Abstract
Long bone development involves the embryonic formation of a primary ossification center (POC) in the incipient diaphysis followed by postnatal development of a secondary ossification center (SOC) at each epiphysis. Studies have elucidated major basic mechanisms of POC development, but relatively little is known about SOC development. To gain insights into SOC formation, we used Col2-Cre Rosa-tdTomato (Col2/Tomato) reporter mice and found that their periarticular region contained numerous Tomato-positive lineage cells expressing much higher Tomato fluorescence (termed TomatoH) than underlying epiphyseal chondrocytes (termed TomatoL). With time, the TomatoH cells became evident at the SOC invagination site and cartilage canal, increased in number in the expanding SOC, and were present as mesenchymal lineage cells in the subchondral bone. These data were verified in two mouse lineage tracing models, Col2-CreER Rosa-tdTomato and Glil-CreER Rosa-tdTomato. In vitro tests showed that the periarticular TomatoH cells from Col2/Tomato mice contained mesenchymal progenitors with multidifferentiation abilities. During canal initiation, the cells expressed vascular endothelial growth factor (VEGF) and migrated into epiphyseal cartilage ahead of individual or clusters of endothelial cells, suggesting a unique role in promoting vasculogenesis. Later during SOC expansion, chondrocytes in epiphyseal cartilage expressed VEGF, and angiogenic blood vessels preceded TomatoH cells. Gene expression analyses of microdissected samples revealed upregulation of MMPs in periarticular cells at the invagination site and suggested potential roles for novel kinase and growth factor signaling pathways in regulating SOC canal initiation. In summary, our data indicate that the periarticular region surrounding epiphyseal cartilage contains mesenchymal progenitors that initiate SOC development and forms subchondral bone.
Keywords: Mesenchymal progenitors, Periarticular layer, Cartilage, Secondary ossification center, Blood vessel
INTRODUCTION
Long bones, like much of the skeleton, are formed through endochondral ossification [1, 2]. This process starts with mesenchymal cell condensations emerging at sites of future skeletal elements [3]. During condensation, the cells alter their adhesiveness to both extracellular matrix (ECM) and one another, migrate toward the center, and exclude vessels. At the boundary of this condensation, a thin layer of loose mesenchymal cells forms while the condensation reaches a critical size. Cells within the boundary then differentiate into chondrocytes, while boundary cells begin to flatten, elongate, and form the perichondrium. As these processes are ongoing, a mesenchymal interzone emerges at the future location of synovial joints and separates neighboring skeletal elements such as the femur and tibia or humerus and ulna. Over time, the interzone cells and their descendants give rise to joint tissues, including articular chondrocytes, with some persisting as periarticular cells [4,5]. The diaphyseal portion of developing long bones is the first in which prehypertrophic and hypertrophic chondrocytes develop. The cells induce perichon-drial osteoprogenitors to undergo differentiation via Indian hedgehog (lhh) and produce a bone collar around the diaphysis, representing nascent cortical bone [6]. The diaphyseal hypertrophic chondrocytes also produce angiogenic factors and matrix metalloproteases (MMPs) that induce and facilitate the invasion of hypertrophic cartilage by blood vessels and osteoprogenitors to establish the primary ossification center (POC). A previous genetic cell tracing study demonstrated that osteoprogenitors, but not osteoblasts, in the perichondrium follow blood vessels during invasion into hypertrophic cartilage, and along with other cells, ultimately establish the POC [7]. Shortly after birth, a secondary ossification center (SOC) forms in the epiphyseal regions, thus defining the boundary of the growth plate between the POC and SOC, as well as the area of articular cartilage.
There are several similarities in the formation of the POC and SOC, including chondrocyte hypertrophy and mineralization, invasion of blood vessels into cartilage matrix, and remodeling of the cartilage area into bone and bone marrow space [8]. However, SOC formation also has interesting and unique features. First, unlike the perichondrial region at the midshaft that forms the mineralized bone collar prior to POC initiation, the periarticular region surrounding epiphyseal cartilage remains as a layer of flattened and fibroblastic-like cells parallel to the surface during the early stage of SOC formation. Second, the SOC starts with invagination of periarticular segments at only a few sites that generate cartilage canals, a special anatomical structure, to carry blood vessels and loose connective tissue into the centrally located cartilage. Third, the cartilage canals initially invade non-mineralized epiphyseal cartilage, a striking contrast to calcified hypertrophic cartilage into which vessels migrate during POC formation. Eventually, when the canals fuse to form the expanding SOC, the local cartilage tissue undergoes hypertrophy, mineralizes and is gradually replaced by endochondral bone.
It has long been assumed that mechanisms underlying POC formation apply, and can be extrapolated to, SOC formation. This is certainly so with reference to important regulators such as metalloproteinases (MMP9, 13, and 14) [9]. Several signaling pathways, including thyroid hormone [10], epidermal growth factor receptor (EGFR) [11], Wnt/β-catenin [12], and insulin-like growth factor 1 [13], have been reported to be required for proper SOC development. The unique features of SOC formation, particularly the initiation stages, indicate that it may require additional mechanisms distinct from those observed in POC formation. In particular, it is currently unclear what mechanisms control periarticular cell migration, tissue invagination and cartilage canal formation, and what may be the source and function of mesenchymal progenitors responsible for SOC formation.
Collagen 2 promoter-driven Cre transgenic mice are often used to activate reporter gene expression or knockout a target floxed gene in cartilage [14]. However, more recent studies have shown that these mice, as well as the inducible Col2-CreER, Sox9-CreER, and Acan-CreER mice, can target and activate reporter gene expression in neighboring mesenchymal cells [15]. In this study, we have exploited these features of Col2-Cre mice to carry out mesenchymal cell labeling during SOC development, focusing on the initiation and expansion of cartilage canals and on mesenchymal progenitors within periarticular region as a possible contributor to SOC development and bone formation. Two other inducible mouse models (Col2-CreER and Glil-CreER) were also used to support our observations and conclusion. Furthermore, gene expression profiling of invaginating cells in comparison with the neighboring periarticular region cells revealed expression of novel genes and signaling pathways, all likely regulating SOC initiation.
MATERIALS AND METHODS
Animals
Col2/Tomato, Col2ER/Tomato, and GlilER/Tomato mice were generated by breeding Rosa-tdTomato mice with Col2-Cre mice, Col2-CreER mice, and Glil-CreER mice, respectively. All mice were obtained from Jackson Laboratory (Bar Harbor, ME). Mice containing CreER received 50 μg tamoxifen in corn oil per mouse or vehicle at P4. Only mice homozygous for Tomato were used for experiments. Knee joints were harvested at various time points after birth. To label newly mineralized sites, mice received 10 mg/kg calcein (Sigma, Allentown, C0875) injection 2 hours before sacrifice. For EdU labeling, mice were injected with 1.6 mg/kg EdU (Invitro-gen, Carlsbad, A10044) 2 days and 2 hours before harvesting, then stained according to the manufacturer’s protocol. All animal work performed in this report was approved by the Institutional Animal Care and Use Committee at the University of Pennsylvania.
Sample Preparation and Imaging
For thin cryosections, mouse knee joints were fixed in 4% paraformaldehyde for 4–6 hours followed by 20% sucrose for 1 day. Tissues were then embedded and sectioned using low-profile blades on a Leica CM3050 cryostat. For whole mount images, samples were treated similarly with the addition of 2% polyvinylpyrrolidone (PVP) during the cryoprotection step and frozen in 8% gelatin (porcine) in the presence of 20% sucrose and 2% PVP. Samples were sectioned at a thickness of 100 μm. Sections were stained with primary antibodies against endomucin (Santa Cruz, Minneapolis, sc-65,495), CD31 (R&D Systems, Pittsburg), matrilin-1 (Abcam, Cambridge, ab106384), CD45 (BioLegend, Dedham), Ter119 (Biolegend, 116201), vascular endothelial growth factor (VEGF; Abcam, ab46154), VEGFR-2 (Abcam, ab10972), and proteoglycan degraded products (gifts from Dr. John S. Mort), followed by Alexa Fluor 488-conjugated donkey anti-rabbit IgG secondary antibody (Thermo Scientific, Philadelphia, A-21206), Alexa Fluor 488-conjugated donkey anti-rat (Thermo Scientific, A21208), Alexa Fluor 647 conjugated goat anti-rabbit 647 (Thermo Scientific, A21246), or Alexa Fluor 647 conjugated goat anti-rat (Thermo Scientific, A21247). Fluorescent signals were acquired from Zeiss LSM-710 laser scanning confocal microscope. Maximum intensity projections were generated using ImageJ. Area was quantified from section with the largest SOC using ImageJ. In figures, a single representative image is shown to represent the average results of at least three independent histological analyses conducted.
