ABSTRACT
Salmonella enterica Javiana is a leading cause of severe foodborne Salmonellosis. Despite its emergence as a major foodborne pathogen, little is known of how S. Javiana interacts with intestinal epithelial cells, or of potential methods for ameliorating the bacterial-host interaction. Using cell-based adhesion, invasion and lactate dehydrogenase release assays, we observed an invasive and cytotoxic effect of S. Javiana on intestinal epithelial cells. We assessed the effect of probiotic species of lactic acid bacteria (LAB) on the S. Javiana-host cell interaction, and hypothesized that LAB would reduce S. Javiana infectivity. Salmonella enterica Javiana invasion was significantly impaired in host cells pre-treated with live Lactobacillus acidophilus and Lactobacillus rhamnosus. In addition, pre-exposure of host cells to live L. acidophilus, L. rhamnosus and L. casei reduced S. Javiana-induced cytotoxicity, while heat-killed LAB cultures had no effect on S. Javiana invasion or cytotoxicity. qRT-PCR analysis revealed that S. Javiana exposed to L. acidophilus and L. rhamnosus exhibited reduced virulence gene expression. Moreover, pre-treating host cells with LAB prior to S. Javiana infection reduced host cell production of inflammatory cytokines. Data suggest a potential protective effect of L. acidophilus, L. rhamnosus and L. casei against intestinal epithelial infection and pathogen-induced damage caused by S. Javiana.
Keywords: Salmonella enterica Javiana, lactic acid bacteria, probiotics, intestinal epithelial cells, invasion, cytotoxicity
Lactic acid bacteria (LAB) decrease S. Javiana virulence and modulate host inflammatory response to infection.
INTRODUCTION
Salmonella is a Gram-negative, facultatively anaerobic bacillus and member of the Enterobacteriaceae family. The Salmonella genus is comprised of two species, Salmonella bongori and Salmonella enterica, the latter of which contains most medically-relevant strains. Salmonella enterica is a highly ubiquitous species containing over 2600 serovars that can be divided into typhoidal and non-typhoidal Salmonella (NTS) (Gal-Mor, Boyle and Grassl 2014). Typhoidal serovars (S. enterica Typhi and S. enterica Paratyphi) cause life-threatening systemic disease, while most NTS serovars cause serious gastroenteritis and other acute infections in humans and animals. NTS strains are the third leading cause of bacterial foodborne illness globally (Majowicz et al. 2010), and cause nearly 94 million illnesses and 155 000 deaths annually worldwide (Kirk et al. 2015). Each year in the U.S., Salmonella causes an estimated 1.2 million illnesses, 23 000 hospitalizations and 450 deaths (Centers for Disease Control and Prevention 2018), and the majority of these infections are attributed to consumption of NTS strains in contaminated food products, including meat, poultry, eggs, cheese, seafood and produce (Jackson et al. 2013). Although more than 2400 serovars of NTS have been identified, most cases of Salmonella-induced gastroenteritis are attributed to five serovars: S. Typhimurium, S. Enteriditis, S. Newport, S. Javiana and S. Heidelberg (Boore et al. 2015).
Of the leading strains of NTS, S. Javiana is the fourth most common (Boore et al. 2015), but is one of the least characterized in terms of its interaction with the host intestinal epithelium. The majority of S. Javiana strains contain Salmonella Pathogenicity Island-1 (SPI-1) and SPI-2 encoded virulence genes (Allard et al. 2013; Mezal, Stefanova and Khan 2013) commonly found in other gastroenteritis-associated NTS serovars, and which enable colonization and invasion of the intestinal epithelium, and intracellular survival, respectively (Zhou 2001; Abrahams and Hensel 2006). In addition, genomic analyses have revealed that S. Javiana strains possess the genes pltA, pltB and cdtB, which together encode the cytolethal distending toxin (CDT), a virulence factor more commonly associated with infection by the typhoidal strain S. Typhi than NTS strains (Mezal, Stefanova and Khan 2013), and which promotes host cell invasion, cell cycle arrest, DNA damage and systemic spread (Williams et al. 2015; Miller et al. 2018). However, few studies have examined the interaction of S. Javiana with intestinal epithelial cells during infection or potential ways to ameliorate the host-pathogen interaction.
NTS are a public health concern not only due to the frequency of infection, but also because of the emergence of antimicrobial resistant strains. In the U.S., there are an estimated 100 000 cases of drug-resistant Salmonellosis each year, over 66 000 of which are caused by multidrug-resistant NTS (Centers for Disease Control and Prevention 2013). Not surprisingly, resistance is rising fastest among the leading NTS strains, including S. Javiana. In 2002 the CDC reported no resistance to commonly used antibiotics among S. Javiana isolates from an outbreak (Centers for Disease Control and Prevention 2002), but more recent studies indicate emergence of ampicillin-, tetracycline- (Mezal, Stefanova and Khan 2013), sulfisoxazole- (Micallef et al. 2012), gentamicin-, streptomycin- and kanamycin-resistance (Santos et al. 2007; Mezal, Stefanova and Khan 2013; Angelo et al. 2016; Nair, Venkitanarayanan and Johny 2018) among S. Javiana isolated from clinical, food and environmental sources. Given the high incidence of foodborne Salmonellosis and rise in drug resistance among NTS serovars, there is urgent need for discovery and development of antibiotic alternatives for prevention or treatment of Salmonella infections, including those caused by S. Javiana.
Probiotics, which are viable microorganisms that upon ingestion have beneficial effects on the host, have gained widespread attention for their anti-infective properties and potential use as non-antibiotic prophylactic or therapeutic agents (Pereira et al. 2018). While a wide variety of bacteria and yeast are reported to have probiotic properties (Surendran Nair, Amalaradjou and Venkitanarayanan 2017), species of lactic acid bacteria (LAB) are well-documented for exhibiting antimicrobial and anti-virulence effects against gastrointestinal (GI) pathogens (Fuller 1992) and for exerting immunoregulatory and homeostatic effects on the host (Lievin-Le Moal and Servin 2014; George et al. 2018; Zhang et al. 2018). Indeed, in vitro and in vivo studies have shown that LAB can limit infectivity of GI pathogens, including NTS serovars (Chen et al. 2012; Lievin-Le Moal and Servin 2014; Dutra et al. 2016; Muyyarikkandy and Amalaradjou 2017). However, since the anti-infective properties of probiotics are often unique to specific strains of probiotics and pathogens (Chen et al. 2012; Campana, van Hemert and Baffone 2017), and since to our knowledge, no studies have examined the impact of probiotics on intestinal epithelial cell infection by S. Javiana, whether LAB might affect S. Javiana interaction with intestinal epithelial cells is not known. Therefore, we sought to use a cell-based infection model to examine the interaction of S. Javiana with intestinal epithelial cells and to investigate the potential for probiotic LAB strains to modulate S. Javiana infection. Here, we report that S. Javiana has an invasive phenotype and cytotoxic effect on the human HT29-MTX intestinal epithelial cell line, and that the LAB species Lactobacillus acidophilus, Lactobacillus rhamnosus and Lactobacillus casei can reduce S. Javiana invasion, limit pathogen-induced cell damage, alter S. Javiana virulence gene expression and modulate the host cell inflammatory response to infection.
MATERIALS AND METHODS
Bacterial strains and growth conditions
All bacterial strains used in this study are listed in Table 1. Salmonella enterica Javiana and Escherichia coli were grown in Luria Bertani (LB) broth, while LAB species L. acidophilus, L. rhamnosus, L. casei, Lactobacillus plantarum and Leuconostoc mesenteroides were grown in deMann-Rogosa-Sharpe (MRS) broth. All bacterial cultures were stored at −80C with addition of 20% (vol/vol) glycerol. Prior to use in experiments, cultures were subcultured onto either LB agar or MRS agar and individual colonies were grown overnight either in LB broth in a shaking incubator at 37°C or in MRS broth in a static incubator at 37°C.
Table 1.
Bacterial strains used in this study.
| Bacterium | Strain description | Source or reference |
|---|---|---|
| Salmonella enterica Javiana | CFSAN1 0 01992, human stool isolate | Obtained from Dr. Marc Allard (U.S. FDA) (2) |
| Escherichia coli | DUP-101 strain, ATCC2 51 739 | ATCC |
| Lactobacillus acidophilus | ARS NRRL3 B1910 | Our collection |
| Lactobacillus casei | KCTC4 3109 | Our collection |
| Leuconostoc mesenteroides | ARS NRRL B-1118, isolated from olives | Our collection |
| Lactobacillus plantarum | ARS NRRL B-4496, ATCC 14 917, isolated from pickled cabbage | Our collection |
| Lactobacillus rhamnosus GG | ATCC 53 103, human stool isolate | Our collection (15) |
Center for Food Safety and Applied Nutrition
American Type Culture Collection
Agricultural Research Service (ARS) Northern Regional Research Laboratory (NRRL) Culture Collection
Korean Collection Type Cultures
Cell culture
The mucus-secreting human colonic cell line HT29-MTX-E12 (HT29-MTX) was purchased from Sigma Aldrich. HT29-MTX cells were cultured in Roswell Park Memorial Institute 1640 (RPMI) medium containing 10% FBS (v/v), 1% GlutaMAX (Life Technologies), 1% HEPES buffer and 1% of a 100X penicillin-streptomycin (VWR). Cells were incubated at 37°C in a humidified 5% (v/v) CO2 atmosphere and used between passages 10 and 20. Cells were seeded into 24-well plates (for adhesion, invasion, cytotoxicity and cytokine assays) at a density of 4.0 × 104 cells per well or into 6-well culture plates (for gene expression studies) at a density of 1.7 × 105 cells per well. Cell medium was changed every two days and medium without antibiotic was used for the last medium change prior to infection assays. Infections were performed on confluent monolayers at 14–21 days post-seeding, to ensure that HT29-MTX cells had reached maturity (Lesuffleur et al. 1993).
