Changes in mechanical load, hormones, or metabolic stress provoke remodeling of the actin-based thin filaments within muscle fibers. Solís and Russell show that several signaling pathways converge at the actin-capping protein CapZ to regulate muscle fiber growth in response to mechanical stiffness and neurohumoral signaling.
Abstract
Muscle adaptation is a response to physiological demand elicited by changes in mechanical load, hormones, or metabolic stress. Cytoskeletal remodeling processes in many cell types are thought to be primarily regulated by thin filament formation due to actin-binding accessory proteins, such as the actin-capping protein. Here, we hypothesize that in muscle, the actin-capping protein (named CapZ) integrates signaling by a variety of pathways, including phosphorylation and phosphatidylinositol 4,5-bisphosphate (PIP2) binding, to regulate muscle fiber growth in response to mechanical load. To test this hypothesis, we assess mechanotransduction signaling that regulates muscle growth using neonatal rat ventricular myocytes cultured on substrates with the stiffness of the healthy myocardium (10 kPa), fibrotic myocardium (100 kPa), or glass. We investigate how PIP2 signaling affects CapZ using the PIP2 sequestering agent neomycin and the effect of PKC-mediated CapZ phosphorylation using the PKC-activating drug phorbol 12-myristate 13-acetate (PMA). Molecular simulations suggest that close interactions between PIP2 and the β-tentacle of CapZ are modified by phosphorylation at T267. Fluorescence recovery after photobleaching (FRAP) demonstrates that the kinetic binding constant of CapZ to sarcomeric thin filaments in living muscle cells increases with stiffness or PMA treatment but is diminished by PIP2 reduction. Furthermore, CapZ with a deletion of the β-tentacle that lacks the phosphorylation site T267 shows increased FRAP kinetics with lack of sensitivity to PMA treatment or PIP2 reduction. Förster resonance energy transfer (FRET) probes the molecular interactions between PIP2 and CapZ, which are decreased by PIP2 availability or by the β-tentacle truncation. These data suggest that CapZ is bound to actin tightly in the idle, locked state, with little phosphorylation or PIP2 binding. However, this tight binding is loosened in growth states triggered by mechanical stimuli such as substrate stiffness, which may have relevance to fibrotic heart disease.
Introduction
Muscle adaptation is a response to physiological demand elicited by changes in mechanical load, hormones, or metabolic stress that can combine to have both acute or long-lasting effects. During striated muscle growth, new sarcomeres are added in parallel to provide additional force or in series to return the sarcomere to its optimal resting length (Russell et al., 2000; Mansour et al., 2004; Yang et al., 2016). Cardiac myocytes integrate the mechanical inputs from the extracellular matrix through cell membrane complexes to the myofibrillar cytoskeleton (Danowski et al., 1992; Samarel et al., 2013). Mechanotransduction signaling to trigger muscle growth is an active area of research.
Cytoskeletal remodeling processes in many cell types are thought to be primarily regulated by thin filament formation due to actin-binding accessory proteins (Dabiri et al., 1997; Pollard, 2016), such as the actin-capping proteins that regulate the polymerization and depolymerization of individual filaments (Edwards et al., 2014). The actin cap is a mushroom-like heterodimeric protein (α and β subunits) that binds to the barbed end of an actin filament. In muscle, the caps are named CapZ, because they are found in the Z-disc, where the thin filament polarity is reversed at each sarcomere. Striated muscle uses a large percentage of the body’s energy and its mass is highly regulated, with the default being atrophy. This “use it or lose it” regulation extends to existing proteins that are only assembled and maintained to meet local tension demands. Therefore, regulation is mainly accomplished at the subcellular level by modulation of existing proteins.
Since CapZ plays such a crucial role in the control of thin filament assembly, it is an obvious choice for study of muscle hypertrophy. Both the α and β subunits interact with the thin filament through two main surfaces: the first with the terminal actin subunits (site 1), and the second with a mobile β-tentacle that consists on the hydrophobic side of an amphipathic α-helix at the β subunit C terminus (site 2; Hug et al., 1992; Kim et al., 2010; Edwards et al., 2014). Recent work has shown that the posttranslational modifications in CapZ modulate its capping dynamics. Ectopic expression of dominant-negative PKCɛ in neonatal rat myocytes blunted the phenylephrine-induced increase in CapZ dynamics and concomitantly reduced phosphorylation and acetylation of CapZβ1 (Lin et al., 2016). While much biochemical detail has been reported, the question of the physiological regulation of this capping and uncapping in muscle is not fully understood.
Here, we propose that CapZ acts as an integration node for signaling pathways through phosphorylation, acetylation, and binding of phosphatidylinositol 4,5-bisphosphate (PIP2), which responds to myocyte mechanical straining or neurohormonal chemical activation (Li and Russell, 2013; Li et al., 2014). Previous work established that PIP2 binds to CapZ in the Z-disc (Heiss and Cooper, 1991; Hartman et al., 2009). The lipid signaling through PIP2 was found to interact with an actin-capping protein, a nonsarcomeric form of CapZ, inhibiting its binding to the F-actin barbed end (Edwards et al., 2014). PIP2 interacts with a cationic-rich patch in the actin-capping protein to the actin binding face, site 1 (Heiss and Cooper, 1991; Kim et al., 2007). Note that muscle is highly evolved in its actin-associated capping proteins; for example, the fission yeast (Schizosaccharomyces pombe) capping protein lacks phosphoinositide response due to decreased number of positively charged residues (Kuhn and Pollard, 2007).
Higher local tension is needed by a muscle working in a stiff fibrotic environment than in a healthy soft one. Substrate stiffness varies from 10 kPa in normal heart tissue to over 100 kPa in diseased rat myocardium (Bhana et al., 2010; Fomovsky and Holmes, 2010). Substrate stiffness regulates cardiac myocyte hypertrophy through PIP2-mediated dynamics on CapZ (Li et al., 2016). We hypothesize that CapZ acts as a destination of multiple signaling pathways (e.g., PIP2 binding and PKC phosphorylation) that ultimately regulate actin dynamics in response to neurohumoral and mechanical stimuli. The approach is to culture neonatal rat ventricular myocytes (NRVMs) on substrata with stiffness that resembles normal or pathological cardiac environments. Docking simulations between CapZ and free molecules of PIP2 led to the hypothesis that the β-tentacle is actively involved in stabilizing PIP2 interactions with CapZ. Also, a phosphorylation site in the β-tentacle affected this binding affinity. To test the hypothesis in vivo, the experimental approach was to infect NRVMs using CapZ-GFP with or without its β-tentacle. Signaling pathways were assessed by treatment with neomycin to reduce the PIP2 lipid level in cells or PMA to activate PKC phosphorylation.
Quantitative biophysical techniques tested for PIP2 regulation of the CapZ interactions. FRAP determined the kinetics of binding of CapZ to sarcomeric actin in living muscle cells. Förster resonance energy transfer (FRET) was also used to examine the nanoscale molecular interactions of CapZ and PIP2. Results suggest a major role for PIP2 binding to the β-tentacle of CapZ that is affected by its phosphorylation. Thus, the lipid signaling pathway may contribute significantly to the regulation of actin assembly, ultimately being integrated with other posttranslational modifications of phosphorylation and acetylation to determine the shape and size of heart cells and the gross morphology of the ventricle.
Materials and methods
PIP2 and CapZ docking simulations
The CapZ (α and β subunit) crystal structure (Protein Data Bank accession no. 1IZN) was represented in an energy-minimized state to obtain a CapZ conformation that aims to represent CapZ structure in solution instead of its crystal structure. Energy minimization with the Molecular Modeling Toolkit in UCSF Chimera (Pettersen et al., 2004) used the AMBER ff14 SB force field and explicit representation of hydrogen bonds and charges (Gasteiger partial charge calculation). Modeling of PIP2 binding to the minimized CapZ structure was conducted using Autodock Vina (Trott and Olson, 2010) and the PyRx working interface (Dallakyan and Olson, 2015). CapZ and a single PIP2 molecule (Protein Data Bank ligand no. PT5) were represented in a grid space of x = 70.6 Å, y = 75.0 Å, z = 79.3 Å using a Universal Force Field (Rappe et al., 1992). Each docking simulation used a single PIP2 molecule and the software selected the top nine PIP2 conformers exhibiting the largest change in free energy of binding. This procedure was repeated three times to derive a total of 27 PIP2 docked structures on CapZ for each condition.
