Abstract
N-glycosylation is one of the predominant modifications of eukaryotic proteins. It is catalyzed by oligosaccharyl transferase (OST), an eight-subunit protein complex in the endoplasmic reticulum membrane. OST transfers the oligosaccharide from a lipid-linked donor (LLO) to the Asn-Xaa-Ser/Thr sequon of nascent polypeptide, usually co-translationally by partnering with the ribosome and the translocon. We and two other groups have recently determined high-resolution cryo-EM structures of the yeast and mammalian OST complexes. In this Structural Snapshot, we describe the molecular mechanism of eukaryotic OST and its interaction with the translocon.
Keywords: Protein N-glycosylation, structural biology, protein structure, glycosyltransferase, cryo-EM, Saccharomyces cerevisiae
Introduction
Protein glycosylation is a process of covalently attaching sugar moieties to proteins. It plays important role in many essential biological processes, such as intercellular recognition, protein folding and stability, protein–protein or protein–ligand interactions, and in many enzyme activities [1]. There are several types of glycosylation, including N-, O-, and C-linked glycosylation, glypiation, and phosphoglycosylation [2]. At the three-dimensional structural level, the vast majority of glycosyltransferases belong to one of three folds: GT-A, which has a Rossmann-like domain; GT-B, with two Rossmann-like domains; and GT-C, containing a transmembrane helical domain and a mixed α/β soluble domain [3, 4]. Whereas O-linked glycosylation is generally catalyzed by enzymes of GT-A and GT-B folds, N-linked glycosylation is exclusively catalyzed by enzymes of GT-C fold.
N-glycosylation is wide spread in eukaryotes [5], but it is also found in some prokaryotes [6–8]. About half of all proteins in a eukaryotic cell are N-glycosylated, which is catalyzed by oligosaccharyl transferase (OST), an eight-subunit protein complex found in the ER membrane [9, 10]. Previous studies revealed that the Saccharomyces cerevisiae OST contains Ost1, 2, 4, and 5; Stt3; Wbp1; Swp1; and either Ost3 or Ost6 [11, 12]. All these subunits have homologs in higher organisms, which express two isoforms, STT3A-OST and STT3B-OST: ribophorin I corresponds to the yeast Ost1; DAD1 to Ost2; MagT1/TUSC3 or DC2/KCP2 to Ost3/6; OST4 to Ost4; TMEM258 to Ost5; OST48 to Wbp1; STT3A/STT3B to Stt3; and ribophorin II to Swp1 [5]. In S. cerevisiae, because Ost3 binds co-translational translocon Sec61, and Ost6 binds the post-translational translocon Ssh1 [11], the Ost3-containing OST is co-translational and the Ost6-containing OST is post-translational. In higher organisms, DC2/KPC2, but not MagT1/TUSC3, interact with the co-translational translocon Sec61 [29]. Therefore, the DC2/KPC2-containing STT3A-OST functions co-translationally, whereas the MagT1/TUSC3-containing STT3B-OST functions post-translationally [5]. N-glycosylation in prokaryotes is catalyzed by a single-subunit enzyme that is homologous to the catalytic subunit Stt3 of the eukaryotic OST complex [13, 14].
The first structural insight into the N-linked glycosyl transfer reaction was derived from the crystal structures of the bacterial PglB and the archaeal AglB, which are homologous to the catalytic Stt3 subunit of OST [15–18]. The structures of eukaryotic OST had been limited to low-resolution EM envelopes (which included a low-resolution shape of the yeast OST and the mapping of the approximate locations of four essential subunits [19]), and three cryo-ET studies on the native mammalian OST−Sec61−ribosome complex at nanometer resolution [20–22]. These works lacked structural detail and did not reveal any clues to the functions of the individual subunits. Recently, our lab at Van Andel Research Institute and the labs of Kasper Locher and Markus Aebi at ETH Zurich determined the first atomic-resolution cryo-EM structures of the yeast OST complex [23, 24]. Around the same time, the labs of Roland Beckman in Munich and Fredrich Förster, now in Utrecht, published cryo-EM maps of several mammalian OST−Sec61−ribosome complexes [25]. These structures are starting to reveal the molecular mechanism of eukaryotic N-glycosylation.
