Abstract
O6-Alkylguanine DNA-alkyltransferase (AGT), a DNA repair protein, can form crosslinks with DNA. The AGT-DNA crosslinks are known to be mutagenic when AGT is heterologously expressed in Escherichia coli, as well as in mammalian cells. To understand the biological consequences, reliable access to AGT-oligonucleotide crosslinks is needed. This unit describes the synthesis and characterization of site-specific AGT-oligonucleotide crosslinks at the N2-position of deoxyguanosine and N6-position of deoxyadenosine. We developed a post-oligomerization strategy for the synthesis of propargyl-modified oligonucleotides. Copper-catalyzed azide-alkyne cycloaddition was used as a key step to obtain the iodoacetamide-linked oligonucleotides, which serve as good electrophiles for the crosslinking reaction with cysteine-145 of the active site of AGT. Trypsinization of AGT and hydrolysis of oligonucleotides, combined with analysis by liquid chromatography-tandem mass spectrometry, was utilized to confirm the nucleobase-adducted peptides. This method provides a useful strategy for the synthesis and characterization of site-specific DPCs, which can be further used to understand proteolytic degradation coupled DNA repair mechanisms.
Keywords: DNA-protein crosslinks, O6-alkylguanine DNA-alkyltransferase, oligonucleotides, post-oligomerization, copper-catalyzed azide-alkyne cycloaddition (CuAAC), proteomics
Introduction
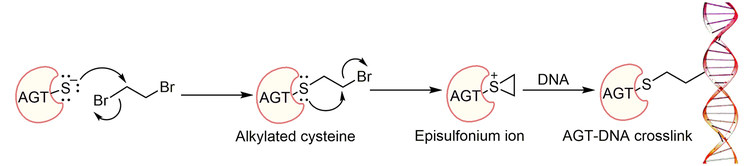
DNA-protein crosslinks (DPCs) are an understudied type of DNA damage formed when a protein becomes covalently bound to DNA. DPCs can be generated in the presence of exogenous and endogenous carcinogenic agents, e.g. ionizing radiation, UV light, bifunctional alkylating agents, aldehydes, and anticancer drugs (Ide et al., 2015). The bulkiness of the DPC adducts can contribute to cell lethality, replication stress and genomic instability by disturbing physiological processes such as replication, transcription, and DNA repair (Barker et al., 2005). DPC repair and its biological consequences are not completely understood, due in part to the lack of efficient synthetic methodologies to obtain site-specific DPCs for further studies. O6-Alkylguanine DNA-alkyltransferase (AGT, MGMT) can crosslink to DNA in the presence of bis-electrophile 1,2-dibromoethane (DBE, or ethylene dibromide, a potent carcinogen), as depicted in Figure 1 (Liu et al., 2002). The AGT active site Cys-145 reacts with DBE and forms a half-mustard, which subsequently forms an episulfonium ion. The AGT, bearing the Cys-145 episulfonium ion, is highly unstable and readily reacts with various nucleophiles including those in DNA (Figure 1). AGT-DNA crosslinks are known to be cytotoxic and mutagenic (Kalapila et al., 2010; Liu et al., 2002). Nuclear proteases such as SPRTN and its yeast homolog Wss1 can degrade the DNA-protein crosslinks (larger than 16 KDa) into small DNA-peptide crosslinks, which can be further processed by a DNA repair pathway such as nucleotide excision repair or bypassed by translesion synthesis (TLS) DNA polymerases (Barker et al., 2005; Fielden et al., 2018; Vaz et al., 2017). Nucleotide excision repair, coupled with proteolytic degradation, can play an important role in DPC repair. The bypass of proteolytically degraded DNA-peptide crosslinks can be mutagenic and, in order to understand the biological consequences, site-specific crosslink substrates are needed. Toward this end, we synthesized site-specific oligonucleotide-AGT crosslinked structural analogues utilizing a copper-catalyzed azide-alkyne cycloaddition (CuAAC, click chemistry) approach. We explored the CuAAC reaction for the synthesis of iodoacetamide-linked oligonucleotides, which serve as good substrates for reaction with Cys-145 of the active site of AGT. This unit outlines detailed protocols for the synthesis of structural analogues of AGT-oligonucleotide crosslinks. Basic Protocol 1 describes the synthesis of an AGT-oligonucleotide crosslink at the N2-position of deoxyguanosine (dG). Basic Protocol 2 highlights the details of synthesis of an AGT crosslink at the N6-position of deoxyadenosine (dA). Basic Protocol 3 describes the validation of AGT-DNA crosslinks at the N2-position of dG and N6-position of dA using a proteomic approach.
Figure 1.
Mechanism of DBE-derived AGT-DNA crosslink formation.
This approach can likely be adapted for the synthesis of other crosslinked proteins in which cysteine is responsible for the crosslink formation. Additionally, the methods presented here can be easily modified to accommodate biologically relevant DPCs involving different DNA binding proteins of interest and at various reactive sites of DNA. Overall, the protocol presented here is a valuable tool for the synthesis of site-specific DPCs which can be further used to understand the complex repair mechanisms and TLS pathway.
BASIC PROTOCOL 1
Synthesis of a Site-Specific AGT- N2-Deoxyguanosine Oligonucleotide Crosslink
This section describes the synthesis of an AGT-oligonucleotide crosslink at the N2-position of dG. It is divided into the two basic procedures: (i) Synthesis, purification, and characterization of an N2-propargyl dG-modified DNA template, and (ii) CuAAC followed by crosslink reaction with AGT.
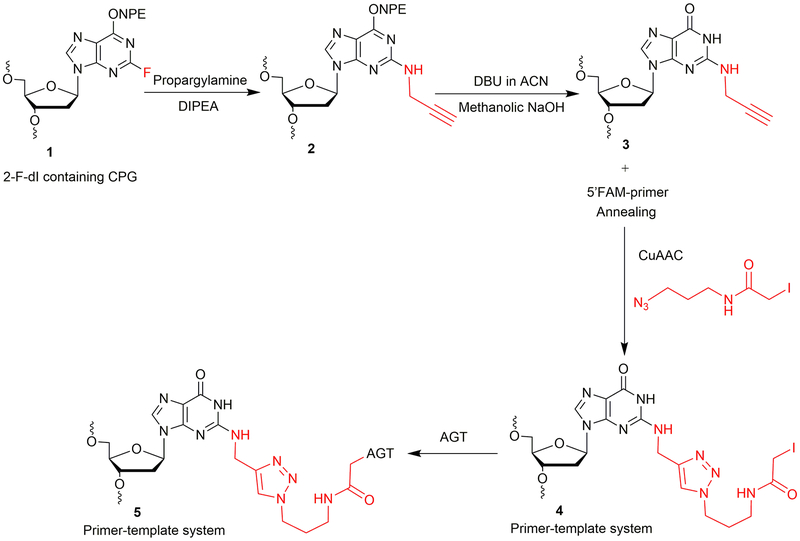
2-Fluoro-2′-deoxyinosine (2-F-dI) phosphoramidite was obtained from Glen Research (Sterling, VA). An oligonucleotide containing a 2-F-dI (1, Figure 2) was synthesized on a PerSeptive Biosystems Model 8909 DNA synthesizer using Ultramild phosphoramidites/solid supports (Glen Research) on a 1-μmol scale, utilizing a standard synthetic protocol (Gruppi et al., 2014). A post-oligomerization approach was developed to achieve the synthesis of the N2-propargyl-dG modified oligonucleotide template, as depicted in Figure 2. The synthesis is carried out at a CPG (controlled pore glass) support level, which involves nucleophilic displacement using propargylamine followed by O6-p-nitrophenylethyl (NPE) deprotection. The final deprotection is carried out to cleave CPG beads and to remove base-protecting groups to obtain oligonucleotide 3 (Figure 2). The N2-propargyl-dG template 3 (Figure 2, Table 1) is annealed with a 6-carboxyfluorescein (FAM)-labeled 12-mer primer. The annealed primer-template system is subjected to CuAAC to obtain the iodoacetamide-linked oligonucleotide system 4 (Figure 2). The crosslinking reaction is then carried out with the AGT protein. This strategy is used to obtain an AGT-crosslinked template-primer system 5 (Figure 2) to perform TLS.
Figure 2.
Synthetic strategy of N2-dG oligonucleotide-AGT crosslink.
Table 1.
