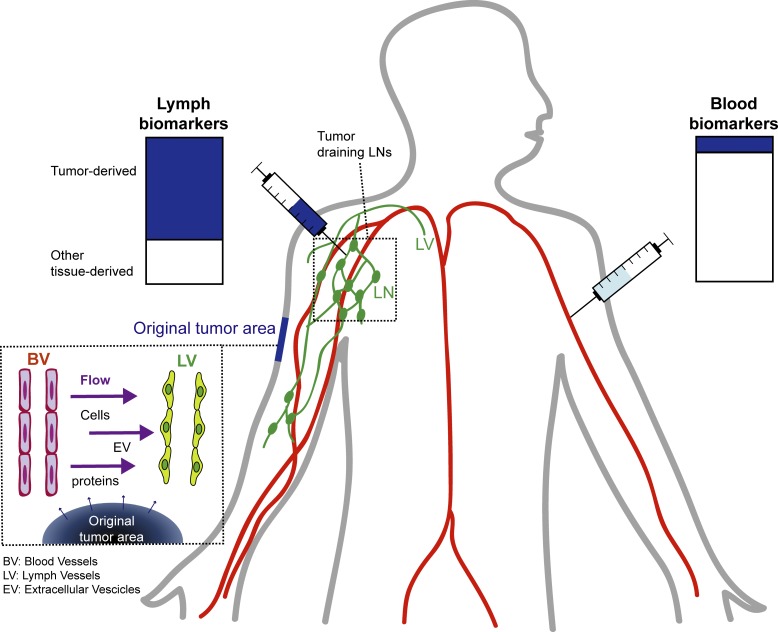
Identifying cancer biomarkers is of great interest for patient prognosis and monitoring. However, tumor-associated factors are diluted in blood and challenging to detect. Broggi et al. show that lymphatic exudate, accessible at the time of lymphadenectomy, is highly enriched in tumor-associated factors and therefore a promising source for biomarker discovery.
Abstract
Liquid biopsies allow monitoring of cancer progression and detection of relapse, but reliable biomarkers in melanoma are lacking. Because secreted factors preferentially drain to lymphatic vessels before dilution in the blood, we hypothesized that lymph should be vastly enriched in cancer biomarkers. We characterized postoperative lymphatic exudate and plasma of metastatic melanoma patients after lymphadenectomy and found a dramatic enrichment in lymphatic exudate of tumor-derived factors and especially extracellular vesicles containing melanoma-associated proteins and miRNAs, with unique protein signatures reflecting early versus advanced metastatic spread. Furthermore, lymphatic exudate was enriched in memory T cells, including tumor-reactive CD137+ and stem cell–like types. In mice, lymph vessels were the major route of extracellular vesicle transport from tumors to the systemic circulation. We suggest that lymphatic exudate provides a rich source of tumor-derived factors for enabling the discovery of novel biomarkers that may reflect disease stage and therapeutic response.
Graphical Abstract
Introduction
Early detection and monitoring of melanoma are crucial to improve patient survival. Melanoma staging considers the extent of dermal invasion, presence of ulceration, proliferation of tumor cells, and presence of loco-regional or distant metastases. In addition to the tumor node metastasis staging system, as new effective therapies are available, there is an urgent need for biomarkers capable of detecting melanoma before distant metastasis formation and assessing the risk of relapse. Good biomarkers should allow not only the monitoring of disease progression and response to treatment, but also the identification of patients at high risk of recurrence who may benefit from closer monitoring and adjuvant therapies early on, at the time of diagnosis. Biomarkers comprise mainly proteins, circulating microRNA (miRNA), exosomes, cell-free tumor DNA, and circulating tumor cells (Huang and Hoon, 2016). While extensive work has been done and multiple biomarkers have been identified, as of now there is no reliable blood test to diagnose melanoma occurrence and recurrence. For example, elevated concentrations of serum lactate dehydrogenase (LDH) and S100B proteins in melanoma patients are associated with worse prognosis and lower response to therapy (Buchbinder and Flaherty, 2016). However, their specificity is low. Therefore, biomarker signatures, comprising a panel of tumor-related factors, might be more robust in predicting disease progression and response to treatment.
Among factors released by the tumor microenvironment, extracellular vesicles (EVs), including exosomes (50–200 nm), have been shown to reflect disease status and to carry proteins and miRNA that could be used as biomarkers. For example, exosomes found in the plasma of melanoma patients were shown to be enriched in the melanoma-specific protein TYRP2, and high levels of this protein in exosomes were predictive of poor prognosis (Peinado et al., 2012). Similarly, S100 protein signatures carried or induced by exosomes could be useful as biomarkers (Bresnick et al., 2015; Hoshino et al., 2015; Alegre et al., 2016). Another study revealed that the inhibition of BRAF alters miRNAs released in EVs by melanoma cells, highlighting the potential of EV miRNA signature measurement to monitor treatment response (Lunavat et al., 2017). Thus, circulating EVs hold great promise as a source for cancer biomarkers, as they are enriched in tumor-relevant factors.
Biomarker levels are typically measured in liquid biopsies from the patient’s blood. The advantages are that analyses can be repeated over time, and blood withdrawal is noninvasive. However, at early stages of metastatic spread, the concentration of circulating biomarkers in the plasma is often very low and therefore undetectable. Proteins and other secreted factors released in the tumor extracellular space enter nearby lymphatic vessels with interstitial flow (Swartz and Lund, 2012) and are transported to the LNs, the thoracic duct, and eventually the blood. Because interstitial fluid flow is necessarily directed toward draining lymphatics, most secreted proteins and factors (e.g., from a tumor) are carried into the lymphatics, where they are exposed to (and can be sampled by) the endothelial and immune cells in the vessels and nodes. In this way, tissue fluid balance is strongly coupled to local immune regulation through antigen transport (Alitalo, 2011; Swartz and Lund, 2012; Trevaskis et al., 2015; Lund et al., 2016; Triacca et al., 2017). Directly sampling the tumor interstitium, the lymph may contain a higher concentration of melanoma biomarkers, possibly reaching detectable levels already at early stages of metastasis. Moreover, it has been reported that premetastatic alterations are already observed in LNs before tumor cell seeding, supporting the idea that secreted factors found in the lymph can possibly serve as biomarkers to detect early tumor spreading (Hood et al., 2011; Liu and Cao, 2016; Peinado et al., 2017).
Proteomic analyses of the lymph reveal that the lymph is not merely a plasma ultrafiltrate (Leak et al., 2004; Dzieciatkowska et al., 2014) and that in physiological conditions, the lymph proteome reflects the tissue it originates from and carries a pool of tissue-derived self-antigens important for immunological tolerance (Clement and Santambrogio, 2013; Clement et al., 2016). Similarly, during inflammatory conditions, the lymph mirrors the tissue inflammatory signature. Therefore, we hypothesized that lymph carries important tumor-derived factors that reflect the tumor microenvironment and may allow patient stratification or help predict response to therapy.
Here we collected postoperative lymphatic leak, hereafter referred to as lymphatic exudate, from metastatic melanoma patients undergoing lymphadenectomy (LAN) following positive LN biopsy and compared its contents to that of the plasma from each patient. We show not only that the lymphatic exudate is drastically enriched in secreted tumor-associated factors, but also that they contain proteomic signatures that may be used to differentiate early from late stages of metastatic spread.
Results
Lymphatic exudate is enriched in melanoma-associated proteins and EVs compared with plasma
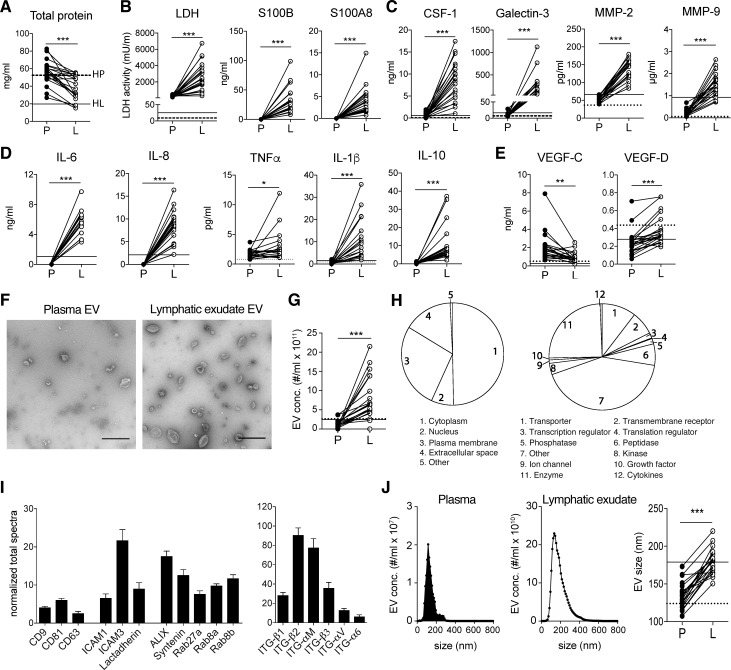
Lymph from healthy donors was collected immediately after dissection of afferent lymphatic vessels using an electrocautery or harmonic scalpel, while in patients undergoing LAN (Table S1 and Fig. S1), lymphatic exudate (composed of both the draining lymph and interstitial lymph) was collected in a suction drain and immediately processed for analysis as previously reported (Rutkowski et al., 2010). We first compared the lymphatic exudate and plasma from these patients for known melanoma biomarkers and other tumor-related factors, as well as the relative abundance of EVs. As previously reported in lymph (Clement et al., 2010, 2013; Dzieciatkowska et al., 2014), lymphatic exudate contained lower overall total protein concentrations compared with plasma (Fig. 1 A). However, the lymphatic exudate was dramatically enriched in melanoma-associated proteins such as LDH, S100B, and S100A8 (Buchbinder and Flaherty, 2016; Huang and Hoon, 2016) compared with plasma, where they were undetectable (Fig. 1 B). Factors known to strongly correlate with metastatic potential in melanoma (Egeblad and Werb, 2002), including colony stimulating factor-1 (CSF-1), galectin-3, and matrix metalloproteinases (MMP)–2 and -9, were also dramatically higher in lymphatic exudate compared with plasma (Fig. 1 C). Likewise, in melanoma patients, we detected an enrichment in lymphatic exudate of cytokines associated with tumor growth and neo-angiogenesis (Colotta et al., 2009)—namely, IL-6, IL-8, TNF-α, and IL-1β—as well as IL-10, which are associated with immunosuppressive tumor-associated macrophage skewing (Kim et al., 2013; Fig. 1 D). Importantly, each of these factors was higher in metastatic melanoma patients compared with healthy controls (HCs), especially in the lymph compartment (Fig. 1, A–D, dotted and solid horizontal lines). Interestingly, the lymphangiogenic growth factor vascular endothelial growth factor (VEGF)–C was less concentrated in lymphatic exudate than in plasma for most patients, while its homologue VEGF-D, present in much lower concentrations, was slightly more elevated in lymphatic exudate compared with plasma (Fig. 1 E). In particular, VEGF-D was detected at similar levels in HCs and patients, suggesting that the expression of this growth factor is not modulated during melanoma metastasis.
Figure 1.