Structures of the yeast OST complex
The two cryo-EM structures of Saccharomyces cerevisiae OST were both derived from the endogenous source, one purified with the detergent digitonin and used directly for EM imaging, and the other purified with a mixture of N-dodecyl-β-D-maltopyranoside and cholesteryl hemisuccinate and further reconstituted in the nanodisc MSPE3D1 [23, 24]. The structures were determined to comparable resolutions of 3.5-Å and 3.3-Å, respectively, in which all eight subunits were unambiguously resolved (Fig. 1A–C). These two structures represent a rare example in which the same structure has been determined in two different environments, in detergent and in nanodisc. The two structures are remarkably similar, with a root-mean-square deviation of 1.78 Å, suggesting that the protein structures are not appreciably altered if mild manipulations are used to extract them from their native lipid bilayers, as in these two cases. This is highly encouraging, because the potential for structural change of membrane proteins outside their native membrane environment is a constant concern.
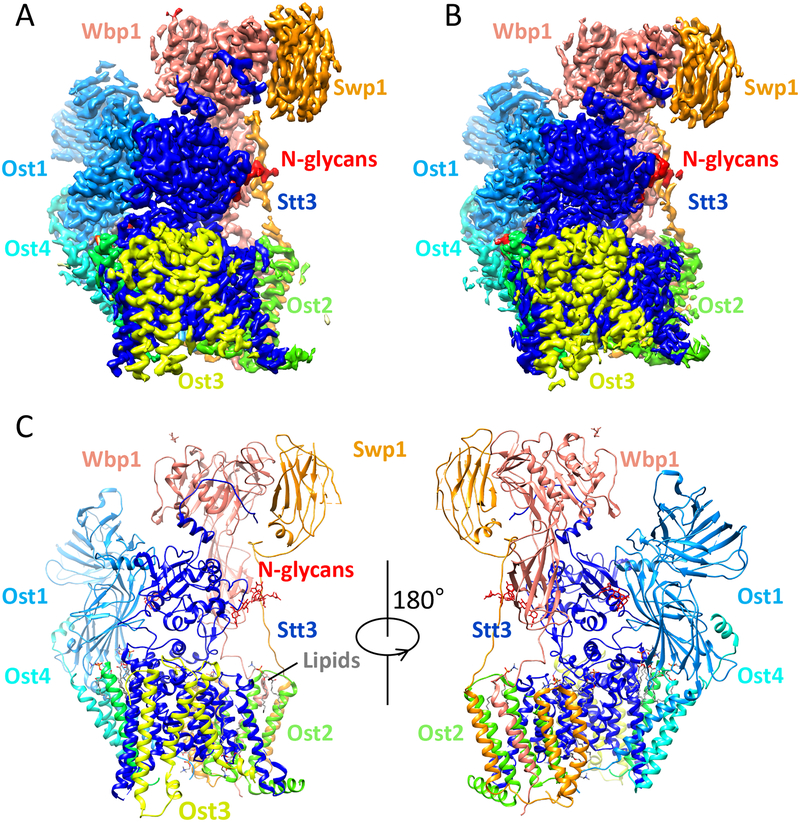
Fig. 1.
Atomic structure of the yeast OST. (A) Cryo-EM 3D map of the OST isolated in digitonin at 3.5 Å resolution. (B) Cryo-EM 3D map of the OST reconstituted in nanodisc at 3.3 Å resolution. (C) The OST structure is shown in cartoons and colored in the same scheme as in panel A. The three N-linked glycans are shown as sticks.
