Enigma Homologue (ENH) is a Z-disc protein whose loss causes cardiomyopathy, but whose function in muscle mechanics is poorly understood. Gregorich et al. show that ENH affects tension redevelopment in mouse myocardium, possibly by altering cross-bridge cycling kinetics or the compliance of the Z-disc.
Abstract
Enigma Homologue (ENH) is a component of the Z-disc, a structure that anchors actin filaments in the contractile unit of muscle, the sarcomere. Cardiac-specific ablation of ENH protein expression causes contractile dysfunction that ultimately culminates in dilated cardiomyopathy. However, whether ENH is involved in the regulation of myocardial contractility is unknown. To determine if ENH is required for the mechanical activity of cardiac muscle, we analyze muscle mechanics of isolated trabeculae from the hearts of ENH+/+ and ENH−/− mice. We detected no differences in steady-state mechanical properties but show that when muscle fibers are allowed to relax and then are restretched, the rate at which tension redevelops is depressed in ENH−/− mouse myocardium relative to that in ENH+/+ myocardium. SDS-PAGE analysis demonstrated that the expression of β-myosin heavy chain is increased in ENH−/− mouse myocardium, which could partially, but not completely, account for the depression in tension redevelopment kinetics. Using top-down proteomics analysis, we found that the expression of other thin/thick filament regulatory proteins is unaltered, although the phosphorylation of a cardiac troponin T isoform, cardiac troponin I, and myosin regulatory light chain is decreased in ENH−/− mouse myocardium. Nevertheless, these alterations are very small and thus insufficient to explain slowed tension redevelopment kinetics in ENH−/− mouse myocardium. These data suggest that the ENH protein influences tension redevelopment kinetics in mouse myocardium, possibly by affecting cross-bridge cycling kinetics. Previous studies also indicate that ablation of specific Z-disc proteins in myocardium slows contraction kinetics, which could also be a contributing factor in this study.
Introduction
In recent years, the importance of Z-discs, the protein-dense structures that anchor the antiparallel actin filaments from adjoining sarcomeres, in muscle has been underscored by the identification of a growing list of mutations in Z-disc proteins in patients with cardiac and skeletal muscle myopathies (Knöll et al., 2011; Hwang and Sykes, 2015). The Enigma Homologue (ENH) protein belongs to the Enigma subfamily of the larger PDZ/LIM family of proteins, which also contains Enigma and Cypher/ZASP/Oracle (Zheng et al., 2010). Among members of this protein subfamily, comparatively little is known about the biological functions of the ENH protein.
To date, nine isoforms, which are produced via alternative splicing of the ENH transcript, have been identified (Kuroda et al., 1996; Nakagawa et al., 2000; Niederländer et al., 2004; Cheng et al., 2010). These isoforms are categorized based on the presence or absence of LIM domains at the C terminus. Whereas all ENH protein isoforms contain a single N-terminal PDZ domain, the “short” isoforms lack three C-terminal LIM domains present in “long” isoforms of the protein (Nakagawa et al., 2000; Cheng et al., 2010). The N-terminal PDZ domain binds α-actinin and promotes localization of ENH protein isoforms to the Z-discs in striated muscle (Nakagawa et al., 2000). Conversely, the LIM domains have been shown to interact with a number of important signaling proteins, including several kinases (Kuroda et al., 1996; Maturana et al., 2008; Yan et al., 2015). Long isoforms of the ENH protein have a ubiquitous tissue distribution (Kuroda et al., 1996; Ueki et al., 1999), whereas the short isoforms are expressed predominantly in striated muscles (Kuroda et al., 1996; Niederländer et al., 2004).
Up to seven ENH isoforms (five long and two short isoforms) may be expressed in the heart (Cheng et al., 2010), with expression patterns that are developmentally regulated (Yamazaki et al., 2010). Long isoforms of ENH are highly expressed early in cardiac development, and the short isoforms predominate in the adult heart (Yamazaki et al., 2010), but reexpression of long ENH isoforms occurs in response to pressure overload (Yamazaki et al., 2010). Moreover, overexpression of ENH1/1a, a long isoform, in neonatal cardiomyocytes promotes the expression of hypertrophic markers and increases cell volume (Yamazaki et al., 2010), implicating the protein in the hypertrophic response. Altered expression of ENH was detected by RNA-seq in transgenic mice overexpressing PLNR9C/+, a model of dilated cardiomyopathy (Burke et al., 2016). However, it was recently shown that cardiac-specific ablation of ENH isoform expression results in impaired myocardial function and dilated cardiomyopathy in the absence of activation of common cardiac stress pathways (Cheng et al., 2010). These findings suggest that the ENH protein may also have a role, either directly or indirectly, in regulating myocardial contractility. The results presented herein suggest that the ENH protein influences tension redevelopment kinetics in mouse myocardium, possibly by affecting cross-bridge cycling kinetics or the compliance of the Z-disc.
Materials and methods
Transgenic mice
Mice lacking cardiac-specific expression of ENH protein isoforms have been described previously (Cheng et al., 2010). ENH−/− mice have a deletion in exon 3, which encodes part of the N-terminal PDZ domain present in all ENH protein isoforms; thus, ENH−/− mice have cardiac-specific ablation of all ENH protein isoforms (Cheng et al., 2010). Two pairs of male and female ENH+/− mice were obtained as a gift from Dr. J. Chen (University of California San Diego, La Jolla, CA). Mice were housed in clear plastic cages on a 12-h/12-h light/dark cycle with access to food and water ad libitum. Initial crossing of ENH+/− mice produced ENH+/+, ENH+/−, and ENH−/− animals at expected Mendelian ratios. ENH−/− mice were obtained by intercrossing the ENH−/− progeny resulting from the initial crosses. ENH+/+ mice were obtained from ENH+/− crosses. Genotypes were determined from toe clips by PCR with the following primer sets: P1 (5′-GCAGGAAGCACACCCAGTAT-3′) versus P2 (5′-TGGTCTCCAACATTTCACCA-3′) to probe for the wild-type allele, and Frtneo (5′-AATGGGCTGACCGCTTCCTCGT-3′) versus P4 (5′-TCGGATGGATGCTCTCTTCT-3′) to probe for the knockout allele as described previously (Cheng et al., 2010). The genotypes of animals used for mechanical analyses were reconfirmed using tail snips after removal of hearts for pulling trabeculae. All procedures involving animal care and handling were reviewed and approved by the University of Wisconsin School of Medicine and Public Health Animal Care and Use Committee.
Solutions
The composition of solutions was calculated using the computer program of Fabiato (1988) and the stability constants (corrected to pH 7.0 and 22°C) listed by Godt and Lindley (1982). Unless otherwise stated, all solutions used for mechanical measurements contained (in mmol/liter) 100 BES, 15 creatine phosphate, 5 dithiothreitol, 1 free Mg2+, and 4 MgATP. In addition, (a) negative log of the calcium concentration (pCa) 9.0 solution contained 7 EGTA and 0.02 CaCl2, (b) pCa 4.5 solution contained 7 EGTA and 7.01 CaCl2, and (c) preactivating solution contained 0.07 EGTA. The ionic strength of all solutions was adjusted to 180 mmol/liter using potassium propionate. A range of solutions containing different amounts of [Ca2+]free were prepared by mixing pCa 9.0 and pCa 4.5 solutions.