For CapZΔC12, amino acids 266–278 corresponding to the β-tentacle in the CapZβ subunit were removed from CapZ, while for CapZΔC26, amino acids 252–278 in CapZβ1 were removed. Phosphorylation of the CapZ in the β-tentacle was introduced in T267 using UCSF Chimera. PIP2 docking simulations with the modified CapZ forms were performed under the same conditions as for full-length CapZ. Since the majority of PIP2 in cells is associated with membranes or liposomes, CapZ simulations are usually done in the presence of a cell membrane (Smith et al., 2006; Kuhn and Pollard, 2007), but here, that restriction was removed. However, actin was not present in the simulation, nor can the noncrystalline amino acids be visualized. Particularly, 6 aa in the C terminus of the CapZβ1 (YIQPDN) subunit were not present in the simulations.
Isolation and culture of neonatal ventricular cardiomyocytes on substrata of varying stiffness, PIP2 levels, and PMA treatment
NRVMs were extracted as described previously (Li et al., 2016). Briefly, hearts obtained from 1–2-d-old Sprague–Dawley rats were minced and then digested with collagenase type II (Worthington). After incubation at 37°C for 10 min with agitation, NRVMs in suspension (10 ml) were collected and added to fetal bovine serum (10 ml). This process was repeated until the tissue was completely digested (usually between five and six repetitions). NRVMs were filtered through a 70-µm nylon sieve, centrifuged, and resuspended in plating media (Louch et al., 2011). Myocytes were plated on glass-bottom dishes modified with fibronectin or on these dishes with a layer of polyacrylamide (PAA) of 10 or 100 kPa that were modified with fibronectin as described previously (Li et al., 2016). Neomycin (500 µM) was applied 30 min before experiments to reduce the PIP2 level (Schacht, 1976; Li and Russell, 2013). PMA (100 nM) was applied 30 min before experiments to activate PKC.
CapZβ1-GFP and CapZβ1ΔC-GFP infection
NRVMs grown in culture for at least 24 h were infected with a CapZβ1-GFP virus or a CapZβ1-GFP virus with its β-tentacle (12 aa in C terminus) deleted (CapZβ1ΔC-GFP). The CapZβ1-GFP and CapZβ1ΔC-GFP DNA constructs were a gift from Dr. Dorothy Shafer (University of Virginia, Charlottesville, VA; Schafer et al., 1995). The CapZβ1-GFP and CapZβ1ΔC-GFP viruses were provided by Dr. Jody Martin (Vector Core, University of Illinois at Chicago, Chicago, IL). Briefly, the plasmids containing the cDNA of CapZβ1-GFP or CapZβ1ΔC-GFP were subcloned into pShuttle-cytomegalovirus AdEasy XL adenoviral vector system (Stratagene) as described previously (Hartman et al., 2009; Lin et al., 2013). Infected NRVMs (multiplicity of infection = 20) were incubated for 1 h at 37°C before substituting the media with maintenance media (Louch et al., 2011). Cells were cultured for 24 h after infection before conducting FRAP or FRET assays.
FRAP
FRAP measurements were made in a confocal light scanning microscope (LSM 880; Carl Zeiss) with controlled temperature (37°C) and CO2 (5%). A single sarcomeric region of interest (ROI) was selected for photobleaching, while a contiguous sarcomere was used as the reference. After an ROI was photobleached, the light intensity in the ROI and the reference were monitored for up to 900 s at 5 frames per second. The characteristic intensity recovery rate, kFRAP, was determined by the equation
| (1) |
where I is the intensity in an reference-weighed ROI, C1 and C2 are preexponential constants, and koff1 and koff2 are the kinetic constants. FRAP kinetics can be modeled with two phases, but only the simple constant was used here to provide the most general information. The amplitude-weighted kinetic constant kFRAP was determined using the following formula:
| (2) |
Immunofluorescence
NRVMs were fixed after extraction of the membrane and cytosol. NRVMs were washed with PBS and treated with the extraction buffer II derived from the ProteoExtract Subcellular Proteome Extraction kit (EMD Millipore) with the addition of protease (no. 539131; Calbiochem) and phosphatase (no. 524625; EMD Millipore) inhibitors. Extraction was conducted for 10 min at 4°C. NRVMs were gently washed with PBS and fixed with a formaldehyde solution (10%) for 10 min at room temperature. Mouse anti-PIP2 (1:100 dilution, no. 2335; Abcam) and rabbit anti-α-actinin (1:50, no. 31346; Cell Signaling Technology) antibodies were incubated overnight at 4°C, while the corresponding anti-mouse AF488 (1:400, no. A21202; ThermoFisher) and anti-rabbit AF594 (1:400, no. 21207; ThermoFisher) were incubated for 1 h at room temperature before applying mounting medium (no. H-1200; Vector Laboratories).
Line scan measurements
Fixed fluorescently labeled NRVMs were analyzed using ImageJ or a customized MATLAB algorithm. Line scans, corresponding to intensity profiles from a straight line along a myofibril, were used to interrogate the relative abundance of PIP2 in the Z-discs. The peak-to-peak intensity ratio was defined as the fluorescence intensity ratio of PIP2 to α-actinin on the Z-discs. Three to five sarcomeres were included in each line scan for a total of three line scans per image.
FRET
FRET interactions between CapZ-GFP and PIP2 were conducted by extracting the cytosol and the membrane and fixing as described above. The primary PIP2 antibody (1:100) was incubated overnight at 4°C, followed by incubation with an antigen-binding fragment F(ab′)2 anti-mouse AF555 (1:400, no. A21425; ThermoFisher) for 1 h at room temperature. This smaller antibody fragment used as the secondary antibody proved to yield a higher and more reproducible FRET response than the larger undigested secondary antibody. After incubation and washing, 10% formaldehyde was applied for 10 min at room temperature before adding the mounting medium. This improved the quality of FRET analysis and the preservation of fluorescent probe localization in the samples. FRET measurements were conducted in a confocal light scanning microscope (LSM 880) at room temperature by the acceptor photobleaching method (König et al., 2006; Mkrtschjan et al., 2018). Briefly, images of the donor (CapZ-GFP) and the acceptor (PIP2 secondary antibody) were captured with alternating, partial photobleaching of the acceptor dye in an ROI of the cell. This ROI constituted approximately one fifth to one half of the cell area, while a reference ROI was concomitantly imaged without alternating acceptor photobleaching. The FRET efficiency (EFRET) was defined by
| (3) |
where D(i), Dr(i), and Db(i) represent the average fluorescence intensity of the donor in the ROI, the reference ROI (acceptor was not photobleached), and the background (no cells present), at a scanning step i, respectively. D(0), Dr(0), and Db(0) represent the fluorescent intensity before the photobleaching sequence began. Extrapolation of EFRET(i) to an acceptor intensity of zero derived the EFRET of the sample.
Statistical analysis
Significance between two groups uses Student’s t tests. Multiple sample comparisons uses ANOVA followed by Fisher’s least significant difference test.