Bacterial adhesion, invasion and intracellular survival assays
All infection assays were performed in RPMI growth medium, at 37°C in 5% CO2 atmosphere. To assess adhesion and invasion of S. Javiana or E. coli (non-pathogenic negative control) to HT29-MTX monolayers, overnight bacterial cultures were washed and resuspended in RPMI medium and then added to HT29-MTX monolayers grown on sterile coverslips in 24-well plates at multiplicity of infection (MOI) of 10. For adhesion assays, at 1 h p.i., monolayers were washed three times with Dulbecco's PBS (D-PBS, Lonza) to remove non-adherent bacteria, then incubated with 0.01% Triton X-100 (Sigma Aldrich) for 5 min to dislodge attached bacteria (Burkholder and Bhunia 2010), and adherent bacteria were enumerated by plating onto LB agar. For invasion assays, at 2 h p.i. infected monolayers were treated with gentamicin (100 µg/mL) for 30 min to kill extracellular bacteria, then washed three times with D-PBS and lysed with 0.1% Triton-X (Burkholder and Bhunia 2009; Burkholder and Bhunia 2010). Intracellular bacteria were enumerated by plating cell lysates onto LB agar.
To examine the impact of LAB pre-treatment on S. Javiana adhesion or invasion, overnight LAB cultures were washed and resuspended in RPMI medium, then added to HT29-MTX monolayers grown on sterile coverslips in 24-well plates at multiplicity of exposure (MOE) of 10. At 1 h post-LAB exposure, non-adherent LAB were removed by washing HT29-MTX cells with D-PBS, S. Javiana was added to cell medium at MOI 10 and infected cells were incubated for 1 h. At 1 h post-infection, S. Javiana adhesion was assessed, and at 2 h post-infection S. Javiana invasion was determined as described above. Adherent and intracellular S. Javiana were enumerated on LB agar, which did not support the growth of any LAB tested (data not shown).
For invasion assays using heat-killed LAB (HK-LAB), overnight LAB cultures were washed, resuspended in sterile water and autoclaved at 121°C for 15 min to kill the bacteria (Wagner et al. 2000). Aliquots of autoclaved cultures were plated on MRS agar to assess loss of viability, and Gram stains were prepared to ensure presence of bacterial cells in the autoclaved suspensions. HK-LAB were pelleted, resuspended in RPMI medium and added to HT29-MTX monolayers at MOE 10. At 1 h post-exposure to HK-LAB, HT29-MTX cells were washed, infected with S. Javiana (MOI 10) and S. Javiana invasion was assessed as described above.
To assess the effect of LAB pre-treatment on the number of intracellular S. Javiana present at 6 h p.i., HT29-MTX cells were either infected with S. Javiana alone (MOI 10) or pre-treated with live L. acidophilus, L. rhamnosus or L. casei (MOE 10) for 1 h prior to S. Javiana infection. At 2 h p.i., gentamicin (100 µg/ml) was added to the cell media to kill extracellular bacteria. At 6 h p.i., epithelial cells were washed three times with D-PBS, lysed with 0.1% Triton-X, and intracellular S. Javiana were enumerated by plating on LB agar.
Cytotoxicity assays
A lactate dehydrogenase (LDH) release assay (Thermo Fisher Scientific) was used to quantify host cell damage induced by S. Javiana (Burkholder and Bhunia 2009). HT29-MTX cells were grown in 24-well plates and infected with S. Javiana or E. coli K12 (non-pathogenic control) as described above for adhesion and invasion assays. Uninfected cells were used as a negative control and cells treated with 0.1% Triton-X (TX) served as a positive control. At 2 h p.i., HT29-MTX supernatants were collected and centrifuged (800 x g for 5 min) to remove bacterial and eukaryotic cells. A 100 ul aliquot of each sample was dispensed into triplicate wells of a 96-well plate and LDH activity was determined spectrophotometrically per manufacturer's protocol, using the formula: % Cytotoxicity of sample = ((AbsTX–Abssample)/(AbsTX–AbsUninfected))*100.
To assess the effect of individual LAB strains on S. Javiana-induced cytotoxicity, overnight LAB or E. coli cultures were added to HT29-MTX monolayers at MOE of 10 as described above. At 1 h post- exposure, non-adherent bacteria were removed by washing HT29-MTX cells with D-PBS, S. Javiana was added to cell medium at MOI 10 and infected cells were incubated for 2 h. At 2 h p.i., HT29-MTX cell supernatants were collected, LDH was quantitated and % cytotoxicity calculated as detailed above. For cytotoxicity assays using HK-LAB, overnight LAB cultures were autoclaved at 121°C for 15 min as described above, and then resuspended in RPMI medium and added to HT29-MTX monolayers at MOE 10 for 1 h prior to infection with S. Javiana. In separate experiments, we also performed an LDH assays with individual LAB strains, in absence of S. Javiana, to ensure that LAB alone did not have cytotoxic effects on HT29-MTX cells (Fig. S1, Supporting Information).
Salmonella viability assay
To assess the impact of LAB on S. Javiana viability, overnight cultures of S. Javiana alone or S. Javiana plus individual LAB strains were diluted 1:50 in RPMI and incubated at 37°C in 5% CO2, to mimic the growth conditions and bacterial concentrations used in infection assays. At 0, 2, 4 and 6 h post-inoculation, aliquots were obtained and plated on LB agar (which allowed enumeration of S. Javiana but not LAB) to quantify S. Javiana viability, and on MRS agar (which allowed enumeration of LAB but not S. Javiana) to confirm viability of LAB strains (MRS data not shown).
Analysis of the effect of LAB on Salmonella virulence gene expression
The effect of L. acidophilus, L. rhamnosus and L. casei on expression of S. Javiana virulence genes invA, prgH, pltA and cdtB was determined by qRT-PCR. These strains of LAB were chosen for these experiments because they were the strains that impaired S. Javiana invasion and S. Javiana-induced cytotoxicity. To mimic conditions used in infection assays, gene expression studies were performed in RPMI medium in the presence of HT29-MTX cells. Briefly, HT29-MTX cells grown in 6-well plates were pre-treated with or without individual strains of L. acidophilus, L. rhamnosus or L. casei (MOE 10). At 1 h post-LAB exposure, S. Javiana was added to the cell medium at MOI 10. At 2 h p.i., HT29-MTX supernatant and monolayers were harvested (to collect extracellular and intracellular S. Javiana), samples were pelleted and pellets were resuspended in Qiagen RNeasy mini kit buffer RLT containing β-mercaptoethanol and added to tubes containing 0.2 mm Rnase-free stainless steel beads (Next Advance). Samples were homogenized using a Bullet Blender cell disruptor (Next Advance) and mRNA was prepared from the homogenate using the Qiagen RNeasy kit according to the manufacturer protocol. Samples were treated with RNase-free DNase I (Qiagen) to remove remaining host cell and bacterial DNA and were analyzed for purity using a NanoDrop spectrophotometer. The resulting RNA was stored at −80°C. cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Life Technologies) and samples were used for RT-PCR with PowerUp Sybr Green Master Mix (Life Technologies) and the primer sequences listed in Table 2. Real time detection and relative quantitation of transcripts were achieved with the StepOne Plus Real-Time PCR System (Life Technologies). Prior to performing qRT-PCR, all primers were validated against DNA from S. Javiana, L. acidophilus, L. rhamnosus, L. casei and HT29-MTX cells to ensure that they amplified only S. Javiana DNA, and that there was no non-specific amplification of LAB or host cell DNA.
Table 2.
Primers used in this study.
| Primera | Sequence (5′–3′) | Source |
|---|---|---|
| cdtB-F primer | ATGTCTTGCGTCCGACAACT | This studyb |
| cdtB-R primer | CGTGCGCTGTCAGAAAAACA | |
| pltA-F primer | TTTACCAGACCTGTTGCGCT | This study |
| pltA-R primer | AGCTTGCTCCCATCCATCAC | |
| invA-F primer | CGCGCTTGATGAGCTTTACC | This study |
| invA-R primer | TCGCTTAACAAACGCTGCAC | |
| prgH-F primer | GGGCGCTCGATGATGTAGAA | This study |
| prgH-R primer | TGGCCTGGGCTCATTTTGAT | |
| 16S-F primer | GGCGCATACAAAGAGAAGCG | This study |
| 16S-R primer | CTCCAATCCGGACTACGACG |
F, forward; R, reverse
All primer sequences based on the Salmonella enterica subsp. enterica serovar Javiana strain CFSAN001992 chromosomal genome sequence (GenBank accession no. NC_02 0307.1)
The effect of LAB on S. Javiana gene expression was determined using the comparative quantification (∆∆C T) method (Livak and Schmittgen 2001; Tranchemontagne et al. 2016). Briefly, the ∆∆CT method compares the threshold cycle (CT) from an experimental sample (from LAB-exposed S. Javiana) with both a calibrator sample (from S. Javiana not exposed to LAB) and a normalizer (S. Javiana-specific 16S rRNA housekeeping gene measured in experimental and calibrator samples). The ∆CT value, representing the difference in threshold cycle between the target and normalizer genes, was determined by subtracting the CT value of the 16S rRNA gene from the CT values for each target gene (invA, prgH, pltA, or cdtB). The ∆∆CT value was derived from the subtraction of the ∆CT of the calibrator sample from the ∆CT of the experimental sample. 2−∆∆CT was expressed as the n-fold difference in gene expression in the experimental sample compared to the calibrator sample at each time-point tested. Genes exhibiting greater than 3-fold change in expression were considered to be altered by LAB exposure.