N2-dG- and N6-dA-modified oligonucleotide templates used here
| Code | Oligonucleotide sequence (5′ to 3′) | Calculated mass [M−H]− | Observed mass [M−H]− |
|---|---|---|---|
| 3 | TCTCXGTTTATGGACCACC (X = N2-propargyl-dG) | 5791.812 | 5792.600 |
| 7 | TCTCXGTTTATGGACCACC (X = N6-propargyl-dA) | 5775.812 | 5778.000 |
Materials
2-F-dI phosphoramidite (Glen Research)
2-F-dI containing modified DNA CPG column (Gruppi et al., 2014)
Nuclease-free water
Argon gas
Screw cap vial
Dimethyl sulfoxide (DMSO, anhydrous, Sigma-Aldrich)
N,N-Diisopropylethylamine (DIPEA, Sigma-Aldrich)
Propargylamine (Sigma-Aldrich)
Acetonitrile (ACN or CH3CN, anhydrous, Sigma-Aldrich)
Vortex mixing device (Fisher Scientific)
Benchtop centrifuge (VWR)
Heating block set to 55 or 95 °C (VWR)
1,8-Diazabicyclo[5.4.0]undec-7-ene (DBU, Sigma-Aldrich)
Shaker
Methanol, anhydrous (Sigma-Aldrich)
0.4 M Methanolic NaOH
Sonicator (Fisher Scientific)
Centrifugal vacuum evaporator (Speed Vac)
HPLC system
C18 HPLC column (Phenomenex; 150 mm × 10 mm, 5 μm or 150 mm × 4.6 mm, 5 μm)
15- and 50-mL centrifuge tubes
Lyophilizer
TEN (Tris (10 mM)-EDTA (1 mM)-NaCl (300 mM)) buffer, pH 8.0)
C18 Sep-Pak column (Waters)
Nanodrop One Spectrophotometer (Thermo Scientific)
HPLC vials
High resolution mass spectrometer (Waters Synapt, quadrupole/OA-TOF)
Hexafluoroisopropanol (HFIP, Sigma-Aldrich)
Triethylamine (Sigma-Aldrich)
Methanol (CH3OH, HPLC grade)
UPLC column (Acquity UPLC BEH C18,1.7 μm, Waters)
1.5 mL Microcentrifuge tubes (low protein binding, Thermo Scientific)
2-[4-(2-Hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (HEPES) buffer (pH 7.3)
Sodium chloride
FAM-labeled 12-mer primer (Integrated DNA Technology)
Copper(II) sulfate pentahydrate (CuSO4•5H2O, Sigma-Aldrich)
Tris-hydroxypropyltriazolylmethylamine (THPTA, Sigma-Aldrich)
Sodium ascorbate (Acros Organics)
Magnetic beads
Stirrer
Black paper or black box
Sodium acetate (Fisher Biotech)
Ethanol (100%, Fisher Chemical)
Centrifuge (4 °C, Thermo Scientific Sorvall Legend X1)
Amicon centrifugal filters, MWCO 10000, 0.5 mL (Merck Millipore)
Tris-HCl buffer (20 mM, pH 7.6)
2X Laemmli sample buffer (Bio-Rad)
NuPAGE mini gels (12-well, 10% Bis-Tris, Invitrogen)
Gel system (Invitrogen Novex mini cell)
2-(N-Morpholino)ethanesulfonic acid (MES) buffer (20X, NuPAGE, Novex)
Loading tips (Gel round tip, 1–200 μL)
Simply Blue Safe Stain (Coomassie Blue stain, Invitrogen)
−20 and −80 °C freezers
Gel Doc EZ imager (Bio-Rad)
Synthesis of N2-propargyl-dG modified oligonucleotide template 3
1. Synthesize 2-F-dI containing oligonucleotide as per reported protocol (Gruppi et al., 2014).
2. Flush the 2-F-dI oligonucleotide-containing CPG column with argon.
3. Weigh 5 mg of 2-F-dI oligonucleotide CPG support in a screw cap vial.
4. Add 86 μL anhydrous DMSO.
5. Add 10 μL of 500 mM propargylamine (final concentration 50 mM).
6. Add 4.4 μL DIPEA (final concentration 250 mM).
7. Heat at 55 °C.
8. Add 10 μL of 500 mM propargylamine after 7 h and continue the reaction for an additional 15 h (total 22 h).
9. After 22 h, spin down the reaction mixture and discard the supernatant carefully using loading tips.
10. Wash the CPG support with anhydrous DMSO (2 × 1 mL) and with anhydrous CH3CN (3 × 1 mL).
11. Air dry the CPG support for 30 min.
12. NPE deprotection: Treat the CPG support twice with 1 mL of 1 M DBU in CH3CN, one hour each time at room temperature (on shaker).
13. After DBU treatment, carefully remove the supernatant using loading tips.
14. Wash the CPG support with anhydrous CH3OH (2 × 1 mL) and with anhydrous CH3CN (3 × 1 mL).
15. Air dry the CPG support for 1 h.
16. Treat the CPG support with 1 mL of 0.4 M methanolic NaOH at room temperature for overnight (shaking is recommended).
17. After completion of reaction, sonicate the CPG support for 5 min.
18. Collect the supernatant and wash the CPG support with water twice (2 × 200 μL).
19. Neutralize the collected supernatant to pH 7 with glacial acetic acid and evaporate it under vacuum with a centrifugal evaporator.
20. Resuspend the DNA pellet in 500 μL of nuclease-free water.
21. Purify the oligonucleotide by HPLC using a Phenomenex Clarity Oligo-RP (C18) column (150 mm × 10 mm, 5 μm) at room temperature. Buffers consist of Mobile Phase A (0.1 M triethylammonium acetate (TEAA), pH 7) and Mobile Phase B (0.1 M triethylammonium acetate and CH3CN, 1:1, v/v). The UV detector is set at 260 nm for detection of the oligonucleotide. The following gradient program (v/v) is used with a flow rate of 3 mL/min: start at 17% B, continue for 5 min, then increase to 40% B over 20 minutes, then increase to 100% B at 21 min, hold at 100% B for 5 min, and re-equilibrate for 4 min at 17% B (all v/v).
22. The desired oligonucleotide elutes at ~ 18 min.
23. Lyophilize the N2-propargyl-dG oligonucleotide fractions.
24. Desalt the N2-propargyl-dG oligonucleotide using a C18 Sep-Pak column.
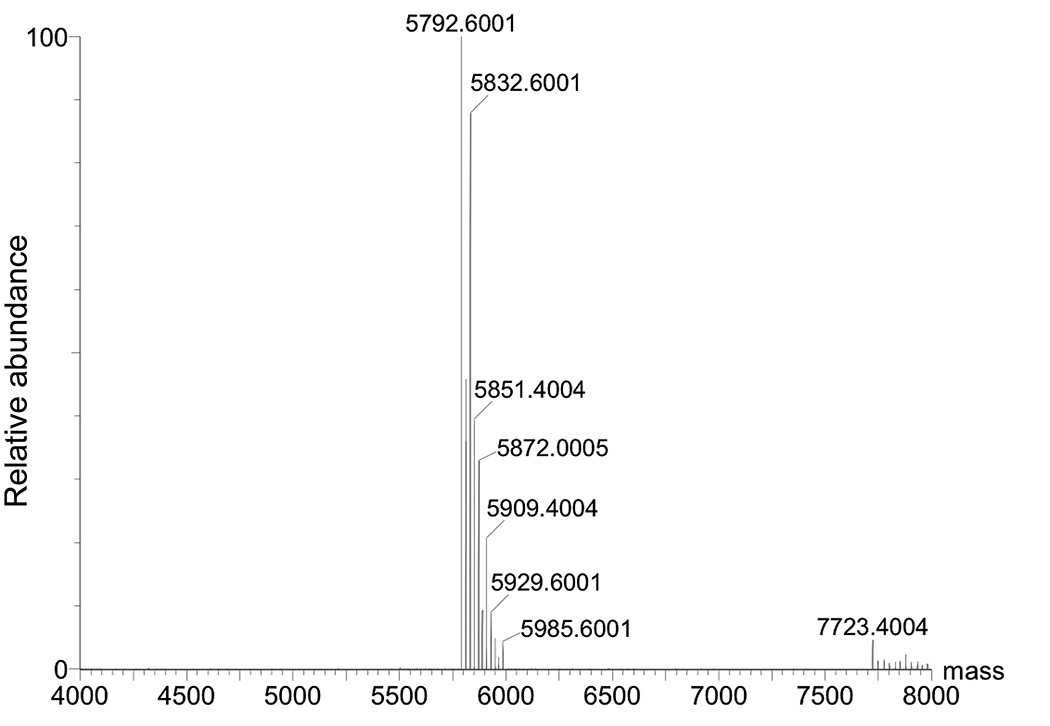
25. Confirm the identity of N2-propargyl-dG oligonucleotide 3 (Figure 2) using negative ion ESI-MS (Figure 3).
Figure 3.
