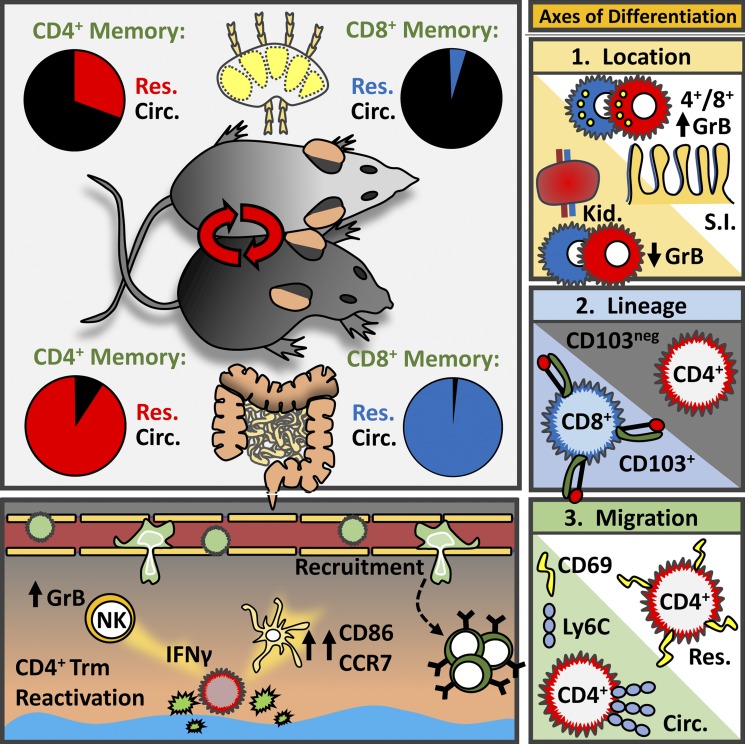
CD4 T cell localization impacts function and differentiation. Beura et al. show that memory CD4+ T cells are largely resident in both lymphoid and non-lymphoid tissues, organize local recall responses, and share overlapping transcriptional and location-specific features with CD8+ TRM.
Abstract
This study examines the extent to which memory CD4+ T cells share immunosurveillance strategies with CD8+ resident memory T cells (TRM). After acute viral infection, memory CD4+ T cells predominantly used residence to survey nonlymphoid tissues, albeit not as stringently as observed for CD8+ T cells. In contrast, memory CD4+ T cells were more likely to be resident within lymphoid organs than CD8+ T cells. Migration properties of memory-phenotype CD4+ T cells in non-SPF parabionts were similar, generalizing these results to diverse infections and conditions. CD4+ and CD8+ TRM shared overlapping transcriptional signatures and location-specific features, such as granzyme B expression in the small intestine, revealing tissue-specific and migration property–specific, in addition to lineage-specific, differentiation programs. Functionally, mucosal CD4+ TRM reactivation locally triggered both chemokine expression and broad immune cell activation. Thus, residence provides a dominant mechanism for regionalizing CD4+ T cell immunity, and location enforces shared transcriptional, phenotypic, and functional properties with CD8+ T cells.
Graphical Abstract
Introduction
Immunosurveillance by naive T cells is biased toward secondary lymphoid organs (SLOs) via a selective program of recirculation that uses blood and lymphatic vessels as conduits. CD8+ memory T cells are typically 103- to 104-fold more abundant than their naive counterparts, which provides the numerical luxury to extend direct immunosurveillance more broadly, including to visceral, mucosal, and barrier organs. Within nonlymphoid tissues (NLTs), CD8+ T cell immunosurveillance is generally dominated by resident populations. Resident memory T cells (TRM) are parked within tissues and do not recirculate through blood and lymphatics like their naive counterparts (Schenkel and Masopust, 2014; Carbone, 2015). CD8+ TRM have also been reported in SLOs, although these are typically rare after systemic primary infections (Schenkel et al., 2014b; Beura et al., 2018).
The extent to which residence contributes to global memory CD4+ T cell surveillance is less clear. First, antiviral antigen-specific memory CD4+ T cells are typically much less abundant than their CD8+ T cell counterparts (Seder and Ahmed, 2003; Surh and Sprent, 2008; Taylor and Jenkins, 2011), and thus may require different strategies for patrolling the organism for evidence of reinfection. Moreover, the proportion of blood-borne memory CD4+ T cells that express an effector memory phenotype is often higher than observed for CD8+ T cells, which may be consistent with nonlymphoid recirculation strategies (Nascimbeni et al., 2004). Moreover, early reports documenting CD8+ TRM in skin highlighted that CD4+ memory T cells were almost entirely comprised of a recirculating population in the skin and reproductive mucosa (Gebhardt et al., 2011), establishing a precedent that CD8+ and CD4+ T cells may obey fundamentally different rules of NLT immunosurveillance. However, firm evidence for CD4+ TRM in the reproductive mucosa has been reported (Iijima and Iwasaki, 2014; Stary et al., 2015). Follow-up studies indicated that memory CD4+ T cells in resting mouse skin equilibrated with circulation, although there was a biased retention of perifollicular CD4+ T cells after herpes simplex virus infection, and inflammation altered the equilibration set-point (Collins et al., 2016). Similarly, after Candida albicans infection, mouse skin was shown to harbor both resident and migratory CD4 memory T cells (Park et al., 2018). In support of recirculation, CD4+ T cells expressing intermediate levels of CCR7 and CD62L have been shown to egress from the skin of specific pathogen–free (SPF) mice (Bromley et al., 2013). In humans, alemtuzumab (anti-CD52) depletes circulating cells, but leaves behind CCR7− CD4+ T cells in skin, supporting that they are resident. However, CD62L+/CCR7+ (central memory T cell [TCM]) and CD62L−/CCR7+ (migratory memory T cell) CD4+ T cells are depleted, indicating skin recirculation (Watanabe et al., 2015). In a separate study, CD4+ T cells that confer protective immunity against Leishmania major were shown to be resident by skin grafting experiments (Glennie et al., 2015). While skin surveillance by memory CD8+ T cells appears dominated by residence, memory CD4+ T cell immunosurveillance may be more complex.
Reports have differed regarding the equilibration of lung memory CD8+ T cells with the circulating population (Wu et al., 2014; Takamura et al., 2016; Slütter et al., 2017). However, several studies indicate the dominant presence of CD4+ TRM in lung or nasal mucosa, where they may be critical for protection (Teijaro et al., 2011; Turner et al., 2014, 2018; Wilk et al., 2017; Allen et al., 2018; Hondowicz et al., 2018; Oja et al., 2018; Smith et al., 2018). Evidence for CD4+ TRM also extends to the small intestine, bone marrow, and liver (Romagnoli et al., 2017; Steinfelder et al., 2017; Benoun et al., 2018; Siracusa et al., 2018).
Many TRM stably express CD69, and most memory CD4+ and CD8+ T cells isolated from lymphoid or NLTs of human cadavers express CD69 (Sathaliyawala et al., 2013; Thome et al., 2014). However, reports in non-SPF mice indicate that CD69 expression may not be sufficient to infer residence (Beura et al., 2018). Indeed, residence can be difficult to quantify. First, there are no perfect markers; unequivocal evaluation really still depends on migration experiments, and experimental approaches have varied considerably. Second, vascular contamination has historically led to overestimation (or complete misinterpretation) of recirculation, although this can be solved with intravascular staining in animal models (Galkina et al., 2005; Anderson et al., 2014). Third, TRM may have very poor isolation efficiency, causing them to be grossly underestimated, sometimes by a factor of >50. Quantitative immunofluorescence microscopy (QIM) enumerates TRM in situ and has shown that cell isolation is subject to sampling bias and overestimations of recirculation (Steinert et al., 2015).
To summarize, among primary memory CD8+ T cells, LN surveillance patterns are typically dominated by recirculation whereas NLTs are dominated by residence. There is less consensus for CD4+ T cells because there have been fewer studies and they have produced varied conclusions or indicated more complexity. It should be noted that CD4+ T cell migration studies have typically focused on a single tissue. To perform a more comprehensive analysis, we interrogated migration properties in several tissues after lymphocytic choriomeningitis virus (LCMV) infection (which we found establishes broadly distributed memory CD4+ T cells), or in “dirty” mice (laboratory mice that have been cohoused with non-SPF pet store mice) that have very diverse microbial experience. Here we report that (1) residence is the dominant mechanism for CD4+ memory T cell immunosurveillance in NLTs and to a substantial extent in lymphoid tissues; (2) CD4+ TRM share phenotypic, transcriptional, and functional properties with CD8+ TRM inhabiting similar locations; and (3) reactivation of mucosal CD4+ TRM triggers local immune activation.
Results
LCMV-Armstrong infection establishes broadly distributed memory CD4+ T cells
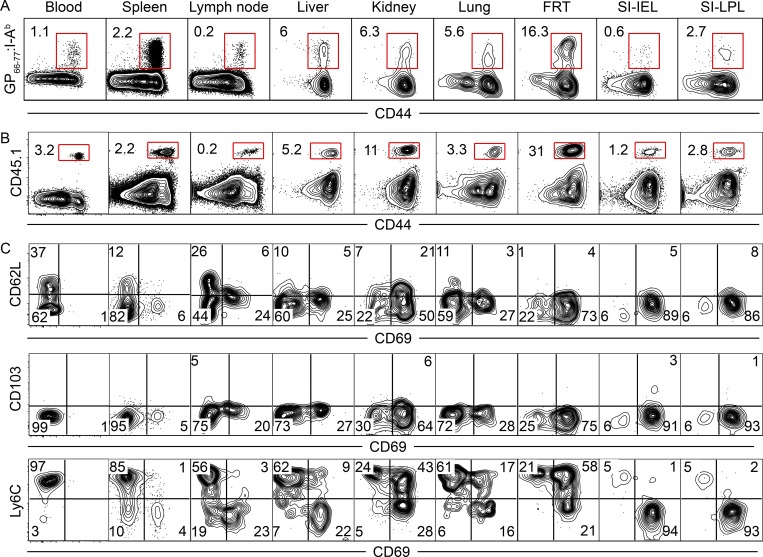
We assessed the distribution of memory CD4+ T cells after a systemic acute viral infection. Adult female C57BL/6J mice were injected with 2 × 105 PFU LCMV (Armstrong strain) i.p., which is putatively cleared about 1 wk after infection (Wherry et al., 2003). GP66-77 represents an immunodominant CD4+ T cell LCMV epitope, and GP66-77:I-Ab MHCII tetramers were constructed as described (Pepper et al., 2011). 50 d after LCMV infection, lymphocytes were isolated from blood, SLOs, and several nonlymphoid organs. Tetramer staining revealed that LCMV-specific memory CD4+ T cells were present in all locations examined (Fig. 1 A). The frequency of CD4+ T cells that stained with GP66-77:I-Ab MHCII tetramers varied considerably among tissues, being lowest in LNs and the epithelium of the small intestine, and highest in the female reproductive tract (FRT) and kidney.
