Summary
Bacterial growth and division require insertion of new peptidoglycan (PG) into the existing cell wall by PG synthase enzymes. Emerging evidence suggests that many PG synthases require activation to function, however it is unclear how activation of division-specific PG synthases occurs. The FtsZ cytoskeleton has been implicated as a regulator of PG synthesis during division, but the mechanisms through which it acts are unknown. Here we show that FzlA, an FtsZ-binding protein and essential regulator of constriction in Caulobacter crescentus, helps link FtsZ to PG synthesis to promote division. We find that hyperactive mutants of the PG synthases FtsW and FtsI specifically render fzlA, but not other division genes, non-essential. However, FzlA is still required to maintain proper constriction rate and efficiency in a hyperactive PG synthase background. Intriguingly, loss of fzlA in the presence of hyperactivated FtsWI causes cells to rotate about the division plane during constriction and sensitizes cells to cell wall-specific antibiotics. We demonstrate that FzlA-dependent signaling to division-specific PG synthesis is conserved in another α-proteobacterium, Agrobacterium tumefaciens. These data establish that FzlA helps link FtsZ to cell wall remodeling, and is required for signaling to both activate and spatially orient PG synthesis during division. Overall, our findings support the paradigm that activation of SEDS-PBP PG synthases is a broadly conserved requirement for bacterial morphogenesis.
eTOC blurb
Lariviere et al. show that in the bacterium Caulobacter crescentus, the constriction regulator FzlA plays an essential role in signaling from FtsZ to peptidoglycan synthesis activation. fzlA, can be deleted upon hyperactivation of FtsWI and regulates cell shape maintenance and resilience to antibiotic stress.
Introduction
Bacterial division is driven by the insertion of new cell wall material at midcell in a tightly regulated manner, allowing for determination of cell shape and maintenance of envelope integrity [1,2]. The cell wall is made of peptidoglycan (PG), a meshwork consisting of glycan strands crosslinked by peptide stems [3,4]. PG synthesis requires the coordination of glycan polymerization and peptide crosslinking by either coupled monofunctional glycosyltransferases (GTases) and transpeptidases (TPases), or bifunctional enzymes that contain both activities, with these proteins being more generally referred to as PG synthases [1].
Monofunctional PG synthase pairs have been implicated as the primary synthetic enzymes of the elongation (elongasome) and division (divisome) machineries. A paradigm has been proposed whereby a shape, elongation, division, and sporulation (SEDS) family GTase is functionally coupled to a penicillin binding protein (PBP) TPase, which together facilitate cell wall synthesis [5–7]. Through characterization of the elongation-specific PG synthases RodA and PBP2 in Escherichia coli, it has been postulated that SEDS-PBP enzymes require activation to function [5]. Specifically, mutations in RodA or PBP2 that increase GTase activity in vitro and PG synthesis in cells render other components of the elongasome non-essential, arguing that their normal function is to activate the RodA-PBP2 complex [5]. Intriguingly, analogous mutations in the division-specific SEDS-PBP enzymes, FtsW and FtsI, allow cells to constrict faster than normal [8], indicating that these mutations promote formation of an activated PG synthase complex [5,9]. However, it is unclear precisely how SEDS-PBP activation normally occurs during division.
Recent studies have established that the conserved cytoskeletal protein FtsZ [10,11], which recruits the division machinery to a ring-like structure at midcell [12–14], is coupled to PG synthesis activation during division. In multiple organisms, the C-terminal linker domain of FtsZ was found to be required for regulating cell wall integrity [15–17] and shape, as well as PG chemistry [16,18]. Moreover, in E. coli and Bacillus subtilis, FtsZ dynamics were demonstrated to drive PG synthase dynamics in both organisms, as well as division site shape in E. coli and constriction rate in B. subtilis [19,20]. Collectively these data indicate that, at least in some organisms, FtsZ acts as a “dynamic scaffold” or “dynamic activator” of PG synthesis likely impinging on FtsWI. However, the signaling pathway connecting these two endpoints remains unresolved.
We previously demonstrated that an essential FtsZ-binding protein, FzlA [21], is required for division and regulates the rate of constriction in the α-proteobacterium Caulobacter crescentus [22]. Mutations in FzlA with diminished affinity for FtsZ were found to have slower constriction rates and altered cell pole shape, indicative of reduced PG synthetic activity during division [22]. We therefore postulated that FzlA facilitates a link between FtsZ and PG synthesis by serving as an upstream activator of PG synthases and, here, set out to test this hypothesis.
Results
fzlA lies upstream of ftsWI in a PG synthesis pathway
We reasoned that if FzlA impacts constriction through PG synthases, it likely acts on the division-specific SEDS family GTase FtsW and/or the monofunctional PBP TPase FtsI. To assess if FzlA is required for activation of FtsWI, we exploited fast-constricting strains containing hyperactive mutant variants of FtsI and/or FtsW termed ftsW**I* [8,9] and ftsW* [9]. ftsW**I* bears the mutations F145L and A246T in FtsW and I45V in FtsI, whereas ftsW* contains only the FtsW A246T mutation [9]. These mutations are thought to stabilize an activated form of the FtsWI complex [5,9], leading to increased rates of constriction [8] via unrestrained PG synthesis.
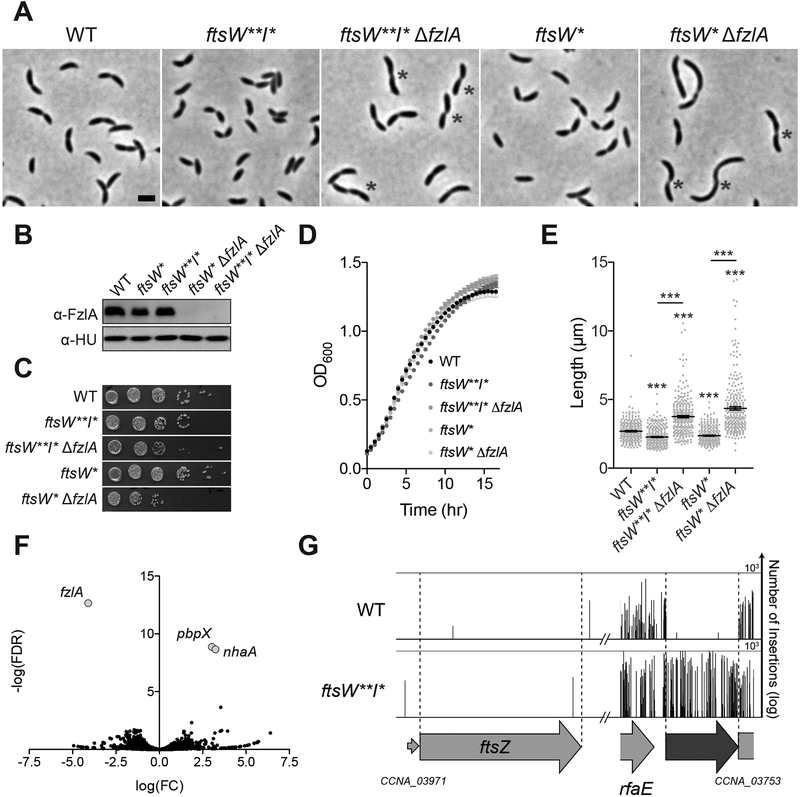
If fzlA lies upstream of ftsWI in a PG synthesis pathway, then the hyperactive variants ftsW**I* and/or ftsW* may bypass the essentiality of fzlA. Accordingly, we found that fzlA could be readily deleted in either the ftsW**I* or ftsW* strain backgrounds (Figure 1A–B). This is a particularly striking finding given that depletion of FzlA in a WT background completely inhibits division and induces cell filamentation and death [21]. Interestingly, a number of ftsW**I*/ftsW*ΔfzlA cells appeared to be “S”-shaped with the direction of curvature in future daughter cells facing opposite directions, as opposed to the characteristic “C”-shape of pre-divisional WT and ftsW**I* Caulobacter cells (Figure 1A, asterisk, discussed further below). Strains lacking fzlA displayed a slight reduction in colony forming units, compared to the corresponding hyperactive PG synthase mutant strains (Figure 1C), whereas growth rate was unaffected (Figure 1D). Additionally, ftsW**I*/ftsW*ΔfzlA cells displayed an increase in length (Figure 1E), suggesting a division defect. Because ftsW*ΔfzlA cells are longer than ftsW**I*ΔfzlA cells, we conclude that ftsW**I* suppresses loss of fzlA better than the single mutant.
Figure 1. Hyperactive ftsWI mutants suppress loss of fzlA.
(A) Micrographs of WT Caulobacter and PG synthase hyperactive mutant cells +/− fzlA. White asterisks mark S-shaped cells. Scale bar: 2 μm.
(B) α-FzlA immunoblot (top) and α-HU immunoblot (bottom, loading control) of the indicated strains.
(C, D) Spot dilutions (diluted ten-fold) (C) and growth curves (D) of the indicated strains.
(E) Lengths of unsynchronized cells from the indicated strains. Mean ± SEM shown. Kruskal-Wallis tests with Dunn’s post-test were performed to compare WT and the indicated strains: ***P≤0.001. From left to right, n = 254, 262, 261, 260, 258.