Cytokine assays
To determine whether LAB exposure would alter host cell cytokine secretion in response to S. Javiana infection, HT29-MTX monolayers were grown in 24-well plates and were either uninfected, exposed to S. Javiana alone (MOI 10), or pre-treated with individual LAB (MOE 10) for 1 h followed by S. Javiana infection (MOI 10). At 2 h p.i., gentamicin (100 µg/mL) was added to the infection medium to kill extracellular bacteria, and monolayers were incubated for an additional 4 h to allow for secretion of cytokines. Infected cell supernatants were collected and analyzed for cytokines using the Human Inflammation Quanitbody array (RayBiotech) according to manufacturer protocol. Completed arrays were shipped to the manufacturer for scanning and cytokine quantitation. The limit of detection for cytokines in this assay is 3 pg/ml (RayBiotech).
Statistical analysis
Differences between treatments were assessed by analysis of variance (ANOVA) using R, version 2.15 (R Development Core Team 2012). Any significant ANOVA tests were further analyzed using Tukey's post hoc pairwise comparisons. We used an alpha value of 0.05, so P values of <0.05 were considered significant. All error bars represent standard deviations.
RESULTS
Adhesion, invasion and cytotoxic effect of S. Javiana during infection of HT29-MTX epithelial cells
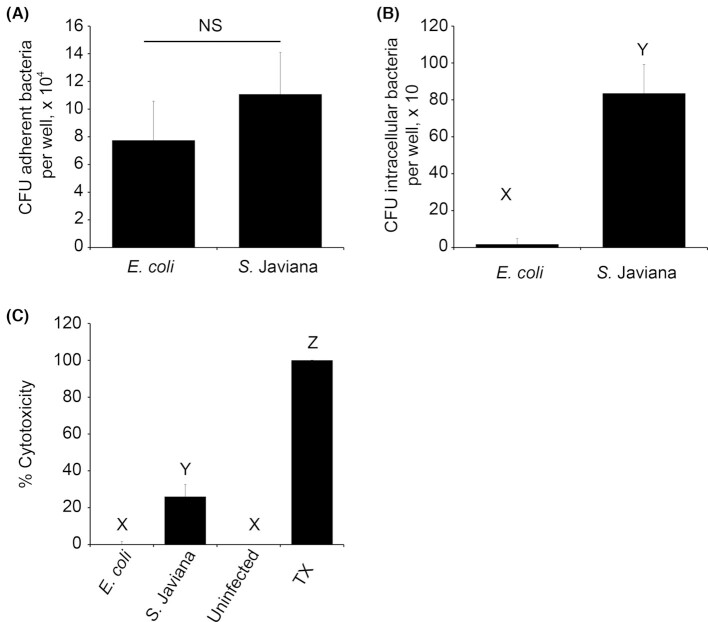
Few studies have examined the interaction of S. Javiana with intestinal epithelial cells. Therefore, we used the human colonic HT29-MTX intestinal epithelial cell line (Gagnon et al. 2013) to examine S. Javiana adhesion, invasion and cytotoxic effect during host cell infection. Nonpathogenic E. coli was used as a control that, although moderately adhesive, exhibits little invasion or cytotoxicity. While S. Javiana adhered to HT29-MTX cells at levels similar to E. coli (Fig. 1A), as expected, S. Javiana exhibited significantly greater invasion than E. coli into host cells (Fig. 1B). LDH release assays revealed significantly greater cytotoxic effect of S. Javiana compared to the nonpathogen (Fig. 1C). Together, these findings confirm that, similar to other gastroenteritis-producing strains of NTS, S. Javiana interacts with, invades and damages epithelial cells during infection (Wallis and Galyov 2000; Bergeron et al. 2009; Burkholder and Bhunia 2009).
Figure 1.
Salmonella Javiana adheres to, invades and exhibits cytotoxic effect on HT29-MTX intestinal epithelial cells. (A, B) HT29-MTX intestinal epithelial cells were inoculated with E. coli (non-pathogenic negative control) or S. Javiana, each at MOI 10. (A) At 1 h p.i., adherent bacteria were enumerated by plating. (B) At 2 h p.i., gentamicin-containing media (100 µg/ml) was added to the monolayers to kill extracellular bacteria, and 30 min later monolayers were washed and lysed with 0.1% Triton-X. Intracellular bacteria were enumerated from lysates by plating. (C) HT29-MTX intestinal epithelial cells were infected with E. coli (non-cytotoxic negative control) or S. Javiana. Uninfected cells served as a negative control, while 0.1% Triton-X (TX) was used as a positive control. At 2 h p.i., LDH was measured in HT29-MTX cell supernatants, and % cytotoxicity was determined as described in methods. In A and B, number of adherent or intracellular bacteria were compared between treatments. In C, % cytotoxicity was compared between treatments. NS indicates no significant difference in bacterial adhesion between treatments, while different letters (X,Y,Z) indicate significant pairwise differences in invasion or cytotoxicity between treatment groups (P < 0.05).
LAB reduce S. Javiana invasion and S. Javiana-induced cytotoxicity during epithelial cell infection
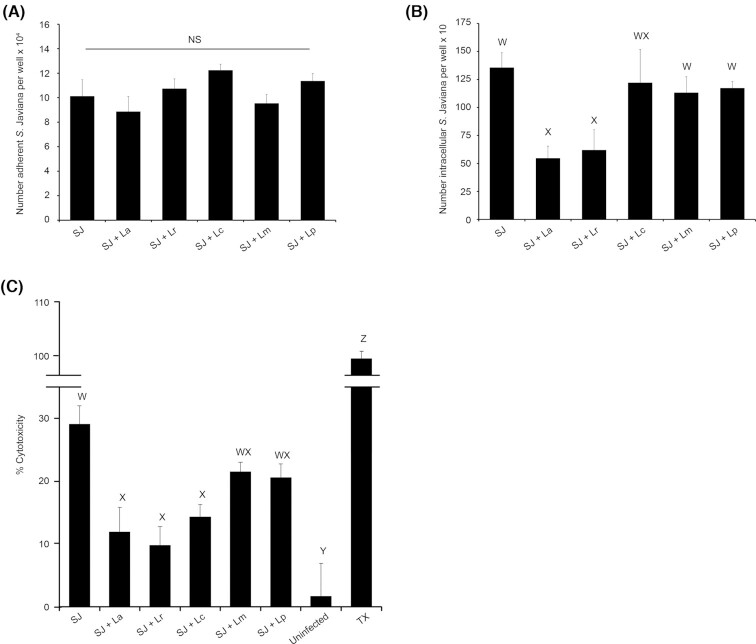
Previous reports from our work and others have shown that LAB can hinder the interaction of Salmonella with host cells and ameliorate Salmonella-induced cell or tissue damage (Burkholder and Bhunia 2009; Chen et al. 2012; Campana, van Hemert and Baffone 2017; Muyyarikkandy and Amalaradjou 2017; Liu et al. 2018). Therefore, we sought to examine, in our HT29-MTX infection model, the effect of LAB species commonly used as probiotics (Burkholder and Bhunia 2009; Campana, van Hemert and Baffone 2017; Choi et al. 2018) on S. Javiana adhesion, invasion and S. Javiana-induced cytotoxicity. We performed adhesion, invasion and LDH release assays in which host cells were either untreated or pre-exposed to L. acidophilus, L. mesenteroides, L. rhamnosus, Lactobacillus plantarum or L. casei (MOE 10) for 1 h prior to infection with S. Javiana (MOI 10). All LAB strains bound to the HT29-MTX cells and were adherent at the time of S. Javiana infection, with L. acidophilus, L. rhamnosus and L. casei exhibiting greatest host cell adhesion (Fig. S2, Supporting Information). While there was no effect of any LAB strain on S. Javiana adhesion to HT29-MTX cells (Fig. 2A), S. Javiana invasion was significantly reduced in host cells pre-exposed to L. acidophilus or L. rhamnosus (Fig. 2B). Similarly, the cytotoxic effect of S. Javiana was significantly decreased in host cells pre-treated with L. acidophilus, L. rhamnosus and L. casei compared to cells infected with S. Javiana alone (Fig. 2C). In contrast, pre-exposing HT29-MTX cells to nonpathogenic E. coli prior to S. Javiana infection had no impact on S. Javiana adhesion, invasion or cytotoxic effect (Fig. S3, Supporting Information). Data suggest that pre-exposing host cells to L. acidophilus, L. rhamnosus and L. casei has some protective effect that can reduce S. Javiana invasion and pathogen-induced host cell damage.
Figure 2.