Because these two structures are both in the apo state − in the absence of LLO and peptide substrate − and nearly identical, we will focus our discussion on the structure determined in digitonin. Yeast OST has a total of 28 predicted transmembrane helices (TMHs), and the 3.5-Å 3D map has densities for 26 of them. The two unresolved TMHs are TMH9 of Stt3 and TMH1 of Ost3, both of which are around the LLO binding site and are likely dynamic in the absence of the large donor substrate. The yeast OST contains five soluble domains. The aqueous domains of Stt3, Ost1, Wbp1, and Swp1 are well resolved in the 3D density map, but the soluble domain of Ost3 is largely absent, visible only in one of the 3D map classes and at a very low display threshold, indicating its high flexibility in the apo state of the enzyme complex. In the OST structure, the catalytic subunit Stt3 forms the core and the other seven subunits assemble around it. Consistent with previous biochemical studies, the eight subunits can be grouped into 3 classes according to their assembly features: Ost1−Ost5, Ost2−Swp1−Wbp1, and Stt3−Ost4−Ost3 [26]. Interestingly, several phospholipid molecules were identified in the subunit interfaces, mediating the OST complex assembly.
The stabilizing function of an N-glycan in the OST structure
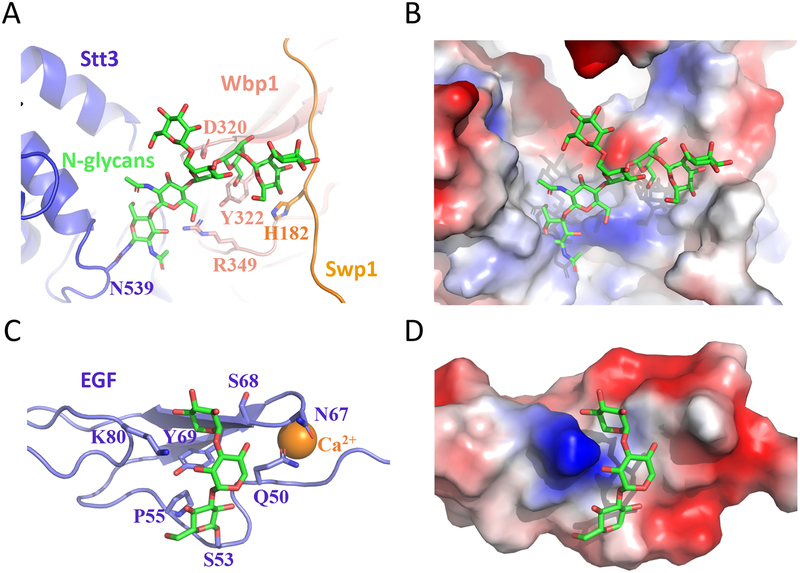
All three predicted glycoproteins in the OST were found to indeed be glycosylated: Asn336 of Ost1, Asn60 of Wbp1, and Asn539 of Stt3. There was enough density on Asn539 for atomic modeling of seven covalently linked sugars for the N-glycan. This glycan directly interacts with Wbp1 and Swp1, suggesting a stabilizing role of N-glycosylation in regulating the enzymatic activity of the N-glycosylation complex OST (Fig. 2A–B). Similarly, we recently found an O-linked trisaccharide that binds and stabilizes the epidermal growth factor-like (EGF) repeat of the human coagulation factor 9 (hFA9) [27] (Fig. 2C–D). Although glycans are usually portrayed like a branch of a tree protruding outwards into the solvent, these observations clearly indicate that glycans can also bind non-covalently to protein via interactions such as hydrogen bonds and participate in protein folding and stability.
Fig. 2.
A glycan can directly interact with the underlying protein to exert a stabilizing effect on the protein structure. (A-B) The 7-sugar N-glycan on Asn539 of Stt3 interacts with and stabilizes neighboring Wbp1 and Swp1. (C-D) The O-linked trisaccharide Xyl−Xyl−Glc on Ser53 of the EGF motif of hFA9 folds back and interacts with a shallow groove to stabilize the structure (PDB ID 5VYG).
