In this review, Aiello and Kang discuss the molecular mechanisms, regulatory networks, and functional consequences of epithelial–mesenchymal transition (EMT) in the context of cancer metastasis, with a particular focus on partial EMT and cellular plasticity.
Abstract
Epithelial–mesenchymal transition (EMT) is a developmental process whereby stationary, adherent cells acquire the ability to migrate. EMT is critical for dramatic cellular movements during embryogenesis; however, tumor cells can reactivate EMT programs, which increases their aggressiveness. In addition to motility, EMT is associated with enhanced stem cell properties and drug resistance; thus it can drive metastasis, tumor recurrence, and therapy resistance in the context of cancer. However, the precise requirements for EMT in metastasis have not been fully delineated, with different tumor types relying on discrete EMT effectors. Most tumor cells do not undergo a full EMT, but rather adopt some qualities of mesenchymal cells and maintain some epithelial characteristics. Emerging evidence suggests that partial EMT can drive distinct migratory properties and enhance the epithelial-mesenchymal plasticity of cancer cells as well as cell fate plasticity. This review discusses the diverse regulatory mechanisms and functional consequences of EMT, with an emphasis on the importance of partial EMT.
Introduction
Epithelial–mesenchymal transition (EMT) is a developmental program that facilitates motility in otherwise adherent epithelial cells. During EMT, an epithelial cell sheds its connections to neighboring cells, converts from apico-basal to front-back polarity, and takes on the properties of a migratory mesenchymal cell (Greenburg and Hay, 1982). Both EMT and its reverse process, mesenchymal–epithelial transition (MET), occur throughout development, wound healing, fibrosis, and tumor progression. During embryogenesis, EMT is required for gastrulation, the stage at which epithelial epiblast-derived cells ingress and transition into mesenchymal cells, forming the three germ layers (Carver et al., 2001). At the onset of gastrulation, fibroblast growth factor signaling in the primitive streak activates the EMT- transcription factor (TF) Snail (SNAI1), which in turn transcriptionally represses E-cadherin, resulting in EMT (Nakaya and Sheng, 2008). A similar paradigm plays out repeatedly through development: EMT also occurs during neural crest cell migration (Cheung et al., 2005), somitogenesis (Dale et al., 2006), and cardiac valve formation (Timmerman et al., 2004). In adult organisms, facets of the EMT program are activated in response to cutaneous injury to facilitate collective migration. Keratinocytes at the edge of the wound respond to epidermal growth factor and TGF-β signaling to activate the EMT-TF Slug (SNAI2), which promotes motility and wound closure (Haensel and Dai, 2018). EMT also occurs in pathological conditions, including fibrosis and cancer. In chronic pulmonary obstructive disease, chronic inflammation leads to small-airway fibrosis that appears to be driven by the EMT of bronchial epithelial cells (Jolly et al., 2018). Similarly, EMT of alveolar epithelial cells has been reported as the source of myofibroblasts in idiopathic pulmonary fibrosis (Kim et al., 2006). Likewise in the kidney, tubular epithelial cells undergo EMT to contribute to renal fibrosis (Iwano et al., 2002). Finally, EMT plays a significant role in tumor progression, where it has been implicated in many of the hallmarks of cancer (Hanahan and Weinberg, 2011), particularly metastasis. This review discusses the evolving definition of EMT in the context of cancer as well as its functional consequences.
Characteristics and functional consequences of cancer EMT
Since EMT enhances cellular mobility, it is no surprise that it has been connected to the dissemination of tumor cells. Indeed, carcinomas often lose epithelial markers or express EMT markers at the invasive front (Brabletz et al., 2001; Vincent et al., 2009; Kahlert et al., 2011; Paterson et al., 2013; Kunita et al., 2018) and in circulating tumor cells (Aktas et al., 2009; Hyun et al., 2016; Lapin et al., 2017), which represent the first steps of the metastatic cascade (invasion and intravasation, respectively). Expression of EMT-TFs correlates with poor clinical outcomes in cholangiocarcinoma, gastric cancer, and breast cancer, among others (Ryu et al., 2012a,b; Jang et al., 2015). This is in part due to EMT’s role in promoting metastasis, which will be discussed in detail in a later section; however, it should be noted that while the primary consequence of EMT is increased motility, the phenomenon is also associated with stemness, therapy resistance, and immune evasion.
The connection between EMT and stem cell properties was first reported in a study by Mani et al. (2008), which demonstrated that mammary epithelial cells and breast cancer cells that have undergone EMT exhibit stem cell markers (CD44hi/CD24lo) and functional characteristics. When EMT was induced through treatment with TGF-β or overexpression of EMT-TFs, the cells formed more mammospheres (an in vitro test for self-renewal) and had an increased ability to repopulate a cleared mammary fat pad (in the case of normal mammary epithelial cells) or form tumors (in the case of breast cancer cells; Mani et al., 2008). Similarly, in prostate cancer, EMT was accompanied by an increase in the expression of embryonic stem cell markers as well as an enhanced ability to form spheres in vitro and tumors in vivo (Kong et al., 2010). In a mouse model of breast cancer recurrence, Snail-driven EMT promoted the regrowth of tumors (Moody et al., 2005), and in patients, residual breast tumors left behind after conventional therapy often exhibit EMT and stem cell features (Creighton et al., 2009). Thus, in addition to stemness (or perhaps because of stemness), EMT is strongly associated with therapy resistance. Tumor cells selected for chemoresistance acquire an EMT phenotype (Shah et al., 2007); conversely, tumor cells that are induced to undergo EMT acquire resistance to chemotherapy (Yin et al., 2007), and inhibition of EMT can increase drug sensitivity (Ren et al., 2013; Fischer et al., 2015; Zheng et al., 2015). EMT-TFs confer chemo- and radioresistance through a number of molecular mechanisms, including resistance to apoptosis, enhanced DNA damage repair and altered drug metabolism (van Staalduinen et al., 2018).
With the advent of cancer immunotherapy, it has become apparent that EMT also protects tumor cells from immune cell–mediated killing. The first clue was the finding that SNAIL-induced EMT promotes melanoma metastasis by induction of regulatory T cell–mediated immunosuppression (Kudo-Saito et al., 2009). Cancer cells that undergo EMT secrete cytokines such as TGF-β, IL-10, and thrombospondin-1 that result in a generally immunosuppressive tumor microenvironment (Yaguchi et al., 2011). Breast cancer cell lines that skew mesenchymal recruit more immunosuppressive T regulatory cells and M2-polarized macrophages and fewer effector and cytotoxic T cells compared with epithelial lines when implanted into an immunocompetent host; moreover, tumors derived from the mesenchymal cell lines are resistant to anti-cytotoxic T lymphocyte–associated protein 4 (CTLA4) immunotherapy (Dongre et al., 2017). EMT has been associated with resistance to cytotoxic T lymphocyte killing due to the interruption of the immunological synapse (Akalay et al., 2013). Another mechanism of EMT-related immune escape is the up-regulation of immune checkpoint proteins on tumor cells such as programmed death ligand-1 (PD-L1), PD-L2, and B7-H3 (Chen et al., 2014; Lou et al., 2016; Noman et al., 2017). With a hand in seemingly every aspect of tumor progression, EMT is a formidable obstacle in the treatment of cancer.
Molecular mechanisms of EMT
Many developmental signal transduction pathways are capable of inducing EMT, including the TGF-β, epidermal growth factor, fibroblast growth factor, hepatocyte growth factor, Wingless/integrated, Sonic hedgehog, and Notch pathways (Li et al., 1994, 2006; Miettinen et al., 1994; Kim et al., 2002; Timmerman et al., 2004). Cytokines such as IL-8, IL-6, and TNF-α, often secreted by tumor stroma, can also promote EMT (Sullivan et al., 2009; Wu et al., 2009; Fernando et al., 2011). Tumor cell interactions with extracellular matrix components can also induce EMT. For example, ovarian and prostate cancer cell lines that come into contact with type I collagen up-regulate EMT-TFs Snail and Slug (Cheng and Leung, 2011). Moreover, EMT programs can be activated through mechanotransduction: matrix stiffness, fluid flow, osmotic pressure, and tissue tension all influence the EMT status of cancer cells (Mihalko and Brown, 2018). In the context of breast cancer, dense collagen fibrils increase matrix stiffness, which in turn promotes the nuclear translocation of the EMT-TF Twist1 (TWIST1; Wei et al., 2015).
But how do epithelial cells respond to these extracellular signals to achieve the dramatic changes necessary to become motile? At the molecular level, cells going through EMT must repress epithelial genes that contribute to cellular adhesion (adherens junctions, tight junctions, and desmosomes), allowing them to detach from their neighbors. The classic epithelial marker E-cadherin (CDH1), a critical component of the adherens junction, is the most prominent target of repression during the EMT process. A number of EMT-TFs including Snail, Slug, and Zinc-finger E-box binding homeoboxes 1 and 2 (ZEB1/2) directly target CDH1 and other epithelial genes for transcriptional repression (Cano et al., 2000; Comijn et al., 2001; Bolós et al., 2003; Shirakihara et al., 2007). EMT-TFs themselves are repressed by epithelial-associated proteins such as ELF5, TFs Grainyhead-like 2 (GRHL2), and Ovo-like zinc fingers 1 and 2 (OVOL1/2), which help maintain an epithelial phenotype and can drive MET (Chakrabarti et al., 2012; Cieply et al., 2012; Roca et al., 2013). EMT-TFs are also negatively regulated by micro-RNAs (miRNAs), including miR-34, which represses SNAI1, and the miR-200 family, which represses ZEB1 (Bracken et al., 2008; Gregory et al., 2008; Korpal et al., 2008; Park et al., 2008; Siemens et al., 2011). The balance between EMT-TFs and their antagonistic miRNAs plays a critical role in determining where a cell falls on the EMT spectrum as well as its potential for plasticity and metastasis (Celià-Terrassa et al., 2018).
