Abstract
Background:
Pleural effusion (PE) has been reported useful in many studies for testing epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer (NSCLC) with variable results. This systematic review and meta-analysis was performed to elucidate whether PE could be used as a surrogate for tumor tissue to detect EGFR mutations.
Methods:
We extracted 2 × 2 diagnostic table from each included study and calculated data on specificity, sensitivity, negative likelihood ratio (NLR), positive likelihood ratio (PLR) ,and diagnostic odds ratio (DOR). We used the area under curve (AUC) and summary receiver operating characteristic curve (SROC) to summarize the overall diagnostic performance and assessed publication bias by Deeks’ funnel plot.
Results:
Our meta-analysis included 15 eligible publications. The following summary estimates for diagnostic parameters of the EGFR mutations detection in PE were made: sensitivity, 0.86 (95%CI 0.83–0.89); specificity, 0.93 (95%CI 0.91–0.95); PLR, 8.53 (95%CI 5,94–12.25); NLR, 0.18 (95%CI 0.13–0.25); DOR, 63.40 (95%CI 38.83–103.51); and AUC, 0.94. Funnel plot indicated publication bias insignificant.
Conclusions:
The meta-analysis suggests that EGFR mutation detecting in PE, especially supernatants, is a promising surrogate for tumor tissue in EGFR mutations testing of patients with NSCLC.
Keywords: diagnosis, epidermal growth factor receptor, meta-analysis, pleural effusion
1. Introduction
Lung cancer is worldwide health problem that places heavy burden on patients and society, of which about 85% of the total cases diagnosed were non-small cell lung cancer (NSCLC).[1] According to reports, 222,500 cases were newly diagnosed and 155,870 cases died of lung cancer in 2017, United States of American.[2] With decades of research and work, the treatment of NSCLC has been improved a lot with significant clinical progress, especially on targeted therapy. NSCLCs with mutation events in the domain of epidermal growth factor receptor (EGFR) tyrosine kinase, which express as a percentage of about 30% to 40% of NSCLC cases in Asian and of about 2% to 8% in Western countries, respond to tyrosine kinase inhibitors (TKIs), for instance, gefitinib, icotinib, and erlotinib.[3,4] For patients with EGFR mutation, TKIs treatment improved the clinical prognosis of such patients with improved quality of life and increased overall survival date.[5] Thus, the determination of EGFR mutation status has an importance role in the NSCLC treatment.
The most used examination sample is the biopsy sample from tumor tissue through closed pleura biopsy or thoracoscopic lung biopsy, and which means invasive procedures and potentially high risk for patients. Many NSCLC are presented with malignant pleural effusion (PE). About 15% of lung cancer patients have PE and more than 10% of the patients have PE when they received a diagnosis.[6] Approximately 50% of NSCLC patients with PE are initially positive cytologically. Although tissue biopsy is recommended for genetic analysis, the risk of biopsy complication, failure to obtain homogeneous tumor samples, and no patient's informed consent can lead to suboptimal mutation detection.[7] Because PE sampling is usually facile, less traumatic and reproducible, further research is necessary to determine whether it can be used for gene testing as a tissue substitute. Growing studies suggest that pleural effusion is a potential sample to determine EGFR status,[8,9] while those studies gave different and discordant results. Thus, this meta-analysis which based current available evidence aimed to evaluate the overall accuracy of PE test in determining the EGFR status of NSCLC patients.
2. Methods
This meta-analysis was based on the guidelines about diagnostic studies and the Preferred Reporting Items for Systematic Reviews and Meta-Analyses report.[10–12] Ethics Committee approval was not required for this retrospective study.
3. Literature search
We searched in PubMed, Embase, Wanfang, Weipu, and CNKI to identify potentially eligible studies. No starting data limit was applied and the last search was performed on December 20, 2017. The following key words and medical subheadings were used as search terms: “pleural effusion” AND “epidermal growth factor receptor” OR “EGFR” AND “non-small cell lung cancer” OR “NSCLC”. Different combinations of the previously mentioned terms were also searched. Articles were identified using the “similar-articles” function in PubMed.
4. Study selection
First, we selected records through the titles and summaries, and then reviewed the full text of the potentially qualified researches. Studies met the following criteria were included:
-
1.
it was an original research paper published in English or Chinese;
-
2.
all patients involved should be diagnosed NSCLC with histopathology or cytology;
-
3.
EGFR mutation status should be detected by matched tumor tissues and PE samples;
-
4.
enough initial data to build the 2 × 2 diagnostic table.
5. Data extraction and quality evaluation
Data were collected including first author, publication year, country, patients number, percentage of female patients, percentage of smokers (ever and current), histologic type, TNM stage, type of PE samples (cell blocks or supernatants), detection methods of EGFR mutation in PE or tumor tissues, true positive (TP), true negative (TN), false negative (FN), and false positive (FP). When EGFR mutation detection was performed using different methods and PE samples, TP, TN, FP, and FN were extracted separately.
QUADAS-2 instrument was used to evaluated the methodological quality of eligible studies.[13] Risk of bias were assessed in 4 domains and applicability concerns in 3 domains within QUADAS-2. Review Manager software (version 5.2, the Cochrane Collaboration) was used to develop QUADAS plot.