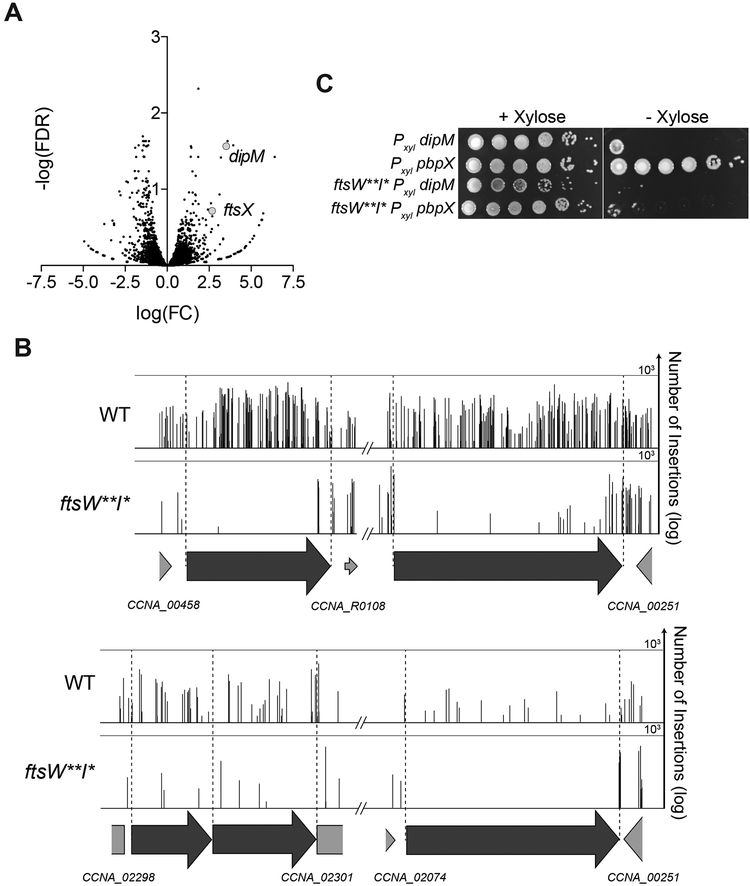
(F) Volcano plot of the negative log10 of the false discovery rate (−log(FDR)) vs. log2 of the fold change of each gene in WT vs. ftsW**I* strains determined by Tn-Seq analysis.
(G) Plot of transposon insertion frequency in essential division genes in WT (top) vs. ftsW**I* (bottom) cells determined by Tn-Seq analysis.
Strain key (Caulobacter): WT (EG865 A-E; EG2366 F-G), ftsW**I*(EG1557), ftsW**I* ΔfzlA (EG2170), ftsW*(EG1556), ftsW*ΔfzlA (EG2166). See also Figures S1–S3 and Table S1.
We also observed that ftsW**I* suppresses length, width, shape, and fitness defects associated with slowly constricting fzlA point mutants fzlANH2 and fzlANH3 (Figure S1), further indicating that hyperactivated ftsWI are dominant to, and likely downstream of fzlA. To determine the contribution of the FtsZ-FzlA interaction to activation of FtsWI, we assessed cell morphology, fitness, and cell length of ftsW**I* strains containing FzlA mutants with decreasing affinity for FtsZ [22] (Figure S2; FzlA > FzlANH2 = FzlANH3 > FzlANH1; FzlANB2, FzlANB1 = no binding). We found that decreased affinity of FzlA towards FtsZ correlated with an increase in cell length (Figure S2E), indicating that high-affinity binding to FtsZ is required for FzlA to contribute to the FtsWI activation pathway. We term this activation pathway the [FtsZ-FzlA]-FtsWI signaling pathway.
We previously showed that mutation of FzlA’s conserved C-terminus inhibits division, but does not affect binding to FtsZ, suggesting the C-terminus might facilitate an essential interaction with another protein [22]. We reasoned that if this other essential interaction is involved in the PG synthesis activation pathway, then FzlA’s C-terminus should be mutable in an ftsW**I*background. Indeed, fzlAUE2, which encodes a charge reversal mutation in the C-terminus of FzlA normally preventing division but not FtsZ-binding, phenocopies loss of fzlA in an ftsW**I*background. ftsW**I* fzlAUE2 cells are slightly elongated and “S”-shaped (Figure S3A), but have normal cell growth, viability, and FzlA levels (Figure S3B–D). Because the 5’ end of fzlAs coding sequence overlaps with the non-essential downstream gene, CCNA_03753, we exogenously expressed fzlAUE2 in an ftsW**I* ΔfzlA background to leave CCNA_03753 intact. Again, cells became slightly elongated and “S”-shaped in the presence of fzlAUE2 (Figure S3E), with no major impact on growth or viability compared to exogenous expression of fzlA (Figure S3F–H), confirming that change in cell shape is due to mutation of FzlA’s C-terminus. Because FzlA’s C-terminus becomes dispensable upon hyperactivation of ftsW**I*, we propose that the C-terminus facilitates interaction with another essential factor in the PG synthesis activation pathway.
ftsW**I* specifically permits loss of fzlA
To assess the specificity of the fzlA-ftsWI genetic interaction and potentially identify additional components of this pathway, we performed comparative transposon sequencing (Tn-Seq) on WT and ftsW**I*strains. Surprisingly, fzlA was the only essential gene to become non-essential in the ftsW**I* background, with few insertions in WT but plentiful insertions in ftsW**I* cells (Figure 1F–G, Table S1). All other known essential division genes, e.g. ftsZ (Figure 1G, Table S1), had few transposon insertions in either background; examination of known division genes’ insertion profiles showed all domains remain essential in ftsW**I*. These data indicate that ftsW**I*specifically rescues loss of fzlA, confirming that FzlA is normally required for activation of FtsWI. We expect that other essential division proteins participate in this pathway too, but that they have additional essential functions.
FzlA contributes to efficient division in a hyperactive PG synthase background
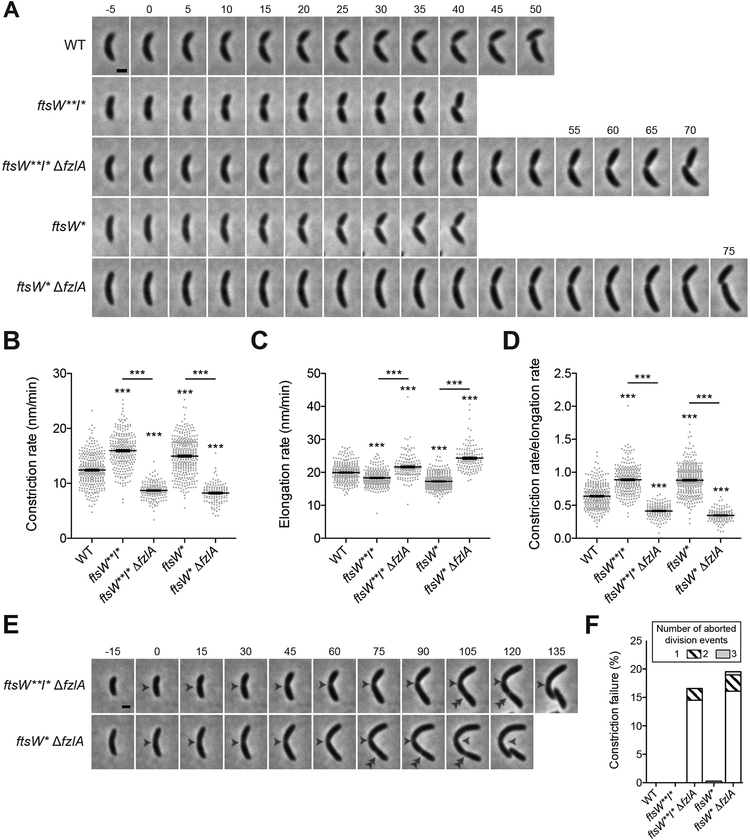
Given that cells lacking fzlA in the hyperactive PG synthase backgrounds were elongated, we assessed constriction rate and division efficiency of ftsW**I* and ftsW* cells ± fzlA using time-lapse microscopy and MicrobeJ to track division [22,23] (Figure 2A, Video S1). Consistent with previous findings [8], ftsW**I* and ftsW* cells constrict more quickly than WT (Figure 2B). Intriguingly, the hyperactive PG synthase strains lacking fzlA constricted significantly more slowly than the corresponding strain with fzlA present, with constriction rates cut nearly in half (Figure 2B). This suggests that hyperactivated FtsWI are not sufficient for efficient division and underscores the importance of FzlA in dictating constriction rate. As with cell length and fitness, ftsW**I* acted as a better suppressor to fzlA deletion, allowing for a faster constriction rate than did ftsW*(Figure 2B).
Figure 2. FzlA contributes to efficient division in a hyperactive PG synthase background.
(A) Time-lapse micrographs of constriction in WT or PG synthase hyperactive mutant cells +/− fzlA. Constriction starts at t=0 and concludes in the last frame upon cell separation. Time relative to constriction initiation (minutes) is indicated. Scale bar: 1 μm. (B, C, D) Plots of constriction rate (B), total elongation rate (C), and ratio of constriction rate to total elongation rate (D) for a population of synchronized cells from each indicated strain, calculated from single cell microscopy data. Mean ± SEM shown. Kruskal-Wallis tests with Dunn’s post-test were performed to compare WT and the indicated strains: ***P≤0.001. From left to right, n = 324, 280, 161, 366, 139 (B) and 321, 280, 161, 363,139 (C, D).