Cell Culture
Periarticular cells were isolated from the distal femurs and proximal tibia epiphyses of P4 mice via transient enzymatic digestion as described previously [16]. Briefly, tissues were incubated with 0.25% trypsin (Invitrogen; wt/vol) for 1 hour, followed by a 2.5 hours digestion with 173 U/ml type 1 collagenase (Worthington Biochemical, Lakewood, NJ). Dissociated cells were seeded on culture dishes precoated with Fibronectin from bovine plasma (Sigma-Aldrich, F1141) at a concentration of 1–5 μg/cm2 for 4 hours, followed by blocking with 3% bovine serum albumin (BSA) for 30 minutes. The attached cells were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum (FBS), 100 μg/ml streptomycin and 100 lU/ml penicillin. Remaining cartilage tissues underwent further digestion in 86 U/ml type 1 collagenase overnight and cells collected were considered as epiphyseal chondrocytes. To eliminate a possible damaging effect of prolonged digestion on cells, periarticular cells collected from the transient digestion were subjected to the same overnight digestion used for collecting epiphyseal chondrocytes before culture. However, we did not observed any difference in the following assays between periarticular cells harvested with or without extra digestion (data not shown).
For colony forming units-fibroblast (CFU-F) assay, cells were seeded in T25 flask at 6,000 cells per flask for 1 week, and then stained with 0.3% crystal violet in methanol. For proliferation assay, cells were seeded in 96-well plate at 5,000 cells per well, and MTT assay was conducted at 1, 3, and 5 days after seeding. For differentiation assays, cells were seeded at 3 × 106 per well in 6-well plate. At 80%‒90% confluence, cells were incubated with either osteogenic medium (αMEM with 10% FBS, 50 μg/ml ascorbic acid, 10 mM β-glycerol phosphate) followed by Alizarin Red staining 2 weeks later or adipogenic medium (αMEM with 10% FBS, 0.5 mM isobuthylmethylxanthine, 10 mM indometha-cin, 1 μM dexamethasone, and 10 μg/ml insulin) followed by Oil red staining 1 week later.
To perform migration assay, 5,000 endothelial progenitor cells (EPCs) were added to the upper parts of a 96-well microchemo-taxis Boyden chamber (Neuro Probe) and conditioned media from TomatoH cells or bone marrow mesenchymal stem cells (MSCs), with or without SU5416 (Selleckchem, Houston, S2845), were added to the lower parts. The chamber was then incubated at 37°C in humidified 5% CO2 for 4 hours. The migrated cells on the lower side of filter were then stained with 0.3% crystal violet in methanol and counted under a microscope. EPCs were generated by flushing bone marrow cells from 2-month-old C57/BI6 mice in M199 media and seeded into Vitronectin-coated 6-well plate [17]. Cells were validated by staining with Dil-Ac-LDL (Biomedical Technologies, Inc., BT 902). Bone marrow MSCs were generated by flushing bone marrow cells from 2-month-old C57/BI6 mice and cultured in the growth media (αMEM with 10% FBS, 1% glutamine) until 80%−90% confluence.
Gene Expression Analyses
To quantify the expression level of individual marker genes, cells were collected in Tri Reagent (Sigma, St. Louis, MO) for RNA purification. A Taqman Reverse Transcription Kit (Applied BioSystems, Inc., Foster City, CA) was used to reverse transcribe mRNA into cDNA. Following this, quantitative realtime PCR (qRT-PCR) was performed using a Power SYBR Green PCR Master Mix Kit (Applied BioSystems, Inc.). The primer sequences for the genes used in this study are listed in Supporting Information Table S1. Gene expression was calculated using the ΔΔCT method after normalizing to β-actin.
For microarray assay, knee joints from P5.5 mice were fresh frozen, embedded and sectioned using the cryo-jane tape system. After dehydration, slides were air-dried and regions of interest from the distal femur were collected using the infrared laser of the ArcturusXT laser capture microdissection (LCM) system (ThermoFisher, Watham, MA) via CapSure Macro LCM caps (Applied Biosystems, Foster City, CA). Samples were pooled for RNA isolation via Qiagen RNeasy micro kit (Gaithersburg, MD). RNA was then subjected to GeneChip WT Pico Reagent Kit (Affymetrix, Santa Clara, CA) followed by Clariom D assay (Thermo Scientific). Data were analyzed by Transciptome Analysis Console 4.0 (TAC, Applied Biosystems). Threshold for a gene being significantly regulated was a fold change greater than 2 with p <.05. GO terms and clusters, as well as KEGG pathway enrichments, were identified using the Database for Annotation, Visualization and Integrated Discovery [18,19]. Genes in KEGG pathways regulated in the canal versus peri were visualized for interactive GO term nodes using ClueGo plug-in (v2.5.0) [20] of cytsocape (v3.5.1, http://www.cytoscape.org/). Heat map and hierarchical clustering was conducted on the 1,200 most regulated genes using ClustVis [21].
Statistical Analyses
Data are expressed as means ±SEM. All statistical analyses were conducted using GraphPad Prism. Comparisons between groups were analyzed by either a Student’s t test (paired or unpaired as stated) or one-way or two-way analysis of variance (with repeated measurements or not as appropriate) with a Bonferroni’s post-test as stated. p values less than .05 were considered to be statistically significant.
RESULTS
Periarticular Surface of Mouse Long Bone Epiphysis Contains Mesenchymal Progenitors that Contribute to SOC Development
Femoral epiphyses in mice remain uniformly cartilaginous until around postnatal day 4 (P4), with initiation of SOC formation becoming apparent around P5-P6. In line with this notion, nearly all epiphyseal chondrocytes in P4 Col2/Tomato pups were reporter positive (Fig. 1Aa, 1Ab). Surprisingly, Tomato fluorescent intensity was not uniform throughout the epiphysis. As shown in Figure 1Ac, 1Ad, cells at the outer epiphyseal perimeter had notably higher Tomato signal (Tomatohlgh cells, TomatoH, hereafter) than those within the cartilage mass (Tomatolow cells, TomatoL, hereafter; Supporting Information Fig. S1). The TomatoH cells were present in ~4–6 cell layers, were dense, elongated, and relatively small cells (Supporting Information Fig. S2A), and lacked matrillin-1 expression (Supporting Information Fig. S2B), whereas the underlying, less dense, and relatively larger chondrocytes were matrilin-1+. Matrilin-1 is normally expressed in epiphyseal chondrocytes that will eventually be replaced by bone and bone marrow after epiphyseal marrow compartment formation [22]. Hence, the TomatoH cells represented periarticular cells at this stage that would likely contribute to articular cartilage development. In addition to the periarticular region (Fig. 1Ba), TomatoH cells were also found in the groove of Ranvier (Fig. 1Bb), the resting zone of growth plate (Fig. 1Bc), and the primary spongiosa (Fig. 1Bd), likely belonging to the osteogenic lineage.
Figure 1.
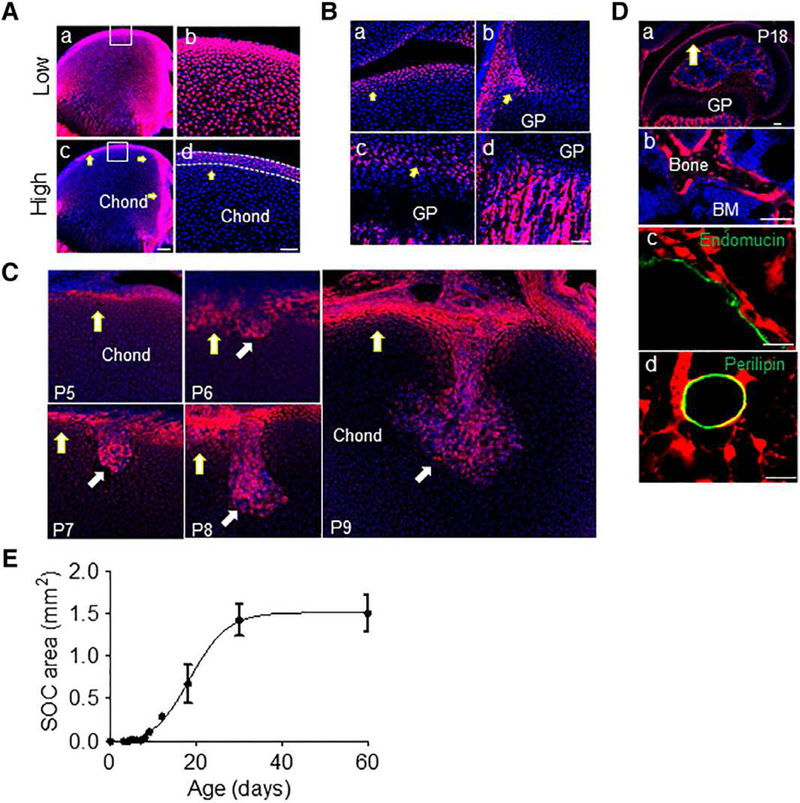
The highly fluorescent periarticular region in Col2/Tomato mice contains multipotent cells that forms the invading canal and reconstitutes subchondral bone. (A): A femur from P4 Col2/Tomato mouse was imaged by confocal microscopy and displayed at low (Aa, Ab) and high (Ac, Ad) intensity threshold. Yellow arrows point to TomatoH cells. The periarticular region was defined as the 4–6 layers of TomatoH cells along the epiphyseal surface (white dotted lines). Blue: DAPI. Bar = 200 μm (Aa, Ac), 50 μm (Ab, Ad). (B): High magnification images of the periarticular region (Ba), groove of Ranvier (Bb), growth plate (arrows point to the resting zone; Bc), and primary spongiosa (Bd) of the same femur showing labeling with high Tomato fluorescence. Bar = 50 μm. (C): Col2/Tomato mice were sacrificed postnatally at days indicated and the forming cartilage canal imaged by confocal microscopy. Bar = 100 μm. (D): By P18 (Da), TomatoH cells were found to give rise to osteoblasts and osteocytes (Db), pericytes along Endomucin+ blood vessels (Dc), and adipocytes (Dd). (Db), (Dc), and (Dd) are high magnification images from (Da). Bar = 200 μm (Da, Db), 25 μm (Dc, Dd). (E): Monitoring of secondary ossification center expansion revealed rapid growth during the invasion and expansion phases, with a maximum epiphyseal marrow area being reached by P30. Values represent averages ± SEM (n = 3–5/age).