All records were independently checked by 2 reviewers and a consensus was reached in case of disagreement. During research selection, data collection or quality evaluation, any differences between the 2 authors (CSP and HWM) were resolved through consultation with a third author (JYQ).
6. Statistical analysis
The EGFR mutation status in tumor tissue samples were used as a reference standard. We used standard methods which was recommended for diagnostic accuracy meta-analysis. And calculated pooled specificity, sensitivity, negative likelihood ratio (NLR), positive likelihood ratio (PLR), and diagnostic odds ratio (DOR) by the data (TP, FP, FN, and TN) retrieved from original studies, together with 95% confidence intervals (95% CIs). Based on summary receiver operating characteristic (SROC) curve, area under the curve (AUC) was calculated. According to the results of heterogeneity tests, either a random-effects model or a fixed-effects model was chosen to calculated related indices across studies.
Chi-Squared and Fisher exact tests were used to detect heterogeneity across studies, and then meta-regression was used to identify potential covariates. Publication bias were detected by Deeks’ funnel plot.[14] Data analyses were carried out with RevMan 5.2 (Cochrane Collaboration, Oxford, UK), Meta-DiSc 1.4 (XI. Cochrane Colloquium, Barcelona, Spain), and STATA 12.0 (Stata Corp., College Station, TX). All tests were double-sided. P < .05 was considered statistically significant.
7. Results
7.1. Study selection and characteristics
After rigorous identification and selection, 15 publications of detecting EGFR mutations in paired PE and tumor tissues of NSCLC patients were eligible for inclusion.[15–29] Major reasons for excluding studies were unpaired PE and tumor tissues and insufficient data. Figure 1 shows the process of selecting of potentially eligible studies. In some publications, EGFR mutations were detected both in cell blocks and supernatants of PE.[15,17–19] Besides, more than 1 assays were used for EGFR-mutation analysis.[16,18,22] Thus, each of these publications was treated as 2 independent studies, giving 24 studies in our meta-analysis altogether.
Figure 1.
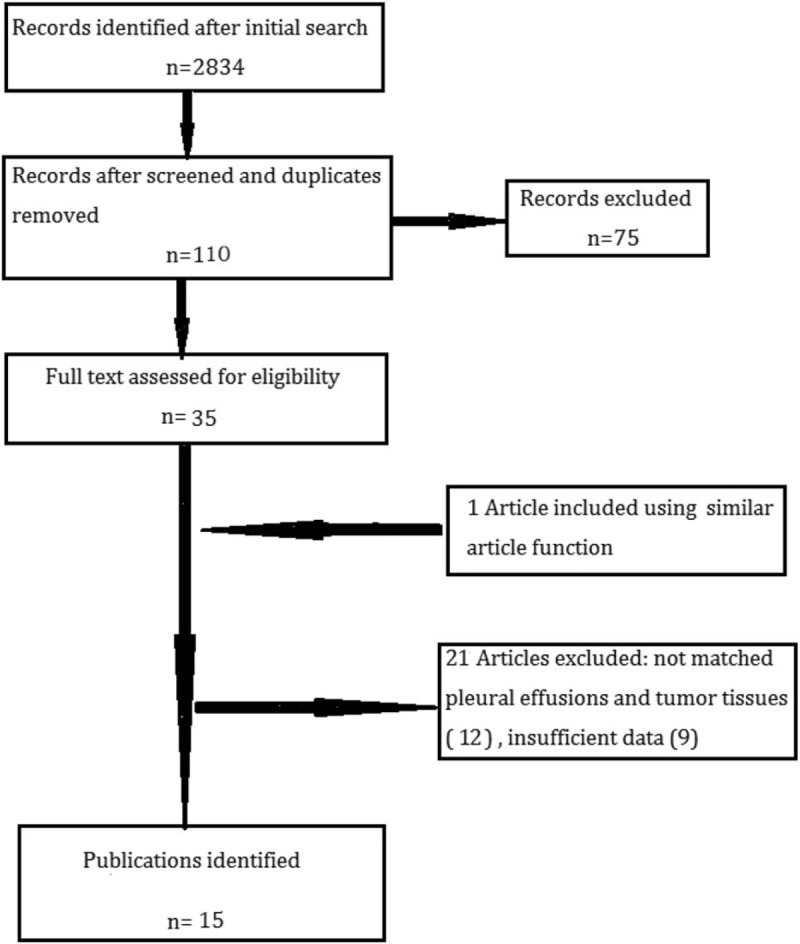
Studies selection process for the meta-analysis.
