Abstract
Accurate diagnosis and stratification of children with irritable bowel syndrome (IBS) remain challenging. Given the central role of recurrent abdominal pain in IBS, we evaluated the relationships of pediatric IBS and abdominal pain with intestinal microbes and fecal metabolites using a comprehensive clinical characterization and multiomics strategy. Using rigorous clinical phenotyping, we identified preadolescent children (aged 7 to 12 years) with Rome III IBS (n = 23) and healthy controls (n = 22) and characterized their fecal microbial communities using whole-genome shotgun metagenomics and global unbiased fecal metabolomic profiling. Correlation-based approaches and machine learning algorithms identified associations between microbes, metabolites, and abdominal pain. IBS cases differed from controls with respect to key bacterial taxa (eg, Flavonifractor plautii and Lachnospiraceae bacterium 7_1_58FAA), metagenomic functions (eg, carbohydrate metabolism and amino acid metabolism), and higher-order metabolites (eg, secondary bile acids, sterols, and steroid-like compounds). Significant associations between abdominal pain frequency and severity and intestinal microbial features were identified. A random forest classifier built on metagenomic and metabolic markers successfully distinguished IBS cases from controls (area under the curve, 0.93). Leveraging multiple lines of evidence, intestinal microbes, genes/pathways, and metabolites were associated with IBS, and these features were capable of distinguishing children with IBS from healthy children. These multi-omics features, and their links to childhood IBS coupled with nutritional interventions, may lead to new microbiome-guided diagnostic and therapeutic strategies.
Irritable bowel syndrome (IBS) is a functional gastrointestinal (GI) tract disorder affecting up to 20% of the world's population.1, 2 IBS symptoms include bloating, altered bowel habits, and abdominal pain, with pain being the most debilitating.3, 4 The etiologies of IBS are multifactorial,5, 6, 7, 8 and increasing evidence supports the involvement of gut microbes in IBS onset and symptoms.9, 10, 11, 12, 13 Relative to healthy controls (HCs), IBS gut microbial communities (child or adult) often display greater heterogeneity and temporal instability7 and shifts in the relative abundances of members of the phyla Bacteroidetes, Proteobacteria (specifically, Gammaproteobacteria), and Firmicutes.9, 14 In other reports, adults with IBS have been found to have altered fecal concentrations of metabolites that may be produced or modified by microbes, including short-chain fatty acids, bile acids, and amino acid degradation products.15, 16, 17
Despite the growing number of IBS microbiome-based studies, little consensus exists with respect to IBS-associated alterations in microbial community structure, function, or associated metabolites. This lack of consensus may be attributed to varying molecular profiling methods, the limited resolution (ie, family or genus level) often associated with partial 16S rRNA gene data, varying definitions of IBS, subtypes evaluated, and/or other physiological factors. In addition, most past studies have relied on subject symptom recall to confirm diagnosis of IBS. However, in both children and adults, recall is unreliable and poorly correlated with prospective IBS symptom diary capture.18, 19 Given this, recent expert guidance recommends the use of prospective 2-week symptom diaries for clinical studies of IBS,20 but prospective diary-based guidance has yet to be implemented in many IBS microbiome studies.
Our previous prospective diary-based characterization of children with IBS suggests that IBS subtype and symptom severity may provide insight regarding the microbial ecology of IBS, as we identified differentially abundant fecal bacterial taxa according to IBS subtype and pain frequency.9 Similarly, recent work in adults reported the association of key microbes with symptom severity and the ability of these taxa to distinguish severe cases of IBS from mild cases or controls with moderate accuracy.12
To date, the roles of gut microbiota composition and function in adult IBS remain poorly understood.10 Data addressing pediatric IBS are even more limited,9, 21 despite pediatric IBS typically being associated with fewer comorbidities. No previous study in children or adults has combined prospective diary-based evaluation of IBS symptoms with shotgun metagenomic sequencing [whole-genome sequencing (WGS)] and global metabolomic profiling to characterize the IBS-associated gut microbiome. Thus, we sought to integrate rigorous, prospective, clinical phenotyping with multiple lines of omics-based evidence to improve our understanding of the interrelationships among IBS, abdominal pain, gut microbes, and microbial metabolites. By focusing on children, our intent was to evaluate microbiome-based contributions to IBS pathogenesis, and by focusing on pain, we addressed a primary component of symptom severity and detractor from quality of life, which affects both children and adults with IBS.3, 4 We hypothesized that microbial signatures and fecal metabolites may be associated with abdominal pain frequency and severity in children with IBS. These microbiome-based features, when taken together, may enable clinical laboratories to refine the diagnosis of IBS and match patients with potentially beneficial treatment strategies.