Results
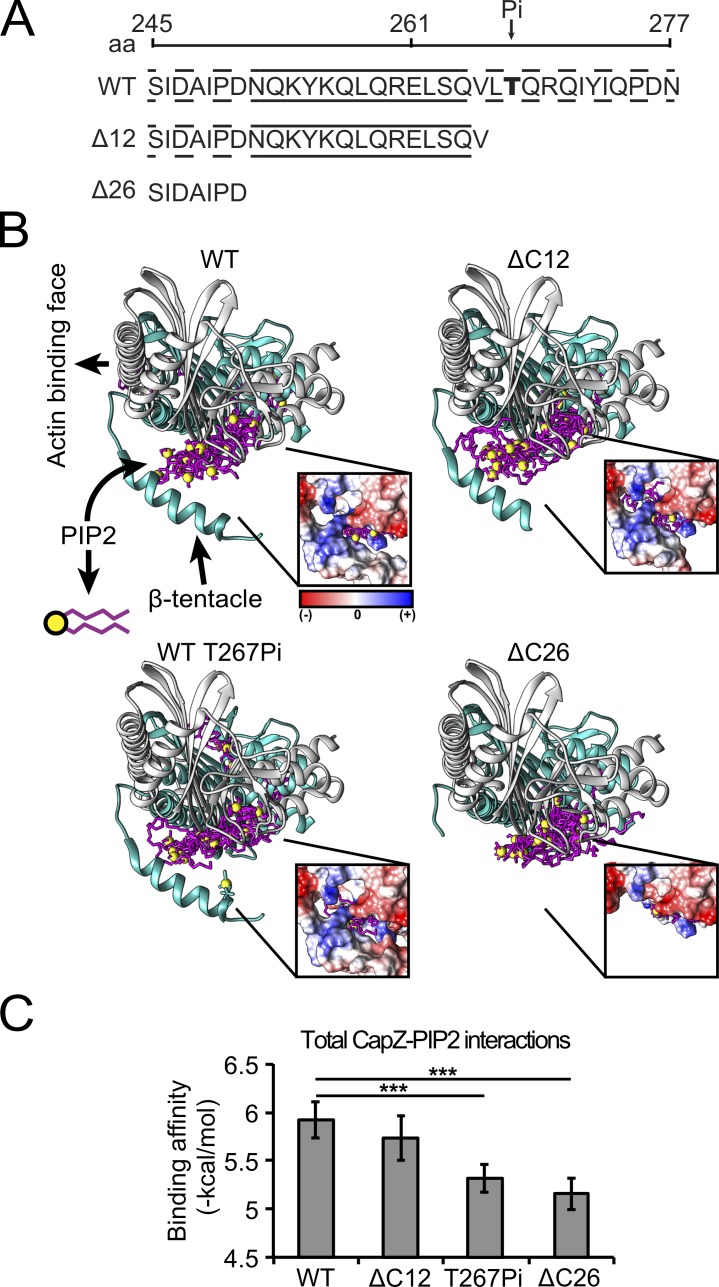
CapZ and PIP2 interactions were studied by docking simulations and alterations of the CapZ structure that include unmodified CapZ (amino acids 1–277), a phosphorylation in T267, and a ΔC12 (amino acids 1–265) and ΔC26 (amino acids 1–251) truncation in the CapZβ1 subunit (Fig. 1 A). CapZΔC26 was used to model the state when the β-tentacle was away from the core structure, which was expected to occur when the β-tentacle is bound to actin (locked state). CapZ and PIP2 docking simulations revealed favorable binding interactions localized in three regions of unmodified CapZ: the actin binding face and two hydrophobic pockets in the α- and β-subunits of CapZ (Fig. 1 B). The most frequent (56%) and energetically favorable interaction was with the aliphatic carbon chains buried in the β-subunit hydrophobic pocket (site 2 of capping protein; Edwards et al., 2014), while the phosphoinositide group interacted with the CapZ–β-tentacle (β-subunit C-terminal region). A positively charged patch at the base of the β-tentacle comprised by K256 and R260 might be involved in interactions with the PIP2 phosphoinositide group (Fig. 1 B, insert). A secondary PIP2-binding cluster was found in the putative actin and PIP2 binding face with less frequency (33%). Incorporation of the phosphate significantly reduced the binding affinity of PIP2 for CapZ (P < 0.001) due to the negative charges that repelled the negatively charged PIP2 molecules (Fig. 1 B, insert). The ΔC26 form, but not the ΔC12 form, significantly reduced the binding affinity of PIP2 to CapZ (P < 0.001). The ΔC26 form eliminates a positively charged patch (K256 and R260) localized at the base of the β-tentacle while in the ΔC12 form is preserved (Fig. 1 B, insert).
Figure 1.
Molecular simulations suggest that close interactions between PIP2 and the β-tentacle of CapZ are modified by phosphorylation at T267. (A) The CapZ C-terminal domain is represented as are a potential T267 phosphorylation site (Pi) and with truncations of the last 12 (ΔC12) or 26 (ΔC26) aa. Across the amino acid sequence, continuous lines represent the α helix, while discontinuous lines are unstructured regions. (B) The CapZ simulation of the crystal structure is oriented with the actin binding face to the left and tentacle below. Actin is not included in the simulation. PIP2 conformers (covalent bonds, magenta; phosphate groups, yellow) were predominantly found in a pocket distal to the actin binding face. This suggested a testable hypothesis that the β-tentacle interacts directly with PIP2. Insert shows the Coulombic surface electrostatic potential of CapZ on the PIP2 and β-tentacle interphase with positive-charge density (blue) and negative-charge density (red). (C) Incorporation of a phosphate on T267 or full deletion of β-tentacle (ΔC26), but not partial deletion of β-tentacle (ΔC12), in the simulation decreased the PIP2-binding affinity to CapZ (mean ± SD, n = 27 PIP2 conformers; ***, P < 0.001).
To test the physiological effects of a CapZ with a β-tentacle deletion (ΔC12 form), NRVMs on glass substrates were infected with viruses encoding CapZβ1-GFP or CapZβ1ΔC-GFP and their dynamics measured by FRAP. Both forms were found to assemble at the Z-disc (Fig. 2 A). Therefore, the β-tentacle does not affect subcellular localization of CapZβ1 as described previously (Lin et al., 2013). The β-tentacle deletion increased CapZβ1 dynamics based on the kinetics of fluorescence recovery (Fig. 2 B). CapZβ1-GFP kFRAP was smaller than CapZβ1ΔC-GFP kFRAP (Fig. 2 C), suggesting that the β-tentacle is required to stabilize CapZ in the Z-disc.
Figure 2.
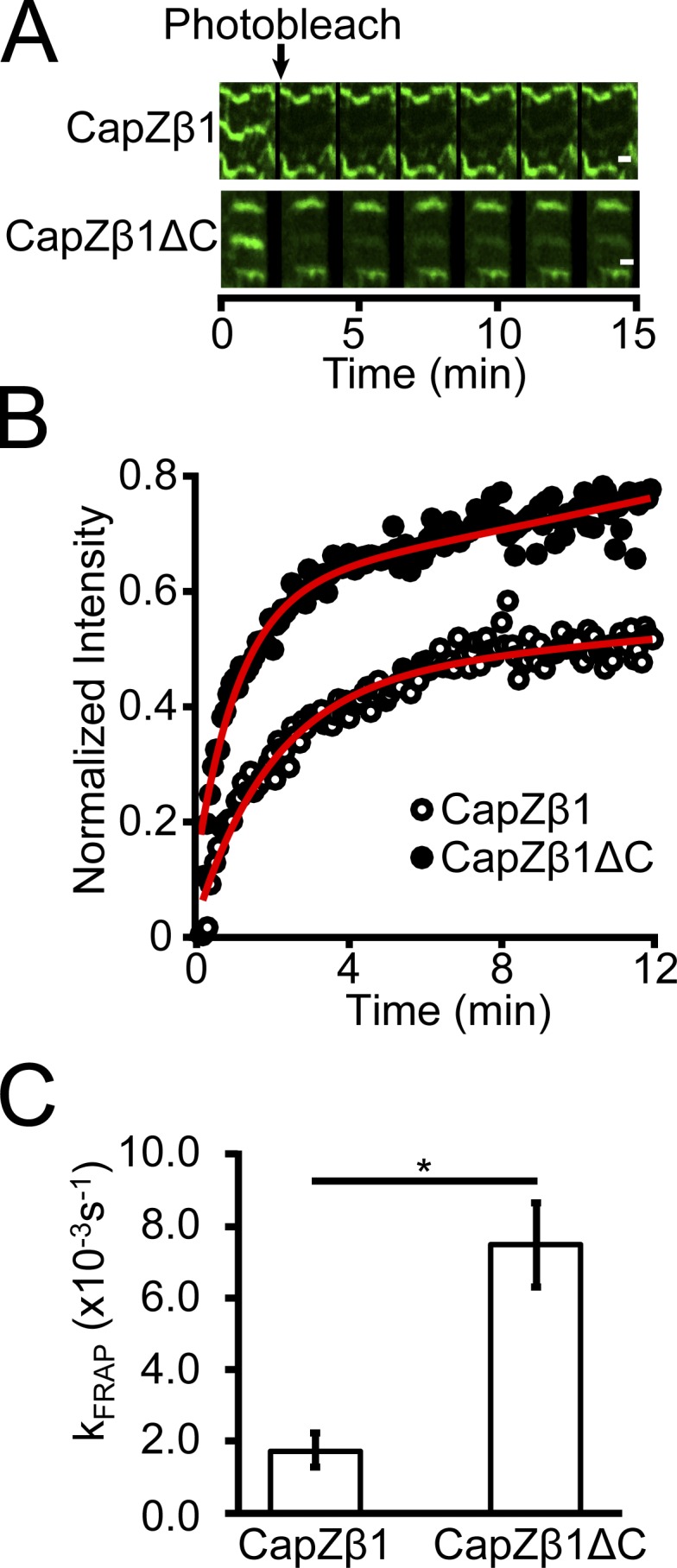
CapZ-GFP dynamics increase with deletion of the β-tentacle determined by the FRAP kinetic constant. (A) Fluorescence recovery over a period of 15 min after photobleaching GFP (green) a single Z-disc in the middle of three sarcomeres in an NRVM grown on glass infected with virus for CapZβ1-GFP (upper) or CapZβ1ΔC-GFP (lower). Scale bar, 1 µm. (B) Fluorescence recovery intensity traces of CapZβ1-GFP or CapZβ1ΔC-GFP are used to calculate the fluorescence recovery rates (kFRAP). (C) Deletion of the β-tentacle increases CapZ-GFP dynamics (mean ± SEM, n = 4; *, P < 0.05).