A cell undergoing EMT must also activate mesenchymal genes to promote the morphological and behavioral transformations necessary to become migratory. EMT-TFs Twist (TWIST1) and Pair related homeobox 1 (PRRX1) are strong promoters of the mesenchymal transcription program (Yang et al., 2004; Ocaña et al., 2012). Although the Snail and Zeb families of EMT-TFs were originally thought to act only as transcriptional repressors, it has been demonstrated that they can also act as transcriptional activators in certain contexts (Wels et al., 2011; Rembold et al., 2014; Lehmann et al., 2016). EMT-TFs promote the expression of crucial mesenchymal genes such as vimentin (VIM), N-cadherin (CDH2), fibronectin (FN1), and fibroblast-specific protein 1 (FSP1). These downstream mesenchymal targets reshape the cytoskeleton and cell membrane to allow for migration.
Beyond transcriptional control of EMT
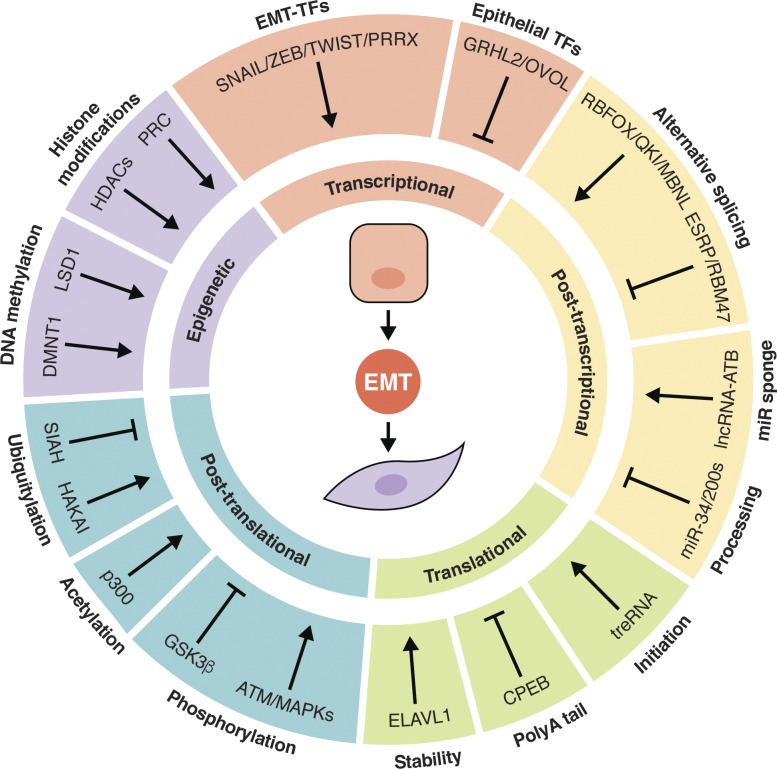
Despite the intense focus within the field on EMT-TFs, it has become increasingly apparent that the mechanisms behind this process are not limited to the transcriptional level. EMT is facilitated through many levels of regulation, from epigenetic to posttranslational modifications and every step in between (Fig. 1). DNA methylation of the CDH1 promoter is one mechanism of E-cadherin repression: ZEB1 interacts with DNA methyltransferase 1 (DNMT1) to accomplish this (Fukagawa et al., 2015). EMT-TFs can also recruit histone-modifying enzymes, including the histone demethylase LSD1, histone deacetylases HDAC1/2, and the polycomb repressive complex (PRC2), to repress the CDH1 promoter (Herranz et al., 2008; von Burstin et al., 2009; Lin et al., 2010; Aghdassi et al., 2012; Skrypek et al., 2017). At the RNA level, alternative splicing by Epithelial splicing regulatory proteins 1 and 2 (ESRP1/2), RNA binding motif protein 47 (RBM47), Quaking, RNA binding fox-1 homologue 2 (RBFOX2), and Muscleblind-like splicing regulator 1 (MBNL1) play a role in regulating EMT. ESRP1/2 and RBM47 promote epithelial-specific splicing, while the latter proteins promote mesenchymal-specific splicing (Shapiro et al., 2011; Venables et al., 2013; Yang et al., 2016; Neumann et al., 2018). At the posttranscriptional level, besides miRNAs, long noncoding RNAs (lncRNAs) also contribute to EMT regulation. For example, lncRNA-activated by TGF-β (lncRNA-ATB) and lncRNA-PNUTS have been suggested to act as sponges for the miR-200 family and miR-205, respectively, sequestering these miRNAs to prevent them from inhibiting EMT-TF transcripts (Yuan et al., 2014; Grelet et al., 2017). Another EMT-promoting lncRNA, translational regulator lncRNA, negatively regulates the translation of CDH1 mRNA (Gumireddy et al., 2013; Dhamija and Diederichs, 2016). Similarly, cytoplasmic polyadenylation element binding protein 1 (CPEB1) promotes the shortening of the polyA tail of matrix metalloprotease 9 (MMP9), which reduces its translation; upon deletion of CPEB1, mammary tumor cells undergo EMT and become more metastatic (Nagaoka et al., 2016). In contrast, embryonic lethal abnormal vision–like RNA binding protein 1 (ELAVL1 or HUR) promotes EMT by stabilizing Snai1 mRNA (Zhou et al., 2016). Posttranslational regulation of EMT-TFs is also an important level of control. SNAI1 and TWIST1 can be acetylated by p300, which modulates their stability, localization, and interactions with other proteins (Shiota et al., 2010; Chang et al., 2017). SNAI1, SNAI2, TWIST1, and ZEB1 can also be phosphorylated, which affects their stabilization/degradation. Glycogen synthase kinase 3 β (GSK3β) and PKD1 phosphorylate Snail and Slug to promote their degradation, while MAPKs and Ataxia-Telangiesctasia mutated serine/threonine kinase phosphorylate TWIST1 and ZEB1, respectively, to stabilize them (Bauer et al., 2009; Hong et al., 2011; Kim et al., 2012; Zhang et al., 2014; Zheng et al., 2014). ZEB1 is regulated by the E3 ubiquitin ligase SIAH as well, which marks it for degradation (Chen et al., 2015). Epithelial proteins can likewise be posttranslationally regulated during EMT. The E3 ubiquitin ligase Hakai ubiquitinates E-cadherin, inducing its endocytosis and destruction (Fujita et al., 2002). During hepatocyte growth factor–mediated EMT, E-cadherin can be phosphorylated by PKCδ, which disrupts its interaction with β-catenin on the cytoplasmic side and with E-cadherin on adjacent cells (Chen et al., 2016). The E-cadherin protein also contains four asparagine (Asn) residues that are N-glycosylated, two of which are critical to its adhesive function: site-directed mutagenesis of Asn-554 and Asn-566 significantly reduced adhesion and enhanced the migratory ability of a breast cancer cell line (Zhao et al., 2008). EMT is a tightly regulated process because the consequences of an aberrant transition are significant, especially in the context of cancer.
Figure 1.
Layers of EMT regulation. EMT is regulated at the epigenetic, transcriptional, posttranscriptional, translational, and posttranslational levels. EMT-TFs recruit DNA methylation and histone modification machinery to stably repress epithelial genes and prevent their transcription. They are opposed by epithelial-associated TFs, which in turn repress EMT-TFs. Both epithelial and mesenchymal transcripts are alternatively spliced and regulated by miRNAs and lncRNAs. Translation initiation, mRNA stability, and polyadenylation affect the translation rate of epithelial and mesenchymal transcripts. Posttranslational modifications such as ubiquitylation, acetylation, and phosphorylation determine the balance between stability and degradation of epithelial and mesenchymal proteins.
EMT/MET model of metastasis
The EMT/MET model of metastatic dissemination attempts to reconcile the seemingly contradictory observation that metastatic lesions tend to have epithelial features, much like the primary tumor they arose from, but epithelial cells are not inherently invasive. The model postulates that cancer cells of epithelial origin undergo EMT to achieve the first steps of the metastatic cascade, including invasion into the tumor stroma, intravasation, and possibly extravasation at distant organs; however, in order to successfully form secondary tumors, cancer cells must undergo the reverse process of MET after reaching the metastatic site. Just like EMT, which is often induced by factors produced by stromal cells at the invasive front, MET is often an active process stimulated by molecular cues from metastatic niches in secondary organ sites (Gao et al., 2012; Del Pozo Martin et al., 2015; Esposito et al., 2019). While there is strong support in the literature for the role of EMT/MET in metastasis, the model continues to be challenged and updated with new research findings. This section presents results from these studies and synthesizes competing views for the EMT/MET model of metastasis.
EMT occurs during the natural history of tumor progression
The first challenge for the EMT/MET model was the difficulty in identifying tumor cells that had undergone EMT in vivo. Pathologists had long noted the mesenchymal morphology of what seemed to be cancer cells at the invasive front of tumors, but the origin of those cells was unclear because a mesenchymal tumor cell is for the most part indistinguishable from a mesenchymal stromal cell. However, with integration of the lineage labeling technique into the cancer field, it became possible to detect tumor cells that had undergone EMT spontaneously in vivo. The first direct evidence for EMT in breast cancer was reported by Trimboli et al. (2008). In this study, whey acidic protein (Wap)–Cre was used to genetically label mammary epithelial cells with LacZ. These strains were crossed to three murine models of breast cancer: Wap-myc, mouse mammary tumor virus (MMTV)–neu, and MMTV-polyoma middle T antigen (PyMT). In Wap-Cre/Wap-myc tumors, LacZ+ cells could be found in the stroma (i.e., cells of epithelial origin that acquired mesenchymal characteristics); however, this was not seen in MMTV-neu or MMTV-PyMT tumors, suggesting that EMT occurs in Myc-driven metastasis but not in Neu- or PyMT-induced metastasis (Trimboli et al., 2008). Rhim et al. (2012) used a similar method to demonstrate that pancreatic cells of epithelial origin undergo EMT and disseminate not only in the context of cancer but even at the preneoplastic stage. In this study, a YFP genetic label was used in conjunction with the KrasG12D/p53fl/+/Pdx1-Cre mouse model (KPCY) of pancreatic ductal adenocarcinoma (PDAC) to show that tumor cells with mesenchymal features can be found in the primary tumor stroma, circulation, and metastatic sites (Rhim et al., 2012). Upon careful inspection of disseminated tumor cells in the liver of this mouse model, it was revealed that EMT features are predominant in small metastatic lesions, but large lesions tend to have epithelial characteristics, consistent with an EMT-to-MET switch (Aiello et al., 2016).