Pre-treating HT29-MTX cells with LAB prior to infection reduces S. Javiana invasion and S. Javiana-induced cytotoxicity. (A, B) HT29-MTX cells were infected with S. Javiana alone (MOI 10) or pre-treated for 1 h with individual LAB (MOE 10) prior to infection with S. Javiana. (A) At 1 h p.i., epithelial cells were washed and adherent S. Javiana were enumerated by plating. (B) At 2 h p.i., monolayers were treated with gentamicin (100 µg/ml) and 30 min later, epithelial cells were lysed and intracellular S. Javiana were enumerated by plating. (C) HT29-MTX cells were pre-treated with LAB (MOE 10) for 1 h prior to infection with S. Javiana (MOI 10) for 2 h. At 2 h p.i., LDH was measured in HT29-MTX cell supernatants and % cytotoxicity was determined. In A and B, number of adherent or intracellular S. Javiana were compared between treatments. In C, % cytotoxicity was compared between treatments. NS indicates no significant difference in S. Javiana adhesion between treatment groups, while different letters (W,X,Y,Z) indicate significant pairwise differences in invasion or cytotoxicity (P < 0.05) between treatments. (SJ = S. Javiana, La = L. acidophilus, Lr = L. rhamnosus, Lc = L. casei, Lm = L. mesenteroides, Lp = L. plantarum, TX = Triton X-treated cells)
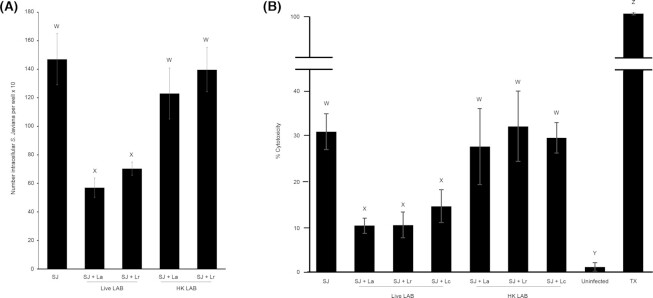
Heat-killed LAB do not affect S. Javiana invasion or S. Javiana-induced cytotoxicity
The mechanism by which probiotic bacteria might alter S. Javiana invasion and cytotoxic effect is unclear. Therefore, we performed similar invasion and cytotoxicity studies using live and HK-LAB, to ascertain whether LAB-mediated perturbation of S. Javiana infection is an active or passive probiotic process. While live LAB altered S. Javiana invasion and cytotoxic effect on host cells (Fig. 3A-B) there was no effect of HK L. acidophilus or L. rhamnosus on S. Javiana invasion (Fig. 3A). Similarly, there was no impact of HK L. acidophilus, L. rhamnosus or L. casei on S. Javiana-induced HT29-MTX cytotoxicity (Fig. 3B). Data indicate that perturbation of S. Javiana interaction with HT29-MTX cells is an active LAB process, and for that reason, all additional experiments were conducted with live LAB cultures.
Figure 3.
Heat-killed LAB do not alter S. Javiana invasion or S. Javiana-induced cytotoxicity. (A) HT29-MTX cells were infected with S. Javiana alone (MOI 10) or pre-treated for 1 h with live or heat-killed L. acidophilus or L. rhamnosus (MOE 10) prior to infection with S. Javiana. At 2 h p.i., monolayers were treated with gentamicin (100 µg/ml) and 30 min later, epithelial cells were lysed and intracellular S. Javiana were enumerated by plating. (B) HT29-MTX cells were either infected with S. Javiana alone (MOI 10) or pre-treated with live or heat-killed L. acidophilus, L. rhamnosus or L. casei (MOE 10) for 1 h prior to infection with S. Javiana. At 2 h p.i., LDH was measured in HT29-MTX cell supernatants and % cytotoxicity was determined. In A, the number of intracellular S. Javiana was compared between each treatment group, and in B the % cytotoxicity was compared between each treatment. Different letters (W,X,Y,Z) indicate significant pairwise differences in intracellular bacteria or cytotoxicity (P < 0.05) between treatments. (SJ = S. Javiana, La = L. acidophilus, Lr = L. rhamnosus, Lc = L. casei, HK = heat-killed, TX = Triton X-treated cells).
LAB do not kill S. Javiana
We next sought to examine potential mechanisms by which LAB might reduce S. Javiana-induced cytotoxicity during HT29-MTX cell infection. Probiotic bacteria are well-known producers of bacteriocins that can kill enteric pathogens including Salmonella (Dobson et al. 2012). To determine whether the LAB used in our study exerted antimicrobial effects against S. Javiana, we grew S. Javiana alone or in co-culture with L. acidophilus, L. rhamnosus, L. casei, L. plantarum or L. mesenteroides and measured S. Javiana viability over time. There was no impact of any LAB species on S. Javiana viability during 6 h of co-culture (Fig. 4), suggesting that in our model, the LAB are not producing compounds that kill S. Javiana.
Figure 4.
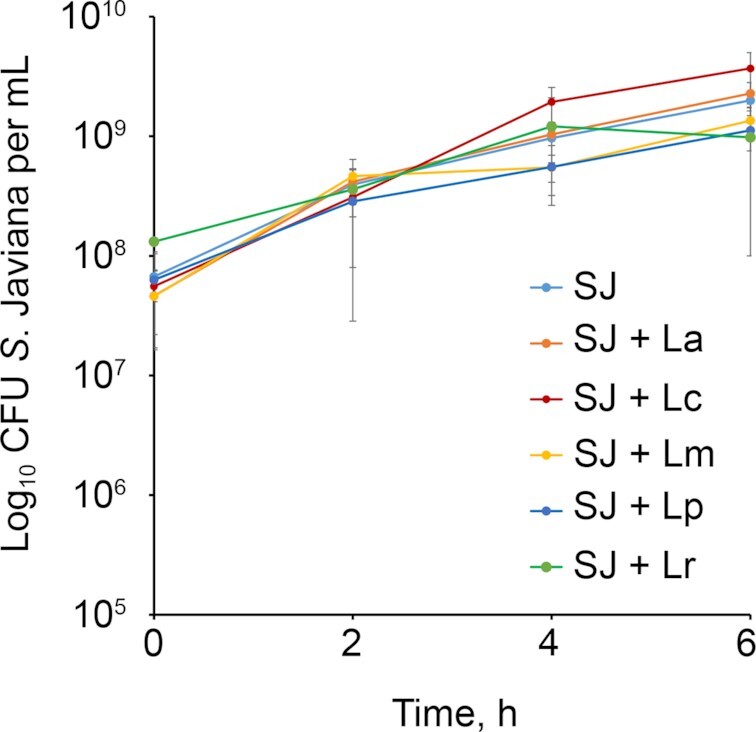
LAB do not kill S. Javiana. S. Javiana (SJ) was incubated in cell medium alone, or in presence of individual LAB strains. At 2, 4 and 6 h, viable S. Javiana were enumerated by plating. (SJ = S. Javiana, La = L. acidophilus, Lc = L. casei, Lm = L. mesenteroides, Lp = L. plantarum, Lr = L. rhamnosus).
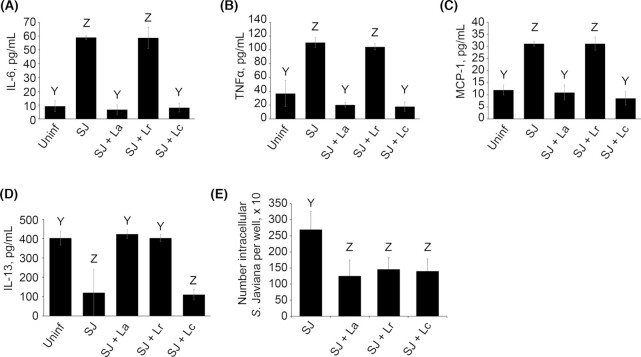
Exposure to LAB alters S. Javiana virulence gene expression during epithelial cell infection
Some strains of probiotics have been shown to exhibit anti-virulence effects on enteric pathogens such as E. coli and Salmonella (Medellin-Pena et al. 2007; Bayoumi and Griffiths 2012; Yang et al. 2014; Muyyarikkandy and Amalaradjou 2017; Peng et al. 2018), where exposure to the probiotic bacterium alters pathogen virulence gene expression. Given the fact that the LAB tested did not kill S. Javiana (Fig. 4), but did alter S. Javiana invasion (Fig. 2B) and its cytotoxic effect on host cells (Fig. 2C), we speculated that the LAB might be impacting expression of S. Javiana virulence traits. Therefore, we sought to examine the effect of L. acidophilus, L. rhamnosus and L. casei on expression of S. Javiana virulence genes invA, prgH, pltA and cdtB. The SPI-1 genes invA and prgH encode key parts of the SPI-1 type three secretion system (TTSS) that delivers effectors into intestinal epithelial cells, and have been shown in other NTS serovars to mediate epithelial invasion and cytotoxic effects on host cells (Galan and Curtiss 1989; Behlau and Miller 1993; Mills, Bajaj and Lee 1995; Collazo and Galan 1996). The cdtB gene also drives host cell invasion (Mezal, Bae and Khan 2014; Miller et al. 2018) and cdtB and pltA promote S. Javiana-induced host cell damage and cell cycle arrest (Mezal, Bae and Khan 2014; Miller and Wiedmann 2016; Miller et al. 2018). We assessed the impact of LAB on S. Javiana invA, prgH, pltA and cdtB expression in the presence of HT29-MTX cells to mimic conditions used in our previous infection assays and to account for the fact that Salmonella TTSS activity requires host cell contact (Galan and Collmer 1999). The HT29-MTX cells were pretreated with or without L. acidophilus, L. rhamnosus or L. casei, then infected with S. Javiana. At 2 h p.i., expression of S. Javiana invA, prgH, pltA and cdtB were assayed via qRT-PCR (Fig. 5). S. Javiana exposed to L. acidophilus and L. rhamnosus exhibited marked (8 to 15-fold) reduction in invA and prgH expression. In addition, S. Javiana exposed to L. acidophilus exhibited a 4.9-fold decrease in pltA and 3.1-fold reduction in cdtB expression, and S. Javiana exposed to L. rhamnosus exhibited a 4.8-fold reduction in pltA expression. We observed no effect of L. casei on expression of any S. Javiana virulence genes tested (Fig. 5). Together, data indicate that in our infection model, L. acidophilus and L. rhamnosus somehow cause a pronounced reduction in S. Javiana expression of key SPI-1 TTSS genes invA and prgH, and a moderate reduction in expression of expression of pltA and cdtB. Such effects on S. Javiana virulence expression might contribute to the anti-invasive and anti-cytotoxic effect of the LAB during S. Javiana infection, although additional studies are needed to confirm the role of invA, prgH, pltA and cdtB during S. Javiana infection of the HT29-MTX cell line.
