Abstract
Recent advances in native mass spectrometry (MS) have enabled the elucidation of how small molecule binding to membrane proteins modulates their structure and function. The protein stabilizing osmolyte, Trimethylamine oxide (TMAO), exhibits attractive properties for native MS studies. Here, we report significant charge reduction, nearly three-fold, for three membrane protein complexes in the presence of this osmolyte without compromising mass spectral resolution. TMAO improves the ability to resolve individual lipid binding events to the ammonia channel (AmtB) by over 200% compared to typical native conditions. The generation of ions with compact structure and access to a larger number of lipid binding events through the incorporation of TMAO increases the utility of IM-MS for structural biology studies.
Graphicl Abstract
Introduction
Native mass spectrometry (MS) in conjunction with ion mobility (IM) is a powerful means to gain insight into the structure and function of protein assemblies, especially for membrane proteins[1–4], which represent one of the most important targets for drug discovery[5]. Recent advances in IM-MS have led to the ability to preserve non-covalent interactions and maintain intact, folded membrane proteins in the gas phase[2, 3, 6] providing information on subunit stoichiometry[7] and noncovalent interactions with lipids and other molecules[6, 8–11], including the measurement of binding affinities and thermodynamics[12]. Recently, native IM-MS has revealed how lipids can selectively stabilize oligomers[13] and allosterically modulate interactions with lipids and proteins[14–16].
Native IM-MS analysis of membrane proteins, typically encapsulated in detergent micelles, involves gentle removal of buffer and detergent molecules from the complex while preserving non-covalent interactions and structure in the mass spectrometer[17]. Removal of detergent from membrane proteins presents a particular challenge and requires careful optimization of mass spectrometer tuning parameters to maintain compact, folded states[18]. Harsh instrument tuning can result in perturbation of protein structure resulting in erroneous binding measurements for noncovalent interactions. A number of detergents, such as dodecylmaltoside (DDM), commonly used for solubilizing membrane proteins prior to IM-MS analysis, give rise to ions that are activated, or non-native, with collision cross section (CCS) measurements deviating far from calculated values[6, 9, 19]. In contrast, the discovery of charge reducing detergents, such as tetraethylene glycol monooctyl ether (C8E4) and n-dodecyl-N,N-dimethylamine-N-oxide (LDAO) (Figure 1a), yields ions with less coulombic repulsion and a lower propensity for unfolding or activation[6]. The ability to produce cleanly resolved mass spectra while simultaneously preserving native-like membrane protein structure, as determined by experimental CCS measurements agreeing with those calculated from crystal structures, has revolutionized the field[6]. The addition of small molecules, such as imidazole, and the use of negative mode ESI to manipulate charge for soluble and membrane proteins has also been explored[19–21]. Unfortunately, imidazole exhibits poor charge reduction (~2–3 charges), especially in cases where non-charge-reducing detergents are used, and often forms adducts[21]. The adducting not only lowers spectral resolution, but it also reduces spectral signal-to-noise and therefore apparent sensitivity. Negative mode ESI has been employed for membrane protein analysis, however, charge reducing phenomena appear to be protein dependent[22]. Despite advances in charge manipulation, there remains an ongoing need to further investigate the manipulation of charge on membrane proteins.