(E) Micrographs of constriction failure at the initial division site (single white arrowhead), then initiation and completion at a second site (double white arrowhead) in ΔfzlA cells. As in (A), constriction initiates at t=0 and concludes in the last frame. Scale bar: 1 μm.
(F) Plot of the constriction failure rate in cells in which constriction initiated, then failed at one division site and subsequently initiated and finished at another division site. “Number of aborted division events” refers to the number of times a cell abandoned division at distinct sites within the cell. The y-axis indicates the percentage of cells out of the whole population that aborted division at least once. From left to right, n = 324, 280, 193, 368, 174.
Strain key (Caulobacter): WT (EG865), ftsW**I* (EG1557), ftsW**I* ΔfzlA (EG2170), ftsW*(EG1556), ftsW*ΔfzlA (EG2166). See also Videos S1–S2.
To ensure that changes in constriction rate were not due to global differences in PG synthesis, we determined elongation rates across strains (Figure 2C), which enabled calculation of the ratio of constriction to elongation rate (Figure 2D). We saw the same trend as for constriction rate itself, with ftsW**I* and ftsW* mutant strains having higher ratios of constriction to elongation and loss of fzlA giving lower ratios (Figure 2D). Interestingly, elongation rate was inversely correlated with constriction rate in all mutant strains (Figure 2C–D), perhaps reflecting competition between the elongasome and divisome for PG precursor substrate [24]. Altogether, these data support the conclusion that alterations to the [FtsZ-FzlA]-FtsWI pathway affect constriction to a greater extent than they affect elongation, with FzlA increasing the constriction rate in both WT and hyperactive PG synthase mutant backgrounds.
While tracking division in ftsW**I*/ftsW*ΔfzlA cells, we noticed that some cells initiated, then aborted division at one location, before successfully dividing at a second (or third or fourth) site (Figure 2E, Video S2). We found that 16.6–19.5% of ftsW*/ftsW**I*ΔfzlA cells aborted division at one site before successfully dividing at another, compared to a 0–0.3% failure rate for WT or hyperactive PG synthase cells with fzlA present (Figure 2F). These data further demonstrate that ftsW**I*are not sufficient for efficient division, and that fzlA is required to ensure division processivity and efficiency.
fzlA is required for maintenance of proper cell shape
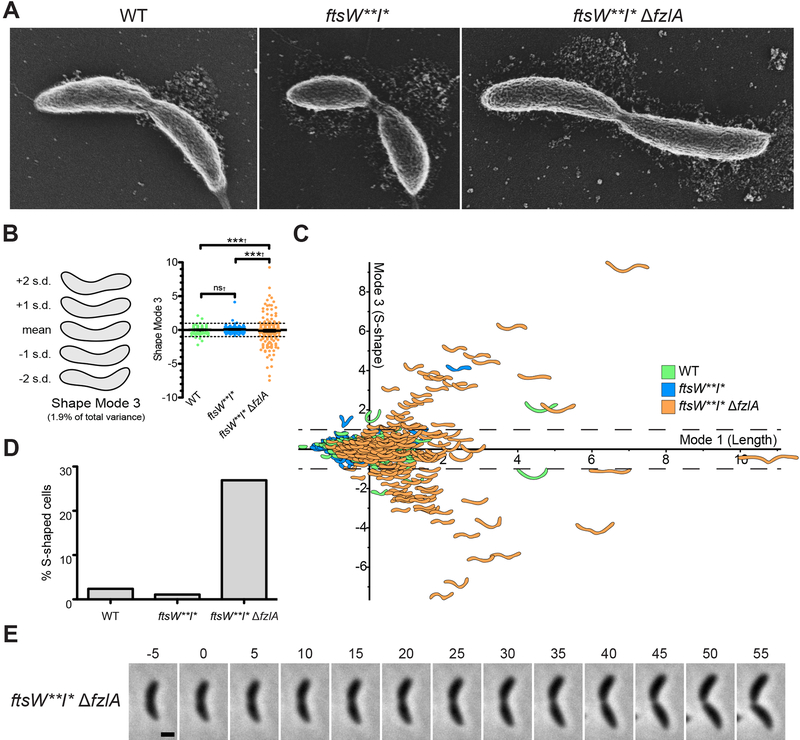
As mentioned earlier, deletion of fzlA in the hyperactive PG synthase backgrounds impacted global cell morphology, with many pre-divisional cells appearing “S-shaped”. By scanning electron microscopy (SEM), we saw a relatively high frequency of S-shaped ftsW**I*ΔfzlA cells, whereas most WT or ftsW**I* cells displayed the typical “C-shaped” morphology characteristic of Caulobacter (Figure 3A). To quantify S-shape frequency, we utilized Celltool to isolate variance in cell shape to shape modes [22,25]. Shape mode 3 captured the variation due to degree of S-versus C-shape and we set a cutoff such that cells with a standard deviation |sd| > 1 from the mean for this shape mode are considered S-shaped (Figure 3B–C). We found a significant difference in variance in degree of S-shape across populations (Figure 3B), corresponding with a large difference in S-shaped cells across strains: 26.9% of dividing ftsW**I*ΔfzlA cells are S-shaped, compared to 2.4% of WT and 1.1% of ftsW**I* cells (Figure 3D).
Figure 3. fzlA is required for global shape maintenance.
(A) SEM images of cells from the indicated strains. Scale bar: 200 nm.
(B) PCA of cell shape in a population of cells that have initiated constriction from the indicated strains. Shape mode 3 approximately captures degree of S-shape in cells. Mean cell contour ± 1 or 2 standard deviations (s.d.) is shown (left). Shape mode values for cells in each strain are plotted and mean ± SEM is shown (right). The dashed line drawn at s.d.=1 indicates the cutoff for S-shaped cells (cells with an |s.d.| ≥ 1 are considered to be S-shaped). A Brown-Forsythe Levene-type test (used in populations not assumed to be normally distributed) was performed using R to compare population variances (†): nsP>0.05, ***P≤0.001. From left to right, n = 292, 279, 290.
(C) Plot of shape mode 3 (S-shape) vs. shape mode 1 (length) values for a population of cells that have initiated constriction from the indicated strains. The dashed lines drawn at |s.d.|=1 indicates the cutoff for S-shaped cells.
(D) Plot of percentage of S-shaped cells present in a population of cells that have initiated constriction from the indicated strains. Cells from (B) with an |s.d.| ≥ 1 are considered S-shaped.
(E) Micrographs of cell twisting during division in an ftsW**I*ΔfzlA cell. Constriction starts at t=0 minutes and concludes in the last frame upon cell separation. Scale bar: 1 μm.
Strain key (Caulobacter): WT (EG865), ftsW**I* (EG1557), ftsW**I* ΔfzlA (EG2170). See also Figures S4–S5 and Video S3.
To shed light on the origin of S-shape, we next asked at what point during growth do ftsW**I*ΔfzlA cells begin to adopt this morphology. Using time-lapse microscopy, we observed that ftsW**I*ΔfzlA cells were C-shaped at the beginning of the cell cycle and began to twist or rotate about the division plane after constriction initiated. S-shape only became apparent in the latter part of constriction, when daughters had rotated ~180° relative to each other (Figure 3E, Video S3). This finding suggests that the [FtsZ-FzlA]-FtsWI pathway determines geometry of PG insertion at the site of division in a manner that influences global cell morphology, normally constraining cells in their characteristic C-shape as constriction progresses.
Changes in division site shape and formation of S-shaped cells have been previously linked to aberrant localization of FtsZ and the elongation factor, MreB, respectively [19,26–28]. However, mNG-FtsZ and Venus-MreB localization in ftsW**I* ΔfzlA cells was comparable to ftsW**I*cells (Figure S4A–B). Additionally, imaging with HADA did not reveal any gross changes in PG synthesis localization (Figure S4C). Together, these findings suggest that cell twisting is likely induced by a finer scale alteration of PG synthesis at the division site due to disruption of the [FtsZ-FzlA]-FtsWI pathway. Finally, we observed that affinity of mutant FzlA for FtsZ was inversely correlated with the frequency of S-shaped cells (Figure S5), demonstrating that the FtsZ-FzlA interaction is important for maintaining proper morphology.