EMT is a driver of metastasis
While these reports are suggestive that EMT and MET are important for metastasis, they do not address whether either is necessary and sufficient for this process. Tsai et al. (2012) elegantly demonstrated that Twist1-mediated EMT in squamous cell carcinoma was sufficient to drive dissemination, but Twist1 had to be down-regulated at metastatic sites for colonization to occur. Using a doxycycline-inducible Twist1 construct to induce EMT at the primary site either locally (by topical application) or systemically (by oral administration), the authors showed that Twist1 expression (and thus EMT) drove dissemination. However, metastatic colonization occurred only in mice that had locally activated Twist1, which could be reversed at metastatic sites that were not exposed to doxycycline (Tsai et al., 2012). Similarly, Snail expression is sufficient to drive breast cancer cells into the circulation, but it must be down-regulated once those cells reach the lung in order for the cells to successfully colonize (Tran et al., 2014). Along those lines, Takano et al. (2016) determined that isoform switching of Prrx1 from the EMT-promoting Prrx1b to the MET-promoting Prrx1a is necessary for liver metastasis in pancreatic cancer. Likewise, EMT driven by loss of p120-catenin accelerates pancreatic tumor progression and distant metastasis; however, p120-catenin–mediated stabilization of E-cadherin (and therefore MET) is required for metastatic colonization of the liver (Reichert et al., 2018). Interestingly, experimental lung metastasis did not require restoration of p-120 catenin, suggesting that MET is not required in this context. Along similar lines of inquiry into the requirements of EMT and MET for metastasis, Title et al. (2018) deleted it in β cells of a mouse model of neuroendocrine cancer to determine whether loss of the miR-200 family is sufficient to drive metastasis. The authors found that although mir-200 ablation increased survival, the resulting tumors metastasized more frequently, presumably due to increased EMT. The authors also deleted the miR-200 sites within the Zeb1 3′ untranslated region to promote EMT and found that this model phenocopies miR-200 ablation. Similar results were also seen in the KPC mouse model of PDAC with miR-200 deletion (Title et al., 2018). While suppression of the miR-200 family can promote dissemination, it is also an important driver of metastatic colonization. In addition to suppressing Zebs, the miR-200s target Sec23 homologue A (Sec23a), a gene important for the secretion of metastasis-suppressive proteins such as Tinagl1 (Shen et al., 2019). Through this dual mechanism, ectopic expression of the miR-200 family suppresses tumor migration and invasion but promotes lung colonization (Korpal et al., 2011). These studies demonstrate that EMT drives dissemination and MET is necessary for metastatic colonization.
Context-dependent requirement of EMT for metastasis
To address the question of whether EMT is required for metastasis, Zheng et al. (2015) generated Snail and Twist knockout mice on the background of the KrasG12D/p53R172H/+/Pdx1-Cre (KPC) PDAC model. Surprisingly, neither Snail nor Twist deletion resulted in a significant decrease in metastasis, suggesting that EMT is not required for pancreatic cancer metastasis (Zheng et al., 2015). However, a similar study in which Zeb1 was deleted in the same mouse model reported a significant reduction in metastasis, reinvigorating the debate over whether EMT is required for metastasis (Krebs et al., 2017). Snail does seem to be critical for breast cancer metastasis, however, as conditional deletion of Snail in the context of the MMTV-PyMT model significantly reduced lung metastasis (Tran et al., 2014). On the other hand, a report that used lineage labeling called into question the role of EMT in breast cancer metastasis. Fischer et al. (2015) used Fsp1-Cre to genetically label MMTV-PyMT breast cancer cells that undergo Fsp1-mediated EMT with GFP and found that most lung metastases did not express GFP, suggesting that EMT is not required in this context (Fischer et al., 2015). Similarly, an EMT lineage label driven by either α-smooth muscle actin–Cre or Fsp1-Cre was not activated in macrometastatic lesions in an Flp-FRT mouse model of PDAC (Chen et al., 2018). However, the results of these two studies contradict an earlier report that used an Fsp1 knock-in GFP reporter and suicide construct (thymidine kinase) on an MMTV-PyMT background (Xue et al., 2003). The resulting metastatic lesions contained cells that were double positive for the mammary cell marker casein and GFP, suggesting that EMT occurred during tumor progression. Moreover, replacement of endogenous Fsp1 with GFP or ablation of FSP1+ cells with the nucleoside analogue ganciclovir both significantly abrogated lung metastasis. Thus, evidence supporting or arguing against an essential role of EMT in metastasis have been presented in different studies (Table 1); however, looking at the question from a contextual perspective can shed some light on the issue.
Table 1. Summary of evidence for/against EMT in metastasis.
| Gene of interest | Approach | Tumor type | Model | Result | Reference |
|---|---|---|---|---|---|
| Twist1 | Conditional/inducible overexpression | SCC | DMBA | Sufficient for dissemination but must be down-regulated at metastatic site | Tsai et al. (2012) |
| Snai1 | Inducible overexpression | BC | MMTV-Neu | Sufficient for dissemination but must be down-regulated at metastatic site | Tran et al. (2014) |
| Prrx1a/b | Inducible overexpression | PDAC | KPC | Isoform B is sufficient for dissemination but must switch with isoform A at metastatic site | Takano et al. (2016) |
| miR-200s | Conditional KO | PNET/PDAC | RT2/KPC | Sufficient to drive metastasis | Title et al. (2018) |
| Snai1 | Conditional KO | BC | MMTV-PyMT | Critical for metastasis | Tran et al. (2014) |
| Snai1/Twist1 | Conditional KO | PDAC | KPC | Dispensable for metastasis | Zheng et al. (2015) |
| Zeb1 | Conditional KO | PDAC | KPC | Critical for metastasis | Krebs et al. (2017) |
| Fsp1 | Lineage tracing and cell ablation | BC | MMTV-PyMT | Labeled cells contribute to and are required for metastasis | Xue et al. (2003) |
| Fsp1 | Lineage tracing | BC | MMTV-PyMT | Labeled cells did not contribute to metastasis | Fischer et al. (2015) |
| Fsp1/αSMA | Lineage tracing | PDAC | KPF | Labeled cells did not contribute to metastasis | Chen et al. (2018) |
BC, breast cancer; DMBA, 7,12-dimethylbenz(a)anthracene; KPF, KrasG12D/p53FRT/+ or p53R172H/+/Pdx1-Flp; Neu, rat Errb2 transgene; PNET, pancreatic neuroendocrine tumor; RT2, Rip-Tag2; SCC, squamous cell carcinoma.
Context matters
There does not appear to be a single unifying molecular definition of EMT, especially in the context of cancer. Carcinomas use diverse programs to achieve the same goal of generating migratory tumor cells with the capability to invade, metastasize, and evade therapy. Cells that have undergone EMT in different tumor types (sometimes even within the same tumor) may look similar morphologically but can have dramatically divergent gene expression profiles. This might explain the apparently incongruous results of the aforementioned studies of EMT’s role in metastasis (Li and Kang, 2016; Aiello et al., 2017; Ye et al., 2017). Context-dependent manifestations of the EMT program were recently covered in an excellent review that discusses the nonredundant functions of various EMT-TFs (Stemmler et al., 2019). Of note, while Twist appears to be a critical EMT-TF in breast cancer (Yang et al., 2004; Mani et al., 2008), its role in PDAC EMT is not strongly supported (Hotz et al., 2007). Instead, Zeb1 seems to the primary EMT-TF responsible for PDAC EMT, which would explain why its deletion significantly reduced EMT and metastasis (Krebs et al., 2017), whereas Twist deletion did not (Zheng et al., 2015). Likewise, while FSP1 is a reliable marker of EMT in the KPC model of PDAC (Aiello et al., 2016), its usefulness as an EMT marker in breast cancer varies by model (Trimboli et al., 2008), which could explain why Fsp1 lineage-labeled tumor cells could not be found in lung metastases (Fischer et al., 2015). Thus it is likely that inconsistent findings within the EMT/metastasis field are due to different requirements for EMT effectors depending on tumor type, or due to an incomplete or partial EMT phenotype that does not involve EMT-TFs.