Figure 5.
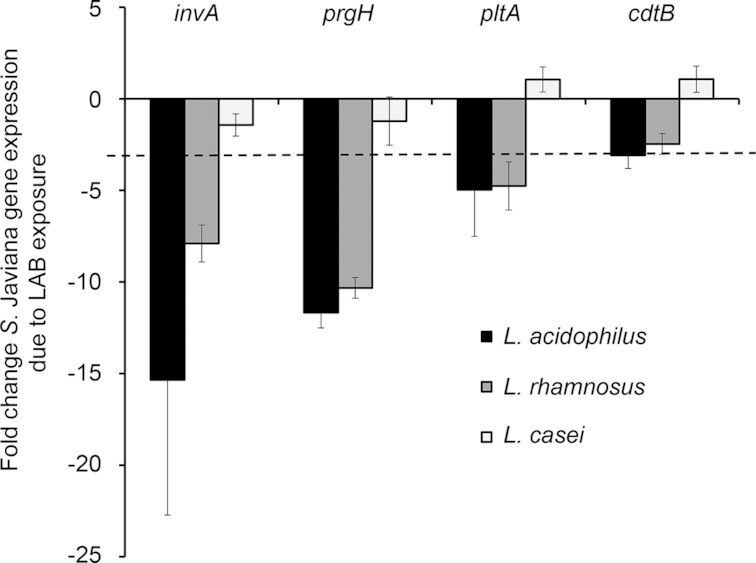
L. acidophilus and L. rhamnosus reduce S. Javiana virulence gene expression. HT29-MTX cells were pre-treated with or without LAB species L. acidophilus, L. rhamnosus or L. casei (MOE 10) for 1 h prior to infection with S. Javiana (MOI 10). At 2 h post-infection, bacteria were harvested, RNA was isolated and expression of S. Javiana invA, prgH, pltA and cdtB was analyzed via qRT-PCR. The effect of LAB exposure on S. Javiana gene expression was determined using the ΔΔCT method. Data are presented as fold change in gene expression in S. Javiana cells exposed to LAB compared to S. Javiana alone. Genes exhibiting > 3-fold change (denoted by dotted line in figure) were considered altered for expression.
LAB alter HT29-MTX cytokine production during S. Javiana infection
Having observed LAB-induced reduction in S. Javiana invasion, S. Javiana-mediated host cell cytotoxicity and virulence gene expression, we next examined whether LAB could alter the host inflammatory response by measuring the impact of LAB pre-treatment on HT29-MTX cytokine production during S. Javiana infection. HT29-MTX cells were either infected with S. Javiana alone or were pre-treated with L. acidophilus, L. rhamnosus or L. casei prior to infecting with S. Javiana for 6 h, after which infected cell supernatants were collected and assayed for inflammatory and anti-inflammatory cytokines using a commercial antibody array (Fig. 6A-D, Full array results in Table S1, Supporting Information). Pre-treating host cells with L. acidophilus and L. casei prior to infection significantly reduced epithelial production of inflammatory cytokines IL-6, TNFα and MCP-1 (Fig. 6A-C). In addition, while S. Javiana infection reduced host cell production of immunoregulatory cytokine IL-13, host cells exposed to L. acidophilus and L. rhamnosus prior to infection exhibited normal IL-13 production (Fig. 6D). We also measured the effect of LAB pre-exposure on S. Javiana intracellular viability at 6 h p.i., which is the same time-point at which cytokine production was assayed, and found fewer S. Javiana in host cells pre-treated with L. acidophilus, L. rhamnosus and L. casei than in untreated cells (Fig. 6E). Together, data point to a potential protective effect of LAB against potential host cell damage, inflammation, and pathogen intracellular survival during S. Javiana infection.
Figure 6.
LAB alter host epithelial cytokine production during S. Javiana infection. (A-D) HT29-MTX cells were either uninfected or pre-treated with or without L. rhamnosus, L. acidophilus or L. casei (MOE 10) for 1 h prior to infection with S. Javiana (MOI 10). At 2 h p.i., gentamicin was added to cell media and monolayers were incubated for 4 h to allow for synthesis and secretion of cytokines. Infected cell supernatants were collected and analyzed for inflammatory and anti-inflammatory cytokines using a commercial antibody array (Ray Biotech). (E) To measure the number of intracellular S. Javiana present at 6 h p.i. (the time-point at which cytokine production was measured), HT29-MTX cells were either infected with S. Javiana alone or pre-treated with L. acidophilus, L. rhamnosus or L. casei for 1 h prior to S. Javiana infection. At 2 h p.i., gentamicin was added to the cell media and at 6 h p.i., epithelial cells were lysed and intracellular S. Javiana were enumerated by plating. In A-D, concentration of individual cytokines was compared between different treatments and in E, number of intracellular S. Javiana were compared between treatments. Different letters (Y,Z) indicate a significant pairwise differences (P < 0.05) in cytokine concentration or S. Javiana intracellular survival between treatments. (Uninf = Uninfected, SJ = S. Javiana, La = L. acidophilus, Lr = L. rhamnosus, Lc = L. casei)
DISCUSSION
Probiotic microorganisms, and LAB in particular, have been shown to exhibit anti-infective properties against GI pathogens, including serovars of NTS (Burkholder and Bhunia 2009; Lievin-Le Moal and Servin 2014). Although precise mechanisms by which probiotic bacteria might limit infection remain inconclusive (Corr, Hill and Gahan 2009; Oelschlaeger 2010), proposed mechanisms are wide ranging and may include direct antimicrobial effects on pathogens (Alvarez-Sieiro et al. 2016), competition with pathogens for resources and colonization space within the host (Deriu et al. 2013; Lievin-Le Moal and Servin 2014), alteration of pathogen virulence (Medellin-Pena et al. 2007; Bayoumi and Griffiths 2010; Bayoumi and Griffiths 2012; Dutra et al. 2016) or modulation of host barriers and defenses (Anderson et al. 2010; Anderson et al. 2010). Moreover, the anti-pathogenic effects of probiotics are often specific to individual probiotic and pathogen strains (Sherman, Ossa and Johnson-Henry 2009; Campana, van Hemert and Baffone 2017). Despite the fact that S. Javiana is one of the leading NTS causes of foodborne gastroenteritis, the interaction of S. Javiana with host cells and the potential impact of LAB on S. Javiana pathogenesis was poorly understood. Using the human intestinal HT29-MTX epithelial cell line as an infection model, we report here that S. Javiana invades and has a cytotoxic effect on intestinal epithelial cells. We also show that three species of LAB—L. acidophilus, L. rhamnosus and L. casei—exhibit anti-pathogenic effects against S. Javiana in our infection model. Pre-treatment of host cells with L. acidophilus or L. rhamnosus reduced S. Javiana invasion, while L. acidophilus, L. rhamnosus and L. casei limited S. Javiana-induced cytotoxicity and intracellular survival. We also demonstrate reduced expression of S. Javiana virulence genes in the presence of L. acidophilus and L. rhamnosus, as well as an altered inflammatory response in host cells pre-treated with L. acidophilus, L. rhamnosus and L. casei prior to S. Javiana infection. Collectively, data suggest that L. acidophilus, L. rhamnosus and L. casei can exert a protective effect against S. Javiana infection, potentially by altering the virulence properties of the pathogen.
Our findings that L. acidophilus and L. rhamnosus reduced S. Javiana invasion are in agreement with previous reports which showed that LAB strains reduced invasion of other NTS serovars both in vitro (Tsai et al. 2005; Lin et al. 2008) and in vivo (Tsai et al. 2005; Lin et al. 2007; Chiu et al. 2008). These data also provide the first evidence of anti-invasive effects of LAB against S. Javiana. Similarly, we and others have shown that LAB can reduce host cell damage induced by enteric pathogens (Sherman et al. 2005), including NTS (Burkholder and Bhunia 2009; Eom, Song and Choi 2015). Although few studies have examined S. Javiana-induced host cell cytotoxicity, one recent study showed that while S. Javiana induced DNA damage and cell cycle arrest during infection of HIEC-6 human intestinal epithelial cells, the bacterium caused little host cell membrane damage or death (Miller et al. 2018). The discrepancies between their study and ours, in which we show a cytotoxic effect of S. Javiana—indicated by host cell release of the intracellular enzyme LDH—is potentially due to the use of different host cell lines. Here, our findings that S. Javiana-induced cytotoxicity was ameliorated in host cells pre-treated with L. acidophilus, L. rhamnosus and L. casei suggest a potential prophylactic effect of these LAB species against S. Javiana-mediated host cell damage.