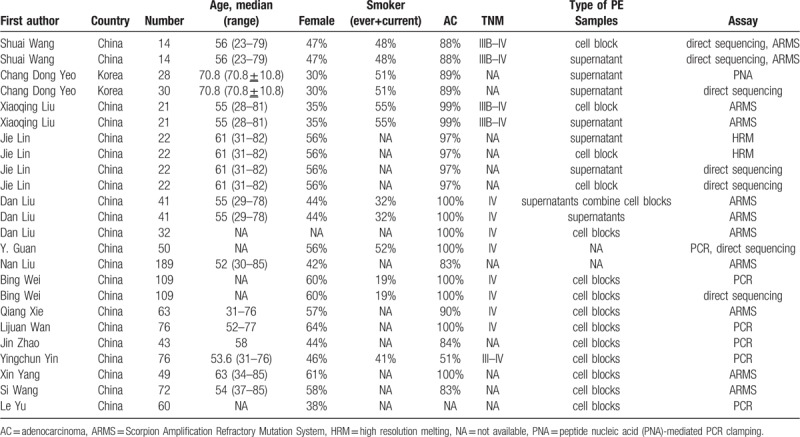
All studies were conducted in Asia, 2 in Korea, the others in China. The average sample size was 50 (range, 14–189). Most patients had been enrolled consecutively and prospectively, except 3 studies were retrospectively.[21,23,24] Up to 80% of the NSCLC participants were adenocarcinoma and all were diagnosed histopathologically or cytologically. The participants in 5 included studies were in stages III to IV,[15,17,26] 8 studies in IV,[19,20,22–24] and others unknown. There were many ways to detect EGFR mutation. Direct sequencing, amplification refractory mutation system (ARMS), and polymerase chain reaction (PCR) were the main methods, which was adopted in 4,[16,18,22] 9[17,19,21,23,27,28], and 5[22,24–26,29] studies, respectively. The details of key characteristics were indicated in Table 1.
Table 1.
Characteristics of eligible studies.
QUADAS-2 was proposed in 2011 and was integrated into RevMan 5.2 in 2012.[10,30] We evaluated the methodological quality in 4 domains (patient selection, index test, reference standard, flow and timing). A response of “Yes” or “Unclear” or “No” was given according to the criterion. These responses for each criterion are then converted into applicability concerns and risk of bias as high, low, or unclear. The original studies quality in our meta-analysis was moderate, most of the included studies did not clearly report whether the results of both tumor tissues and PE samples were blinded from each other and whether a threshold was prespecified.[15–20] Thus, the index text and reference standard remain unclear risk and unclear concern, respectively. However, in patient selection and flow and timing, these studies did generally good. Except 1 was judged to have high risk of bias,[15] related to do not avoid a case-control design, and inappropriate exclusion. Figure 2 shows the summary quality of studies included.
Figure 2.
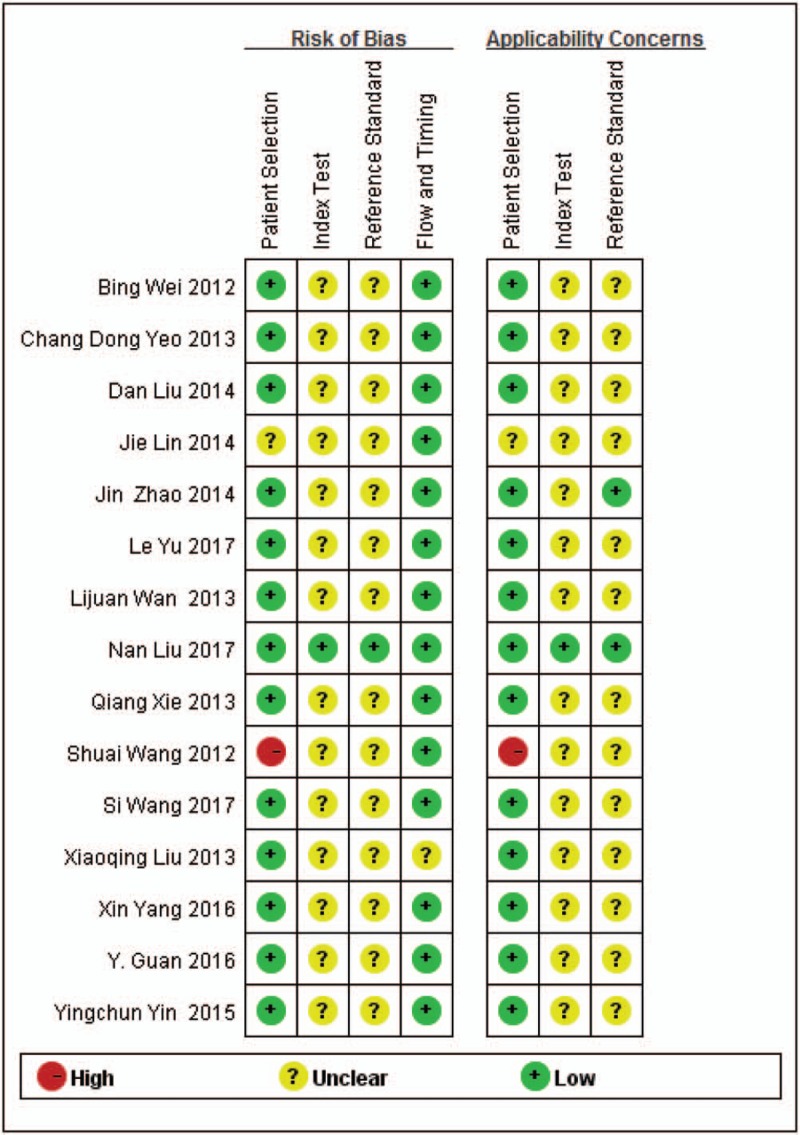
Summary of QUADAS-2 assessments of included studies. QUADAS-2: Quality Assessment of Diagnostic Accuracy Studies-2.