Deconvoluted mass spectrum of N2-propargyl-dG modified oligonucleotide. Expected mass [M−H]− 5791.812, Found [M−H]− 5792.6001.
26. ESI-MS method (negative mode): Waters Synapt; Column: Acquity UPLC BEH C18 1.7 μm 2.1 mm × 100 mm; Solvent A: 200 mM HFIP + 8.1 mM TEA at pH 7.7; Solvent B: CH3OH; Flow: 0.2 mL/min; gradient program (v/v): start at 5% B then increase to 35% B over 10 min; then increase to 80% B over 13 min; re-equilibrate for 4 min; stop time: 17 min; injection volume: 5 μL; sample temperature: 4 °C; column temperature: 40 °C. The deconvoluted mass spectrum of N2-propargyl-dG oligonucleotide 3 is shown in Figure 3.
Note: For NPE deprotection using DBU in ACN (Figure 2, Step 2), a Glen Research Protocol can be used (http://www.glenresearch.com/Technical/TB_2-F-dI.pdf). For post-oligomerization, reaction in multiple batches is recommended.
CuAAC reaction
Perform the CuAAC reaction to produce iodoacetamide linked-oligonucleotide system 4 (Figure 2). An azide linker (N-(3-azidopropyl)-2-iodoacetamide, Figure 2) was prepared according to a reported procedure (Kotoku et al., 2014). The CuAAC reaction was performed according to a reported protocol (Presolski et al., 2011).
27. Anneal the N2-propargyl-dG modified oligonucleotide template 3 (Table 1) with 12-mer FAM-labeled DNA primer.
The N2-propargyl-dG DNA template (3.5 nmol) is dissolved in 20 μL of nuclease-free water.
12-mer FAM labeled DNA primer (3.5 nmol) is dissolved in 20 μL of nuclease-free water. Mix with the template DNA.
Add 5 μL of 500 mM NaCl (to a final concentration of 50 mM).
Add 5 μL 500 mM HEPES buffer (pH 7.3, to a final concentration of 50 mM).
Total 50 μL volume: anneal at 95 °C for 5 min and slowly cool to room temperature (~ 3 h).
28. Add 36 μL of nuclease-free water to the annealed primer-template system (final concentration of template and primer for CuAAC reaction is 35 μM for each).
29. Add 5 μL of 500 mM HEPES buffer (pH 7.3) to the annealed template-primer system.
30. Add 2 μL of azide linker (final concentration 2 mM).
31. Add a premixed solution of CuSO4 (1 μL, final concentration 0.1 mM) and ligand THPTA (1 μL, final concentration 0.5 mM) to the reaction mixture and deaerate it with argon by sparging for 40 seconds.
32. Add 5 μL of 100 mM sodium ascorbate (final concentration 5 mM).
33. Stir the reaction mixture for 4 h (slowest speed) at room temperature in the dark, covering the reaction tube with black paper.
34. After 4 h, precipitate the reaction mixture using 3 M sodium acetate (pH 5.2).
Add 19 μL of 3 M sodium acetate (pH 5.2) and 650 μL of cold ethanol. Mix well.
Store in a −80 °C freezer for overnight.
Centrifuge the sample (21,000 × g for 30 min) and decant the supernatant to obtain an iodoacetamide-linked oligonucleotide pellet.
Wash the iodoacetamide-linked oligonucleotide pellet with 70% cold ethanol (200 μL).
Centrifuge the iodoacetamide-linked oligonucleotide pellet (21,000 × g for 20 min) and remove the ethanol carefully. Air dry the DNA pellet in the dark to obtain the iodoacetamide linked-oligonucleotide system 4 (Figure 2).
Crosslink reaction of iodoacetamide-linked oligonucleotide with AGT
AGT was expressed and purified as described previously (Chowdhury et al., 2013) and used further for the crosslinking reactions.
35. Perform the crosslink reaction to get the AGT-oligonucleotide crosslink complex 5 (Figure 2) as follows.
Buffer exchange the AGT protein (7 nmol) with 20 mM Tris-HCl (pH 7.6, 500 μL) using an Amicon centrifugal filter, MWCO 10000.
Add 30 μL of buffer-exchanged AGT to the iodoacetamide-linked oligonucleotide pellet from CuAAC reaction (from Step 34e).
Mix it briefly and keep it on shaker for 3 h at room temperature (in the dark using a black box).
After completion of reaction, store the reaction mixture at 4 °C.
Analyze the crosslink reaction by separation via 10% Bis-Tris SDS-PAGE gel (12 well, 120 V for 1.5 h) using MES buffer (Figure 4).
Load the samples on SDS-PAGE gel using 2X Laemmli sample buffer.
After electrophoresis, stain the gel using Coomassie Blue.
Wash the gel with water to remove excess stain and image it using Gel Doc EZ imager to analyze the AGT-oligonucleotide crosslink complex 5 (Figure 2).
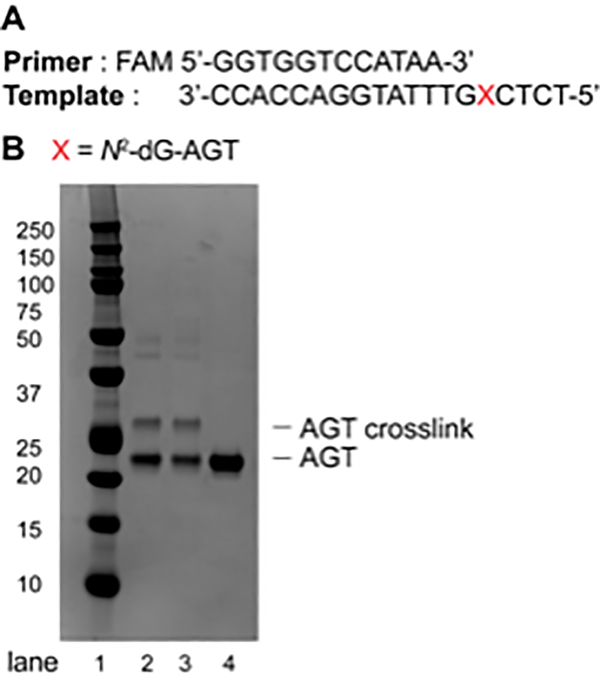
Figure 4.
(A) Sequence of the primer (FAM-labeled, 12-mer) and the template (19-mer); (B) SDS-PAGE (10%, Bis-Tris) of AGT-oligonucleotide crosslink reaction at the N2-position of dG. Lane 1: Precision Plus Protein dual color standard; Lane 2: AGT-oligonucleotide crosslink reaction after 1 h; Lane 3: AGT-oligonucleotide crosslink reaction after 3 h; Lane 4: AGT protein (21.9 KDa).
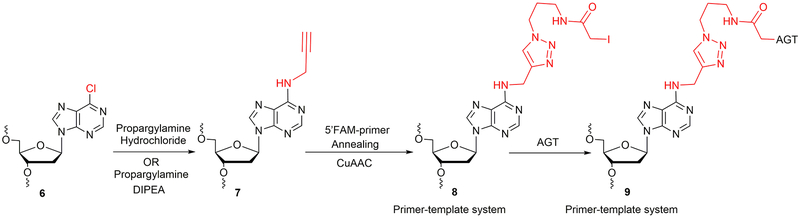
BASIC PROTOCOL 2
Synthesis of Site-Specific AGT-oligonucleotide Crosslink at N6-Deoxyadenosine
This section describes the synthesis of AGT-oligonucleotide crosslink at the N6-position of dA utilizing 6-chloropurine containing oligonucleotide. It is divided into two basic procedures: (i) Synthesis, purification and characterization of N6-propargyl-dA-modified oligonucleotide template 7 (Table 1, Figure 5), and (ii) CuAAC using N6-propargyl-dA and followed by crosslink reaction with AGT (Figure 5). The 6-chloropurine phosphoramidite (Wang et al., 2006) was a generous gift from Prof. Carmelo J. Rizzo (Vanderbilt University, Nashville, TN). The oligonucleotide containing a 6-chloropurine was synthesized on a PerSeptive Biosystems Model 8909 DNA synthesizer using Ultramild phosphoramidites/solid supports (Glen Research) on a 1-μmol scale, utilizing a standard synthetic protocol (Wang et al., 2006). The synthesis of N6-propargyl-dA modified oligonucleotide template was carried out in-solution as well as at the CPG level. For in-solution synthesis, the deprotection, HPLC purification of 6-chloropurine oligonucleotide 6 was carried out using a reported protocol (Wang et al., 2006). Further, the identity of oligonucleotide 6 was confirmed by ESI-MS. The oligonucleotide 6 was subjected to a nucleophilic displacement reaction with propargylamine hydrochloride to obtain N6-propargyl-dA-modified oligonucleotide template 7 (Figure 5).
