefully, with a median follow-up time of 15.7 mo. Routinely, surveillance was recommended every 3–4 mo. “No evidence of disease” was defined as no recurrence of disease as determined by LN palpation and imaging according to national recommendations and “alive with disease” as relapse of disease on treatment. The characteristics of the patients in the different cohorts are summarized in Tables S1, S2, and S12. All samples were collected using clinical study protocols approved for this purpose by the Instituto de Salud Carlos III Committee for Ethical Research. All individuals provided informed consent for blood and seroma donation. ES samples were collected 24–48 h after radical lymphadenectomy from routinely used, sucking postoperative drainages; the localization of the LN where the drainage was placed, and where the ES was obtained, is specified in Table S12. After collection, samples (5–10 ml) were centrifuged for 10 min at 500 × g at room temperature and incubated with red blood cell lysing buffer for 10 min on ice. Samples were again centrifuged and supernatant was immediately frozen at −80°C.
Plasma samples (initial volume ranging from 2 to 5 ml) were obtained by venipuncture from arterial blood in the upper side of the arm. Whole blood was collected in EDTA tubes, inverted for 8 to 10 times, and centrifuged for 10 min at 1,100 × g at room temperature using a swing-out bucket rotor. Then, the plasma fraction (supernatant) was poured into the collection tube and immediately frozen at −80°C.
Cell lines
Primary melanocyte cultures and melanoma cell lines (WM164, SK-MEL-28 and SK-MEL-103) were kindly provided by Dr. M. Soengas (Spanish National Cancer Research Centre, Madrid, Spain). All melanoma cell lines were grown in high glucose DMEM (Lonza) supplemented with 10% fetal bovine serum (Hyclone), 2 mM glutamine and 20 μg/ml gentamicin. Primary melanocytes were culture in 254CF medium (Gibco) supplemented with human melanocyte growth supplement (Gibco). All cells were grown at 37ºC in a humidified 5% CO2 atmosphere. For exosome isolation, cells were cultured during 72 h in medium supplemented with 10% exosome-depleted fetal bovine serum (Hyclone).
Exosome isolation
Purification of exosomes from plasma and ES was performed in the same way after thawing the samples at 37°C for 3–5 min. First, samples were centrifuged at 3,000 × g for 20 min, followed by further centrifugation of the supernatants at 12,000 × g for 20 min. The exosomes were subsequently harvested by centrifugation at 100,000 × g for 70 min. The exosome pellet was resuspended in PBS and collected by a second ultracentrifugation at 100,000 × g for 70 min. For isolation of exosomes from cell cultures, supernatant fractions were collected after 72 h and centrifuged at 500 × g for 10 min followed by centrifugations at 12,000 × g for 20 min and 100,000 × g for 70 min. Finally, the exosome pellet was washed with PBS and collected by another ultracentrifugation at 100,000 × g for 70 min. All centrifugations were performed at 10°C using a Beckman Optima X100 centrifuge with a Beckman 50.4Ti or 70.1Ti rotor. Exosomes were resuspended in PBS and the protein content was measured by bicinchoninic acid (BCA) assay (Pierce). Particle content was measured from an aliquot of 1 µl of plasma or ES exosomes diluted in 1 ml of PBS using NTA (NanoSight; Malvern) equipped with a blue laser (405 nm). For density gradient analysis, EVs isolated in the 100,000 × g pellet were loaded on top of a discontinuous iodixanol gradient prepared by diluting a stock solution of OptiPrep (60% wt/vol) with 0.25 M sucrose/10 mM Tris (pH 7.5) to generate 40%, 20%, 10%, and 5% wt/vol iodixanol solutions. A discontinuous iodixanol gradient was generated by sequentially layering 2 ml each of the 40%, 20%, 10%, and 5% (wt/vol) iodixanol solutions. A 100-µl volume of PBS containing exosomes was overlaid onto the discontinuous iodixanol gradient and centrifuged using a Beckman Optima X100 with a 70.1Ti rotor at 10°C for 16 h. Fractions of 1 ml were collected from the top of the gradient and analyzed for particle concentration by NTA. Positive fractions (5, 6, and 7) were diluted in 20 ml in PBS and centrifuged at 100,000 × g for 70 min at 10°C using a Type 70 Ti rotor. The resulting pellets were resuspended in 100 µl PBS.