At P5-P6 when the SOC initiates, incipient invagination sites from the periarticular region were enriched with TomatoH cells (Fig. 1C). At P7-P8, newly formed cartilage canals contained numerous TomatoH cells, while the surrounding cartilage cells were TomatoL. Typically, there were 1–2, but no more than 3, canals per distal femur and those canals originate randomly from the anterior and posterior sides of periarticular surface excluding the articular surface facing the tibia. When subchondral trabecular bone structure was established around P18, it became evident that all osteoblasts and osteocytes were TomatoH (Fig. 1Da, 1Db). Other TomatoH cells included pericytes, considered primitive mesenchymal progenitors or MSCs (Fig. 1Dc), as well as adipocytes (Fig. 1Dd). Quantification revealed that the SOC area undergoes drastic growth from initiation stage (P5-P6) to the cartilage canal advancement stage (P7-P8) and expansion stage (P9 and after), finally reaching a mature stage around P30 (Fig. 1E). The dynamic distribution of TomatoH cells during these stages provided the first evidence suggesting that mesenchymal progenitors within the SOC originate from the periarticular TomatoH cell population.
To further support this conclusion, we performed pulse-chase experiments using inducible Col2ER/Tomato mice. Without tamoxifen injection, epiphyseal cartilage in P5 mice exhibited sporadic TomatoH cells only (Fig. 2Aa, 2Ab, 2B), suggesting minor, leaky expression in this model. A single injection of tamoxifen at P4 elicited creation of many TomatoH cells largely around the periarticular region by P5 (Fig. 2Ac, 2Ad, 2B). Later, similar to Col2/Tomato pups, the tamoxifen-induced Col2ER/Tomato mice exhibited many TomatoH osteoblasts, osteocytes, perivascular cells and bone marrow adipocytes at P60 (Fig. 2C), indicating that periarticular TomatoH cells contain multipotent mesenchymal progenitors for SOC formation.
Figure 2.
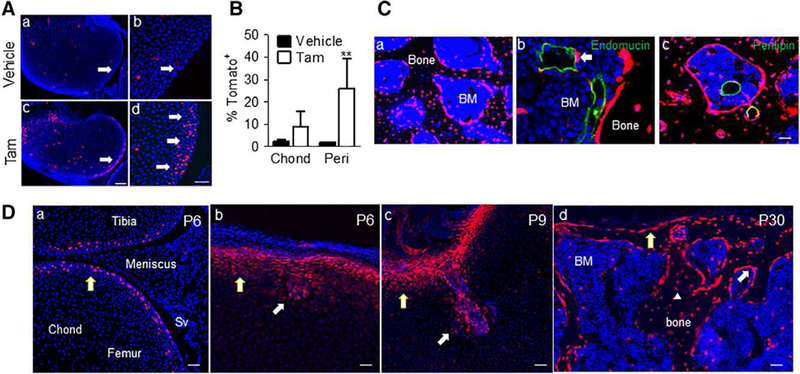
Lineage tracing specifically labels periarticular cells which give rise to the invading canal and reconstitutes subchondral bone. (A): Tomato+ cells were predominantly found at the periarticular region (pointed by arrows) in Col2ER/Tomato mice at P5 after tamoxifen injection at P4. (Ab) and (Ad) are magnified images from (Aa) and (Ac), respectively. Bar = 200 μm (Aa and Ac), 50 μm (Ab and Ad). (B): Quantification of the number of Tomato+ cells in the chond and peri regions following tamoxifen injection; n = 3. Values represent average values ± SEM. **, p <.01 relative to vehicle control by two-way analysis of variance with Bonferroni’s post-test. (C): These mice were also found to have Tomato+ osteoblasts, osteocytes (Ca), perictyes (Cb), and adipocytes (Cc) at P60. Bar = 50 μm. (D): Gli1-CreER/ Tomato mice were injected with tamoxifen at P4 and sacrificed during canal initiation (P6), canal invasion (P9) and following secondary ossification center (SOC) formation (P30). Tomato signal was found to specifically label periarticular cells, the cartilage canal and mesenchymal lineage cells within the bone marrow following SOC formation, consistent with the TomatoH cells in the Col2/Tomato mice. Bar = 50 μm. Abbreviations: BM, bone marrow; Chond, Epiphyseal chondrocytes; Peri, periarticular; SV, synovium.
Due to the existence of TomatoL cells in the epiphyseal cartilage, the above experiments cannot completely exclude the possibility that TomatoL cells might become TomatoH cells and contribute to SOC formation. To address this concern, we analyzed SOC development in Gli1ER/Tomato mice. In this mouse model, when injected at P4,Tomato+ cells were found exclusively in the periarticular cells but not underlying epiphyseal chondrocytes and other joint tissues including the meniscus and synovium (Fig. 2Da). When the SOC initiated, these Tomato+ cells were enriched at the invagination site (Fig. 2Db) and in the invading cartilage canal (Fig. 2Dc). Later, numerous osteoblasts and osteocytes within the SOC were found to be Tomato+ (Fig. 2Dd). This result further supports a direct contribution of periarticular cells to the establishment of the mesenchymal lineage cells within the epiphyseal bone compartment.
A Subset of Periarticular TomatoH Cells Displays Mesenchymal Progenitor Properties
To characterize the phenotypic properties of TomatoH and TomatoL cells, we isolated these two cell populations from the long bone epiphyses of P4 Col2/Tomato pups prior to SOC initiation. After a brief enzymatic digestion, the periarticular layer was depleted and most TomatoH cells were released (Fig. 3A). Subsequent longer digestion released TomatoL cells from the inner portion of cartilage. Once adhered, TomatoH cells exhibited higher fluorescent intensity than TomatoL cells and had an elongated and fibroblastic shape similar to bone marrow mesenchymal progenitors, while the majority of TomatoL cells were polygonal, a typical chondrocyte morphology (Fig. 3B). Gene expression analysis showed that TomatoH cells expressed much lower levels of chondrogenic markers Sox9, Col2a1 and Col10a1 than TomatoL cells (Fig. 3C), indicating that the Toma-toH population represented largely undifferentiated cells. MTT assays implied a greater proliferation ability in TomatoH cells than TomatoL cells (Fig. 3D). When seeded at a low density, TomatoH cells generated 10-fold more colonies than TomatoL cells (Fig. 3E). Another key feature of mesenchymal progenitors is a strong migratory ability. Trans-well assays revealed a 4-fold increased migration in TomatoH cells compared with TomatoL cells (Fig. 3F).
Figure 3.