Skinned right ventricular trabeculae
Right ventricular trabeculae (n = 15 for ENH+/+; n = 20 for ENH−/−) were isolated as described previously (Olsson et al., 2004; Patel et al., 2012). Briefly, hearts were excised from ENH+/+ (n = 11) and ENH−/− (n = 12) mice of either sex (3–6 mo of age) previously injected (intraperitoneal injection) with 5,000 U heparin/kg body weight, anesthetized with isoflurane, and pinned down to the base of a dissecting dish filled with modified Ringer’s solution (in mmol/liter: 120 NaCl, 19 NaHCO3, 1.2 Na2HPO4, 1.2 MgSO4, 5 KCl, 1 CaCl2, and 10 glucose, pH 7.4, 22°C) preequilibrated with 95% O2/5% CO2. The right ventricles were cut open and exposed to fresh Ringer’s solution containing 20 mmol/liter 2,3-butanedione monoxime for 30 min (1× solution change). The unbranched right ventricular trabeculae were then dissected free, tied to sticks to hold muscle length fixed, and transferred to relaxing solution (in mmol/liter: 100 KCl, 20 imidazole, 7 MgCl2, 2 EGTA, and 4 ATP, pH 7.0, 4°C) containing 1% Triton X-100 and 0.25 g/ml saponin. After skinning overnight, the trabeculae were washed in fresh relaxing solution (∼1 h) and then stored at −20°C in relaxing solution containing glycerol (50:50 vol/vol). The left and right ventricles were separated at the septum, frozen in liquid nitrogen, and stored at −80°C for later analysis by top-down proteomics.
Experimental apparatus and protocols
On the day of an experiment, skinned trabeculae were washed in relaxing solution for 30 min before cutting them free from the sticks and trimming their ends. The trimmed trabeculae were then transferred to a stainless steel experimental chamber containing pCa 9.0 solution (Moss et al., 1983). The ends of each trabecula were attached to the arms of a motor (model 312B; Aurora Scientific) and force transducer (model 403; Aurora Scientific), as described earlier (Moss et al., 1983). The chamber assembly was then placed on the stage of an inverted microscope (Olympus) fitted with a 40× objective and a CCTV camera (model WV-BL600; Panasonic). The light from a halogen lamp was used to illuminate the skinned preparations. Bitmap images of the preparations were acquired using an AGP 4X/2X graphics card, as well as associated software (ATI Technologies), and were used to assess mean sarcomere length (SL) during the course of each experiment. Changes in tension and motor position were sampled (16-bit resolution, DAP5216a; Microstar Laboratories) at 2.0 kHz using SLControl software developed in this laboratory (http://www.slcontrol.com; Campbell and Moss, 2003). Data were saved to computer files for later analysis.
Active tension–pCa and rate constant of tension redevelopment (ktr)–pCa/tension relationships were established at SL 2.2 µm as described previously (Patel et al., 2012). Briefly, the skinned trabeculae were stretched to a mean SL of ∼2.2 µm in pCa 9.0 solution. After measuring length and width, the preparations were transferred first to preactivating solution, then to Ca2+ activating solution, and finally back to pCa 9.0 solution. Once in Ca2+ activating solution, steady-state tension and ktr were measured simultaneously using the modified multistep protocol developed by Brenner and Eisenberg (1986), described in detail previously (Patel et al., 2001), and illustrated in Fig. 1 A. Briefly, after tension reached a steady level in activating solution (pCa 6.2–4.5), the length of the preparation was rapidly reduced by ∼20%, held for ∼14 ms, and then restretched back to its original length. As a result of restretch, there was an initial transient increase, followed by a decrease in tension (seen as a spike in the tension trace) and subsequent slow recovery of tension nearing the initial steady-state level. The ktr reported in the present study is the rate constant of tension redevelopment after the spike. The drop in tension recorded in solution of pCa 9.0 was taken as passive tension and was therefore subtracted from the drop in total tension at each pCa to yield the Ca2+-activated tension (P). The protocol was repeated to establish active tension–pCa and ktr–pCa/relative active tension relationships. Upon completing mechanical measurements, the trabeculae were cut free at the points of attachment, placed in urea/thiourea SDS sample buffer, and stored at −80°C until subsequent protein analysis.
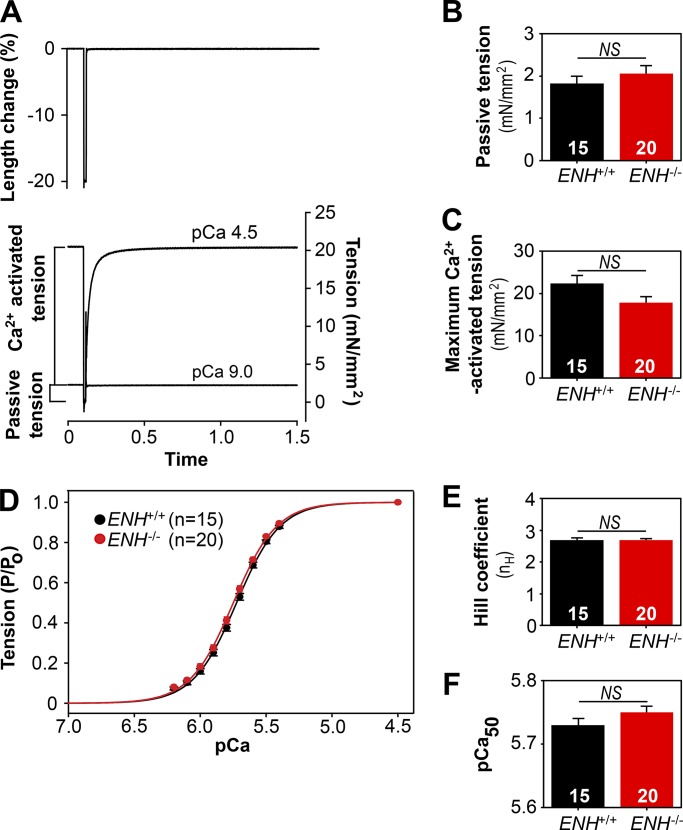
Figure 1.
Loss of ENH protein expression does not alter steady-state mechanical properties of mouse myocardium. (A) Experimental protocol for simultaneous determination of Ca2+-activated tension and ktr. (B and C) The passive (B) and maximum (C) Ca2+-activated tension generated by skinned right ventricular trabeculae isolated from the hearts of ENH+/+ and ENH−/− mice. (D) Tension–pCa relationships established in ENH+/+ and ENH−/− trabeculae. (E and F) The Hill coefficient (E) and pCa50 (F) values established in ENH+/+ and ENH−/− trabeculae. Each data point is the mean and error bar the SEM.
To ensure that the differences in mechanical properties between ENH+/+ and ENH−/− myocardium were not confounded by the differential expression of key contractile proteins, the trabeculae stored in sample buffer were electrophoresed using 5% and 12.5% Tris-HCl Precast Criterion gels (Bio-Rad) as described previously (Warren and Greaser, 2003). The gels were silver stained using methods described previously (Shevchenko et al., 1996) with minor modifications (Stelzer et al., 2006b) as follows: the gels were (a) incubated for 20 min in fixing solution containing 50% methanol and 10% acetic acid, (b) washed for 20 min (4× ddH2O changes) with ddH2O, (c) incubated for 1.5 min in 0.01% sodium thiosulfate solution and then rinsed 4× with ddH2O, (d) incubated for 20 min in 0.09% silver nitrate solution and then rinsed 4× with ddH2O, (e) incubated in developing solution containing 0.0004% sodium thiosulfate, 2% potassium carbonate, and 0.0068% formaldehyde until proteins were visible and then rinsed 4× with ddH2O, (f) incubated for 20 min in destaining solution containing 10% methanol and 10% acetic acid, rinsed 4× in ddH2O, and (g) finally washed for 30 min (6× ddH2O changes) with ddH2O. The gels were then scanned and protein bands quantified using a Bio-Rad Chemi Doc MP Imaging System.