Partial EMT
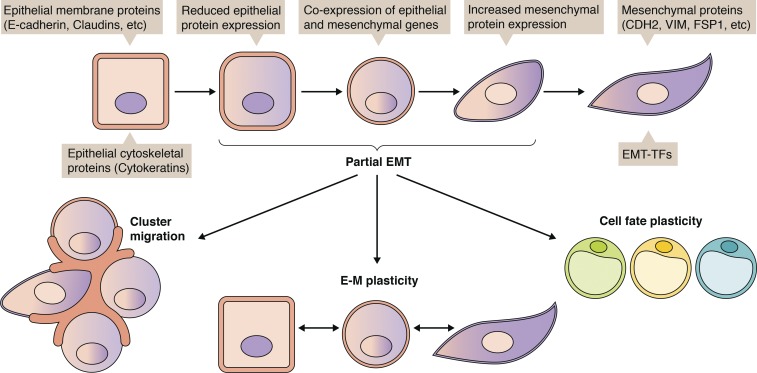
Emerging evidence suggests that EMT is a spectrum, and cancer cells often fall somewhere between fully epithelial and fully mesenchymal (Nieto et al., 2016). Tumor cells rarely commit to full EMT except in rare cases such as in hereditary diffuse gastric cancer and hereditary lobular breast cancer, where germline CDH1 mutations and subsequent epigenetic inactivation results in irreversible EMT (Barber et al., 2008; Dossus and Benusiglio, 2015). More often, tumor cells exhibit partial EMT, which could manifest as the coexpression of epithelial and mesenchymal markers or the loss of epithelial markers without gain of mesenchymal markers. Partial EMT appears to confer tumor cells with enhanced epithelial–mesenchymal plasticity, which is imperative for metastasis, tumor recurrence, and therapy resistance (Fig. 2). Therefore, it is critical to acknowledge and investigate the myriad ways tumor cells move through the EMT spectrum to address the issue in the clinic.
Figure 2.
Partial EMT: Heterogeneity and functional consequences. EMT is a spectrum of epithelial and mesenchymal phenotypes. Partial EMT, which typically involves a combination of epithelial and mesenchymal gene expression, facilitates cluster migration/dissemination, plasticity between epithelial and mesenchymal states, and even plasticity in cell fate (i.e., transdifferentiation to adipocytes).
A recent study reported that a partial EMT phenotype was predominant in the KPCY mouse model of PDAC, especially in association with the classic subtype (Aiello et al., 2018). Using a lineage-labeling strategy, CDH1+ epithelial and CDH1− mesenchymal tumor cells were isolated from autochthonous PDAC tumors and subjected to RNA sequencing, which revealed that in a majority of tumors, Cdh1 mRNA was maintained in cells that had lost it at the protein level. In contrast to the common EMT mechanism of EMT-TFs transcriptionally repressing the epithelial program, epithelial proteins like CDH1 were internalized in RAB11+ recycling endosomes. This potentially results in a cell poised for MET: indeed, CDH1− cells derived from tumors that undergo partial EMT were found to be more capable of generating CDH1+ cells compared with cells from tumors that typically undergo full EMT. Interestingly, partial EMT tumor cell lines engaged in collective migration in vitro and generated more circulating tumor clusters in vivo compared with full EMT tumor lines, which disseminated as single cells in vitro and in vivo. Moreover, a partial EMT phenotype was detected in a number of human pancreatic, breast, and colon cancer cell lines, suggesting that partial EMT is a common feature of carcinomas.
Single-cell RNA sequencing (scRNA-seq) is a powerful technique that overcomes the ambiguity of population transcriptomics. Recently, this method has been used to demonstrate the coexpression of both epithelial and mesenchymal genes at single-cell resolution during development and tumor progression, revealing that partial EMT occurs naturally in vivo. During murine organogenesis, scRNA-seq transcriptional profiling has identified intestine, liver, and lung cells with a partial EMT phenotype at embryonic day 9.5–11.5 (Dong et al., 2018). These cells express epithelial markers, including CDH1, EPCAM, claudins, and cytokeratins, as well as mesenchymal markers VIM, FN1, and SPARC. Interestingly, partial EMT cells had very low expression of classic EMT-TFs (SNAI1/2, ZEB1/2, and TWIST1/2), which is consistent with the expression of epithelial genes but suggests alternative mechanisms for inducing mesenchymal transcription. Partial EMT cells were not observed in adult intestine, liver, or lung; therefore, they probably represent a transient population during development. Dong et al. (2018) also investigated two previously published scRNA-seq datasets in primary breast cancer and lung adenocarcinoma patient-derived xenografts for evidence of partial EMT and found tumor cells that coexpressed VIM, FN1, EPCAM, and CDH1. Whether tumor cells arrive at partial EMT the same way that cells do during organogenesis remains an open question. In the same vein, Puram et al. (2017) surveyed primary and metastatic head and neck squamous cell carcinomas (HNSCCs) using scRNA-seq and identified a subset of malignant cells with a partial EMT signature. These cells bore some classic features of EMT, including expression of VIM, TGFβ-induced (TGFBI), and extracellular matrix genes, but expression of EMT-TFs was notably low, and consequently, epithelial gene expression was maintained. Using TGFBI expression to isolate partial EMT cells from an HNSCC cell line, the authors demonstrated that partial EMT cells are invasive and highly plastic. In situ, partial EMT cells were found at the leading edge of tumors near cancer-associated fibroblasts. Moreover, the partial EMT signature correlated with a malignant basal HNSCC subtype and lymph node metastasis, suggesting that partial EMT promotes loco-regional invasion.
Pastushenko et al. (2018) investigated the EMT spectrum in lineage-labeled primary murine skin and mammary tumors using a panel of cell surface markers to identify cells in intermediate EMT states. Using different combinations of the markers CD106, CD51, and CD61, the authors distinguished six populations within the YFP+/EPCAM− (presumably mesenchymal) compartment located throughout the EMT spectrum. Immunostaining revealed that these populations all expressed the mesenchymal marker VIM but varied in their expression of the epithelial marker cytokeratin-14. scRNA-seq further demonstrated the heterogeneity of EMT features among the six EPCAM− populations. Transplantation assays revealed that all EPCAM− populations had higher tumor-initiating capacity compared with EPCAM+ cells, but in vitro experiments showed that they varied in plasticity. Tumor cells in the middle of the EMT spectrum exhibited the most plasticity (i.e., were able to generate cells of all six subpopulations after sorting). Interestingly, although cells with a partial EMT phenotype were readily able to disseminate hematogenously, they did not have an increased ability to colonize the lung, suggesting that factors besides MET contribute to metastatic ability in this context (Pastushenko et al., 2018). This study paints a picture of the remarkably heterogeneous nature of EMT within tumors and begs the question of how it is established.
Ishay-Ronen et al. (2019) recently demonstrated that the partial EMT state makes breast cancer cells conducive to transdifferentiation into adipocytes. In this study, combination treatment with rosiglitazone (a peroxisome proliferator-activated receptor-γ agonist to induce adipocyte differentiation) and trametinib (a kinase inhibitor to block TGF-β signaling) was sufficient to convert partial EMT breast cancer cells into benign, cell cycle–arrested adipocytes, significantly reducing lung metastasis (Ishay-Ronen et al., 2019). This intriguing report raises the possibility that cancer cell plasticity, which seems to be at its peak during partial EMT, could be exploited therapeutically.
Perspectives and future directions
EMT is a dynamic, highly regulated process that occurs during embryogenesis and tumor progression to endow epithelial cells with motility, stemness, and therapy resistance. Its role in metastasis has been hotly debated, with recent reports both refuting and supporting the requirement for EMT. This is likely due to context-dependent EMT mechanisms, but further investigation of EMT effectors will be necessary to clarify the situation. Partial EMT is emerging as a common manifestation of the EMT program in tumors and can bestow cancer cells with increased plasticity.
At this point, the EMT field has collected an impressive body of knowledge on the signals that can promote EMT as well as effector proteins and their regulators. However, it remains unclear under which contexts these molecular players are involved in the natural history of tumor progression. Considering the emerging evidence that tumor types vary widely in their requirements for EMT-TFs and effectors in order to undergo EMT (or partial EMT), a systematic investigation of the necessity for each EMT driver in different in vivo tumor models would significantly enhance our understanding of how EMT actually happens in a living tumor. Moreover, this information would inform future experiments to determine whether EMT is required for metastasis: a clearer picture of which EMT programs are active in a given tumor type will be critical to successfully block EMT or eliminate cells that have undergone EMT. Only then will it be possible to definitively answer the question of whether EMT is crucial for metastasis in different cancer types and subtypes.
In the context of cancer, full EMT is rarely achieved, but rather partial EMT is more common, with cancer cells falling along a spectrum of epithelial and mesenchymal traits. What are the molecular mechanisms that cause a cell to begin down the path to EMT and stop partway there? It probably depends on a combination of cell-autonomous and non–cell-autonomous factors: perhaps microenvironmental cues, the transcriptomic landscape of the cell, or chromatin accessibility (which could be attributed to the cell of origin). What are the functional consequences of partial EMT? Recent reports suggest it pushes cells toward collective migration, but how does partial EMT relate to other facets of the EMT program such as stemness? Recent studies also started to appreciate the importance of EMT dynamics in mediating its biological impact in stemness and metastasis (Celià-Terrassa et al., 2018), an area that will benefit from systems and computational biology approaches of investigation. Epithelial–mesenchymal plasticity seems to be important for allowing tumor cells to adapt to their ever-changing microenvironment, whether they find themselves in a distant organ or bombarded with a new therapy. Partial EMT appears to bestow dramatic plasticity on tumor cells, giving them the ability to transdifferentiate into an entirely different cell type, according to one recent report (Ishay-Ronen et al., 2019). Therapeutically targeting partial EMT, and thus cellular plasticity, could prove to be efficacious; however, the molecular mechanisms governing partial EMT must be parsed first.