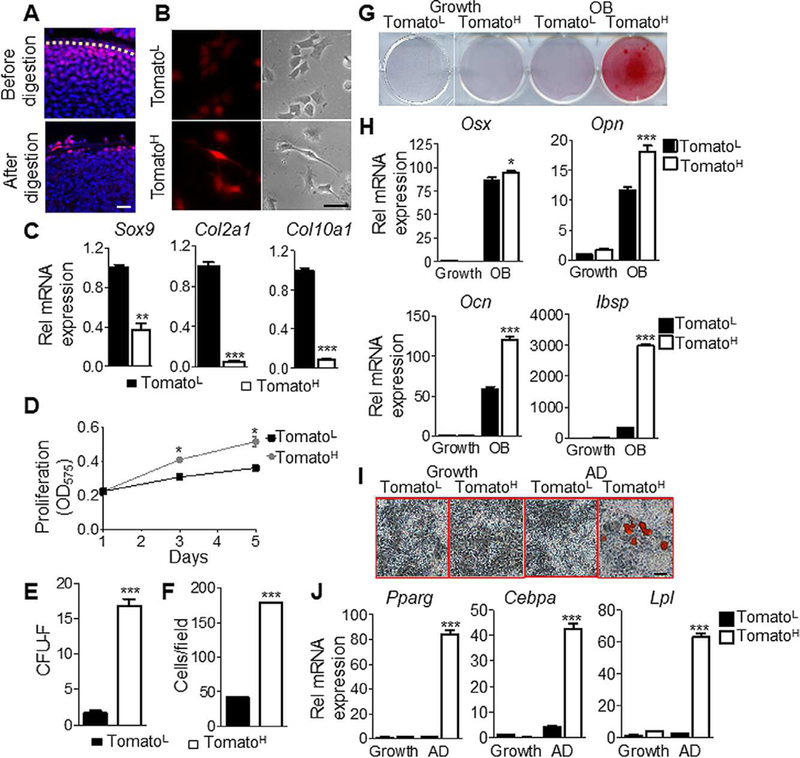
The periarticular region contains mesenchymal progenitors with increased osteogenic and adipogenic potential. (A): TomatoH and TomatoL cells were isolated by short-term or overnight enzymatic digestion, respectively. Bar = 50 μm. (B): Morphologically, TomatoL cells appear more rounded, characteristic of chondrocytes, whereas TomatoH cells appear more fibroblastic and elongated with increased fluorescence following adhesion. Bar = 10 μm. (C): Gene expression confirmed TomatoL cells expressed elevated levels of the chondro-genic markers Sox9, Col2, and Col10, relative to TomatoH cells. (D): TomatoH cells possessed significantly greater proliferation, as assessed by MTT assays. (E): TomatoH possess an increased colony forming ability. (F): TomatoH cells show increased migration rates, as assessed by trans-well assays. (G): TomatoH cells showed increased mineralization under osteogenic differentiation conditions. (H): TomatoH cells showed increased expression of the osteogenic markers Osx, Opn, Ocn, and Ibsp, relative to TomatoL cells, under osteogenic differentiation conditions. (I): Under adipogenic differentiation conditions, TomatoH cells show increased staining for lipid accumulation. Bar = 10 μm. (J): TomatoH cells expressed significantly elevated levels of the adipogenic markers Pparγ, Cebpa, and Lpl when cultured under adipogenic conditions. Graphs represent average values ±SEM. Data was analyzed by paired t test (C-F) or by repeated measures two-way analysis of variance with Bonferroni’s post-test (*, p <.05; **, p <.01; ***, p <.001; n = 3).
Next, we investigated whether TomatoH cells can differentiate into osteoblasts and adipocytes. Compared with TomatoL cells, TomatoH cells displayed higher osteogenic potential, as indicated by alizarin red staining (Fig. 3G) and expression of osteogenic marker genes osterix (Osx), osteopontin (Opn), osteocalcin (Ocn), and integrin binding sialoprotein (Ibsp; Fig. 3H). TomatoH cells also displayed higher adipogenic potential as revealed by Oil Red O staining (Fig. 3I) and expression of adipogenic markers peroxisome proliferator-activated receptor γ (Pparγ), CCAAT/en-hancer binding protein α (Cebpα) and lipoprotein lipase (Lpl; Fig. 3J).
In addition, we performed EdU labeling to study the proliferative ability of TomatoH cells during SOC development in vivo (Supporting Information Fig. S3A). Quantification of EdU+ cells among the TomatoH cell population at different locations showed that the proliferation ability of TomatoH cells increased drastically as they migrated from the periarticular region into the cartilage canal and the expanding SOC (Supporting Information Fig. S3B). Taken together, the above in vitro and in vivo observations indicate that the periarticular TomatoH cells represent mesenchymal progenitors able to produce various cell types within epiphyseal bone and to increase their proliferation when moving into epiphyseal cartilage to establish the SOC.
The Invaginating Canal Recruits Endothelial Cells and Is Preceded by Chondrocyte ECM Degradation
Cartilage canals start off as invaginations of the periarticular region at P5-P6 in mice. Strikingly, at this stage, we observed an influx of TomatoH cells penetrating into epiphyseal cartilage from discrete surface sites, followed by Endomucin+ endothelial cells, many of which exhibited an elongated shape or were in clusters but had not formed lumenized vessels yet (Fig. 4A, P5). Later, cartilage canals initiating from those discrete surface sites became more evident and rich in TomatoH cells at the front edge that made contact with chondrocytes and endothelial cells lagging behind these TomatoH cells by P7 (Fig. 4A, P6). The cartilage canals had two important features. First, TomatoH cells were always located at the front edge of the advancing canal and preceded other types of cells including endothelial cells. Second, inside the canals, many endothelial cells were loosely connected and did not form lumenized vessels. These data suggest that vasculogenesis, the de novo process of vessel formation, contributed to SOC initiation.
Figure 4.
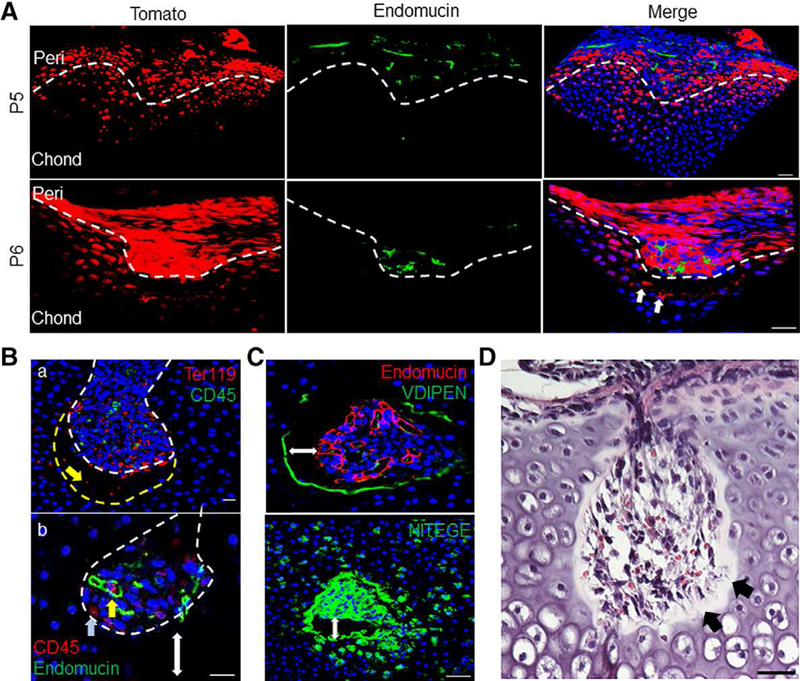
Canal invasion precedes endothelial cell recruitment and is led by chondrocyte extracellular matrix (ECM) degradation. (A): Canal initiation was led by TomatoH cells (white arrows) and trailed by blood vessels stained with the endothelial marker endomucin. Bar = 50 μm. (Ba): Canal invasion was preceded by an “empty space” between the canal front (dotted white line) and the surrounding chondrocytes (dotted yellow line), containing only Ter119+ erythroid lineage cells (yellow arrow). Bar = 20 μm. (Bb): Within the canal, CD45+ hematopoietic cells were either occasionally associated with immature vasculature (yellow arrow) or away from endothelial cells (blue arrow). Double-headed arrow shows gap between canal front and epiphyseal chondrocytes. Bar = 20 μm. (C): Immunofluorescent images show intense staining for chondrocyte ECM degradation around the invading canal. Double-headed arrow marks gap between canal and epiphyseal chondrocytes. Bar = 50 μm. (D): Staining with H&E confirmed a cell-free gap, with residual fiber or matrix components in the observed empty space (black arrows). Bar = 50 μm.
In addition to TomatoH and endothelial cells, the developing cartilage canals contained short blood vessels, erythrocytes (Ter 119+) and hematopoietic cells (CD45+) either loosely associated with vessels or away from vessels (Fig. 4B). Interestingly, DAPI staining revealed that within the cartilage canal, between the front edge and dense underlying chondrocytes, there was an “empty” space approximately 30 μm in length, where only erythrocytes but no nucleated cells were detected (Fig. 4B). The canal wall, including the one surrounding the dense cell cluster and the front edge, was distinctly labeled by antibodies against the aggrecan-degradation products VDIPEN and NITEGE (Fig. 4C), suggesting that the cartilage matrix was being degraded during the advancement of the cartilage canals. H&E staining of paraffin sections confirmed that such space was composed of matrix and remnants of surrounding dead chondrocytes (Fig. 4D). Thus, this “empty” space appears to represent a transitional zone of cartilage matrix degraded by neighboring chondrocytes into which canal cells migrate.