Lymphatic exudate is enriched dramatically in tumor-secreted factors compared with plasma. Plasma (P) was compared with lymphatic exudate (L) from each metastatic melanoma patient. Average of expression in lymph from healthy donors (n ≥ 3) is shown, when detectable, in each graph for lymph (HL, solid line) and plasma (HP, dotted line). (A) Total protein content. (B) Melanoma biomarkers: LDH activity and melanoma proteins S100B and S100A8. (C) Metastasis-related proteins: CSF-1, galectin-3, and MMP-2 and -9. (D) Inflammatory cytokines: IL-6, -8, -1β, -4, and -10, and TNF-α. (E) Lymphatic growth factors VEGF-C and VEGF-D. (F) TEM analysis of EVs isolated from plasma and lymphatic exudate (bars, 500 nm). (G) EV density in plasma compared with lymph. (H) Subcellular localization and function of exosomal proteins analyzed by mass spectrometry. (I) Detection of exosomal markers tetraspanins (CD9, CD63, and CD81) adhesion molecules (ICAM1, ICAM3, and lactadherin), ALIX, syntenin, Rab proteins, and integrins on lymphatic exudate EVs of melanoma patients. Data shown as mean ± SEM. (J) Size comparison of EVs from plasma versus lymphatic exudate. n ≥ 17; *, P < 0.05; **, P < 0.01; ***, P < 0.001 by paired Student’s t test.
We next purified and examined the EVs from lymphatic exudate and found that while they shared the typical cup-shaped morphology of plasma-derived exosomes (Fig. 1 F), they were present at dramatically higher densities than in the plasma of nearly all patients, with a fivefold average increase (Fig. 1 G). Proteomic profiles of lymphatic exudate EVs revealed that most of the proteins were derived from the cytoplasm, the plasma membrane, and the nucleus, and included mainly enzymes, transporters, receptors, peptidases, and transcriptional regulators (Fig. 1 H). We also detected a few proteins normally found in the extracellular space, suggesting either that they are bound to the EV surface or that they were contained in other types of released vesicles contaminating the EV fraction. Lymphatic exudate EVs contained common exosomal markers, such as tetraspanins (cluster of differentiation [CD]–9, CD81, and CD63), adhesion molecules (intercellular adhesion molecule [ICAM]–1, ICAM-3, and lactadherin), ALIX, syntenin, Rab proteins (Rab27a, Rab8a, and Rab8b), and integrins (ITG-β1, ITG-β2, ITG-αM, ITG-β3, ITG-αV, and ITG-α6; Fig. 1 I). Interestingly, the average size of EVs was ∼30% larger in lymphatic exudate versus plasma (Fig. 1 J).
Lymphatic exudate is enriched in melanoma-associated miRNAs, which are mostly carried in EVs
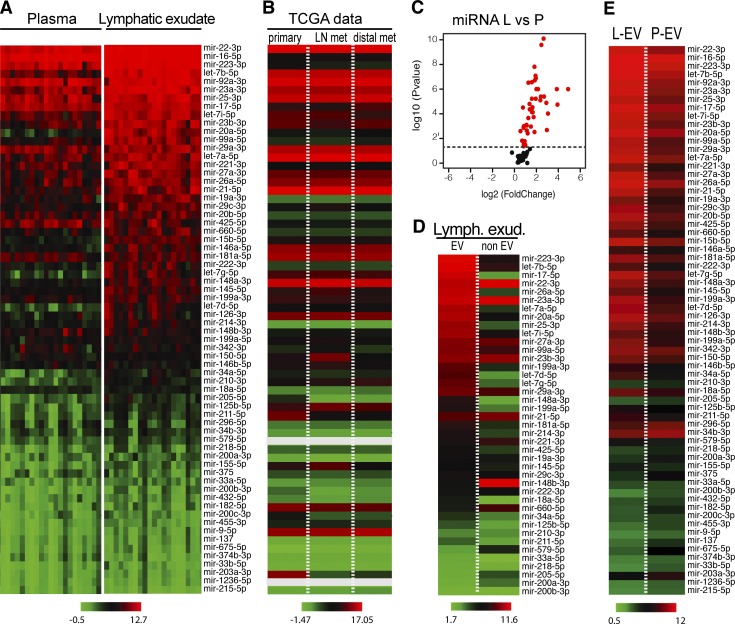
Several miRNAs have been shown to be potential mediators of metastasis and involved in the formation of the premetastatic niche, and, after literature research, we focused on 66 miRNAs that had been shown to be associated with metastatic melanoma (Table S1). We next assessed the lymphatic exudate and plasma for expression of these 66 miRNAs using a custom miRNA array and found that most were at significantly higher concentrations in the lymphatic exudate than in the plasma (Fig. 2 A and Fig. S2 A). Interestingly, these roughly correlated with miRNA expression levels reported in The Cancer Genome Atlas (TCGA) datasets for cutaneous metastatic melanoma, both in primary and metastatic tumors (Fig. 2 B).
Figure 2.
Lymphatic exudate is dramatically enriched compared with plasma in melanoma-relevant miRNAs, which mostly associate with EVs. (A) Heat map of melanoma-associated miRNAs contained in plasma and lymphatic exudate of metastatic melanoma patients (n = 12, data sorted by average expression levels in lymph). (B) TCGA data showing average expression levels of the same miRNAs, in the same order, in primary melanoma (n = 104) and at metastatic sites (regional LN metastasis, n = 226; distal metastasis, n = 67). Gray bars reflect miRNAs that were not reported. (C) Volcano plot of miRNA expression in lymphatic exudate (L) relative to that in plasma (P; each miRNA averaged across all patients, with red indicating the miRNAs with P < 0.05). (D) Within the lymphatic exudate (Lymph. exud.), content of lymph-enriched melanoma–associated miRNAs in EV and non-EV fractions of metastatic melanoma patients. (E) Comparative average miRNA expression in the EV fraction from lymphatic exudate (L-EV) or plasma (P-EV; n = 11, data sorted by average expression of miRNAs in EV fraction, raw data in Table S1).
It has been shown that tumor-secreted miRNAs that circulate and target the premetastatic niche are often contained within EVs (Peinado et al., 2017). Therefore, we also analyzed the miRNA that were enriched in the lymphatic exudate for their content in the EV and non-EV fractions (Fig. 2 C). As expected, most of the miRNAs were found primarily within the EV compartment, although interestingly, some were more abundant in the non-EV compartment, including mir-148-3b, mir-660-5p, and mir-579 (Fig. 2 D). The miRNA content of lymphatic exudate EVs was also increased compared with that in plasma EVs (Fig. 2 E). Taken together, melanoma-associated miRNAs, probably coming from residual tumor cells or cells from the tumor microenvironment, were dramatically enriched in the lymphatic exudate and were mostly circulating in association with EVs.
Proteomic analysis of lymphatic exudate reveals distinct signatures related to nodal metastatic spread
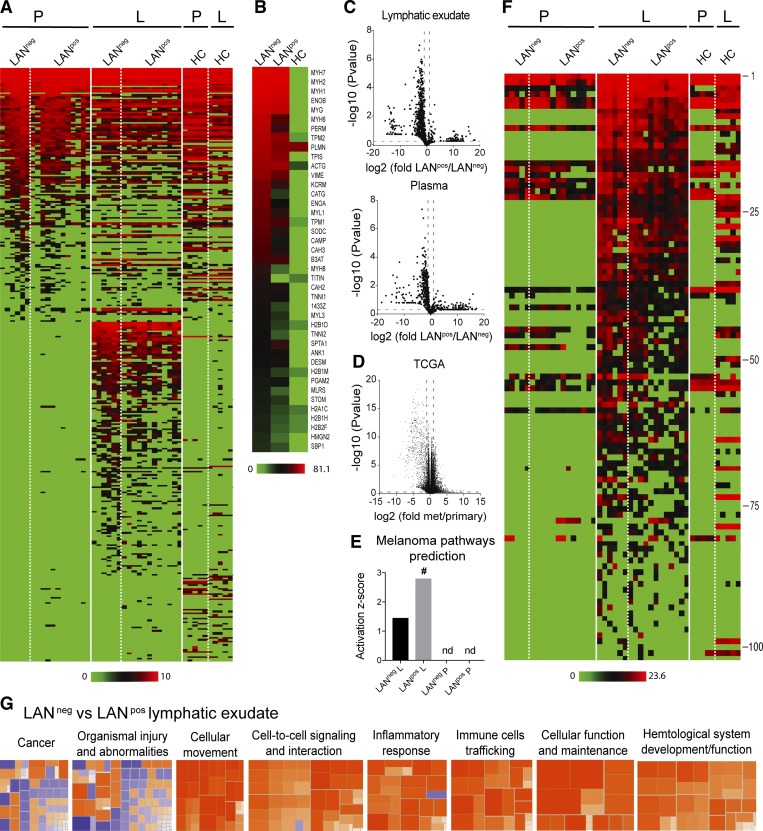
As the samples were collected from stage IIIA-C melanoma patients undergoing radical LAN following a positive sentinel LN, we could divide patients into two groups based on the nodal metastatic spread at the time of lymphatic exudate collection (Fig. S2 B). Patients with no evidence of further nodal spread beyond the sentinel LN were defined throughout the study as LAN-negative (LANneg), correlating with a less advanced disease, whereas patients with nodal involvement were defined as LAN-positive (LANpos), correlating with a more advanced disease.
We next performed an in-depth comparative proteomic analysis of the lymphatic exudate and plasma from LANneg and LANpos metastatic melanoma patients, as well as lymph from HCs for comparison (noting the important differences in collection methods). First, comparing lymphatic exudate versus plasma, we found a distinct protein signature in the lymphatic exudate that was absent in the plasma as well as in HCs (Fig. 3 A and Table S1). The 40 most highly expressed proteins found in the lymphatic exudate of melanoma patients were mainly related to cytoskeletal rearrangement and adhesion (myosin, actin, tropomyosin, troponin, and integrins), matrix remodeling (vimentin, enolases, and cathepsins), histone variants, and glycolytic enzymes (triosephosphate isomerase; Fig. 3 B). These processes are known to be differentially regulated in tumor progression and metastasis (Wilson et al., 2010; Hsiao et al., 2013); however, they may also be altered in other processes such as wound healing.
Figure 3.
Lymphatic exudate contains distinct protein signatures from plasma that better predict cancer pathways and that correlate with degree of metastatic spread. Patients were grouped as LAN negative or positive, according to the absence (LANneg) or presence (LANpos) of metastases in one or more of the resected LNs; HCs are shown for comparison. (A) Unbiased heat map highlighting global proteomic profiling of lymphatic exudate (L) and plasma (P) from LANneg and LANpos patients; data sorted by average expression of proteins in plasma and then lymphatic exudate. (B) Averaged expression profiles of the 40 most abundant proteins in the lymphatic exudate from each group (ranked by averaged expression in both LANneg and LANpos patients). (C) Volcano plot depictions of global proteomics profiling comparison of lymphatic exudate (top) and plasma (bottom) from LANneg and LANpos patients. (D) Volcano plot depiction of TCGA mRNA profiling of metastatic versus primary melanoma tumors. Dotted lines indicate limits where P value < 0.05 and ± twofold changes. (E) Activation scores of the melanoma-specific pathway prediction in lymphatic exudate versus plasma of patients; # indicates significant increase (P < 0.05); nd, not determined. (F) Heat map of 102 melanoma-associated proteins (described in the publicly available “Human Protein Atlas”) analyzed by LFQ proteomics and sorted from highest to lowest levels in lymphatic exudate (protein names and raw data are found in Table S1). (G) Activation z-scores of pathways that are up-regulated (>2, orange) or down-regulated (< −2, blue) in lymphatic exudate of LANneg versus LANpos patients. All data represent n = 6 for LANneg, n = 12 for LANpos, and n = 5 for HC. Raw data in Table S1.