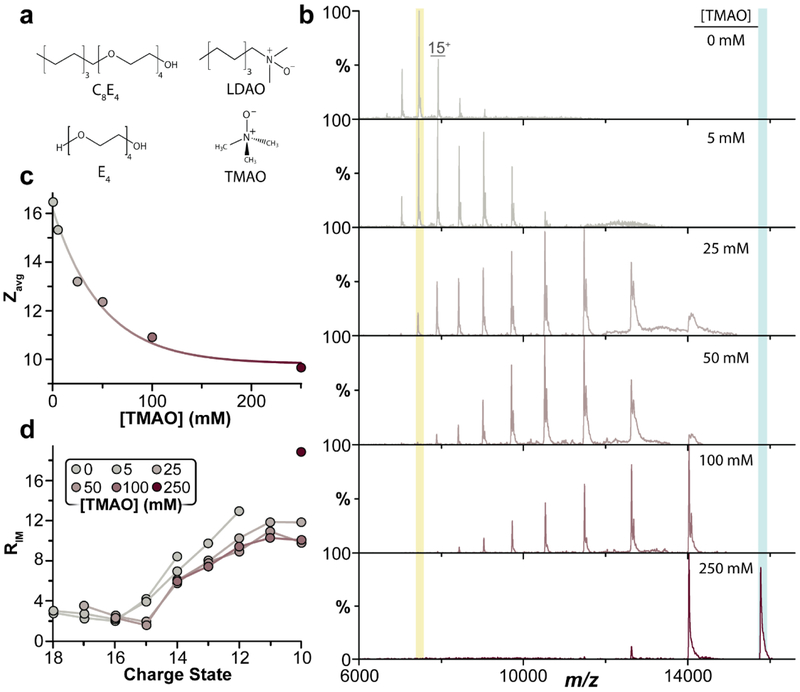
Figure 1.
Trimethylamine oxide (TMAO) charge reduces and preserves compact, folded states of the ammonium channel (AmtB). a) Chemical structures of tetraethylene glycol monooctyl ether (C8E4) and ndodecyl-N,N-dimethylamine-N-oxide (LDAO) and their respective head groups: tetraethylene glycol (E4) and TMAO. b) Native mass spectra of AmtB in 200mM ammonium acetate (AA) with 2x critical micelle concentration (CMC) C8E4 and increasing amounts of TMAO. The final concentration of TMAO is listed for each mass spectrum. c) Plotted is the average charge state as a function of TMAO concentration. TMAO exhibits a concentration dependent charge reduction. d) IM resolving power increases with lower charge states accessed with TMAO.
Trimethylamine oxide (TMAO) is an intracellular osmolyte with properties that stabilize and protect proteins against extreme conditions, such as high urea concentrations[23, 24]. Inspired by these observations, we sought to evaluate the ability of TMAO to also preserve membrane protein structure from solution into the gas phase. Membrane proteins are ionized using nano electrospray ionization (nESI) where droplets are produced that undergo cooling as well as evaporative processes that increase local salt/buffer ion concentrations, exclude water, and crowd molecules in the droplets[25, 26]. Here, we report the unique properties of TMAO to charge reduce membrane proteins ionized by nESI, which is consistent with a recent report showing significant charge reduction of soluble proteins[27]. Beyond charge reduction, IM measurements provide evidence of protein structuring in the presence of TMAO.
Methods
Protein expression and purification.
The Ammonia channel (AmtB) from Escherichia coli was expressed and purified as previously described[15]. Aquaporin Z (AqpZ) and Mechanosenstive Channel of Large Conductance (MscL) were expressed and purified as previously described.[6] Purified membrane proteins were loaded onto a Superdex 200 Increase 10/300 GL column (GE Healthcare) equilibrated in GF buffer (130 mM sodium chloride, 10% glycerol and 50 mM TRIS, pH7.4 at room temperature) supplemented with 2x CMC detergent. Peak fractions containing AqpZ were pooled, concentrated, flash frozen in liquid nitrogen, and stored at −80 °C.
Native mass spectrometry analysis.
Native mass spectrometry (MS) was performed on a Synapt G1 HDMS instrument (Waters corporation) equipped with a 32k RF generator. Nano electrospray ionization was performed using gold coated capillaries prepared in house as previously described[17]. Purified protein was buffer exchanged into ammonium acetate buffer containing 0.5% C8E4(v/v) for native mass MS experiments as previously described[6]. Stock solutions of trimethylamine oxide (TMAO) (Sigma Aldrich, St. Louis, MO) were prepared in water and diluted accordingly into MS buffer and used fresh daily.