TomatoH Cells Secret VEGF to Stimulate Endothelial Cell Migration
The above observations led us to hypothesize that periarticular TomatoH mesenchymal progenitors play a leading role in initiating cartilage canal formation by promoting migration of endothelial cells into the epiphyseal cartilage. To further test this possibility, we determined the distribution of VEGF in P5 samples. Interestingly, TomatoH cells located at sites of invagination strongly expressed VEGF (Fig. 5A) and topographically, were distributed ahead of VEGFR-2+ endothelial cells migrating behind them (Fig. 5B). Neighboring epiphyseal chondrocytes did not have detectable levels of VEGF expression at this stage, while TomatoH cells at the front edge and inside the canals remained VEGF+ at later stages (Fig. 5C). In vitro, we found that culture media conditioned by TomatoH cells markedly stimulated the migration of bone marrow endothelial progenitors as seen with bone marrow mesenchymal progenitors. This stimulatory effect was greatly attenuated by SU5416, a VEGFR-2 inhibitor (Fig. 5D). Taken together, our results suggest that TomatoH mesenchymal progenitors originating in the periarticular region stimulate migration of endothelial progenitors and formation of cartilage canals, with VEGF as a candidate mediator.
Figure 5.
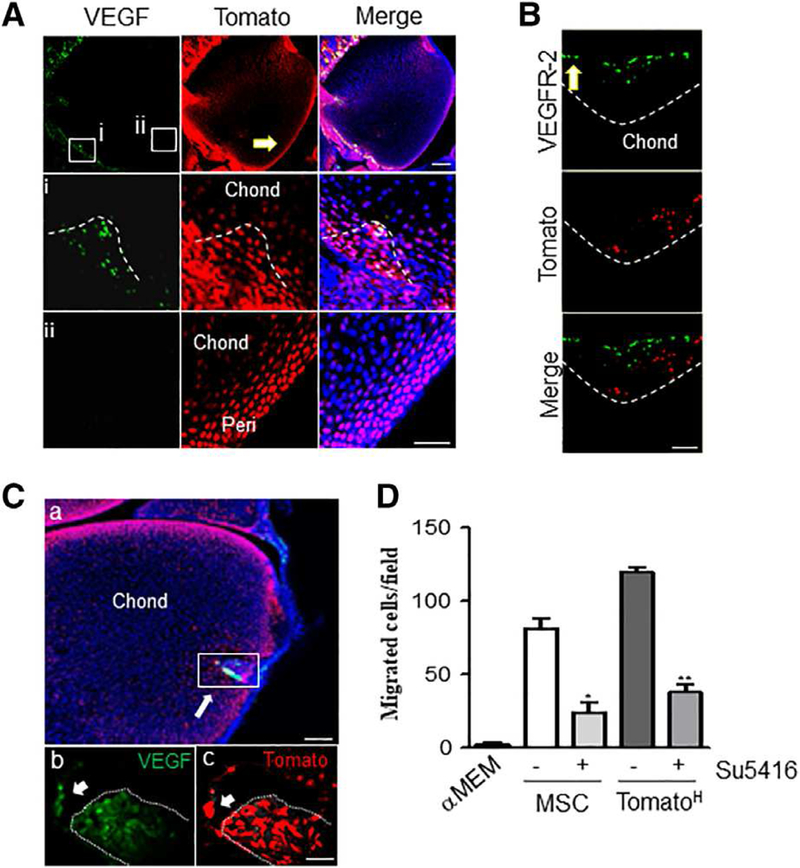
Secondary ossification center canal initiation and progression is led by perichondrial-derived Tomato cells independent of epiphyseal chondrocyte-derived vascular endothelial growth factor (VEGF) expression. (A): At P5, VEGF expression is confined to cells forming the canal and the neighboring periarticular region (Ai) while no signal was detected along the periarticular surface away from the canal (Aii). Dotted line depicts canal invasion site. Bar = 100 μm. (B): These leading cells are closely followed by a wave of VEGFR-2+ cells. Dotted line depicts canal front. Bar = 100 μm. (Ca): At P8, VEGF is expressed robustly by most cells of the canal, as well as a few chondrocytes in front of the advancing canal. Bar = 200 μm. (Cb, Cc): Magnified insert from (Ca). Arrow denotes TomatoH cells in front of canal positive for VEGF staining. Bar = 20 μm. (D): Trans-well migration of endothelial cells was conducted using conditioned media from mesenchymal stem cells orTomatoH cells and treatment with the VEGFR-2 inhibitor Su5416. TomatoH cells were found to induce endothelial cell migration in a VEGR-2-dependent manner. Graphs represent average values ± SEM. Data analyzed by repeat measures two-way analysis of variance with Bonferroni’s post-test (*, p <.05; **, p <.01; n = 3).
Angiogenic Blood Vessels Precede Mesenchymal Progenitors During SOC Expansion
A mature vascular network within the SOC became evident at P9 when the canals reached the center of the epiphyseal cartilage (Fig. 6A). Interestingly, and in contrast to previous stages, vessels preceded TomatoH cells in invading neighboring cartilage during SOC expansion at P12 (Fig. 6B). These intruding vessels at the junction between SOC marrow space and epiphyseal cartilage contained numerous tip cells with characteristic filopodia visualized by CD31 staining (Fig. 6C). These data suggested that angiogenesis played a dominant role in forming new vessels and leading mesenchymal progenitors to migrate at this late stage, similar to what is known to occur in POC formation. Consistent with these changes, we observed that chondrocytes at the canal front edge started to express VEGF at these late stages (Fig. 6D) and surrounding hypertrophic chondrocytes expressed a detectable level of VEGF during SOC expansion (Fig. 6E). It is therefore possible that this switch in VEGF expression pattern contributes to the reversed migration sequence of mesenchymal progenitors and endothelial cells.
Figure 6.
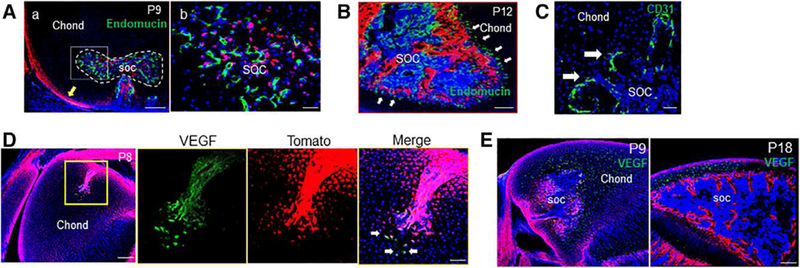
The transition from canal invasion to secondary ossification center (SOC) expansion is associated with altered sources of vascular endothelial growth factor (VEGF) and a spatial reorganization of blood vessels. (A): By P9, some mature vessels were visualized and present along the front of the expanding SOC canal (dotted line). Yellow arrow denotes periarticular surface. Bar = 200 μm (Aa), 50 μm (Ab). (B): By P12, endothelial cells were found to be ahead of the expanding TomatoH bone compartment (arrows). Bar = 200 μm. (C): These vessels were associated with a high density of angiogenic sprouts (arrows) along the invading front. Bar = 50 μm. (D): This change to a blood vessel-led SOC expansion was associated with a change in VEGF expression, with a few TomatoH and TomatoL cells out front of the invading canal (arrows) visibly expressing VEGF at P8. Bar = 200 μm, 50 μm. (E): At P9 and P18, VEGF production was detected by the majority of hypertrophic chondrocytes throughout the remaining epiphyseal chondrocytes. Bar = 200 μm.
Chondrocyte Mineralization Occurs During SOC Expansion, but Not During Canal Invasion
Traditional immunohistochemistry requires decalcification of bone tissues. However, whole mount staining can be performed on undecalcified samples, allowing us to visualize newly mineralized tissue and antibody staining simultaneously through cal-cein labeling. Using this approach, we analyzed the relationship among tissue mineralization, vessel formation, and mesenchymal progenitors during SOC formation. We observed mineralization inside the expanding SOC, but not inside or around the cartilage canal (Supporting Information Fig. S4A), indicating that TomatoH cells remained as progenitors during early stages of SOC development and that neither TomatoH cells nor chondrocytes surrounding the canals had terminally differentiated yet. During SOC expansion, we found that hypertrophic chondrocytes immediately next to the SOC junction were positive for calcein staining along their longitudinal intercellular axis and that staining extended inside the SOC in roughly parallel shapes (Supporting Information Fig. S4B). Interestingly, at P12, vessels were growing next to these mineralized surfaces but always lagged behind, suggesting that mineralization patterns have roles in vessel growth directionality. Consistent with the aforementioned data, TomatoH cells further lagged behind vessels. The same pattern was also observed at the junction of growth plate and primary spongiosa during POC. Later, the longitudinal calcein labeling at the SOC junction was gradually shortened and eventually, around P18, turned into a transverse line that appeared to completely block further ingression of vessels (Supporting Information Fig. S4C), resulting in the completion of SOC expansion and establishment of the articular cartilage.