EMT contributes to nearly all of the hallmarks of cancer and continues to be an attractive target for cancer therapy. However, because the reverse process of MET appears to be critical for metastatic colonization, there has been justifiable hesitation to translate EMT inhibitors into the clinic for fear of stabilizing micrometastases. Moreover, it would be a challenge to address the problem of intra- and intertumor variation in EMT mechanisms. Nevertheless, a thoughtfully designed anti-EMT therapy combined with chemo-, radio-, and/or immunotherapy could optimize treatment outcome. Potential anti-EMT strategies could include reversing EMT, directly targeting cells that have undergone EMT, or inducing transdifferentiation of EMT cells into a harmless cell type as undertaken in Ishay-Ronen et al. (2019). Direct targeting of EMT tumor cells has proved challenging due to their general drug resistance and the difficulty of drugging TFs such as the EMT-TFs. Targeting the downstream effectors of EMT/MET or the overall plasticity of cancer cells might be a more precise alternative for therapeutic development. For example, Tinagl1 is a secreted metastasis-inhibitory protein that is down-regulated by miR-200s during MET. Therapeutic treatment with recombinant Tinagl1 reduces tumor progression and metastasis while avoiding the induction of EMT by targeting miR-200s directly (Shen et al., 2019). Finally, the transdifferentiation strategy would be valuable at any stage, perhaps even more so in the context of metastasis, since these lesions are especially refractory to treatment (Ishay-Ronen et al., 2019). These approaches should be combined with other established treatments for maximum effect. For example, since EMT is associated with immunosuppression, anti-EMT therapy could precede immunotherapy to sensitize the tumor to treatment. As the spectrum of EMT continues to be refined, hopefully novel druggable targets will be identified to augment current treatments.
Acknowledgments
We thank members of our laboratories for helpful discussions. We also apologize to the many investigators whose important studies could not be cited directly here owing to space limitations.
The work was supported by grants from the Brewster Foundation, the Breast Cancer Research Foundation, the Susan G. Komen Foundation, US Department of Defense, and the National Institutes of Health to Y. Kang and a US Department of Defense Breast Cancer Research Program Fellowship (BC170055) to N.M. Aiello.
The authors declare no competing financial interests.
Author contributions: The manuscript was written and edited by N.M. Aiello and Y. Kang.
References
- Aghdassi A., Sendler M., Guenther A., Mayerle J., Behn C.O., Heidecke C.D., Friess H., Büchler M., Evert M., Lerch M.M., and Weiss F.U.. 2012. Recruitment of histone deacetylases HDAC1 and HDAC2 by the transcriptional repressor ZEB1 downregulates E-cadherin expression in pancreatic cancer. Gut. 61:439–448. 10.1136/gutjnl-2011-300060 [DOI] [PubMed] [Google Scholar]
- Aiello N.M., Bajor D.L., Norgard R.J., Sahmoud A., Bhagwat N., Pham M.N., Cornish T.C., Iacobuzio-Donahue C.A., Vonderheide R.H., and Stanger B.Z.. 2016. Metastatic progression is associated with dynamic changes in the local microenvironment. Nat. Commun. 7:12819 10.1038/ncomms12819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello N.M., Brabletz T., Kang Y., Nieto M.A., Weinberg R.A., and Stanger B.Z.. 2017. Upholding a role for EMT in pancreatic cancer metastasis. Nature. 547:E7–E8. 10.1038/nature22963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello N.M., Maddipati R., Norgard R.J., Balli D., Li J., Yuan S., Yamazoe T., Black T., Sahmoud A., Furth E.E., et al. 2018. EMT Subtype Influences Epithelial Plasticity and Mode of Cell Migration. Dev. Cell. 45:681–695.e4. 10.1016/j.devcel.2018.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akalay I., Janji B., Hasmim M., Noman M.Z., André F., De Cremoux P., Bertheau P., Badoual C., Vielh P., Larsen A.K., et al. 2013. Epithelial-to-mesenchymal transition and autophagy induction in breast carcinoma promote escape from T-cell-mediated lysis. Cancer Res. 73:2418–2427. 10.1158/0008-5472.CAN-12-2432 [DOI] [PubMed] [Google Scholar]
- Aktas B., Tewes M., Fehm T., Hauch S., Kimmig R., and Kasimir-Bauer S.. 2009. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 11:R46 10.1186/bcr2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber M., Murrell A., Ito Y., Maia A.T., Hyland S., Oliveira C., Save V., Carneiro F., Paterson A.L., Grehan N., et al. 2008. Mechanisms and sequelae of E-cadherin silencing in hereditary diffuse gastric cancer. J. Pathol. 216:295–306. 10.1002/path.2426 [DOI] [PubMed] [Google Scholar]
- Bauer K., Dowejko A., Bosserhoff A.K., Reichert T.E., and Bauer R.J.. 2009. P-cadherin induces an epithelial-like phenotype in oral squamous cell carcinoma by GSK-3beta-mediated Snail phosphorylation. Carcinogenesis. 30:1781–1788. 10.1093/carcin/bgp175 [DOI] [PubMed] [Google Scholar]
- Bolós V., Peinado H., Pérez-Moreno M.A., Fraga M.F., Esteller M., and Cano A.. 2003. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J. Cell Sci. 116:499–511. 10.1242/jcs.00224 [DOI] [PubMed] [Google Scholar]
- Brabletz T., Jung A., Reu S., Porzner M., Hlubek F., Kunz-Schughart L.A., Knuechel R., and Kirchner T.. 2001. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc. Natl. Acad. Sci. USA. 98:10356–10361. 10.1073/pnas.171610498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken C.P., Gregory P.A., Kolesnikoff N., Bert A.G., Wang J., Shannon M.F., and Goodall G.J.. 2008. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 68:7846–7854. 10.1158/0008-5472.CAN-08-1942 [DOI] [PubMed] [Google Scholar]
- Cano A., Pérez-Moreno M.A., Rodrigo I., Locascio A., Blanco M.J., del Barrio M.G., Portillo F., and Nieto M.A.. 2000. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2:76–83. 10.1038/35000025 [DOI] [PubMed] [Google Scholar]
- Carver E.A., Jiang R., Lan Y., Oram K.F., and Gridley T.. 2001. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol. Cell. Biol. 21:8184–8188. 10.1128/MCB.21.23.8184-8188.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celià-Terrassa T., Bastian C., Liu D.D., Ell B., Aiello N.M., Wei Y., Zamalloa J., Blanco A.M., Hang X., Kunisky D., et al. 2018. Hysteresis control of epithelial-mesenchymal transition dynamics conveys a distinct program with enhanced metastatic ability. Nat. Commun. 9:5005 10.1038/s41467-018-07538-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti R., Hwang J., Andres Blanco M., Wei Y., Lukačišin M., Romano R.A., Smalley K., Liu S., Yang Q., Ibrahim T., et al. 2012. Elf5 inhibits the epithelial-mesenchymal transition in mammary gland development and breast cancer metastasis by transcriptionally repressing Snail2. Nat. Cell Biol. 14:1212–1222. 10.1038/ncb2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang R., Zhang Y., Zhang P., and Zhou Q.. 2017. Snail acetylation by histone acetyltransferase p300 in lung cancer. Thorac. Cancer. 8:131–137. 10.1111/1759-7714.12408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A., Wong C.S.F., Liu M.C.P., House C.M., Sceneay J., Bowtell D.D., Thompson E.W., and Möller A.. 2015. The ubiquitin ligase Siah is a novel regulator of Zeb1 in breast cancer. Oncotarget. 6:862–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.L., Wang S.H., Chan P.C., Shen M.R., and Chen H.C.. 2016. Phosphorylation of E-cadherin at threonine 790 by protein kinase Cδ reduces β-catenin binding and suppresses the function of E-cadherin. Oncotarget. 7:37260–37276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Gibbons D.L., Goswami S., Cortez M.A., Ahn Y.H., Byers L.A., Zhang X., Yi X., Dwyer D., Lin W., et al. 2014. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 5:5241 10.1038/ncomms6241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., LeBleu V.S., Carstens J.L., Sugimoto H., Zheng X., Malasi S., Saur D., and Kalluri R.. 2018. Dual reporter genetic mouse models of pancreatic cancer identify an epithelial-to-mesenchymal transition-independent metastasis program. EMBO Mol. Med. 10:e9085 10.15252/emmm.201809085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J.C., and Leung P.C.K.. 2011. Type I collagen down-regulates E-cadherin expression by increasing PI3KCA in cancer cells. Cancer Lett. 304:107–116. 10.1016/j.canlet.2011.02.008 [DOI] [PubMed] [Google Scholar]
- Cheung M., Chaboissier M.C., Mynett A., Hirst E., Schedl A., and Briscoe J.. 2005. The transcriptional control of trunk neural crest induction, survival, and delamination. Dev. Cell. 8:179–192. 10.1016/j.devcel.2004.12.010 [DOI] [PubMed] [Google Scholar]
- Cieply B., Riley P. IV, Pifer P.M., Widmeyer J., Addison J.B., Ivanov A.V., Denvir J., and Frisch S.M.. 2012. Suppression of the epithelial-mesenchymal transition by Grainyhead-like-2. Cancer Res. 72:2440–2453. 10.1158/0008-5472.CAN-11-4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comijn J., Berx G., Vermassen P., Verschueren K., van Grunsven L., Bruyneel E., Mareel M., Huylebroeck D., and van Roy F.. 2001. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell. 7:1267–1278. 10.1016/S1097-2765(01)00260-X [DOI] [PubMed] [Google Scholar]