Figure 5.
Synthetic strategy of N6-dA oligonucleotide-AGT crosslink.
For Materials, see Basic Protocol 1
6-Chloropurine phosphoramidite
6-Chloropurine-containing DNA CPG column (Wang et al., 2006)
Propargylamine hydrochloride (Sigma-Aldrich)
1,4 Dioxane (Sigma-Aldrich)
Synthesis of N6-propargyl-dA modified oligonucleotide template 7: In-solution
1. Synthesize, deprotect and purify the 6-chloropurine-containing oligonucleotide as per reported protocol (Wang et al., 2006).
2. Dissolve 6-chloropurine oligonucleotide pellet (3.5 nmol) in 65 μL of nuclease-free water.
3. Add 10 μL of 500 mM propargylamine hydrochloride (final concentration 50 mM).
4. Add 25 μL 1 M DIPEA in dioxane (250 mM final concentration). Total volume 100 μL.
5. Heat at 55 °C for 22 h in a heating block.
6. Desalt the reaction mixture using a Sep-Pak column and confirm the identity of the N6-propargyl-dA oligonucleotide 7 (Figure 5, Table 1) by negative ion ESI-MS, without further purification (Figure 6).
Figure 6.
Deconvoluted mass spectrum of N6-propargyl-dA-modified oligonucleotide. Expected mass [M−H]− 5775.812, found [M−H]− 5778.0005.
7. The deconvoluted mass spectrum of N6-propargyl-dA oligonucleotide 7 is shown in Figure 6.
CuAAC and crosslink reaction
8. Perform the CuAAC and crosslink reaction as per Basic Protocol 1 to obtain AGT-oligonucleotide crosslink at the N6-position of dA.
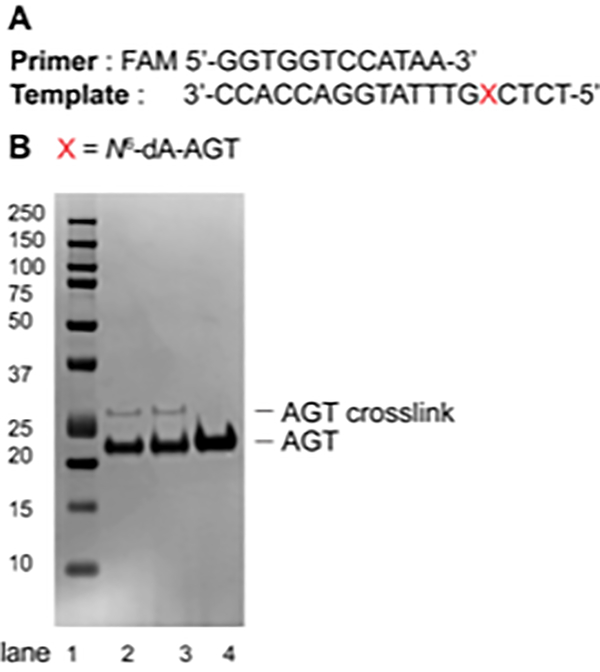
Analyze the crosslink reaction on a 10% Bis-Tris SDS-PAGE gel (12 well) using MES buffer (Figure 7).
Figure 7.
(A) Sequence of the primer (12-mer) and the template (19-mer); (B) SDS-PAGE (10%, Bis-Tris) of AGT-cross-link reaction at the N6-position of dA. Lane 1: Protein Kaleidoscope standard; Lane 2: AGT-oligonucleotide crosslink reaction after 1 h; Lane 3: AGT-oligonucleotide crosslink reaction after 3 h; Lane 4: AGT protein (21.9 KDa).
ALTERNATE PROTOCOL 2
Synthesis of N6-propargyl-dA modified oligonucleotide template 7: CPG support level
Post-oligomerization can be done in solution or at the CPG bead level (Wickramaratne et al., 2015). For the synthesis of N6-propargyl-dA template, post-oligomerization reaction was carried out in solution as mentioned in Basic Protocol 2. To avoid oligonucleotide degradation and to improve the yield of modified oligonucleotide 7 by reducing the number of HPLC purifications, we also focused on the post-oligomerization reaction at the CPG bead level. In this Alternate Protocol 2, we have discussed only the synthesis of N6-propargyl-dA oligonucleotide 7 (Figure 5) at the CPG support level and its HPLC purification. Further, the N6-propargyl-dA oligonucleotide 7 (Figure 5) was also explored to obtain AGT-oligonucleotide crosslink.
1. Flush the 6-chloropurine oligonucleotide containing CPG column with argon.
2. Weigh 5 mg of 6-chloropurine oligonucleotide CPG into a screw cap vial.
3. Perform the nucleophilic displacement reaction using propargylamine at CPG level as described in steps 4 to 11 of Basic Protocol 1.
4. Cleave the oligo from CPG support as described in steps 16 to 20 of Basic Protocol 1.
5. Purify, desalt, and characterize the N6-propargyl-dA oligonucleotide 7 (Figure 5, Table 1) using HPLC and ESI-MS as described in steps 20 to 25 of Basic Protocol 1.
6. The N6-propargyl-dA oligonucleotide 7 (Figure 5) is eluted at ~ 18.3 min.
7. Follow the Basic Protocol 1 to obtain AGT-oligonucleotide crosslink.
BASIC PROTOCOL 3
Proteomic analysis: Trypsin digestion, oligonucleotide hydrolysis, and LC-MS/MS analysis of AGT-DNA crosslinks
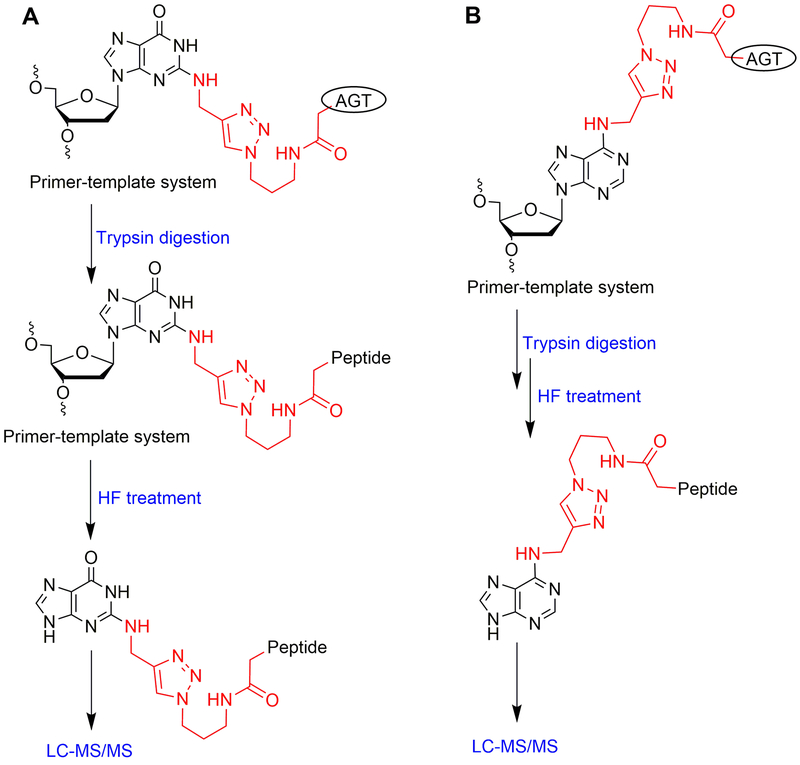
The following is the standard protocol that is used for the detection of the crosslink at Cys-145 of AGT using a proteomic approach. This section describes the detailed identification (proteomic analysis) of an AGT-oligonucleotide crosslink at the N2 position of dG and N6 position of dA. In the method presented here, the AGT protein bearing crosslinks was digested with trypsin (Shevchenko et al., 2007) and further oligonucleotide hydrolysis was carried out with 48% hydrofluoric acid (Geyer et al., 2004). The detailed flow chart for the proteomic analysis of N2-dG and N6-dA oligonucleotide-AGT crosslinks is outlined in Figure 8.