Sample preparation for proteomics analysis
Frozen left ventricular myocardium from ENH+/+ (n = 8) and ENH−/− (n = 13) mice was homogenized in ∼25× (wt/vol) HEPES extraction buffer containing (in mmol/liter) 25 HEPES, pH 7.5, 100 sodium fluoride, 5 sodium pyrophosphate, 50 β-glycerophosphate, 5 Na3VO4, 2.5 EDTA, and 1× protease inhibitor cocktail (Sigma) using a Polytron electric homogenizer (Model PRO200; PRO Scientific) on ice with 3–4-s bursts. The resulting homogenates were centrifuged at 21,000 g for 20 min at 4°C, and the supernatants were discarded. The insoluble pellets were subsequently rehomogenized in 3× (wt/vol) trifluoroacetic acid (TFA) solution (containing 1% TFA and 10 mmol/liter Tris(2-carboxyethyl)phosphine [TCEP]) to extract a protein fraction enriched in sarcomeric proteins. After centrifugation (21,000 g, 4°C, 30 min), the supernatants were transferred to new microcentrifuge tubes, taking care not to disturb the pellet, and centrifuged an additional time (21,000 g, 4°C, 90 min) to ensure complete removal of all particulate matter from the samples. After careful collection of the supernatant, the protein concentration of individual samples was determined using the Pierce Coomassie Plus (Bradford) Assay Kit (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions. A single ENH+/+ sample was randomly chosen, and six dilutions were prepared with the following total protein concentrations (in ng): 50, 100, 250, 400, 500, and 750. These samples were used to verify that the measured protein intensities fell within the linear response range for the mass spectrometer (Fig. S1). The remaining samples were diluted to a final total protein concentration of 400 ng. All samples were diluted in HPLC-grade water containing 0.1% formic acid and 10 mmol/liter TCEP.
Top-down proteomics
Diluted protein extracts prepared from the left ventricular myocardium of ENH+/+ (n = 8) and ENH−/− (n = 13) mice were separated using a NanoAcquity ultra-high pressure liquid chromatography system (Waters), equipped with a home-packed PLRP column (PLRP-S, 250 mm × 500 µm, 10-µm particle size, 1,000-Å pore size; Varian), coupled directly online with a high-resolution Impact II Q-TOF mass spectrometer (Bruker Daltonics). Proteins were eluted using a gradient going from 5% B to 95% B over 55 min (solvent A: 0.10% formic acid in water; solvent B: 0.10% formic acid in a 50:50 mixture of acetonitrile and ethanol) at a flow rate of 8 µl/min. Spectra were collected using a scan rate of 1 Hz over a 500–3,000 m/z range. Chromatographic separation and protein signal intensities (Fig. S2), as well as the relative intensity ratios of unmodified and modified protein forms (Fig. S3), were highly reproducible across replicates.
Data analysis
The cross-sectional areas of skinned trabeculae were calculated by assuming that the trabeculae were cylindrical and by equating the width, measured from video images of the mounted preparations, to diameter. Each tension (P) at pCa between 6.2 and 5.4 was expressed as a fraction of the maximum Ca2+-activated tension (Po) developed by the same preparations at pCa 4.5, i.e., P/Po. To determine the concentration of Ca2+ at half-maximal activation (pCa50), tension–pCa data were fitted with the Hill equation: P/Po = [Ca2+]n/(kn + [Ca2+]n)], where n is the slope (Hill coefficient; nH) of the fitted curve and k is the pCa50. ktr was determined by linear transformation of the half-time of tension recovery (ktr = −ln 0.5 × (t1/2)−1), as described previously (Chase et al., 1994; Patel et al., 2001).
All MS data were analyzed using DataAnalysis (Bruker Daltonics). To quantify the relative abundances of unmodified and modified protein forms, spectra were averaged across the entire width of the chromatographic peaks corresponding to elution of the protein of interest, deconvoluted using Maximum Entropy, and processed using the SNAP function in DataAnalysis, which yielded a mass list containing intensity values for each detected protein species in the specified chromatographic time frame. The total protein population intensity was determined by summing the intensity values for all unmodified and modified species corresponding to the same protein. Subsequently, the relative abundance (expressed as a percentage of the total population intensity) of each individual form of that specific protein was determined by dividing the summed intensities of all species corresponding to a specific protein form by the total population intensity for that protein and multiplying by 100%. Protein phosphorylation levels were determined using the relative abundance values calculated for all differentially phosphorylated species from the same protein using the following equation: Ptotal = (%Pmono + (2 × %Pbis) + (3 × %Ptris) + …)/100, as described previously (Dong et al., 2012; Peng et al., 2014; Gregorich et al., 2015).
For the MS-based quantification of sarcomeric protein expression levels in ENH+/+ and ENH−/− mouse myocardium, extracted ion chromatograms (EICs) were generated for individual sarcomeric proteins from each sample in DataAnalysis by inputting all protein forms corresponding to a specific protein of interest for the top five most abundant charge states. A line was then manually drawn under the EIC generated for a given sarcomeric protein using the Chromatogram Compounds Manually function in DataAnalysis. This procedure yielded an area value corresponding to the area under the curve, which represented the relative expression of the specific protein.
All data are presented as means ± SEM. Data were not normally distributed; thus, statistical significance was assessed using the nonparametric two-tailed Mann–Whitney U test. P values <0.05 were considered significant.
Online supplemental material
Also included are the linear response ranges for individual sarcomeric proteins (Fig. S1), the reproducibility of the chromatographic separation and sarcomeric protein mass spectrometry signal intensities (Fig. S2), the reproducibility of protein phosphorylation measurements by top-down proteomics (Fig. S3), and SDS-PAGE analysis of thin/thick filament regulatory protein expression in ENH+/+ and ENH−/− trabeculae (Fig. S4).
Results
Loss of ENH protein expression does not alter the steady-state contraction or mechanical properties of mouse myocardium
To determine whether ablation of ENH protein expression affects the mechanical properties of mouse myocardium, trabeculae were isolated from the hearts of ENH+/+ and ENH−/− mice and skinned, and the steady-state tension and ktr were measured simultaneously using the modified multistep protocol developed by Brenner and Eisenberg (1986), described in detail previously (Fig. 1 A; Patel et al., 2001). Although skinned trabeculae from ENH+/+ and ENH−/− mice generated similar amounts of passive tension (pCa 9.0; Fig. 1 B and Table 1), there was a trend toward decreased maximum Ca2+-activated tension (pCa 4.5) in ENH−/− trabeculae, although this trend did not reach statistical significance (P = 0.051; Fig. 1 C and Table 1). The tension–pCa relationships measured in ENH+/+ and ENH−/− trabeculae were sigmoidal and were fitted with a Hill equation (as described in Materials and methods) to yield nH, an index of apparent cooperativity in the activation of tension, and pCa50, a measure of the Ca2+ sensitivity of tension. The obtained tension–pCa relations, as well as the derived values for nH and pCa50, did not differ between ENH+/+ and ENH−/− trabeculae (Fig. 1, D–F; and Table 1).
Table 1. Summary of mechanical properties of right ventricular trabeculae isolated from the hearts of ENH+/+ and ENH−/− mice.
| Number of trabeculae | Passive tension (mN/mm2) | Maximum Ca2+ activated tension (mN/mm2) | Hill coefficient (nH) | Ca2+ sensitivity of tension (pCa50) | Maximum rate of tension redevelopment (s−1) |
|---|---|---|---|---|---|
| ENH+/+, n = 15 | 1.83 ± 0.17 | 22.34 ± 1.85 | 2.69 ± 0.06 | 5.73 ± 0.01 | 32.33 ± 1.94 |
| ENH−/−, n = 20 | 2.06 ± 0.18 | 17.85 ± 1.48 | 2.69 ± 0.04 | 5.75 ± 0.01 | 19.80 ± 1.17a |
The mechanical properties of skinned trabeculae isolated from the right ventricles of ENH+/+ (n = 11) and ENH−/− (n = 12) mice were examined at SL 2.2 µm and 22°C. Passive tension was measured at pCa 9.0. Po and ktr were measured at pCa 4.5. pCa50 and nH values were derived by fitting the tension–pCa relationships to a Hill equation as described in Materials and methods.
The measured values were significantly different (P < 0.05) from values recorded in ENH+/+ trabeculae.