Interestingly, by comparing the whole proteomic profile of both lymphatic exudate and plasma of LANpos versus LANneg patients, we detected an overall reduction of the proteins in LANpos patients (Fig. 3 C). This was in line with previous observations that increased metastatic potential correlates with down-regulation of gene and protein expression overall (Duffy, 1992; Olson and Joyce, 2015; Wojtukiewicz et al., 2015), as well as data from the TCGA showing more genes are down-regulated in metastatic lesions compared with primary tumor in the cutaneous metastatic melanoma dataset (Fig. 3 D).
Using the Ingenuity pathway analysis (IPA), we found that the overall proteomic signature predicted to be specific for melanoma was uniquely associated with the lymphatic exudate, but not the plasma (Fig. 3 E). Interestingly, the LANpos patients had a higher activation z-score than LANneg patients (Fig. 3 E and Table S1). Other pathways, including cancer, cellular movement, cell–cell signaling and interaction, and cell death and survival, were all quantitatively more predicted in the lymphatic exudate than in the plasma (Table S1). However, since the analyzed lymphatic exudate was collected after primary tumor excision, the protein signatures could partially reflect the wounding response to the LAN; therefore, we narrowed our analysis to proteins that have been reported in the publicly available Human Protein Atlas database to be up- or down-regulated specifically in melanoma as compared with healthy tissue. As before, we found that in most cases, expression of these proteins was unique to the lymphatic exudate (Fig. 3 F and Table S1), with decreased levels overall in the lymphatic exudate of patients with bulky nodal disease (LANpos) compared with LANneg (Fig. S2 B).
We next performed IPA analysis on the enriched melanoma-associated proteins found in the lymphatic exudate of patients and compared them with proteins in the HC lymph. Similar protein changes were detected in lymphatic exudate of LANneg and LANpos patients (Fig. S3, A and B; and Table S1); however, the melanoma-associated proteome of LANpos patients was predicted to have a higher (greater than +2.8) z-score for pathways associated with tumor cellular movement and cellular apoptosis, overall indicating a more invasive tumor phenotype (Fig. S3 C). In particular, the proteins filamin-A, fibronectin-1, granulin, laminin subunit-β, galectin-1, melanotransferrin, serpin family A member 1, and pigment epithelium-derived factor precursor predicted tumor cell migration in LANpos patients (Fig. S3 D). Moreover, the IPA analysis predicted TGF-β1 and KRAS, both known to be involved in tumorigenesis and metastasis, as principal upstream master gene regulators for the main protein hits in LANpos patients (Fig. S3 E).
When directly comparing the LANneg versus LANpos lymph proteomes, LANneg lymphatic exudate displayed an up-regulation of pathways involved in cellular movement, inflammation, and cellular maintenance, while the proteome of LANpos patients showed up-regulated pathways associated with morbidity and mortality, indicating a more advanced disease stage (Fig. 3 G and Table S1). Overall, these results point to the lymphatic exudate as the biological fluid that, as compared with the plasma, carries a more predictive signature of tumor state, as it is richer in cancer-associated proteins, and allows the differentiation of LANpos from LANneg patients.
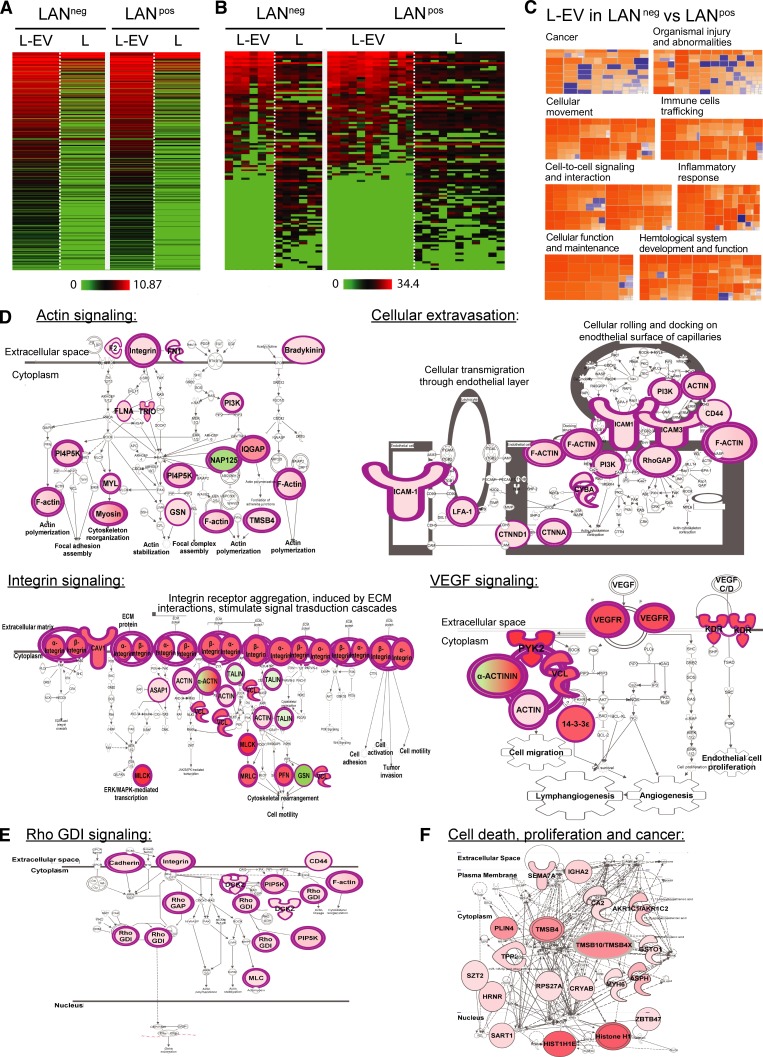
Lymphatic exudate EVs are enriched in tumor-related proteins that differentiate LANneg from LANpos patients
We next wanted to assess whether proteins derived from remaining tumor cells (in LANpos patients) or derived from the original tumor microenvironment (LANneg patients) might be carried by EVs, similarly to miRNAs. Thus, we characterized the proteomic content of lymphatic exudate EVs (Fig. 4 A and Table S1). Interestingly, around half of the melanoma-associated proteins, which we previously detected in whole lymphatic exudate (Fig. 3 F), were specifically enriched in EV fractions (Fig. 4 B). To determine the predicted pathways associated with the EV proteomic profile, we performed a comparative IPA analysis of the whole protein content of lymphatic exudate EVs purified from LANneg and LANpos patients. Similarly, to the whole lymphatic exudate proteome, the proteome of EVs derived from LANneg patients had a different signature from that of LANpos patients. In particular, EVs from LANneg patients presented a higher activation z-score (greater than +2.5) for several cellular functions including actin signaling, cellular extravasation, integrin signaling, and VEGF pathways (Fig. 4, C and D). Notably, the pathways up-regulated in LANneg patients are suggestive of an earlier disease state, where cytoskeleton rearrangement, cellular movement and adhesion, and lymphatic and blood vessel neogenesis are occurring. On the other hand, in EVs purified from LANpos patients, we observed a down-regulation of the Rho GDP dissociation inhibitor signaling pathway (Fig. 4 E), known to be a signature of highly metastatic breast cancer (Bozza et al., 2015), and an up-regulation of pathways associated with cell death, proliferation, and cancer-associated pathways (Fig. 4 F). These data suggest that EVs of the two patient subsets express protein signatures that predict pathways correlated with different stages of the metastatic process.
Figure 4.
Lymph-derived EVs are enriched in tumor-related proteins that may differentiate LANneg from LANpos melanoma patients. (A and B) Unbiased heat map highlighting the averaged protein concentration profiles (A) and expression profiles of melanoma-associated proteins (B; specified in Table S1) from lymph-derived EVs (L-EV) versus whole lymphatic exudate (L). (C–F) Proteomic profile comparisons using IPA. (C) Activation z-scores for the pathways up-regulated (orange, z-score >2) or down-regulated (blue, z-score < −2) in the lymphatic exudate EVs of LANneg versus LANpos patients. (D) Top up-regulated pathways in LANneg versus LANpos lymphatic exudate EVs. ECM, extracellular matrix. (E and F) Top down-regulated (E) or top up-regulated (F) pathways in LANpos versus LANneg lymphatic exudate EVs. n = 6 for LANneg and n = 11 for LANpos. Rho GDI, Rho GDP dissociation inhibitor. Raw data in Table S1.
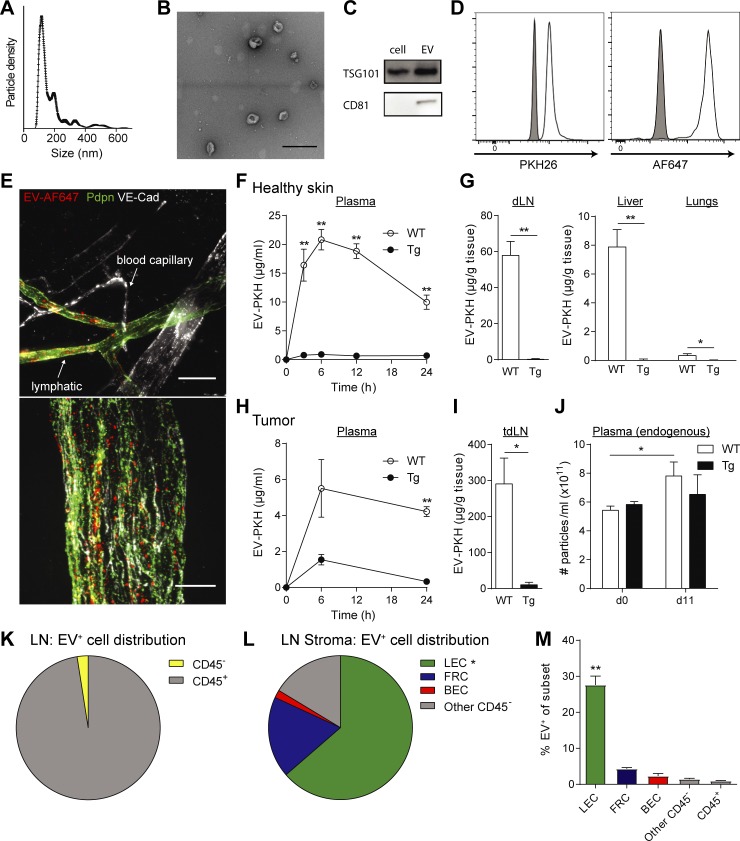
Lymphatic vessels are required for systemic distribution of EVs in mice
Lymphatic endothelial cells (LECs) have been shown to actively up-regulate macromolecular transport from the interstitial space (Triacca et al., 2017), and recent work in mice highlighted the role of lymphatics in the transport of EVs from the periphery to draining LNs (dLNs; Hood et al., 2011; Pucci et al., 2016; Srinivasan et al., 2016). However, the extent to which lymphatics are needed for the systemic distribution of EVs remains unclear. We purified and fluorescently labeled EVs from in vitro cultures of the mouse melanoma cell line B16-F10 (Fig. 5, A–D). After injection into the mouse ear dermis, fluorescent EVs were observed in draining lymphatic vessels, but not in surrounding blood capillaries, suggesting that lymphatics constitute an important route for EV transport (Fig. 5 E). To determine the relative contribution of lymphatic vessels to EV distribution to distant sites, we used K14-VEGFR-3-Ig (transgenic [Tg]) mice lacking dermal lymphatics (Mäkinen et al., 2001) and performed a biodistribution study 24 h after intradermal (i.d.) injection of labeled EVs into mouse hocks. We observed that while EVs injected i.d. into WT mice were detected in dLNs, plasma, lungs, and liver, they were nearly undetectable in these tissues after i.d. injection into Tg mice (Fig. 5, F and G). Next, we injected labeled EVs directly into implanted B16-F10/VEGF-C melanomas, where again they were seen abundantly in WT mice and to a much lesser extent in the dLNs and plasma of Tg mice. To confirm that lymphatic vessels played a role in endogenous EV distribution from melanoma tumors, we analyzed the plasma of mice before tumor inoculation (day 0) and after B16-F10/VEGF-C tumor inoculation and growth (day 11). We observed that while the quantity of endogenous (nonlabeled) EVs increased upon tumor growth in WT mice, no significant increase in EV levels was observed in Tg mice (Fig. 5 J). Last, in WT mice, we observed that LECs were the main CD45neg stromal cells taking up EVs in the tumor-draining LN (Fig. 5, K–M). Together, these findings demonstrate that lymphatics represent a major route of EV trafficking from the interstitial space into the lymph, LN, and systemic circulation.