7.2. Diagnostic accuracy
As shown in Figure 3, Sensitivity of PE samples of EGFR mutation detection ranged from 0.64 to 1.00, specificity ranged from 0.75 to 1.00. Compared with tumor tissues, the pooled sensitivity of NSCLC PE samples was 0.86 (95%CI: 0.83–0.89, I2 = 40.9%) and pooled specificity was 0.93 (95%CI: 0.91–0.95). The PLR was 8.53 (95%CI: 5.94–12.25) and NLR was 0.18 (95%CI: 0.13–0.25). The DOR was 63.40 (95%CI: 38.83–103.51). Chi-Squared values of these parameters were as follows: sensitivity, 50.12 (P < .001); specificity, 62.28 (P < .001); PLR, 32.01 (P = .1); NLR, 53.44 (P < .001); and DOR, 28.07 (P = .21), indicating that there was significant heterogeneity among studies (Table 2). The overall diagnostic performance was evaluated by calculating SROC curves and the corresponding AUC. The SROC curve was positioned close to the ideal upper left corner of the plot. Figure 4 showed the SROC with AUC of 0.94 (SEM = 0.01), and Q value for sensitivity and specificity was 0.88 (SEM = 0.01) indicating a high discriminatory ability for PE EGFR mutation test.
Figure 3.
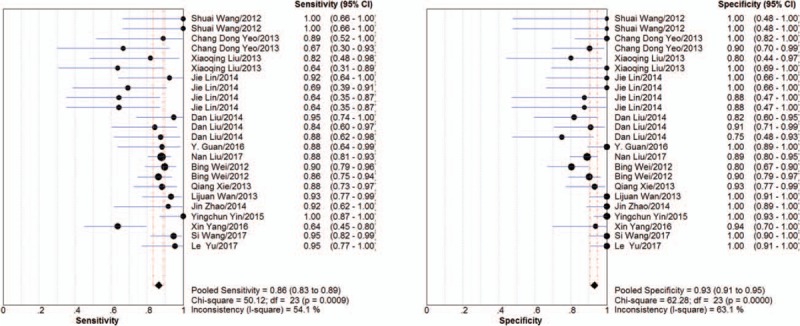
Forest plot of the summary sensitivity and specificity of pleural effusion. The sensitivity/specificity of individual study is represented by a circle, through which runs a horizontal line (95% CI). The diamond at the bottom represents the pooled sensitivity/specificity from the studies. df = degrees of freedom.
Table 2.
Meta-analyses results.
Figure 4.
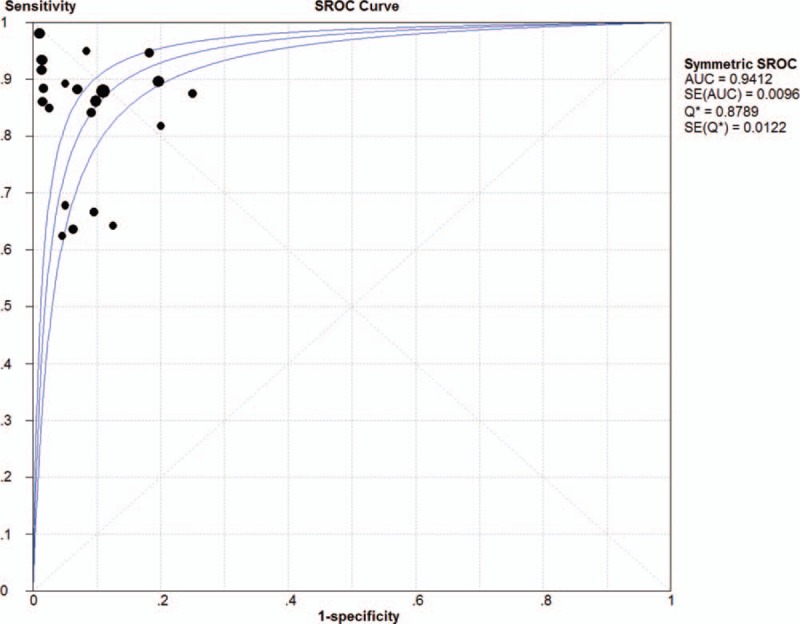
Summary receiver operating characteristic (SROC) curve of pleural effusion. AUC = area under the curve.
7.3. Sub-group analysis and publication bias
We performed sub-group analyses to exam the influence of type of PE samples, countries, detection methods, and TNM stages. We compared 3 main methods used for EGFR mutation test: ARMS, direct sequencing, polymerase chain reaction (PCR), and high resolution melting (HRM). HRM have a perfect specificity of 1.00 (95%CI: 0.81–1.00), while PCR seems to have the optimal sensitivity 0.93 (95%CI: 0.88–0.97), and the highest DOR of 418.04 (95%CI: 49.50–3530.8), indicating PCR and HRM performed better than the other 2 methods in PE EGFR mutation detection. When considering the type of PE samples, supernatants have a more excellent specificity than cell blocks (0.95 vs 0.94), however, a suboptimal sensitivity than cell blocks (0.80 vs 0.87). AUC of supernatants were higher than cell blocks (0.96 vs 0.94), indicating supernatants may have better diagnostic performance than cell blocks. Table 2 shows the details.