The [FtsZ-FzlA]-FtsWI pathway contributes to resistance to PBP-targeting antibiotics
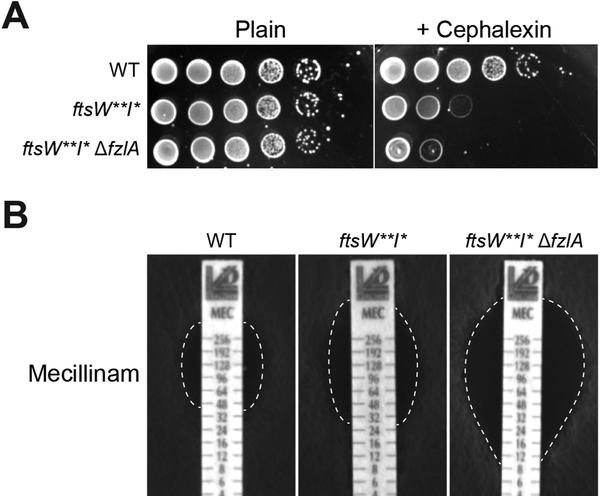
Because FzlA is important for regulation of PG synthesis in the context of determining constriction rate and cell shape, we hypothesized that it might also contribute to resistance to cell wall-targeting antibiotics. ftsW**I*, as has been previously shown [9], displayed sensitivity to cephalexin (Figure 4A), which inhibits FtsI and other penicillin-binding proteins in Caulobacter [29,30]. Interestingly, deletion of fzlA in the ftsW**I* background exacerbated sensitivity to cephalexin (Figure 4A). We found a similar trend upon treatment with mecillinam, which targets the elongation-specific PG synthase PBP2 [31,32], whereby the minimum inhibitory concentration (MIC) for ftsW**I* cells was decreased compared to WT, with deletion of fzlA further lowering the MIC (Figure 4B). However, neither ftsW**I*nor ftsW**I*ΔfzlA cells displayed sensitivity to the β-lactam ampicillin or antibiotics that block other aspects of cell wall synthesis (Figure S6A). fzlA therefore supports robust cell wall synthesis in the presence of PBP-inactivating drugs, perhaps by compensating for partial PBP-inactivation by boosting the intensity of cell wall synthesis activation signaling. To determine if the interaction between FtsZ and FzlA is important for maintaining cell wall integrity, we assessed sensitivity to cephalexin using the panel of fzlA mutants which display varying affinities towards FtsZ (Figure S6B). We found that mutants with decreased FzlA affinity towards FtsZ in fact became more sensitive to cephalexin (Figure S6B), demonstrating that the entire [FtsZFzlA]-FtsWI pathway is required for promoting cell wall integrity during antibiotic treatment.
Figure 4. Loss of fzlA leads to increased cell wall antibiotic sensitivity.
(A) Spot dilutions (diluted ten-fold) of the indicated strains plated on PYE ± cephalexin (6 μg/ml).
(B) Plates of the indicated strains grown in the presence of mecillinam minimum inhibitory concentration (MIC) test strips, with antibiotic concentration decreasing from top to bottom. The zone of clearance is highlighted in white (dashed line).
Strain key (Caulobacter): WT (EG865), ftsW**I*(EG1557), ftsW**I*ΔfzlA (EG2170). See also Figure S6.
Since ftsW**I* cells are more sensitive to perturbation of other PG synthetic activities even when fzlA is present, we asked if any normally non-essential division genes become more important for fitness in an ftsW**I* background, as they might help bolster resistance to assaults on PG synthesis. Examination of the ftsW**I* Tn-Seq data indicated that pbpX (encoding a bifunctional PG synthase) [33,34], and to a lesser extent ftsX (encoding a cell separation factor) [35] and dipM (encoding an envelope maintenance/cell separation factor) [35–38], had fewer transposon insertions in an ftsW**I* background than in WT (Figures 1F and 5A–B). Depletion of the otherwise nonessential DipM or PbpX completely inhibited growth of ftsW**I* cells (Figure 5C), validating our Tn-Seq results. Because ftsW**I* cells have misregulated division site PG synthase activity, we suspect that DipM and PbpX become important for ensuring robust PG synthesis during constriction. Surprisingly, the normally non-essential nhaA locus, coding for a putative sodium-proton antiporter [39], was also predicted by Tn-Seq to become essential in ftsW**I*cells (Figures 1F and 5A–B). Disruption of nhaA in the presence of sucrose has been shown to arrest division [39], suggesting nhaA may be important for osmoregulation during division under certain conditions, possibly explaining its apparent synthetic lethality with ftsW**I*.
Figure 5. Multiple non-essential division genes become essential in a hyperactive PG synthase background.
(A) Volcano plot of the negative log10 of the false discovery rate (−log(FDR)) vs. log2 of the fold change of each gene in WT vs. ftsW**I* strains determined by Tn-Seq analysis. This is a zoomed in and cropped view of the volcano plot from Figure 1F.
(B) Plot of transposon insertion frequency in essential division genes in WT (top) vs. ftsW**I* (bottom) cells. Genetic loci are annotated below the plot. Number of reads is displayed on a logarithmic scale.
(C) Spot dilutions (diluted ten-fold) of the indicated strains grown +/− xylose (to induce gene expression) or +/− glucose (non-inducer control).
Strain key (Caulobacter): WT (EG2366), ftsW**I*(EG1557), Pxyl dipM (EG2771), Pxyl pbpX (EG2772), ftsW**I* dipMΔC Pxyl dipM (EG2779), ftsW**I*pbpXΔC Pxyl pbpX (EG2781). See also Table S1.
The fzlA-ftsW genetic interaction is conserved in diverse α-proteobacteria
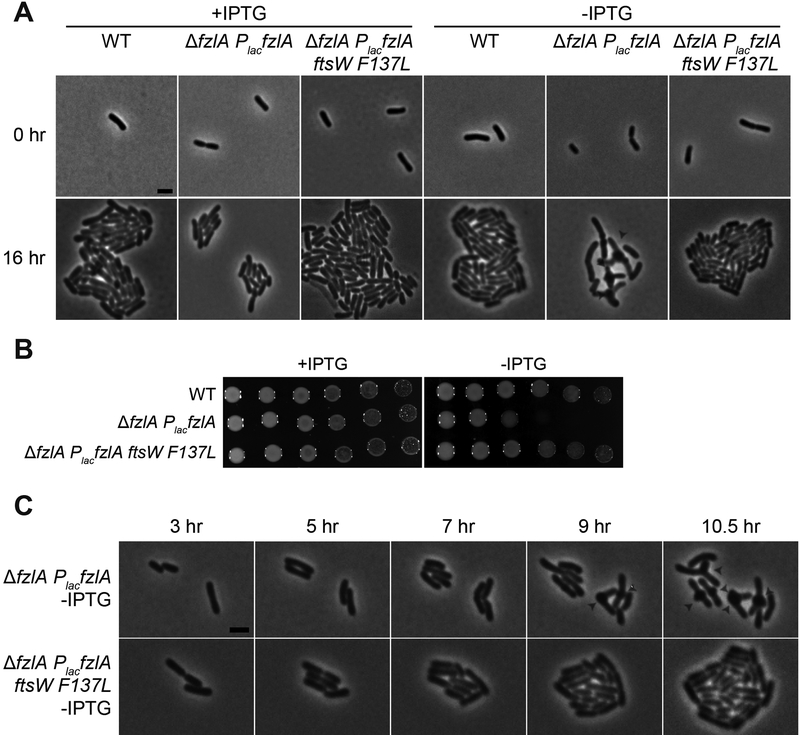
To assess the conservation of FzlA’s role in regulating PG synthesis, we sought to characterize the genetic interaction between fzlA and PG synthases in another α-proteobacterium, Agrobacterium tumefaciens. While A. tumefaciens and Caulobacter display disparate growth patterns [40–43], the components of the division machinery are largely conserved. To test if the genetic interaction between fzlA and ftsW is conserved, we made an IPTG-dependent FzlA depletion construct in a WT background or in a background with a single hyperactivating mutation in A. tumefaciens ftsW (F137L, equivalent to Caulobacter FtsW F145L) at the ftsW locus. Depletion of FzlA in a WT background resulted in reduced viability, division arrest, and ectopic pole formation at midcell (Figure 6), reminiscent of FtsW depletion in A. tumefaciens [18]. Importantly, we found that the decrease in viability and morphology defects associated with depletion of FzlA were rescued by ftsWF137L (Figure 6). These data indicate that FzlA’s essential role in regulating division-specific PG synthesis is conserved in another α-proteobacterium and further highlight the importance of FzlA as a key regulator of constriction and cell morphology.
Figure 6. The ability of hyperactive ftsW to suppress loss of fzlA is conserved.
(A) Micrographs of PG synthase hyperactive mutant cells ± FzlA in Agrobacterium tumefaciens. fzlA was induced where indicated with IPTG and depleted where indicated upon removal of IPTG, then grown for 16 hours on agarose pads. White arrowheads mark ectopic poles at midcell. Scale bar: 2 μm.
(B) Spot dilutions (diluted ten-fold) of the indicated strains grown in the presence or absence of IPTG to control fzlA expression, in A. tumefaciens.
(C) Phase contrast time-lapse microscopy images depicting WT and PG synthase hyperactive mutant cells depleted of FzlA over time. White arrowheads mark ectopic poles at midcell. Scale bar: 2 μm.
Strain key (A. tumefaciens): WT (PBA44)[55]; AfzlA PlacfzlA (PBA199); AfzlA PlacfzlA ftsWF137L (PBA232).