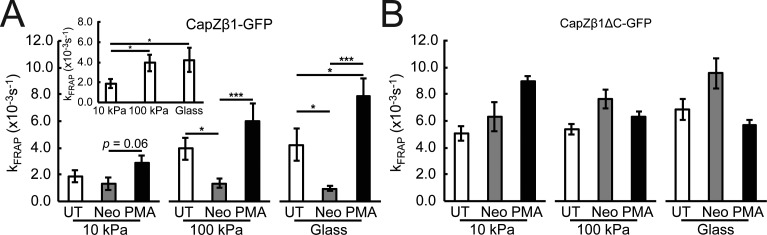
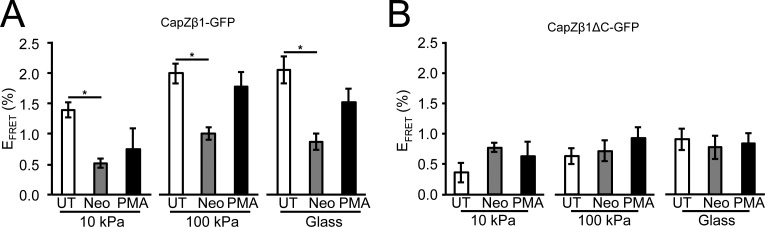
To determine whether mechanical properties affected CapZ dynamics, FRAP was measured on CapZ β1-GFP–infected cells grown on substrata with different stiffness. Results show that kFRAP at 10 kPa was reduced relative to 100 kPa and glass (P < 0.05, P < 0.05, respectively), suggesting that stiffness affects CapZ dynamics (Fig. 3 A, insert). Acute doses of neomycin to cells infected with CapZβ1-GFP reduced kFRAP on 100 kPa and glass, but not on 10 kPa (Fig. 3 A), possibly due to the relatively low baseline level of CapZ dynamics at 10 kPa. Acute exposure to PMA increased CapZβ1-GFP dynamics on glass (pathological stiffness) relative to untreated conditions, while PMA activation was significant for 100 kPa and glass with neomycin. This was consistent with previous observations of increasing NRVM cytoskeletal remodeling by PMA under stiff substrates, but not soft substrates (Pandey et al., 2018). Acute exposure of NRVMs infected with CapZβ1ΔC-GFP to substrata with different stiffness, neomycin, or PMA was insufficient to modify its FRAP dynamics (Fig. 3 B), suggesting that the β-tentacle end region (amino acids 266–277) is sensitive to PIP2 and PKC signaling.
Figure 3.
Stiffness and PKC signaling increase CapZ FRAP dynamics that is diminished by PIP2 reduction, while β-tentacle-truncated CapZ is irresponsive to stimuli. (A and B) FRAP kinetics were measured on NRVMs plated on PAA substrates (10 kPa and 100 kPa) or glass and infected with CapZβ1-GFP (A) or CapZβ1ΔC-GFP virus (B). FRAP kinetics in untreated (UT; white bars) NRVMs or NRVMs treated with 500 µM neomycin for 30 min to reduce the PIP2 level (Neo; gray bars) or PMA for 30 min (black bars). (A) kFRAP is higher on 100-kPa substrates and glass relative to 10-kPa substrates (insert). kFRAP was low on 10-kPa substrates and not reduced further by neomycin. However, kFRAP on stiffer substrates (100 kPa) or glass were initially higher but significantly reduced by neomycin. PMA increased kFRAP on stiff substrates. (B) CapZβ1ΔC-GFP dynamics is insensitive to stiffness, neomycin, and PMA treatments (mean ± SEM, n = 6–16 in A and n = 4–9 in B; *, P < 0.05, ***, P < 0.01).
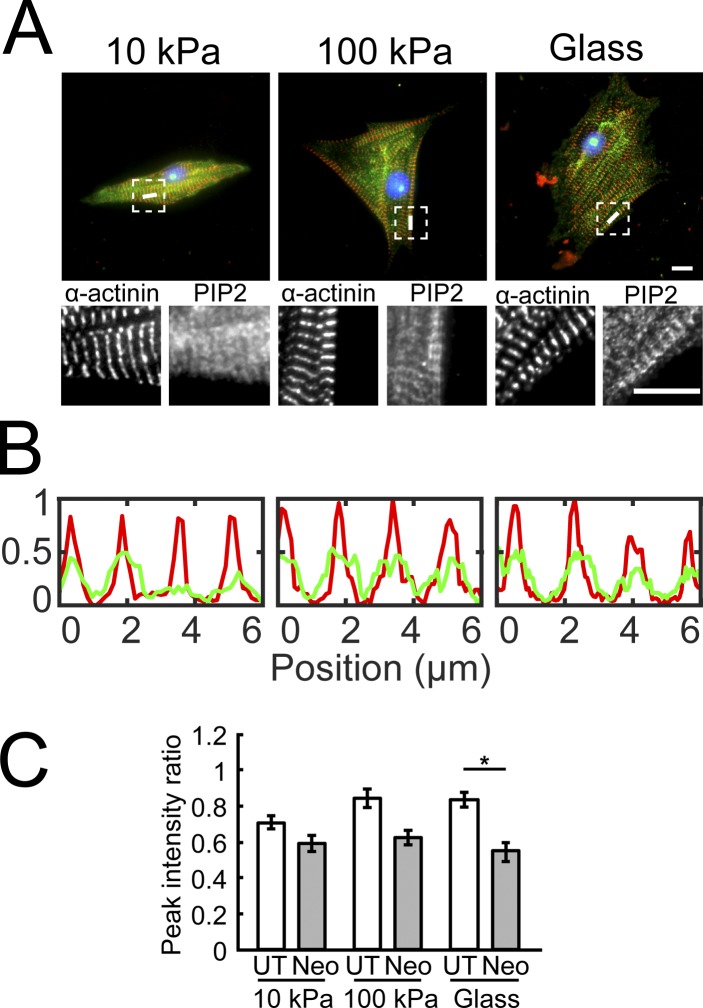
Immunostaining quantified the effects of neomycin and stiffness on PIP2 localization in the Z-discs. Z-disc (α-actinin) and PIP2 localization are shown with immunostaining, line scans, and quantification (Fig. 4, A and B). PIP2 to α-actinin intensity ratios at the Z-discs revealed significant reductions (P < 0.05) only on glass (Fig. 4 C). This suggested the PIP2 levels across Z-discs might change. However, changes cannot distinguish the localization of PIP2 proximal to CapZ or the localization of PIP2 in other Z-disc regions, such as α-actinin (Ribeiro et al., 2014), and costameric assemblies, such as vinculin (Chinthalapudi et al., 2016). Therefore, FRET studies were done to attain molecular specificity. However, note that FRET was done on fixed tissue, which may cause unknown redistribution.