ENH ablation slows kinetics in mouse myocardium, an effect that involves changes in the rates of cross-bridge attachment and detachment
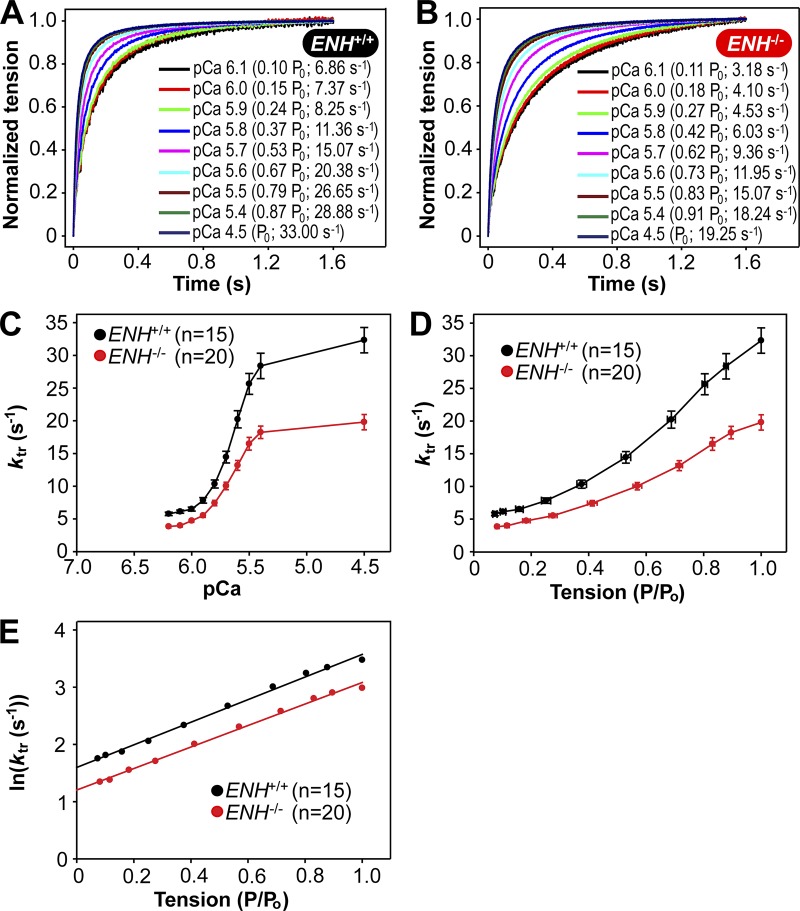
Trabeculae from ENH+/+ and ENH−/− mice exhibited [Ca2+]free-dependent (and tension-dependent) changes in ktr. To illustrate this, records of tension redevelopment in ENH+/+ and ENH−/− trabeculae at various levels of [Ca2+]free (and tension) are shown in Fig. 2, A and B, where steady-state tension at each pCa was normalized to 1.0 to provide better visualization of changes in the kinetics of tension redevelopment. Compared with ENH+/+ trabeculae, ENH−/− trabeculae redeveloped tensions at a significantly slower rate at both submaximal and maximal [Ca2+]free (Fig. 2, A–C; and Table 1). When ktr values were replotted against steady-state isometric tension (as a function of maximum tension), the curvilinear ktr–relative tension relationships measured in ENH−/− trabeculae were shifted to the right (to higher [Ca2+]free) relative to those established in ENH+/+ trabeculae (Fig. 2 D).
Figure 2.
Ablation of ENH protein expression slows tension redevelopment kinetics in mouse myocardium. (A and B) Representative tension redevelopment traces at different [Ca2+]free in skinned right ventricular trabeculae isolated from the hearts of ENH+/+ (A) and ENH−/− (B) mice. (C) ktr–pCa relationships established in ENH+/+ and ENH−/− trabeculae. (D) ktr–relative tension relationship established in ENH+/+ and ENH−/− trabeculae. (E) In(ktr)–relative tension relationships established in ENH+/+ and ENH−/− trabeculae. The natural logarithm of mean ktr values were plotted against relative tension, and the regression fitted to the data yielded y intercepts of 1.60 (gapp = 4.94 s−1) and 1.14 (gapp = 3.33 s−1) for ENH+/+ and ENH−/− trabeculae, respectively. Each data point is the mean and error bar the SEM.
The deletion of ENH-induced reduction in ktr values may involve a decrease in either the rate of attachment of myosin heads with actin (fapp) or the rate of detachment of myosin head from actin (gapp), or both. Considering the two-state cross-bridge model suggesting that myosin cross-bridges are either attached to the thin filament and generating tension (in which case ktr = fapp + gapp) or detached and not generating tension (in which case ktr = gapp); it is possible to estimate gapp by extrapolating either the curvilinear ktr–tension relationship (Fig. 2 D) or the ln[ktr]–tension relationship to zero tension, i.e., to the y axis (Brenner and Eisenberg, 1986; Brenner, 1988). Here we chose the latter method to determine whether ENH deletion affects gapp (Fig. 2 E). The natural logarithms of mean ktr values were plotted against relative tensions and extrapolated with a linear regression fit to the y axis. The gapp values thus determined for ENH+/+ and ENH−/− trabeculae were 4.94 and 3.33 s−1, respectively. Knowing the maximum value of ktr (Table 1) and gapp for ENH+/+ and ENH−/− trabeculae, we calculated fapp using the equation ktr = fapp + gapp to be 27.39 s−1 for ENH+/+ trabeculae and 16.47 s−1 for ENH−/− trabeculae. These results suggest that deletion of ENH protein in mouse myocardium is likely to reduce the rate of cross-bridge attachment by as much as ∼40% and rate of detachment by ∼33%.
The relative expression of β-myosin heavy chain (β-MHC), but not other thin/thick filament regulatory proteins, is altered in the myocardium of ENH−/− mice relative to that in ENH+/+ mice
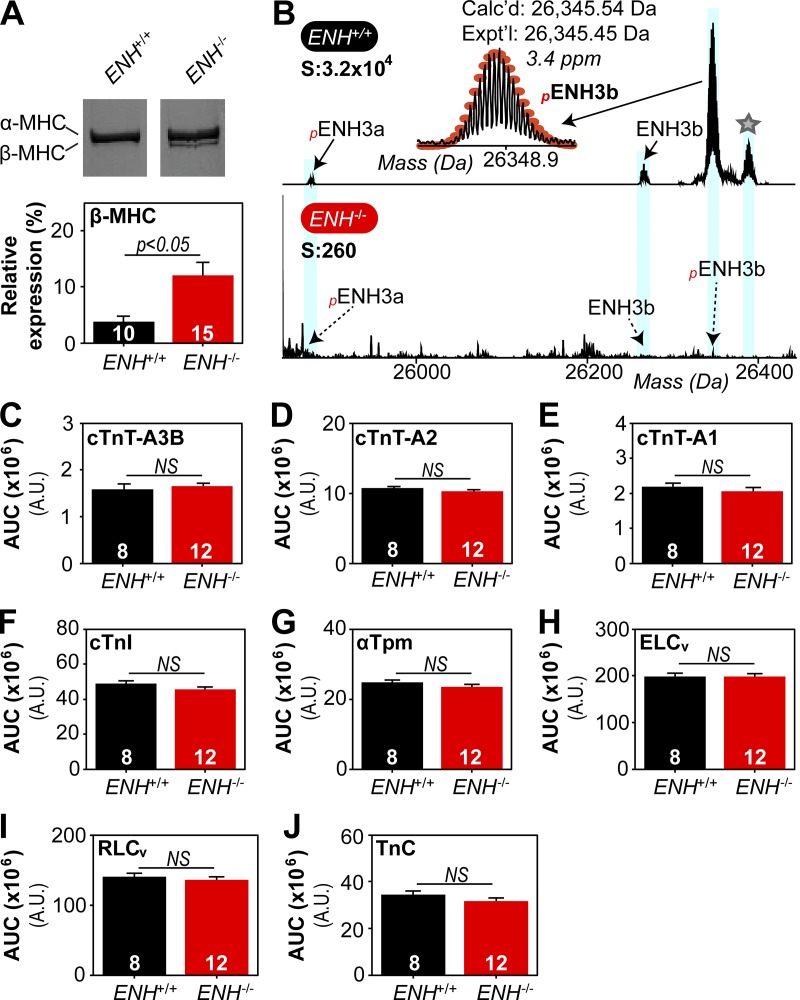
Tension redevelopment kinetics are determined fundamentally by the expression of MHC isoforms (Locher et al., 2009; Ford and Chandra, 2012), and thus we assessed the relative expression of α- and β-MHC in trabeculae from ENH+/+ and ENH−/− mice. In ENH+/+ trabeculae, α-MHC and β-MHC accounted for 96.2 ± 1.1% and 3.8 ± 1.1%, respectively, of the total MHC; while in ENH−/− trabeculae these isoforms accounted for 87.9 ± 2.3% and 12.1 ± 2.3%, respectively, of total MHC (Fig. 3 A). The expression of the slow isoform (β-MHC) was increased by ∼8% in trabeculae from ENH−/− compared with ENH+/+ trabeculae (P < 0.05; Fig. 3 A).