Instrument parameters were tuned to maximize ion intensity but simultaneously preserve the native-like state of proteins as determined by IM. The instrument was set to a capillary voltage of 1.7 kV, sampling cone voltage of 100–200 V, extraction cone voltage of 10 V, trap collision energy 50–150 V, transfer collision energy 10 – 50 V, and argon flow rate at 7 ml/min (5.2 × 10−2 mbar). The trap (5.25 × 10−2 mbar) and TWIM (5.33 × 10−1 mbar) devices were filled with agron and nitrogen, respectively. The T-wave settings for trap (300 ms-1/2.0 V), IMS (300 ms-1/20–24 V) and transfer (100 ms-1/10 V), source temperature (90 °C) and trap bias (25–35 V) were optimized accordingly.
Recorded spectra were deconvoluted using MassLynx 4.1 (Waters Corp.), UniDec[28], and Pulsar[29]. The arrival time distributions for the main peaks of each charge state were fit using Gaussian populations in Pulsar to determine RIM.
Results and Discussion
We first titrated TMAO into a sample of Ammonia Channel (AmtB), a trimeric integral membrane protein complex from E. coli, encapsulated in the charge-reducing C8E4 detergent and recorded their mass spectra (Figure 1b). In the absence of TMAO, the average charge state (Zavg) was 16 (Figure 1b, top panel). Interestingly, the addition of 250 mM TMAO (Figure 1B, bottom) significantly reduced the Zavg to 8.5. This effect is even more dramatic when considering the reported Zavg of 24 for AmtB in the non-charge-reducing detergent DDM. We also note that charge reduction does not arise only from a synergistic effect between TMAO and C8E4, since TMAO reduces AmtB in the non-charge-reducing nonyl glucoside (NG) by 11 charges (Figure S1). Importantly, high concentrations of TMAO did not significantly compromise mass spectral resolution indicating TMAO easily releases from the protein during desolvation (Figure S2) unlike other charge-reducing small molecules, such as E4, that result in peak broadening owing to non-specific adduction to the protein[19]. Titration of TMAO reveals a concentration dependent charge reduction (Figure 1C) similar to that shown for soluble proteins[27]. A compaction of the charge state distribution was also observed from six down to three charge states at the highest concentration of TMAO. To the best of our knowledge, TMAO is the most potent charge-reducing molecule for membrane protein complexes reported to date.
Given the ability of TMAO to charge reduce without compromising MS resolution, the IM measurements for different charge states in the presence of TMAO were analyzed and the IM resolving power (RIM, t/Δt) for each peak was calculated to assess the impact of TMAO on peak width (Figure 1d and S3). We acknowledge that IM peaks will inherently widen as drift time increases if RIM is constant. However, comparison of RIM informs on the narrowing or widening of peaks in the presence of additives. The addition of TMAO resulted in charge reduced ions that retain a compact arrival time distributions (ATD) (Figure 1D and 2). For the 8+ ion of AmtB, representing the most charge reduced ion, RIM increased nearly two-fold compared to the 14+ charge state. At present, it is not possible to generate the 8+ ion in the absence of TMAO thus direct comparison of the peak width with and without an osmolyte present is not feasible. Overall, RIM gradually increased for charge reduced ions or narrowing of the ATD (Figure 1d) consistent with the established concentration dependence of TMAO impacting protein structure[23]. While RIM alone does not directly inform on protein structure an increase in resolving power upon osmolyte addition does suggest that either i) a reduced number of structures are adopted by the protein or ii) the reduced surface charge on the protein allows for trapping in wells along the potential energy surface therefore reducing the overall broadness of the population. These results demonstrate the remarkable ability of TMAO to yield charge reduced species with while maintaining relatively narrow peak widths in terms of arrival time and charge state distributions. Molecular modeling of protein ions in the presence and absence of osmolyte ions may reveal greater insight into the effects of small molecules on protein folding landscapes.