Figure 4.
PIP2 localization in the Z-lines in neonatal myocytes grown on varying stiffness and with neomycin treatment. (A) NRVMs plated on PAA substrates (10 kPa and 100 kPa) or glass were fixed and stained for α-actinin (red) and PIP2 (green). Dashed squares are enlarged in lower panels to show separate α-actinin and PIP2 fluorescence images. Scale bars, 10 µm. (B) Line scans corresponding to white lines in A depict PIP2 (green) localization in the Z-lines represented by α-actinin (red). (C) Peak PIP2 to α-actinin intensity ratio quantifies the effect of neomycin and stiffness on PIP2 localization in the Z-lines (mean ± SEM, n = 15–21; *, P < 0.05).
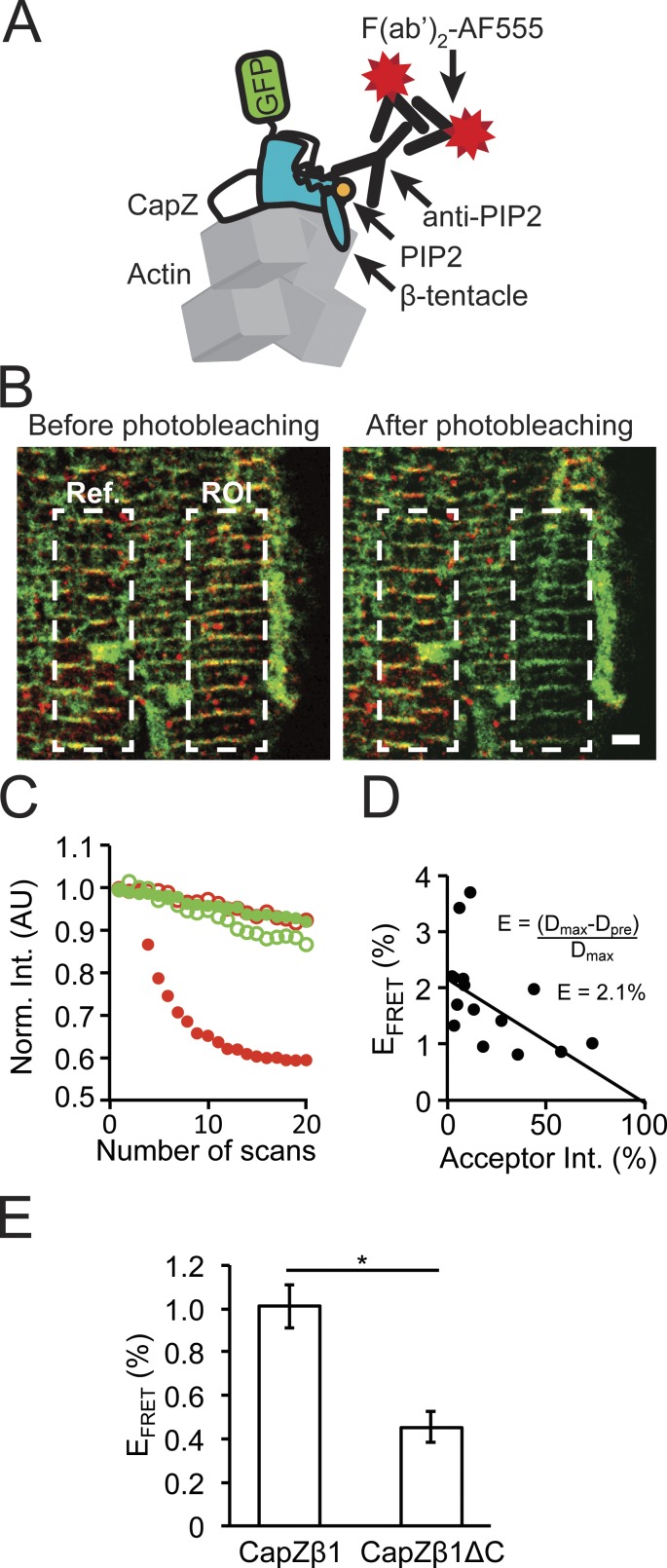
A FRET assay tested the hypothesis that the β-tentacle was required to stabilize CapZ and PIP2 interactions. Using the GFP tag on CapZ as the FRET donor fluorophore and a set of primary and secondary antibodies to PIP2 as the FRET acceptor fluorophore, the FRET response was quantified in NRVMs infected with CapZβ1-GFP or CapZβ1ΔC-GFP (Fig. 5 A). After photobleaching the acceptor signal in a ROI in fixed NRVMs (Fig. 5 B), the donor fluorescence decay in the ROI recovery was assessed as a function of the acceptor fluorescence relative to the reference region that was not photobleached (Fig. 5 C). The FRET response showed a linear trend with the intercept corresponding to the recovered EFRET percentage (Fig. 5 D). Deletion of the β-tentacle decreased CapZ and PIP2 interactions, as suggested by the decrease in EFRET (Fig. 5 E; P < 0.05).
Figure 5.
Direct molecular interaction of PIP2 with CapZ in the Z-disc confirmed by FRET. (A) NRVMs grown on glass were infected with a CapZβ1-GFP or a CapZβ1ΔC-GFP virus, where the GFP (green) acts as the FRET donor. Demembranated, fixed cells were treated with protease and phosphatase inhibitors and a primary to PIP2 antibody and then fluorescently labeled with short secondary antibody (red) acting as the FRET acceptor. (B) Confocal images of NRVMs containing donor (green) and acceptor (red) before (left) and immediately after (right) photobleaching. Dashed lines correspond to the photobleached (ROI) and reference (Ref) zones, respectively. Scale bar, 2 µm. (C) Fluorescence emission traces of donor (green, filled), acceptor (red, filled), donor reference (green, open) and acceptor reference (red, open) during the acceptor photobleaching sequence (20 scans over a period of 2 min). AU, arbitrary units. (D) Linear fit of an example of acceptor intensity to FRET efficiency, where the intercept (2.1%) represents the recovered transfer efficiency (EFRET). (E) Deletion of the β-tentacle decreases CapZ and PIP2 interactions (mean ± SEM, n = 10; *, P < 0.05).
To resolve the relationship between PIP2 and CapZ interactions as a function of substrate stiffness, FRET assays were done on NRVMs plated on 10 kPa, 100 kPa, or glass. EFRET between different substrates were not significantly different (Fig. 6 A). However, acute treatment with neomycin significantly reduced EFRET in all substrates (P < 0.05 10 kPa, 100 kPa, and glass). Treatment with PMA did not change EFRET relative to untreated samples at any stiffness. No change was seen in CapZβ1ΔC-GFP with stiffness, neomycin, or PMA treatments (Fig. 6 B). These results suggest that PIP2 interacts with CapZ located in the Z-disc and that the β-tentacle was involved in stabilizing the CapZ–PIP2 interactions.
Figure 6.
The molecular interactions between PIP2 and CapZ determined by FRET are decreased by PIP2 availability but become nonresponsive with β-tentacle truncation. EFRET of CapZ and PIP2 interactions were determined for NRVMs grown on varying stiffness substrata, untreated (UT; white bars) NRVMs, and NRVMs treated with neomycin to scavenge PIP2 (Neo; gray bars), or treated with PMA to activate PKC (black bars). (A) Stiffness or PMA had no significant effect on EFRET, but neomycin significantly decreased EFRET in all the substrates. (B) The EFRET for CapZβ1ΔC-GFP was low and did not change with stiffness, PIP2 decrease, or PMA treatments (mean ± SEM, n = 12–40 for A and n = 9–21 for B; *, P < 0.05).