EM
For negative staining, purified exosome fractions were applied onto freshly glow-discharged, carbon-coated, 400-mesh copper EM grids at a concentration of 4 × 107 particles/ml and incubated for 5 min at room temperature. The grids were placed consecutively on top of three distinct 50-µl drops of MilliQ water, rinsed gently for 2 s, laid on the top of two different 50-µl drops of 1% uranyl acetate, and stained for 1 min. Finally, the grids were rinsed gently for 5 s and air dried. Grid visualization was performed on a Tecnai 12 transmission electron microscope (Thermo Fisher Scientific) with a lanthanum hexaboride cathode operated at 120 keV. Images were recorded at 21,900 nominal magnification with a 4k×4k TemCam-F416 CMOS camera (TVIPS).
Immunoblotting
Exosomal protein content was measured by BCA, and aliquots of 10 µg were used for Western blot analysis. Exosomes were lysed in Laemmli buffer at 95°C for 5 min, and the protein extracts were resolved by SDS-PAGE and probed using antibodies against HSP90 (1:1,000, SMC-107; Stressmarq Bioscience), TRP-2 (1:1,000; PEP8, kindly supplied by Dr. V.J. Hearing, National Institutes of Health, Bethesda, MD), β-actin (1:10,000, no. A5441; Sigma-Aldrich), GADPH (1:7,000, no. ab8245; Abcam), CD-9 (1:1,000, no. 92726; Abcam). Anti-CD63 and anti-CD81 hybridomas were kindly supplied by Dr. M. Yáñez-Mo (Molecular Biology Centre, Madrid, Spain). Peroxidase-conjugated AffinityPure donkey anti-rabbit or anti-mouse (1:5,000; Jackson ImmunoResearch) were used as secondary antibodies and their binding was detected using ECL Western Blotting Substrate kit (GE Healthcare). The intensity of the immunoreactive bands was quantified by densitometry using ImageJ software (National Institutes of Health).
Sample preparation for proteomic analysis
The protein content of ES- and plasma-derived exosomes isolated as described above was solubilized using 8 M urea in 100 mM Tris-HCl (pH 8.0). Samples (7.5 µg) were digested using the standard filter-aided sample preparation protocol. Briefly, proteins were reduced (10 mM dithiothreitol, 30 min, room temperature), alkylated (55 mM iodoacetamide, 20 min in the dark, at room temperature), and sequentially digested with Lys-C (protein-to-enzyme ratio, 1:50; overnight at room temperature; Wako) and with trypsin (protein-to-enzyme ratio, 1:100; 6 h at 37°C; Promega). Resulting peptides were desalted using C18 stage-tips.
Mass spectrometry
For the proteomic analysis of matched ES- and plasma-derived exosomes, LC-MS/MS was performed by coupling an Ultimate 3000 RSLCnano System (Dionex) with a Q-Exactive Plus mass spectrometer (Thermo Scientific). Peptides were loaded into a trap column (Acclaim PepMap 100; 100 µm × 2 cm; Thermo Scientific) over 3 min at a flow rate of 10 µl/min in 0.1% formic acid (FA). Then peptides were transferred to an analytical column (PepMap rapid separation liquid chromatography C18; 2 µm, 75 µm × 50 cm; Thermo Scientific) and separated using a 120-min effective linear gradient (buffer A: 0.1% FA; buffer B: 100% acetonitrile, 0.1% FA) at a flow rate of 250 nl/min. The gradient used was as follows: 0–5 min 2% B, 5–7 min 5% B, 7–100 min 20% B, 100–127 min 38% B, 127–137 min 98% B, and 137–145 min 2% B. The peptides were electrosprayed (2.1 keV) into the mass spectrometer through a heated capillary at 300°C and an S-Lens radio frequency (RF) level of 50%. The mass spectrometer was operated in a data-dependent mode, with an automatic switch between the MS and MS/MS scans using a top 15 method (minimum automatic gain control target, 3E3) and a dynamic exclusion time of 32 s MS (350–1,400 m/z), and MS/MS spectra were acquired with a resolution of 70,000 and 17,500 full width at half maximum (FWHM; 200 m/z), respectively. Peptides were isolated using a 2 Thompson unit (Th) window and fragmented using higher-energy collisional dissociation at 27% normalized collision energy. The ion target values were 3E6 for MS (25-ms maximum injection time) and 1E5 for MS/MS (45-ms maximum injection time). Samples were analyzed twice.