Figure 1.
LCMV-Armstrong infection establishes broadly distributed memory CD4+ T cells. (A) C57BL/6J mice were infected with 2 × 105 PFU LCMV-Armstrong. Distribution of endogenous (GP66-77:I-Ab MHCII tetramer+) memory CD4+ T cells within blood, SLOs, and NLTs was assessed 50 d after infection. (B and C) CD45.1+ SMARTA CD4+ T cells were transferred to C57BL/6J mice 1 d before infection with LCMV-Armstrong. Mice were analyzed 64 d after infection. Distribution (B) and phenotype (C) of SMARTA CD4+ memory T cells within blood, SLOs, and NLTs are shown. Data are representative of three independent experiments with n = 4 mice per experiment (A) or three separate experiments with n = 3 mice per experiment (B and C). SI, small intestine; IEL, intraepithelial lymphocytes; LPL, lamina propria lymphocytes.
CD45.1+ naive SMARTA transgenic CD4+ T cells specific for GP66-77 were transferred to naive adult female C57BL/6J mice the day before LCMV infection, then analyzed 64 d later. Similar to the endogenous GP66-77-specific CD4+ T cell population (Fig. 1 A), SMARTA CD4+ memory T cells were isolated from all locations examined, and were least abundant in LNs and small intestine epithelium and most frequent in the FRT and kidney (Fig. 1 B). Further phenotypic analysis via flow cytometry revealed CD62L expression was restricted to blood and secondary lymphoid populations (Fig. 1 C). CD69 expression was abundant in NLTs, was expressed by a modest portion of CD62L− SMARTA CD4+ T cells in LNs and spleen, and was absent from blood (Fig. 1 C). Ly6C expression varied widely with location, whereas CD103 was not expressed anywhere (with the exception of a very small SMARTA population isolated from small intestine epithelium and lamina propria; Fig. 1 C). The adoption of the CD69+ CD62L− Ly6Clo phenotype on gut SMARTA CD4+ T cells was gradual (Fig. S1). These results indicate that both polyclonal endogenous and SMARTA TCR transgenic CD4+ T cells are broadly distributed after LCMV-Armstrong infection of SPF mice.
Residence is a dominant feature of memory CD4+ T cells in NLTs
CD69 expression was stably maintained on LCMV-specific memory CD4+ T cells long after viral clearance, suggesting that they could be TRM (Fig. 1 and data not shown). Previous studies on CD8+ T cell migration properties demonstrated that the isolation efficiency of TRM is poor relative to recirculating cells, an issue that may be related to survival (Steinert et al., 2015; Fernandez-Ruiz et al., 2016; Georgiev et al., 2018). Accordingly, immunohistological analyses that relied on imaging rather than cell extraction provided more accurate measurements of the relative abundance of TRM versus recirculating cells (Steinert et al., 2015).
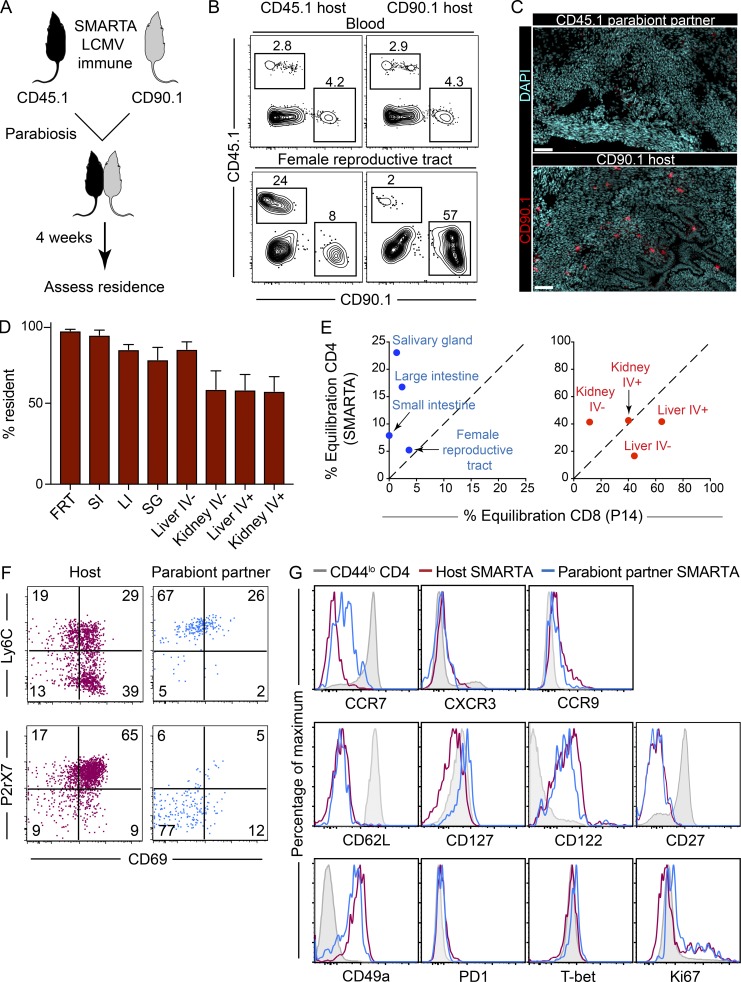
SMARTA transgenic T cells have an advantage over endogenous CD4+ T cells because they can be easily measured by both flow cytometry and immunofluorescence via staining for a readily identifiable congenic marker (either CD45.1 or CD90.1). To examine the recirculation properties of CD4+ memory T cells after LCMV infection, we transferred either CD45.1+ or CD90.1+ SMARTA CD4+ T cells to naive mice. The following day, both mice received LCMV-Armstrong. 45–60 d later, the vasculature of the two mice was conjoined via parabiotic surgery (Fig. 2 A). 4 wk later, tissues were analyzed by flow cytometry (Fig. 2 B) or by QIM (Fig. 2, C and D), as previously described (Steinert et al., 2015). Each NLT that we examined was populated by SMARTA CD4+ memory T cells that failed to equilibrate, and were thus resident for at least the 4-wk duration of the experiment. Interestingly, even the vascular contiguous compartments of liver and kidney had more TRM than recirculating memory CD4+ T cells (Fig. 2 D). We previously used QIM to evaluate LCMV-specific memory CD8+ T cells in parabionts, which provided an opportunity to compare CD4+ and CD8+ T cell equilibration in the same infection model and in diverse anatomical locations. As shown in Fig. 2 E, while memory CD4+ T cells predominantly used residence to survey NLTs, they did so with less stringency than has been observed for CD8+ T cells. However, memory CD4+ T cells were equally resident, if not more so, than memory CD8+ T cells within the compartments of kidney and liver that were permissive to intravascular staining (e.g., sinusoids and glomeruli).
Figure 2.
Residence is a dominant feature of memory CD4+ T cells in NLTs. (A) CD90.1+ and CD45.1+ SMARTA memory immune chimeras were conjoined via parabiosis 45–60 d after LCMV-Armstrong infection. Parabiont pairs were analyzed 4 wk after parabiosis. (B) Frequency of host- and parabiont partner–derived SMARTA cells in blood and FRT were determined by flow cytometric analysis. Plots are gated on total live CD4+ T cells. (C) FRT of both mice stained with DAPI (cyan) and anti-CD90.1 (to label SMARTA cells; red). Bars, 50 µm. (D) Number of CD45.1+ and CD90.1+ SMARTA CD4+ T cells in both parabiont partners in indicated tissues was calculated by QIM. Percentage of CD4+ residence was determined by the formula described previously (Steinert et al., 2015). IV+ indicates cells labeled by anti-CD45 antibody injected i.v. 3 min before sacrifice. LI, large intestine; SG, salivary gland. (E) Equilibration of CD4+ SMARTA cells and CD8+ P14 cells in NLTs and vascular compartments following LCMV-Armstrong infection (as determined by QIM). (F and G) Phenotypic comparison of host (CD90.1+, maroon)– and partner (CD45.1+, blue)–derived SMARTA CD4+ T cells in FRT. CD44lo naive CD4+ T cells are shown in gray. Data are representative of two separate experiments with a total of 12 individual mice. Bars represent mean ± SEM. SI, small intestine.
Consistent with the interpretation that NLTs were predominantly surveyed by resident populations of SMARTA CD4+ memory T cells, we found that the phenotype of host cells was distinct from migrated SMARTA cells (derived from the parabiont partner) that were recovered from the FRT, with a CD69+, P2rX7+, and Ly6C+/− phenotype being associated with TRM. In contrast, migrated populations were primarily CD69−, P2rX7−, and Ly6C+ (Fig. 2 F). Further characterization revealed that migrating cells also expressed higher levels of CCR7 and CD127, and marginally distinct levels of CD49a, T-bet, and CD122, but also exhibited many similarities with TRM (Fig. 2 G).
Taken together, these data indicate that after acute LCMV infection of SPF mice, TRM were the dominant surveyor of all NLTs examined, and they exhibited unique phenotypic characteristics when compared with their migratory counterparts.
Abundant CD4+ TRM in SLOs
In Fig. 1, we noted that a substantive fraction of SMARTA CD4+ memory T cells expressed CD69 in SLOs, particularly within LNs. Most TRM express CD69; however, CD69 expression is insufficient to infer stable residence, at least by memory CD8+ T cells within SLOs (Beura et al., 2018). Moreover, activated effector and CD4+ follicular helper T cells (TFH) also express CD69 presumably in response to recent TCR stimulation. We next asked if SLOs were surveyed by bona fide CD4+ TRM following acute LCMV infection.