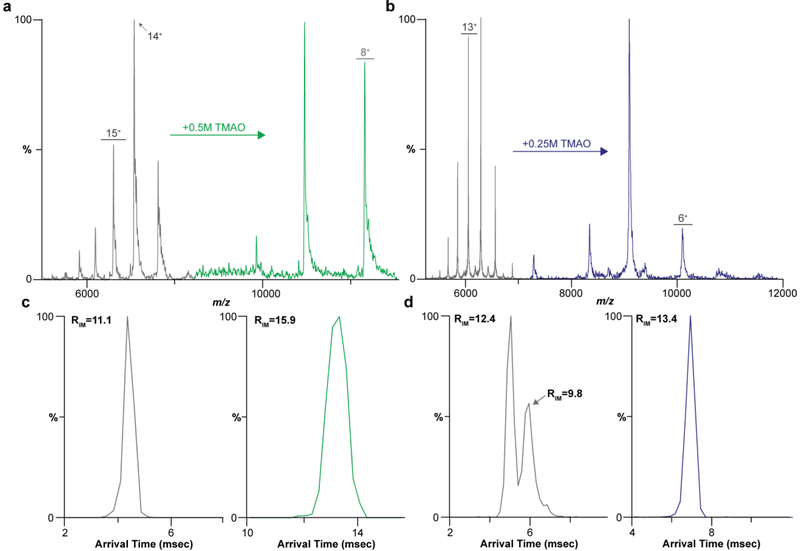
Figure 2.
Charge reduction and reduced conformation heterogeneity for aquaporin Z (AqpZ) from E. coli and mechanosensitive channel of large conductance (MscL) from M. tuberculosis with TMAO. a) ApqZ in 200 mm AA with 2 × CMC C8E4 (black) and doped with 500 mM TMAO (green). b) Mass spectra of MscL in 200 mm AA with 2 × CMC C8E4 (black) and 250 mM TMAO (blue). c) Arrival time distributions (ATD) for the 12+ and 9+ of AqpZ and their respective RIM. d) ATD for the 12+ and 7+ IM of MscL and their respective RIM.
To determine whether TMAO charge reduction is universal, we investigated two additional integral membrane proteins (Figure 2). The first was Aquaporin Z (AqpZ) from E. coli, a tetrameric integral membrane protein[30]. AqpZ has a charge state distribution centered on the 14+ charge state in C8E4 (Figure 2a). The addition of 500 mM TMAO reduced the Zavg by 6 charges (Figure 2a). For reference, in non-charge-reducing detergents, such as octyl glucoside, the reported Zavg for AqpZ is centered around the 18+ charge state[19]. As observed for AmtB, concomitant with charge reduction was a decrease in peak width as observed in the ATD (Figure 2c). The 12+ charge state of AqpZ produced without TMAO contained the most compact, “native-like” ATD of the charge states observed with an RIM of 11.1 (Figure 2C, left panel). In contrast, the 9+ ion produced using TMAO exhibited an RIM of 15.9, corresponding to a 143% increase in resolving power (Figure 2C, right panel). This observed narrowing in peak width suggests that even the highest charge states produced using TMAO sample fewer conformations. Moreover, the charge-reduced ions have similar Rm/z indicated that they have do not have fewer adducts, such as salts or solvent, then the most native of charge states that were previously accessible using charge reducing detergents such as C8E4.
In some cases, more activating, higher energy regimes might be necessary to remove detergents and adducts from the membrane protein complex to obtain a resolved mass spectrum. To demonstrate this effect, we purposely tuned the instrument to yield slightly unfolded charge states using a sample of the mechanosensitive channel of large conductance (MscL) prepared from M. tuberculosis in the charge-reducing detergent C8E4 (Figure 2b). Here, the charge state distribution for MscL is centered on the 14+ charge state and shifts to 7+ upon addition of 250mM TMAO (Figure 2b). Two peaks are observed in the ATD for the 12+ charge state, selected as the most abundant of the “native-like” charge states, with the first having an RIM of 12.1 and 9.8 for the second, consistent with slightly activating conditions (Figure 2d) typically employed during native MS analysis of membrane protein complexes. Strikingly, at higher collision energies (>100 V Trap CE) only one compact, folded conformer is recorded for the ions reduced with 250mM TMAO indicating preservation of native-like structure with an RIM for the 7+ charge state increasing to 13.4 (Figure 2d). We also tuned the instrument for conditions to slightly unfold AmtB and Aqpz and observed, similarly to MscL, upon addition of TMAO the ATD narrowed corresponding to a more compact, folded structure (Figure S3). The addition of TMAO has the potential to preserve protein structure while affording the use of higher collision energies for ion activation.