The N-glycosyltransferase reaction mechanism is likely conserved
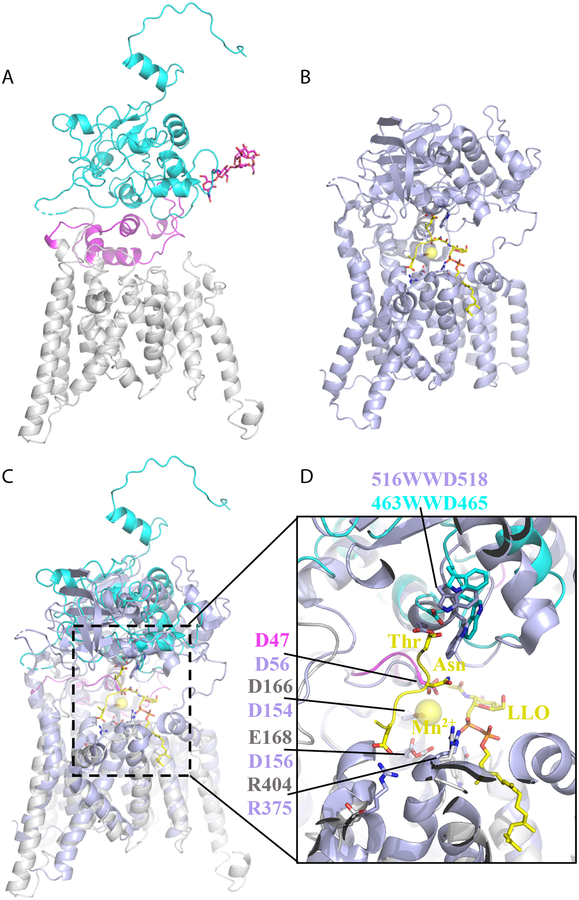
The catalytic subunit Stt3 has 13 TMHs, a lumenal domain, and an α-helical accessory domain that is formed by external loop 1 (EL1) (Fig. 3A), but the structure is in the apo state and does not contain substrates. However, structures of the bacterial homolog PglB have been determined either in complex with Mn2+ and a peptide acceptor or in complex with Mn2+, an LLO analog, and a peptide acceptor (Fig. 3B) [17, 18]. Structural alignment showed high similarity between Stt3 and PglB (Fig. 3C). In fact, many catalytically important residues and structural motifs are conserved and share the same positions in the two structures (Fig. 3D). For examples, the conserved glycosylation-sequon-binding WWD motif is located at the top of the active site pocket of Stt3; residues that are expected to coordinate Mn2+ and LLO, such as D47, D166, E168, W208, and R404, are superimposable with their counterparts in PglB. These observations suggest that Stt3 and PglB share the same catalytic mechanism. In other words, the mechanism of glycosylation sequon recognition and LLO binding, as proposed for PglB structure, will likely apply to the eukaryotic Stt3.
Fig. 3.
Structural comparison between the yeast Stt3 and the bacterial PglB. (A) Atomic structure of Stt3 in carton view. TMHs are in grey, carboxyl terminal domain (CTD) in cyan, EL1 in magenta, and the carboxyl terminal extension (CTE) in red. (B) Atomic structure of PglB in complex with Mn2+, an acceptor peptide, and a nonhydrolyzable LLO donor analog. (C) Superimposition of Stt3 and PglB. (D) A close-up view of the active site of Stt3 aligned with that of PglB.
Interface between OST and the translocon
The determination of the yeast OST structure made it feasible to interrogate the interface between the OST and the translocon, given the available crystal structure of the yeast Sec61 and the low-resolution cryo-ET map of the mammalian OST–translocon–ribosome. Our lab and the labs of Kasper Locher and Markus Aebi realized that Ost3 of the yeast OST (DC2/KCP2 in the mammalian OST) mediated the physical interaction with the co-translocon [23, 24].