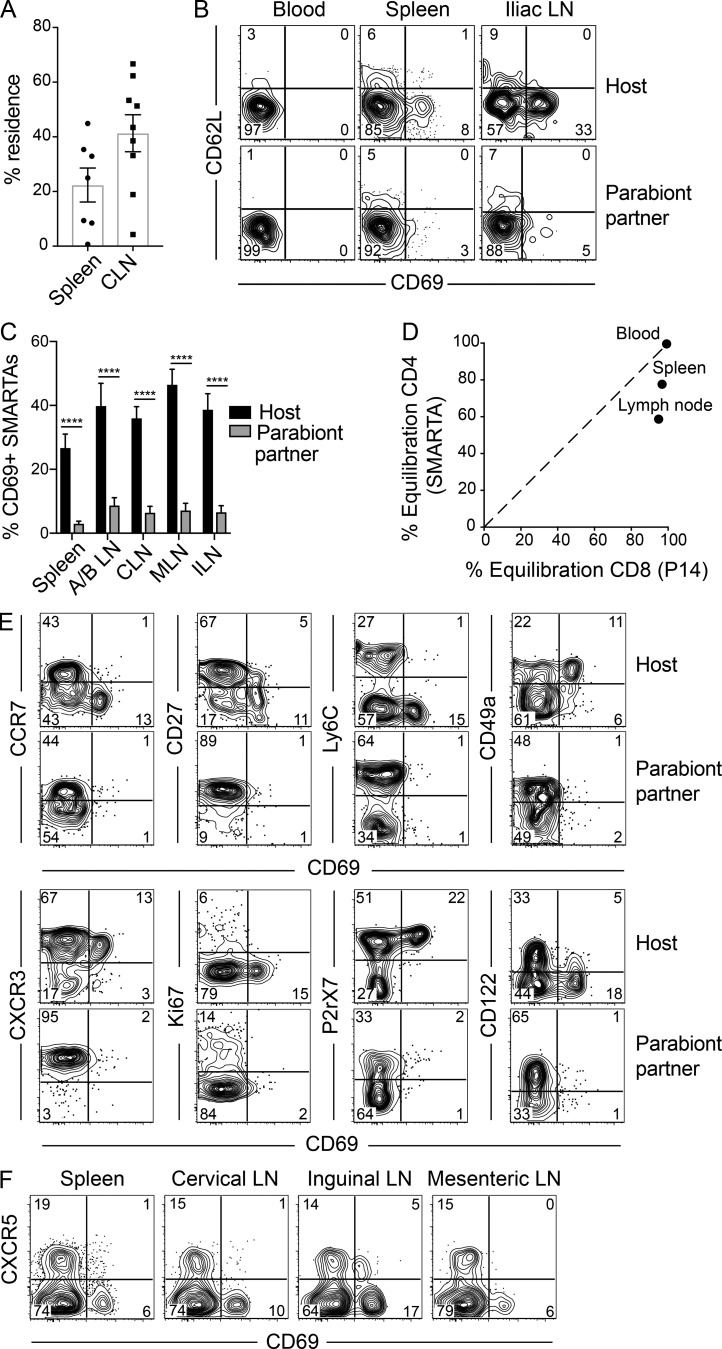
Parabiosis experiments and QIM analysis, as in Fig. 2, revealed a bias toward SMARTA CD4+ memory T cells of host origin in spleen and LNs, which supported the interpretation of stable residence, for at least the 28-d duration of conjoining (cervical LN shown, plotted as percent resident in Fig. 3 A). Phenotypic profiling indicated that CD69+ SMARTA CD4+ memory T cells were particularly refractory to equilibration between parabiont pairs (Fig. 3, B and C). This suggests that TRM may be enriched within the CD69+ population, although this distinction was not absolute because we did observe some equilibration among CD69+ cells. Notably, after LCMV infection, antigen-specific memory CD4+ T cells were more likely to be resident than memory CD8+ T cells within SLOs (Fig. 3 D).
Figure 3.
Abundant CD4+ TRM cells in SLOs. (A–D) LCMV-immune SMARTA parabionts were generated as described in Fig. 2 A. (A) Percent residence of SMARTA CD4+ T cells in spleen and cervical LNs (CLN) as determined by QIM. (B) CD69 and CD62L expression on host- and partner-derived SMARTA CD4+ T cells in blood, spleen, and iliac LNs. (C) Percent CD69+ SMARTA cells from spleen and LNs (combined axillary/brachial [A/B LN], cervical, mesenteric [MLN], and iliac [ILN]) of host and parabiont partner as determined by flow cytometry. (D) Equilibration of CD4+ SMARTA cells and CD8+ P14 cells in spleen and cervical LN following LCMV-Armstrong infection (as determined by QIM). (E) Phenotypic analysis of host- and partner-derived SMARTA cells isolated from spleen 28 d after parabiosis. (F) CD69 and CXCR5 expression on SMARTA CD4+ T cells of SMARTA immune chimeras 75 d after LCMV-Armstrong infection. Data are representative of two separate experiments with a total of 12 individual mice (A–D) or two independent experiment with n = 4 mice per experiment (E). ****, P < 0.0001. Two-way ANOVA with Sidak’s multiple comparison test (C). Bars represent mean ± SEM.
In comparing the phenotypes of host- and partner-derived SMARTA CD4+ T cells isolated from spleen, we found that host SMARTA cells were phenotypically distinct from SMARTA cells derived from the parabiont partner (Fig. 3 E). Host SMARTA cells displayed a phenotypic profile associated with TRM (CD69+, P2rX7+), whereas migrated populations were primarily CD69− and P2rX7−. TRM cells in spleen expressed high levels of CD49a but low levels of Ly6C and CD27, suggesting similarities with NLT TRM.
It should be noted that effector and certain memory CD4+ TFH have been shown to express CD69 (Fazilleau et al., 2009; Asrir et al., 2017). However, when we analyzed SMARTA CD4+ memory T cells 75 d after LCMV infection, we failed to detect cells expressing the CD69+ CXCR5+ phenotype that would otherwise indicate the presence of TFH cells within our analyzed SLO TRM population (Fig. 3 F). Moreover, previous studies showed waning of TFH populations after effector time points in acute LCMV infection (Hale et al., 2013). In summary, we identified a population of virus-specific memory CD4+ T cells in SLO that are bona fide residents, share some phenotypic signatures with nonlymphoid TRM, and are distinct from TFH.
Comparison of mucosal memory CD4+ and CD8+ T cells
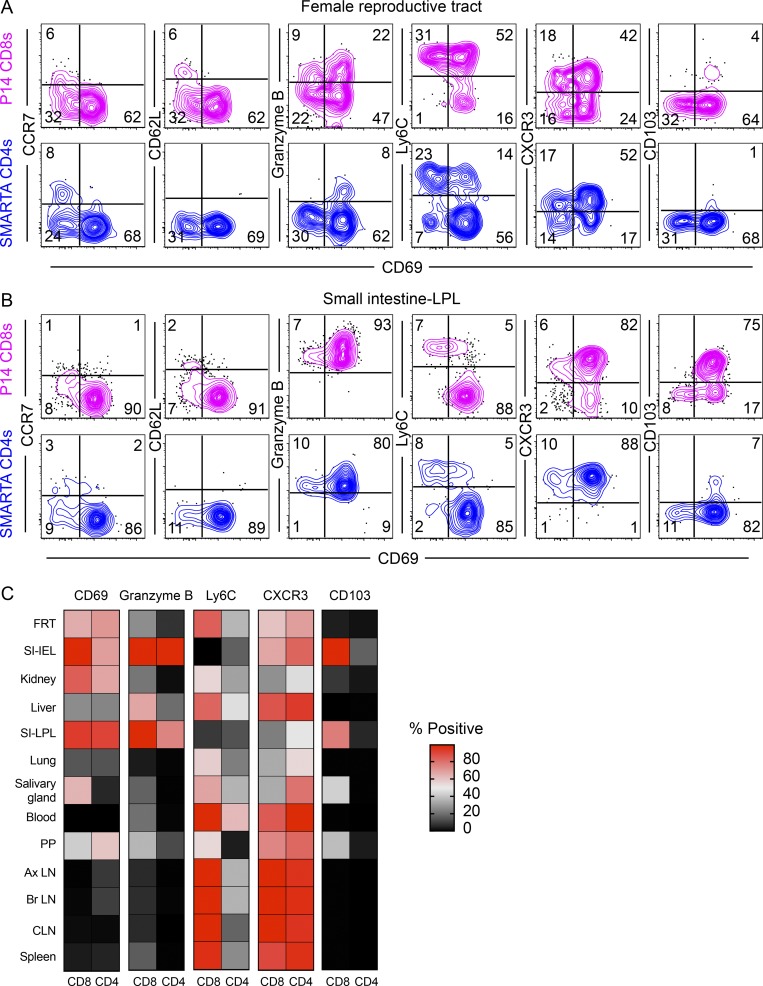
It has been hypothesized that tissue microenvironments shape memory T cell phenotypes. To explore this concept further, we wanted to compare both CD4+ and CD8+ memory T cells (including TRM) that were specific for the same virus and isolated from the same tissue. P14 TCR transgenic CD8+ T cells, specific for the H-2Db–restricted LCMV GP33-41 epitope, were transferred to naive mice, which were then infected with LCMV-Armstrong 1 d later. 40 d after infection, memory P14 CD8+ T cells were isolated and assessed by flow cytometry. For comparison, we examined SMARTA transgenic CD4+ T cells, also isolated 40 d after LCMV infection. Fig. 4, A and B, shows data from the FRT and the small intestine lamina propria, two tissues where LCMV-specific CD8+ TRM have been previously well characterized, and that also highlight tissue-specific differences. A broader tissue survey is summarized in Fig. 4 C. With respect to many phenotypic markers, memory CD4+ and CD8+ T cells were most aligned based on location rather than lineage. To highlight key lineage-specific differences, memory CD4+ T cells were more likely to express CD69 within SLOs, did not express CD103 regardless of anatomical compartment, and exhibited lower Ly6C expression than their CD8+ tissue counterparts. Interestingly, granzyme B expression by resting memory CD4+ and CD8+ T cells was quite similar, and expression was constitutively maintained particularly within the small intestine epithelium and lamina propria.
Figure 4.
Comparison of mucosal memory CD4+ and CD8+ T cells after LCMV infection. SMARTA and P14 immune chimeras were prepared by transferring naive SMARTA CD4+ and P14 CD8+ T cells to C57BL/6J mice and infecting the recipients with LCMV-Armstrong 1 d after. (A and B) Phenotypic comparison between memory P14 CD8+ T cells and SMARTA CD4+ T cells (40 d after infection) from (A) FRT and (B) small intestine lamina propria (SI-LPL) are shown. (C) Heatmap showing comparative expression of indicated markers between P14 CD8+ and SMARTA CD4+ T cells in various NLTs and SLOs. Data are representative of two independent experiments with n = 4 mice per treatment/experiment. CLN, cervical LN; Ax LN, axillary LN; Br LN, brachial LN; PP, Peyer's patches.