Discussion
The goal of this work was to determine how several different mechanotransduction signaling pathways were integrated to cap or uncap CapZ from the actin filament. Molecular simulations suggested that close interactions between PIP2 and the β-tentacle of CapZ were modified by phosphorylation at T267. This was confirmed experimentally by CapZ-GFP dynamics, which increased with a 12-aa deletion of the β-tentacle including T267, as determined by the FRAP kinetic constant. Stiffness increased CapZ FRAP dynamics, but that was diminished by PIP2 reduction Furthermore, at all stiffnesses, the FRAP kinetic constant was higher with activated PKC (PMA treatment) than with low PIP2 levels (neomycin treatment). Interestingly, the β-tentacle–truncated CapZ was irresponsive to substrate stiffness stimuli, PIP2 levels, or activation of phosphorylation. kFRAP on stiffer 100 kPa or glass were initially higher than on 10 kPa and were significantly reduced by neomycin. The immunofluorescent approach for PIP2 localization in the Z-lines in neonatal myocytes was not sensitive enough to detect changes on varying substrate stiffness and with neomycin treatment. Therefore, direct molecular interaction of PIP2 with CapZ in the Z-disc was confirmed by FRET. Interactions were decreased by PIP2 availability but became nonresponsive with β-tentacle truncation. Surprisingly, PKC activation did not increase the molecular interaction of PIP2 and CapZ, which was predicted by the simulation to decrease because the T267 phosphate would repel PIP2. This discrepancy could be due to the lack of free motion of the β-tentacle or the absence of actin during the PIP2 docking simulations. Alternatively, FRET was done on fixed tissue with potential issues. The FRAP data were physiological from living myocytes and suffer from fewer limitations.
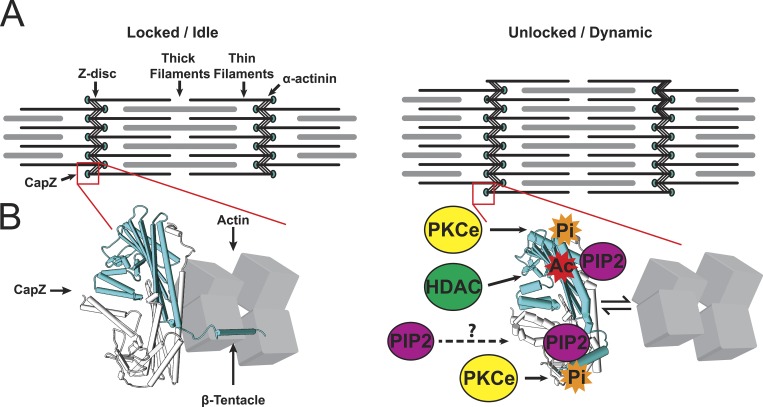
A complex mechanism is proposed for regulation of actin assembly where the CapZ acts as a destination to integrate several signaling pathways by phosphorylation, acetylation, and PIP2 binding. A cartoon (Fig. 7) suggests that the actin binding face (site 1) and the β-tentacle (site 2) cooperate to regulate CapZ docking to the actin filament. This model is based on the new results, other investigations (Kim et al., 2007; Kuhn and Pollard, 2007; Smith et al., 2006), and our prior work (Hartman et al., 2009; Li and Russell, 2013; Lin et al., 2013, 2015, 2016; Li et al., 2014, 2016; Mkrtschjan et al., 2018). We propose the tight binding is loosened in growth states of hypertrophy triggered by stimuli such as mechanical flexing, loading, stiffer substrate, angiotensin II, and phenylephrine. Signaling pathways from these stimuli modify CapZ at the actin binding face (site 1) by PKC phosphorylation (S204), acetylation (K199), or PIP2 binding. Additionally, PIP2 and T267 phosphorylation interact near the β-tentacle (site 2) to modulate CapZ dynamics. Growth may only occur when the CapZ is in unlocked to the dynamic state by a combination of several signaling pathways at both the β-tentacle and the actin binding face.
Figure 7.
Proposed mechanism for regulation of actin assembly by CapZ acting as a destination to integrate several signaling pathways by phosphorylation, acetylation, and PIP2 binding. (A) CapZ caps the actin of the thin filament in the Z disc, shown in a stable sarcomere (left) or one undergoing hypertrophy (right). (B) Based on studies from many groups (Kim et al., 2007; Kuhn and Pollard, 2007; Smith et al., 2006) and the FRET and FRAP data in this paper, CapZ is shown at the molecular scale as bound to actin tightly in the locked/idle state, with little phosphorylation or PIP2 binding. However, this tight binding is loosened in growth states of hypertrophy triggered by stimuli such as mechanical flexing, loading, stiffer substrate, angiotensin II, and phenylephrine. Signaling pathways from these stimuli modify CapZ at the actin binding face by PKC phosphorylation (S204), acetylation (K199), or PIP2 binding. Additionally, PIP2 and T267 phosphorylation interact near the β-tentacle to modulate CapZ dynamics. Growth only occurs when the CapZ is in the unlocked/dynamic state by a combination of several signaling pathways at both the actin binding face and the β-tentacle.
Distribution of PIP2
CapZ modulation presumably involves single PIP2 molecules interacting with CapZ at the barbed end of the actin filament. The source of the PIP2 is shown as a dashed arrow (Fig. 7), because it is controversial. Clearly, single PIP2 molecules are not found in aqueous solution and must be associated with a membrane or liposome or shuttled in hydrophobic pockets of unknown proteins. It is highly unlikely that lipid micelles could be situated within the Z-disc itself due to its complex ultrastructural architecture (Pyle and Solaro, 2004; Lange et al., 2006; Burgoyne et al., 2015). A more likely explanation for PIP2 in the Z-disc is that lipids bind to hydrophobic pockets in many proteins, obviating the need for a lipid bilayer nearby. Other structural proteins are now known to interact with PIP2 at specific domains. In the Z-disc, PIP2 regulated binding of actin and titin Z-repeats simultaneously to α-actinin-2 by interacting with three closely located arginines (Ribeiro et al., 2014). This raises the question of whether a common domain arises from these structures that is selective to binding PIP2 molecules away from cell membranes. This would require a comprehensive study since a vast number of candidate domains could be targets for PIP2 (Chen et al., 2012). However, there are common arginine and lysine-containing motifs that are proposed as PIP2-interacting regions for capping protein (Kim et al., 2007), gelsolin (Cunningham et al., 2001), and villin and profilin (Janmey et al., 1999).
The β-tentacle and PIP2
Prior biochemical and computational work has assumed that PIP2 interacts while bound to a membrane (Smith et al., 2006; Kim et al., 2007; Kuhn and Pollard, 2007). However, in this proposed mechanism, single PIP2 molecules can bind to CapZ bound to actin. Biochemical assays assume that PIP2 is reconstituted into liposomes, and dispersion of single PIP2 molecules is not considered. Tryptophan quenching assays showed that truncating the 34 C-terminal amino acids of actin-capping protein (nonsarcomeric form of CapZβ1) had no effect on its ability to bind PIP2. However, our simulations using a ΔC26 truncation show that PIP2 binds to this region but with the binding affinity reduced by 12%. Experimental findings here appear to contradict our earlier study, which suggested that removing the C-terminal β-tentacle enhances CapZβ1 interaction with PIP2 (Li and Russell, 2013). However, those studies were biochemical, using whole lysate and pull-down of the readily solubilized fractions, which could perhaps be enhanced in the unlocked state after mechanical strain or also in the artifactual unlocked state when the β-tentacle can no longer bind to actin.
The proposed mechanism (Fig. 7) does not invalidate the uncapping mechanism by “wobbling” of the core region while the β-tentacle is still bound to actin (Kim et al., 2007). This is because PIP2 binding to the hinge composed by the CapZ core region and the β-tentacle might facilitate “wobbling” of the core region, which assumes that the β-tentacle remained bound to actin, by stabilizing the unlocked state of CapZ. PIP2 binding to the actin binding face of CapZ is still necessary to stabilize the CapZ core region detached from F-actin while the β-tentacle presumably maintains contact with F-actin. FRAP data using a CapZβ1ΔC-GFP showed that this mutant was insensitive to reduced PIP2 levels with neomycin treatment. Thus, it is possible that the PIP2-induced binding of the CapZ core region relative to the β-tentacle is necessary to initiate uncapping and subsequently to enable PIP2 to bind to the actin face and stabilize the uncapped state.