Gene Expression Profiling of Canal Invasion Shows Altered Regulation of Genes Involved in Cell Migration and Differentiation
To identify regulatory pathways involved in canal initiation and progression, we isolated the invagination sites/early cartilage canals (Canal), chondrocytes neighboring the canal (CNC), the neighboring periarticular region (Peri), and the underlying epiphyseal chondrocytes (Chond), respectively, from P5.5 pups via LCM for gene profiling (Fig. 7A). Successful separation of tissues was confirmed by expression of matrilin-1, an epiphyseal chondrocyte marker, along with mature chondrocyte genes such as Col2/9/11, Aggrecan, and Hpalnl in Chond, while Peri expressed elevated levels of the matrix markers Col1/3/8/14/15, Fn1, and Eln (Supporting Information Table S2). Pairwise comparison of the Peri population with Canal, CNC or Chond populations revealed a total of 1,881 genes up or downregulated in the latter three groups relative to Peri (Fig. 7B). The large number of regulated genes in Canal relative to Peri pointed to extensive changes in periarticular cells at this early stage of canal initiation. Principle component analyses confirmed clustering of sample replicas and indicated much more changes occurred in Canal versus Peri than CNC versus Chond (Fig. 7C). Hierarchical clustering analysis of the most highly regulated genes revealed that transition from periarticular region to canal was associated with strong upregula-tion of many genes (Fig. 7D). Differential gene expression analyses for CNC relative to Chond (Supporting Information Table S3) revealed significant upregulation of several MMPs, specifically MMP9 and 13, and a drastic downregulation of articular cartilage surface marker PRG4 (Fig. 7E). Gene analysis, along with GO term enrichment (Fig. 7F) suggested that a primary role for CNC in SOC canal progression was to limit cell proliferation and differentiation, degrade the ECM and prepare for apoptosis to make way for the advancing canal.
Figure 7.
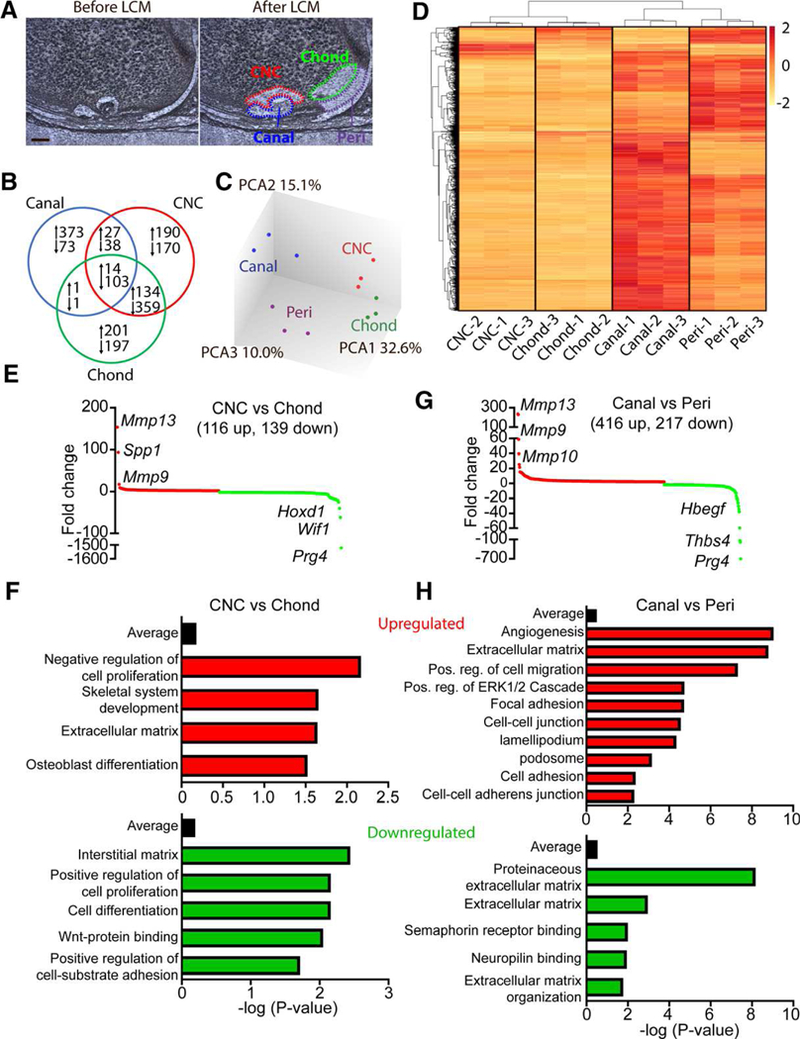
Gene expression analyses associated with canal initiation and progression. (A): Laser capture microdissection was used to dissect the canal, chondrocytes near the canal (CNC), periarticular region (peri) and underlying chondrocytes (chond). Bar = 50 μm. (B): Gene expression analyses, with peri as the control tissue, showed 1,881 significantly regulated genes across the three isolated tissues. (C): PCA confirmed clustering of individual samples and showed notable changes between the peri and canal whereas more modest changes were observed between the CNC and chond. (D): Heat map of the 1,200 most regulated genes show robust gene expression changes during canal formation. (E): Differential expression profiles of significantly regulated genes from CNC relative to epiphyseal chondrocytes. Red denotes upregulated genes whereas green represents downregulated genes. Some of the most highly regulated genes have been listed. (F): Enrichment analysis of GO terms up and down-regulated in the CNC versus epiphyseal chondrocytes. (G): Differential expression profiles of significantly regulated genes from the canal relative to the periarticular region. Red denotes upregulated genes whereas green represents downregulated genes. Some of the most highly regulated genes have been listed. (H): Enrichment analysis of GO terms up and downregulated in the canal versus the periarticular region (n = 3).
Like the CNC, the most regulated genes within the Canal versus Peri remained MMPs and Prg4 (Fig. 7G; Supporting Information Table S4). However, unlike the CNC, GO terms enriched for in the canal showed strong upregulation of angiogenesis, cell adhesion and migration, and kinase signaling, and downregulation of semaphorin and neuropilin signaling (Fig. 7H). The presence of ECM GO terms in both up and downregulated groups suggests that matrix remodeling and protein content may play an important role in regulating canal progression and cell fate within the canal. Genes associated with significantly regulated KEGG pathways in the canal, relative to the periarticular region, were further analyzed for linked GO term clusters (Supporting Information Fig. S5A). Large GO term clusters were observed for myeloid lineage cell regulation and vascular development, consistent with our immunofluorescent results. In addition, several GO term clusters were observed in the regulation of cell migration, adhesion, and cytoskeletal reorganization, mesenchymal cell development, and PDGFR signaling, along with upregulation of several kinase pathways, including Akt, MAPK, PI3K and ERK½ signaling cascades. Immunofluorescence confirmed a robust upre-gulation of Akt activation (p-Akt) specifically in the initiating SOC canal (Supporting Information Fig. S5B). These extensive pathway modifications suggest the possibility that endogenous signals drive periarticular cells to invade and form the canal rather than external signals derived from underlying chondrocytes to attract or initiate periarticular cell invasion. A search for potential signaling pathways which could regulate canal initiation and invasion revealed several key genes in the EGFR, semaphorin, TGFβ, WNT, and delta/notch pathways significantly regulated in the canal versus the periarticular region (Supporting Information Table S5).
DISCUSSION
It is well accepted that POC formation is initiated by hypertrophic chondrocytes in the diaphysis that secrete VEGF for recruiting blood vessels accompanied by osteoprogenitors. Using animal models that differentially mark periarticular chondrocytes and underlying epiphyseal chondrocytes, we provide evidence here that SOC initiation may occur via distinct mechanisms. Our data suggest that the periarticular surface contains undifferentiated mesenchymal progenitors eventually forming epiphyseal bone. SOC development is initiated by invasion of these progenitors into underlying cartilage rather than vascular invasion as in POC formation. At the time of SOC initiation, those progenitors, but not surrounding chondrocytes, express VEGF that would be important to recruit individual endothelial cells (vasculogenesis) and existing vessels (angiogenesis) and establish cartilage canals through which mesenchymal progenitors, vessels, and hematopoietic cells migrate into the center of epiphyseal cartilage. Later, during cartilage canal advancement and SOC expansion, these periarticular mesenchymal progenitors undergo proliferation, eventually giving rise to all cell types of the mesenchymal lineage within the forming epiphyseal bone compartment.