Figure 3.
The expression of MHC isoforms, but not other thin/thick filament regulatory proteins, is altered in ENH−/− mouse myocardium. (A) Representative silver-stained gels showing expression profile of myosin heavy chain isoforms (top) and associated quantification (bottom) in ENH+/+ and ENH−/− trabeculae. (B) Zoomed-in deconvoluted mass spectrum showing ENH protein species detected in ENH+/+, but not ENH−/−, mouse myocardium. Star denotes potential acetylated form of pENH3b (ΔM = 42.00 Da). (C–J) Top-down proteomics quantification of thin/thick filament protein expression in the ventricular myocardium of ENH+/+ and ENH−/− mice. Each data point is the mean and error bar the SEM.
We next determined whether the expression of other thin/thick filament regulatory proteins was altered in the myocardium of ENH−/− mice. Quantitative top-down proteomics was employed to assess the relative expression of these proteins in the myocardium of ENH+/+ and ENH−/− mice. Proteomics analysis of sarcomere protein-enriched extracts permitted highly reproducible quantification of the expression of thin/thick filament regulatory proteins, including cardiac troponin T (cTnT) isoforms, cardiac troponin I (cTnI), troponin C, α-tropomyosin (αTpm), myosin essential light chain (ELCv), and myosin regulatory light chain (RLCv), in the myocardium of ENH+/+ and ENH−/− mice (Fig. S2). Top-down proteomics analysis also enabled the detection of two short isoforms of ENH (ENH3a and ENH3b; Cheng et al., 2010) in mouse myocardium and confirmed ablation of ENH isoform expression in ENH−/− mouse myocardium (Fig. 3 B). No differences in the expression of thin/thick filament regulatory proteins were detected in ENH+/+ and ENH−/− mouse myocardium by top-down proteomics analysis (Fig. 3, C–J). The top-down proteomics data were corroborated by results from SDS-PAGE analysis of thin/thick filament regulatory protein expression in trabeculae from ENH+/+ and ENH−/− mice (Fig. S4).
The phosphorylations of a cTnT isoform, cTnI, and RLCv are reduced in the myocardium of ENH−/− mice relative to ENH+/+ mouse myocardium
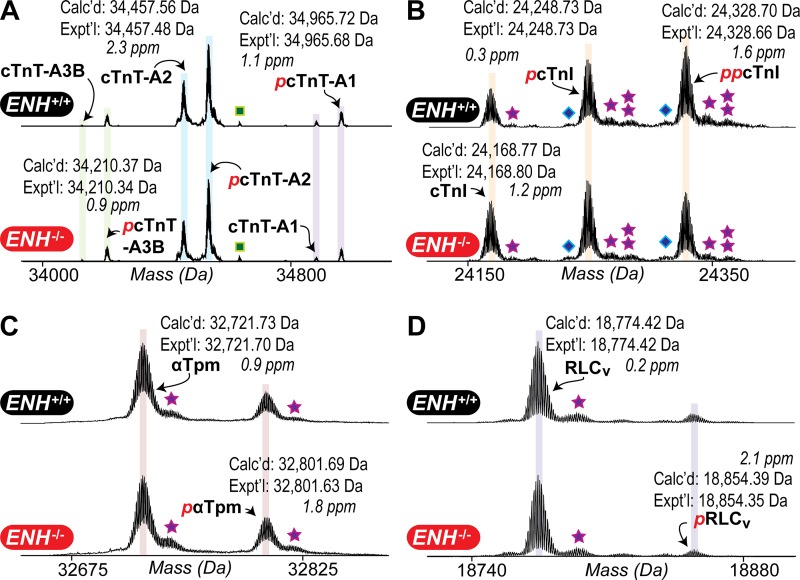
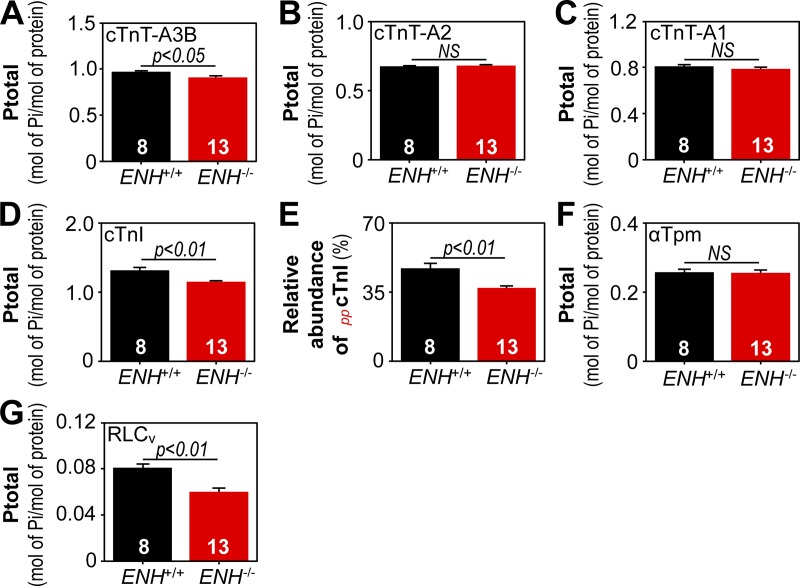
Since contractile protein phosphorylation can also regulate tension redevelopment kinetics (Korte et al., 2003; Stelzer et al., 2006a,b; Mamidi et al., 2016; Toepfer et al., 2016), the phosphorylation status of thin/thick filament regulatory proteins in the ventricular myocardium of ENH+/+ and ENH−/− mice was also reproducibly determined using top-down proteomics (Fig. S3). Representative zoomed-in deconvoluted mass spectra with detected cTnT, cTnI, αTpm, and RLCv protein species in ENH+/+ and ENH−/− mouse myocardium are shown in Fig. 4. Analysis of sarcomeric protein phosphorylation in ENH+/+ and ENH−/− mouse myocardium showed that, while the phosphorylation of cTnT isoforms A2 (cTnT-A2) and A1 (cTnT-A1), as well as αTpm, did not differ, the phosphorylation of cTnT isoform A3B (cTnT-A3B), cTnI, and RLCv were significantly decreased in the myocardium of ENH−/− mice (relative to that in ENH+/+ mouse myocardium; Fig. 5). Specifically, the phosphorylations of cTnT-A3B, cTnI, and RLCv were decreased by ∼6% (P < 0.05), 12% (P < 0.01), and 25% (P < 0.01), respectively, in ENH−/− mouse myocardium (Fig. 5). The relative abundance of the bis-phosphorylated form of cTnI (ppcTnI) was decreased by ∼10% in the myocardium of ENH−/− mice (P < 0.01 versus relative abundance in ENH+/+ mouse myocardium; Fig. 5 E).