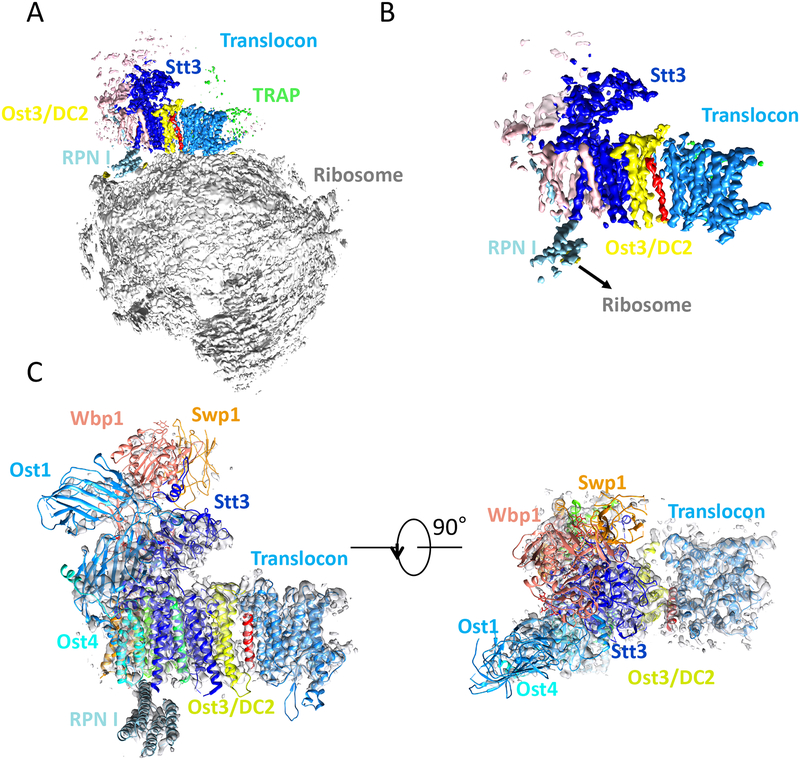
The architectural difference between the mammalian STT3A-OST and STT3B-OST in which DC2/KCP2 − but not MagT1/TUSC3 − physically binding to the translocon [28, 29], was elegantly confirmed in the labs of Fredrich Förster and Roland Beckmann. They showed that the translocon was associated with the OST in the cryo-EM map derived from an STT3B-knockout cell line and that the OST was absent in the density map of the translocon derived from an STT3A-knockout cell line [25]. Their single-particle cryo-EM efforts led to an improved 3D map of the OST–Sec61–ribosome in which the translocon had a resolution of 3.5–4.5 Å and the OST had a resolution of 4.5–5.5 Å in the transmembrane region (the resolution was lower at the lumenal soluble region of the OST). With the improved 3D map, the Förster and Beckmann labs also realized that DC2/KCP2, equivalent to Ost3/6, was responsible for interfacing with the translocon [25].
To provide a better view of the interface between OST and translocon, we docked the yeast OST structure into the improved 3D map of the mammalian OST−translocon−ribosome (Fig. 4A–C). The fact that Ost3/6 interfaces with the translocon explains why two yeast OST isoforms, the Ost3-containing or the Ost6-containing, exist in yeast and their distinct association with the Sec61 or Ssh1 translocon, respectively [11]. The OST/translocon interface is also in agreement with previous biochemical studies demonstrating that DC2/KCP2 mediates the interaction between the human OST and the translocon [29]. DC2 seems to maintain tight binding with the translocon, suggesting an essential role in co-translational N-glycosylation. Interestingly, there is an interaction between the ribosome and the cytosolic RPN1 domain of ribophorin I (Ost1 in yeast). STT3B possesses a unique 47-amino-acid aqueous domain directly beneath the RPN1 domain and thus interferes with ribosome binding, potentially explaining the why STT3B-OST does not function co-translationally.
Fig. 4.
Structure of the mammalian OST−translocon−ribosome complex. (A) Cryo-EM 3D map at 4.2-Å overall resolution determined in the Forster and Beckmann labs. (B) A close-up view of the translocon region using a high surface-rendering threshold as depicted in (A). (C) Docking of the yeast OST structure into the 3D map of the mammalian OST−translocon−ribosome. The identity of the red TMH is currently unknown.