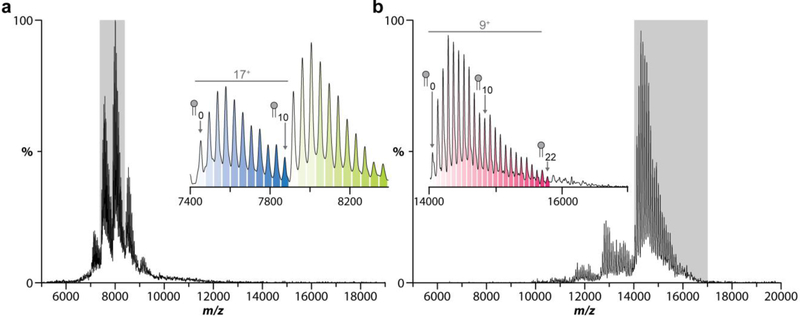
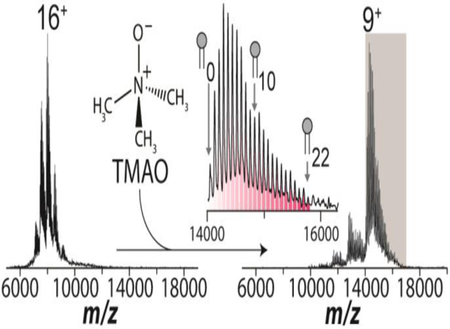
Despite the utility of charge-reducing detergents, there remains a numerical limit to the number of lipid binding events that can be captured before significant overlap occurs with neighboring ions. Overlap of charge states can negatively impact deconvolution programs used to obtain fractional abundances thwarting the ability to obtain quantitative information. While higher mass resolution instruments like orbitraps can mitigate this problem, they do not completely remove overlap between charge states (Figure S4B-D). The native mass spectrum for AmtB doped with 10 equivalents of POPE reveals a distribution of lipids bound to the complex (Figure 3). Importantly, up to ten POPE molecules can be resolved between charge states in correspondence with the predicted mass spectrum (Figure 3a, inset and S4). The addition of TMAO to AmtB mixed with 25 equivalents of POPE increased the amount of resolved lipid binding events to AmtB by two-fold (Figure 3b and S5) before potential overlap with adjacent charge states (Figure 3b, inset) owing to the inherent increase in m/z space between each charge state, which increases as the number of charges decreases. Notably, the presence of TMAO did not interfere with lipid binding. In short, TMAO opens new opportunities to investigate how larger number of lipid binding events, such as the annular belt of lipids immediately surrounding membrane proteins, modulates the protein structure and function.
Figure 3.
Addition of TMAO captures a large number of lipid binding events to AmtB. a) Mass spectrum of 4 μM AmtB doped with 40μM POPE. Inset displays the 16+ and 17+ charge states with a maximum of 10 POPE lipids bound before overlap with neighboring ions. b) Mass spectrum of AmtB doped with 100μM POPE and 250 mM TMAO. Inset displays the 9+ charge state with 22 POPE lipids bound between the 9+ and 8+ charge states.
Activation of ions, observed as unfolding events in the gas phase, severely diminishes the ability of native IM-MS to report on physicochemical properties such as binding affinity and oligomeric state. In some cases, the optimized instrument conditions for longer acyl chain and other detergents, which require more energy to release from the protein[19], may contain IM profiles containing native-like state and partially activated states for lower and higher charge states, respectively. A balance is often struck between ejecting the membrane protein complex from the detergent micelle and minimizing collisional activation.
While C8E4 is an effective charge-reducing detergent for membrane proteins[6], in many cases the highest charge state(s) of the charge-reduced membrane protein complex under optimized instrument settings exhibit minor unfolding as a result of activation, such as seen for AqpZ (Figure S3). The addition of TMAO further reduces charge for membrane proteins in charge-reducing detergents enabling the instrument to be tuned under higher energy regimes while not perturbing protein structure to yield cleanly resolved ions across the charge state distribution. This added benefit by TMAO will be most useful when ejecting membrane protein complexes from longer acyl chain detergents.