Chi-Squared values for sensitivity (P < .001), specificity (P < .001), and NLR (P < .001) suggested significant heterogeneity. Then a meta-regression was performed to identify possible sources. However, we found diagnostic accuracy of EGFR mutation detection in PE did not depended on detection methods, type of PE samples, TNM stages, proportion of female and smoker, or on countries. We found that none of these covariates was source of significant heterogeneity (all P > .05). Table 3 shows the results of the RDOR analysis.
Table 3.
Meta-regression of EGFR mutation detection in PE.
Deeks’ funnel plot tested the publication bias. The P value associated with Deeks test was not significant (P = .59). Figure 5 suggests insignificant publication bias.
Figure 5.
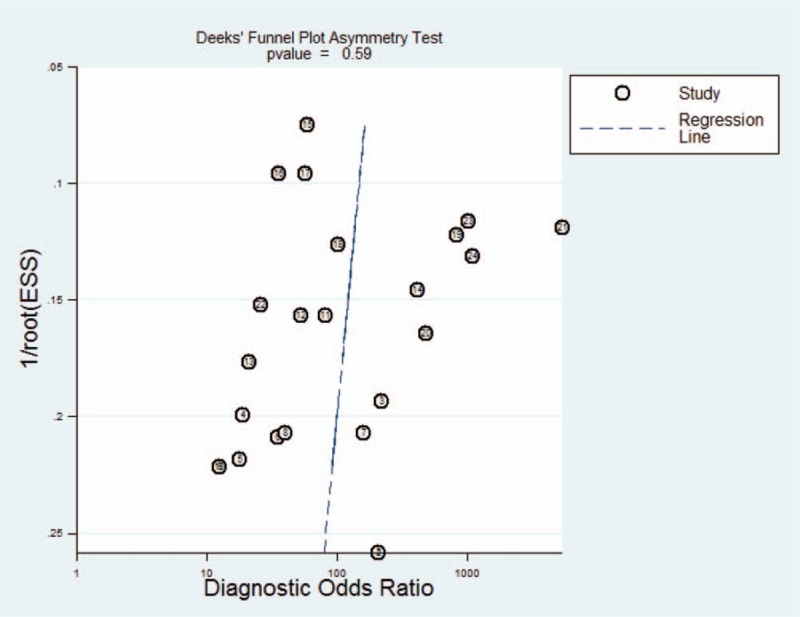
Deeks's funnel plot to assess the likelihood of publication bias.
8. Discussion
Targeted therapy is irreplaceable in the NSCLC treatment, especially in adenocarcinoma. Mutations in EGFR gene are related to the objective responsiveness of tumor to targeted therapy of EGFR TKIs.[31,32] Needless to say, a highly feasible sample is essential to perform EGFR mutations test. Although detection the mutations in tumor tissue plays an vital role in guiding treatment with EGFR TKIs,[33,34] its restriction is obvious, such as inadequate tissue acquisition and non-ideal tissue positions.[35] Thus, researchers have focused on blood and PE samples to find substitutes for tumor tissues.[36–38] Previous studies have suggested that serum is a good alternative when tumor tissue is unavailable or insufficient for EGFR mutations detection.[39] To our knowledge, this is the first meta-analysis to comprehensively assess the overall accuracy of EGFR mutations test within PE samples.
Our meta-analysis of the available evidence showed a pooled sensitivity of 0.86 and specificity of 0.93. The relatively high specificity indicates that a low rate of misdiagnosis (7%), which is more important than the rate of missed diagnosis in recommendation of EGFR TKIs treatment. However, the suboptimal sensitivity indicates a relatively high rate of missed diagnoses (14%). At the same time, our meta-analysis calculated an AUC of 0.94 for the SROC curve. Since an AUC of 1.0 (100%) indicates spotless discriminating ability, our meta-analysis suggests a relatively high level of overall diagnostic accuracy. With 1 accord, a pooled DOR of 63.40 was calculated indicated a competitive discriminatory performance. The optimal DOR and AUC indicate that PE might be a suitable screening samples for detection of EGFR mutation. However, pooled PLR and NLR were moderate in our study. Our meta-analysis indicated a pooled PLR of 8.53. This is dismal for clinical use to some extent. Similarly, the pooled NLR was 0.18, which is not low enough to draw a diagnosis of exclusion in the clinic.
Variant types of mutations in EGFR of NSCLC were detected in PE, most mutations focus on exon 19, 20, and 21 regardless of the sample type, Such as exon 19 del, exon 20 (T790 M), and exon 21 (L858R). About 90% of these mutations are deletions of exon 19 and point mutations of exon 21, which are known to be sensitive for TKIs therapy. T790 M mutation of exon 20 is responsible to TKIs resistance. Types of EGFR mutations, including secondary mutations and resistance mutations, can be detected in PE samples. Our work is not so much about the specific mutation type, because most of original publications did not pay attention to the topic.