Concluding remarks
The atomic structure of the yeast OST and the near-atomic model of the mammalian OST−translocon−ribosome complex have revealed the architecture of eukaryotic OST, the likely conserved glycosyl transfer reaction mechanism of the eukaryotic Stt3, and the physical interactions between the OST and the translocon−ribosome. However, many key questions remain. For example, how does the eukaryotic OST accommodate and recognize its donor substrate, which is considerably larger than the prokaryotic donors? Is an atomic-resolution structure of the mammalian OST on the horizon that would clarify several mammal-specific features of the enzyme complex? How do the noncatalytic subunits contribute to N-glycosylation? Last but not the least, what are the structures and functions of the dozen or so enzymes that sequentially synthesize the LLO donor for the OST to transfer en bloc onto the nascent peptide chain? We look forward to answering these questions in the coming years.
Abbreviations
- AglB
archaeal glycosylation B
- cryo-EM
cryo-electron microscopy
- cryo-ET
cryo-electron tomography
- EGF
epidermal growth factor-like repeat
- EL1
external loop 1
- hFA9
human coagulation factor 9
- LLO
lipid-linked oligosaccharide
- OST
oligosaccharyl transferase
- PglB
protein glycosylation locus B
- TMH
transmembrane helix
- WWD motif
tryptophan-tryptophan-aspartate motif
REFERENCES
- 1.Helenius A & Aebi M (2004) Roles of N-linked glycans in the endoplasmic reticulum, Annu Rev Biochem. 73, 1019–49. [DOI] [PubMed] [Google Scholar]
- 2.Spiro RG (2002) Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds, Glycobiology. 12, 43R–56R. [DOI] [PubMed] [Google Scholar]
- 3.Lairson LL, Henrissat B, Davies GJ & Withers SG (2008) Glycosyltransferases: structures, functions, and mechanisms, Annu Rev Biochem. 77, 521–55. [DOI] [PubMed] [Google Scholar]
- 4.Hurtado-Guerrero R & Davies GJ (2012) Recent structural and mechanistic insights into post-translational enzymatic glycosylation, Curr Opin Chem Biol. 16, 479–87. [DOI] [PubMed] [Google Scholar]
- 5.Shrimal S, Cherepanova NA & Gilmore R (2015) Cotranslational and posttranslocational N-glycosylation of proteins in the endoplasmic reticulum, Semin Cell Dev Biol. 41, 71–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mescher MF & Strominger JL (1976) Purification and characterization of a prokaryotic glycoprotein from the cell envelope of Halobacterium salinarium, J Biol Chem. 251, 2005–14. [PubMed] [Google Scholar]
- 7.Szymanski CM, Yao R, Ewing CP, Trust TJ & Guerry P (1999) Evidence for a system of general protein glycosylation in Campylobacter jejuni, Mol Microbiol. 32, 1022–30. [DOI] [PubMed] [Google Scholar]
- 8.Larkin A & Imperiali B (2011) The expanding horizons of asparagine-linked glycosylation, Biochemistry-Us. 50, 4411–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwarz F & Aebi M (2011) Mechanisms and principles of N-linked protein glycosylation, Curr Opin Struct Biol. 21, 576–82. [DOI] [PubMed] [Google Scholar]
- 10.Apweiler R, Hermjakob H & Sharon N (1999) On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database, BBA-Gen Subjects. 1473, 4–8. [DOI] [PubMed] [Google Scholar]
- 11.Yan AX & Lennarz WJ (2005) Two oligosaccharyl transferase complexes exist in yeast and associate with two different translocons, Glycobiology. 15, 1407–1415. [DOI] [PubMed] [Google Scholar]
- 12.Cherepanova N, Shrimal S & Gilmore R (2016) N-linked glycosylation and homeostasis of the endoplasmic reticulum, Curr Opin Cell Biol. 41, 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nothaft H & Szymanski CM (2010) Protein glycosylation in bacteria: sweeter than ever, Nat Rev Microbiol. 8, 765–78. [DOI] [PubMed] [Google Scholar]