Common transcriptional signature of resident T cells
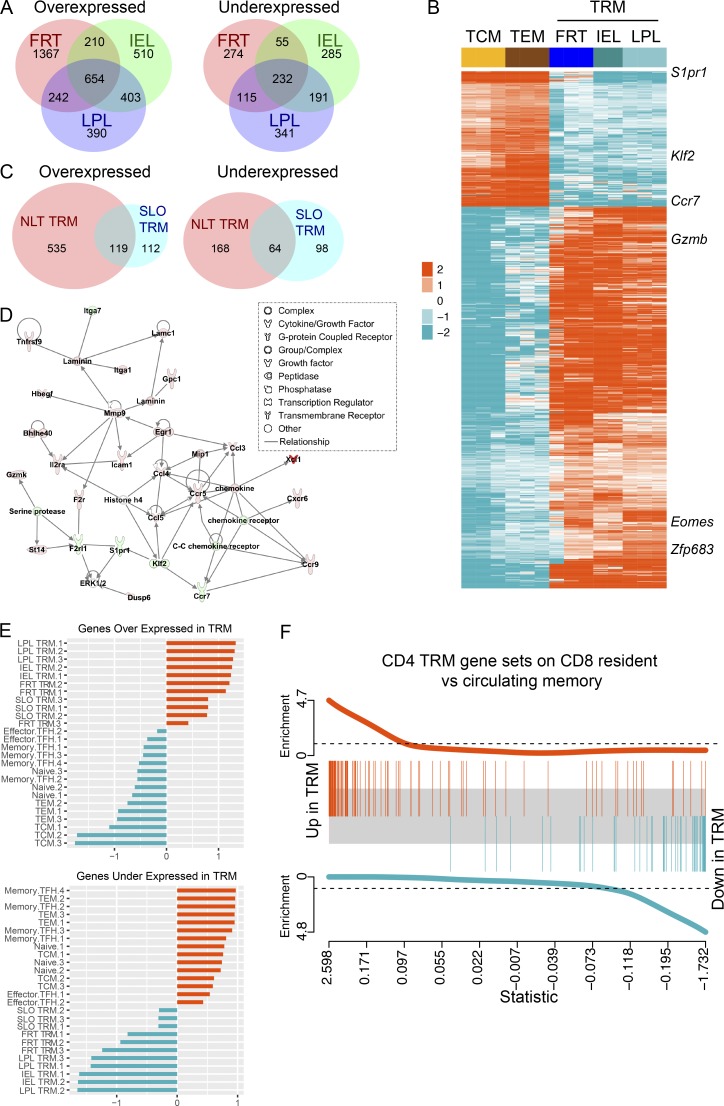
We next assessed the extent to which TRM gene signatures are shared among different mucosal compartments. Here, we used RNA sequencing (RNA-seq) transcriptional profiling, as flow cytometry (Figs. 1, 2, 3, and 4) is limited to only a few molecules. 54 d after LCMV infection, CD69+ SMARTA CD4+ memory T cells were isolated and sorted from the FRT, small intestine epithelium (IEL), and small intestine lamina propria. Sort-purified CD62L+ (TCM) and CD62L−/CD69− (effector memory T cell [TEM]) SMARTA CD4+ T cells were isolated from spleen. Differential gene expression (DGE) analysis was performed between each NLT CD69+ SMARTA CD4+ T cell population and circulating TCM and TEM (criteria for significance: false discovery rate [FDR] ≤0.05 and absolute fold change value ≥2). Here, we observed transcriptional heterogeneity among TRM isolated from different mucosal tissues, particularly the FRT (Fig. 5 A and Table S2). Nonetheless, we identified a common set of 654 overexpressed and 232 underexpressed genes in all NLT TRM compared with TCM and TEM (Fig. 5, A and B; and Table S2). The common gene sets included transcripts that were previously identified to be associated with TRM cells, including underexpression of S1pr1, Klf2, and Ccr7 and overexpression of Zfp683 and Gzmb (Fig. 5 B and Table S2). Consistent with our flow cytometric observation and previously reported human and mouse CD4+ TRM datasets (Thome et al., 2014; Turner et al., 2014; Kumar et al., 2017), Itgae (which encodes CD103) was not differentially expressed in CD4+ NLT TRM versus recirculating subsets. Pathway enrichment analysis highlighted both tissue-specific and shared TRM pathways. Pathways represented within a refined core resident gene signature include those involved in cell communication, cell junction organization, nuclear receptor signaling, and Eph-ephrin signaling (Fig. S2 A).
Figure 5.
Common transcriptional signature of resident memory T cells. NLT CD69+ SMARTA memory CD4+ T cells were isolated and sorted from the FRT, small intestine epithelium (IEL), and small intestine lamina propria (LPL). TCM (CD62L+), TEM (CD62L−/CD69−), and SLO TRM (CD69+CD62L−) SMARTA CD4+ T cells were isolated and sorted from spleen. Cells were obtained 54 d after LCMV infection. (A) Venn diagrams showing the number of unique and common DEGs between NLT CD69+ SMARTA TRM cells and TCM and TEM (criteria for significance: FDR ≤0.05 and absolute value of fold change ≥2). Full lists of common and unique DEGs are provided in Table S2. (B) Heatmap generated from clustering analysis with normalized expression of genes that showed differential expression in all NLT TRM as identified in A. Several known resident memory genes are highlighted. (C) Venn diagram showing the overlap of overexpressed and underexpressed genes between NLT CD69+ SMARTA TRM cells and SLO TRM cells relative to circulating TCM and TEM cells. A full list of DEGs is provided in Table S2. (D) Most enriched gene network. IPA was used to generate the network from the genes shared by both SLO TRM and NLT TRM (identified in Fig. 5 C) and the average fold change value obtained from averaging the absolute fold change from all comparisons of TRM to circulating TCM and TEM cells. Edges (lines and arrows) represent direct interactions as supported by information in the IPA database. Genes included in the TRM gene list have a colored node. Node color indicates up-regulated genes (orange) and down-regulated genes (blue) in TRM. Node shapes represent functional classes of gene product. (E) Comparison of CD4+ datasets with a previously published dataset containing transcript profiles of mouse effector and memory TFH from an LCMV infection model (GSE43863; Hale et al., 2013). Gene set summary values were calculated for each sample. Z scores of gene set summary scores are plotted. (F) GSEA plot. A CD8+ ranked gene list was obtained from the output of a GSEA enrichment test comparing previously published gut TRM to circulating memory samples (GSE47045; Mackay et al., 2013). The limma barcode plot function was used to plot enrichment of the overexpressed and underexpressed CD4+ TRM gene sets in the CD8+ ranked gene list.
Thus far, our data raised the hypothesis that there were three broad axes of regulation that contributed to T cell gene expression: NLT versus SLO distribution, resident versus recirculating migration pattern, and CD8+ versus CD4+ memory lineage. To explore this more deeply, we first compared gene expression by TRM isolated from spleen to the core TRM signature common to all NLT TRM populations. Indeed, principal component analysis (PCA) highlighted that many gene expression patterns are uncoupled from residence, and align with location (Fig. S2 B). Using the same criteria for significance as in Fig. 5 A, we identified 393 differentially expressed genes (DEGs) in SLO TRM (from sort-purified CD69+ CD62L− SMARTA CD4+ T cells isolated from spleen) relative to both TCM and TEM. Of these genes, 119 over- and 64 underexpressed genes were shared with the core NLT TRM gene signature (Fig. 5 C and Table S2). Ingenuity Pathway Analysis (IPA) was used to identify canonical pathways and gene networks for this core set of TRM genes (data not shown). Leukocyte extravasation signaling, neuroinflammation signaling, IL-8 signaling, and integrin signaling pathways were all up-regulated in TRM relative to circulating memory cells (data not shown). The top two most significant gene networks contained 48 and 46 genes, respectively, from the core set of TRM genes, and were enriched for genes involved in cell-mediated immune response, cellular movement, and hematological system development and function (Fig. 5 D and Fig. S2 C). The most significant network included Ccl4, Ccl5, Ccl3l3, Ccr5, Ccr7, Ccr9, Gzmk, Klf2, Xcl1, Icam1, Itga7m, Itga1, Il2ra, and S1pr1 (Fig. 5 D).
We then used this core set of TRM genes to determine whether SLO TRM cells are transcriptionally distinct from CD4+ T follicular helper cells (TFH), which comprise a separate resident SLO population. To answer this question, we compared our gene expression data from the aforementioned CD4+ T cell subsets to a previously published dataset containing transcriptional profiles of mouse naive SMARTA CD4+ T and effector and memory TFH cells from an LCMV infection model (GSE43863; Hale et al., 2013). We first calculated gene set summary values for each sample (see Materials and methods) and then calculated a Z score from the summary values. We found that gene set summary value Z scores for the NLT and SLO TRM samples move in the same direction, while the circulating and TFH sample Z scores move in the opposite direction (Fig. 5 E). These data indicate that SLO TRM are transcriptionally distinct from TFH.
Last, we tested the extent to which our core CD4+ TRM gene expression signature (shared by SLO, FRT, IEL, and lamina propria TRM) would overlap with a previously reported core gene signature shared by CD8+ TRM (skin, IEL, and lung TRM; Mackay et al., 2013). We found a remarkable degree of enrichment of our CD4+ TRM genes in the CD8+ TRM dataset (Fig. 5 F). Taken together, these results reveal a shared gene signature of tissue residence that transcends anatomical location and T cell lineage, yet discriminates TRM from TFH as well as TCM and TEM.
Sensing and alarm function of mucosal CD4+ TRM
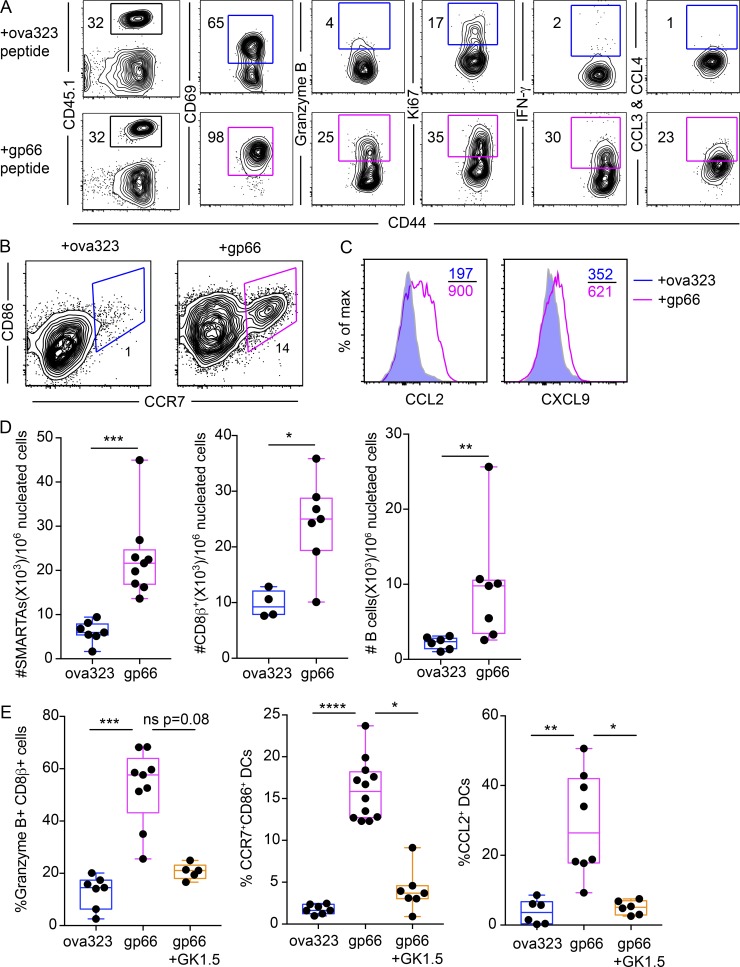
Because CD4+ TRM are the dominant surveyor of frontline mucosal organs, we next interrogated their function in the event of reinfection. To this end, we established SMARTA memory immune chimeras as described earlier, then reactivated SMARTA CD4+ TRM cells in the FRT by depositing LCMV-gp66 peptide in the cervical lumen (trans-cervical delivery [t.c.]) as previously described (Schenkel et al., 2013). In response to cognate antigen-specific reactivation, SMARTA CD4+ T cells in FRT became uniformly CD69+, and a significant fraction of them also up-regulated the cytotoxic molecule granzyme B, the cytokine IFN-γ, and the chemokines CCL3 and CCL4 (Fig. 6 A). Ki67, a marker associated with cell cycle entry, also increased in response to antigen-specific recall compared with ova323 control peptide (Fig. 6 A).