Figure 5.
Tumor EVs are taken up and transported by lymphatic vessels. (A) Size distribution of purified EVs from cultured B16-F10 melanoma cells, as determined by nanoparticle tracking analysis. (B) TEM image of B16-F10 cell-secreted EVs (bar, 200 µm). (C) Western blot analysis of exosomal markers CD81 and TSG101 in cell lysates and EVs (20 µg protein). (D) Bead-bound, fluorescently labeled EVs analyzed by flow cytometry for labels (PKH26, Alexa Fluor 647). (E) EV uptake by lymphatic (VE-cadherin+ [VE-Cad+] and podoplanin+ [Pdpn+]), but not blood (VE-Cad+Pdpn−) endothelium, in the mouse ear dermis 30 min after i.d. injection of AF647-labeled EVs (EV-AF647). Bars, 50 μm (top) and 10 μm (bottom). (F and G) Biodistribution of PKH26-labeled EVs (EV-PKH) after i.d. injection into WT mice and mice lacking dermal lymphatics (K14-VEGFR-3-Ig mice, Tg) in plasma (F) and dLN (G), liver, and lungs after 24 h. (H–J) EV biodistribution after intratumoral injections into B16-F10/VEGF-C tumors, (H) in plasma and (I) in dLN after 24 h. (J) Total endogenous particle concentration in plasma before tumor inoculation (day 0) and after 11 d as determined by nanoparticle tracking analysis. (K–M) Distribution of EV-positive cells among (K) total LN and (L) LN stromal cell (CD45−) populations 24 h after intratumoral injection. (M) Percentage of EV-positive cells within each cell subtype among LN cells. For all data shown, n ≥ 4 per group in at least two independent experiments; *, P < 0.05, **, P < 0.01 by two-tailed unpaired Student’s t test. Data in F–J and M shown as mean ± SEM.
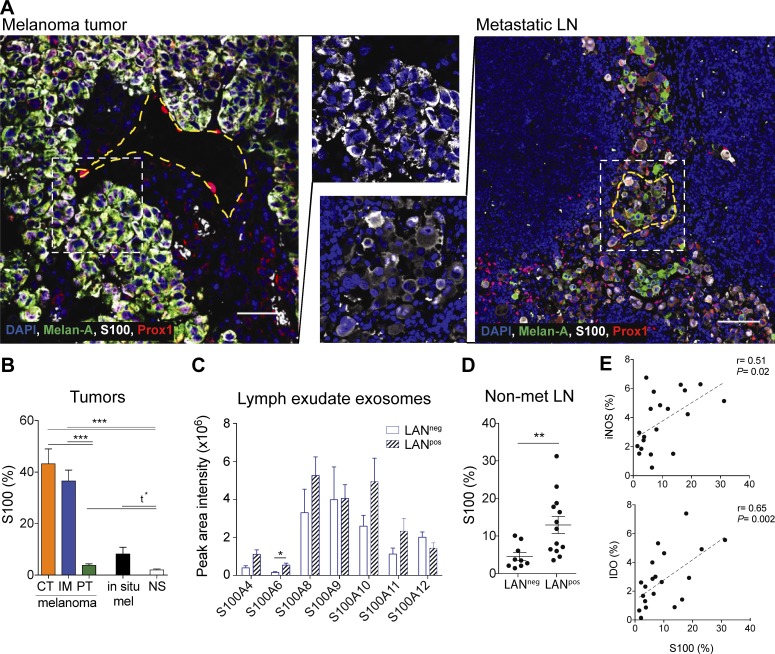
S100 proteins are expressed in melanoma tumors, lymphatic exudate EVs, and premetastatic LNs
Knowing that lymph can transport exosomes to the dLNs and to distal sites, we were interested in characterizing the role of tumor-derived EVs in the formation of the premetastatic niche. Therefore, we investigated the transport of the inflammatory protein S100 that is also a well-known melanoma antigen and expressed in the premetastatic niche (Hiratsuka et al., 2008; Peinado et al., 2012; Bresnick et al., 2015; Costa-Silva et al., 2015; Hoshino et al., 2015). Immunostaining of human primary melanoma and metastatic LNs showed that S100 staining was specific to tumor cells, which were heterogeneous in their expression of the tumor antigen Melan-A (Fig. 6 A). As reported earlier, metastatic melanoma cells also displayed low levels of cytoplasmic Prospero homeobox 1 (Prox-1; Bordry et al., 2018), which can be differentiated from the nuclear Prox-1 expression in LECs (Fig. 6 A). S100, which is known to be up-regulated in premetastatic LNs (Hiratsuka et al., 2008; Peinado et al., 2012; Bresnick et al., 2015; Costa-Silva et al., 2015; Hoshino et al., 2015), was highly up-regulated in tumor-rich areas (tumor center and invasive margin) and to a lesser extent in the peritumoral region of primary tumors, as compared with normal skin (Fig. 6 B). Non-invasive in situ melanomas expressed lower levels of S100 (Fig. 6 B), consistent with the notion that the S100 antigen is associated with highly aggressive melanoma tumors.
Figure 6.
Premetastatic LNs express S100 proteins that are found in lymphatic exudate EVs of melanoma patients. (A) Primary melanoma (left; bar, 50 µm) and metastatic LN (right; bar, 200 µm) analyzed by immunofluorescence staining. Yellow dotted lines indicate the perimeter of lymphatic vessels (identified by nuclear Prox-1 staining) while the white dotted lines indicate the zoomed areas shown in the center. (B) Quantification of S100 (% of positive pixels) in different regions of the melanoma (CT, center of the tumor; IM, invasive margin; PT, peritumoral region; n = 14) compared with in situ melanoma (mel; also called stage 0 melanoma, which is confined to the epidermis; n = 14) and normal skin (NS, n = 12). (C) Expression of S100 proteins in EVs isolated from lymphatic exudate of melanoma patients as analyzed by mass spectrometry. (D) S100 expression in whole sections of tumor-free (nonmetastatic [non-met]) LNs of LANneg (n = 10) and LANpos (n = 12) patients. Data shown as mean ± SEM. (E) Correlation of S100 expression with iNOS and IDO expression in the tumor-free LNs of both LANpos and LANneg patients (n = 23).
Next, we confirmed the presence of the S100 isoforms (S100A4, S100A6, S100A8, S100A9, S100A10, S100A11, and S100A12) in lymphatic exudate EVs of both LANneg and LANpos patients (Fig. 6 C). By comparing nonmetastatic LNs of both patient subsets, we found that the highly metastatic LANpos patients expressed significantly more S100 in the tumor-free LNs compared with low metastatic LANneg patients (Fig. 6 D). Moreover, S100 expression correlated with the expression of the suppressive molecules inducible nitric oxide synthase (iNOS) and indoleamine-pyrrole 2,3-dioxygenase (IDO) in tumor-free LNs (Fig. 6 E). This suggests that the tumor-free LNs in LANpos patients were undergoing more changes than those in LANneg patients.
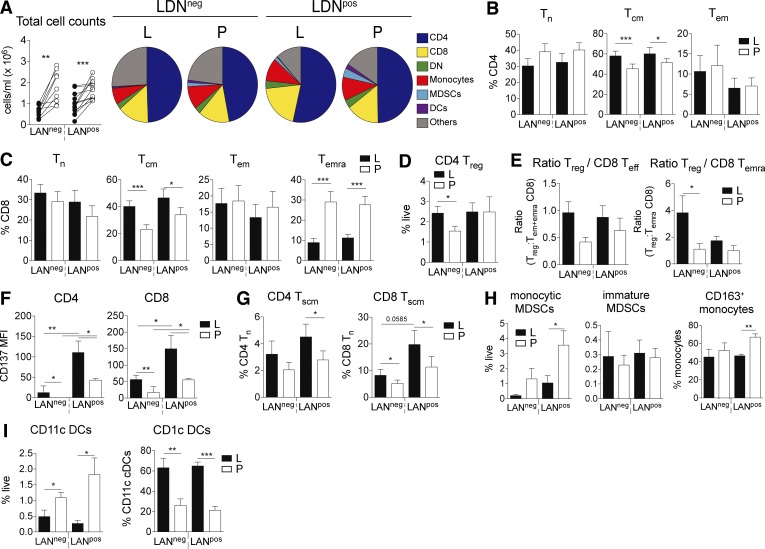
Lymphatic exudate is enriched in central memory and stem cell–like memory T cells
Immune cells can also travel from the tumor microenvironment or metastatic LNs to other secondary lymphoid tissues through lymphatic vessels. Therefore, we used flow cytometry to characterize the cellular composition of the lymphatic exudate of LANneg and LANpos patients and compared it to the one of the plasma. Although we found less cells circulating in the lymphatic exudate, the overall composition of immune cells in lymphatic exudate and plasma was similar (Fig. 7 A). Central memory T cells (TCM) of both CD4+ and CD8+ subsets were enriched in lymphatic exudate compared with plasma (Fig. 7, B and C; and Fig. S4 A). In contrast, the terminally differentiated effector memory CD8+ T cell population was enriched in plasma of metastatic patients (Fig. 7 C). LANneg patients showed more circulating regulatory T (T reg) cells (Fig. 7 D), and interestingly, the ratio of T reg cells to effector cells, particularly for the terminally differentiated effector memory (TEMRA) subset, was higher in the lymphatic exudate (Fig. 7 E). The activation marker CD137 is significantly more expressed on T cells isolated from the lymphatic exudate, and at highest level in LANpos patients (Fig. 7 F). Given the observation that lymphatic exudate contained more memory T cells, we next investigated the presence of another memory T cell phenotype that characterizes stem cell memory T cells (TSCM) and is defined within the naive cell subset by the high expression of CD45RA, CCR7, IL-7R, and CD95 (Fig. S4 B). We found that lymphatic exudate is enriched in both CD4+ and CD8+ TSCM cells especially in highly metastatic patients (Fig. 7 G). Lastly, monocytic myeloid-derived suppressor cells (MDSCs) and CD163+ monocytes (Fig. S4 C) were enriched in LANpos plasma (Fig. 7 H). While the total dendritic cell (DC) content was increased in plasma, the CD1c+ DC subset was significantly enriched in lymphatic exudate of metastatic melanoma patients (Fig. 7 I). Taken together, our data suggest that memory and CD137+ tumor antigen-experienced T cells preferentially recirculate into the lymphatics.