- 14.Szymanski CM & Wren BW (2005) Protein glycosylation in bacterial mucosal pathogens, Nat Rev Microbiol. 3, 225–37. [DOI] [PubMed] [Google Scholar]
- 15.Matsumoto S, Taguchi Y, Shimada A, Igura M & Kohda D (2017) Tethering an N-Glycosylation Sequon-Containing Peptide Creates a Catalytically Competent Oligosaccharyltransferase Complex, Biochemistry. 56, 602–611. [DOI] [PubMed] [Google Scholar]
- 16.Matsumoto S, Shimada A, Nyirenda J, Igura M, Kawano Y & Kohda D (2013) Crystal structures of an archaeal oligosaccharyltransferase provide insights into the catalytic cycle of N-linked protein glycosylation, P Natl Acad Sci USA. 110, 17868–17873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Napiorkowska M, Boilevin J, Sovdat T, Darbre T, Reymond JL, Aebi M & Locher KP (2017) Molecular basis of lipid-linked oligosaccharide recognition and processing by bacterial oligosaccharyltransferase, Nat Struct Mol Biol. [DOI] [PubMed] [Google Scholar]
- 18.Lizak C, Gerber S, Numao S, Aebi M & Locher KP (2011) X-ray structure of a bacterial oligosaccharyltransferase, Nature. 474, 350–5. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Chavan M, Schindelin H, Lennarz WJ & Li HL (2008) Structure of the oligosaccharyl transferase complex at 12 angstrom resolution, Structure. 16, 432–440. [DOI] [PubMed] [Google Scholar]
- 20.Pfeffer S, Dudek J, Schaffer M, Ng BG, Albert S, Plitzko JM, Baumeister W, Zimmermann R, Freeze HH, Engel BD & Forster F (2017) Dissecting the molecular organization of the translocon-associated protein complex, Nat Commun. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfeffer S, Dudek J, Gogala M, Schorr S, Linxweiler J, Lang S, Becker T, Beckmann R, Zimmermann R & Forster F (2014) Structure of the mammalian oligosaccharyltransferase complex in the native ER protein translocon, Nat Commun. 5. [DOI] [PubMed] [Google Scholar]
- 22.Pfeffer S, Burbaum L, Unverdorben P, Pech M, Chen YX, Zimmermann R, Beckmann R & Forster F (2015) Structure of the native Sec61 protein-conducting channel, Nat Commun. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wild R, Kowal J, Eyring J, Ngwa EM, Aebi M & Locher KP (2018) Structure of the yeast oligosaccharyltransferase complex gives insight into eukaryotic N-glycosylation, Science. 359, 545–550. [DOI] [PubMed] [Google Scholar]
- 24.Bai L, Wang T, Zhao G, Kovach A & Li H (2018) The atomic structure of a eukaryotic oligosaccharyltransferase complex, Nature. 555, 328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braunger K, Pfeffer S, Shrimal S, Gilmore R, Berninghausen O, Mandon EC, Becker T, Forster F & Beckmann R (2018) Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum, Science. 360, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan A, Ahmed E, Yan Q & Lennarz WJ (2003) New findings on interactions among the yeast oligosaccharyl transferase subunits using a chemical cross-linker, J Biol Chem. 278, 33078–87. [DOI] [PubMed] [Google Scholar]
- 27.Takeuchi H, Yu H, Hao H, Takeuchi M, Ito A, Li H & Haltiwanger RS (2017) O-Glycosylation modulates the stability of epidermal growth factor-like repeats and thereby regulates Notch trafficking, J Biol Chem. 292, 15964–15973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelleher DJ, Karaoglu D, Mandon EC & Gilmore R (2003) Oligosaccharyltransferase isoforms that contain different catalytic STT3 subunits have distinct enzymatic properties, Mol Cell. 12, 101–11. [DOI] [PubMed] [Google Scholar]
- 29.Shrimal S, Cherepanova NA & Gilmore R (2017) DC2 and KCP2 mediate the interaction between the oligosaccharyltransferase and the ER translocon, J Cell Biol. 216, 3625–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]