Figure 8.
Flow chart of proteomics sample preparation: (A) for N2-dG oligonucleotide-AGT crosslink and (B) for N6-dA oligonucleotide-AGT crosslink.
Note: For iron-IMAC purification please refer to the Sigma protocol provided with SigmaPrep Spin columns.
Materials
Nuclease-free water
Acetonitrile (ACN or CH3CN, anhydrous, Sigma-Aldrich)
Vortex mixing device (Fisher Scientific)
Benchtop centrifuge (VWR)
Heating block set to 37 °C (VWR)
Shaker
Methanol (CH3OH, anhydrous, Sigma-Aldrich)
Centrifugal vacuum evaporator (Speed Vac)
Microcentrifuge tubes (1.5 mL, low protein binding, Thermo Scientific)
NuPAGE mini gels (10-well, 10% Bis-Tris, Invitrogen)
Gel system (Invitrogen Novex mini cell)
2-(N-Morpholino)ethanesulfonic acid (MES) buffer (20X, NuPAGE, Novex)
Loading tips (Gel round tip, 1–200 μL)
Simply Blue Safe Stain (Coomassie Blue stain, Invitrogen)
−20 and −80 °C freezers
4X LDS sample buffer (NuPAGE, Invitrogen)
Razor blades
Dithiothreitol (DTT, Research Products International)
Urea (Sigma-Aldrich)
2-Iodoacetamide (Sigma-Aldrich)
Trypsin (sequencing grade, Promega)
Water bath (37 °C)
Ammonium bicarbonate (Fluka Analytical)
PHOS-select iron-affinity slurry (Fe-IMAC slurry, Sigma-Aldrich)
SigmaPrep Spin columns (Sigma-Aldrich)
Centrifuge (IEC micromax)
End-over-end rotator
Glacial acetic acid (Fisher)
Ammonium hydroxide solution (30–33% w/v, Sigma-Aldrich)
Formic acid (HCO2H, LC-MS grade, Thermo Scientific)
Hydrofluoric acid (HF, 48%, Sigma-Aldrich)
Nitrogen gas
In-solution digestion of AGT-oligonucleotide crosslinks
Note: This is a common method for both N2-dG and N6-dA oligonucleotide-AGT crosslinks.
1. Reduction (of disulfide bridges)
To 10 μL (50 μg of protein) of crosslink reaction mixture (in 20 mM Tris-HCl, pH 7.6; in low binding tube), add 30 μL of 8 M urea (final concentration 6 M) and 1 μL of 500 mM DTT (final concentration 10 mM).
Heat at 37 °C for 30 min (in heating block).
2. Alkylation (of free thiol groups of cysteines)
Add 5 μL of 250 mM iodoacetamide to the above tube (final concentration 25 mM).
Shake for 30 min in the dark at room temperature.
3. Quenching of remaining iodoacetamide
Add 4 μL of 500 mM DTT to the above tube.
Heat at 37 °C for 10 min (in heating block).
4. Total reaction volume from steps 1 to 3 is 50 μL; add 450 μL of 20 mM Tris-HCl (pH 7.6).
5. Trypsin digestion: Add trypsin (50:1 protein:trypsin, w/w) and incubate in water bath at 37 °C overnight or for 20 h.
6. After complete digestion, add 10 μL of glacial acetic acid to adjust the pH to 2 – 3.
7. Purify the digested peptides using an Fe-IMAC column:
Carefully mix the iron affinity slurry and add 80 μL of the slurry into a clean SigmaPrep spin column using a wide orifice pipette tip.
Add 500 μL of equilibration solution to the spin column, vortex, and centrifuge it for 30 s at 8,200 × g.
Discard the flow-through liquid and repeat the above step b twice.
Place an end cap onto the equilibrated spin column outlet and add 500 μL of the digested solution (from step 6). Incubate for 30 min with mixing (end-over-end rotation). After incubation, remove the end cap and centrifuge as described in step b.
Wash the slurry with 500 μL of equilibration solution, vortex, and centrifuge as described in step b.
Wash the slurry with 500 μL of water and centrifuge as described in step b.
Elute the digested DNA-peptide crosslink: place an end cap onto the column outlet and add 250 to 500 μL of elution solution. Incubate for ~ 5 min with mixing (end-over-end rotation).
After incubation, remove the end cap and centrifuge as described in step b and evaporate the eluent under vacuum with a centrifugal evaporator.
8. Perform DNA hydrolysis:
Treat the dry sample (from step h) with 50 μL of 48% HF at 4 °C for 14 h to hydrolyze the DNA.
Remove the HF under a stream of nitrogen.
Dissolve the resulting product in 50 μL of anhydrous CH3OH and dry under a stream of nitrogen.
Repeat above step c.
Dissolve the sample in 20 μL of 0.1% formic acid (v/v) and shake for 10 min.
Centrifuge at 21,000 × g for 5 min.
Analyze the sample using LC-MS/MS.
In-gel digestion of AGT-oligonucleotide crosslinks
Note: This is a common method for both the N2-dG and N6-dA oligonucleotide-AGT crosslinks.
9. Reduction (of disulfide bridges)
Aliquot 10 μL of crosslink reaction mixture (in 20 mM Tris-HCl, pH 7.6; in low binding tube), add 1 μL of 100 mM DTT (final concentration 10 mM) and 1 μL of 10% SDS (w/v).
Heat at 37 °C for 30 min (in heating block).
10. Alkylation (of free thiol groups of cysteines)
Add 2 μL of 250 mM iodoacetamide to the above tube (final concentration 25 mM).
Shake for 30 min in the dark at room temperature.
11. Quenching of residual iodoacetamide
Add 2 μL of 100 mM DTT to the above tube.
Heat at 37 °C for 10 min (in heating block).
12. Total reaction volume from steps 1 to 3 is 16 μL; add 6 μL of 4X LDS loading dye and (without heating) load on the SDS gel (10% Bis-tris, 10-well); run the gel for 1.5 h in MES buffer and at 120 V.
13. Stain gel with Coomassie blue stain (Simply Blue Safe Stain).
14. Wash the gel with water to remove excess stain.
15. Carefully excise the band corresponding to the crosslink from the gel (Figures 4 and 7, respectively) using a razor blade and collect into a low-binding tube (1 mm3 gel pieces; avoid keratin contamination by using disposable gloves).
16. Add 1 mL of destaining solution to the gel pieces and shake at room temperature for 30 min.
17. After 30 min, carefully remove the destaining solution using loading tips.
18. Dehydrate the gel pieces with CH3CN (3 × 1 mL). After dehydration with CH3CN, centrifuge the mixture and remove the traces of CH3CN using loading tips. The gel should be completely dehydrated.
19. Add freshly prepared trypsin solution (10 ng/μL, 10 mM ammonium bicarbonate) to completely cover the rehydrated gel pieces and incubate in water bath overnight at 37 °C.
20. After overnight incubation, add extraction solution (5% formic acid in 2:1 CH3CN:H2O) in a 1:1 ratio (v/v) to the previously added trypsin solution (step 19).
21. Shake for 15 min.
22. Remove the entire volume (using loading tips; try to avoid gel pieces) into a new tube.
23. Add the same volume of extraction solution to the gel pieces for a second extraction and shake for 15 min.
24. Remove all volume and pool both extractions together (from step 22).
25. Evaporate it under vacuum with a centrifugal evaporator (heating on).
26. Add 50 μL of 48% HF to the dry sample (from step 25) and incubate at 4 °C for 14 h.
27. Remove the HF under a stream of nitrogen.
28. Dissolve the resulting product in 50 μL of anhydrous CH3OH and dry the sample under a stream of nitrogen.
29. Repeat step 28.
30. Dissolve the dry sample in 0.1% aq. formic acid (20 μL) and centrifuge at 21,000 × g for 5 min (~ 10 pmol/μL)
31. Analyze using LC-MS/MS.
Proteomics methods: LC-MS/MS analysis
32. Analyze the nucleobase adducted-peptides using following LC-MS/MS parameters:
NanoLC Ultra system (Eksigent Technologies, Dublin, CA) interfaced with an LTQ Orbitrap XL mass spectrometer (Thermo Scientific, San Jose, CA).