Effects of stiffness
Substrate stiffness varies in health and disease (Bhana et al., 2010; Fomovsky and Holmes, 2010), and stiffness has been shown to regulate actin assembly and cell size (Li et al., 2016; Mkrtschjan et al., 2018). FRAP data (Fig. 3) showed that at 10 kPa, PIP2 sequestration did not decrease CapZ dynamics, whereas at 100 kPa and glass, PIP2 sequestration did alter CapZ dynamics. The FRET results demonstrated that PIP2 interacted similarly with CapZ for NRVMs grown on all stiffness levels, and neomycin reduced the interaction significantly at 10 kPa and 100 kPa and on glass (Fig. 6). One possible explanation for this discrepancy between the FRET and FRAP data are that posttranslational modifications in CapZ such as phosphorylation and acetylation are involved in modulating CapZ dynamics with stiffness, as well as with acute doses of phenylephrine (Lin et al., 2016). It has been shown that PKCε activity on CapZ was increased at 100 kPa and glass relative to 10 kPa (Mkrtschjan et al., 2018). Furthermore, broad nonspecific drugs like neomycin and PMA are useful but have limits, and there can be cross-talk. Neomycin scavenging can have side effects on PLC/PKC signaling cascades. High-dose neomycin might have side effects other than PIP2 chelation, such as inhibition of the ryanodine receptor (Mead and Williams, 2002). Even a low dose of 1 to 10 µM has affinity for PIP2 (Gabev et al., 1989). Thus, it is possible that increased levels of phosphorylation and acetylation in CapZ on stiff substrates work cooperatively with PIP2 binding to increase CapZ dynamics.
CapZ isoforms
In striated cardiac muscle, there is also a nonsarcomeric CapZ isoform, CapZβ2. The major difference between the sarcomeric CapZβ1 and CapZβ2 is that the latter has a 5-aa longer tentacle with unknown function. Interestingly CapZβ2 is costameric (membrane bound), but when CapZβ2 was overexpressed in transgenic mice, it was also found in the Z-disc with myofibrillar disarray, and the mice suffer cardiomyopathy and heart failure (Hart and Cooper, 1999). Overexpression of CapZβ2 has been shown to decrease myofibrillar PKCε levels and increase myofibrillar Ca2+ sensitivity upon acute stimulus with phenylephrine or endothelin (Pyle et al., 2002, 2006). These results point to a critical role for sarcomere function and development from the localization of the appropriate CapZ isoform.
In summary, the metabolic rate by resting and active cardiac muscle is one of the highest of any tissue, and the default is to disassemble any nonessential myofibrils to conserve energy. The regulation of actin assembly by the barbed-end capping proteins is complex and highly evolved. From many studies, including this paper, a mechanism for integration of multiple signaling pathways is proposed by the actin-capping protein CapZ. To date, there are no human mutations of CapZ, perhaps supporting its essential function for maintenance of thin filament assembly for appropriate control of normal cardiac cell size and heart function. PIP2 binding to CapZ contributes to regulatory pathways of muscle remodeling that depend on mechanical stimuli, which may have relevance to stiffness in heart disease.
Acknowledgments
The authors thank Dr. Dorothy Schafer for providing the CapZβ1-GFP and CapZβ1Δ12C constructs and Dr. Jody Martin for introducing the constructs into viral vectors.
This work was supported by the National Institutes of Health (grant HL-62426 to B. Russell, Project 2).
The authors declare no competing financial interests.
Author contributions: C. Solís and B. Russell designed the study. C. Solís performed experiments and analyzed data. C. Solís and B. Russell wrote the manuscript.
Henk L. Granzier served as editor.
Footnotes
This work is part of a special collection on myofilament function.
References
- Bhana B., Iyer R.K., Chen W.L., Zhao R., Sider K.L., Likhitpanichkul M., Simmons C.A., and Radisic M.. 2010. Influence of substrate stiffness on the phenotype of heart cells. Biotechnol. Bioeng. 105:1148–1160. [DOI] [PubMed] [Google Scholar]
- Burgoyne T., Morris E.P., and Luther P.K.. 2015. Three-Dimensional Structure of Vertebrate Muscle Z-Band: The Small-Square Lattice Z-Band in Rat Cardiac Muscle. J. Mol. Biol. 427:3527–3537. 10.1016/j.jmb.2015.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Sheng R., Kallberg M., Silkov A., Tun M.P., Bhardwaj N., Kurilova S., Hall R.A., Honig B., Lu H., and Cho W.. 2012. Genome-wide Functional Annotation of Dual-Specificity Protein- and Lipid-Binding Modules that Regulate Protein Interactions. Mol. Cell. 46:226–237. 10.1016/j.molcel.2012.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinthalapudi K., Rangarajan E.S., Brown D.T., and Izard T.. 2016. Differential lipid binding of vinculin isoforms promotes quasi-equivalent dimerization. Proc. Natl. Acad. Sci. USA. 113:9539–9544. 10.1073/pnas.1600702113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham M.L., Waldo G.L., Hollinger S., Hepler J.R., and Harden T.K.. 2001. Protein kinase C phosphorylates RGS2 and modulates its capacity for negative regulation of Galpha 11 signaling. J. Biol. Chem. 276:5438–5444. 10.1074/jbc.M007699200 [DOI] [PubMed] [Google Scholar]
- Dabiri G.A., Turnacioglu K.K., Sanger J.M., and Sanger J.W.. 1997. Myofibrillogenesis visualized in living embryonic cardiomyocytes. Proc. Natl. Acad. Sci. USA. 94:9493–9498. 10.1073/pnas.94.17.9493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallakyan S., and Olson A.J.. 2015. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 1263:243–250. 10.1007/978-1-4939-2269-7_19 [DOI] [PubMed] [Google Scholar]
- Danowski B.A., Imanaka-Yoshida K., Sanger J.M., and Sanger J.W.. 1992. Costameres are sites of force transmission to the substratum in adult rat cardiomyocytes. J. Cell Biol. 118:1411–1420. 10.1083/jcb.118.6.1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M., Zwolak A., Schafer D.A., Sept D., Dominguez R., and Cooper J.A.. 2014. Capping protein regulators fine-tune actin assembly dynamics. Nat. Rev. Mol. Cell Biol. 15:677–689. 10.1038/nrm3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomovsky G.M., and Holmes J.W.. 2010. Evolution of scar structure, mechanics, and ventricular function after myocardial infarction in the rat. Am. J. Physiol. Heart Circ. Physiol. 298:H221–H228. 10.1152/ajpheart.00495.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabev E., Kasianowicz J., Abbott T., and McLaughlin S.. 1989. Binding of neomycin to phosphatidylinositol 4,5-bisphosphate (PIP2). Biochim. Biophys. Acta. 979:105–112. 10.1016/0005-2736(89)90529-4 [DOI] [PubMed] [Google Scholar]
- Hart M.C., and Cooper J.A.. 1999. Vertebrate isoforms of actin capping protein beta have distinct functions In vivo. J. Cell Biol. 147:1287–1298. 10.1083/jcb.147.6.1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman T.J., Martin J.L., Solaro R.J., Samarel A.M., and Russell B.. 2009. CapZ dynamics are altered by endothelin-1 and phenylephrine via PIP2- and PKC-dependent mechanisms. Am. J. Physiol. Cell Physiol. 296:C1034–C1039. 10.1152/ajpcell.00544.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss S.G., and Cooper J.A.. 1991. Regulation of CapZ, an actin capping protein of chicken muscle, by anionic phospholipids. Biochemistry. 30:8753–8758. 10.1021/bi00100a006 [DOI] [PubMed] [Google Scholar]
- Hug C., Miller T.M., Torres M.A., Casella J.F., and Cooper J.A.. 1992. Identification and characterization of an actin-binding site of CapZ. J. Cell Biol. 116:923–931. 10.1083/jcb.116.4.923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey P.A., Xian W., and Flanagan L.A.. 1999. Controlling cytoskeleton structure by phosphoinositide-protein interactions: phosphoinositide binding protein domains and effects of lipid packing. Chem. Phys. Lipids. 101:93–107. 10.1016/S0009-3084(99)00058-4 [DOI] [PubMed] [Google Scholar]