Several assays were used in our included studies. ARMS, HRM, PCR, and direct sequencing were used at a relatively high frequency. Thus, we compared the pooled diagnostic accuracy of these 4 methods. Direct sequencing is a historical method used to detect EGFR mutations and provides detailed mutation information. However, detecting mutations in this way requires at least 30% of the mutant DNA within the sample, which can lead to less satisfactory sensitivity (0.78 in our meta-analysis).[40] Due to the pessimistic sensitivity of direct sequencing, patients with EGFR mutations may be missed as not having any mutations.[41] Our results suggest PCR has the highest diagnostic performance, which is consistent with previous study.[9] PCR had an excellent specificity of 0.95, which means when a patient is diagnosed with EGFR mutations, there is only 5% chance of misdiagnosed. This was satisfactory if it triggers invasive diagnostic procedures or targeted treatment.[42] Another method, HRM, is a powerful method that judges the existence of mutations without detailed mutation information. In spite of its optimal specificity (1.00) and DOR (75.47), it still cannot completely replace the sequencing method. Further study is needed to make up for its imperfections in clarifying the mutation type and gene sequencing. However, it should be mentioned that not only the assay but also reagents, DNA quality, software, and crucially primer design and amplicon size have impact on mutation detection potential. A standardised method of detecting EGFR mutations in PE samples is needed urgently.
Two types of PE samples were tested in our included studies. We made a comparison and our results showed that supernatants have a more optimal diagnostic performance than cell blocks (AUC: 0.96 vs 0.94). We conclude that cell-free supernatants might be a better resource for mutation detection than cell pellets. It is in keeping with previous studies.[18,19] The probably reason is tumor DNA may be released into PE through the apoptosis or necrosis of disseminated tumor cells. And supernatants has more dissolved tumor DNA. Although the tumor cells in PE are fewer than that in tumor tissues, we can more or less make up for deficiencies by extracting the supernatants part for detection as far as possible. Besides, the test procedure of supernatants EGFR mutation is simplified by eliminating the need of embedding. It is easier to purify DNA from PE supernatants than from tumor tissues. Examination of PE is rapid, efficient, and minimally invasive. Supernatants of PE might be a more suitable substitute of tumor tissues than cell blocks.
A meta-analysis by Luo et al assessed the diagnostic performance of cfDNA, compared with tissues.[43] The pooled specificity was 0.935 and pooled sensitivity was 0.674. Another meta-analysis by Chen Mao et al evaluated the accuracy of EGFR mutation testing in blood against that in tumor tissues, and drew the conclusion that serum showed lower sensitivity (0.56 vs 0.65) but higher specificity (0.95 vs 0.85) than plasma.[39] The pooled results in our meta-analysis indicate a relatively higher sensitivity of 0.86, and an optimal specificity of 0.93. It seems that EGFR mutations detection in PE samples performs better than that in blood samples. Maybe the local site influence can explain this. PE is concentrated at the site of tumor lesion, such as pleural cavity, while peripheral blood is relatively far away from tumor lesion.
Several limitations should be discussed. First, only 15 publications were included after the strict search strategy and study selection. The statistical power maybe inadequate for drawing definitive conclusions about the ability of detecting EGFR mutation in PE. The relatively low prevalence of NSCLC complicated with PE may contribute to the limited original research.[44] Lacking of studies which compare the EGFR mutation test results in paired tumor tissues and PE samples urgently calls for further studies on a large scale. Second, although the publication bias were not significant in our meta-analysis, we have to mention that we only include articles written in English and Chinese because of language restrictions. Moreover, all of the included studies were conducted in Asia, although the mutation frequency of Asians is significantly higher than that of Caucasians (30–40% vs 10%),[45] this may lead to bias. Third, the methodological quality of included studies were modest, based on QUADAS-2 assessment. This was mainly due to the lack of key information. Although substantial heterogeneity was detected across the included studies using meta-regression, we were unable to identify causes. Future research should be more rigorous in order to reduce the risk of bias.
In conclusions, the present meta-analysis suggests that PE, especially supernatants, is a promising substitute for tumor tissue in EGFR mutations testing of patients with NSCLC. Its relatively high specificity suggests that EGFR mutation positivity in PE could be used to recommend EGFR TKIs treatment. Further study and optimization are required to improve the sensitivity and identify the best testing methods.
Acknowledgments
The funders had no role in design, data collection and analysis for this study, decision to publish, or preparation of the manuscript.
Author contributions
Data curation: Jiangyue Qin, Sixiong Wang.
Methodology: Caishuang Pang, Huiwen Ma, Jiangyue Qin, Sixiong Wang, Ting Yang.
Supervision: Yongchun Shen, Donglin Wang.
Validation: Yongchun Shen, Donglin Wang.
Writing – original draft: Caishuang Pang, Huiwen Ma, Chun Wan.
Writing – review & editing: Caishuang Pang.
Footnotes
Abbreviations: ARMS = amplification refractory mutation system, AUC = area under the curve, DOR = diagnostic odds ratio, EGFR = epidermal growth factor receptor, FN = false negative, FP = false positive, HRM = high resolution melting, NLR = negative likelihood ratio, NSCLC = non-small cell lung cancer, PCR = polymerase chain reaction, PE = pleural effusion, PLR = positive likelihood ratio, SROC = summary receiver operating characteristic, TKIs = tyrosine kinase inhibitors, TN = true negative, TP = true positive.
Institutional review board approval was not required for this retrospective meta-analysis.
Informed consent was not required for this retrospective meta-analysis.
This work was supported by National Key Research and Development Program (2016YFC1304500 and 2016YFC0903600), the Science and Technology Pillar Program of the Department of Science and Technology of Sichuan province (2015SZ0151), and Medical scientific research project of Chongqing Municipal Health and Family Planning Commission (2015ZDXM040).