Figure 4.
Top-down proteomic analysis of contractile protein phosphorylation in ENH+/+ and ENH−/− mouse myocardium. (A–D) Representative zoomed-in deconvoluted mass spectra showing the relative intensities of unmodified and modified forms of cTnT isoforms (A), cTnI (B), αTpm (C), and RLCv (D) in ENH+/+ and ENH−/− mouse myocardium. Reported masses are the most abundant masses. Squares, stars, and diamonds denote peaks corresponding to proteins associated noncovalently with phosphoric acid (+H3PO4), proteins associated noncovalently with sodium ions (+Na+), and proteins displaying a 62-Da mass increase relative to the unmodified and modified protein species, respectively.
Figure 5.
The phosphorylation of cTnT-A3B, cTnI, and RLCv is decreased in ENH−/− mouse myocardium. (A–D) Quantification of cTnT isoform (A–C) and cTnI (D) phosphorylation in the ventricular myocardium of ENH+/+ and ENH−/− mice. (E) Relative abundance of ppcTnI in ENH+/+ and ENH−/− mouse myocardium. (F and G) Quantification of αTpm (F) and RLCv (G) phosphorylation in ENH+/+ and ENH−/− mouse myocardium. Each data point is the mean and error bar the SEM.
Discussion
The results of the present study demonstrate that ablation of ENH protein expression slows tension redevelopment kinetics in mouse myocardium. In mammalian cardiac muscle, tension redevelopment kinetics are determined fundamentally by the expression of MHC isoforms (Locher et al., 2009; Ford and Chandra, 2012). The relative expression of the slow isoform, β-MHC, was significantly increased in ENH−/− trabeculae. This finding is in agreement with previous results showing an up-regulation of β-MHC at the messenger RNA level in ENH−/− mouse myocardium (Cheng et al., 2010) and suggests that increased expression of β-MHC contributes to slowed tension redevelopment kinetics in the myocardium of these mice. Prior studies have demonstrated that a linear relationship exists between the expression of β-MHC and ktr in rat and mouse ventricular myocardium (Locher et al., 2009; Ford and Chandra, 2012). Based on the linear relationships established in those studies, the 8% increase in β-MHC in ENH−/− mouse myocardium would be expected to reduce ktr by ∼8–10%, indicating that the observed increase in β-MHC in ENH−/− mouse myocardium contributes to slowed contraction kinetics but is insufficient to explain the ∼40% decrease in ktr. Furthermore, such low-level expression of β-MHC only affects tension redevelopment kinetics at high levels of activation (unpublished observations) and, thus, a modest increase in β-MHC expression cannot explain the pronounced reduction in ktr at low levels of [Ca2+]free observed here. Collectively, this information argues against increased expression of β-MHC as a major factor underlying the decreased ktr in ENH−/− mouse myocardium. Since the expression of other thin/thick filament regulatory proteins did not differ in ENH+/+ and ENH−/− mouse myocardium, the ENH deletion-induced decrease in ktr is unlikely to be due to changes in the expression of other contractile proteins.
Tension redevelopment kinetics can also be modulated by the phosphorylation of contractile proteins such as cMyBP-C (Korte et al., 2003; Stelzer et al., 2006a; Mamidi et al., 2016), cTnT (Gollapudi et al., 2012; Michael et al., 2014), and RLCv (Olsson et al., 2004; Stelzer et al., 2006b; Toepfer et al., 2016). Using top-down proteomics, we identified significant reductions in the phosphorylation of cTnT-A3B (∼6%), cTnI (∼12%), and RLCv (∼25%) in ENH−/− mouse myocardium. Based on prior studies (Ayaz-Guner et al., 2009; Scruggs et al., 2010; Zhang et al., 2011), reduced phosphorylation of cTnT-A3B, cTnI, and RLCv likely occurs at Ser1, Ser22/23, and Ser14/15, respectively. Reconstitution of mouse cardiac muscle fibers with pseudo-phosphorylated cTnT (T203E) has previously been shown to reduce ktr by decreasing gapp in an α-MHC background (Michael et al., 2014). Therefore, reduced phosphorylation of cTnT in ENH−/− myocardium would be expected to increase ktr (by increasing gapp), arguing against a role for reduced cTnT phosphorylation in slowed tension redevelopment kinetics in the myocardium of ENH−/− mice. The observed decrease in cTnI phosphorylation in ENH−/− myocardium would not be expected to affect ktr (Stelzer et al., 2007), although an increase in the Ca2+ sensitivity of tension would be anticipated. A possible explanation for unaltered Ca2+ sensitivity of tension in ENH−/− trabeculae is that the effect of reduced cTnI phosphorylation may have been counteracted by an effect of reduced RLCv phosphorylation (i.e., decreased Ca2+ sensitivity of tension; Olsson et al., 2004; Stelzer et al., 2006b; Toepfer et al., 2016), which was also observed in these mice. In contrast to alterations in cTnT-A3B and cTnI phosphorylation, decreased RLCv phosphorylation (∼25%) in ENH−/− mouse myocardium could contribute to slowed tension redevelopment kinetics in the myocardium of ENH−/− mice (Olsson et al., 2004; Stelzer et al., 2006b; Toepfer et al., 2016). Yet, decreased RLCv phosphorylation should also produce concomitant decreases in the Ca2+ sensitivity of tension and maximal tension (Olsson et al., 2004; Stelzer et al., 2006b; Toepfer et al., 2016). While decreased Ca2+ sensitivity of tension may be offset by the effect of reduced cTnI phosphorylation as suggested above, a significant decrease in maximal tension was not detected in ENH−/− trabeculae. Together, this information indicates that aside from decreased RLCv phosphorylation, which could contribute to decreased ktr, the observed changes in contractile protein phosphorylation do not explain slowed tension redevelopment kinetics in ENH−/− mouse myocardium. The possibility that alterations in the expression or phosphorylation of other sarcomeric proteins (i.e., those not analyzed in this study) contribute to slowed tension redevelopment kinetics in ENH−/− mouse myocardium must be acknowledged; however, the findings of the present study indicate that removal of the ENH protein from the Z-disc directly underlies depressed tension redevelopment kinetics in ENH−/− mouse myocardium. Moreover, our ktr data suggest that removal of the ENH protein slows tension redevelopment kinetics by decreasing cross-bridge cycling kinetics.
An alternative possibility is that loss of ENH expression slows tension redevelopment kinetics by increasing the compliance of sarcomeres. Prior studies have shown that cardiac-specific ablation of MLP, a LIM domain–containing protein that localizes to the Z-disc, increases the width of Z-disc (Knöll et al., 2002) and reduces passive tension. Even though a similar increase in the width of Z-discs in the myocardium of ENH−/− mice has been reported previously (Cheng et al., 2010), we did not observe a decrease in passive force in the present study. A more recent study of MLP−/− mice also reported no change in passive tension, a decreased rate of cross-bridge detachment (gapp), and no change in the Ca2+ sensitivity of tension; however, a decrease in maximal tension was also noted (Li et al., 2018). Although we did not detect a significant change in maximal tension in ENH−/− myocardium, the possibility that removal of the ENH protein introduces a compliance within the sarcomeres, similar to that induced by MLP removal, cannot be excluded.