- Kim K., McCully M.E., Bhattacharya N., Butler B., Sept D., and Cooper J.A.. 2007. Structure/function analysis of the interaction of phosphatidylinositol 4,5-bisphosphate with actin-capping protein: implications for how capping protein binds the actin filament. J. Biol. Chem. 282:5871–5879. 10.1074/jbc.M609850200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T., Cooper J.A., and Sept D.. 2010. The interaction of capping protein with the barbed end of the actin filament. J. Mol. Biol. 404:794–802. 10.1016/j.jmb.2010.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König P., Krasteva G., Tag C., König I.R., Arens C., and Kummer W.. 2006. FRET-CLSM and double-labeling indirect immunofluorescence to detect close association of proteins in tissue sections. Lab. Invest. 86:853–864. 10.1038/labinvest.3700443 [DOI] [PubMed] [Google Scholar]
- Kuhn J.R., and Pollard T.D.. 2007. Single molecule kinetic analysis of actin filament capping. Polyphosphoinositides do not dissociate capping proteins. J. Biol. Chem. 282:28014–28024. 10.1074/jbc.M705287200 [DOI] [PubMed] [Google Scholar]
- Lange S., Ehler E., and Gautel M.. 2006. From A to Z and back? Multicompartment proteins in the sarcomere. Trends Cell Biol. 16:11–18. 10.1016/j.tcb.2005.11.007 [DOI] [PubMed] [Google Scholar]
- Li J., and Russell B.. 2013. Phosphatidylinositol 4,5-bisphosphate regulates CapZβ1 and actin dynamics in response to mechanical strain. Am. J. Physiol. Heart Circ. Physiol. 305:H1614–H1623. 10.1152/ajpheart.00477.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Tanhehco E.J., and Russell B.. 2014. Actin dynamics is rapidly regulated by the PTEN and PIP2 signaling pathways leading to myocyte hypertrophy. Am. J. Physiol. Heart Circ. Physiol. 307:H1618–H1625. 10.1152/ajpheart.00393.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Mkrtschjan M.A., Lin Y.H., and Russell B.. 2016. Variation in stiffness regulates cardiac myocyte hypertrophy via signaling pathways. Can. J. Physiol. Pharmacol. 94:1178–1186. 10.1139/cjpp-2015-0578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.H., Li J., Swanson E.R., and Russell B.. 2013. CapZ and actin capping dynamics increase in myocytes after a bout of exercise and abates in hours after stimulation ends. J. Appl. Physiol. 114:1603–1609. 10.1152/japplphysiol.01283.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.H., Swanson E.R., Li J., Mkrtschjan M.A., and Russell B.. 2015. Cyclic mechanical strain of myocytes modifies CapZβ1 post translationally via PKCε. J. Muscle Res. Cell Motil. 36:329–337. 10.1007/s10974-015-9420-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.H., Warren C.M., Li J., McKinsey T.A., and Russell B.. 2016. Myofibril growth during cardiac hypertrophy is regulated through dual phosphorylation and acetylation of the actin capping protein CapZ. Cell. Signal. 28:1015–1024. 10.1016/j.cellsig.2016.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louch W.E., Sheehan K.A., and Wolska B.M.. 2011. Methods in cardiomyocyte isolation, culture, and gene transfer. J. Mol. Cell. Cardiol. 51:288–298. 10.1016/j.yjmcc.2011.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour H., de Tombe P.P., Samarel A.M., and Russell B.. 2004. Restoration of resting sarcomere length after uniaxial static strain is regulated by protein kinase Cepsilon and focal adhesion kinase. Circ. Res. 94:642–649. 10.1161/01.RES.0000121101.32286.C8 [DOI] [PubMed] [Google Scholar]
- Mead F., and Williams A.J.. 2002. Ryanodine-induced structural alterations in the RyR channel suggested by neomycin block. Biophys. J. 82:1964–1974. 10.1016/S0006-3495(02)75545-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mkrtschjan M.A., Solís C., Wondmagegn A.Y., Majithia J., and Russell B.. 2018. PKC epsilon signaling effect on actin assembly is diminished in cardiomyocytes when challenged to additional work in a stiff microenvironment. Cytoskeleton (Hoboken). 75:363–371. 10.1002/cm.21472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P., Hawkes W., Hu J., Megone W.V., Gautrot J., Anilkumar N., Zhang M., Hirvonen L., Cox S., Ehler E., et al. 2018. Cardiomyocytes Sense Matrix Rigidity through a Combination of Muscle and Non-muscle Myosin Contractions. Dev. Cell. 44:326–336.e3. 10.1016/j.devcel.2017.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., and Ferrin T.E.. 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Pollard T.D. 2016. Actin and Actin-Binding Proteins. Cold Spring Harb. Perspect. Biol. 8:a018226 10.1101/cshperspect.a018226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle W.G., and Solaro R.J.. 2004. At the crossroads of myocardial signaling: the role of Z-discs in intracellular signaling and cardiac function. Circ. Res. 94:296–305. 10.1161/01.RES.0000116143.74830.A9 [DOI] [PubMed] [Google Scholar]
- Pyle W.G., Hart M.C., Cooper J.A., Sumandea M.P., de Tombe P.P., and Solaro R.J.. 2002. Actin capping protein: an essential element in protein kinase signaling to the myofilaments. Circ. Res. 90:1299–1306. 10.1161/01.RES.0000024389.03152.22 [DOI] [PubMed] [Google Scholar]
- Pyle W.G., La Rotta G., de Tombe P.P., Sumandea M.P., and Solaro R.J.. 2006. Control of cardiac myofilament activation and PKC-betaII signaling through the actin capping protein, CapZ. J. Mol. Cell. Cardiol. 41:537–543. 10.1016/j.yjmcc.2006.06.006 [DOI] [PubMed] [Google Scholar]
- Rappe A.K., Casewit C.J., Colwell K.S., Goddard W.A., and Skiff W.M.. 1992. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J. Am. Chem. Soc. 114:10024–10035. 10.1021/ja00051a040 [DOI] [Google Scholar]
- Ribeiro E.A. Jr., Pinotsis N., Ghisleni A., Salmazo A., Konarev P.V., Kostan J., Sjöblom B., Schreiner C., Polyansky A.A., Gkougkoulia E.A., et al. 2014. The structure and regulation of human muscle α-actinin. Cell. 159:1447–1460. 10.1016/j.cell.2014.10.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell B., Motlagh D., and Ashley W.W.. 2000. Form follows function: how muscle shape is regulated by work. J. Appl. Physiol. 88:1127–1132. 10.1152/jappl.2000.88.3.1127 [DOI] [PubMed] [Google Scholar]
- Samarel A.M., Koshman Y., Swanson E.R., and Russell B.. 2013. Biophysical forces modulate the costamere and Z-disc for sarcomere remodeling in heart failure. In Biophysics of the Failing Heart: Physics and Biology of Heart Muscle. R.J. Solaro and J.C. Tardiff, editors. Springer, New York. 141–174. 10.1007/978-1-4614-7678-8_7 [DOI] [Google Scholar]
- Schacht J. 1976. Inhibition by neomycin of polyphosphoinositide turnover in subcellular fractions of guinea-pig cerebral cortex in vitro. J. Neurochem. 27:1119–1124. 10.1111/j.1471-4159.1976.tb00318.x [DOI] [PubMed] [Google Scholar]
- Schafer D.A., Hug C., and Cooper J.A.. 1995. Inhibition of CapZ during myofibrillogenesis alters assembly of actin filaments. J. Cell Biol. 128:61–70. 10.1083/jcb.128.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J., Diez G., Klemm A.H., Schewkunow V., and Goldmann W.H.. 2006. CapZ-lipid membrane interactions: a computer analysis. Theor. Biol. Med. Model. 3:30 10.1186/1742-4682-3-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., and Olson A.J.. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Schmidt L.P., Wang Z., Yang X., Shao Y., Borg T.K., Markwald R., Runyan R., and Gao B.Z.. 2016. Dynamic Myofibrillar Remodeling in Live Cardiomyocytes under Static Stretch. Sci. Rep. 6:20674 10.1038/srep20674 [DOI] [PMC free article] [PubMed] [Google Scholar]