The authors declare that they have no competing interests.
References
- [1].Inamura K. Lung cancer: understanding its molecular pathology and the 2015 WHO clasification. Front Oncol 2017;7:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. A Cancer J Clin 2017;67:7–30. [DOI] [PubMed] [Google Scholar]
- [3].Brueckl WM. Treatment choice in EGFR-mutant non-small-cell lung cancer. Lancet Oncol 2017;18:1425–6. [DOI] [PubMed] [Google Scholar]
- [4].Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299–311. [DOI] [PubMed] [Google Scholar]
- [5].Lee CK, Davies L, Wu YL, et al. Gefitinib or erlotinib vs chemotherapy for egfr mutation-positive lung cancer: individual patient data meta-analysis of overall survival. J Natl Cancer Inst 2017;109(6.): doi:10.1093/jnci/djw279. [DOI] [PubMed] [Google Scholar]
- [6].Fenton KN, Richardson JD. Diagnosis and management of malignant pleural effusions. Am J Surg 1995;170:69–74. [DOI] [PubMed] [Google Scholar]
- [7].Eberhard DA, Giaccone G, Johnson BE. Non-small-cell lung cancer working group. Biomarkers of response to epidermal growth factor receptor inhibitors in non-small-cell lung cancer working group: standardization for use in the clinical. J Clin Oncol 2008;26:983–94. doi: 10.1200/JCO.2007.12.9858. [DOI] [PubMed] [Google Scholar]
- [8].Xu HM, Sun WY, Zhang G, et al. Detection of epidermal growth factor receptor mutation in non-small-cell lung carcinoma using cytological and histological specimens. JBUON 2015;20:142–5. [PubMed] [Google Scholar]
- [9].Zhong J, Li X, Bai H, et al. Malignant pleural effusion cell blocks are substitutes for tissue in EML4-ALK rearrangement detection in patients with advanced non-small-cell lung cancer. Cytopathology 2016;27:433–43. doi:10.1111/cyt. 12322. [DOI] [PubMed] [Google Scholar]
- [10].Moher D, Liberati A, Tetzlaff J, et al. PRISMA Group. Preferred reporting items for systematic reviews and meta analyses: the PRISMA statement. PLoS Med 2009;6:e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Leeflang MM, Deeks JJ, Takwoingi Y, et al. Cochrane diagnostic test accuracy reviews. Syst Rev 2013;2:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Leeflang MM, Deeks JJ, Gatsonis C, et al. Systematic reviews of diagnostic test accuracy. Ann Intern Med 2008;149:889–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011;155:529–36. [DOI] [PubMed] [Google Scholar]
- [14].Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol 2005;58:882–93. [DOI] [PubMed] [Google Scholar]
- [15].Wang S, Han XH, Li JL, et al. Detection of epidermal growth factor receptor gene mutations in advanced non-small cell lung cancer. Chin J Pathol 2012;41:530–3. [DOI] [PubMed] [Google Scholar]
- [16].Yeo CD, Kim JW, Kim KH, et al. Detection and comparison of EGFR mutations in matched tumor tissues, cell blocks, pleural effusions, and sera from patients with NSCLC with malignant pleural effusion, by PNA clamping and direct sequencing. Lung Cancer 2013;81:207–12. [DOI] [PubMed] [Google Scholar]
- [17].Liu X, Lu Y, Zhu G, et al. The diagnostic accuracy of pleural effusion and plasma samples versus tumor tissue for detection of EGFR mutation in patients with advanced non-small cell lung cancer: comparison of methodologies. J Clin Pathol 2013;66:1065–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lin J, Gu Y, Du R, et al. Detection of EGFR mutation in supernatant, cell pellets of pleural effusion and tumor tissues from non-small cell lung cancer patients by high resolution melting analysis and sequencing. Int J Clin Exp Pathol 2014;7:8813–22. [PMC free article] [PubMed] [Google Scholar]
- [19].Liu D, Lu Y, Hu Z, et al. Malignant pleural effusion supernatants are substitutes for metastatic pleural tumor tissues in EGFR mutation test in patients with advanced lung adenocarcinoma. PLoS One 2014;9:e89946doi:10.1371/journal. pone. 0089946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Guan Y, Wang ZJ, Wang LQ, et al. Comparison of EGFR mutation rates in lung adenocarcinoma tissue and pleural effusion samples. Genet Mol Res 2016;15(2.): doi: 10.4238/gmr.15027001. [DOI] [PubMed] [Google Scholar]
- [21].Liu N, Sun RZ, Du J, et al. Comparison of epidermal growth factor receptor gene mutations identified using pleural effusion and primary tumor tissue samples in non-small cell lung cancer. Appl Immunohistochem Mol Morphol 2018;26:e44–51. doi:10.1097/PAI. 00000000 00000543. [DOI] [PubMed] [Google Scholar]
- [22].Wei B, Ma J, Ma ZY, et al. The study of detection EGFR mutation in pleural effusion of lung adenocacinoma. J Clin Exp pathol 2012;28:323–5. [Google Scholar]
- [23].Xie Q, Chen Q, Shi Q, et al. Detection of EGFR gene mutations in thoracoscopy biopsy and pleural effusion in advanced non small cell lung cancer. J Clin Exp Pathol 2013;29:881–3. [Google Scholar]