Figure 7.
Lymphatic exudate is enriched in memory and antigen-experienced CD137+ T cells compared with plasma. (A) Total cell counts and pie charts of composition of circulating immune cells as analyzed by flow cytometry. DN, double-negative (CD4− CD8−) T cells. (B–F) CD4+ T cell (B) and CD8+ T cell subsets (C; naive [TN]; central memory [TCM], and effector memory [TEM]), CD4+ T reg cells (D), and ratio of CD4+ T reg to total CD8+ effector (TEFF = TEM + TEMRA) or to CD8+ TEMRA (E), stem cell–like memory CD4+ and CD8+ T cells (F; TSCM). L, lymphatic exudate; P, plasma. (G–I) CD137 expression on CD4+ and CD8+ T cells (G), MDSC subtypes (H), and DCs (I). T cells were gated after live/dead exclusion on CD3+ and defined as follows: TCM, CD45RA− CCR7+; TN, CD45RA+ CCR7+; TEMRA, CD45RA+ CCR7−; TEM, effector, CD45RA− CCR7−; and TSCM, defined as CD95hi and CD127hi T cells. Monocytic cells were gated according to the gating strategy described in Fig. S4. n = 10 for LANneg and n = 11 for LANpos melanoma patients. Data shown as mean ± SEM; *, P < 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 compared to plasma using a paired Student's t test (A) or one-way ANOVA followed by Tukey's multiple comparison test (B–I).
Discussion
The ability to predict metastatic spread is still a major goal in cancer research, and liquid biopsies are a major focus for monitoring disease progression. Liquid biopsies are considered minimally invasive, and have the potential to provide information about the tumor at very early stages as well as allow the detection of novel biomarkers to guide the design of personalized treatments and monitor response to therapy (Mandal and Chan, 2016; Wan et al., 2017). Here, we demonstrate that lymph, which can be collected as lymphatic exudate after LAN, is a uniquely rich source of tumor-derived factors that may be useful for biomarker discovery.
The lymphatic exudate analyzed here, from patients undergoing LAN, was collected after the removal of the primary melanoma, and thus did not directly drain from the primary tumor. However, the fact that this lymphatic exudate was dramatically enriched in known melanoma biomarkers suggests that these factors remained in the tumor site after tumor resection. Furthermore, LECs are known to actively take up proteins and vesicles (Hirosue et al., 2014) and can archive them over weeks (Tamburini et al., 2014). Undoubtedly, the composition of the lymphatic exudate would be altered by the surgery and wound-healing response there; indeed, some of the up-regulated pathways (such as cellular movement and endothelial cell proliferation) are associated with both wound healing and metastasis. Therefore, we first restricted our analysis on melanoma-associated proteins found exclusively in the lymphatic exudate, and then directly compared LANneg with LANpos patients, which both underwent the same surgical procedure.
It is well appreciated that metastasis formation is a complex process initiated by the primary tumor and involving tumor cell transition into an invasive phenotype, remodeling of the tumor microenvironment, and promoting changes in distant organs that will support later metastatic tumors (Jiang et al., 2015). The transformation of the distant sites, referred to as premetastatic niches, is initiated by tumor-derived EVs containing factors secreted by both tumor cells and immune cells of the tumor microenvironment, including antigens, integrins, immunomodulatory factors, and miRNA (Peinado et al., 2012; Costa-Silva et al., 2015; Hoshino et al., 2015). Therefore, EVs are considered potential biomarkers for early detection of tumors (Kahlert and Kalluri, 2013). Since the interstitial fluid of the tumor flows directly into draining lymphatics, and since EVs are large enough that diffusion is negligible compared with convection for their interstitial transport, we were not surprised to find dramatically higher EV densities in the lymphatic exudate versus plasma of melanoma patients. Our finding that EVs in the lymphatic exudate were on average substantially larger than those in plasma may possibly reflect the notion that tumor and immune cells in the tumor microenvironment produce larger EVs, or that the plasma EVs arise from a wider variety of sources, including platelets. Interestingly, the proteomic profile of EVs from LANneg patients’ lymphatic exudate was associated with pathways involved in cellular movement, vascularization, extravasation, and adhesion, which correlate with early stages of metastasis. In contrast, the proteomic profile of EVs isolated from LANpos patients showed up-regulated signaling pathways connected to cell death, proliferation, and cancer, suggestive of more advanced disease. Importantly, these results are consistent with those independently obtained by García-Silva et al. in this issue.
Metastasis-related miRNAs were specifically expressed in EVs of metastatic melanoma patients, which were dramatically enriched in the lymphatic exudate compared with plasma. However, these miRNAs were present at similar levels in LANpos versus LANneg patients, possibly suggesting that the prometastatic miRNAs released in EVs by melanoma cells are independent of tumor stage. However, further analyses are needed to assess the contribution and the biomarker potential of EV-bound miRNA isolated from lymphatic exudate in metastatic patients, including a more extensive miRNA analysis.
Our data provides evidence in mice that tumor-derived EVs are primarily taken up by LECs and transported by lymphatic vessels, and that EVs require lymphatics in order to reach distant sites. This is consistent with recent studies showing that EVs are delivered to the LN via lymphatic vessels (Pucci et al., 2016; Srinivasan et al., 2016). Moreover, we found that lymphatic exudate EVs of melanoma patients contain S100, which is known to be up-regulated in premetastatic LNs (Hiratsuka et al., 2008; Peinado et al., 2012; Bresnick et al., 2015; Costa-Silva et al., 2015; Hoshino et al., 2015). Since LECs can archive antigens during viral infections (Tamburini et al., 2014), it is interesting to speculate whether LN LECs directly contribute to premetastatic niche formation by taking up and storing S100-containing tumor-derived EVs.
Finally, we found that although lymphatic exudate contains an overall lower density of immune cells compared with plasma, specific populations were enriched in lymph of melanoma patients. CD4+ and CD8+ T cells in the lymphatic exudate versus plasma expressed higher levels of the costimulatory molecule CD137, which is expressed on tumor-reactive T cells and widely used as a marker to enrich and expand T cells for immunotherapy (Zhu and Chen, 2014). Terminally differentiated CD8+ T cells were significantly higher in the plasma, consistent with their lack of lymphoid homing chemokine receptors, while central memory CD4+ and CD8+ T cells, which migrate to and among secondary lymphatic organs, were enriched in the lymph. Interestingly, we found, particularly in LANpos patients, an enrichment of so-called TSCM cells: CD45RA−, CCR7+, IL-7R+ T cells expressing the activation marker CD95. TSCM cells have been described as a primordial memory subset with self-renewing capacity and strong anti-tumor activity (Gattinoni et al., 2011).
One limitation of introducing lymph as a liquid biopsy is that lymph collection is to date technically challenging. Lymph can be collected after sentinel node biopsy during the surgical procedure or in already diagnosed metastatic melanoma patients who undergo a radical LAN, where additional LNs surrounding the sentinel node are removed. During the latter procedure, large numbers or lymphatic vessels are excised, and both circulating lymph and interstitial lymph (lymphatic exudate) can be easily collected after surgery using a surgical drain. The main advantages of collecting lymphatic exudate compared with cannulated circulating lymph is that the procedure does not delay the surgery time, and much larger volumes of material can be analyzed, allowing a more complete screening. On the other hand, lymphatic exudate cannot be analyzed from healthy individuals, where circulating lymph is collected by cannulation (Clement et al., 2010). Nonetheless, the analyses presented here can be performed with much smaller volumes, and thus collecting lymph from patients undergoing primary tumor and sentinel node dissection at the same time (i.e., in breast cancer patients) would allow a more comprehensive analysis of specific tumor-predictive biomarker signatures. Furthermore, the relevance of collecting lymph during primary tumor or sentinel node dissection is strengthened by the recent decision to remove radical LAN from standard clinical protocols on metastatic melanoma patients following a recent multicenter selective lymphadenectomy trial II study showing no survival benefit (Faries et al., 2017).
Taken together, our data suggest that lymph from cancer patients is a rich source of tumor-derived factors and may provide a highly valuable source for identifying protein, miRNA, EV, and cellular fingerprints of each patient’s cancer. As such, lymph should be taken into consideration during the development of diagnostic tests to predict organ-specific metastasis and therapies to halt metastatic spread. This proteomic and miRNAomic analysis, together with the immune cell signature, could be pivotal to the phenotyping of cancer at very early stages as well as its interaction with the immune system in the so called “cancer immunogram” (Blank et al., 2016). Further analysis on lymphatic exudate or lymph collected from a larger cohort of melanoma patients, including at a wider range of disease state, will be required to fully explore its potential for biomarker discovery that may guide the design of personalized therapies.
Materials and methods
Patients
This study investigated lymphatic exudate and plasma of 22 patients with stage IIIA to IIIC cutaneous melanoma (Table S1) who were treated at the Lausanne University Hospital, Lausanne, Switzerland. All the studies were conducted according to the relevant regulatory standards, upon approval by Swissmedic and the Commission d’Éthique de la Recherche Clinique de la Faculté de Biologie et de Médecine, Université de Lausanne.
Lymphatic exudate and lymph collection
Newly diagnosed melanoma patients with a positive sentinel LN underwent radical LAN. Out of all the patients, 10 had a negative LAN (LANneg), meaning that all the LNs removed during the surgery were tumor-free, and 11 had a positive LAN (LANpos; Fig. S1). During the surgery, lymphatic vessels were dissected by using electrocautery or harmonic scalpel. Postoperative lymphatic leak was collected in a suction drain for 2 d following the surgery (collected daily). Immediately after collection, lymphatic exudate was supplemented with a cocktail of protease/phosphatase inhibitors (Roche) and immediately used for further analysis as previously described (Rutkowski et al., 2010). At the same time as lymphatic exudate collection, plasma was also collected and used for comparative analysis. Given their similarity, in all the analyses, the expression of day 1 and day 2 of both lymphatic exudate and plasma was averaged and considered as n = 1. The study was approved by the Commission Cantonale d’Éthique de la Recherche sur l’Être Humain and performed according to the relevant regulatory standards. HCs used for lymphatic exudate and plasma were collected from the laboratory of L. Santambrogio as previously described (Clement et al., 2010). Briefly, prenodal peripheral lymph of healthy subjects was collected from the subcutaneous lymphatic vessel 20 cm above the ankle, through lymphatic cannulation performed with polyethylene tubing (IntramedicH PE60; 427416; Becton Dickinson).