Column: Use a 360 μm outer diameter/100 μm inner diameter microcapillary analytical column packed with Jupiter octadecylsilane (C18) (3 μm, 300 AÅ, Phenomenex) and equipped with an integrated electrospray emitter tip.
Sample loading: load approximately 10 pmol of nucleobase-adducted peptides (1.5 μL min−1).
Peptide separation conditions: linear gradient formed with 0.1% HCO2H in H2O (solvent A) and 0.1% HCO2H in CH3CN (solvent B) (all v/v) by increasing from 2–45% solvent B (v/v) over a period of 0–45 min at a flow rate of 500 nL min−1.
Mass spectrometer settings: Spray voltage = 2.3 kV, capillary temperature = 200 °C. Higher energy collisional dissociation (HCD) and collision-induced dissociation (CID) MS/MS spectra were collected in a data-dependent mode using Top 2 for each fragmentation mode. MS1 spectra were collected with a resolution of 70,000, an automatic gain control (AGC) target of 1e6, and a mass range from m/z 300 to 1,500. HCD MS/MS spectra were acquired with a resolution of 7,500, an AGC target of 1e5, and normalized collision energy (NCE) of 35. CID MS/MS spectra were acquired with an NCE of 35 and 50 ms max injection time.
If the crosslinked peptide intensity is too low or there is interference, a targeted multiple reaction monitoring (MRM) approach may be required for positive MS/MS identification.
Note: These settings apply for an LTQ Orbitrap XL mass spectrometer. It is not necessary to follow the steps form this protocol. Parameters may change depending on the instrument. The identified adducted peptides in IDPicker can be alternatively confirmed using Skyline software. These parameters were used for the analysis of both N2-dG and N6-dA oligonucleotide-AGT crosslinks. The analysis was carried out in the positive ion mode.
Peptide data analysis
33. Raw data files were searched using MyriMatch software Version 2.2.140 (Tabb et al., 2007) against a forward and reverse Uniprot/Swissprot human proteome database with only reviewed proteins included (Version 20170202—20,165 entries).
34. Trypsin (specific cleavage) was used as the enzyme search parameter. The number of missed cleavages permitted was two, the precursor ion mass tolerance was set at 10 ppm, and a fragmentation tolerance of 20 ppm was used for the database search for HCD fragmentation; 0.5 Da was used for CID fragmentation searches.
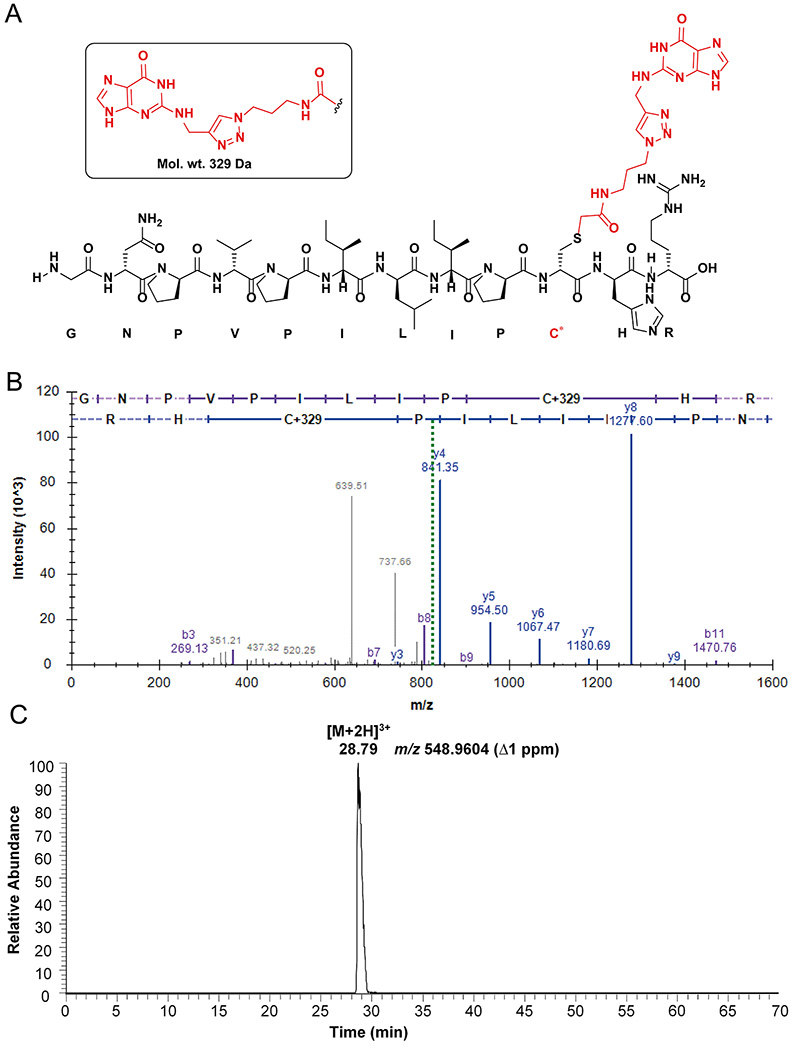
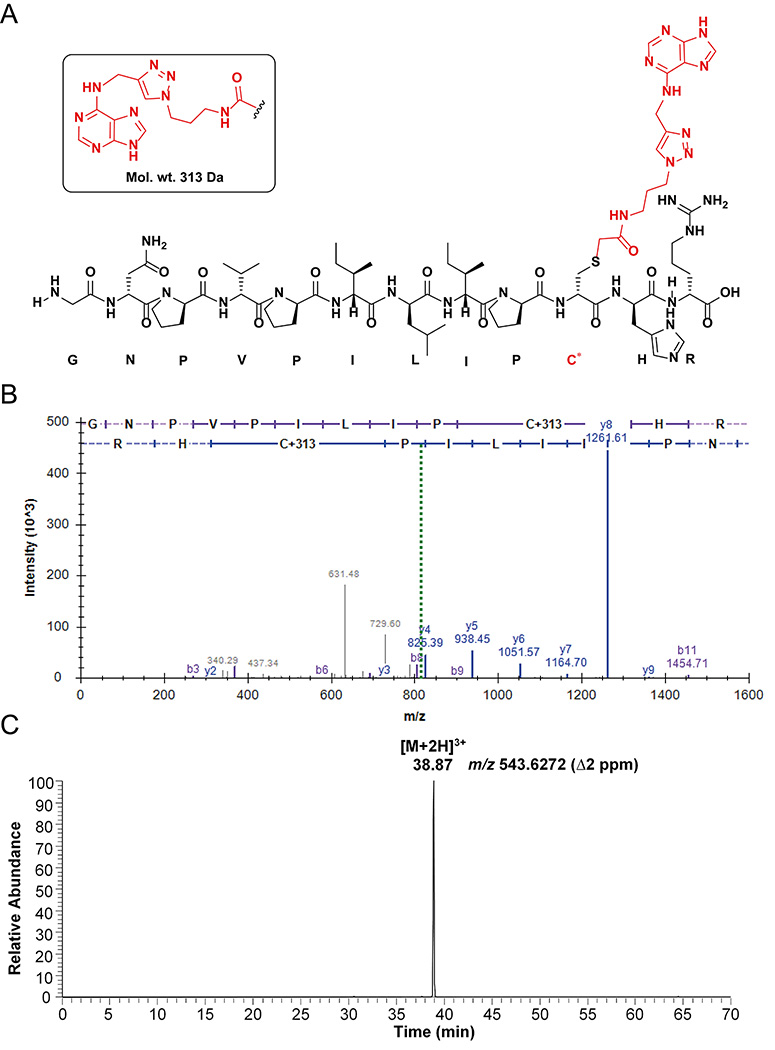
35. Cysteine modifications were included as variable search modifications. The guanine adduct mass was +329 Da (Figure 9A) and adenine adduct mass was +313 Da (Figure 10A).
Figure 9.
Peptide analysis of AGT-N2-dG oligonucleotide crosslink: (A) Observed structure of dodecamer guanine-adducted peptide (GNPVPILIPC*R) from the active site of AGT; (B) Annotated spectra of CID MS/MS of the [M+2H]2+ ion at m/z 822.9362 of the guanine-adducted peptide; (C) Extracted ion chromatogram of the guanine-adducted peptide for the [M+3H]3+ ion at m/z 548.9604. (C* = guanine-adducted Cys-145, observed an adduct with the guanine base only, due to the loss of deoxyribose).
Figure 10.