- Creighton C.J., Li X., Landis M., Dixon J.M., Neumeister V.M., Sjolund A., Rimm D.L., Wong H., Rodriguez A., Herschkowitz J.I., et al. 2009. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc. Natl. Acad. Sci. USA. 106:13820–13825. 10.1073/pnas.0905718106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale J.K., Malapert P., Chal J., Vilhais-Neto G., Maroto M., Johnson T., Jayasinghe S., Trainor P., Herrmann B., and Pourquié O.. 2006. Oscillations of the snail genes in the presomitic mesoderm coordinate segmental patterning and morphogenesis in vertebrate somitogenesis. Dev. Cell. 10:355–366. 10.1016/j.devcel.2006.02.011 [DOI] [PubMed] [Google Scholar]
- Del Pozo Martin Y., Park D., Ramachandran A., Ombrato L., Calvo F., Chakravarty P., Spencer-Dene B., Derzsi S., Hill C.S., Sahai E., and Malanchi I.. 2015. Mesenchymal Cancer Cell-Stroma Crosstalk Promotes Niche Activation, Epithelial Reversion, and Metastatic Colonization. Cell Reports. 13:2456–2469. 10.1016/j.celrep.2015.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhamija S., and Diederichs S.. 2016. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int. J. Cancer. 139:269–280. 10.1002/ijc.30039 [DOI] [PubMed] [Google Scholar]
- Dong J., Hu Y., Fan X., Wu X., Mao Y., Hu B., Guo H., Wen L., and Tang F.. 2018. Single-cell RNA-seq analysis unveils a prevalent epithelial/mesenchymal hybrid state during mouse organogenesis. Genome Biol. 19:31 10.1186/s13059-018-1416-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongre A., Rashidian M., Reinhardt F., Bagnato A., Keckesova Z., Ploegh H.L., and Weinberg R.A.. 2017. Epithelial-to-Mesenchymal Transition Contributes to Immunosuppression in Breast Carcinomas. Cancer Res. 77:3982–3989. 10.1158/0008-5472.CAN-16-3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossus L., and Benusiglio P.R.. 2015. Lobular breast cancer: incidence and genetic and non-genetic risk factors. Breast Cancer Res. 17:37 10.1186/s13058-015-0546-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito M., Mondal N., Greco T.M., Wei Y., Spadazzi C., Lin S.-C., Zheng H., Cheung C., Magnani J.L., Lin S.-H., et al. 2019. Bone vascular niche E-selectin induces mesenchymal-epithelial transition and Wnt activation in cancer cells to promote bone metastasis. Nat. Cell Biol. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando R.I., Castillo M.D., Litzinger M., Hamilton D.H., and Palena C.. 2011. IL-8 signaling plays a critical role in the epithelial-mesenchymal transition of human carcinoma cells. Cancer Res. 71:5296–5306. 10.1158/0008-5472.CAN-11-0156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K.R., Durrans A., Lee S., Sheng J., Li F., Wong S.T.C., Choi H., El Rayes T., Ryu S., Troeger J., et al. 2015. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 527:472–476. 10.1038/nature15748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Krause G., Scheffner M., Zechner D., Leddy H.E.M., Behrens J., Sommer T., and Birchmeier W.. 2002. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat. Cell Biol. 4:222–231. 10.1038/ncb758 [DOI] [PubMed] [Google Scholar]
- Fukagawa A., Ishii H., Miyazawa K., and Saitoh M.. 2015. δEF1 associates with DNMT1 and maintains DNA methylation of the E-cadherin promoter in breast cancer cells. Cancer Med. 4:125–135. 10.1002/cam4.347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D., Joshi N., Choi H., Ryu S., Hahn M., Catena R., Sadik H., Argani P., Wagner P., Vahdat L.T., et al. 2012. Myeloid progenitor cells in the premetastatic lung promote metastases by inducing mesenchymal to epithelial transition. Cancer Res. 72:1384–1394. 10.1158/0008-5472.CAN-11-2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenburg G., and Hay E.D.. 1982. Epithelia suspended in collagen gels can lose polarity and express characteristics of migrating mesenchymal cells. J. Cell Biol. 95:333–339. 10.1083/jcb.95.1.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory P.A., Bert A.G., Paterson E.L., Barry S.C., Tsykin A., Farshid G., Vadas M.A., Khew-Goodall Y., and Goodall G.J.. 2008. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10:593–601. 10.1038/ncb1722 [DOI] [PubMed] [Google Scholar]
- Grelet S., Link L.A., Howley B., Obellianne C., Palanisamy V., Gangaraju V.K., Diehl J.A., and Howe P.H.. 2017. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol. 19:1105–1115. 10.1038/ncb3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumireddy K., Li A., Yan J., Setoyama T., Johannes G.J., Orom U.A., Tchou J., Liu Q., Zhang L., Speicher D.W., et al. 2013. Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. EMBO J. 32:2672–2684. 10.1038/emboj.2013.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haensel D., and Dai X.. 2018. Epithelial-to-mesenchymal transition in cutaneous wound healing: Where we are and where we are heading. Dev. Dyn. 247:473–480. 10.1002/dvdy.24561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., and Weinberg R.A.. 2011. Hallmarks of cancer: the next generation. Cell. 144:646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Herranz N., Pasini D., Díaz V.M., Francí C., Gutierrez A., Dave N., Escrivà M., Hernandez-Muñoz I., Di Croce L., Helin K., et al. 2008. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol. Cell. Biol. 28:4772–4781. 10.1128/MCB.00323-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J., Zhou J., Fu J., He T., Qin J., Wang L., Liao L., and Xu J.. 2011. Phosphorylation of serine 68 of Twist1 by MAPKs stabilizes Twist1 protein and promotes breast cancer cell invasiveness. Cancer Res. 71:3980–3990. 10.1158/0008-5472.CAN-10-2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotz B., Arndt M., Dullat S., Bhargava S., Buhr H.J., and Hotz H.G.. 2007. Epithelial to mesenchymal transition: expression of the regulators snail, slug, and twist in pancreatic cancer. Clin. Cancer Res. 13:4769–4776. 10.1158/1078-0432.CCR-06-2926 [DOI] [PubMed] [Google Scholar]
- Hyun K.A., Koo G.B., Han H., Sohn J., Choi W., Kim S.I., Jung H.I., and Kim Y.S.. 2016. Epithelial-to-mesenchymal transition leads to loss of EpCAM and different physical properties in circulating tumor cells from metastatic breast cancer. Oncotarget. 7:24677–24687. 10.18632/oncotarget.8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishay-Ronen D., Diepenbruck M., Kalathur R.K.R., Sugiyama N., Tiede S., Ivanek R., Bantug G., Morini M.F., Wang J., Hess C., and Christofori G.. 2019. Gain Fat-Lose Metastasis: Converting Invasive Breast Cancer Cells into Adipocytes Inhibits Cancer Metastasis. Cancer Cell. 35:17–32.e6. 10.1016/j.ccell.2018.12.002 [DOI] [PubMed] [Google Scholar]
- Iwano M., Plieth D., Danoff T.M., Xue C., Okada H., and Neilson E.G.. 2002. Evidence that fibroblasts derive from epithelium during tissue fibrosis. J. Clin. Invest. 110:341–350. 10.1172/JCI0215518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang M.H., Kim H.J., Kim E.J., Chung Y.R., and Park S.Y.. 2015. Expression of epithelial-mesenchymal transition-related markers in triple-negative breast cancer: ZEB1 as a potential biomarker for poor clinical outcome. Hum. Pathol. 46:1267–1274. 10.1016/j.humpath.2015.05.010 [DOI] [PubMed] [Google Scholar]
- Jolly M.K., Ward C., Eapen M.S., Myers S., Hallgren O., Levine H., and Sohal S.S.. 2018. Epithelial-mesenchymal transition, a spectrum of states: Role in lung development, homeostasis, and disease. Dev. Dyn. 247:346–358. 10.1002/dvdy.24541 [DOI] [PubMed] [Google Scholar]
- Kahlert C., Lahes S., Radhakrishnan P., Dutta S., Mogler C., Herpel E., Brand K., Steinert G., Schneider M., Mollenhauer M., et al. 2011. Overexpression of ZEB2 at the invasion front of colorectal cancer is an independent prognostic marker and regulates tumor invasion in vitro. Clin. Cancer Res. 17:7654–7663. 10.1158/1078-0432.CCR-10-2816 [DOI] [PubMed] [Google Scholar]
- Kim J.Y., Kim Y.M., Yang C.H., Cho S.K., Lee J.W., and Cho M.. 2012. Functional regulation of Slug/Snail2 is dependent on GSK-3β-mediated phosphorylation. FEBS J. 279:2929–2939. 10.1111/j.1742-4658.2012.08674.x [DOI] [PubMed] [Google Scholar]
- Kim K., Lu Z., and Hay E.D.. 2002. Direct evidence for a role of beta-catenin/LEF-1 signaling pathway in induction of EMT. Cell Biol. Int. 26:463–476. 10.1006/cbir.2002.0901 [DOI] [PubMed] [Google Scholar]
- Kim K.K., Kugler M.C., Wolters P.J., Robillard L., Galvez M.G., Brumwell A.N., Sheppard D., and Chapman H.A.. 2006. Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc. Natl. Acad. Sci. USA. 103:13180–13185. 10.1073/pnas.0605669103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D., Banerjee S., Ahmad A., Li Y., Wang Z., Sethi S., and Sarkar F.H.. 2010. Epithelial to mesenchymal transition is mechanistically linked with stem cell signatures in prostate cancer cells. PLoS One. 5:e12445 10.1371/journal.pone.0012445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M., Lee E.S., Hu G., and Kang Y.. 2008. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 283:14910–14914. 10.1074/jbc.C800074200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M., Ell B.J., Buffa F.M., Ibrahim T., Blanco M.A., Celià-Terrassa T., Mercatali L., Khan Z., Goodarzi H., Hua Y., et al. 2011. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat. Med. 17:1101–1108. 10.1038/nm.2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs A.M., Mitschke J., Lasierra Losada M., Schmalhofer O., Boerries M., Busch H., Boettcher M., Mougiakakos D., Reichardt W., Bronsert P., et al. 2017. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 19:518–529. 10.1038/ncb3513 [DOI] [PubMed] [Google Scholar]