Materials and Methods
Subject Recruitment and Classification
Controls and children with IBS (aged 7 to 12 years) were recruited from a large health care network based in Houston, TX.9 Patients and HCs were age matched. Informed consent was obtained from parents, assent was obtained from children, and all recruitment and study procedures were approved by the Baylor College of Medicine Institutional Review Board (H-25184).
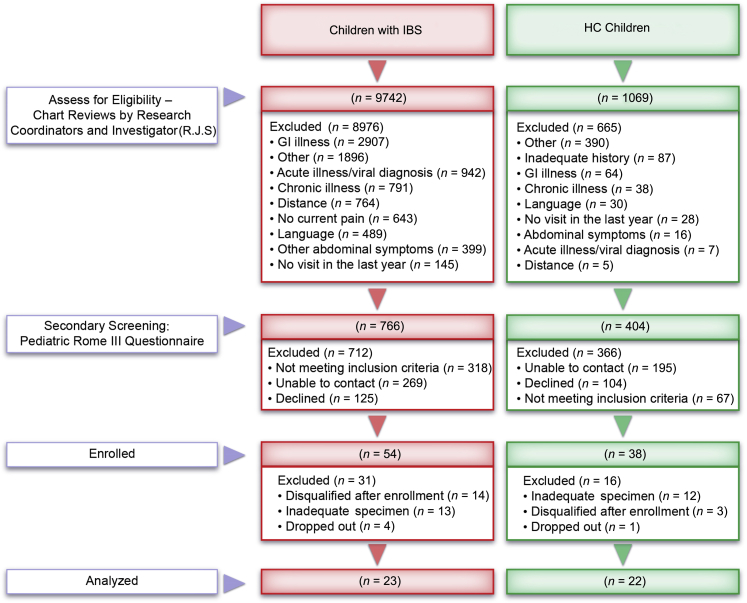
Participants were rigorously classified as IBS cases or HCs (Figure 1). Medical charts were reviewed by trained research coordinators examining International Classification of Diseases, Ninth Revision (ICD-9) codes for abdominal pain and IBS or well-child visits. Potential study participants completed a physician visit within the prior year for assessment of symptoms or well-child care. Secondary review of medical and laboratory records was performed by an investigator (R.J.S.), and those who passed secondary review were screened via telephone and initially classified (potential IBS or potential HC) using a modified pediatric Rome III questionnaire.1, 22 Children were excluded if chart review or screening revealed a significant chronic medical/pain condition (eg, diabetes), chronic vomiting, unexplained weight loss, hematochezia, major GI tract surgery, significant developmental delay, an organic GI tract disorder, use of antibiotics within the prior month or probiotics within the prior 6 months, and/or menarche. Detailed inclusion and exclusion criteria and subject metadata are archived in the Database of Genotypes and Phenotypes (https://www.ncbi.nlm.nih.gov/gap; accession number phs000265.v3.p1).
Figure 1.
Study flowchart outlining subject recruitment, participation, and classification. GI, gastrointestinal.