In addition to TMAO, other approaches have also been developed to achieve the same level of charge reduction for soluble proteins. Electron transfer dissociation has been implemented to reduce the charge state of intact protein complexes post nESI as low as a singly charged species[31, 32]. Cation-to-anion protein transfer reactions (CAPTR) is another post ionization technique to reduce the charge state of proteins[33–35]. For example, application of CAPTR has revealed that collision cross section of charge-reduced ions generated under denaturing conditions are significantly larger than those generated under native conditions[36]. Lastly, by coupling a corona discharge probe with nESI intact protein complexes could be significantly charge reduced[37]. Although these methods are effective at charge reduction, the addition of TMAO is much easier and more accessible to researchers.
Conclusion
The unique properties of TMAO make it an attractive additive for native IM-MS studies of intact membrane protein complexes. The charge reduction properties of TMAO facilitate generation of ions with reduced charge making them ideal for native analyses. Increased activation energy regimes were able to be used without significant perturbation to protein arrival time distributions. TMAO has recently been shown to counteract urea denaturation on a soluble protein[27], implying the osmolyte could be useful for both soluble and membrane protein complexes. IM-MS has revealed that ions generated using TMAO are compact and display narrow arrival time distributions. Moreover, we observe significant IM peak broadening for AmtB and AqpZ whereas for MscL no significant broadening occurred. These observations suggest that the solvent accessible surface area on AmtB and AqpZ is inherently different than MscL and thus different structures are populated which account for the broad ATD. The addition of TMAO appears to promote fewer structure(s) thus yielding a tighter ATD consistent with the increase in mobility resolution. However, further investigations are warranted to probe protein structure upon binding of lipids and other molecules to membrane protein complexes, such as G protein coupled receptors (GPCR’s), in the presence of TMAO[11, 38]. Charge reduction with TMAO provides an attractive route to elucidate the structural effects that arise from a larger number of bound lipid at the resolution of individual binding events. This will provide the opportunity to move towards more complex lipid mixtures and decipher the intricate roles they have on membrane protein structure and function at unprecedented detail.
Supplementary Material
References
- 1.Loo JA: Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom Rev. 16, 1–23 (1997) [DOI] [PubMed] [Google Scholar]
- 2.Hilton GR, Benesch JL: Two decades of studying non-covalent biomolecular assemblies by means of electrospray ionization mass spectrometry. J R Soc Interface. 9, 801–816 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robinson CV: From molecular chaperones to membrane motors: through the lens of a mass spectrometrist. Biochem Soc Trans. 45, 251–260 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calabrese AN, Radford SE: Mass spectrometry-enabled structural biology of membrane proteins. Methods. (2018) [DOI] [PubMed] [Google Scholar]
- 5.Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M: Drug-target network. Nature biotechnology. 25, 1119–1126 (2007) [DOI] [PubMed] [Google Scholar]