For the proteomic analysis of ES exosomes derived from patients with different LN involvement, LC-MS/MS was performed by coupling a nanoLC-Ultra 1D+ system (Eksigent) with an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) via a Nanospray Flex source (Thermo Fisher Scientific). Peptides were loaded into a trap column (NS-MP-10 BioSphere C18 5 µm, 20-mm length; Nanoseparations) over 10 min at a flow rate of 2.5 µl/min in 0.1% FA. Then, peptides were transferred to an analytical column (ReproSil Pur C18-AQ 2.4 µm, 500-mm length and 0.075-mm internal diameter [ID]) and separated using a 120-min linear gradient (buffer A: 4% ACN; 0.1% FA; buffer B: 100% ACN; 0.1% FA) at a flow rate of 250 nl/min. The gradient used was as follows: 0–2 min 6% B, 2–103 min 30% B, 103–113 min 98% B, and 113–120 min 2% B. The peptides were electrosprayed (1.8 keV) into the mass spectrometer with a PicoTip emitter (360/20 Tube OD/ID µm, tip ID 10 µm; New Objective), using a heated capillary temperature of 325°C and an S-Lens RF level of 60%. The mass spectrometer was operated in a data-dependent mode, with an automatic switch between the MS and MS/MS scans using a top 15 method (threshold signal ≥800 counts and dynamic exclusion of 60 s). MS spectra (350–1,500 m/z) were acquired in the Orbitrap at a resolution of 60,000 FWHM (400 m/z). Peptides were isolated using a 1.5-Th window and fragmented using collision-induced dissociation with a linear ion trap read-out at an normalized collision energy of 35% (0.25 Q-value and 10-ms activation time). The ion target values were 1E6 for MS (500-ms maximum injection time) and 5,000 for MS/MS (100-ms maximum injection time). Samples were run in duplicate.
Proteomic data analysis
Raw files were processed with MaxQuant (v 1.5.1.2) using the standard settings against a human protein database (UniProtKB/Swiss-Prot, August 2014, 20,187 sequences) supplemented with contaminants. Label-free quantification was done with match between runs (match window of 0.7 min and alignment window of 20 min). Carbamidomethylation of cysteines was set as a fixed modification, whereas methionine oxidation and N-term acetylation were variable protein modifications. The minimal peptide length was set to seven amino acids, and a maximum of two tryptic missed-cleavages was allowed. The results were filtered at 0.01 false discovery rate (FDR; peptide and protein level) and subsequently the “proteinGroup.txt” file was loaded in Perseus (v1.5.1.6) for further statistical analysis. A minimum of 50% label-free quantification valid values per group was required for quantification. Missing values were imputed from the observed normal distribution of intensities. Then, a two-sample Student’s t test with a permutation-based FDR was performed. Only proteins with a q-value <0.05 and log2 ratio >1.32 or < −1.32 were considered as regulated.
Analysis of GOterms and Reactome pathways was performed using the ClueGO plug-in v2.5.1 run on Cytoscape v3.6.0. The human ExoCarta database was considered as the reference set list, and a right-sided hypergeometric test was selected for enrichment analysis. A Bonferroni stepdown test was applied, and only GOterms and pathways with at least P < 0.05 were considered for analysis.
The MS proteomic data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD009505 and PXD012042.
Analysis of BRAF mutations in ES-derived vesicles
All patient samples were analyzed using the ExoDx BRAF V600E/K test by Exosome Diagnostics. In summary, 1 ml of fluid from each sample was used for co-isolation of exosomal RNA and DNA along with any cfDNA present using the ExoLution Plus extraction technology (Exosome Diagnostics), followed by nucleic acid purification. The total nucleic acids were subjected to reverse transcription and a highly sensitive quantitative PCR analysis of BRAFWT as well as BRAFV600E and BRAFV600K.
DNA extraction from paraffin tissue for BRAF analysis
DNA was isolated from paraffin tissue sections with a Maxwell RSC DNA FFPE kit (Promega) and quantified using a Quantus Fluorometer (Promega). DNA from tumor specimens was analyzed using the Cobas 4800 BRAF V600 Mutation test (Roche) following the manufacturer’s instructions.