- [24].Wan LJ, Zou JT, Luo H, et al. Detection EGFR mutation in pleural effusion and blood of NSCLC. Shandong Med Pharm 2013;53:53–5. [Google Scholar]
- [25].Zhao J, Tian QM, Wu BP. Comparison of EGFR mutation in pleural effusion and biopsies of NSCLC. Heilongjiang Med Pharm 2014;37:109–10. [Google Scholar]
- [26].Yang YC, Wang XM, Li L, et al. Detection of EGFR K-ras mutations and ELM4-alk fusion gene in non-small cell lung cancer using cytological specimen materials and their clinical pathology significance. Chin J Clin Thorac Cardiovasc Surg 2015;22:870–4. [Google Scholar]
- [27].Yang X, Wang QS, X HL. Analysis of cells blocks EGFR mutation in pleural effusion of lung adenocarcinoma. J Clin Exp Pathol 2016;32:61–3. [Google Scholar]
- [28].Wang S, Li M, Wang L, et al. The study of detection EGFR gene mutation in non small cell lung cancer tissue and matched pleural effusion. Modern Oncol 2017;25:1729–31. [Google Scholar]
- [29].Yu L, Zhu QG, Yang LM, et al. Clinical significance of EGFR gene mutation in pleural effusion cell blocks in non-small cell lung cancer. J Shanxi Med Univ 2017;48:462–6. [Google Scholar]
- [30].Review Manager (RevMan) [Computer program] Version 5.2. Copenhagen: The Nordic Cochrane Centre. 2012;The Cochrane Collaboration. [Google Scholar]
- [31].Huang SF, Liu HP, Li LH, et al. High frequency of epidermal growth factor receptor mutations with complex patterns in non-small cell lung cancers related to gefitinib responsiveness in Taiwan. Clin Cancer Res 2004;10:8195–203. [DOI] [PubMed] [Google Scholar]
- [32].To kumo M, Toyooka S, Kiura K, et al. The relationship between epidermal growth factor receptor mutations and clinicopathologic features in non-small cell lung cancers. Clin Cancer Res 2005;11:1167–73. [PubMed] [Google Scholar]
- [33].Bentel JM, Thomas MA, Sadowska A, et al. From theory to practice: EGFR mutation testing in current practice and roadblocks to its implementation. J OncoPathol 2013;1:93–102. [Google Scholar]
- [34].Lozano MD, Zulueta JJ, Cheveste JI, et al. Assessment of epidermal growth factor receptor and K-ras mutation status in cytological stained smears of non-small cell lung cancer patients: correlation with clinical outcomes. Oncologist 2011;16:877–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Simone G, Mangia A, Malfettone A, et al. Chromogenic in situ hybridization to detect EGFR gene copy number in cell blocks from fine-needle aspirates of non small cell lung carcinomas and lung metastases from colo-rectal cancer. J Exp Clin Cancer Res 2010;29:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Smits AJ, Kummer JA, Hinrichs JW, et al. EGFR and KRAS mutations in lung carcinomas in the Dutch population: increased EGFR mutation frequency in malignant pleural effusion of lung adenocarcinoma. Cell Oncol (Dordr) 2012;35:189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Nakamura T, Sueoka-Aragane N, Iwanaga K, et al. Application of a highly sensitive detection system for epidermal growth factor receptor mutations in plasma DNA. J Thorac Oncol 2012;7:1369–81. [DOI] [PubMed] [Google Scholar]
- [38].Kim ST, Jung HY, Sung JS, et al. Can serum be used for analyzing the EGFR mutation status in patients with advanced non-small cell lung cancer? Am J Clin Oncol 2013;36:57–63. [DOI] [PubMed] [Google Scholar]
- [39].Mao C, Yuan JQ, Yang ZY, et al. Blood as a substitute for tumor tissue in detecting EGFR mutations for guiding EGFR TKIs treatment of nonsmall cell lung cancer. Medicine 2015;94:e775doi: 10.1097/MD. 0000000000000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Fan J, Zhang Q, Tochio H, et al. Structural basis of diverse sequence-dependent target recognition by the 8 kDa dynein light chain. J Mol Biol 2001;306:97–108. [DOI] [PubMed] [Google Scholar]
- [41].Zhang X, Zhao Y, Wang M, et al. Detection and comparison of epidermal growth factor receptor mutations in cells and fluid of malignant pleural effusion in non-small cell lung cancer. Lung Cancer 2008;60:175–82. [DOI] [PubMed] [Google Scholar]
- [42].Brawley OW, Kramer BS. Cancer screening in theory and in practice. J Clin Oncol 2005;23:293–300. [DOI] [PubMed] [Google Scholar]
- [43].Luo J, Shen L, Zheng D. Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: a systematic review and meta-analysis. Sci Rep 2014;4:6269doi:10.1038/srep06269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lippincott Williams and Wilkins, Pass HI, Johnson DH, Minna JD, et al. Lung cancer: principles and practice. 3rd ed.2005. [Google Scholar]
- [45].Zhang X, Chang A. Somatic mutations of the epidermal growth factor receptor and non-small-cell lung cancer. J Med Genet 2007;44:166–72. [DOI] [PMC free article] [PubMed] [Google Scholar]