- 6.Laganowsky A, Reading E, Allison TM, Ulmschneider MB, Degiacomi MT, Baldwin AJ, Robinson CV: Membrane proteins bind lipids selectively to modulate their structure and function. Nature. 510, 172–175 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrera NP, Isaacson SC, Zhou M, Bavro VN, Welch A, Schaedler TA, Seeger MA, Miguel RN, Korkhov VM, van Veen HW, Venter H, Walmsley AR, Tate CG, Robinson CV: Mass spectrometry of membrane transporters reveals subunit stoichiometry and interactions. Nat Methods. 6, 585–587 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou M, Morgner N, Barrera NP, Politis A, Isaacson SC, Matak-Vinkovic D, Murata T, Bernal RA, Stock D, Robinson CV: Mass spectrometry of intact V-type ATPases reveals bound lipids and the effects of nucleotide binding. Science. 334, 380–385 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcoux J, Wang SC, Politis A, Reading E, Ma J, Biggin PC, Zhou M, Tao H, Zhang Q, Chang G, Morgner N, Robinson CV: Mass spectrometry reveals synergistic effects of nucleotides, lipids, and drugs binding to a multidrug resistance efflux pump. Proceedings of the National Academy of Sciences of the United States of America. 110, 9704–9709 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Housden NG, Hopper JT, Lukoyanova N, Rodriguez-Larrea D, Wojdyla JA, Klein A, Kaminska R, Bayley H, Saibil HR, Robinson CV, Kleanthous C: Intrinsically disordered protein threads through the bacterial outer-membrane porin OmpF. Science. 340, 1570–1574 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gault J, Donlan JA, Liko I, Hopper JT, Gupta K, Housden NG, Struwe WB, Marty MT, Mize T, Bechara C, Zhu Y, Wu B, Kleanthous C, Belov M, Damoc E, Makarov A, Robinson CV: High-resolution mass spectrometry of small molecules bound to membrane proteins. Nat Methods. 13, 333–336 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cong X, Liu Y, Liu W, Liang X, Russell DH, Laganowsky A: Determining Membrane Protein-Lipid Binding Thermodynamics Using Native Mass Spectrometry. J Am Chem Soc. 138, 4346–4349 (2016) [DOI] [PubMed] [Google Scholar]
- 13.Gupta K, Donlan JAC, Hopper JTS, Uzdavinys P, Landreh M, Struwe WB, Drew D, Baldwin AJ, Stansfeld PJ, Robinson CV: The role of interfacial lipids in stabilizing membrane protein oligomers. Nature. 541, 421–424 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cong X, Liu Y, Liu W, Liang X, Laganowsky A: Allosteric modulation of protein-protein interactions by individual lipid binding events. Nat Commun. 8, 2203 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patrick JW, Boone CD, Liu W, Conover GM, Liu Y, Cong X, Laganowsky A: Allostery revealed within lipid binding events to membrane proteins. Proceedings of the National Academy of Sciences of the United States of America. 115, 2976–2981 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yen HY, Hoi KK, Liko I, Hedger G, Horrell MR, Song W, Wu D, Heine P, Warne T, Lee Y, Carpenter B, Pluckthun A, Tate CG, Sansom MSP, Robinson CV: PtdIns(4,5)P2 stabilizes active states of GPCRs and enhances selectivity of G-protein coupling. Nature. 559, 423–427 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laganowsky A, Reading E, Hopper JT, Robinson CV: Mass spectrometry of intact membrane protein complexes. Nat Protoc. 8, 639–651 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen SH, Russell DH: How Closely Related Are Conformations of Protein Ions Sampled by IMMS to Native Solution Structures? J Am Soc Mass Spectrom. 26, 1433–1443 (2015) [DOI] [PubMed] [Google Scholar]
- 19.Reading E, Liko I, Allison TM, Benesch JL, Laganowsky A, Robinson CV: The role of the detergent micelle in preserving the structure of membrane proteins in the gas phase. Angew Chem Int Ed Engl. 54, 4577–4581 (2015) [DOI] [PubMed] [Google Scholar]
- 20.Hopper JT, Sokratous K, Oldham NJ: Charge state and adduct reduction in electrospray ionization-mass spectrometry using solvent vapor exposure. Anal Biochem. 421, 788–790 (2012) [DOI] [PubMed] [Google Scholar]