For DNA obtained from LN samples, the target region was amplified using a Qiagen Multiplex PCR kit (Qiagen) and PCR was performed with BRAF specific primers (forward 5′-TGCTTGCTCTGATAGGAAAATGAGA-3′ and reverse 5′-CCATCCACAAAATGGATCCAGACA-3′) under the following cycling conditions: 15 min at 95°C, 5 cycles of 64°C, 5 cycles of 62°C, and 35 cycles of 60°C. The samples were then prepared for sequencing using the 16S Metagenomic Sequencing Library Preparation kit for the Illumina MiSeq System (Illumina) following the manufacturer’s protocol.
cfDNA isolation
cfDNA was extracted from ES samples using a QIAamp Circulating Nucleic Acid Kit (Qiagen) according to the manufacturer's protocol. Samples were centrifuged at 16,000 × g for 10 min before starting the extraction, and the supernatant was recovered and subjected to cfDNA extraction. cfDNA concentration was measured by a Qubit 2.0 Fluorometer (Thermo Fisher Scientific).
Statistical analysis
The error bars in the graphical data represent the means ± SEM. When appropriate, statistical significance was determined with a paired or unpaired two-tailed Student’s t test or one-way ANOVA or Mantel-Cox log-rank test for survival curves using GraphPad Prism software. For GOTerms and pathway analysis, group-corrected P values were obtained applying Bonferroni stepdown test using the ClueGO plug-in.
Online supplemental material
Fig. S1 shows characterization of ES-derived exosomes. Fig. S2 shows proteomic analysis of ES- and melanoma-derived exosomes. Fig. S3 shows detection of the BRAFV600E mutation in melanoma patient samples. Table S1 shows clinicopathologic characteristics of melanoma patients for matched plasma and ES samples. Table S2 shows clinicopathologic characteristics of melanoma patients for independent plasma and ES samples. Table S3 presents proteins identified by LC/MS-MS in EVs from plasma and ES paired samples. Table S4 shows proteins identified in melanoma cell line–derived exosomes. Table S5 is an analysis of proteins differentially expressed between melanoma- and melanocyte-derived exosomes. Table S6 shows clinicopathologic characteristics of the melanoma patients included in the proteomic profile of ES samples with different LN involvement. Table S7 presents proteins found in melanoma patient ES-derived exosomes. Table S8 shows proteins found in melanoma patient ES-derived exosomes not found in human lymph datasets. Table S9 shows proteins upregulated in ES-derived exosomes from N3 versus N1a melanoma patients. Table S10 shows proteins differentially expressed in melanoma AJCC stage III LNs found in melanoma patient–derived ES exosomes identified in Mactier et al. (2014). Table S11 shows prognostic markers in melanoma AJCC stage III found in melanoma patient–derived ES exosomes identified in Mactier et al. (2014). Table S12 shows details of the cohort of patients included in the proteomic analysis for different LN involvement and the analysis of BRAFV600E in EV-NAs.
Supplementary Material
Acknowledgments
We thank the members of Dr. H. Peinado’s, Dr. M. Soengas’, Dr. M. Yañez-Mo’s, and Dr. D. Lyden’s laboratories for reagents and discussions.
This work was supported by the US National Institutes of Health (R01-CA169416), the Feldstein Foundation, the Starr Cancer Consortium, Ministerio de Economía y Competitividad, the Ramón y Cajal Program, Atresmedia-Fundación AXA, "La Caixa" Foundation (ID 100010434, Fellowship LCF/BQ/ES17/11600007), Fundación Científica Asociación Española Contra el Cáncer, Fundación de Investigación Oncológica, Ministerio de Economía y Competitividad-Red de Excelencia en Investigación e Innovación en Exosomas, the Ramón Areces Foundation, and Comunidad Autónoma de Madrid.
K. Brinkmann, L. Meyer, J. Skog, and M. Noerholm are employees of Exosome Diagnostics. P. Rutkowski has served as a member of the Advisory Board for Novartis, Amgen, Merck Sharp & Dohme, Roche, Bristol-Myers Squibb, Bayer, and Blueprint Medicine and has received honoraria for lectures from Novartis, Pfizer, Merck Sharp & Dohme, Bristol-Myers Squibb, and Roche.
The other authors declare no competing financial interests.