Figure 6.
Mucosal CD4+ TRM execute sensing and alarm functions. SMARTA memory immune chimeras were challenged transcervically with ova323 (control) or gp66 peptide 35–65 d after LCMV-Armstrong infection. (A) Phenotypic analysis of SMARTA CD4+ T cells in FRT 16 h following reactivation. Left-most plots are gated on total live CD4+ T cells. All other plots are gated on CD45.1+ SMARTA CD4+ T cells. (B and C) Expression of maturation markers CCR7, CD86 (B) and chemokines (C) on CD11chi MHCIIhi dendritic cells in the FRT 16 h following peptide challenge. (D) Enumeration of SMARTA CD4+ T cells, CD8β+ T cells, and B cells in the FRT 48 h after recall by QIM. (E) SMARTA memory immune chimeras were injected with either CD4-depleting antibody (GK1.5) or PBS. Treated mice were t.c. challenged with indicated peptides. CD11chi MHCIIhi dendritic cell maturation and CD8+ T cell activation (granzyme B upregulation) in the FRT were assessed 16 h after challenge by flow cytometry. Data are representative of three separate experiments with n = 3 mice/group per experiment (A–D) or two independent experiments with n = 3 mice/group per experiment (E). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant. Mann-Whitney U test (D), Kruskal Wallis one-way ANOVA with Dunn’s multiple comparison test (E). Box plots indicate medians (center lines), 25th and 75th percentiles (bottom and top box edges, respectively), minima and maxima (whiskers), and individual data points (circles).
In response to memory SMARTA reactivation, the classical dendritic cells (CD11chi MHCIIhi) in the FRT increased their surface expression of CD86 and CCR7, markers associated with dendritic cell maturation (Fig. 6 B). Expression of CCL2 and CXCL9 chemokines was also induced on dendritic cells (Fig. 6 C), and SMARTA CD4+ T cells, host CD8+ T cells, and host B cells accumulated within the FRT by 48 h after CD4+ TRM reactivation (Fig. 6 D). The accumulated CD8+ T cells up-regulated granzyme B and to a lesser extent, Ki67, in response to CD4+ T cell activation (Fig. S3 A). To ensure that this process was indeed CD4+ T cell–dependent, we depleted CD4+ T cells in vivo (using the CD4+ T cell depleting antibody GK1.5; see Fig. S3, B and C) before challenging with gp66 peptide. Here, we no longer observed CD8+ T cell activation and dendritic cell maturation (Fig. 6 E). To confirm this rapid elaboration of effector function by local CD4+ T cells is not an artifact of SMARTA transgenic cells, we repeated the local recall in LCMV-endogenous immune mice (no transgenic T cell transfer). To this end, C57BL/6J mice were infected with LCMV-Armstrong, and 63 d after infection, the mice received either gp66 or ova323 control peptide (t.c.). After 16 h, GP66-77:I-Ab tetramer+ T cells were found to up-regulate IFN-γ (Fig. S3 D), as we observed with SMARTA CD4+ T cells. CD4+ TRM reactivation was not limited to peptide challenge. Indeed, when we transcervically rechallenged SMARTA immune chimeras with a virus engineered to express cognate antigen (VV-gp61), SMARTA T cells up-regulated both IFN-γ and granzyme B 24 h after recall (Fig. S3 E).
To determine whether the observed sensing and alarm functions were indeed attributable to resident CD4+ T cells, we established SMARTA memory immune chimeras, then paired them with naive mice via parabiosis. After equilibration of SMARTA cells in the blood, both mice were contemporaneously challenged with LCMV-gp66 peptide transcervically (Fig. S4, A and B). Here we found that granzyme B expression by CD8+ T cells and natural killer (NK) cells, and CD11chi MHCIIhi classical dendritic cell maturation (as assessed by CD86 up-regulation), were magnified in the SMARTA memory parabiont compared with the naive parabiont (Fig. S4, C–E), which supports a specific role for tissue-resident memory in these processes. We further confirmed these findings by selective depletion of circulating SMARTA cells while preferentially preserving a TRM population in the FRT. Here, we administered α-CD90.1 depleting antibody to CD90.1+ SMARTA immune chimeras before transcervical challenge with gp66 peptide (Fig. S5). Taken together, these results suggest that local CD4+ TRM reactivation triggers rapid innate and adaptive immune responses, providing protection from reinfection at mucosal barrier surfaces.
Residence is the dominant mechanism of immune surveillance by CD4+ T cells in non-SPF (dirty) mice
Our findings showed that after acute LCMV infection, CD4+ TRM dominate all nonvascular NLTs that we examined. It was unclear whether these results were generalizable to memory CD4+ T cells in mice that have been exposed to a diverse array of natural mouse pathogens acquired via physiological routes. Moreover, residence may be overestimated in an SPF environment which may be unphysiologically quiescent.
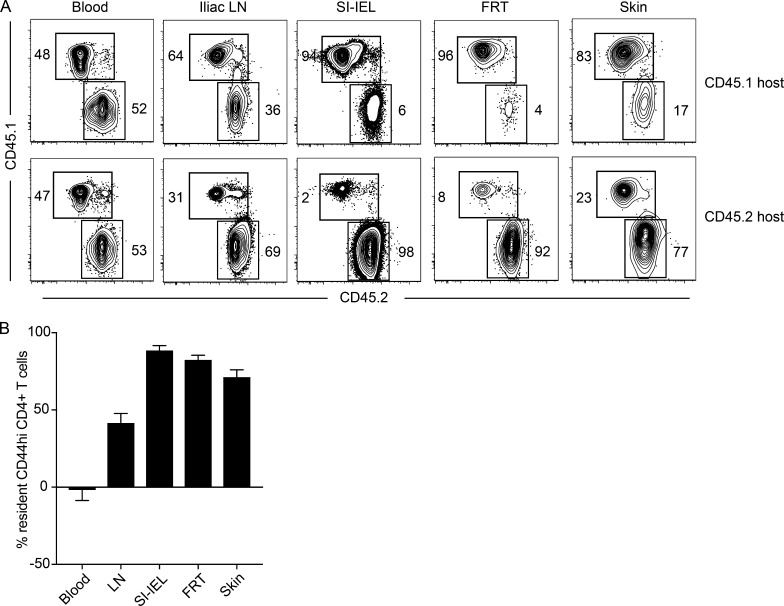
To address these issues, we used a non-SPF dirty mouse model, in which laboratory mice are cohoused with pet shop mice in order to expose them to a diverse spectrum of microbes naturally found in mice. We have previously shown that cohoused dirty laboratory mice are well populated by CD4+ T cells in NLT (Beura et al., 2016). We cohoused both CD45.1+ and CD45.2+ SPF laboratory mice with pet store mice. 60 d later, dirty CD45.1+ and CD45.2+ laboratory mice were removed from cohousing and underwent parabiosis. 4 wk after parabiosis, blood and various tissues were analyzed for memory CD4+ T cells. While the memory CD4+ T cells of both hosts were equally represented in blood, there was significant bias toward host-derived cells in most NLTs (Fig. 7, A and B). Similar to our LCMV-specific findings, 40% of CD44hi CD4+ T cells in the iliac LN of dirty mice were resident (Fig. 7 B). Overall these observations suggest that among CD4+ T cells, TRM are the predominant surveyors of NLTs after diverse infections, and comprise a substantial population in SLOs. These results extend our findings beyond acute LCMV infection and indicate that the dominance of TRM is not an artifact of SPF housing.
Figure 7.
Residence is the dominant mechanism of immune surveillance by CD4+ T cells in non-SPF (dirty) mice. Congenically distinct SPF C57BL/6J mice (CD45.1+ and CD45.2+) and pet store mice were cohoused for 60 d to generate dirty mice. Dirty CD45.1+ and CD45.2+ mice were conjoined via parabiosis for 4 wk before analysis by flow cytometry and QIM. (A) Frequency of host- and partner-derived CD4+ T cells in indicated tissues. Plots are gated on CD44hi CD4+ live T cells. (B) Host- and partner-derived cells were enumerated by flow cytometry–based counting in indicated tissues, and the percent residence of CD44hi CD4 T cells was calculated by the formula described previously (Steinert et al., 2015). SI, small intestine. Bars represent mean ± SEM. Data are representative of three separate experiments with at least three parabiont pairs per experiment.
Discussion
This study sought to help bring forth a generalizable conclusion on residence among memory CD4+ T cells, doing so by investigating migration in many tissues after a single acute viral infection (LCMV) or after a more diverse and agnostic microbial exposure (after cohousing laboratory mice with pet shop mice). Here we found that CD4+ TRM dominated surveillance of all NLTs investigated under these diverse conditions, and surprisingly TRM constituted 30% of the LN CD4+ T cell population as well, which is much larger than what has been observed for CD8+ T cells. Local reactivation of CD4+ TRM triggered accumulation of circulating lymphocytes and local dendritic cell maturation, indicating that CD4+ TRM execute MHCII-initiated sensing and alarm functions within mucosal tissues. Core transcriptional signatures revealed tissue-specific, lineage-specific, and migration property–specific axes of differentiation. These results highlight TRM heterogeneity, and ontogeny models will have to incorporate this newfound complexity.