It was recently demonstrated that the giant protein nebulin, which is anchored in the Z-disc but spans the length of the thin filament, has a role in determining the rate of cross-bridge cycling, as well as the Ca2+ sensitivity and cooperativity of activation, in skeletal muscle (Chandra et al., 2009). Notably, the width of Z-discs is also increased in skeletal muscles lacking nebulin (Tonino et al., 2010). However, the effects of nebulin on contractility are likely mediated by direct modulation of actin–myosin interactions by the portion of the protein that extends into the A band (Chandra et al., 2009). Supporting this conclusion is the fact that ablation of nebulette, the short cardiac-specific isoform of nebulin, which is also anchored in the Z-disc but does not span the thin filament, does not alter the steady-state or dynamic mechanical properties of mouse myocardium (Mastrototaro et al., 2015). Therefore, it is unclear at this point how a protein such as ENH, which is within the Z-disc (Nakagawa et al., 2000), could influence actin–myosin interactions in the A band. One possible explanation is that the structural changes occurring within the Z-disc upon ENH removal (Cheng et al., 2010) alter the spatial orientation of the actin filaments with respect to the myosin filaments. As a result, the myosin heads may exhibit slower attachment to and/or detachment from binding sites on the thin filament, thereby slowing cross-bridge cycling kinetics. Although our findings provide the first evidence that the ENH protein influences tension redevelopment kinetics in mouse myocardium, further studies such as sinusoidal analysis and tension–velocity curves will nonetheless be necessary to fully understand the role of ENH as a determinant of myocardial contractility.
Supplementary Material
Acknowledgments
The authors thank Dr. Ju Chen for generously providing transgenic mice heterozygous for the ENH protein.
Financial support for this work was provided by National Institutes of Health F31 HL128086 (to Z.R. Gregorich), R37 HL82900 (to R.L. Moss), and R01 HL109810 and R01 HL096971 (to Y. Ge). Y. Ge also acknowledges National Institutes of Health R01 GM117058 and S10 OD018475.
The authors declare no competing financial interests.
Author contributions: Z.R. Gregorich, J.R. Patel, R.L. Moss, and Y. Ge designed research; Z.R. Gregorich, J.R. Patel, W. Cai, Z. Lin, and R. Heurer performed research; Z.R. Gregorich, J.R. Patel, W. Cai, Z. Lin, R. Heurer, D.P. Fitzsimons, R.L. Moss, and Y. Ge analyzed data; Z.R. Gregorich, J.R. Patel, D.P. Fitzsimons, R.L. Moss, and Y. Ge wrote the paper.
Henk L. Granzier served as editor.
Footnotes
This work is part of a special collection on myofilament function.
References
- Ayaz-Guner S., Zhang J., Li L., Walker J.W., and Ge Y.. 2009. In vivo phosphorylation site mapping in mouse cardiac troponin I by high resolution top-down electron capture dissociation mass spectrometry: Ser22/23 are the only sites basally phosphorylated. Biochemistry. 48:8161–8170. 10.1021/bi900739f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner B. 1988. Effect of Ca2+ on cross-bridge turnover kinetics in skinned single rabbit psoas fibers: implications for regulation of muscle contraction. Proc. Natl. Acad. Sci. USA. 85:3265–3269. 10.1073/pnas.85.9.3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner B., and Eisenberg E.. 1986. Rate of force generation in muscle: correlation with actomyosin ATPase activity in solution. Proc. Natl. Acad. Sci. USA. 83:3542–3546. 10.1073/pnas.83.10.3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke M.A., Chang S., Wakimoto H., Gorham J.M., Conner D.A., Christodoulou D.C., Parfenov M.G., DePalma S.R., Eminaga S., Konno T., et al. 2016. Molecular profiling of dilated cardiomyopathy that progresses to heart failure. JCI Insight. 1:e86898 10.1172/jci.insight.86898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell K.S., and Moss R.L.. 2003. SLControl: PC-based data acquisition and analysis for muscle mechanics. Am. J. Physiol. Heart Circ. Physiol. 285:H2857–H2864. 10.1152/ajpheart.00295.2003 [DOI] [PubMed] [Google Scholar]
- Chandra M., Mamidi R., Ford S., Hidalgo C., Witt C., Ottenheijm C., Labeit S., and Granzier H.. 2009. Nebulin alters cross-bridge cycling kinetics and increases thin filament activation: a novel mechanism for increasing tension and reducing tension cost. J. Biol. Chem. 284:30889–30896. 10.1074/jbc.M109.049718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase P.B., Martyn D.A., and Hannon J.D.. 1994. Isometric force redevelopment of skinned muscle fibers from rabbit activated with and without Ca2+. Biophys. J. 67:1994–2001. 10.1016/S0006-3495(94)80682-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Kimura K., Peter A.K., Cui L., Ouyang K., Shen T., Liu Y., Gu Y., Dalton N.D., Evans S.M., et al. 2010. Loss of enigma homolog protein results in dilated cardiomyopathy. Circ. Res. 107:348–356. 10.1161/CIRCRESAHA.110.218735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Sumandea C.A., Chen Y.C., Garcia-Cazarin M.L., Zhang J., Balke C.W., Sumandea M.P., and Ge Y.. 2012. Augmented phosphorylation of cardiac troponin I in hypertensive heart failure. J. Biol. Chem. 287:848–857. 10.1074/jbc.M111.293258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabiato A. 1988. Computer programs for calculating total from specified free or free from specified total ionic concentrations in aqueous solutions containing multiple metals and ligands. Methods Enzymol. 157:378–417. 10.1016/0076-6879(88)57093-3 [DOI] [PubMed] [Google Scholar]
- Ford S.J., and Chandra M.. 2012. The effects of slow skeletal troponin I expression in the murine myocardium are influenced by development-related shifts in myosin heavy chain isoform. J. Physiol. 590:6047–6063. 10.1113/jphysiol.2012.240085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godt R.E., and Lindley B.D.. 1982. Influence of temperature upon contractile activation and isometric force production in mechanically skinned muscle fibers of the frog. J. Gen. Physiol. 80:279–297. 10.1085/jgp.80.2.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollapudi S.K., Mamidi R., Mallampalli S.L., and Chandra M.. 2012. The N-terminal extension of cardiac troponin T stabilizes the blocked state of cardiac thin filament. Biophys. J. 103:940–948. 10.1016/j.bpj.2012.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorich Z.R., Peng Y., Lane N.M., Wolff J.J., Wang S., Guo W., Guner H., Doop J., Hacker T.A., and Ge Y.. 2015. Comprehensive assessment of chamber-specific and transmural heterogeneity in myofilament protein phosphorylation by top-down mass spectrometry. J. Mol. Cell. Cardiol. 87:102–112. 10.1016/j.yjmcc.2015.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang P.M., and Sykes B.D.. 2015. Targeting the sarcomere to correct muscle function. Nat. Rev. Drug Discov. 14:313–328. 10.1038/nrd4554 [DOI] [PubMed] [Google Scholar]
- Knöll R., Hoshijima M., Hoffman H.M., Person V., Lorenzen-Schmidt I., Bang M.L., Hayashi T., Shiga N., Yasukawa H., Schaper W., et al. 2002. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 111:943–955. 10.1016/S0092-8674(02)01226-6 [DOI] [PubMed] [Google Scholar]
- Knöll R., Buyandelger B., and Lab M.. 2011. The sarcomeric Z-disc and Z-discopathies. J. Biomed. Biotechnol. 2011:569628 10.1155/2011/569628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte F.S., McDonald K.S., Harris S.P., and Moss R.L.. 2003. Loaded shortening, power output, and rate of force redevelopment are increased with knockout of cardiac myosin binding protein-C. Circ. Res. 93:752–758. 10.1161/01.RES.0000096363.85588.9A [DOI] [PubMed] [Google Scholar]