Discussion
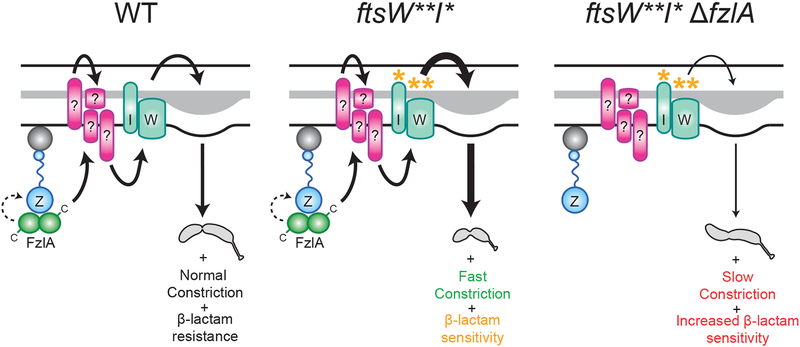
Here we have described a conserved PG synthesis activation pathway in which FtsZ and FzlA, likely via its C-terminus, regulates PG synthesis during division in α-proteobacteria by signaling to FtsWI (solid arrow) and/or possibly by influencing FtsZ dynamics (dashed arrow) (Figure 7, left). Specifically, the [FtsZ-FzlA]-FtsWI pathway determines geometry of cell wall insertion at the site of division, sets the constriction rate, and promotes cell wall integrity (Figure 7, left). FtsW**I* can still receive input from FzlA which, in combination with their intrinsic hyperactivity, leads to shorter, faster-constricting cells with sensitivity to cell wall antibiotics (Figure 7, middle). In the absence of fzlA, ftsW**I* cells lose critical regulation of PG synthesis, leading to rotation during division, slower constriction, and increased sensitivity to cell wall antibiotics (Figure 7, right). We establish FzlA as a critical participant in signaling from FtsZ to FtsWI and demonstrate that this division-specific SEDS-PBP pair require FzlA-dependent activation for normal division.
Figure 7. FzlA is required for activation of FtsWI and regulates the geometry of PG insertion.
FzlA is required for activation of FtsWI and likely signals (solid arrow) through unidentified intermediate factor(s), possibly via its C-terminus, in a manner dependent on interaction with FtsZ to effect normal cell shape, normal constriction rate, and antibiotic resistance (left). FzlA may also or alternatively contribute to cell wall synthesis activation by influencing FtsZ dynamics (dashed arrow, left). FtsW**I* can still receive input from FzlA (either through signaling, influence of FtsZ dynamics or some combination of both), which in addition to its own hyperactivity, leads to faster constriction and antibiotic sensitivity likely associated with positive misregulation of PG insertion (middle). FtsW**I* can function in the absence of FzlA, but with misregulated activity, leading to twisting during constriction, slower constriction speed, and increased antibiotic sensitivity (right).
On a molecular level, how does FzlA provide input into the PG synthesis process? We can envision at least two potential mechanisms. In the first, FzlA influences PG synthesis by regulating FtsZ superstructure or dynamics. According to this model, FzlA would function analogously to ZipA or FtsA, which have been proposed to contribute to constriction, at least in part, by influencing FtsZ assembly properties [44–49]. FtsW**I* hyperactivation could suppress loss of fzlA by overcoming the requirement for regulation by FtsZ dynamics or, perhaps less likely, by restoring the requisite dynamic properties of FtsZ. We previously demonstrated that FzlA can alter FtsZ superstructure and reduce its turnover in vitro under certain biochemical conditions [22], which would be consistent with this model. However, our prior identification of FzlA mutants (e.g. FzlANH3) that fail to alter FtsZ superstructure or dynamic properties in vitro but that, nevertheless, function in division suggests that these effects on FtsZ assembly properties are not required for activation of constriction [22]. We cannot, however, rule out the possibility that FzlA contributes to PG synthesis activation by influencing FtsZ dynamics.
We favor a model in which FzlA serves as a bridge between FtsZ and the conserved core PG synthesis activation pathway, transducing a cell wall synthesis activation signal. We have no evidence that FzlA and FtsWI directly interact, and hypothesize that other intermediary factor(s) transduce the activation signal from FzlA to FtsWI. Our finding that FzlA’s normally essential C-terminus, which is not involved in FtsZ binding [22], becomes non-essential in an ftsW**I* background implicates this region of the protein as the likely interface with the downstream division activation pathway. Genetic studies in E. coli have implicated a number of divisome proteins as part of the core divisome activation machinery, including FtsA, FtsN, FtsK, and FtsQLB [47,50–52], each of which is essential and conserved in Caulobacter. FtsN is proposed to signal to FtsQLB, either directly or through FtsA, to lead to FtsWI activation [46,47,50], with FtsK also playing a role in constriction licensing [51,52]. According to our signal transduction model, FzlA contributes to this activation signal by providing regulatory input via its C-terminus into the core conserved signaling pathway. The two models described above are not mutually exclusive; indeed, the two may work in concert (Figure 7). Future work will clarify how FzlA contributes to PG synthesis activation.
That cells become “S”-shaped upon deletion of fzlA and hyperactivation of ftsW**I* suggests that regulation of cell wall synthesis during division is required to globally maintain cell shape. Although further studies will be required to inform the precise mechanism of cell twisting, we propose a few potential models. If FzlA influences FtsZ dynamics, then loss of fzlA could result in aberrant FtsZ movement and misdirection of PG synthesis dynamics that ultimately induces rotation, somewhat reminiscent of the D158A and D212G ftsZ mutants in E. coli [19]. In those mutants, FtsZ forms helical structures that induce helical septa. For a similar model to hold in Caulobacter, the scale on which dynamics are altered would need to be significantly more finely focused, since FtsZ localization and septum morphology is obviously aberrant in the E. coli ftsZ mutants, but not in ftsW**I* ΔfzlA cells. Alternatively, loss of fzlA may impact PG synthase dynamics independent of FtsZ dynamics, effectively uncoupling the two, but still leading to aberrant movement of PG synthases and altered patterning of PG insertion. Finally, FzlA may be required for balanced transpeptidation and transglycosylation activities, thereby impacting PG chemistry and cell shape, as is the case for maintenance of helical shape in Helicobacter pylori [53].
More broadly, our model advances the idea that regulation of SEDS-PBP pairs for growth and division is conserved at numerous levels. The finding that FzlA governs division-specific PG synthesis in both Caulobacter and A. tumefaciens argues that α-proteobacteria require FzlA as a part of a conserved and dedicated pathway to activate FtsWI. FzlA is absent outside of this clade, however, so we propose that other divisome components play equivalent roles in contributing to FtsWI activation in other organisms. On a greater scale, our findings expand the paradigm for PG synthesis by SEDS-PBP PG synthase pairs in bacteria and provide evidence that the requirement for PG synthase activation is conserved. Elongation is facilitated by the coordination of the SEDS family GTase RodA and the monofunctional TPase PBP2, orthologs to FtsW and FtsI, respectively [5]. During elongation activation (in E. coli) these PG synthases are activated by another protein, MreC, forming an activated complex that in turn regulates assembly and directional motion of the polymerizing scaffold MreB [5]. In this system, hyperactivating mutations in RodA or PBP2 allow for bypass of the activator, MreC, similar to our finding that FtsW**I* can bypass the activator FzlA. Our data provide experimental support for the proposal that the requirement for activation of the SEDSPBP pair of PG synthases is generally conserved for elongation and division.
There are prominent differences between the models for elongasome and divisome activation, however. Containing fewer proteins than the divisome, the elongasome is akin to a stripped down version of the divisome [54], with hyperactive RodA or PBP2 rendering dispensable all elongasome components save MreB and the PG synthases [5]. Thus for elongation, the cell needs an activated SEDS-PBP pair and a spatial regulator to orient their motion [5]. Conversely, hyperactive FtsWI in Caulobacter only allows for disruption of FzlA, with the rest of the divisome remaining essential. Since division requires invagination and fission of all layers of the envelope in coordination with DNA segregation and cell cycle progression, this added complexity necessitates functions beyond PG synthesis and remodeling. Further, whereas PG synthesis during elongation comprises insertion of new PG in the same plane as old cell wall material, PG synthesis during constriction requires a lasting, directional change to shape the new cell poles. So while there are key similarities in the paradigm of PG synthase activation, regulation of division likely requires a more complicated network of inputs to manage the additional outputs and constraints discussed above. In summation, this work provides evidence that the requirement for SEDS-PBP activation is conserved across multiple modes of PG synthesis, which has broad implications for determining the speed of division, cell shape, and cell wall robustness.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Erin Goley (egoley1@jhmi.edu).
Experimental Model and Subject Details
Strains
Strains (Table S2) used in this study can be found in the supplemental information. Caulobacter crescentus strains were derived from the NA1000 WT strain. Unless otherwise indicated, Caulobacter colonies were isolated from solid 1.5% agar peptone yeast extract (PYE) plates grown at 30 °C and cells were grown in liquid culture in PYE shaking at 30 °C. Where indicated, Caulobacter cells were treated with 6 μg/ml of cephalexin. Antibiotic MIC analysis was performed using antibiotic test strips (Liofilchem), which include a concentration gradient of 0.016–256 mg/L for all antibiotics tested. Where indicated, cells were treated with 0.3% xylose to drive inducible gene expression or with 0.3% glucose as a non-inducer control.
A. tumefaciens were grown in A. tumefaciens glucose and (NH4)2S04 (ATGN) minimal medium [56], with 0.5% glucose at 28°C. E. coli strains were grown in LB medium at 37°C. IPTG was added at a concentration of 1 mM when necessary. To make the ΔfzlA PlacfzlA strain (PBA199), first, a mini-Tn7 vector containing IPTG inducible fzlA, along with the pTNS3 helper plasmid, were introduced into ΔtetRA a-attTn7 cells (PBA44) via electroporation as previously described [55]. Deletion of fzlA (for PBA199) and allelic exchange of ftsW (for PBA232) were subsequently performed by transferring the corresponding suicide vector to A. tumefaciens via conjugation with E. coli S17.