Authors contributions: S. García-Silva, A. Benito-Martín, S. Sánchez-Redondo, A. Hernández-Barranco, P. Ximénez-Embún, L. Nogués, M.S. Mazariegos, K. Brinkmann, A. Amor López, L. Meyer, C. Rodríguez, C. García-Martín, J. Boskovic, R. Letón, C. Montero, and Y. Ruano performed the experiments. P. Rutkowski, J.L. Rodríguez-Peralto, P.L. Ortiz-Romero, L. Santambrogio, M.S. Brady, I. Kalinowska, and A. Szumera-Ciećkiewicz provided the patient samples and/or clinical advice. J. Skog, M. Noerholm, J. Boskovic, M. Robledo, and J. Muñoz provided the experimental facilities. S. García-Silva, P. Ximénez-Embún, and J. Muñoz performed the proteomic data analysis. M. Robledo, J. Skog, M. Noerholm, and H. Peinado performed the genomic data analysis. H. Peinado conceived the study. S. García-Silva and H. Peinado planned the experiments and wrote the manuscript. All authors contributed to and approved the final version of the manuscript.
References
- Allenson K., Castillo J., San Lucas F.A., Scelo G., Kim D.U., Bernard V., Davis G., Kumar T., Katz M., Overman M.J., et al. 2017. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann. Oncol. 28:741–747. 10.1093/annonc/mdx542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman R.L., Wang Q., Carro A., Verhaak R.G., and Squatrito M.. 2017. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro-oncol. 19:139–141. 10.1093/neuonc/now247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broggi M.A.S., Maillat L., Clement C.C., Bordry N., Corthésy-Henrioud P., Auger A., Matter M., Hamelin R., Potin L., Demurtas D., et al. 2019. Tumor-associated factors are enriched in lymphatic exudate compared to plasma in metastatic melanoma patients. J. Exp. Med. 10.1084/jem.20181618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M., Johnson L.A., Leone D.A., Majek P., Vaahtomeri K., Senfter D., Bukosza N., Schachner H., Asfour G., Langer B., et al. 2018. Lymphatic exosomes promote dendritic cell migration along guidance cues. J. Cell Biol. 217:2205–2221. 10.1083/jcb.201612051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calapre L., Warburton L., Millward M., Ziman M., and Gray E.S.. 2017. Circulating tumour DNA (ctDNA) as a liquid biopsy for melanoma. Cancer Lett. 404:62–69. 10.1016/j.canlet.2017.06.030 [DOI] [PubMed] [Google Scholar]
- Castellanos-Rizaldos E., Grimm D.G., Tadigotla V., Hurley J., Healy J., Neal P.L., Sher M., Venkatesan R., Karlovich C., Raponi M., et al. 2018. Exosome-based detection of EGFR T790M in plasma from non-small cell lung cancer patients. Clin. Cancer Res. 24:2944–2950. 10.1158/1078-0432.CCR-17-3369 [DOI] [PubMed] [Google Scholar]
- Figueroa J.M., Skog J., Akers J., Li H., Komotar R., Jensen R., Ringel F., Yang I., Kalkanis S., Thompson R., et al. 2017. Detection of wild-type EGFR amplification and EGFRvIII mutation in CSF-derived extracellular vesicles of glioblastoma patients. Neuro-oncol. 19:1494–1502. 10.1093/neuonc/nox085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K.C., D’Alessandro A., Clement C.C., and Santambrogio L.. 2015. Lymph formation, composition and circulation: a proteomics perspective. Int. Immunol. 27:219–227. 10.1093/intimm/dxv012 [DOI] [PubMed] [Google Scholar]
- Hood J.L., San R.S., and Wickline S.A.. 2011. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 71:3792–3801. 10.1158/0008-5472.CAN-10-4455 [DOI] [PubMed] [Google Scholar]
- Hoshino A., Costa-Silva B., Shen T.L., Rodrigues G., Hashimoto A., Tesic Mark M., Molina H., Kohsaka S., Di Giannatale A., Ceder S., et al. 2015. Tumour exosome integrins determine organotropic metastasis. Nature. 527:329–335. 10.1038/nature15756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaman S., and Detmar M.. 2014. Mechanisms of lymphatic metastasis. J. Clin. Invest. 124:922–928. 10.1172/JCI71606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke J.J., Flaherty K.T., Ribas A., and Long G.V.. 2017. Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nat. Rev. Clin. Oncol. 14:463–482. 10.1038/nrclinonc.2017.43 [DOI] [PubMed] [Google Scholar]