These periarticular cells, hereby identified by their higher Tomato fluorescence (TomatoH) compared with epiphyseal chondrocytes (TomatoL), express minimal levels of chondrogenic or osteogenic markers both in vivo and in vitro. Using high resolution confocal microscopy on thick sections, we observed that TomatoH cells are the first to migrate inside cartilage at invagination sites and represent the majority of proliferating cells migrating into the center of cartilage during cartilage canal advancement. Interestingly, in contrast to TomatoL cells, the TomatoH cells clearly show multilineage potential in culture. In vivo, all mesenchymal lineage cells, including osteoblasts, osteo-cytes, and adipocytes in subchondral bone region are TomatoH, suggesting that they are likely derived from TomatoH periarticular cells. Although tamoxifen-induced Col2ER/Tomato and GlilER/ Tomato mouse models further strengthen this conclusion, we still cannot completely rule out that progenitors from neighboring joint tissues or circulation migrate into cartilage canal and eventually reconstitute the entire mesenchymal lineage cells in the subchondral bone. Future in vivo longitudinal fluorescence microscopy could clarify this issue. Note that TomatoH cells also existed in the groove of Ranvier and the resting zone of growth plate, two sites that are known to harbor long-lived mesenchymal progenitors during development [23,24]. Considering that mesenchymal lineage cells in the metaphysis come from perichondrium during POC formation, it is reasonable to assume a similar location of source cells in SOC formation.
One question remaining is why our Col2/Tomato mice, while labeling all cells of the epiphyseal region, showed increased fluorescent intensity in sites known to harbor progenitor cells, including the periarticular region. One possibility is the altered metabolic activity of these progenitors, resulting in either greater Tomato protein production or a reduced protein catabolism. Also, likely a contributing factor is the relatively small cell size and high cell density present at the periarticular region relative to the underlying hypertrophic chondrocytes. These factors could concentrate the fluorescent signal into a smaller region, thus increasing the detectable Tomato fluorescence. Interestingly, we observed a similar phenomenon in the growth plate where resting chondrocytes are TomatoH but the rest of the growth plate chondrocytes are TomatoL. Another possible explanation is that epigenetic changes in chondrocytes result in different expression patterns of Tomato in those cells.
Mesenchymal condensation occurs when previously dispersed mesenchymal cells gather together to differentiate into a single tissue type. Although this process is critical for the formation of many organs including tooth, cartilage, bone, muscle, tendon, kidney, and lung, the boundary cells seem to be mostly studied in skeletal development. Although all periarticular cells surrounding epiphyseal cartilage have high Tomato fluorescence, we normally only observed 2–3 cartilage canals per bone. There are two likely explanations for this phenomenon. One is that only a small portion of TomatoH periarticular cells are mesenchymal progenitors; the cells would not be evenly distributed across the periarticular region, but would rather be enriched at certain locations that initiate invagination. The other explanation is that all TomatoH cells in periarticular region have the same progenitor ability, but unknown stochastic events would activate only certain cells and at only certain locations to initiate or inhibit cartilage canal formation. Future studies are required to distinguish between these two possibilities and understand the signaling mechanisms that initiate SOC formation.
The crosstalk between stem cells and blood vessels plays a fundamental role in tissue development and regeneration. In POC formation, invading vessels express MMPs that degrade chondrocyte ECM and recruit hematopoietic cells to establish marrow cavity [25,26]. Although mature Col1-expressing osteoblasts remain outside the neo-forming POC and mineralize the bone collar, Osx-expressing osteoprogenitors move into the incipient POC by association with invading vessels or migration along tracks created by vessel-derived MMP action [7]. In contrast to the POC, initiation of SOC formation appears to be initiated and driven by expansion and invasion of undifferentiated periarticular TomatoH cells. VEGF is known for its crucial function in SOC formation [27]. Interestingly, these TomatoH mesenchymal progenitors are a primary source of VEGF expression and are likely responsible for early vessel infiltration during invagination and cartilage canal initiation. Our results are consistent with a previous finding that VEGF is mainly detected at the periarticular region at the invagination site and in certain areas of resting cartilage immediately below it, but not in the center of epiphyseal cartilage [28]. At that time, it was concluded that resting chondrocytes in close proximity to the apical tip of canals express VEGF to drive vessel infiltration. Our data indicate that instead, periarticular mesenchymal progenitors express VEGF, move into the resting cartilage and promote vascular formation in the canals. Of course, we cannot completely rule out the possibility that VEGF is expressed in underlying epiphyseal chondrocytes but not detectable by our staining procedures. Hypertrophic chondrocytes at subsequent stages would become the major source of VEGF production for further cartilage canal advancement and SOC expansion. Although our data suggest that these periarticular cells are responsible for the initial invasion of the SOC canal, we also cannot discount a potential role for the overlying vasculature in canal initiation, possibly through secreted factors which could help distinguish those periarticular regions destined for canal formation from those remaining on the surface. Furthermore, functional assays using conditional knockout mouse models are required to elucidate the differential roles of mesenchymal progenitors and vessels in early stages of canal formation.
In addition to angiogenesis, our data suggest that vasculo-genesis also takes place during SOC development, involving individual endothelial cells or clusters that enter the incipient canals and give rise to luminized vessels de novo. Vasculogen-esis is the formation of blood vessels by differentiation of (hem)angioblasts and occurs in embryos, in the uterus during menstruation cycle and in wound repair [29–31]. Additional work is required to confirm these results and explore whether this process is unique to SOC or whether vasculoneogenesis may also occur during POC formation as well.
Recently, studies based on lineage tracing revealed that at least some of the terminally differentiated, hypertrophic chondrocytes within the growth plate could escape death and trans-differentiate into osteoblasts and osteocytes in the primary spongiosa [32–34]. In line with this idea, a recent study proposed that epiphyseal growth plate chondrocytes are able to transdif-ferentiate into osteoblasts in the resulting marrow space using inducible Col2-CreER reporter mice [35]. However, our results here, as well as those in published reports [15,36], have shown that the Col2-CreER system also labels multipotent progenitors. Moreover, our GliER/Tomato mice in which epiphyseal chondrocytes are not labeled, clearly display numerous Tomato+ bone forming cells in subchondral bone. Also of note is the presence of a preresorption gap between invading canal and underlying resting chondrocytes with no nucleated cells at an early canal invasion stage. Morphologically, we observe that the surrounding resting chondrocytes undergo apoptosis, a conclusion that has been proposed in previous reports using caspase 3 staining [28] and electron microscopic examination [37]. The space barrier, together with the death of surrounding chondrocytes, strongly suggests that transdifferentiation from chondrocytes to mesenchymal progenitors or osteoblasts is an unlikely event.
To contribute to a better understanding of signaling pathways regulating SOC invasion, we conducted gene expression analyses on LCM samples. Canal initiation could conceivably be regulated from two distinct mechanisms. The first one is that internal signals within the periarticular surface trigger cell proliferation and invasion. The second one is that signals from the underlying chondrocytes stimulate invasion of periarticular cells into the underlying tissue. Although we cannot discount a minor role of the underlying chondrocytes, cells at the invagination site showed robust transcriptional changes compared with their periarticular donors, strongly suggesting that an intrinsic signal from within the periarticular region is associated with canal initiation.
MMPs play a critical role in SOC formation. Interestingly, our data showed that Mmp14 is expressed at uniformly high levels throughout the four regions. In contrast, Mmp9 and Mmp13 were expressed at relatively low levels in the periarticular region and epiphyseal chondrocytes but was prominently upregulated 81- and 235-fold, respectively, in the CNC and cartilage canal, during SOC initiation. These data provide an explanation for previous reports showing that SOC formation was completely blocked in Mmp14 null mice [38,39], was severely delayed in Mmp9 and 13 double-knockout mice, but was not altered in Mmp9 or 13 single knockout mice [40]. Our results also bring up an interesting role for cell migration, adhesion and communication. Several genes involved with these pathways, such as Cxcr4, vinculin (Vd), and Notch3, were found to be significantly upregulated within the canal. Finally, due to limitations in the timing of canal collection for LCM experiments, the importance of those novel signaling pathways identified in our assay remains to be elucidated in terms of whether they function in invagination, early canal progression or both. Among them, EGFR, TGFp, Semaphorin, Wnt, and delta-notch pathways warrant further scrutiny for their potential involvement in periarticular canal formation and progression.
CONCLUSION
Our data provide strong evidence that the epiphyseal periarticular region contains undifferentiated, multipotent progenitors that are responsible for initiating SOC canal formation ahead of the invading vasculature. In past decades, MSCs have rapidly emerged as an investigational therapy for treating cartilage degeneration diseases such as osteoarthritis [41]. Since periarticular mesenchymal progenitors are located at future articular cartilage site, it would be intriguing to test their potential therapeutic application in this common joint disease and compare efficacies with MSC from other tissues, such as adipose and bone marrow. It would also be interesting to establish what relationships exist between these progenitors and the joint interzone cells responsible for generating articular cartilage [4], providing further insights into their repair and regenerative potentials.