Method Details
Plasmid construction
Plasmids (Table S3) used in this study can be found in the supplemental information. Plasmids were created using standard molecular cloning procedures including PCR, restriction digestion, and ligation. Mutagenesis of ftsW for A. tumefaciens was performed using a QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Agilent Genomics), with primers designed using Agilent’s QuikChange Primer Design Program and the manufacturer’s protocol. pEG1345 was constructed using an NEBuilder HiFi DNA Assembly Cloning Kit (NEB).
Spot dilutions, growth analysis, and synchrony
Spot dilutions for Caulobacter were performed by serially diluting cells at the indicated fraction (1/10 or 1/2), before plating. Growth rates were obtained by measuring optical density at 600 nm (OD600) values of cells every 30 minutes. Spot dilutions and growth assays were performed in triplicate. Caulobacter cell synchrony was performed as previously described [12]. Briefly, log phase cells were washed with M2 salts (6.1 mM Na2HPO4, 3.9 mM KH2PO4, 9.3 mM NH4Cl) [57], resuspended in 1:1 M2:Percoll, then centrifuged at 11,200 x g. The swarmer band was isolated, and cells were subsequently washed twice in M2, then resuspended in PYE.
For A. tumefaciens spot dilutions, cells were grown overnight in ATGN minimal medium in the presence of IPTG at 1 mM concentration, washed, then pre-depleted of IPTG for 16 hours where indicated. Cells were then serially diluted (ten-fold) and spotted on ATGN minimal medium with the presence or absence of IPTG.
Light microscopy imaging and analysis
Images of log phase Caulobacter cells were obtained using either phase contrast microscopy, with cells grown on either 1% agarose PYE pads or 1% agarose dH2O pads, or, when indicated, fluorescence microscopy, with cells grown on 1% agarose dH2O pads. For fluorescence microscopy, mNG-FtsZ expression was induced for 1 hour with xylose then imaged through the GFP filter and venus-MreB expression was induced for 2 hours with xylose then imaged through the YFP filter. For determination of PG incorporation localization, cells were pulsed with 0.82 mM HADA for 5 minutes, washed twice with PBS, then visualized through the DAPI filter. For time-lapse imaging, synchronized cells were placed on 1% agarose PYE pads and imaged using phase contrast microscopy at room temperature (RT), with images being acquired at 5 minute intervals at 100x. Imaging of Caulobacter cells was performed using a Nikon Eclipse Ti inverted microscope with a Nikon Plan Fluor × 100 (numeric aperture 1.30) oil Ph3 objective and Photometrics CoolSNAP HQ2 cooled CCD (charge-coupled device) camera. For A. tumefaciens phase contrast microscopy, exponentially growing cells were spotted on 1% agarose ATGN pads as previously described [58], then imaged. For A. tumefaciens time-lapse microscopy, images were collected every ten minutes. Microscopy of A. tumefaciens cells was performed with an inverted Nikon Eclipse TiE with a QImaging Rolera em-c2 1K EMCCD camera and Nikon Elements Imaging Software.
For determination of dimensions of log phase cells, cell length and width were measured using FIJI [59] and MicrobeJ software, similar to as previously described [22]. Constriction rate and elongation rate were also determined using MicrobeJ [23] after blinding image file names. Briefly, MicrobeJ software allowed for tracking of cells imaged by time-lapse microscopy throughout the division process, with automatic detection of constriction initiation and manual determination of cell separation. Cell length was found for cells at each time point, cell width was found at the site of constriction, and constriction time was calculated by multiplying the number of frames in which constriction was detected by 5 (since images were acquired every 5 minutes), allowing for calculation of constriction and elongation rates. Constriction failure rate was determined by counting the number of cells which initiated constriction at one division site, failed, then ultimately divided at a separate site. Prism was used for graphing and statistical analysis of calculated terms. All cells that were in focus were included in these analyses.
Cell shape analysis
For cell shape analysis, binary masks of phase contrast images of log phase cells were inputted into Celltool [25], allowing for creation of cell contours, similar to as previously described [22]. Following alignment of cell contours (not allowing for reflection), a model of cell shape was created. The shape modes of interest were either plotted as histograms displaying the cell shape across two dimensions, or as single data points. R software was used to perform statistical analyses to compare population variances in shape modes across strains. Prism was used for graphing calculated terms. All cells that were in focus were included in these analyses except where noted otherwise in the text.
Scanning electron microscopy sample preparation and imaging
For SEM, log phase cells were incubated on poly-lysine (1:10) coated glass cover slip for 15 minutes, then fixed for 1 hour using fixation buffer (1% glutaraldehyde, 0.02 M cacodylate, and 3 mM MgCl2). Cells were gradually dried by washing 3 times with wash buffer (3% sucrose, 0.02 M cacodylate, and 3 mM MgCl2), twice with dH2O, once each with 30%, 50%, 70%, 90%, 100% ethanol, once with 1:1 ethanol:hexamethyldisiloxane (HMDS), and once with HMDS at 5 minute intervals each, before desiccation overnight. Cover slips were mounted, then coated with a 15 nm gold palladium sputter coat. Samples were then imaged with a LEO/Zeiss Field-emission SEM. Representative images are presented.
Immunoblot analysis
Immunoblot analysis was performed using standard procedures, with a 1:5,000 −1:6,666 dilution of α-FzlA primary antibody [21], a 1:50,000 dilution of α-HU primary antibody [60], and/or 1:10,000 of HRP-labeled α-rabbit secondary antibody (PerkinElmer) on nitrocellulose membranes. Chemiluminescent substrate (PerkinElmer) was added to facilitate protein visualization via an Amersham Imager 600 RGB gel and membrane imager (GE).
Transposon library preparation, sequencing, and analysis
Wild type Caulobacter crescentus NA1000 (EG2366) or ftsW**I*triple mutant (EG1557) cells were grown in a large culture (1 liter) to mid-log (0.4–0.6), washed of excess Mg2+ with 10% glycerol, and mutagenized with the Ez-Tn5 <Kan-2> transposome (Epicentre). Cells recovered by shaking at 30 °C for 90 minutes, then plated on kanamycin containing plates for 3 days at 30 °C in order to yield roughly 100–500 colonies per plate. Libraries were grown at 30 °C and comprised ~100,000–200,000 colonies each. Mutants were pooled into one library by scraping colonies from the surface of the agar and added into ~25–40 mL PYE. Pooled libraries were shaken to yield a homogenous slurry and sterile glycerol was added to 20%. Libraries were then frozen in liquid nitrogen and stored at −80 °C. Two libraries of each genetic background were prepared individually and compared as biological replicates.
Genomic DNA was extracted from one aliquot of each library using DNeasy Blood and Tissue Kit (Qiagen). Tn-Seq libraries were prepared for Illumina Next-Generation sequencing through sequential PCR amplifications using arbitrary hexamer primers and Tn5-specific primer facing outward for the first round, and indexing primer sets that include unique molecular identifier to filter artifacts arising from PCR duplicates for the second round. Libraries were then pooled and sequenced at the University of Massachusetts Amherst Genomics Core Facility on the NextSeq 550 (Illumina).
For analyses, reads were demultiplexed by index, then each sample Tn-Seq library was concatenated and clipped of the unique molecular identifier linker from the second PCR using Je [61] and the following command:
java -jar /je_1.2/je_1.2_bundle.jar clip F1=compiled.gz LEN=6
Clipped reads were then mapped back to the Caulobacter NA1000 genome (NCBI Reference Sequence: NC_011916.1) using BWA [62] and sorted using Samtools [63]:
bwa mem -t2 clipped.gz | samtools sort -@2 - > sorted.bam
Duplicate reads were removed using Je [61] and indexed with Samtools [63] using the following command:
java -jar /je_1.2/je_1.2_bundle.jar markdupes I=sorted.bam
O=marked.bam M=METRICS.txt MM=0 REMOVE_DUPLICATES=TRUE
samtools index marked.bam
5’ sites of inserted transposons from each library were converted into .wig files containing counts per position and viewed using Integrative Genomics Viewer [64,65]. Coverage and insertion frequency using a bedfile containing all open reading frames from NC_011916.1 with the outer 20% of each gene removed were determined using BEDTools [66] and the following commands:
bedtools genomecov −5 -bg marked.bam > marked.bed
bedtools map -a NA1000.txt -b marked.bed -c 4 > output.txt
Comparison of transposon insertions was performed using the edgeR package in the Bioconductor suite [67,68] using a quasi-likelihood F-test (glmQLFit) to determine the false discovery rate adjusted p-values reported here.
Quantification and Statistical Analysis
Constriction analysis was performed using FIJI and MicrobeJ, as indicated in the Method Details section. Cell shape analysis was performed using Cellool, as indicated in the Method Details section. Tn-Seq analysis using Je, BWA, Samtools, BEDTools, Integrative Genomics Viewer, and the edgeR package in the Bioconductor suite, as indicated in the Method Details section.
Prism GraphPad, and where indicated, R, were used perform statistical analyses. Information regarding individual statistical test parameters can be found in the figure legends.
Data and Software Availability
The Tn-Seq data have been deposited on BioProject (ID: PRJNA526509) and can be accessed at https://www.nchi.nlm.nih.gov/bioproject/7term=PRJNA526509.