Conclusions regarding the extent to which CD4+ T cells are resident have been varied. Here, we conjoined dirty mice with diverse microbial experience and confirmed that residence dominated surveillance of all NLT tissues examined, including skin, where several reports have emphasized the presence of recirculating CD4+ T cells (Gebhardt et al., 2011; Bromley et al., 2013; Watanabe et al., 2015; Collins et al., 2016). This suggests that residence is likely the norm, but not absolute. Indeed, we observed some equilibration in most NLTs under investigation, so while it appears that CD4+ T cells favored residence for the duration of our experiments (28 d parabiosis), perhaps there are unknown conditions that promote a more rapidly equilibrating population in certain models. One important issue that we did not address is the longevity of CD4+ TRM, and how that might compare to the durability of recirculating memory CD4+ T cells (which have been proposed to be quite short-lived after acute, quickly resolving infections, at least in mice; Seder and Ahmed, 2003; Surh and Sprent, 2008; Taylor and Jenkins, 2011).
Our work identified a core CD4+ TRM transcriptional signature shared among SLO, FRT, IEL, and lamina propria TRM. Importantly, this core signature differentiated CD4+ TRM from TFH, which comprise a distinct subset of CD4+ T cells that transiently take up residence following infection. Based on our data, we propose at least three axes of differentiation: one driven by the tissue microenvironment (e.g., the maintenance of granzyme B expression by memory CD8+ and CD4+ T cells in the intestine), one driven by lineage (CD4+ vs. CD8+ T cell, e.g., with respect to CD103 expression), and one coupled with migration properties (resident vs. recirculating lifestyle). These axes likely intersect with separate subsetting strategies that relate directly to priming events, including Th1 cells versus Th2 cells (or Th17 cells, T reg cells, etc.), and T cell exhaustion or anergy. Nevertheless, the broad question that remains is what determines programming of memory CD4+ T cell residence. Related queries include regulation of retention and survival strategies, how location relates to CD4+ TRM metabolism, and how metabolism in turn relates to function.
Interest in barrier CD4+ T cell functions stems from their critical role in control of many human infections, especially phagosomal infections such as tuberculosis. CD4+ T cells are also the principal target of HIV, and reactivation of latently infected CD4+ T cells is responsible for sustaining the chronic infection and perpetuating the viral reservoir (Cohn et al., 2018). Previous work indicates that CD8+ TRM defend barrier tissues against infections, in part through elaboration of cytokines, activation of dendritic cells (DCs), and recruitment of B and T cells (Schenkel et al., 2013, 2014a). Our work shows that CD4+ TRM trigger similar sensing and alarm functions after reactivation, corroborating observations that CD4+ TRM accelerate protective immunity against HSV-2, L. major, C. albicans, influenza virus, and Bordetella pertussis infections at barrier tissues (Teijaro et al., 2011; Iijima and Iwasaki, 2014; Glennie et al., 2015; Wilk et al., 2017; Park et al., 2018). As we now show that reactivated CD4+ TRM recruit lymphocytes to the mucosa, the fate of recruited cells should be examined. For example, do recruited lymphocytes remain in the site of infection long term and potentially contribute to local immunosurveillance? If so, it suggests that CD4+ TRM activation triggers durable and broad changes in local immune cell composition.
This study highlights the dominance of resident-mediated CD4+ T cell immunosurveillance, indicating that a further understanding of CD4+ TRM biology is likely to help inform immune responses in protection and disease throughout the body.
Materials and methods
Mice
C57BL/6J (B6) female mice were from The Jackson Laboratory and were maintained in SPF conditions at the University of Minnesota, Minneapolis, MN. CD90.1+ SMARTA, CD45.1+ SMARTA, and CD45.1+ P14 mice were fully backcrossed to C57BL/6J mice and maintained in our animal colony. To generate dirty mice, pet store mice were purchased from various Twin Cities area pet stores. Male or female pet store mice were introduced into the cages of 6–8-wk-old C57BL/6N and B6.SJL-PtprcaPepcb/BoyJ (CD45.1) mice of the same sex purchased from the National Cancer Institute. Co-housing occurred within a BSL-3 facility as described previously (Beura et al., 2016). All mice used in experiments were 6–20 wk of age. All mice were used in accordance with the Institutional Animal Care and Use Committees guidelines at the University of Minnesota.
Adoptive transfers and infections
We generated immune chimeras by transferring 5 × 104 SMARTA CD4+ T cells or P14 CD8+ T cells into naive C57BL/6J mice and then infecting those mice with 2 × 105 PFU of LCMV-Armstrong i.p. 1 d later. For endogenous CD4+ T cell studies, we infected naive C57BL/6J mice with 2 × 105 PFU of LCMV-Armstrong i.p.
Intravascular staining, lymphocyte isolation, and phenotyping
We used an intravascular staining method to discriminate cells present in the vasculature from cells in the tissue parenchyma, as described (Anderson et al., 2014). We injected mice i.v. with biotin/fluorochrome–conjugated anti-CD45 (30-F11) through the tail vein. 3 min after the injection, we sacrificed the animals and harvested tissues as described (Thompson et al., 2016). Isolated mouse cells were surface-stained with antibodies against CD3 (145-2C11), CD11b (M1/70), CD11c (N418), MHCII I-A/I-E (M5/114.15.2), CD90.1 (OX-7), CD45.1 (A20), CD8β (YTS156.7.7), CD45.2 (104), CD4 (RM4-5), CD62L (MEL-14), CD44 (IM7), CD69 (H1.2F3), CD103 (M290), B220 (RA3-6B2), CD86 (GL1), Ly6C (HK1.4), Ly6G (1A8), CD49a (Ha31/8), CD27 (LG.3A10), CD127 (A7R34), CCR7 (4B12), CXCR3 (CXCR3-173), CD122 (TM-b1), P2rX7 (1F11), and CXCR5 (L138D7). Intracellular staining was performed using the Cytofix/Cytoperm method (BD Bioscience) or the Foxp3/Transcription Factor Staining Buffer Kit (Tonbo Biosciences) with antibodies against CXCL9 (polyclonal), CCL2 (polyclonal), CCL3 (polyclonal), CCL4 (polyclonal), Ki67 (SolA15), IFN-γ (XMG1.2), and granzyme B (GB12). Antibodies were purchased from BD Biosciences, BioLegend, Affymetrix eBioscience, or R&D Systems. Cell viability was determined with Ghost Dye 780 (Tonbo Biosciences). For detecting endogenous gp66-specific CD4 T cells, lymphocytes were stained with a GP66-77:I-Ab MHCII tetramer reagent as described earlier (Beura et al., 2015). The stained samples were acquired with LSRII or LSR Fortessa flow cytometers (BD Biosciences) and analyzed with FlowJo software (Treestar).
Tissue freezing, immunofluorescence, and microscopy
Harvested mouse tissues were fixed in 2% paraformaldehyde for 2 h before being treated with 30% sucrose overnight for cryoprotection. The sucrose-treated tissue was embedded in OCT tissue-freezing medium and frozen in a 2-methyl butane liquid bath. Frozen blocks were processed, stained, imaged, and enumerated by QIM as described (Steinert et al., 2015). Tissues were stained with antibodies to the following markers: CD4 (GK1.5; BD Biosciences), CD90.1 (OX-7; BD Biosciences), CD45.1 (A20; BioLegend), B220 (RA3-6B2; BioLegend), CD8β (YTS156.7.7; BioLegend), and collagen-IV (goat anti-mouse polyclonal; Millipore). We counterstained with DAPI or SYTOX Green (Thermo Fisher Scientific) to detect nuclei. The following secondary antibodies were from Jackson ImmunoResearch: bovine anti-goat (polyclonal) and donkey anti-rat (polyclonal).
In vivo antibody treatment and local TRM cell reactivation
We depleted CD4+ T cells by injecting (i.p.) 250–500 µg of anti-CD4 (GK1.5; Bioxcell) 7 d before recall. To deplete circulating CD90.1+ cells, we injected (i.p.) 0.5–1 µg of anti-CD90.1 antibody (HIS51; Thermo Scientific) 4 d before recall. For local FRT TRM cell rechallenge experiments involving peptides, 50 µg of the indicated peptides (New England Peptides) was delivered trans-cervically as described (Schenkel et al., 2013) in a volume of 30 µl delivered by modified gel-loading pipette. For local FRT TRM cell rechallenge experiments involving virus, 4 × 106 PFU of VV-OVA or VV-gp61 was delivered trans-cervically.
Parabiotic surgery
Parabiotic surgery was done as described (Schenkel et al., 2013). Briefly, mice were anesthetized with ketamine and xylazine. Lateral skin was shaved and disinfected before matching incisions were made from the olecranon to the knee joint of each mouse. The skins were apposed by continuous staples. Parabionts were then allowed to rest for 28–40 d before experiments. Equilibration was confirmed in the peripheral blood before tissue analysis.
RNA-seq data analyses
Cell sorting and RNA preparation
CD4+ CD45.1+ SMARTA TCM (CD62L+ CD69−), TEM (CD62L− CD69−), and SLO TRM (CD62L− CD69+) populations were sorted from spleen of LCMV immune chimeras using a BD FACSAria II. Isolation of TRM cells from the FRT and small intestine epithelial and lamina propria compartment was done by enzymatic digestion as described earlier (Thompson et al., 2016). The resulting single-cell suspensions from the three different tissues were used as a source for sorting CD69+ C62L− TRM cells. Sorted cells were homogenized using QIAshredder columns (QIAGEN), and RNA was extracted using an RNeasy micro kit (QIAGEN) per the manufacturer’s instructions. RNA integrity was assessed using capillary electrophoresis with the Agilent 2100 BioAnalyzer system (Agilent Technologies), which generated an RNA Integrity Number. Samples with an RNA Integrity Number >7 were included in this study.
Library preparation and RNA-seq
Sequencing libraries were prepared using the Clontech SMARTer Stranded Total RNA-Seq Kit v2—Pico Input Mammalian Kit. RNA-seq (50-bp single-end) with the HiSeq 2500 Illumina was done at the University of Minnesota Genomics Center.
RNA-seq data processing
Cutadapt was used to remove the first three nucleotides of the sequencing read, which were derived from the Clontech template-switching oligo (Martin, 2011). Initial quality control analysis of RNA-seq (FASTQ) data for each sample was performed using the FastQC software (version 0.11.5; Andrews, 2010). Trimmomatic (version 0.33) was used to trim FASTQ data (Bolger et al., 2014). UCSC version mm10 (GRCm38; Genome Reference Consortium Mouse Build 38; GCA_000001635.2) was used as the mouse reference genome. HISAT2 (version 2.0.2-β) was used to map single-end reads (Kim et al., 2015). Insertion size metrics were calculated for each sample using Picard software (version 1.126; http://picard.sourceforge.net). Mapping statistics are summarized in Table S1. Samtools (version 1.3_BCFTools_HTSLib) was used to sort and index the bam files (Li et al., 2009). Gene abundance estimates were generated using the Rsubread featureCounts program (Liao et al., 2014). Genes <200 bp were removed before analyses. RNA-seq data from the various CD4 memory T cell populations are available at the National Center for Biotechnology Information Gene Expression Omnibus database under accession no. GSE128197.