Previous reports have shown that probiotics can interfere with infectious processes via active or passive mechanisms and, therefore, probiotic viability is not always a requirement for inhibition of infection or host immunomodulation (Sherman et al. 2005; Kataria et al. 2009). Some studies have demonstrated that killed preparations of probiotic strains can protect against pathogen binding (Hirano et al. 2017) or pathogen-induced epithelial damage (Popovic et al. 2019), and can alter the host response to pathogen challenge (Lopez et al. 2008; Popovic et al. 2019). In contrast, other reports have shown that probiotic viability is necessary for anti-infective properties. For example, Sherman et al (Sherman et al. 2005) showed that viable, but not HK, LAB prevented binding of enterohemorrhagic and enteropathogenic E. coli strains to T84 intestinal epithelial cells and prevented pathogen-induced alterations of epithelial barrier function. In addition, Roselli et al (Roselli et al. 2006) showed that live, but not HK, Bifidobacterium animalis and L. rhamnosus protected Caco-2 monolayers from inflammatory effects of enterotoxigenic E. coli infection. In this study, we demonstrate that only live LAB reduce S. Javiana invasion and cytotoxicity to host cells, as the anti-invasive effects of L. acidophilus and L. rhamnosus and the anti-cytotoxic effects of L. acidophilus, L. rhamnosus and L. casei were abolished when HT29-MTX cells were pre-exposed to HK LAB. This finding suggests that these LAB are somehow actively interfering with S. Javiana epithelial cell infection. The requirement for probiotic viability is likely specific for, and dependent on, the anti-infective mechanisms involved in individual probiotic-pathogen interactions, and the elucidation of mechanisms underlying the effects of LAB strains on S. Javiana epithelial infection will be the focus of future studies.
Several studies have reported that probiotic bacteria can exert anti-pathogenic effects by reducing pathogen virulence gene expression (Medellin-Pena et al. 2007; Bayoumi and Griffiths 2010; Bayoumi and Griffiths 2012; Yang et al. 2014; Younes et al. 2016; Muyyarikkandy and Amalaradjou 2017; Kiymaci et al. 2018; Zhao et al. 2018). Muyyarikkandy and Amalaradjou showed that the exposure of NTS serovars S. Typhimurium, S. Enteriditis and S. Heidelberg to LAB strains L. rhamnosus, Lactobacillus bulgaricus and Lactobacillus paracasei resulted in decreased pathogen expression of motility genes, as well as SPI-1- and SPI-2-encoded genes for epithelial invasion, modulation of host actin cytoskeleton and evasion of macrophage intracellular defenses (Muyyarikkandy and Amalaradjou 2017). In that study, the LAB strains also impacted pathogen phenotype, decreasing NTS motility, invasion and intramacrophage survival. Another report demonstrated that individual and mixed LAB strains isolated from chicken intestinal contents decreased S. Typhimurium expression of SPI-1 virulence genes both in vitro and in an in vivo infection model, and the same LAB strains decreased S. Typhimurium extraintestinal translocation in vivo (Yang et al. 2014). Our finding that S. Javiana exposed to L. acidophilus and L. rhamnosus exhibited an 8–15-fold reduction in invA and prgH expression, and more moderate reductions in pltA and cdtB expression demonstrates that these LAB can modulate S. Javiana virulence. We speculate that LAB-mediated alteration of virulence expression could contribute to the anti-invasive and anti-cytotoxic effect of L. acidophilus and L. rhamnosus we observed in our invasion and cytotoxicity assays. However, these findings are a first step toward understanding how LAB may impact S. Javiana pathogenesis, and additional studies are needed to confirm the role of specific S. Javiana virulence genes, and the impact of LAB-mediated alterations in expression of those genes, during intestinal epithelial infection. In addition, it is possible that the influence of LAB on expression of specific virulence genes is the result of a broader effect of LAB on Salmonella virulence regulators (Bayoumi and Griffiths 2010; Muyyarikkandy and Amalaradjou 2017) such as hilA, the general regulator of SPI1 (Bajaj, Hwang and Lee 1995; Altier et al. 2000) and ssrB, the response regulator of SPI2 (Feng et al. 2004). Future studies will examine the impact of LAB strains and products on S. Javiana virulence regulation.
Probiotics can interfere with pathogen virulence expression in multiple ways that appear to be specific to individual probiotic and pathogen strains (Campana, van Hemert and Baffone 2017). For example, some probiotic bacteria secrete soluble compounds that interact with pathogen receptors (Medellin-Pena et al. 2007; Bayoumi and Griffiths 2012; Yang et al. 2014; Muyyarikkandy and Amalaradjou 2017), while others may require direct cell-to-cell contact with the pathogen to influence its virulence (Lievin-Le Moal and Servin 2014; Younes et al. 2016). In addition, recent studies have indicated that some LAB products alter virulence via disrupting pathogen quorum sensing signaling pathways (Li et al. 2011; Kiymaci et al. 2018; Zhao et al. 2018). Since our experiments were performed with a co-culture of LAB and S. Javiana in the context of host cell infection, we cannot ascertain whether the LAB signal to S. Javiana via a secreted compound or whether virulence inhibition requires direct probiotic-S. Javiana contact. Ongoing studies in our lab aim to characterize the nature of the LAB anti-virulence products and mechanisms by which such products alter S. Javiana virulence.
Although the finding that LAB can influence S. Javiana virulence gene expression is compelling, the underlying mechanisms by which LAB hinder S. Javiana invasion and host cell damage are likely complex and multifaceted. This is particularly true given the fact that L. casei pre-exposure reduced S. Javiana-induced cytotoxicity and intracellular survival, but had no effect on expression of the virulence genes tested. It is possible that L. casei could alter expression of other S. Javiana virulence determinants not tested here, such as SPI-2-encoded genes that govern intracellular survival and pathogenesis, and such effects could manifest as decreased cytotoxic effect or reduced pathogen survival inside host cells. In addition, probiotics do exert biological effects on host cells as well as on pathogens, and such effects on the host can impact the outcome of infection (Lebeer, Vanderleyden and De Keersmaecker 2010). L. casei, L.acidophilus or L. rhamnosus could trigger defense processes in the host cell that contribute to reduced susceptibility to pathogen-induced damage and intracellular survival. Additional studies are warranted to elucidate the potentially distinct mechanisms by which these LAB species perturb the S. Javiana-host cell interaction.
Because probiotics form close associations with the intestinal epithelium, they can interact with host pattern recognition receptors (PRRs) and modulate inflammatory signaling (Lebeer, Vanderleyden and De Keersmaecker 2010; Kanmani and Kim 2018). Since NTS are known inducers of intestinal inflammation (Eckmann, Kagnoff and Fierer 1993; Jung et al. 1995; Eaves-Pyles et al. 2001; Lebeer, Vanderleyden and De Keersmaecker 2010), we examined the effect of L. acidophilus, L. rhamnosus and L. casei on HT29-MTX cytokine production in response to S. Javiana infection, and found that pre-treating host cells with L. acidophilus and L. casei significantly decreased production of inflammatory cytokines IL-6, TNFα and MCP-1, while L. acidophilus and L. rhamnosus prevented pathogen-induced reduction of anti-inflammatory IL-13. The effects of L. acidophilus, L. rhamnosus and L. casei on cytokine production by infected host cells correlate with our observation that S. Javiana intracellular viability at 6 h p.i. was reduced in host cells pre-exposed to those LAB strains. These data also concur with others who reported decreased inflammatory response in host cells and tissues pre-exposed to probiotic strains before infection with S. Typhimurium (Huang et al. 2015; Huang and Huang 2016; Yu et al. 2017), S. Infantis (Yang et al. 2017), and Helicobacter pylori (Lee et al. 2010; Yang et al. 2012), and indicate that L. acidophilus, L. rhamnosus and L. casei may elicit an immunomodulatory effect on host cells infected by S. Javiana.
Collectively, our findings demonstrate that S. Javiana exhibits an invasive and cytotoxic phenotype during infection of HT29-MTX intestinal epithelial cells, and that pre-exposing host cells to L. acidophilus, L. rhamnosus and L. casei can ameliorate S. Javiana virulence and alter the host cell inflammatory response. Since our experiments involved pre-treating host cells with LAB strains prior to infection, these data suggest a potential prophylactic effect of the LAB against S. Javiana infection. Indeed, others have shown that probiotics can have therapeutic, as well as prophylactic effects on infection (Lievin-Le Moal and Servin 2014). Additional studies are warranted to determine if these LAB strains could be effective if added at the same time or even after S. Javiana infection is initiated, to mimic a therapeutic model of probiotic use. Understanding how S. Javiana and host cells respond to LAB may inform future efforts to design functional probiotics to limit infection by this important NTS serovar.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Marc Allard (U.S. Food and Drug Administration) for generously providing S. Javiana CFSAN 001992 for this study. We also thank Dr. Zachary Olson (University of New England) for assistance with statistical analyses, and Ryan Camire for technical assistance.
FUNDING
Funding for this work was provided by University of New England (UNE) start-up funds and a grant from the UNE Office of Research and Scholarship to KMB. LG was funded by a UNE College of Westbrook Health Professions undergraduate research fellowship. AK was supported by an Institutional Developmental Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103423.
Conflicts of interest
Authors declare No conflict of interests.