- Kuroda S., Tokunaga C., Kiyohara Y., Higuchi O., Konishi H., Mizuno K., Gill G.N., and Kikkawa U.. 1996. Protein-protein interaction of zinc finger LIM domains with protein kinase C. J. Biol. Chem. 271:31029–31032. 10.1074/jbc.271.49.31029 [DOI] [PubMed] [Google Scholar]
- Li J., Gresham K.S., Mamidi R., Doh C.Y., Wan X., Deschenes I., and Stelzer J.E.. 2018. Sarcomere-based genetic enhancement of systolic cardiac function in a murine model of dilated cardiomyopathy. Int. J. Cardiol. 273:168–176. 10.1016/j.ijcard.2018.09.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locher M.R., Razumova M.V., Stelzer J.E., Norman H.S., Patel J.R., and Moss R.L.. 2009. Determination of rate constants for turnover of myosin isoforms in rat myocardium: implications for in vivo contractile kinetics. Am. J. Physiol. Heart Circ. Physiol. 297:H247–H256. 10.1152/ajpheart.00922.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamidi R., Gresham K.S., Verma S., and Stelzer J.E.. 2016. Cardiac myosin binding protein-C phosphorylation modulates myofilament length-dependent activation. Front. Physiol. 7:38 10.3389/fphys.2016.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrototaro G., Liang X., Li X., Carullo P., Piroddi N., Tesi C., Gu Y., Dalton N.D., Peterson K.L., Poggesi C., et al. 2015. Nebulette knockout mice have normal cardiac function, but show Z-line widening and up-regulation of cardiac stress markers. Cardiovasc. Res. 107:216–225. 10.1093/cvr/cvv156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maturana A.D., Wälchli S., Iwata M., Ryser S., Van Lint J., Hoshijima M., Schlegel W., Ikeda Y., Tanizawa K., and Kuroda S.. 2008. Enigma homolog 1 scaffolds protein kinase D1 to regulate the activity of the cardiac L-type voltage-gated calcium channel. Cardiovasc. Res. 78:458–465. 10.1093/cvr/cvn052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael J.J., Gollapudi S.K., and Chandra M.. 2014. Effects of pseudo-phosphorylated rat cardiac troponin T are differently modulated by α- and β-myosin heavy chain isoforms. Basic Res. Cardiol. 109:442 10.1007/s00395-014-0442-9 [DOI] [PubMed] [Google Scholar]
- Moss R.L., Swinford A.E., and Greaser M.L.. 1983. Alterations in the Ca2+ sensitivity of tension development by single skeletal muscle fibers at stretched lengths. Biophys. J. 43:115–119. 10.1016/S0006-3495(83)84329-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa N., Hoshijima M., Oyasu M., Saito N., Tanizawa K., and Kuroda S.. 2000. ENH, containing PDZ and LIM domains, heart/skeletal muscle-specific protein, associates with cytoskeletal proteins through the PDZ domain. Biochem. Biophys. Res. Commun. 272:505–512. 10.1006/bbrc.2000.2787 [DOI] [PubMed] [Google Scholar]
- Niederländer N., Fayein N.A., Auffray C., and Pomiès P.. 2004. Characterization of a new human isoform of the enigma homolog family specifically expressed in skeletal muscle. Biochem. Biophys. Res. Commun. 325:1304–1311. 10.1016/j.bbrc.2004.10.178 [DOI] [PubMed] [Google Scholar]
- Olsson M.C., Patel J.R., Fitzsimons D.P., Walker J.W., and Moss R.L.. 2004. Basal myosin light chain phosphorylation is a determinant of Ca2+ sensitivity of force and activation dependence of the kinetics of myocardial force development. Am. J. Physiol. Heart Circ. Physiol. 287:H2712–H2718. 10.1152/ajpheart.01067.2003 [DOI] [PubMed] [Google Scholar]
- Patel J.R., Fitzsimons D.P., Buck S.H., Muthuchamy M., Wieczorek D.F., and Moss R.L.. 2001. PKA accelerates rate of force development in murine skinned myocardium expressing alpha- or beta-tropomyosin. Am. J. Physiol. Heart Circ. Physiol. 280:H2732–H2739. 10.1152/ajpheart.2001.280.6.H2732 [DOI] [PubMed] [Google Scholar]
- Patel J.R., Pleitner J.M., Moss R.L., and Greaser M.L.. 2012. Magnitude of length-dependent changes in contractile properties varies with titin isoform in rat ventricles. Am. J. Physiol. Heart Circ. Physiol. 302:H697–H708. 10.1152/ajpheart.00800.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Gregorich Z.R., Valeja S.G., Zhang H., Cai W., Chen Y.-C., Guner H., Chen A.J., Schwahn D.J., Hacker T.A., et al. 2014. Top-down proteomics reveals concerted reductions in myofilament and Z-disc protein phosphorylation after acute myocardial infarction. Mol. Cell. Proteomics. 13:2752–2764. 10.1074/mcp.M114.040675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scruggs S.B., Reisdorph R., Armstrong M.L., Warren C.M., Reisdorph N., Solaro R.J., and Buttrick P.M.. 2010. A novel, in-solution separation of endogenous cardiac sarcomeric proteins and identification of distinct charged variants of regulatory light chain. Mol. Cell. Proteomics. 9:1804–1818. 10.1074/mcp.M110.000075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., and Mann M.. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850–858. 10.1021/ac950914h [DOI] [PubMed] [Google Scholar]
- Stelzer J.E., Fitzsimons D.P., and Moss R.L.. 2006a Ablation of myosin-binding protein-C accelerates force development in mouse myocardium. Biophys. J. 90:4119–4127. 10.1529/biophysj.105.078147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer J.E., Patel J.R., and Moss R.L.. 2006b Acceleration of stretch activation in murine myocardium due to phosphorylation of myosin regulatory light chain. J. Gen. Physiol. 128:261–272. 10.1085/jgp.200609547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer J.E., Patel J.R., Walker J.W., and Moss R.L.. 2007. Differential roles of cardiac myosin-binding protein C and cardiac troponin I in the myofibrillar force responses to protein kinase A phosphorylation. Circ. Res. 101:503–511. 10.1161/CIRCRESAHA.107.153650 [DOI] [PubMed] [Google Scholar]
- Toepfer C.N., West T.G., and Ferenczi M.A.. 2016. Revisiting Frank-Starling: regulatory light chain phosphorylation alters the rate of force redevelopment (ktr) in a length-dependent fashion. J. Physiol. 594:5237–5254. 10.1113/JP272441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonino P., Pappas C.T., Hudson B.D., Labeit S., Gregorio C.C., and Granzier H.. 2010. Reduced myofibrillar connectivity and increased Z-disk width in nebulin-deficient skeletal muscle. J. Cell Sci. 123:384–391. 10.1242/jcs.042234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki N., Seki N., Yano K., Masuho Y., Saito T., and Muramatsu M.. 1999. Isolation, tissue expression, and chromosomal assignment of a human LIM protein gene, showing homology to rat enigma homologue (ENH). J. Hum. Genet. 44:256–260. 10.1007/s100380050155 [DOI] [PubMed] [Google Scholar]
- Warren C.M., and Greaser M.L.. 2003. Method for cardiac myosin heavy chain separation by sodium dodecyl sulfate gel electrophoresis. Anal. Biochem. 320:149–151. 10.1016/S0003-2697(03)00350-6 [DOI] [PubMed] [Google Scholar]
- Yamazaki T., Wälchli S., Fujita T., Ryser S., Hoshijima M., Schlegel W., Kuroda S., and Maturana A.D.. 2010. Splice variants of enigma homolog, differentially expressed during heart development, promote or prevent hypertrophy. Cardiovasc. Res. 86:374–382. 10.1093/cvr/cvq023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Tsukamoto O., Nakano A., Kato H., Kioka H., Ito N., Higo S., Yamazaki S., Shintani Y., Matsuoka K., et al. 2015. Augmented AMPK activity inhibits cell migration by phosphorylating the novel substrate Pdlim5. Nat. Commun. 6:6137 10.1038/ncomms7137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhang H., Ayaz-Guner S., Chen Y.C., Dong X., Xu Q., and Ge Y.. 2011. Phosphorylation, but not alternative splicing or proteolytic degradation, is conserved in human and mouse cardiac troponin T. Biochemistry. 50:6081–6092. 10.1021/bi2006256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Cheng H., Banerjee I., and Chen J.. 2010. ALP/Enigma PDZ-LIM domain proteins in the heart. J. Mol. Cell Biol. 2:96–102. 10.1093/jmcb/mjp038 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.