DGE analyses
DGE analysis was performed with CLC Genomics Workbench’s (v. 10.1.1; CLC Bio-Qiagen) empirical analysis of DGE (EDGE) tool, which implements the “Exact Test” for our two-group pairwise comparisons of CD4+ TRM (any tissue site) versus SMARTA TCM and CD4+ TRM (any tissue site) versus SMARTA TEM (Robinson and Smyth, 2007; Robinson et al., 2010). EdgeR output results for each pairwise comparison provided in Table S2. Genes were considered to be differentially expressed if they had an FDR P value ≤0.05 and an absolute fold change ≥2. The VennDiagram-package was used to create and draw Venn diagrams of DEG lists (Chen and Boutros, 2011). Log transformed count per million values for DEGs were used for clustering analysis and visualization with the ComplexHeatmap package (Gu et al., 2016).
PCA
To remove unexpressed genes in the dataset, genes with a mean <1 across all of the samples were removed before analysis. The prcomp and autoplot functions in RStudio were used to perform the PCA and plot the first two principal components, respectively.
Pathway analyses
The clusterCompare R package was used to analyze and visualize enriched pathways for shared and unique TRM DEGs (Yu et al., 2012). Gene network analyses for the core set of TRM genes was performed with QIAGEN’s IPA. The reference set for all IPA analyses was the Ingenuity Knowledge Base (genes only), and only direct relationships were considered. Mouse Entrez gene names were used as the IPA output format.
Enrichment analyses
A CD8 ranked gene list was obtained from the output of a gene set enrichment analysis (GSEA) test (GSEA v3.0 [build:0160]) comparing previously published gut TRM to circulating memory samples (GSE47045; Subramanian et al., 2005; Mackay et al., 2013). The limma barcodeplot function was used to plot enrichment of the overexpressed and underexpressed CD4 TRM gene sets in the CD8 ranked gene list.
Comparison of RNA-seq data to previously published TFH dataset
We identified a microarray expression dataset from mouse effector and memory TFH cells in the LCMV infection model (GSE43863; Hale et al., 2013). The GSE43863 array probes were summarized to gene symbols. The summarized array values from the GSE43863 dataset and gene count values of our RNA-seq dataset were both separately trimmed mean of M-values normalized. Genes overexpressed in TRM were considered as one gene set while genes underexpressed in TRM were considered as another gene set. Of the core TRM gene signature, 84 of the 119 overexpressed and 40 of the 64 underexpressed were present on the array. For each sample, a summary score for each of these gene sets was obtained. The sample summary scores were then Z score normalized.
Statistical analysis
If the samples followed Gaussian distribution, then parametric tests (unpaired two-tailed Student’s t test for two groups and one-way ANOVA with Tukey’s multiple comparison test for more than two groups) were used. If the samples deviated from a Gaussian distribution, nonparametric tests (Mann–Whitney U test for two groups, Kruskal–Wallis with Dunn’s multiple comparison test for more than two groups) were used unless otherwise stated. The D’Agostino and Pearson omnibus normality test was used to determine whether samples adhered to Gaussian distribution or not. Variances between groups were compared using an F test and found to be equal. P < 0.05 was considered significant. All statistical analysis was done in GraphPad Prism (GraphPad Software). Sample size was chosen on the basis of previous experience. No sample exclusion criteria were applied. No method of randomization was used during group allocation, and investigators were not blinded.
Online supplemental material
Fig. S1 depicts the gradual acquisition of resident memory phenotype by CD4+ T cells in small intestine lamina propria following LCMV-Armstrong infection. Fig. S2 summarizes the enriched pathways and gene networks identified from gene expression analyses of circulating memory CD4 T cells, SLO CD4 TRM, and NLT CD4 TRM. Fig. S3 shows how local CD4+ TRM reactivation induces activation of other immune cells in the FRT. Fig. S4 summarizes data from parabiosis experiments showing that local CD4+ TRM reactivation is responsible for the activation of other immune cells (CD8 T cells, NK cells, and DCs) in the FRT. Fig. S5 depicts data from selective depletion experiments demonstrating that FRT-resident CD4+ T cells are sufficient to induce local immune activation after reencountering their cognate antigen. Table S1 shows the mapping statistics for each sample generated and used in this study. Table S2 lists the shared and unique TRM DEGs from pairwise comparisons between circulating memory CD4 T cells, SLO TRM, and NLT TRM populations.
Supplementary Material
Acknowledgments
We thank Dr. Thamotharampillai Dileepan and Dr. Marc Jenkins (University of Minnesota, Minneapolis, MN) for providing MHCII tetramer reagents. We also thank the Minnesota Supercomputing Institute for computing time and storage space, the University of Minnesota Genomics Center for sequencing services, and the University of Minnesota Flow Cytometry Resource for cell sorting services. We thank Dr. J. Michael Stolley (University of Minnesota) for help with the graphical abstract design.
This work was funded by the Howard Hughes Medical Institute Faculty Scholars program and National Institutes of Health grants R01AI111671 and R01AI084913 (to D. Masopust).
The authors declare no competing financial interests.
Author contributions: L.K. Beura, N.J. Fares-Frederickson, E.M. Steinert, E.A. Thompson, K.A. Fraser, J.M. Schenkel, and V. Vezys performed the experiments; M.C. Scott performed the bioinformatics analysis; L.K. Beura, N.J. Fares-Frederickson, M.C. Scott, and D. Masopust wrote the manuscript; and D. Masopust was responsible for research supervision, coordination, and strategy.
References
- Allen A.C., Wilk M.M., Misiak A., Borkner L., Murphy D., and Mills K.H.G.. 2018. Sustained protective immunity against Bordetella pertussis nasal colonization by intranasal immunization with a vaccine-adjuvant combination that induces IL-17-secreting TRM cells. Mucosal Immunol. 11:1763–1776. 10.1038/s41385-018-0080-x [DOI] [PubMed] [Google Scholar]
- Anderson K.G., Mayer-Barber K., Sung H., Beura L., James B.R., Taylor J.J., Qunaj L., Griffith T.S., Vezys V., Barber D.L., and Masopust D.. 2014. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat. Protoc. 9:209–222. 10.1038/nprot.2014.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S.2010. FastQC A Quality Control tool for High Throughput Sequence Data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Asrir A., Aloulou M., Gador M., Pérals C., and Fazilleau N.. 2017. Interconnected subsets of memory follicular helper T cells have different effector functions. Nat. Commun. 8:847 10.1038/s41467-017-00843-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoun J.M., Peres N.G., Wang N., Pham O.H., Rudisill V.L., Fogassy Z.N., Whitney P.G., Fernandez-Ruiz D., Gebhardt T., Pham Q.-M., et al. 2018. Optimal protection against Salmonella infection requires noncirculating memory. Proc. Natl. Acad. Sci. USA. 115:10416–10421. 10.1073/pnas.1808339115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura L.K., Anderson K.G., Schenkel J.M., Locquiao J.J., Fraser K.A., Vezys V., Pepper M., and Masopust D.. 2015. Lymphocytic choriomeningitis virus persistence promotes effector-like memory differentiation and enhances mucosal T cell distribution. J. Leukoc. Biol. 97:217–225. 10.1189/jlb.1HI0314-154R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura L.K., Hamilton S.E., Bi K., Schenkel J.M., Odumade O.A., Casey K.A., Thompson E.A., Fraser K.A., Rosato P.C., Filali-Mouhim A., et al. 2016. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature. 532:512–516. 10.1038/nature17655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura L.K., Wijeyesinghe S., Thompson E.A., Macchietto M.G., Rosato P.C., Pierson M.J., Schenkel J.M., Mitchell J.S., Vezys V., Fife B.T., et al. 2018. T Cells in Nonlymphoid Tissues Give Rise to Lymph-Node-Resident Memory T Cells. Immunity. 48:327–338.e5. 10.1016/j.immuni.2018.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., and Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromley S.K., Yan S., Tomura M., Kanagawa O., and Luster A.D.. 2013. Recirculating memory T cells are a unique subset of CD4+ T cells with a distinct phenotype and migratory pattern. J. Immunol. 190:970–976. 10.4049/jimmunol.1202805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone F.R. 2015. Tissue-Resident Memory T Cells and Fixed Immune Surveillance in Nonlymphoid Organs. J. Immunol. 195:17–22. 10.4049/jimmunol.1500515 [DOI] [PubMed] [Google Scholar]
- Chen H., and Boutros P.C.. 2011. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 12:35 10.1186/1471-2105-12-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn L.B., da Silva I.T., Valieris R., Huang A.S., Lorenzi J.C.C., Cohen Y.Z., Pai J.A., Butler A.L., Caskey M., Jankovic M., and Nussenzweig M.C.. 2018. Clonal CD4+ T cells in the HIV-1 latent reservoir display a distinct gene profile upon reactivation. Nat. Med. 24:604–609. 10.1038/s41591-018-0017-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins N., Jiang X., Zaid A., Macleod B.L., Li J., Park C.O., Haque A., Bedoui S., Heath W.R., Mueller S.N., et al. 2016. Skin CD4(+) memory T cells exhibit combined cluster-mediated retention and equilibration with the circulation. Nat. Commun. 7:11514 10.1038/ncomms11514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazilleau N., McHeyzer-Williams L.J., Rosen H., and McHeyzer-Williams M.G.. 2009. The function of follicular helper T cells is regulated by the strength of T cell antigen receptor binding. Nat. Immunol. 10:375–384. 10.1038/ni.1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Ruiz D., Ng W.Y., Holz L.E., Ma J.Z., Zaid A., Wong Y.C., Lau L.S., Mollard V., Cozijnsen A., Collins N., et al. 2016. Liver-Resident Memory CD8+ T Cells Form a Front-Line Defense against Malaria Liver-Stage Infection. Immunity. 45:889–902. 10.1016/j.immuni.2016.08.011 [DOI] [PubMed] [Google Scholar]
- Galkina E., Thatte J., Dabak V., Williams M.B., Ley K., and Braciale T.J.. 2005. Preferential migration of effector CD8+ T cells into the interstitium of the normal lung. J. Clin. Invest. 115:3473–3483. 10.1172/JCI24482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt T., Whitney P.G., Zaid A., Mackay L.K., Brooks A.G., Heath W.R., Carbone F.R., and Mueller S.N.. 2011. Different patterns of peripheral migration by memory CD4+ and CD8+ T cells. Nature. 477:216–219. 10.1038/nature10339 [DOI] [PubMed] [Google Scholar]