Supplementary Material
Table S1. Tn-seq data and analysis for WT vs. ftsWI**I* Caulobacter genes. Related to Figures 1 and 5. Tn-Seq data and analysis for WT vs. ftsW**I* Caulobacter genes. Columns WT 1, WT 2, WI 1, and WI 2 contain the number of unique transposon insertions in each gene in each replicate of WT or ftsW**I* (WI) strain transposon insertion library. These values were used to determine the log2 fold-change (logFC), log counts per million reads (logCPM), PValue, false-discovery rate (FDR), and negative log10 of the FDR for each gene in WT versus ftsW**I*. Genes are ordered by significance (neglog(FDR)).
Table S2. Strain list. Related to STAR Methods. List of strains used in this study.
Table S3. Plasmid list. Related to STAR Methods. List of plasmids used in this study.
Video S1. Time-lapse microscopy of division in WT or PG synthase hyperactive mutant cells with and without fzlA. Related to Figure 2.
Phase contrast time-lapse microscopy movies depicting division in WT or PG synthase hyperactive mutant cells with and without fzlA. As indicated, 5 minutes elapse between frames. Video playback is 10 frames per second.
Strain key (Caulobacter crescentus): WT (EG865), ftsW**I*(EG1557), ftsW**I*ΔfzlA (EG2170), ftsW* (EG1556), ftsW*ΔfzlA (EG2166).
Video S2. Time-lapse microscopy of constriction failure in ΔfzlA cells. Related to Figure 2.
Phase contrast time-lapse microscopy movies depicting examples of constriction failure at the initial division site then initiation and completion at a subsequent site in ΔfzlA cells. As indicated, 5 minutes elapse between frames. Video playback is 10 frames per second. Strain key (Caulobacter crescentus): ftsW**I*ΔfzlA (EG2170), ftsW*ΔfzlA (EG2166).
Video S3. Time-lapse microscopy of cell twisting during division in ftsW**I* ΔfzlA cells. Related to Figure 3.
Phase contrast microscopy movies depicting examples of cell twisting during division in multiple ftsW**I*ΔfzlA cells. As indicated, 5 minutes elapse between frames. Video playback is 10 frames per second. Strain key (Caulobacter crescentus): ftsW**I* ΔfzlA (EG2170).
Highlights.
Hyperactivation of FtsWI allows for deletion of fzlA, but not other essential genes
FzlA participates in signaling from FtsZ to peptidoglycan synthesis activation
Cells lacking FzlA rotate about the division plane during division
FzlA-mediated peptidoglycan synthesis promotes resilience against antibiotic stress
Acknowledgements
We would like to thank Josh Modell and Mike Laub for providing strains; the Manley lab, especially Ambroise Lambert, and members of the Xiao and Goley labs for helpful discussions; the Goley lab for feedback on the manuscript; Anant Bhargava and Adrien Ducret for help with time-lapse analysis; Mike Delannoy, Barbara Smith, and Selam Woldemeskel for developing the SEM protocol, and Mike and Barbara for training on SEM equipment; and Brandon King for providing support with experiments. This work funded in part by the National Institutes of Health, National Institute of General Medical Sciences through R01GM108640 (EDG and PJL), T32GM007445 (training grant support of PJL), R01GM111706 (PC and RZ), R25GM056901(training support of GS-C), and T32GM08515 (training grant support of RZ). PB and MH were supported by the National Science Foundation, I0S1557806.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests
References
- 1.Cabeen MT, and Jacobs-Wagner C (2005). Bacterial cell shape. Nat. Rev. Microbiol 3, 601–610. [DOI] [PubMed] [Google Scholar]
- 2.Woldemeskel SA, and Goley ED (2017). Shapeshifting to Survive: Shape Determination and Regulation in Caulobacter crescentus. Trends Microbiol. 25, 673–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gan L, Chen S, and Jensen GJ (2008). Molecular organization of Gram-negative peptidoglycan. Proc. Natl. Acad. Sci 105, 18953–18957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang KC, Mukhopadhyay R, Wen B, Gitai Z, and Wingreen NS (2008). Cell shape and cell-wall organization in Gram-negative bacteria. Proc. Natl. Acad. Sci, pnas.0805309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rohs PDA, Buss J, Sim SI, Squyres GR, Srisuknimit V, Smith M, Cho H, Sjodt M, Kruse AC, Garner EC, et al. (2018). A central role for PBP2 in the activation of peptidoglycan polymerization by the bacterial cell elongation machinery. PLOS Genet. 14, e1007726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meeske AJ, Riley EP, Robins WP, Uehara T, Mekalanos JJ, Kahne D, Walker S, Kruse AC, Bernhardt TG, and Rudner DZ (2016). SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537, 634–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taguchi A, Welsh MA, Marmont LS, Lee W, Sjodt M, Kruse AC, Kahne D, Bernhardt TG, and Walker S (2019). FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat. Microbiol, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambert A, Vanhecke A, Archetti A, Holden S, Schaber F, Pincus Z, Laub MT, Goley E, and Manley S (2018). Constriction Rate Modulation Can Drive Cell Size Control and Homeostasis in C. crescentus. iScience 4, 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Modell JW, Kambara TK, Perchuk BS, and Laub MT (2014). A DNA Damage-Induced, SOS-Independent Checkpoint Regulates Cell Division in Caulobacter crescentus. PLOS Biol 12, e1001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaughan S, Wickstead B, Gull K, and Addinall SG (2004). Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J. Mol. Evol 58, 19–29. [DOI] [PubMed] [Google Scholar]
- 11.Sundararajan K, and Goley ED (2017). Cytoskeletal Proteins in Caulobacter crescentus: Spatial Orchestrators of Cell Cycle Progression, Development, and Cell Shape In Prokaryotic Cytoskeletons Subcellular Biochemistry. (Springer, Cham: ), pp. 103–137. Available at: https://link.springer.com/chapter/10.1007/978-3-319-53047-5_4 [Accessed September 19, 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goley ED, Yeh Y-C, Hong S-H, Fero MJ, Abeliuk E, McAdams HH, and Shapiro L (2011). Assembly of the Caulobacter cell division machine. Mol. Microbiol 80, 1680–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holden SJ, Pengo T, Meibom KL, Fernandez CF, Collier J, and Manley S (2014). High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. Proc. Natl. Acad. Sci 111, 4566–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu G, Huang T, Buss J, Coltharp C, Hensel Z, and Xiao J (2010). In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM). PloS One 5, e12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buske PJ, and Levin PA (2013). A flexible C-terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo. Mol. Microbiol 89, 249–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sundararajan K, Miguel A, Desmarais SM, Meier EL, Huang KC, and Goley ED (2015). The bacterial tubulin FtsZ requires its intrinsically disordered linker to direct robust cell wall construction. Nat. Commun 6, 7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardner KAJA, Moore DA, and Erickson HP (2013). The C-terminal linker of Escherichia coli FtsZ functions as an intrinsically disordered peptide. Mol. Microbiol 89, 264–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howell M, Aliashkevich A, Sundararajan K, Daniel JJ, Lariviere PJ, Goley ED, Cava F, and Brown PJB (2019). Agrobacterium tumefaciens divisome proteins regulate the transition from polar growth to cell division. Mol. Microbiol Available at: https://onlinelibrary.wiley.com/doi/abs/10.1111/mmi.14212 [AccessedJanuary 31, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang X, Lyu Z, Miguel A, McQuillen R, Huang KC, and Xiao J (2017). GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell 355, 744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bisson-Filho AW, Hsu Y-P, Squyres GR, Kuru E, Wu F, Jukes C, Sun Y, Dekker C, Holden S, VanNieuwenhze MS, et al. (2017). Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355, 739–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goley ED, Dye NA, Werner JN, Gitai Z, and Shapiro L (2010). Imaging based identification of a critical regulator of FtsZ protofilament curvature in Caulobacter. Mol. Cell 39, 975–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lariviere PJ, Szwedziak P, Mahone CR, Löwe J, and Goley ED (2018). FzlA, an essential regulator of FtsZ filament curvature, controls constriction rate during Caulobacter division. Mol. Microbiol 107, 180–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ducret A, Quardokus EM, and Brun YV (2016). MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol 1, 16077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coltharp C, Buss J, Plumer TM, and Xiao J (2016). Defining the rate-limiting processes of bacterial cytokinesis. Proc. Natl. Acad. Sci 113, E1044–E1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pincus Z, and Theriot JA (2007). Comparison of quantitative methods for cell-shape analysis. J. Microsc 227, 140–156. [DOI] [PubMed] [Google Scholar]
- 26.Charbon G, Cabeen MT, and Jacobs-Wagner C (2009). Bacterial intermediate filaments: in vivo assembly, organization, and dynamics of crescentin. Genes Dev. 23, 1131–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bi E, and Lutkenhaus J (1992). Isolation and characterization of ftsZ alleles that affect septal morphology. J. Bacteriol 174, 5414–5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Addinall SG, and Lutkenhaus J (1996). FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol. Microbiol 22, 231–237. [DOI] [PubMed] [Google Scholar]