- Kudo-Saito C., Shirako H., Takeuchi T., and Kawakami Y.. 2009. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell. 15:195–206. 10.1016/j.ccr.2009.01.023 [DOI] [PubMed] [Google Scholar]
- Kunita A., Baeriswyl V., Meda C., Cabuy E., Takeshita K., Giraudo E., Wicki A., Fukayama M., and Christofori G.. 2018. Inflammatory Cytokines Induce Podoplanin Expression at the Tumor Invasive Front. Am. J. Pathol. 188:1276–1288. 10.1016/j.ajpath.2018.01.016 [DOI] [PubMed] [Google Scholar]
- Lapin M., Tjensvoll K., Oltedal S., Javle M., Smaaland R., Gilje B., and Nordgård O.. 2017. Single-cell mRNA profiling reveals transcriptional heterogeneity among pancreatic circulating tumour cells. BMC Cancer. 17:390 10.1186/s12885-017-3385-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann W., Mossmann D., Kleemann J., Mock K., Meisinger C., Brummer T., Herr R., Brabletz S., Stemmler M.P., and Brabletz T.. 2016. ZEB1 turns into a transcriptional activator by interacting with YAP1 in aggressive cancer types. Nat. Commun. 7:10498 10.1038/ncomms10498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., and Kang Y.. 2016. Probing the Fifty Shades of EMT in Metastasis. Trends Cancer. 2:65–67. 10.1016/j.trecan.2016.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Deng W., Nail C.D., Bailey S.K., Kraus M.H., Ruppert J.M., and Lobo-Ruppert S.M.. 2006. Snail induction is an early response to Gli1 that determines the efficiency of epithelial transformation. Oncogene. 25:609–621. 10.1038/sj.onc.1209077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Bhargava M.M., Joseph A., Jin L., Rosen E.M., and Goldberg I.D.. 1994. Effect of hepatocyte growth factor/scatter factor and other growth factors on motility and morphology of non-tumorigenic and tumor cells. In Vitro Cell. Dev. Biol. Anim. 30A:105–110. 10.1007/BF02631401 [DOI] [PubMed] [Google Scholar]
- Lin T., Ponn A., Hu X., Law B.K., and Lu J.. 2010. Requirement of the histone demethylase LSD1 in Snai1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene. 29:4896–4904. 10.1038/onc.2010.234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Y., Diao L., Cuentas E.R.P., Denning W.L., Chen L., Fan Y.H., Byers L.A., Wang J., Papadimitrakopoulou V.A., Behrens C., et al. 2016. Epithelial-Mesenchymal Transition Is Associated with a Distinct Tumor Microenvironment Including Elevation of Inflammatory Signals and Multiple Immune Checkpoints in Lung Adenocarcinoma. Clin. Cancer Res. 22:3630–3642. 10.1158/1078-0432.CCR-15-1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M., et al. 2008. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 133:704–715. 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen P.J., Ebner R., Lopez A.R., and Derynck R.. 1994. TGF-beta induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J. Cell Biol. 127:2021–2036. 10.1083/jcb.127.6.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalko E.P., and Brown A.C.. 2018. Material Strategies for Modulating Epithelial to Mesenchymal Transitions. ACS Biomater. Sci. Eng. 4:1149–1161. 10.1021/acsbiomaterials.6b00751 [DOI] [PubMed] [Google Scholar]
- Moody S.E., Perez D., Pan T.C., Sarkisian C.J., Portocarrero C.P., Sterner C.J., Notorfrancesco K.L., Cardiff R.D., and Chodosh L.A.. 2005. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 8:197–209. 10.1016/j.ccr.2005.07.009 [DOI] [PubMed] [Google Scholar]
- Nagaoka K., Fujii K., Zhang H., Usuda K., Watanabe G., Ivshina M., and Richter J.D.. 2016. CPEB1 mediates epithelial-to-mesenchyme transition and breast cancer metastasis. Oncogene. 35:2893–2901. 10.1038/onc.2015.350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya Y., and Sheng G.. 2008. Epithelial to mesenchymal transition during gastrulation: an embryological view. Dev. Growth Differ. 50:755–766. 10.1111/j.1440-169X.2008.01070.x [DOI] [PubMed] [Google Scholar]
- Neumann D.P., Goodall G.J., and Gregory P.A.. 2018. Regulation of splicing and circularisation of RNA in epithelial mesenchymal plasticity. Semin. Cell Dev. Biol. 75:50–60. 10.1016/j.semcdb.2017.08.008 [DOI] [PubMed] [Google Scholar]
- Nieto M.A., Huang R.Y.J., Jackson R.A., and Thiery J.P.. 2016. EMT: 2016. Cell. 166:21–45. 10.1016/j.cell.2016.06.028 [DOI] [PubMed] [Google Scholar]
- Noman M.Z., Janji B., Abdou A., Hasmim M., Terry S., Tan T.Z., Mami-Chouaib F., Thiery J.P., and Chouaib S.. 2017. The immune checkpoint ligand PD-L1 is upregulated in EMT-activated human breast cancer cells by a mechanism involving ZEB-1 and miR-200. OncoImmunology. 6:e1263412 10.1080/2162402X.2016.1263412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocaña O.H., Córcoles R., Fabra A., Moreno-Bueno G., Acloque H., Vega S., Barrallo-Gimeno A., Cano A., and Nieto M.A.. 2012. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell. 22:709–724. 10.1016/j.ccr.2012.10.012 [DOI] [PubMed] [Google Scholar]
- Park S.M., Gaur A.B., Lengyel E., and Peter M.E.. 2008. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22:894–907. 10.1101/gad.1640608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastushenko I., Brisebarre A., Sifrim A., Fioramonti M., Revenco T., Boumahdi S., Van Keymeulen A., Brown D., Moers V., Lemaire S., et al. 2018. Identification of the tumour transition states occurring during EMT. Nature. 556:463–468. 10.1038/s41586-018-0040-3 [DOI] [PubMed] [Google Scholar]
- Paterson E.L., Kazenwadel J., Bert A.G., Khew-Goodall Y., Ruszkiewicz A., and Goodall G.J.. 2013. Down-regulation of the miRNA-200 family at the invasive front of colorectal cancers with degraded basement membrane indicates EMT is involved in cancer progression. Neoplasia. 15:180–191. 10.1593/neo.121828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puram S.V., Tirosh I., Parikh A.S., Patel A.P., Yizhak K., Gillespie S., Rodman C., Luo C.L., Mroz E.A., Emerick K.S., et al. 2017. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell. 171:1611–1624.e24. 10.1016/j.cell.2017.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert M., Bakir B., Moreira L., Pitarresi J.R., Feldmann K., Simon L., Suzuki K., Maddipati R., Rhim A.D., Schlitter A.M., et al. 2018. Regulation of Epithelial Plasticity Determines Metastatic Organotropism in Pancreatic Cancer. Dev. Cell. 45:696–711.e8. 10.1016/j.devcel.2018.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rembold M., Ciglar L., Yáñez-Cuna J.O., Zinzen R.P., Girardot C., Jain A., Welte M.A., Stark A., Leptin M., and Furlong E.E.M.. 2014. A conserved role for Snail as a potentiator of active transcription. Genes Dev. 28:167–181. 10.1101/gad.230953.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Chen Y., Song H., Chen L., and Wang R.. 2013. Inhibition of ZEB1 reverses EMT and chemoresistance in docetaxel-resistant human lung adenocarcinoma cell line. J. Cell. Biochem. 114:1395–1403. 10.1002/jcb.24481 [DOI] [PubMed] [Google Scholar]
- Rhim A.D., Mirek E.T., Aiello N.M., Maitra A., Bailey J.M., McAllister F., Reichert M., Beatty G.L., Rustgi A.K., Vonderheide R.H., et al. 2012. EMT and dissemination precede pancreatic tumor formation. Cell. 148:349–361. 10.1016/j.cell.2011.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca H., Hernandez J., Weidner S., McEachin R.C., Fuller D., Sud S., Schumann T., Wilkinson J.E., Zaslavsky A., Li H., et al. 2013. Transcription factors OVOL1 and OVOL2 induce the mesenchymal to epithelial transition in human cancer. PLoS One. 8:e76773 10.1371/journal.pone.0076773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H.S., Chung J.H., Lee K., Shin E., Jing J., Choe G., Kim H., Xu X., Lee H.E., Kim D.G., et al. 2012a Overexpression of epithelial-mesenchymal transition-related markers according to cell dedifferentiation: clinical implications as an independent predictor of poor prognosis in cholangiocarcinoma. Hum. Pathol. 43:2360–2370. 10.1016/j.humpath.2012.07.004 [DOI] [PubMed] [Google Scholar]
- Ryu H.S., Park D.J., Kim H.H., Kim W.H., and Lee H.S.. 2012b Combination of epithelial-mesenchymal transition and cancer stem cell-like phenotypes has independent prognostic value in gastric cancer. Hum. Pathol. 43:520–528. 10.1016/j.humpath.2011.07.003 [DOI] [PubMed] [Google Scholar]
- Shah A.N., Summy J.M., Zhang J., Park S.I., Parikh N.U., and Gallick G.E.. 2007. Development and characterization of gemcitabine-resistant pancreatic tumor cells. Ann. Surg. Oncol. 14:3629–3637. 10.1245/s10434-007-9583-5 [DOI] [PubMed] [Google Scholar]