- Mactier S., Kaufman K.L., Wang P., Crossett B., Pupo G.M., Kohnke P.L., Thompson J.F., Scolyer R.A., Yang J.Y., Mann G.J., and Christopherson R.I.. 2014. Protein signatures correspond to survival outcomes of AJCC stage III melanoma patients. Pigment Cell Melanoma Res. 27:1106–1116. 10.1111/pcmr.12290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möhrmann L., Huang H.J., Hong D.S., Tsimberidou A.M., Fu S., Piha-Paul S.A., Subbiah V., Karp D.D., Naing A., Krug A., et al. 2018. Liquid biopsies using plasma exosomal nucleic acids and plasma cell-free DNA compared with clinical outcomes of patients with advanced cancers. Clin. Cancer Res. 24:181–188. 10.1158/1078-0432.CCR-17-2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H., Alečković M., Lavotshkin S., Matei I., Costa-Silva B., Moreno-Bueno G., Hergueta-Redondo M., Williams C., García-Santos G., Ghajar C., et al. 2012. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 18:883–891. 10.1038/nm.2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H., Zhang H., Matei I.R., Costa-Silva B., Hoshino A., Rodrigues G., Psaila B., Kaplan R.N., Bromberg J.F., Kang Y., et al. 2017. Pre-metastatic niches: organ-specific homes for metastases. Nat. Rev. Cancer. 17:302–317. 10.1038/nrc.2017.6 [DOI] [PubMed] [Google Scholar]
- Pucci F., Garris C., Lai C.P., Newton A., Pfirschke C., Engblom C., Alvarez D., Sprachman M., Evavold C., Magnuson A., et al. 2016. SCS macrophages suppress melanoma by restricting tumor-derived vesicle-B cell interactions. Science. 352:242–246. 10.1126/science.aaf1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski P., Nowecki Z.I., Kulik J., Ruka W., and Siedlecki J.A.. 2008. Molecular staging by multimarker reverse transcriptase-polymerase chain reaction assay of lymphatic drainage and blood from melanoma patients after lymph node dissection. Melanoma Res. 18:246–252. 10.1097/CMR.0b013e328307bf3f [DOI] [PubMed] [Google Scholar]
- Santiago-Walker A., Gagnon R., Mazumdar J., Casey M., Long G.V., Schadendorf D., Flaherty K., Kefford R., Hauschild A., Hwu P., et al. 2016. Correlation of BRAF mutation status in circulating-free DNA and tumor and association with clinical outcome across four BRAFi and MEKi clinical trials. Clin. Cancer Res. 22:567–574. 10.1158/1078-0432.CCR-15-0321 [DOI] [PubMed] [Google Scholar]
- Simpson R.J., Kalra H., and Mathivanan S.. 2012. ExoCarta as a resource for exosomal research. J. Extracell. Vesicles. 1:18374 10.3402/jev.v1i0.18374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S., Vannberg F.O., and Dixon J.B.. 2016. Lymphatic transport of exosomes as a rapid route of information dissemination to the lymph node. Sci. Rep. 6:24436 10.1038/srep24436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur B.K., Zhang H., Becker A., Matei I., Huang Y., Costa-Silva B., Zheng Y., Hoshino A., Brazier H., Xiang J., et al. 2014. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 24:766–769. 10.1038/cr.2014.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Włodzimierz R., Rutkowski P., Nowecki Z.I., Kulik J., Nasierowska-Guttmejer A., and Siedlecki J.A.. 2004. Detection of melanoma cells in the lymphatic drainage after lymph node dissection in melanoma patients by using two-marker reverse transcriptase-polymerase chain reaction assay. Ann. Surg. Oncol. 11:988–997. 10.1245/ASO.2004.03.023 [DOI] [PubMed] [Google Scholar]
- Yang S., Che S.P., Kurywchak P., Tavormina J.L., Gansmo L.B., Correa de Sampaio P., Tachezy M., Bockhorn M., Gebauer F., Haltom A.R., et al. 2017. Detection of mutant KRAS and TP53 DNA in circulating exosomes from healthy individuals and patients with pancreatic cancer. Cancer Biol. Ther. 18:158–165. 10.1080/15384047.2017.1281499 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.