- 29.Pogliano J, Pogliano K, Weiss DS, Losick R, and Beckwith J (1997). Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc. Natl. Acad. Sci. U. S. A 94, 559–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costa T, Priyadarshini R, and Jacobs-Wagner C (2008). Localization of PBP3 in Caulobacter crescentus is highly dynamic and largely relies on its functional transpeptidase domain. Mol. Microbiol 70, 634–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spratt BG (1977). The mechanism of action of mecillinam. J. Antimicrob. Chemother 3, 13–19. [DOI] [PubMed] [Google Scholar]
- 32.Spratt BG, and Pardee AB (1975). Penicillin-binding proteins and cell shape in E. coli. Nature 254, 516–517. [DOI] [PubMed] [Google Scholar]
- 33.Yakhnina AA, and Gitai Z (2013). Diverse Functions for Six Glycosyltransferases in Caulobacter crescentus Cell Wall Assembly. J. Bacteriol 195, 4527–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strobel W, Möll A, Kiekebusch D, Klein KE, and Thanbichler M (2014). Function and Localization Dynamics of Bifunctional Penicillin-Binding Proteins in Caulobacter crescentus. J. Bacteriol 196, 1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meier EL, Daitch AK, Yao Q, Bhargava A, Jensen GJ, and Goley ED (2017). FtsEX-mediated regulation of the final stages of cell division reveals morphogenetic plasticity in Caulobacter crescentus. PLOS Genet. 13, e1006999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goley ED, Comolli LR, Fero KE, Downing KH, and Shapiro L (2010). DipM links peptidoglycan remodelling to outer membrane organization inCaulobacter. Mol. Microbiol 77, 56–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Möll A, Schlimpert S, Briegel A, Jensen GJ, and Thanbichler M (2010). DipM, a new factor required for peptidoglycan remodelling during cell division in Caulobacter crescentus. Mol. Microbiol 77, 90–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zielińska A, Billini M, Möll A, Kremer K, Briegel A, Izquierdo Martinez A, Jensen GJ, and Thanbichler M (2017). LytM factors affect the recruitment of autolysins to the cell division site in Caulobacter crescentus. Mol. Microbiol 106, 419–438. [DOI] [PubMed] [Google Scholar]
- 39.Zuleta LFG, Italiani VCS, and Marques MV (2003). Isolation and Characterization of NaCl-Sensitive Mutants of Caulobacter crescentus. Appl Env. Microbiol 69, 3029–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuru E, Velocity Hughes H, Brown PJ, Hall E, Tekkam S, Cava F, de Pedro MA, Brun YV, and VanNieuwenhze MS (2012). In situ Probing of Newly Synthesized Peptidoglycan in Live Bacteria with Fluorescent D-Amino Acids. Angew. Chem. Int. Ed Engl. 51, 12519–12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, and Jacobs-Wagner C (2007). The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol. Microbiol 64, 938–952. [DOI] [PubMed] [Google Scholar]
- 42.Brown PJB, Pedro M.A. de, Kysela DT, Henst CV der, Kim J, Bolle XD, Fuqua C, and Brun YV (2012). Polar growth in the Alphaproteobacterial order Rhizobiales. Proc. Natl. Acad. Sci 109, 1697–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Figueroa-Cuilan WM, and Brown PJB (2018). Cell Wall Biogenesis During Elongation and Division in the Plant Pathogen Agrobacterium tumefaciens. Curr. Top. Microbiol. Immunol [DOI] [PubMed] [Google Scholar]
- 44.Hale CA, Rhee AC, and de Boer PA (2000). ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J. Bacteriol 182, 5153–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hale CA, and de Boer PA (1997). Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88, 175–185. [DOI] [PubMed] [Google Scholar]
- 46.Du S, Pichoff S, and Lutkenhaus J (2016). FtsEX acts on FtsA to regulate divisome assembly and activity. Proc. Natl. Acad. Sci 113, E5052–E5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsang M-J, and Bernhardt TG (2015). A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Mol. Microbiol 95, 925–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schoenemann KM, Krupka M, Rowlett VW, Distelhorst SL, Hu B, and Margolin W (2018). Gain-of-function variants of FtsA form diverse oligomeric structures on lipids and enhance FtsZ protofilament bundling. Mol. Microbiol 109, 676–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haeusser DP, Rowlett VW, and Margolin W (2015). A mutation in Escherichia coli ftsZ bypasses the requirement for the essential division gene zipA and confers resistance to FtsZ assembly inhibitors by stabilizing protofilament bundling. Mol. Microbiol 97, 988–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu B, Persons L, Lee L, and de Boer PAJ (2015). Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Mol. Microbiol 95, 945–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geissler B, and Margolin W (2005). Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK. Mol. Microbiol 58, 596–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grainge I (2010). FtsK--a bacterial cell division checkpoint? Mol. Microbiol 78, 1055–1057. [DOI] [PubMed] [Google Scholar]
- 53.Sycuro LK, Pincus Z, Gutierrez KD, Biboy J, Stern CA, Vollmer W, and Salama NR (2010). Peptidoglycan crosslinking relaxation promotes Helicobacter pylori’s helical shape and stomach colonization. Cell 141, 822–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szwedziak P, and Löwe J (2013). Do the divisome and elongasome share a common evolutionary past? Curr. Opin. Microbiol 16, 745–751. [DOI] [PubMed] [Google Scholar]
- 55.Figueroa-Cuilan W, Daniel JJ, Howell M, Sulaiman A, and Brown PJB (2016). Mini-Tn7 Insertion in an Artificial attTn7 Site Enables Depletion of the Essential Master Regulator CtrA in the Phytopathogen Agrobacterium tumefaciens. Appl. Environ. Microbiol 82, 5015–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morton ER, and Fuqua C (2012). UNIT 3D.1 Laboratory Maintenance of Agrobacterium Curr. Protoc. Microbiol CHAPTER, Unit3D.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hottes AK, Meewan M, Yang D, Arana N, Romero P, McAdams HH, and Stephens C (2004). Transcriptional Profiling of Caulobacter crescentus during Growth on Complex and Minimal Media. J. Bacteriol 186, 1448–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Howell M, Daniel JJ, and Brown PJB (2017). Live Cell Fluorescence Microscopy to Observe Essential Processes During Microbial Cell Growth. J. Vis. Exp. JoVE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fiji: an open-source platform for biological-image analysis | Nature Methods Available at: https://www.nature.com/articles/nmeth.2019 [Accessed March 24, 2019]. [DOI] [PMC free article] [PubMed]
- 60.Bowman GR, Comolli LR, Gaietta GM, Fero M, Hong S-H, Jones Y, Lee JH, Downing KH, Ellisman MH, McAdams HH, et al. (2010). Caulobacter PopZ forms a polar subdomain dictating sequential changes in pole composition and function. Mol. Microbiol 76, 173–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Girardot C, Scholtalbers J, Sauer S, Su S-Y, and Furlong EEM (2016). Je, a versatile suite to handle multiplexed NGS libraries with unique molecular identifiers. BMC Bioinformatics 17, 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, and Durbin R (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinforma. Oxf. Engl 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinforma. Oxf. Engl 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Biotechnol 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thorvaldsdóttir H, Robinson JT, and Mesirov JP (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinforma. Oxf. Engl 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McCarthy DJ, Chen Y, and Smyth GK (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Tn-seq data and analysis for WT vs. ftsWI**I* Caulobacter genes. Related to Figures 1 and 5. Tn-Seq data and analysis for WT vs. ftsW**I* Caulobacter genes. Columns WT 1, WT 2, WI 1, and WI 2 contain the number of unique transposon insertions in each gene in each replicate of WT or ftsW**I* (WI) strain transposon insertion library. These values were used to determine the log2 fold-change (logFC), log counts per million reads (logCPM), PValue, false-discovery rate (FDR), and negative log10 of the FDR for each gene in WT versus ftsW**I*. Genes are ordered by significance (neglog(FDR)).
Table S2. Strain list. Related to STAR Methods. List of strains used in this study.
Table S3. Plasmid list. Related to STAR Methods. List of plasmids used in this study.
Video S1. Time-lapse microscopy of division in WT or PG synthase hyperactive mutant cells with and without fzlA. Related to Figure 2.
Phase contrast time-lapse microscopy movies depicting division in WT or PG synthase hyperactive mutant cells with and without fzlA. As indicated, 5 minutes elapse between frames. Video playback is 10 frames per second.
Strain key (Caulobacter crescentus): WT (EG865), ftsW**I*(EG1557), ftsW**I*ΔfzlA (EG2170), ftsW* (EG1556), ftsW*ΔfzlA (EG2166).
Video S2. Time-lapse microscopy of constriction failure in ΔfzlA cells. Related to Figure 2.
Phase contrast time-lapse microscopy movies depicting examples of constriction failure at the initial division site then initiation and completion at a subsequent site in ΔfzlA cells. As indicated, 5 minutes elapse between frames. Video playback is 10 frames per second. Strain key (Caulobacter crescentus): ftsW**I*ΔfzlA (EG2170), ftsW*ΔfzlA (EG2166).
Video S3. Time-lapse microscopy of cell twisting during division in ftsW**I* ΔfzlA cells. Related to Figure 3.
Phase contrast microscopy movies depicting examples of cell twisting during division in multiple ftsW**I*ΔfzlA cells. As indicated, 5 minutes elapse between frames. Video playback is 10 frames per second. Strain key (Caulobacter crescentus): ftsW**I* ΔfzlA (EG2170).
Data Availability Statement
The Tn-Seq data have been deposited on BioProject (ID: PRJNA526509) and can be accessed at https://www.nchi.nlm.nih.gov/bioproject/7term=PRJNA526509.