Cytokine analysis
Total protein content of lymphatic exudate and plasma was assessed by bicinchoninic acid (BCA) assay (Thermo Fisher Scientific). Most of the cytokines were assessed by ELISA analysis according to the manufacturer’s instructions. ELISAs used were the following: galectin-3 ELISA (DGAL30; R&D Systems), M-CSF (CSF-1) ELISA (DMC00B; R&D Systems), S100B ELISA (KA0037; Abnova), S100A8 (DY4570; R&D Systems), MMP-2 ELISA (DY902; R&D Systems), MMP-9 ELISA (BMS2016/2; Affimetrix), VEGF-C (DuoSet DY293B; R&D Systems), and VEGF-D (DuoSet DY622; R&D Systems). Lactate dehydrogenase activity was measured using the LDH Activity Assay Kit (MAK066; Sigma-Aldrich). IL-6, IL-8, TNF-α, IL-1β, IL-4, and IL-10 were analyzed using the Proinflammatory Panel 1 Kits of Mesoscale (MSD Multi-Spot Assay System).
EV isolation
EVs were isolated with ultracentrifugation technique as described in Théry et al. (2002). Briefly, upon receipt, lymphatic exudate or plasma was centrifuged to remove cells, and proteinase/phosphatase inhibitor tablets were added. The supernatant was then diluted with an equal amount of PBS and centrifuged at 2,000 g for 30 min to remove dead cells and then at 12,000 g for 45 min to remove cell debris. The supernatant was further centrifuged for 2 h at 110,000 g. The pellet containing the EV fraction was washed twice at 110,000 g for 70 min, whereas the supernatant was considered the “non-EV fraction” (containing proteins not present in EVs). The EV fraction was resuspended in 70 µl of PBS. Both fractions were stored at −80°C until analysis. Before proteomic analysis, total protein content was assessed by bicinchoninic acid assay. For in vivo mouse experiments, EVs were purified from 2- to 3-d cell–conditioned medium by concentration and size-exclusion separation. Briefly, cells and debris were cleared from supernatant by serial centrifugations 10 min at 300 g, 10 min at 2,000 g, and 20 min at 10,000 g. Then, the supernatant was concentrated to 500 µl using Amicon Ultra-15 Centrifugal Filter Units, and EVs were separated from free proteins using qEVoriginal Size Exclusion Columns (Izon). For EV tracking experiments, EV membranes were labeled with a PKH26 kit (Sigma-Aldrich), and EV surface proteins were labeled with the Alexa Fluor 647 Protein Labeling Kit (Invitrogen) according to the manufacturer’s protocols and purified from unbound dye using a qEVoriginal Size Exclusion Column.
Nanosight analysis of EVs
EV density and size distribution were assessed by nanoparticle tracking analysis (NanoSight; Malvern Instruments). 1 µl of isolated EVs was diluted in 1 ml of PBS to achieve a uniform particle distribution that was analyzed in three to five sequential measurements at 25°C.
Western blot analysis
20 µg of EVs or cell lysates was mixed with Laemmli SDS sample buffer (Alfa Aesar), incubated 10 min at 95°C, and cooled to 4°C. Electrophoresis was performed on Mini-PROTEAN TGX Gels (Bio-Rad). Proteins were transferred to a polyvinylide difluoride membrane (Bio-Rad) preactivated in 100% methanol. Blocking was performed overnight at 4°C in 5% Tris-buffered saline (TBS) milk, primary in 1–5% TBS milk, 1 h at room temperature (RT; shaking), and secondary (HRP-conjugated) in 1–5% TBS milk, 1 h at RT (shaking). The following antibodies were used: anti-CD9 (1:500; C9993; Sigma-Aldrich), anti-CD63 (1:200; SC-15363; Santa Cruz Biotechnology), anti-CD81 (1:1,000; SAB 3500454; Sigma-Aldrich), and TSG101 (1:1,000; T5701; Sigma-Aldrich).
Transmission electron microscope (TEM) analysis of EVs
Negative stain was performed in the following way: 5 µl sample solution was adsorbed to a glow-discharged carbon-coated copper grid (Canemco & Marivac), washed with deionized water, and stained with 5 µl 2% uranyl acetate. The samples were imaged at RT using a Tecnai Spirit electron microscope equipped with an LaB6 filament and operated at an acceleration voltage of 80 kV. Images were taken at a magnification from 30,000× to 50,000× and recorded with an Eagle 4,098 × 4,098 pixel camera (FEI).
Liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis
To facilitate the identification of the low abundance proteins, both lymphatic exudate and plasma were IgG- and albumin-depleted with Aurum Serum Spin Colums (Bio-Rad) as previously described (Clement et al., 2013). EV fractions were analyzed without prior depletion. 4 µg of total protein was subjected to an “in solution” tryptic digest. 2 µg of each trypsinized sample was used for the nanoLC-MS/MS analysis on an Orbitrap Elite mass spectrometer. Prior to the injection into the mass spectrometer, peptides were desalted on C18 stageTips (Rappsilber et al., 2007), dried down by vacuum centrifugation, and separated by reversed phase chromatography on a Dionex Ultimate 3000 RSLCnano UPLC system connected in-line with an Orbitrap Elite (Thermo Fisher Scientific).
Protein identification
Raw files from each technical and biological replicate were filtered, de novo sequenced, and assigned with protein ID using Peaks 8.0 software (Bioinformatics Solutions) by searching against the human Swiss-Prot database (January 2017; 158,154 entries). The following search parameters were applied for label-free quantitative (LFQ) analysis: trypsin restriction for enzyme and one allowed missed cleaved at one peptide end. The parent mass tolerance was set to 15 ppm using monoisotopic mass, and fragment ion mass tolerance was set to 0.1 daltons. Carbamidomethyl cysteine (+57.0215 on C) was specified in PEAKS 8.0 as a fixed modification. Methionine, lysine, proline, arginine, cysteine, and asparagine oxidations (+15.99 on CKMNPR), deamidation of asparagine and glutamine (NQ-0.98), and pyro-Glu from glutamine (Q-18.01 N-term) were set as variable modifications. Data were validated using the false discovery rate (FDR) method built in PEAKS 8.0, and protein identifications were accepted with a confidence score (−10lgP) >15 for peptides and (−10lgP) >15 for proteins; a minimum of one peptide per protein was allowed after data were filtered for <0.8% FDR for peptides and <1% FDR for proteins identifications (P < 0.05). An independent validation of the MS/MS-based peptides and protein identification was performed using Scaffold (version Scaffold_4.7.3, Proteome Software). The Scaffold built-in option “MuDPIT” was used to combine multiple files from technical replicates of each sample. Peptide identifications were set at >95.0% probability by the Peptide Prophet algorithm with Scaffold delta-mass correction. Protein identifications were set at >90.0% probability and contained at least one identified peptide. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony as described originally (Nesvizhskii et al., 2003).
Label-free relative peptide quantification
LFQ methods (spectral counting and precursor intensity) were used to analyze and contrast the proteomic profiles of lymph, plasma, and EVs from each melanoma patient. LFQ based on the precursor intensity was performed using the quantification algorithm supported by the PEAKS Q module (version 8.0; Bioinformatics Solutions). MS/MS spectra were then extracted using the same software and used to search against the target-decoy database containing all SwissProt entries for Homo sapiens (January 2017; 158,154 entries) as described above. The Quantitative module (Q) of PEAKS 8.0 was used to perform the LFQ analysis using the precursor ion (MS1) quantification built-in algorithm. The relative protein abundance was displayed as heat maps including representative proteins of each protein group after normalization of the corresponding averaged areas (abundances) with respect to the total ion current. The un-clustered heat maps were generated using the CIMminer free software provided by the National Institutes of Health (https://discover.nci.nih.gov/cimminer/) using log2 transformed quantitative values of MS1. The positive values reflect fold increases (red color), and negative values reflect fold decreases (green color). Only proteins that passed a selected significance statistical threshold (ANOVA, P < 0.05 and FDR <1% for protein and peptide expression) and had been identified with at least threefold differential expression across all samples are shown in the representative heat maps. An additional LFQ analysis using the spectral counts intensity (MS/MS or MS2) was employed for the same set of lymph, plasma, and EVs samples using the normalized weighted spectrum count option provided in “perSPECtives” (version 2.0.6; Proteome Software). The corresponding “.mzIdentML” files generated in Scaffold (version 4.7.3) for each set of sample from LANneg, LANpos, and HCs were imported in “perSPECtives” where the threshold of one unique peptide/sample and the significance level of P < 0.05 were set up throughout the control of FDR with the standard Benjamini–Hochberg procedure.
Gene ontology, molecular, and cellular pathways enrichment analysis
Networks, functional analyses, biochemical, and cellular pathways were generated using IPA (Ingenuity Systems) and the list of proteins extracted from LFQ analyses. Specifically, the experimentally determined protein ratios were used to calculate the experimental fold changes by rescaling their values using a log2 transformation, such that positive values reflected fold increases while the negative values reflected fold decreases. For network generation, datasets containing gene identifiers (gene symbols) for the LANneg, LANpos, and HCs were uploaded into the IPA application together with their rescaled log2 transformation of the protein’s area ratios. For all quantitative IPA analysis, we used datasets identified to represent less than threefold differentially abundant expressed proteins across all analyzed samples. These molecules were overlaid onto a global molecular network contained in the Ingenuity Knowledge Base. The networks were then algorithmically generated based on their connectivity index using the built-in IPA algorithm. The probability of having a relationship between each IPA indexed biological function and the experimentally determined genes was calculated by a right-tailed Fisher’s exact test. The level of significance was set to P < 0.05. Accordingly, the IPA analysis identified the molecular and cellular pathways from the IPA library of canonical pathways that were most significant to the dataset (−log (P value) > 2.0). For the quantitative analysis of the expression profiles, IPA assigned the z-score function to all eligible canonical and cellular pathways (where z < −2 represents significant down-regulation while z > 2.0 represents a significant up-regulation of the selected pathways). The specific melanoma-associated proteins were assigned to each proteome dataset using the cancer-related database reported in the Human Protein Atlas (http://www.proteinatlas.org/cancer) and are presented in Table S1.
miRNA analysis
35 µl each of EV and non-EV fractions from lymphatic exudate and plasma were analyzed for their miRNA content with the FirePlex miRNA Assay from Biofluids (Abcam), which allows the detection of up to 68 miRNAs in customized panel using a low sample volume. Samples were shipped to Abcam and profiled (detailed protocol published in Royo et al., 2016). Results were analyzed using the FirePlex Analysis Workbench that allowed geNorm normalization of the samples. Data were presented as log2 transformed after the addition of one pseudoread.
Cell lines
B16-F10 melanoma cells (American Type Culture Collection) were maintained in high-glucose DMEM (10569044; Invitrogen) supplemented with 10% heat-inactivated FBS (A3840002; Invitrogen) and used for in vitro EV production in DMEM 2% heat-inactivated, exosome-depleted FBS (A2720801; Invitrogen). B16-F10 cells were transfected with a Vegfc lentivirus (B16-F10/VEGF-C) and used to induce lymphangiogenic tumors in mice as described previously (Fankhauser et al., 2017).