REFERENCES
- Abrahams GL, Hensel M. Manipulating cellular transport and immune responses: dynamic interactions between intracellular Salmonella enterica and its host cells. Cell Microbiol. 2006;8:728–37. [DOI] [PubMed] [Google Scholar]
- Allard MW, Muruvanda T, Strain Eet al.. Fully assembled genome sequence for Salmonella enterica subsp. enterica Serovar Javiana CFSAN001992. Genome Announc. 2013;1:e0008113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altier C, Suyemoto M, Ruiz AIet al.. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol Microbiol. 2000;35:635–46. [DOI] [PubMed] [Google Scholar]
- Alvarez-Sieiro P, Montalban-Lopez M, Mu Det al.. Bacteriocins of lactic acid bacteria: extending the family. Appl Microbiol Biotechnol. 2016;100:2939–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RC, Cookson AL, McNabb WCet al.. Lactobacillus plantarum DSM 2648 is a potential probiotic that enhances intestinal barrier function. FEMS Microbiol Lett. 2010;309:184–92. [DOI] [PubMed] [Google Scholar]
- Anderson RC, Cookson AL, McNabb WCet al.. Lactobacillus plantarum MB452 enhances the function of the intestinal barrier by increasing the expression levels of genes involved in tight junction formation. BMC Microbiol. 2010;10:316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo KM, Reynolds J, Karp BEet al.. Antimicrobial resistance among nontyphoidal salmonella isolated from blood in the United States, 2003–2013. J Infect Dis. 2016;214:1565–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj V, Hwang C, Lee CA. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol. 1995;18:715–27. [DOI] [PubMed] [Google Scholar]
- Bayoumi MA, Griffiths MW. Probiotics down-regulate genes in Salmonella enterica serovar typhimurium pathogenicity islands 1 and 2. J Food Prot. 2010;73:452–60. [DOI] [PubMed] [Google Scholar]
- Bayoumi MA, Griffiths MW. In vitro inhibition of expression of virulence genes responsible for colonization and systemic spread of enteric pathogens using Bifidobacterium bifidum secreted molecules. Int J Food Microbiol. 2012;156:255–63. [DOI] [PubMed] [Google Scholar]
- Behlau I, Miller SI. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J Bacteriol. 1993;175:4475–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron N, Corriveau J, Letellier Aet al.. Interaction between host cells and septicemic Salmonella enterica serovar typhimurium isolates from pigs. J Clin Microbiol. 2009;47:3413–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore AL, Hoekstra RM, Iwamoto Met al.. Salmonella enterica infections in the United States and assessment of coefficients of variation: A novel approach to identify epidemiologic characteristics of individual serotypes, 1996–2011. PLoS One. 2015;10:e0145416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkholder KM, Bhunia AK. Salmonella enterica serovar Typhimurium adhesion and cytotoxicity during epithelial cell stress is reduced by Lactobacillus rhamnosus GG. Gut Pathog. 2009;1:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkholder KM, Bhunia AK. Listeria monocytogenes uses Listeria adhesion protein (LAP) to promote bacterial transepithelial translocation and induces expression of LAP receptor Hsp60. Infect Immun. 2010;78:5062–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campana R, van Hemert S, Baffone W. Strain-specific probiotic properties of lactic acid bacteria and their interference with human intestinal pathogens invasion. Gut Pathog. 2017;9:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Outbreak of Salmonella serotype Javiana infection in Orlando, Florida, Centers for Disease Control and Prevention, Atlanta, GA. Morbidity and Mortality Weekly Report. 51;2002, 683–4. [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States. http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf(12 December 2018, date last accessed); 2013.
- Centers for Disease Control and Prevention. Salmonella. http://www.cdc.gov/salmonella/(21 December 2018, date last accessed); 2018.
- Chen CY, Tsen HY, Lin CLet al.. Oral administration of a combination of select lactic acid bacteria strains to reduce the Salmonella invasion and inflammation of broiler chicks. Poult Sci. 2012;91:2139–47. [DOI] [PubMed] [Google Scholar]
- Chiu HH, Tsai CC, Hsih HYet al.. Screening from pickled vegetables the potential probiotic strains of lactic acid bacteria able to inhibit the Salmonella invasion in mice. J Appl Microbiol. 2008;104:605–12. [DOI] [PubMed] [Google Scholar]
- Choi AR, Patra JK, Kim WJet al.. Antagonistic activities and probiotic potential of lactic acid bacteria derived from a plant-based fermented food. Front Microbiol. 2018;9:1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collazo CM, Galan JE. Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect Immun. 1996;64:3524–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corr SC, Hill C, Gahan CG. Understanding the mechanisms by which probiotics inhibit gastrointestinal pathogens. Adv Food Nutr Res. 2009;56:1–15. [DOI] [PubMed] [Google Scholar]
- Deriu E, Liu JZ, Pezeshki Met al.. Probiotic bacteria reduce salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe. 2013;14:26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson A, Cotter PD, Ross RPet al.. Bacteriocin production: a probiotic trait?. Appl Environ Microbiol. 2012;78:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutra V, Silva AC, Cabrita Pet al.. Lactobacillus plantarum LB95 impairs the virulence potential of Gram-positive and Gram-negative food-borne pathogens in HT-29 and Vero cell cultures. J Med Microbiol. 2016;65:28–35. [DOI] [PubMed] [Google Scholar]
- Eaves-Pyles T, Murthy K, Liaudet Let al.. Flagellin, a novel mediator of Salmonella-induced epithelial activation and systemic inflammation: I kappa B alpha degradation, induction of nitric oxide synthase, induction of proinflammatory mediators, and cardiovascular dysfunction. J Immunol. 2001;166:1248–60. [DOI] [PubMed] [Google Scholar]
- Eckmann L, Kagnoff MF, Fierer J. Epithelial cells secrete the chemokine interleukin-8 in response to bacterial entry. Infect Immun. 1993;61:4569–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom JS, Song J, Choi HS. Protective effects of a novel probiotic strain of lactobacillus plantarum JSA22 from traditional fermented soybean food against infection by salmonella enterica serovar typhimurium. J Microbiol Biotechnol. 2015;25:479–91. [DOI] [PubMed] [Google Scholar]
- Feng X, Walthers D, Oropeza Ret al.. The response regulator SsrB activates transcription and binds to a region overlapping OmpR binding sites at Salmonella pathogenicity island 2. Mol Microbiol. 2004;54:823–35. [DOI] [PubMed] [Google Scholar]
- Fuller R. History and Development of Probiotics. The Netherlands; Springer, 1992. [Google Scholar]
- Gagnon M, Zihler Berner A, Chervet Net al.. Comparison of the Caco-2, HT-29 and the mucus-secreting HT29-MTX intestinal cell models to investigate Salmonella adhesion and invasion. J Microbiol Methods. 2013;94:274–9. [DOI] [PubMed] [Google Scholar]
- Gal-Mor O, Boyle EC, Grassl GA. Same species, different diseases: How and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front Microbiol. 2014;5:391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JE, Curtiss R 3rd. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci U S A. 1989;86:6383–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JE, Collmer A. Type III secretion machines: Bacterial devices for protein delivery into host cells. Science. 1999;284:1322–8. [DOI] [PubMed] [Google Scholar]
- George F, Daniel C, Thomas Met al.. Occurrence and dynamism of lactic acid bacteria in distinct ecological niches: A multifaceted functional health perspective. Front Microbiol. 2018;9:2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano S, Yokota Y, Eda Met al.. Effect of Lactobacillus plantarum Tennozu-SU2 on salmonella typhimurium infection in human enterocyte-like HT-29-Luc cells and BALB/c mice. Probiotics Antimicrob Proteins. 2017;9:64–70. [DOI] [PubMed] [Google Scholar]
- Huang FC, Huang SC. The different effects of probiotics treatment on Salmonella-induced interleukin-8 response in intestinal epithelia cells via PI3K/Akt and NOD2 expression. Benef Microbes. 2016;7:739–48. [DOI] [PubMed] [Google Scholar]
- Huang IF, Lin IC, Liu PFet al.. Lactobacillus acidophilus attenuates Salmonella-induced intestinal inflammation via TGF-beta signaling. BMC Microbiol. 2015;15:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson BR, Griffin PM, Cole Det al.. Outbreak-associated Salmonella enterica serotypes and food Commodities, United States, 1998–2008. Emerg Infect Dis. 2013;19:1239–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HC, Eckmann L, Yang SKet al.. A distinct array of proinflammatory cytokines is expressed in human colon epithelial cells in response to bacterial invasion. J Clin Invest. 1995;95:55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanmani P, Kim H. Functional capabilities of probiotic strains on attenuation of intestinal epithelial cell inflammatory response induced by TLR4 stimuli. Biofactors. 2018;45:223–235. [DOI] [PubMed] [Google Scholar]
- Kataria J, Li N, Wynn JLet al.. Probiotic microbes: Do they need to be alive to be beneficial?. Nutr Rev. 2009;67:546–50. [DOI] [PubMed] [Google Scholar]
- Kirk MD, Pires SM, Black REet al.. World health organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: A data synthesis. PLoS Med. 2015;12:e1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiymaci ME, Altanlar N, Gumustas Met al.. Quorum sensing signals and related virulence inhibition of Pseudomonas aeruginosa by a potential probiotic strain's organic acid. Microb Pathog. 2018;121:190–7. [DOI] [PubMed] [Google Scholar]