- Georgiev H., Ravens I., Papadogianni G., Malissen B., Förster R., and Bernhardt G.. 2018. Blocking the ART2.2/P2X7-system is essential to avoid a detrimental bias in functional CD4 T cell studies. Eur. J. Immunol. 48:1078–1081. 10.1002/eji.201747420 [DOI] [PubMed] [Google Scholar]
- Glennie N.D., Yeramilli V.A., Beiting D.P., Volk S.W., Weaver C.T., and Scott P.. 2015. Skin-resident memory CD4+ T cells enhance protection against Leishmania major infection. J. Exp. Med. 212:1405–1414. 10.1084/jem.20142101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Eils R., and Schlesner M.. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 32:2847–2849. 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- Hale J.S., Youngblood B., Latner D.R., Mohammed A.U.R., Ye L., Akondy R.S., Wu T., Iyer S.S., and Ahmed R.. 2013. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity. 38:805–817. 10.1016/j.immuni.2013.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondowicz B.D., Kim K.S., Ruterbusch M.J., Keitany G.J., and Pepper M.. 2018. IL-2 is required for the generation of viral-specific CD4+ Th1 tissue-resident memory cells and B cells are essential for maintenance in the lung. Eur. J. Immunol. 48:80–86. 10.1002/eji.201746928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima N., and Iwasaki A.. 2014. T cell memory. A local macrophage chemokine network sustains protective tissue-resident memory CD4 T cells. Science. 346:93–98. 10.1126/science.1257530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., and Salzberg S.L.. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 12:357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar B.V., Ma W., Miron M., Granot T., Guyer R.S., Carpenter D.J., Senda T., Sun X., Ho S.-H., Lerner H., et al. 2017. Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites. Cell Reports. 20:2921–2934. 10.1016/j.celrep.2017.08.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., and Shi W.. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Mackay L.K., Rahimpour A., Ma J.Z., Collins N., Stock A.T., Hafon M.-L., Vega-Ramos J., Lauzurica P., Mueller S.N., Stefanovic T., et al. 2013. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat. Immunol. 14:1294–1301. 10.1038/ni.2744 [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 17:10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Nascimbeni M., Shin E.-C., Chiriboga L., Kleiner D.E., and Rehermann B.. 2004. Peripheral CD4(+)CD8(+) T cells are differentiated effector memory cells with antiviral functions. Blood. 104:478–486. 10.1182/blood-2003-12-4395 [DOI] [PubMed] [Google Scholar]
- Oja A.E., Piet B., Helbig C., Stark R., van der Zwan D., Blaauwgeers H., Remmerswaal E.B.M., Amsen D., Jonkers R.E., Moerland P.D., et al. 2018. Trigger-happy resident memory CD4+ T cells inhabit the human lungs. Mucosal Immunol. 11:654–667. 10.1038/mi.2017.94 [DOI] [PubMed] [Google Scholar]
- Park C.O., Fu X., Jiang X., Pan Y., Teague J.E., Collins N., Tian T., O’Malley J.T., Emerson R.O., Kim J.H., et al. 2018. Staged development of long-lived T-cell receptor αβ TH17 resident memory T-cell population to Candida albicans after skin infection. J. Allergy Clin. Immunol. 142:647–662. 10.1016/j.jaci.2017.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper M., Pagán A.J., Igyártó B.Z., Taylor J.J., and Jenkins M.K.. 2011. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 35:583–595. 10.1016/j.immuni.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., and Smyth G.K.. 2007. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics. 23:2881–2887. 10.1093/bioinformatics/btm453 [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romagnoli P.A., Fu H.H., Qiu Z., Khairallah C., Pham Q.M., Puddington L., Khanna K.M., Lefrançois L., and Sheridan B.S.. 2017. Differentiation of distinct long-lived memory CD4 T cells in intestinal tissues after oral Listeria monocytogenes infection. Mucosal Immunol. 10:520–530. 10.1038/mi.2016.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathaliyawala T., Kubota M., Yudanin N., Turner D., Camp P., Thome J.J.C., Bickham K.L., Lerner H., Goldstein M., Sykes M., et al. 2013. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity. 38:187–197. 10.1016/j.immuni.2012.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel J.M., and Masopust D.. 2014. Tissue-resident memory T cells. Immunity. 41:886–897. 10.1016/j.immuni.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel J.M., Fraser K.A., Vezys V., and Masopust D.. 2013. Sensing and alarm function of resident memory CD8+ T cells. Nat. Immunol. 14:509–513. 10.1038/ni.2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel J.M., Fraser K.A., Beura L.K., Pauken K.E., Vezys V., and Masopust D.. 2014a T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science. 346:98–101. 10.1126/science.1254536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel J.M., Fraser K.A., and Masopust D.. 2014b Cutting edge: resident memory CD8 T cells occupy frontline niches in secondary lymphoid organs. J. Immunol. 192:2961–2964. 10.4049/jimmunol.1400003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seder R.A., and Ahmed R.. 2003. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat. Immunol. 4:835–842. 10.1038/ni969 [DOI] [PubMed] [Google Scholar]
- Siracusa F., McGrath M.A., Maschmeyer P., Bardua M., Lehmann K., Heinz G., Durek P., Heinrich F.F., Mashreghi M.-F., Chang H.-D., et al. 2018. Nonfollicular reactivation of bone marrow resident memory CD4 T cells in immune clusters of the bone marrow. Proc. Natl. Acad. Sci. USA. 115:1334–1339. 10.1073/pnas.1715618115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slütter B., Van Braeckel-Budimir N., Abboud G., Varga S.M., Salek-Ardakani S., and Harty J.T.. 2017. Dynamics of influenza-induced lung-resident memory T cells underlie waning heterosubtypic immunity. Sci. Immunol. 2:eaag2031 10.1126/sciimmunol.aag2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N.M., Wasserman G.A., Coleman F.T., Hilliard K.L., Yamamoto K., Lipsitz E., Malley R., Dooms H., Jones M.R., Quinton L.J., and Mizgerd J.P.. 2018. Regionally compartmentalized resident memory T cells mediate naturally acquired protection against pneumococcal pneumonia. Mucosal Immunol. 11:220–235. 10.1038/mi.2017.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stary G., Olive A., Radovic-Moreno A.F., Gondek D., Alvarez D., Basto P.A., Perro M., Vrbanac V.D., Tager A.M., Shi J., et al. 2015. VACCINES. A mucosal vaccine against Chlamydia trachomatis generates two waves of protective memory T cells. Science. 348:aaa8205 10.1126/science.aaa8205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert E.M., Schenkel J.M., Fraser K.A., Beura L.K., Manlove L.S., Igyártó B.Z., Southern P.J., and Masopust D.. 2015. Quantifying Memory CD8 T Cells Reveals Regionalization of Immunosurveillance. Cell. 161:737–749. 10.1016/j.cell.2015.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinfelder S., Rausch S., Michael D., Kühl A.A., and Hartmann S.. 2017. Intestinal helminth infection induces highly functional resident memory CD4+ T cells in mice. Eur. J. Immunol. 47:353–363. 10.1002/eji.201646575 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surh C.D., and Sprent J.. 2008. Homeostasis of naive and memory T cells. Immunity. 29:848–862. 10.1016/j.immuni.2008.11.002 [DOI] [PubMed] [Google Scholar]
- Takamura S., Yagi H., Hakata Y., Motozono C., McMaster S.R., Masumoto T., Fujisawa M., Chikaishi T., Komeda J., Itoh J., et al. 2016. Specific niches for lung-resident memory CD8+ T cells at the site of tissue regeneration enable CD69-independent maintenance. J. Exp. Med. 213:3057–3073. 10.1084/jem.20160938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.J., and Jenkins M.K.. 2011. CD4+ memory T cell survival. Curr. Opin. Immunol. 23:319–323. 10.1016/j.coi.2011.03.010 [DOI] [PubMed] [Google Scholar]
- Teijaro J.R., Turner D., Pham Q., Wherry E.J., Lefrançois L., and Farber D.L.. 2011. Cutting edge: Tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J. Immunol. 187:5510–5514. 10.4049/jimmunol.1102243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome J.J.C., Yudanin N., Ohmura Y., Kubota M., Grinshpun B., Sathaliyawala T., Kato T., Lerner H., Shen Y., and Farber D.L.. 2014. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell. 159:814–828. 10.1016/j.cell.2014.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson E.A., Beura L.K., Nelson C.E., Anderson K.G., and Vezys V.. 2016. Shortened Intervals during Heterologous Boosting Preserve Memory CD8 T Cell Function but Compromise Longevity. J. Immunol. 196:3054–3063. 10.4049/jimmunol.1501797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D.L., Bickham K.L., Thome J.J., Kim C.Y., D’Ovidio F., Wherry E.J., and Farber D.L.. 2014. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol. 7:501–510. 10.1038/mi.2013.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D.L., Goldklang M., Cvetkovski F., Paik D., Trischler J., Barahona J., Cao M., Dave R., Tanna N., D’Armiento J.M., and Farber D.L.. 2018. Biased Generation and In Situ Activation of Lung Tissue-Resident Memory CD4 T Cells in the Pathogenesis of Allergic Asthma. J. Immunol. 200:1561–1569. 10.4049/JIMMUNOL.1700257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe R., Gehad A., Yang C., Scott L.L., Teague J.E., Schlapbach C., Elco C.P., Huang V., Matos T.R., Kupper T.S., and Clark R.A.. 2015. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Sci. Transl. Med. 7:279ra39 10.1126/scitranslmed.3010302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry E.J., Blattman J.N., Murali-Krishna K., van der Most R., and Ahmed R.. 2003. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J. Virol. 77:4911–4927. 10.1128/JVI.77.8.4911-4927.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk M.M., Misiak A., McManus R.M., Allen A.C., Lynch M.A., and Mills K.H.G.. 2017. Lung CD4 Tissue-Resident Memory T Cells Mediate Adaptive Immunity Induced by Previous Infection of Mice with Bordetella pertussis. J. Immunol. 199:233–243. 10.4049/jimmunol.1602051 [DOI] [PubMed] [Google Scholar]
- Wu T., Hu Y., Lee Y.-T., Bouchard K.R., Benechet A., Khanna K., and Cauley L.S.. 2014. Lung-resident memory CD8 T cells (TRM) are indispensable for optimal cross-protection against pulmonary virus infection. J. Leukoc. Biol. 95:215–224. 10.1189/jlb.0313180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., Han Y., and He Q.-Y.. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 16:284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.