Mice and tumor inoculation
K14-VEGFR3-Ig mice previously described (Mäkinen et al., 2001) were maintained on a C57Bl/6 background and crossbred with WT C57Bl/6 mice (The Jackson Laboratory). 10–16-wk-old K14-VEGFR3-Ig mice and WT littermates of both sexes were used. For tumor distribution experiments, 250,000 B16-F10/VEGF-C cells in 30 µl of PBS were inoculated intradermally into the mouse back flanks. Tumor growth was monitored daily with calipers. All experiments were performed with approval from the Veterinary Authority of the Institutional Animal Care and Use Committee at the University of Chicago under protocol no. 72530.
In vivo EV distribution
To assess drainage from healthy skin, 5–10 µg EVs in 10 µl was injected intradermally into each mouse hock (the lateral tarsal region just above the ankle). After 24 h, mice were sacrificed. Organs were homogenized in radioimmunoprecipitation assay buffer (Sigma-Aldrich) and centrifuged at 10,000 g for 10 min, and supernatant fluorescence was read by plate reader. To assess EV distribution from tumors, B16-F10 VEGF-C tumors were inoculated at day 0, 20–40 µg labeled EVs were injected intratumorally in 20 µl at day 11, and mice were sacrificed at day 12 (24 h after labeled EV injection). Tumors and dLNs were digested according to Broggi et al. (2014) and Fankhauser et al. (2017), and cells were stained with CD31-APC (561814; BD Pharmingen), gp38-PE-Cy7 (127412; BioLegend), and CD45-APC-Cy7 (103116; BioLegend), and analyzed by flow cytometry using a BD LSRFortessa. LECs were defined as CD45−CD31+gp38+, blood endothelial cells as CD45−CD31+gp38−, cancer-associated fibroblasts and fibroblastic reticular cells as CD45−CD31−gp38+, and other CD45− as CD45−CD31−gp38−.
EV drainage in ear dermis and whole mount imaging
1 µl of labeled EVs was injected into mouse dorsal ear dermis. After 30 min, the mouse was sacrificed and perfused with Ringer’s solution followed by zinc fixative (4.5 mM CaCl2, 52 mM ZnCl2, 32 mM Zn(CF3COO)2, 2 mM Tris, and 38 mM glycine, pH 6.5, 340 mOsm/liter). Whole mount staining of the ear was performed as described previously (Kilarski et al., 2013). Ears were cut and fixed for 24 h in zinc fixative and 1% Triton X-100. The dorsal skin was separated from the rest of the ear, washed in TBS, and blocked for 1 h in TBS 0.5% casein. The dorsal skin was incubated with anti-VE-cadherin antibody (550548; BD Biosciences) and anti-podoplanin antibody (AF3244; R&D Systems) for 24 h, washed in TBS 0.1% Tween, and incubated with secondary antibodies for 24 h. After washing in TBS Tween 0.1%, the tissue was dehydrated with 70% ethanol and 100% ethanol and cleared and mounted on a glass slide in 2:1 benzyl benzoate/benzyl alcohol solution. Fluorescence images were acquired with an Olympus IX2-DSU fluorescence microscope and a 63× lens. Image stacks were processed with ImageJ (National Institutes of Health).
Immunofluorescence imaging
Quantification of S100 in different regions of primary tumor and staining of formalin-fixed tumors and LNs was performed as described previously (Bordry et al., 2018). Staining of human LECs monolayer after cocultures was done after 15 min of fixation in 2% paraformaldehyde, RT, and stained for CD31-FITC (Ancell; 180–040), S100 (Novacastra; NCL-L-S100p), and Prox-1 (AF2727; R&D Systems). The fluorescent secondary antibodies were provided by Molecular Probes (Invitrogen).
Analysis of TCGA data
TCGA level 3 gene expression data for skin cutaneous primary melanoma tumors and metastatic sites were downloaded from the Broad GDAC Firehose database (http://gdac.broadinstitute.org/) and analyzed as described in Fankhauser et al. (2017). Volcano plots were generated using RStudio (Version 3.3.2).
Statistics
Unless otherwise specified, the following statistical analyses were performed: paired Student’s t test was used to compare lymphatic exudate and plasma content within the same patient, unpaired t test to compare two groups, and one-way ANOVA followed by Tukey’s multiple comparison test when more than two groups were compared. Non-parametric Spearman’s t test was used for correlations. For each patient, data from two time points (day 1 and day 2) were averaged. Statistical significance is shown as *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Online supplemental material
Fig. S1 provides a schematic of how the lymphatic exudate samples were collected. Fig. S2 shows the enrichment of miRNAs metastatic melanoma–associated proteins in the lymphatic exudate. Fig. S3 shows the cellular and molecular networks and listed melanoma-associated proteins that are mostly up- or down-regulated in lymphatic exudate of LANneg and LANpos patients. Fig. S4 shows the gating strategy for flow cytometric analysis of immune cells in lymphatic exudate and plasma. Table S1 provides the clinical details of the analyzed and all the raw data for the protein and miRNA analysis shown.
Supplementary Material
Acknowledgments
The authors are grateful to the patients that participated in this study. They are grateful to Samia Abed-Maillard, Eleftheria Anastasiu, Hélène Maby-El Hajjami, Paula Marcos Mondéjar, and Loredana Leyvraz for clinical sample handling and information; Witold Kilarski, Yassin Ben Saida, Petra Baumgartner, and Anne-Christine Thierry for technical assistance; Tera Lavoie (Advanced Electron Microscopy Facility, University of Chicago) for assistance with TEM sample preparation and imaging; Kari Alitalo (Translational Biology Laboratory, University of Helsinki, Helsinki, Finland) for the K14-VEGFR3-Ig mice; the École Polytechnique Fédérale de Lausanne Flow Cytometry and Bioimaging core facilities; and Douglas Hanahan, Olivier Michielin, Manuel Fankhauser, and Sachiko Hirosue for helpful discussions and advice.
This work was funded by a grant from SwissTransMed (35/2013).
The authors declare no competing financial interests.
Author contributions: M.A.S. Broggi and M.A. Swartz conceived the ideas, designed the experiments, directed the study, and analyzed the data. M.A.S. Broggi performed the plasma and lymphatic exudate analysis, isolated the EVs, and analyzed the data. L. Maillat and L. Potin performed the EV transport and uptake studies. R. Hamelin ran the proteomics on the samples, while C.C. Clement and L. Santambrogio analyzed the proteomic data. D. Demurtas performed the electron microscopy analysis of lymphatic exudate EVs. M. Matter collected the lymphatic exudate during surgery and, together with E. Romano and D.E. Speiser, wrote the institutional review board protocol. N. Bordry and D.E. Speiser provided clinical vaccine data and S100 staining of primary tumor and LNs. P. Corthésy and L. Maillat helped in analysis and EV extraction of lymph. A. Auger and A. Harari performed flow cytometry analysis. M.A.S. Broggi, L. Maillat, C.C. Clement, L. Santambrogio, and M.A. Swartz wrote and edited the paper. All authors have reviewed and approved the manuscript.
References
- Alegre E., Zubiri L., Perez-Gracia J.L., González-Cao M., Soria L., Martín-Algarra S., and González A.. 2016. Circulating melanoma exosomes as diagnostic and prognosis biomarkers. Clin. Chim. Acta. 454:28–32. 10.1016/j.cca.2015.12.031 [DOI] [PubMed] [Google Scholar]
- Alitalo K. 2011. The lymphatic vasculature in disease. Nat. Med. 17:1371–1380. 10.1038/nm.2545 [DOI] [PubMed] [Google Scholar]
- Blank C.U., Haanen J.B., Ribas A., and Schumacher T.N.. 2016. The “cancer immunogram”. Science. 352:658–660. 10.1126/science.aaf2834 [DOI] [PubMed] [Google Scholar]
- Bordry N., Broggi M.A.S., de Jonge K., Schaeuble K., Gannon P.O., Foukas P.G., Danenberg E., Romano E., Baumgaertner P., Fankhauser M., et al. 2018. Lymphatic vessel density is associated with CD8+ T cell infiltration and immunosuppressive factors in human melanoma. OncoImmunology. 7:e1462878 10.1080/2162402X.2018.1462878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozza W.P., Zhang Y., Hallet K., Rivera Rosado L.A., Hallet K., and Zhang B.. 2015. RhoGDI deficiency induces constitutive activation of Rho GTPases and COX-2 pathways in association with breast cancer progression. Oncotarget. 6:32723–32736. 10.18632/oncotarget.5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnick A.R., Weber D.J., and Zimmer D.B.. 2015. S100 proteins in cancer. Nat. Rev. Cancer. 15:96–109. 10.1038/nrc3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broggi M.A., Schmaler M., Lagarde N., and Rossi S.W.. 2014. Isolation of murine lymph node stromal cells. J. Vis. Exp. (90):e51803 10.3791/51803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchbinder E.I., and Flaherty K.T.. 2016. Biomarkers in melanoma: Lessons from translational medicine. Trends Cancer. 2:305–312. 10.1016/j.trecan.2016.05.003 [DOI] [PubMed] [Google Scholar]
- Clement C.C., and Santambrogio L.. 2013. The lymph self-antigen repertoire. Front. Immunol. 4:424 10.3389/fimmu.2013.00424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement C.C., Cannizzo E.S., Nastke M.D., Sahu R., Olszewski W., Miller N.E., Stern L.J., and Santambrogio L.. 2010. An expanded self-antigen peptidome is carried by the human lymph as compared to the plasma. PLoS One. 5:e9863 10.1371/journal.pone.0009863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement C.C., Aphkhazava D., Nieves E., Callaway M., Olszewski W., Rotzschke O., and Santambrogio L.. 2013. Protein expression profiles of human lymph and plasma mapped by 2D-DIGE and 1D SDS-PAGE coupled with nanoLC-ESI-MS/MS bottom-up proteomics. J. Proteomics. 78:172–187. 10.1016/j.jprot.2012.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement C.C., Becerra A., Yin L., Zolla V., Huang L., Merlin S., Follenzi A., Shaffer S.A., Stern L.J., and Santambrogio L.. 2016. The dendritic cell major histocompatibility complex II (MHC II) peptidome derives from a variety of processing pathways and includes peptides with a broad spectrum of HLA-DM sensitivity. J. Biol. Chem. 291:5576–5595. 10.1074/jbc.M115.655738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colotta F., Allavena P., Sica A., Garlanda C., and Mantovani A.. 2009. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 30:1073–1081. 10.1093/carcin/bgp127 [DOI] [PubMed] [Google Scholar]
- Costa-Silva B., Aiello N.M., Ocean A.J., Singh S., Zhang H., Thakur B.K., Becker A., Hoshino A., Mark M.T., Molina H., et al. 2015. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 17:816–826. 10.1038/ncb3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy M.J. 1992. The role of proteolytic enzymes in cancer invasion and metastasis. Clin. Exp. Metastasis. 10:145–155. 10.1007/BF00132746 [DOI] [PubMed] [Google Scholar]