- Shapiro I.M., Cheng A.W., Flytzanis N.C., Balsamo M., Condeelis J.S., Oktay M.H., Burge C.B., and Gertler F.B.. 2011. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 7:e1002218 10.1371/journal.pgen.1002218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M., Jiang Y.Z., Wei Y., Ell B., Sheng X., Esposito M., Kang J., Hang X., Zheng H., Rowicki M., et al. 2019. Tinagl1 suppresses triple-negative breast cancer progression and metastasis by simultaneously inhibiting integrin/FAK and EGFR signaling. Cancer Cell. 35:64–80.e7. 10.1016/j.ccell.2018.11.016 [DOI] [PubMed] [Google Scholar]
- Shiota M., Yokomizo A., Tada Y., Uchiumi T., Inokuchi J., Tatsugami K., Kuroiwa K., Yamamoto K., Seki N., and Naito S.. 2010. P300/CBP-associated factor regulates Y-box binding protein-1 expression and promotes cancer cell growth, cancer invasion and drug resistance. Cancer Sci. 101:1797–1806. 10.1111/j.1349-7006.2010.01598.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakihara T., Saitoh M., and Miyazono K.. 2007. Differential regulation of epithelial and mesenchymal markers by deltaEF1 proteins in epithelial mesenchymal transition induced by TGF-beta. Mol. Biol. Cell. 18:3533–3544. 10.1091/mbc.e07-03-0249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemens H., Jackstadt R., Hünten S., Kaller M., Menssen A., Götz U., and Hermeking H.. 2011. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 10:4256–4271. 10.4161/cc.10.24.18552 [DOI] [PubMed] [Google Scholar]
- Skrypek N., Goossens S., De Smedt E., Vandamme N., and Berx G.. 2017. Epithelial-to-Mesenchymal Transition: Epigenetic Reprogramming Driving Cellular Plasticity. Trends Genet. 33:943–959. 10.1016/j.tig.2017.08.004 [DOI] [PubMed] [Google Scholar]
- Stemmler M.P., Eccles R.L., Brabletz S., and Brabletz T.. 2019. Non-redundant functions of EMT transcription factors. Nat. Cell Biol. 21:102–112. 10.1038/s41556-018-0196-y [DOI] [PubMed] [Google Scholar]
- Sullivan N.J., Sasser A.K., Axel A.E., Vesuna F., Raman V., Ramirez N., Oberyszyn T.M., and Hall B.M.. 2009. Interleukin-6 induces an epithelial-mesenchymal transition phenotype in human breast cancer cells. Oncogene. 28:2940–2947. 10.1038/onc.2009.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano S., Reichert M., Bakir B., Das K.K., Nishida T., Miyazaki M., Heeg S., Collins M.A., Marchand B., Hicks P.D., et al. 2016. Prrx1 isoform switching regulates pancreatic cancer invasion and metastatic colonization. Genes Dev. 30:233–247. 10.1101/gad.263327.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman L.A., Grego-Bessa J., Raya A., Bertrán E., Pérez-Pomares J.M., Díez J., Aranda S., Palomo S., McCormick F., Izpisúa-Belmonte J.C., and de la Pompa J.L.. 2004. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 18:99–115. 10.1101/gad.276304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Title A.C., Hong S.J., Pires N.D., Hasenöhrl L., Godbersen S., Stokar-Regenscheit N., Bartel D.P., and Stoffel M.. 2018. Genetic dissection of the miR-200-Zeb1 axis reveals its importance in tumor differentiation and invasion. Nat. Commun. 9:4671 10.1038/s41467-018-07130-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H.D., Luitel K., Kim M., Zhang K., Longmore G.D., and Tran D.D.. 2014. Transient SNAIL1 expression is necessary for metastatic competence in breast cancer. Cancer Res. 74:6330–6340. 10.1158/0008-5472.CAN-14-0923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimboli A.J., Fukino K., de Bruin A., Wei G., Shen L., Tanner S.M., Creasap N., Rosol T.J., Robinson M.L., Eng C., et al. 2008. Direct evidence for epithelial-mesenchymal transitions in breast cancer. Cancer Res. 68:937–945. 10.1158/0008-5472.CAN-07-2148 [DOI] [PubMed] [Google Scholar]
- Tsai J.H., Donaher J.L., Murphy D.A., Chau S., and Yang J.. 2012. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 22:725–736. 10.1016/j.ccr.2012.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Staalduinen J., Baker D., Ten Dijke P., and van Dam H.. 2018. Epithelial-mesenchymal-transition-inducing transcription factors: new targets for tackling chemoresistance in cancer? Oncogene. 37:6195–6211. 10.1038/s41388-018-0378-x [DOI] [PubMed] [Google Scholar]
- Venables J.P., Brosseau J.P., Gadea G., Klinck R., Prinos P., Beaulieu J.F., Lapointe E., Durand M., Thibault P., Tremblay K., et al. 2013. RBFOX2 is an important regulator of mesenchymal tissue-specific splicing in both normal and cancer tissues. Mol. Cell. Biol. 33:396–405. 10.1128/MCB.01174-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent T., Neve E.P.A., Johnson J.R., Kukalev A., Rojo F., Albanell J., Pietras K., Virtanen I., Philipson L., Leopold P.L., et al. 2009. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial-mesenchymal transition. Nat. Cell Biol. 11:943–950. 10.1038/ncb1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Burstin J., Eser S., Paul M.C., Seidler B., Brandl M., Messer M., von Werder A., Schmidt A., Mages J., Pagel P., et al. 2009. E-cadherin regulates metastasis of pancreatic cancer in vivo and is suppressed by a SNAIL/HDAC1/HDAC2 repressor complex. Gastroenterology. 137:361–371: 371.e1–371.e5. 10.1053/j.gastro.2009.04.004 [DOI] [PubMed] [Google Scholar]
- Wei S.C., Fattet L., Tsai J.H., Guo Y., Pai V.H., Majeski H.E., Chen A.C., Sah R.L., Taylor S.S., Engler A.J., and Yang J.. 2015. Matrix stiffness drives epithelial-mesenchymal transition and tumour metastasis through a TWIST1-G3BP2 mechanotransduction pathway. Nat. Cell Biol. 17:678–688. 10.1038/ncb3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wels C., Joshi S., Koefinger P., Bergler H., and Schaider H.. 2011. Transcriptional activation of ZEB1 by Slug leads to cooperative regulation of the epithelial-mesenchymal transition-like phenotype in melanoma. J. Invest. Dermatol. 131:1877–1885. 10.1038/jid.2011.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Deng J., Rychahou P.G., Qiu S., Evers B.M., and Zhou B.P.H.. 2009. Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion. Cancer Cell. 15:416–428. 10.1016/j.ccr.2009.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C., Plieth D., Venkov C., Xu C., and Neilson E.G.. 2003. The gatekeeper effect of epithelial-mesenchymal transition regulates the frequency of breast cancer metastasis. Cancer Res. 63:3386–3394. [PubMed] [Google Scholar]
- Yaguchi T., Sumimoto H., Kudo-Saito C., Tsukamoto N., Ueda R., Iwata-Kajihara T., Nishio H., Kawamura N., and Kawakami Y.. 2011. The mechanisms of cancer immunoescape and development of overcoming strategies. Int. J. Hematol. 93:294–300. 10.1007/s12185-011-0799-6 [DOI] [PubMed] [Google Scholar]
- Yang J., Mani S.A., Donaher J.L., Ramaswamy S., Itzykson R.A., Come C., Savagner P., Gitelman I., Richardson A., and Weinberg R.A.. 2004. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 117:927–939. 10.1016/j.cell.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Yang Y., Park J.W., Bebee T.W., Warzecha C.C., Guo Y., Shang X., Xing Y., and Carstens R.P.. 2016. Determination of a Comprehensive Alternative Splicing Regulatory Network and Combinatorial Regulation by Key Factors during the Epithelial-to-Mesenchymal Transition. Mol. Cell. Biol. 36:1704–1719. 10.1128/MCB.00019-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Brabletz T., Kang Y., Longmore G.D., Nieto M.A., Stanger B.Z., Yang J., and Weinberg R.A.. 2017. Upholding a role for EMT in breast cancer metastasis. Nature. 547:E1–E3. 10.1038/nature22816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin T., Wang C., Liu T., Zhao G., Zha Y., and Yang M.. 2007. Expression of snail in pancreatic cancer promotes metastasis and chemoresistance. J. Surg. Res. 141:196–203. 10.1016/j.jss.2006.09.027 [DOI] [PubMed] [Google Scholar]
- Yuan J.H., Yang F., Wang F., Ma J.Z., Guo Y.J., Tao Q.F., Liu F., Pan W., Wang T.T., Zhou C.C., et al. 2014. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 25:666–681. 10.1016/j.ccr.2014.03.010 [DOI] [PubMed] [Google Scholar]
- Zhang P., Wei Y., Wang L., Debeb B.G., Yuan Y., Zhang J., Yuan J., Wang M., Chen D., Sun Y., et al. 2014. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat. Cell Biol. 16:864–875. 10.1038/ncb3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Liang Y., Xu Z., Wang L., Zhou F., Li Z., Jin J., Yang Y., Fang Z., Hu Y., et al. 2008. N-glycosylation affects the adhesive function of E-Cadherin through modifying the composition of adherens junctions (AJs) in human breast carcinoma cell line MDA-MB-435. J. Cell. Biochem. 104:162–175. 10.1002/jcb.21608 [DOI] [PubMed] [Google Scholar]
- Zheng H., Shen M., Zha Y.L., Li W., Wei Y., Blanco M.A., Ren G., Zhou T., Storz P., Wang H.Y., and Kang Y.. 2014. PKD1 phosphorylation-dependent degradation of SNAIL by SCF-FBXO11 regulates epithelial-mesenchymal transition and metastasis. Cancer Cell. 26:358–373. 10.1016/j.ccr.2014.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Carstens J.L., Kim J., Scheible M., Kaye J., Sugimoto H., Wu C.C., LeBleu V.S., and Kalluri R.. 2015. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 527:525–530. 10.1038/nature16064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Chang R., Ji W., Wang N., Qi M., Xu Y., Guo J., and Zhan L.. 2016. Loss of Scribble Promotes Snail Translation through Translocation of HuR and Enhances Cancer Drug Resistance. J. Biol. Chem. 291:291–302. 10.1074/jbc.M115.693853 [DOI] [PMC free article] [PubMed] [Google Scholar]