- 21.Mehmood S, Marcoux J, Hopper JT, Allison TM, Liko I, Borysik AJ, Robinson CV: Charge reduction stabilizes intact membrane protein complexes for mass spectrometry. J Am Chem Soc. 136, 17010–17012 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liko I, Hopper JT, Allison TM, Benesch JL, Robinson CV: Negative Ions Enhance Survival of Membrane Protein Complexes. J Am Soc Mass Spectrom. 27, 1099–1104 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fonin AV, Uversky VN, Kuznetsova IM, Turoverov KK: [Protein Folding and Stability in the Presence of Osmolytes]. Biofizika. 61, 222–230 (2016) [PubMed] [Google Scholar]
- 24.Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN: Living with water stress: evolution of osmolyte systems. Science. 217, 1214–1222 (1982) [DOI] [PubMed] [Google Scholar]
- 25.Konermann L, Ahadi E, Rodriguez AD, Vahidi S: Unraveling the mechanism of electrospray ionization. Anal Chem. 85, 2–9 (2013) [DOI] [PubMed] [Google Scholar]
- 26.Kim D, Wagner N, Wooding K, Clemmer DE, Russell DH: Ions from Solution to the Gas Phase: A Molecular Dynamics Simulation of the Structural Evolution of Substance P during Desolvation of Charged Nanodroplets Generated by Electrospray Ionization. Journal of the American Chemical Society. 139, 2981–2988 (2017) [DOI] [PubMed] [Google Scholar]
- 27.Gault J, Lianoudaki D, Kaldmae M, Kronqvist N, Rising A, Johansson J, Lohkamp B, Lain S, Allison TM, Lane DP, Marklund EG, Landreh M: Mass Spectrometry Reveals the Direct Action of a Chemical Chaperone. J Phys Chem Lett. 9, 4082–4086 (2018) [DOI] [PubMed] [Google Scholar]
- 28.Marty MT, Baldwin AJ, Marklund EG, Hochberg GK, Benesch JL, Robinson CV: Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles. Anal Chem. 87, 4370–4376 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allison TM, Reading E, Liko I, Baldwin AJ, Laganowsky A, Robinson CV: Quantifying the stabilizing effects of protein-ligand interactions in the gas phase. Nat Commun. 6, 8551 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savage DF, Egea PF, Robles-Colmenares Y, O’Connell JD 3rd, Stroud RM: Architecture and selectivity in aquaporins: 2.5 a X-ray structure of aquaporin Z. PLoS Biol. 1, E72 (2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lermyte F, Lacki MK, Valkenborg D, Gambin A, Sobott F: Conformational Space and Stability of ETD Charge Reduction Products of Ubiquitin. J Am Soc Mass Spectrom. 28, 69–76 (2017) [DOI] [PubMed] [Google Scholar]
- 32.Lermyte F, Williams JP, Brown JM, Martin EM, Sobott F: Extensive Charge Reduction and Dissociation of Intact Protein Complexes Following Electron Transfer on a Quadrupole-Ion Mobility-Time-of-Flight MS. J Am Soc Mass Spectrom. 26, 1068–1076 (2015) [DOI] [PubMed] [Google Scholar]
- 33.Laszlo KJ, Bush MF: Analysis of Native-Like Proteins and Protein Complexes Using Cation to Anion Proton Transfer Reactions (CAPTR). J Am Soc Mass Spectrom. 26, 2152–2161 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laszlo KJ, Bush MF: Interpreting the Collision Cross Sections of Native-like Protein Ions: Insights from Cation-to-Anion Proton-Transfer Reactions. Anal Chem. 89, 7607–7614 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laszlo KJ, Munger EB, Bush MF: Folding of Protein Ions in the Gas Phase after Cation-to-Anion Proton-Transfer Reactions. J Am Chem Soc. 138, 9581–9588 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gadzuk-Shea MM, Bush MF: Effects of Charge State on the Structures of Serum Albumin Ions in the Gas Phase: Insights from Cation-to-Anion Proton-Transfer Reactions, Ion Mobility, and Mass Spectrometry. J Phys Chem B. 122, 9947–9955 (2018) [DOI] [PubMed] [Google Scholar]
- 37.Campuzano IDG, Schnier PD: Coupling electrospray corona discharge, charge reduction and ion mobility mass spectrometry: From peptides to large macromolecular protein complexes. International Journal for Ion Mobility Spectrometry. 16, 51–60 (2013) [Google Scholar]
- 38.Poltash ML, McCabe JW, Patrick JW, Laganowsky A, Russell DH: Development and Evaluation of a Reverse-Entry Ion Source Orbitrap Mass Spectrometer. Journal of The American Society for Mass Spectrometry. (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.