- Lebeer S, Vanderleyden J, De Keersmaecker SC. Host interactions of probiotic bacterial surface molecules: Comparison with commensals and pathogens. Nat Rev Microbiol. 2010;8:171–84. [DOI] [PubMed] [Google Scholar]
- Lee JS, Paek NS, Kwon OSet al.. Anti-inflammatory actions of probiotics through activating suppressor of cytokine signaling (SOCS) expression and signaling in Helicobacter pylori infection: A novel mechanism. J Gastroenterol Hepatol. 2010;25:194–202. [DOI] [PubMed] [Google Scholar]
- Lesuffleur T, Porchet N, Aubert JPet al.. Differential expression of the human mucin genes MUC1 to MUC5 in relation to growth and differentiation of different mucus-secreting HT-29 cell subpopulations. J Cell Sci. 1993;106(Pt 3):771–83. [DOI] [PubMed] [Google Scholar]
- Li J, Wang W, Xu SXet al.. Lactobacillus reuteri-produced cyclic dipeptides quench agr-mediated expression of toxic shock syndrome toxin-1 in staphylococci. Proc Natl Acad Sci U S A. 2011;108:3360–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievin-Le Moal V, Servin AL. Anti-infective activities of lactobacillus strains in the human intestinal microbiota: From probiotics to gastrointestinal anti-infectious biotherapeutic agents. Clin Microbiol Rev. 2014;27:167–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CK, Tsai HC, Lin PPet al.. Lactobacillus acidophilus LAP5 able to inhibit the Salmonella choleraesuis invasion to the human Caco-2 epithelial cell. Anaerobe. 2008;14:251–5. [DOI] [PubMed] [Google Scholar]
- Lin WH, Yu B, Lin CKet al.. Immune effect of heat-killed multistrain of Lactobacillus acidophilus against Salmonella typhimurium invasion to mice. J Appl Microbiol. 2007;102:22–31. [DOI] [PubMed] [Google Scholar]
- Liu J, Hu D, Chen Yet al.. Strain-specific properties of Lactobacillus plantarum for prevention of Salmonella infection. Food Funct. 2018;9:3673–82. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- Lopez M, Li N, Kataria Jet al.. Live and ultraviolet-inactivated Lactobacillus rhamnosus GG decrease flagellin-induced interleukin-8 production in Caco-2 cells. J Nutr. 2008;138:2264–8. [DOI] [PubMed] [Google Scholar]
- Majowicz SE, Musto J, Scallan Eet al.. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010;50:882–9. [DOI] [PubMed] [Google Scholar]
- Medellin-Pena MJ, Wang H, Johnson Ret al.. Probiotics affect virulence-related gene expression in Escherichia coli O157:H7. Appl Environ Microbiol. 2007;73:4259–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezal EH, Stefanova R, Khan AA. Isolation and molecular characterization of Salmonella enterica serovar Javiana from food, environmental and clinical samples. Int J Food Microbiol. 2013;164:113–8. [DOI] [PubMed] [Google Scholar]
- Mezal EH, Bae D, Khan AA. Detection and functionality of the CdtB, PltA, and PltB from Salmonella enterica serovar Javiana. Pathog Dis. 2014;72:95–103. [DOI] [PubMed] [Google Scholar]
- Micallef SA, Rosenberg Goldstein RE, George Aet al.. Occurrence and antibiotic resistance of multiple Salmonella serotypes recovered from water, sediment and soil on mid-Atlantic tomato farms. Environ Res. 2012;114:31–39. [DOI] [PubMed] [Google Scholar]
- Miller RA, Wiedmann M. The cytolethal distending toxin produced by nontyphoidal salmonella serotypes javiana, montevideo, oranienburg, and mississippi induces DNA damage in a manner similar to that of serotype typhi. MBio. 2016;7:e02109–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RA, Betteken MI, Guo Xet al.. The typhoid toxin produced by the nontyphoidal salmonella enterica serotype javiana is required for induction of a DNA damage response in vitro and systemic spread in vivo. MBio. 2018;9, e00467–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills DM, Bajaj V, Lee CA. A 40 kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol Microbiol. 1995;15:749–59. [DOI] [PubMed] [Google Scholar]
- Muyyarikkandy MS, Amalaradjou MA. Lactobacillus bulgaricus, lactobacillus rhamnosus and lactobacillus paracasei attenuate salmonella enteritidis, salmonella heidelberg and salmonella typhimurium colonization and virulence gene expression in vitro. Int J Mol Sci. 2017;18, 2381–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair DVT, Venkitanarayanan K, Johny AK. Antibiotic-resistant Salmonella in the food supply and the potential role of antibiotic alternatives for control. Foods. 2018;7, 167–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelschlaeger TA. Mechanisms of probiotic actions - A review. Int J Med Microbiol. 2010;300:57–62. [DOI] [PubMed] [Google Scholar]
- Peng M, Tabashsum Z, Patel Pet al.. Linoleic acids overproducing lactobacillus casei limits growth, survival, and virulence of salmonella typhimurium and enterohaemorrhagic Escherichia coli. Front Microbiol. 2018;9:2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira GVdM, Coelho BdO, Junior AIMet al.. How to select a probiotic? A review and update of methods and criteria. Biotechnol Adv. 2018;36:2060–76. [DOI] [PubMed] [Google Scholar]
- Popovic N, Djokic J, Brdaric Eet al.. The influence of heat-killed Enterococcus faecium BGPAS1-3 on the tight-junction protein expression and immune function in differentiated Caco-2 cells infected with Listeria monocytogenes ATCC 19111. Frontiers in Microbiology. 2019;10:412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria, 2012. [Google Scholar]
- Roselli M, Finamore A, Britti MSet al.. Probiotic bacteria Bifidobacterium animalis MB5 and Lactobacillus rhamnosus GG protect intestinal Caco-2 cells from the inflammation-associated response induced by enterotoxigenic Escherichia coli K88. Br J Nutr. 2006;95:1177–84. [DOI] [PubMed] [Google Scholar]
- Santos FB, Dsouza DH, Jaykus Let al.. Genotypes, serotypes, and antibiotic resistance profiles of Salmonella isolated from commercial North Carolina turkey farms. J Food Prot. 2007;70:1328–33. [DOI] [PubMed] [Google Scholar]
- Sherman PM, Ossa JC, Johnson-Henry K. Unraveling mechanisms of action of probiotics. Nutr Clin Pract. 2009;24:10–14. [DOI] [PubMed] [Google Scholar]
- Sherman PM, Johnson-Henry KC, Yeung HPet al.. Probiotics reduce enterohemorrhagic Escherichia coli O157:H7- and enteropathogenic E. coli O127:H6-induced changes in polarized T84 epithelial cell monolayers by reducing bacterial adhesion and cytoskeletal rearrangements. Infect Immun. 2005;73:5183–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surendran Nair M, Amalaradjou MAR, Venkitanarayanan K. Antivirulence properties of probiotics in combating microbial pathogenesis. Adv Appl Microbiol. 2017;98:1–29. [DOI] [PubMed] [Google Scholar]
- Tranchemontagne ZR, Camire RB, O'Donnell VJet al.. Staphylococcus aureus strain USA300 perturbs acquisition of lysosomal enzymes and requires phagosomal acidification for survival inside macrophages. Infect Immun. 2016;84:241–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CC, Hsih HY, Chiu HHet al.. Antagonistic activity against Salmonella infection in vitro and in vivo for two Lactobacillus strains from swine and poultry. Int J Food Microbiol. 2005;102:185–94. [DOI] [PubMed] [Google Scholar]
- Wagner RD, Pierson C, Warner Tet al.. Probiotic effects of feeding heat-killed Lactobacillus acidophilus and Lactobacillus casei to Candida albicans-colonized immunodeficient mice. J Food Prot. 2000;63:638–44. [DOI] [PubMed] [Google Scholar]
- Wallis TS, Galyov EE. Molecular basis of Salmonella-induced enteritis. Mol Microbiol. 2000;36:997–1005. [DOI] [PubMed] [Google Scholar]
- Williams K, Gokulan K, Shelman Det al.. Cytotoxic mechanism of cytolethal distending toxin in nontyphoidal Salmonella serovar (Salmonella Javiana) during macrophage infection. DNA Cell Biol. 2015;34:113–24. [DOI] [PubMed] [Google Scholar]
- Yang GY, Yu J, Su JHet al.. Oral administration of Lactobacillus rhamnosus GG ameliorates salmonella infantis-induced inflammation in a pig model via activation of the IL-22BP/IL-22/STAT3 Pathway. Front Cell Infect Microbiol. 2017;7:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Brisbin J, Yu Het al.. Selected lactic acid-producing bacterial isolates with the capacity to reduce Salmonella translocation and virulence gene expression in chickens. PLoS One. 2014;9:e93022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YJ, Chuang CC, Yang HBet al.. Lactobacillus acidophilus ameliorates H. pylori-induced gastric inflammation by inactivating the Smad7 and NFkappaB pathways. BMC Microbiol. 2012;12:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younes JA, Reid G, van der Mei HCet al.. Lactobacilli require physical contact to reduce staphylococcal TSST-1 secretion and vaginal epithelial inflammatory response. Pathog Dis. 2016;74:ftw029. [DOI] [PubMed] [Google Scholar]
- Yu J, Zhu YH, Yang GYet al.. Anti-inflammatory capacity of Lactobacillus rhamnosus GG in monophasic variant Salmonella infected piglets is correlated with impeding NLRP6-mediated host inflammatory responses. Vet Microbiol. 2017;210:91–100. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lv J, Pan Let al.. Roles and applications of probiotic Lactobacillus strains. Appl Microbiol Biotechnol. 2018;102:8135–43. [DOI] [PubMed] [Google Scholar]
- Zhao W, Yuan T, Piva Cet al.. The probiotic bacterium, Phaeobacter inhibens, down-regulates virulence factor transcription in the shellfish pathogen, Vibrio coralliilyticus, by N-acyl homoserine lactone production. Appl Environ Microbiol. 2018;85:e01545–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D. Collective efforts to modulate the host actin cytoskeleton by Salmonella type III-secreted effector proteins. Trends Microbiol. 2001;9:567–9.; discussion 569–570. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.