- Dzieciatkowska M., D’Alessandro A., Moore E.E., Wohlauer M., Banerjee A., Silliman C.C., and Hansen K.C.. 2014. Lymph is not a plasma ultrafiltrate: a proteomic analysis of injured patients. Shock. 42:485–498. 10.1097/SHK.0000000000000249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egeblad M., and Werb Z.. 2002. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer. 2:161–174. 10.1038/nrc745 [DOI] [PubMed] [Google Scholar]
- Fankhauser M., Broggi M.A.S., Potin L., Bordry N., Jeanbart L., Lund A.W., Da Costa E., Hauert S., Rincon-Restrepo M., Tremblay C., et al. 2017. Tumor lymphangiogenesis promotes T cell infiltration and potentiates immunotherapy in melanoma. Sci. Transl. Med. 9:eaal4712 10.1126/scitranslmed.aal4712 [DOI] [PubMed] [Google Scholar]
- Faries M.B., Thompson J.F., Cochran A.J., Andtbacka R.H., Mozzillo N., Zager J.S., Jahkola T., Bowles T.L., Testori A., Beitsch P.D., et al. 2017. Completion dissection or observation for sentinel-node metastasis in melanoma. N. Engl. J. Med. 376:2211–2222. 10.1056/NEJMoa1613210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Silva S., Benito-Martin A., Sanchez-Redondo S., Hernández-Barranco A., Ximénez-Embún P., Nogués L., Mazariegos M.S., Brinkmann K., Amor López A., Meyer L., et al. 2019. Use of extracellular vesicles from lymphatic drainage as surrogate markers of melanoma progression and BRAFV600E mutation. J. Exp. Med. 10.1084/jem.20181522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni L., Lugli E., Ji Y., Pos Z., Paulos C.M., Quigley M.F., Almeida J.R., Gostick E., Yu Z., Carpenito C., et al. 2011. A human memory T cell subset with stem cell-like properties. Nat. Med. 17:1290–1297. 10.1038/nm.2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka S., Watanabe A., Sakurai Y., Akashi-Takamura S., Ishibashi S., Miyake K., Shibuya M., Akira S., Aburatani H., and Maru Y.. 2008. The S100A8-serum amyloid A3-TLR4 paracrine cascade establishes a pre-metastatic phase. Nat. Cell Biol. 10:1349–1355. 10.1038/ncb1794 [DOI] [PubMed] [Google Scholar]
- Hirosue S., Vokali E., Raghavan V.R., Rincon-Restrepo M., Lund A.W., Corthésy-Henrioud P., Capotosti F., Halin Winter C., Hugues S., and Swartz M.A.. 2014. Steady-state antigen scavenging, cross-presentation, and CD8+ T cell priming: a new role for lymphatic endothelial cells. J. Immunol. 192:5002–5011. 10.4049/jimmunol.1302492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood J.L., San R.S., and Wickline S.A.. 2011. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 71:3792–3801. 10.1158/0008-5472.CAN-10-4455 [DOI] [PubMed] [Google Scholar]
- Hoshino A., Costa-Silva B., Shen T.-L., Rodrigues G., Hashimoto A., Tesic Mark M., Molina H., Kohsaka S., Di Giannatale A., Ceder S., et al. 2015. Tumour exosome integrins determine organotropic metastasis. Nature. 527:329–335. 10.1038/nature15756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao K.-C., Shih N.-Y., Fang H.-L., Huang T.-S., Kuo C.-C., Chu P.-Y., Hung Y.-M., Chou S.-W., Yang Y.-Y., Chang G.-C., and Liu K.-J.. 2013. Surface α-enolase promotes extracellular matrix degradation and tumor metastasis and represents a new therapeutic target. PLoS One. 8:e69354 10.1371/journal.pone.0069354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.K., and Hoon D.S.B.. 2016. Liquid biopsy utility for the surveillance of cutaneous malignant melanoma patients. Mol. Oncol. 10:450–463. 10.1016/j.molonc.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W.G., Sanders A.J., Katoh M., Ungefroren H., Gieseler F., Prince M., Thompson S.K., Zollo M., Spano D., Dhawan P., et al. 2015. Tissue invasion and metastasis: Molecular, biological and clinical perspectives. Semin. Cancer Biol. 35(Suppl):S244–S275. 10.1016/j.semcancer.2015.03.008 [DOI] [PubMed] [Google Scholar]
- Kahlert C., and Kalluri R.. 2013. Exosomes in tumor microenvironment influence cancer progression and metastasis. J. Mol. Med. (Berl.). 91:431–437. 10.1007/s00109-013-1020-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilarski W.W., Güç E., Teo J.C.M., Oliver S.R., Lund A.W., and Swartz M.A.. 2013. Intravital immunofluorescence for visualizing the microcirculatory and immune microenvironments in the mouse ear dermis. PLoS One. 8:e57135 10.1371/journal.pone.0057135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim O.-H., Kang G.-H., Noh H., Cha J.-Y., Lee H.-J., Yoon J.-H., Mamura M., Nam J.-S., Lee D.H., Kim Y.A., et al. 2013. Proangiogenic TIE2(+)/CD31 (+) macrophages are the predominant population of tumor-associated macrophages infiltrating metastatic lymph nodes. Mol. Cells. 36:432–438. 10.1007/s10059-013-0194-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leak L.V., Liotta L.A., Krutzsch H., Jones M., Fusaro V.A., Ross S.J., Zhao Y., and Petricoin E.F. III. 2004. Proteomic analysis of lymph. Proteomics. 4:753–765. 10.1002/pmic.200300573 [DOI] [PubMed] [Google Scholar]
- Liu Y., and Cao X.. 2016. Characteristics and Significance of the Pre-metastatic Niche. Cancer Cell. 30:668–681. 10.1016/j.ccell.2016.09.011 [DOI] [PubMed] [Google Scholar]
- Lunavat T.R., Cheng L., Einarsdottir B.O., Olofsson Bagge R., Veppil Muralidharan S., Sharples R.A., Lässer C., Gho Y.S., Hill A.F., Nilsson J.A., and Lötvall J.. 2017. BRAFV600 inhibition alters the microRNA cargo in the vesicular secretome of malignant melanoma cells. Proc. Natl. Acad. Sci. USA. 114:E5930–E5939. 10.1073/pnas.1705206114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund A.W., Wagner M., Fankhauser M., Steinskog E.S., Broggi M.A., Spranger S., Gajewski T.F., Alitalo K., Eikesdal H.P., Wiig H., and Swartz M.A.. 2016. Lymphatic vessels regulate immune microenvironments in human and murine melanoma. J. Clin. Invest. 126:3389–3402. 10.1172/JCI79434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkinen T., Jussila L., Veikkola T., Karpanen T., Kettunen M.I., Pulkkanen K.J., Kauppinen R., Jackson D.G., Kubo H., Nishikawa S., et al. 2001. Inhibition of lymphangiogenesis with resulting lymphedema in transgenic mice expressing soluble VEGF receptor-3. Nat. Med. 7:199–205. 10.1038/84651 [DOI] [PubMed] [Google Scholar]
- Mandal R., and Chan T.A.. 2016. Personalized oncology meets immunology: The path toward precision immunotherapy. Cancer Discov. 6:703–713. 10.1158/2159-8290.CD-16-0146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii A.I., Keller A., Kolker E., and Aebersold R.. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75:4646–4658. 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- Olson O.C., and Joyce J.A.. 2015. Cysteine cathepsin proteases: regulators of cancer progression and therapeutic response. Nat. Rev. Cancer. 15:712–729. 10.1038/nrc4027 [DOI] [PubMed] [Google Scholar]
- Peinado H., Alečković M., Lavotshkin S., Matei I., Costa-Silva B., Moreno-Bueno G., Hergueta-Redondo M., Williams C., García-Santos G., Ghajar C., et al. 2012. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 18:883–891. 10.1038/nm.2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H., Zhang H., Matei I.R., Costa-Silva B., Hoshino A., Rodrigues G., Psaila B., Kaplan R.N., Bromberg J.F., Kang Y., et al. 2017. Pre-metastatic niches: organ-specific homes for metastases. Nat. Rev. Cancer. 17:302–317. 10.1038/nrc.2017.6 [DOI] [PubMed] [Google Scholar]
- Pucci F., Garris C., Lai C.P., Newton A., Pfirschke C., Engblom C., Alvarez D., Sprachman M., Evavold C., Magnuson A., et al. 2016. SCS macrophages suppress melanoma by restricting tumor-derived vesicle-B cell interactions. Science. 352:242–246. 10.1126/science.aaf1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Mann M., and Ishihama Y.. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2:1896–1906. 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- Royo F., Diwan I., Tackett M.R., Zuñiga P., Sanchez-Mosquera P., Loizaga-Iriarte A., Ugalde-Olano A., Lacasa I., Perez A., Unda M., et al. 2016. Comparative miRNA analysis of urine extracellular vesicles isolated through five different methods. Cancers (Basel). 8:112 10.3390/cancers8120112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski P., Nowecki Z.I., van Akkooi A.C.J., Kulik J., Wanda M., Siedlecki J.A., Eggermont A.M.M., and Ruka W.. 2010. Multimarker reverse transcriptase-polymerase chain reaction assay in lymphatic drainage and sentinel node tumor burden. Ann. Surg. Oncol. 17:3314–3323. 10.1245/s10434-010-1142-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S., Vannberg F.O., and Dixon J.B.. 2016. Lymphatic transport of exosomes as a rapid route of information dissemination to the lymph node. Sci. Rep. 6:24436 10.1038/srep24436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz M.A., and Lund A.W.. 2012. Lymphatic and interstitial flow in the tumour microenvironment: linking mechanobiology with immunity. Nat. Rev. Cancer. 12:210–219. 10.1038/nrc3186 [DOI] [PubMed] [Google Scholar]
- Tamburini B.A., Burchill M.A., and Kedl R.M.. 2014. Antigen capture and archiving by lymphatic endothelial cells following vaccination or viral infection. Nat. Commun. 5:3989 10.1038/ncomms4989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C., Zitvogel L., and Amigorena S.. 2002. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2:569–579. 10.1038/nri855 [DOI] [PubMed] [Google Scholar]
- Trevaskis N.L., Kaminskas L.M., and Porter C.J.. 2015. From sewer to saviour - targeting the lymphatic system to promote drug exposure and activity. Nat. Rev. Drug Discov. 14:781–803. 10.1038/nrd4608 [DOI] [PubMed] [Google Scholar]
- Triacca V., Güç E., Kilarski W.W., Pisano M., and Swartz M.A.. 2017. Transcellular pathways in lymphatic endothelial cells regulate changes in solute transport by fluid stress. Circ. Res. 120:1440–1452. 10.1161/CIRCRESAHA.116.309828 [DOI] [PubMed] [Google Scholar]
- Wan J.C.M., Massie C., Garcia-Corbacho J., Mouliere F., Brenton J.D., Caldas C., Pacey S., Baird R., and Rosenfeld N.. 2017. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer. 17:223–238. 10.1038/nrc.2017.7 [DOI] [PubMed] [Google Scholar]
- Wilson T.J., Nannuru K.C., Futakuchi M., and Singh R.K.. 2010. Cathepsin G-mediated enhanced TGF-beta signaling promotes angiogenesis via upregulation of VEGF and MCP-1. Cancer Lett. 288:162–169. 10.1016/j.canlet.2009.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtukiewicz M.Z., Hempel D., Sierko E., Tucker S.C., and Honn K.V.. 2015. Protease-activated receptors (PARs)--biology and role in cancer invasion and metastasis. Cancer Metastasis Rev. 34:775–796. 10.1007/s10555-015-9599-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., and Chen L.. 2014. CD137 as a biomarker for tumor-reactive T cells: finding gold in the desert. Clin. Cancer Res. 20:3–5. 10.1158/1078-0432.CCR-13-2573 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.