Peptide analysis of AGT N6-dA-oligonucleotide crosslink: (A) Observed structure of dodecamer adenine-adducted peptide (GNPVPILIPC*HR) from active site of AGT; (B) Annotated spectra of HCD MS/MS of the [M+2H]2+ ion at m/z 814.939 of adenine-adducted peptide; (C) Extracted ion chromatogram of adenine-adducted peptide for the [M+2H]3+ ion at m/z 543.627. (C* = adenine-adducted Cys-145, observed an adduct with the adenine base only, due to the loss of deoxyribose).
36. After MyriMatch identified peptide spectrum matches (PSMs), IDPicker software Version 3.1.642.0 (Holman et al., 2012) was used to rank the PSMs. False discovery rate (FDR) Q values of PSMs were adjusted to achieve a peptide FDR ≤10%.
37. The identified adducted peptides were confirmed in IDPicker as mentioned in Figures 9B and 10B. The [M+2H]2+ ions of guanine and adenine adducts were observed at m/z 822.936 and 814.939 respectively (Figure 9B and 10B).
38. The chromatogram was searched in QualBrowser using a 5 ppm mass window for the extracted ion chromatogram. The extracted ion chromatograms are shown for [M+2H]3+ ions of guanine and adenine adducts at m/z 548.960 and 543.628 respectively (Figures 9C and 10C).
REAGENTS AND SOLUTIONS
Prepare the stock solutions for Basic Protocols 1 to 2:
Propargylamine: 500 mM stock in anhydrous DMSO
DBU: 1 M stock in anhydrous CH3CN
0.4 M Methanolic NaOH (5 mL): 1 mL of 2 M NaOH in 4 mL of anhydrous CH3OH
Propargylamine hydrochloride: 500 mM stock in water.
DIPEA: 1 M stock in 1,4-dioxane.
Azide linker: 100 mM in 10% CH3CN (stored at −20 °C and protected from light in an amber vial)
CuSO4: 10 mM stock in water
THPTA: 50 mM stock in water (stored at −20 °C).
Sodium ascorbate: 100 mM stock in water
Prepare the stock solutions for Basic Protocol 3:
DTT: 500- and 100-mM stocks in water.
Urea: 8 M stock in water.
SDS: 10% (w/v) in water.
Iodoacetamide: 250 mM stock in water.
Trypsin: 10 ng/μL in 10 mM ammonium bicarbonate (pH 7.8).
Equilibration solution: 250 mM acetic acid with 30% CH3CN (v/v).
Elution solution: 400 mM ammonium hydroxide in water.
Destaining solution: 100 mM ammonium bicarbonate with 33% CH3CN (v/v).
5% Formic acid in 2:1 CH3CN:H2O (v/v).
0.1% aq. formic acid (v/v).
Note: All stocks should be prepared fresh.
Commentary
Background information
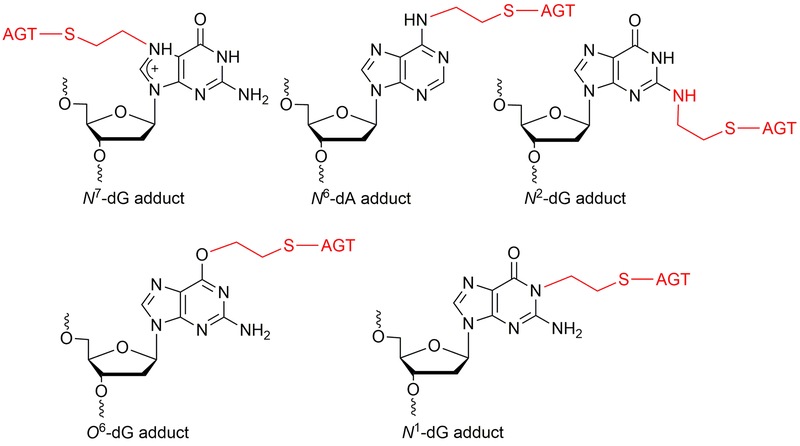
DNA protein crosslinks (DPCs) are very bulky DNA lesions formed by exogeneous (e.g. ultraviolet light, ionizing radiation, cisplatin, dibromoethane) and endogenous (topoisomerases, formaldehyde, abasic sites) agents (Barker et al., 2005). The DPCs can generate genomic instability by affecting physiological processes, including replication and transcription. The DPCs can be repaired by nucleotide excision repair (NER) and homologous recombination (HR) repair pathways (Reardon et al., 2006a, 2006b). In addition to this, DPCs may be digested using proteases before they can be processed by NER pathway. The O6-alkylguanine DNA-alkyltransferase protein is known to form crosslinks with DNA in the presence of 1,2-dibromoethane, resulting in mutations in bacterial as well as in mammalian cells (Kalapila et al., 2010; Liu et al., 2004; Liu et al., 2002). G:C to T:A transversion mutations were observed in mammalian (Chinese hamster ovary, CHO) cells when AGT-expressing cells were treated with dibromoalkanes. (Kalapila et al., 2010). Among the various positions in the DNA, non-labile AGT-DNA crosslinks form notably at the N2-position of dG, O6-position of dG, N1-position of dG, and N6-position of dA (Figure 11) (Chowdhury et al., 2013). Our hypothesis is that AGT-DNA crosslinks formed are processed by nuclear proteases such as SPRTN (Lopez-Mosqueda et al., 2016; Stingele et al., 2016; Vaz et al., 2016) to yield DNA-peptide crosslinks that are small enough to be bypassed by human TLS DNA polymerases. To test this hypothesis, an efficient methodology is needed for the synthesis of oligonucleotide-protein crosslink substrates. In this unit, we developed a protocol for the synthesis and characterization of AGT-oligonucleotide crosslinks at the N2-dG and N6-dA sites of DNA. These DNA-protein crosslinks serve as good substrates to understand the role of translesion polymerases and nuclear proteases as well as the consequences of mutations generated by AGT-DNA crosslinks. Overall, we explored post-oligomerization and CuAAC approaches for the synthesis of site-specific DPCs which are structural analogues of AGT-DNA crosslinks. Further, we confirmed these nucleobase adducted-peptide crosslinks using LC-MS/MS. Predominantly, the dodecamer nucleobase adducted-peptides were observed for N2-dG and N6-dA oligonucleotide-AGT crosslinks from the active site of protein.
Figure 11.
Structures of known AGT-DNA crosslinks
Critical parameters
For successful DPC synthesis the following points need to be considered.
The convertible nucleosides containing DNAs have intrinsic reactivity towards various nucleophiles. Phenoxyacetyl (PAC)-protected phosphoramidites are needed for the solid phase synthesis of convertible nucleoside containing oligonucleotides, which are compatible with ultra-mild deprotection conditions. Post-oligomerization reactions can be carried out at the CPG support level as well as in solution. For the successful post-oligomerization, reactions must be carried out at the CPG support level to reduce the oligonucleotide degradation. Reaction at the CPG support level also improves the yield of modified oligonucleotide by reducing the number of HPLC purifications. For the CuAAC reaction, the stock solutions need to be prepared fresh. The azide linker must be stored in an amber vial. The CuAAC reaction should be carried out in dark conditions. Pre-mixing of CuSO4 and THPTA ligand is very important before addition to the reaction mixture. The oligonucleotide precipitation step also needs to carried out in dark conditions because the iodoacetamide-linked oligonucleotide is light sensitive. For crosslinking reactions, buffer exchange of AGT protein must be carried out at pH 7.6 because the AGT protein is highly sensitive and will precipitate at lower pH values. The crosslink reaction needs to be done in dark conditions using low binding tubes. For proteomics, the use of freshly prepared stock solutions, fresh tips, and low binding tubes is necessary. Also, we emphasize the need to avoid keratin contamination while cutting the crosslink band from the gel for in-gel digestion. For in-solution digestion, the iron-IMAC purification of oligonucleotide-peptide heteroconjugates is a crucial step. The peptide solution needs to be acidified before applying to the iron-IMAC column. Hydrofluoric acid should be handled carefully in a well-ventilated fume hood.