Supplementary Material
SIGNIFICANCE STATEMENT.
Long bone development involves the embryonic formation of a primary ossification center (POC) in the incipient diaphysis followed by postnatal development of secondary ossification centers (SOCs) at each epiphysis. In the presented work, genetically modified fluorescent reporter mice were used to show a layer of highly fluorescent cells along the epiphyseal surface contain mesenchymal stem cells responsible for initiating SOC formation, recruiting surrounding vessels and establishing all mesenchymal lineage cells within this bone compartment. The finding reveals an important role of the periarticular region during skeletal development and highlights its potential use as a source of multipotent progenitor cells for therapeutic treatment.
ACKNOWLEDGEMENTS
We are grateful to Raymond Galante and Xiaofeng Guo from the Center for Sleep and Circadian Neurobiology for the use of their LCM system and Xi Jiang and Nathaniel Dyment from the Department of Orthopedic Surgery for their technical expertise in LCM use and gene analysis. We are also grateful to Hetty Rodriguez from the Molecular Profiling Center for her help in performing the microarray experiment. We also thank Dr. John S. Mort at Shriners Hospital for Children (Montreal, Canada) for kindly providing anti- aggrecan cleavage fragment antibodies. This study was supported by NIH grants NIH/NIAMS R01AR066098, R01DK095803 (to L.Q.), P30 AR069619 (to Penn Center for Musculoskeletal Disorders), and NSFC grant 81702157 (to WT).
REFERENCES
- 1.Kronenberg HM Developmental regulation of the growth plate. Nature 2003;423: 332–336. [DOI] [PubMed] [Google Scholar]
- 2.Long F, Ornitz DM Development of the endochondral skeleton. Cold Spring Harb Per-spect Biol 2013;5:a008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall BK, Miyake T All for one and one for all: Condensations and the initiation of skeletal development. Bioessays 2000;22:138–147. [DOI] [PubMed] [Google Scholar]
- 4.Decker RS, Koyama E, Pacifici M Articular cartilage: Structural and developmental intricacies and questions. Curr Osteoporos Rep 2015; 13:407–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pacifici M, Koyama E, Iwamoto M et al. Development of articular cartilage: What do we know about it and how may it occur? Connect Tissue Res 2000;41:175–184. [DOI] [PubMed] [Google Scholar]
- 6.Long F, Chung UI, Ohba S et al. Ihh signaling is directly required for the osteoblast lineage in the endochondral skeleton. Development 2004;131:1309–1318. [DOI] [PubMed] [Google Scholar]
- 7.Maes C, Kobayashi T, Selig MK et al. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev Cell 2010;19:329–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blumer MJ, Longato S, Fritsch H Structure, formation and role of cartilage canals in the developing bone. Ann Anat 2008;190:305–315. [DOI] [PubMed] [Google Scholar]
- 9.Ortega N, Behonick DJ, Werb Z Matrix remodeling during endochondral ossification. Trends Cell Biol 2004;14:86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xing W, Cheng S, Wergedal J et al. Epiphyseal chondrocyte secondary ossification centers require thyroid hormone activation of Indian hedgehog and osterix signaling. J Bone Miner Res 2014;29:2262–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Zhu J, Li Y et al. Epidermal growth factor receptor (EGFR) signaling regulates epiph¬yseal cartilage development through beta-cate-nin-dependent and -independent pathways. J Biol Chem 2013;288:32229–32240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dao DY, Jonason JH, Zhang Y et al. Cartilage-specific beta-catenin signaling regulates chondrocyte maturation, generation of ossification centers, and perichondrial bone formation during skeletal development. J Bone Miner Res 2012;27:1680–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Menendez A, Fong C et al. IGF-I signaling in osterix-expressing cells regulates secondary ossification center formation, growth plate maturation, and metaphyseal formation during postnatal bone development. J Bone Miner Res 2015;30:2239–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ovchinnikov DA, Deng JM, Ogunrinu G et al. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis 2000;26:145–146. [PubMed] [Google Scholar]
- 15.Ono N, Ono W, Nagasawa T et al. A subset of chondrogenic cells provides early mesenchymal progenitors in growing bones. Nat Cell Biol 2014;16:1157–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yasuhara R, Ohta Y, Yuasa T et al. Roles of beta-catenin signaling in phenotypic expression and proliferation of articular cartilage superficial zone cells. Lab Invest 2011;91:1739–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao Y, Zhang P, Yuan B et al. Pravastatin protects against avascular necrosis of femoral head via autophagy. Front Physiol 2018;9:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang da W, Sherman BT, Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 19.Huang da W, Sherman BT, Lempicki RA Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bindea G, Mlecnik B, Hackl H et al. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009;25:1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metsalu T, Vilo J ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heat-map. Nucleic Acids Res 2015;43:W566–W570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kavanagh E, Ashhurst DE Development and aging of the articular cartilage of the rabbit knee joint: Distribution of biglycan, dec-orin, and matrilin-1. J Histochem Cytochem 1999;47:1603–1616. [DOI] [PubMed] [Google Scholar]
- 23.Karlsson C, Thornemo M, Henriksson HB et al. Identification of a stem cell niche in the zone of Ranvier within the knee joint. J Anat 2009;215:355–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abad V, Meyers JL, Weise M et al. The role of the resting zone in growth plate chondrogenesis. Endocrinology 2002;143:1851–1857. [DOI] [PubMed] [Google Scholar]
- 25.SivarajKK Adams RH. Blood vessel formation and function in bone. Development 2016;143:2706–2715. [DOI] [PubMed] [Google Scholar]
- 26.Ortega N, Behonick DJ, Werb Z Matrix remodeling during endochondral ossification. Trends Cell Biol 2004;14:86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maes C, Stockmans I, Moermans K et al. Soluble VEGF isoforms are essential for establishing epiphyseal vascularization and regulating chondrocyte development and survival. J Clin Invest 2004;113:188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allerstorfer D, Longato S, Schwarzer C et al. VEGF and its role in the early development of the long bone epiphysis. J Anat 2010;216: 611–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Risau W, Sariola H, Zerwes HG et al. Vascu-logenesis and angiogenesis in embryonic-stemcell-derived embryoid bodies. Development 1988;102:471–478. [DOI] [PubMed] [Google Scholar]
- 30.Ferguson JE 3rd, Kelley RW, Patterson C Mechanisms of endothelial differentiation in embryonic vasculogenesis. Arterioscler Thromb Vasc Biol 2005;25:2246–2254. [DOI] [PubMed] [Google Scholar]
- 31.Tonnesen MG, Feng X, Clark RA Angio-genesis in wound healing. J Investig Dermatol Symp Proc 2000;5:40–46. [DOI] [PubMed] [Google Scholar]
- 32.Yang G, Zhu L, Hou N et al. Osteogenic fate of hypertrophic chondrocytes. Cell Res 2014;24:1266–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang L, Tsang KY, Tang HC et al. Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc Natl Acad Sci USA 2014;111:12097–12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou X, von der Mark K, Henry S et al. Chondrocytes transdifferentiate into osteoblasts in endochondral bone during development, postnatal growth and fracture healing in mice. PLoS Genet 2014;10:e1004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aghajanian P, Xing W, Cheng S et al. Epiphyseal bone formation occurs via thyroid hormone regulation of chondrocyte to osteoblast transdifferentiation. Sci Rep 2017;7:10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chandra A, Lin T, Young T et al. Suppression of sclerostin alleviates radiation-induced bone loss by protecting bone-forming cells and their progenitors through distinct mechanisms. J Bone Miner Res 2017;32:360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roach HI, Clarke NM “Cell paralysis” as an intermediate stage in the programmed cell death of epiphyseal chondrocytes during development. J Bone Miner Res 1999;14:1367–1378. [DOI] [PubMed] [Google Scholar]
- 38.Holmbeck K, Bianco P, Caterina J et al. MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell 1999;99:81–92. [DOI] [PubMed] [Google Scholar]
- 39.Zhou Z, Apte SS, Soininen R et al. Impaired endochondral ossification and angiogenesis in mice deficient in membrane-type matrix metal-loproteinase I. Proc Natl Acad Sci USA 2000;97: 4052–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stickens D, Behonick DJ, Ortega N et al. Altered endochondral bone development in matrix metalloproteinase 13-deficient mice. Development 2004;131:5883–5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong L, Zheng LZ, Qin L et al. Role of mes-enchymal stem cells in osteoarthritis treatment. J Orthop Translat 2017;9:89–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