Trouble shooting
Modified oligonucleotide synthesis requires prior experience using an oligonucleotide synthesizer and HPLC purification. Deprotection of modified oligonucleotides needs careful optimization to avoid degradation of oligonucleotides and CPG beads. For NPE deprotection, a Glen Research Protocol can be used (http://www.glenresearch.com/Technical/TB_2-F-dI.pdf). The use of an optimal concentration of NaOH for the deprotection of oligonucleotides is necessary. We prefer 0.4 M NaOH treatment for the deprotection oligonucleotide, which helps to give consistently good yields (Glen Research Protocol: http://www.glenresearch.com/Technical/TB_NaOH_Deprotection.pdf). The boiling point of propargylamine is low (83 °C), and additional equivalents are required for the synthesis of propargyl modified DNAs. The optimal use of copper and ligand is necessary for CuAAC reactions because DNA can bind to copper, which isolates the copper from the alkyne and azide reactants. In addition to this, copper(I) can also cause DNA damage. Precipitation of the CuAAC reaction mixture before the crosslink reaction with AGT is very important because AGT protein has a (His)6 tag on it. The (His)6 tag can bind to the copper, which might affect the crosslink reaction. The loading of AGT-oligonucleotide crosslinks samples on the SDS-PAGE gel can be done using either SDS or LDS loading buffer. In our experience, SDS loading buffer works better than LDS for the AGT protein, but results may vary among proteins. The use of fresh HF and low binding tubes for oligonucleotide hydrolysis prevents contamination by polyethylene glycol (PEG) in the proteomics samples.
Understanding results
For the post-oligomerization reactions, anhydrous conditions are very important to produce the propargyl bearing oligonucleotides in good yield. Also, post-oligomerization should be carried out at the CPG level in multiple batches to obtain the desired oligonucleotide in large scale. The displacement of the iodine of the iodoacetamide-linked oligonucleotide with the Cys-145 of AGT is very crucial to obtain the crosslink in quantitative yield.
Time considerations
In Basic Protocol 1, the 2-F-dI-containing oligonucleotide can be prepared in one day. The nucleophilic displacement, NPE deprotection, and final deprotection of CPG and base protecting groups can be performed in two days. The HPLC purification and desalting of N2-propargyl-dG can be done in two days. The CuAAC and crosslink reactions are easy to perform. Overall, the synthesis of N2-dG-oligonucleotide AGT crosslink can be accomplished in two weeks.
In Basic Protocol 2, the 6-chloropurine-containing oligonucleotide can be prepared in one day. The nucleophilic displacement reaction can be done at the CPG level or in solution. The final deprotection of CPG and base protecting groups can be achieved in 12 h. Overall, synthesis of N6-dA-oligonucleotide AGT crosslink can be accomplished in two weeks.
In Basic Protocol 3, the proteomics sample preparation can be accomplished within three days. The LC MS/MS analysis can be done in two days. Overall, proteomic analysis of N2-dG and N6-dA oligonucleotide-AGT crosslinks can be accomplished in one week.
Acknowledgements
We thank Prof. Carmelo J. Rizzo for the generous gift of 6-chloropurine phosphoramidite and also for providing access to the DNA synthesizer. We thank Dr. Chanchal Malik for help with oligonucleotide synthesis. P.P.G. thanks Carl A. Sedgeman for help with AGT protein expression and purification. This work was supported by a grant from the National Institutes of Health (R01 ES010546).
Literature cited:
- Barker S, Weinfeld M, & Murray D (2005). DNA–protein crosslinks: their induction, repair, and biological consequences. Mutat. Res. Rev. Mutat. Res, 589, 111–135. [DOI] [PubMed] [Google Scholar]
- Chowdhury G, Cho S-H, Pegg AE, & Guengerich FP (2013). Detection and characterization of ethylene dibromide-derived DNA-crosslinks formed with O6-alkylguanine-DNA alkyltransferase. Angew. Chem. Int. Ed, 52, 12879–12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielden J, Ruggiano A, Popović M, & Ramadan K (2018). DNA protein crosslink proteolysis repair: From yeast to premature ageing and cancer in humans. DNA Repair, 71, 198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer H, Geyer R, & Pingoud V (2004). A novel strategy for the identification of protein–DNA contacts by photocrosslinking and mass spectrometry. Nucleic Acids Res, 32, e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruppi F, Salyard TLJ, & Rizzo CJ (2014). Synthesis of G-N2-(CH2)3-N2-G trimethylene DNA interstrand cross-links. Curr. Protoc. Nucleic Acid Chem, 56, 5.14.11–15.14.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman JD, Ma Z-Q, & Tabb DL (2012). Identifying proteomic LC-MS/MS data sets with Bumbershoot and IDPicker. Curr. Protoc. Bioinformatics, 37, 13.17.11–13.17.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide H, Nakano T, Shoulkamy MI, & Salem AMH (2015). Formation, repair, and biological effects of DNA–protein cross-link damage. In Advances in DNA Repair (pp. 43–80). [Google Scholar]
- Kalapila AG, & Pegg AE (2010). Alkyltransferase-mediated toxicity of bis-electrophiles in mammalian cells. Mutat. Res, 684, 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotoku N, Nakata C, Kawachi T, Sato T, Guo X-H, Ito A, Sumii Y, Arai M, & Kobayashi M (2014). Synthesis and evaluation of effective photoaffinity probe molecule of furospinosulin-1, a hypoxia-selective growth inhibitor. Bioorg. Med. Chem, 22, 2102–2112. [DOI] [PubMed] [Google Scholar]
- Liu L, Hachey DL, Valadez G, Williams KM, Guengerich FP, Loktionova NA, Kanugula S, & Pegg AE (2004). Characterization of a mutagenic DNA adduct formed from 1,2-dibromoethane by O6-alkylguanine-DNA alkyltransferase. J. Biol. Chem, 279, 4250–4259. [DOI] [PubMed] [Google Scholar]
- Liu L, Pegg AE, Williams KM, & Guengerich FP (2002). Paradoxical enhancement of the toxicity of 1,2-dibromoethane by O6-alkylguanine-DNA alkyltransferase. J. Biol. Chem, 277, 37920–37928. [DOI] [PubMed] [Google Scholar]
- Lopez-Mosqueda J, Maddi K, Prgomet S, Kalayil S, Marinovic-Terzic I, Terzic J, & Dikic I (2016). SPRTN is a mammalian DNA-binding metalloprotease that resolves DNA-protein crosslinks. ELife, 5, e21491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presolski SI, Hong VP, & Finn MG (2011). Copper-catalyzed azide–alkyne click chemistry for bioconjugation. Curr. Protoc. Chem. Biol, 3, 153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon JT, & Sancar A (2006a). Purification and characterization of Escherichia coli and human nucleotide excision repair enzyme systems. Methods Enzymol. 408, 189–213). [DOI] [PubMed] [Google Scholar]
- Reardon JT, & Sancar A (2006b). Repair of DNA–polypeptide crosslinks by human excision nuclease. Proc. Natl. Acad. Sci. U.S.A, 103, 4056–4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havli J, Olsen JV, & Mann M (2007). In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc, 1, 2856–2860. [DOI] [PubMed] [Google Scholar]
- Stingele J, Bellelli R, Alte F, Hewitt G, Sarek G, Maslen SL, Tsutakawa SE, Borg A, Kjær S, Tainer JA, Skehel JM, Groll M, & Boulton SJ (2016). Mechanism and regulation of DNA-protein crosslink repair by the DNA-dependent metalloprotease SPRTN. Mol. Cell, 64, 688–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, Fernando CG, & Chambers MC (2007). MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res, 6, 654–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz B, Popovic M, Newman JA, Fielden J, Aitkenhead H, Halder S, Singh AN, Vendrell I, Fischer R, Torrecilla I, Drobnitzky N, Freire R, Amor DJ, Lockhart PJ, Kessler BM, McKenna GW, Gileadi O, & Ramadan K (2016). Metalloprotease SPRTN/DVC1 orchestrates replication-coupled DNA-protein crosslink repair. Mol. Cell, 64, 704–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz B, Popovic M, & Ramadan K (2017). DNA–Protein crosslink proteolysis repair. Trends Biochem Sci, 42, 483–495. [DOI] [PubMed] [Google Scholar]
- Wang H, Kozekov ID, Kozekova A, Tamura PJ, Marnett LJ, Harris TM, & Rizzo CJ (2006). Site-specific synthesis of oligonucleotides containing malondialdehyde adducts of deoxyguanosine and deoxyadenosine via a postsynthetic modification strategy. Chem. Res. Toxicol, 19, 1467–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramaratne S, Seiler CL, & Tretyakova NY (2015). Synthesis of DNA oligodeoxynucleotides containing site-specific 1,3-butadiene-deoxyadenosine lesions. Curr. Protoc. Nucleic Acid Chem, 61, 4.61.61–64.61.22. [DOI] [PMC free article] [PubMed] [Google Scholar]