Study participants fulfilling these screening criteria maintained daily pain and stool diaries for 2 weeks.23 Children rated pain using a validated, 10-point scale24 and recorded Bristol stool form.25 After completion of the 2-week diary, clinical phenotype was determined. HCs fulfilled control criteria based on the Rome III–based telephone screening questionnaire and reported ≤1 day with abdominal pain in the 2-week diary.23 IBS cases fulfilled pediatric Rome III IBS criteria based on the questionnaire and reported ≥2 days of pain in the diary. In addition, to ensure that children had IBS, rather than functional abdominal pain or dyspepsia, a published algorithm22 was applied to each subject's 2-week diary. Based on stool form, IBS-confirmed cases were subtyped as follows: IBS constipation predominant (hard stools ≥ 25% of the time); IBS diarrhea predominant (loose stools ≥ 25% of the time); IBS mixed (hard or loose stools ≥ 25% of the time); or IBS unsubtyped (meets none of the definitions above).25
A subset of participants (n = 20 with IBS and 11 HCs) completed 24-hour food diaries along with their stool collection. These data were analyzed for total caloric intake and calories derived from protein, fat, or carbohydrate using Nutritionist Pro (Axxya Systems LLC, Woodinville, WA).
Demographic data, clinical variables, and dietary information were compared using t-tests, U-tests, or χ2 tests, as appropriate.
Stool Collection, DNA Extraction, and Sequencing
In conjunction with the pain and stool diaries, participants submitted a stool sample that was collected in a sterile, self-sealing container, maintained at −20°C, and transferred to the Texas Children's Microbiome Center via courier within 24 hours of collection. Samples were stored at −80°C thereafter.
Stool DNA was extracted using the PowerSoil DNA Isolation kit (MO BIO Laboratories, Carlsbad, CA), with modifications.26 DNA quality and yield were evaluated via agarose gel, NanoDrop 1000 spectrophotometer (NanoDrop, Wilmington, DE), and Qubit fluorometer (Life Technologies Corp., Carlsbad, CA). WGS libraries were generated using 100-bp paired-end libraries and the HiSeq2000 platform (Illumina, Inc., San Diego, CA).26 WGS libraries were quality filtered using the fastq-mcf feature in the ea-utils package version 1.0.4 (https://github.com/ExpressionAnalysis/ea-utils, last accessed August 22, 2018) with a minimum quality score of 20, a minimum length of 75 bp, and removal of duplicate reads. Host-derived sequences were identified by mapping reads to a human reference genome (hg19) using Bowtie2 version 2.0.627 in sensitive mode. Those that mapped successfully were removed from downstream analysis. Taxonomic and functional profiles were generated using the default settings of HUMAnN2 version 0.4.0,28 a pipeline that incorporates MetaPhlAn229 for taxonomic profiling and uses a combination of reference genome mapping and translated search30 of the UniRef50 database31 for functional annotation. Functional profiles are presented in the context of MetaCyc32 pathways and enzymatic reactions. Sequence data associated with this study are available in the National Center for Biotechnology Information Sequence Read archive (https://www.ncbi.nlm.nih.gov/bioproject; accession number PRJNA46339).
Metabolite Extraction and Profiling
Stool samples were prepared for metabolomic profiling and submitted to Metabolon (Durham, NC) for characterization. Protein fractions were removed and small molecules were retained using a series of proprietary organic and aqueous extractions. The small-molecule fractions were divided into two aliquots and further processed using liquid chromatography/tandem mass spectrometry (MS/MS) or gas chromatography/MS. The liquid chromatography/MS/MS platform consisted of an ACQUITY ultraperformance liquid chromatography system (Waters Corp., Milford, MA) coupled with an LTQFT mass spectrometer (Thermo-Finnigan, San Jose, CA), which had a linear ion-trap front end and a Fourier transform ion cyclotron resonance mass spectrometer back end. Fractions characterized using liquid chromatography/MS/MS were analyzed using both acidic positive ion– and basic negative ion–optimized conditions. For quality control purposes, 11 standards of fixed concentration were added before injection into the instrument. The gas chromatography/MS platform had a 5% phenyl column with a temperature ramp of 40°C to 300°C in a 16-minute period. This was coupled with a Trace DSQ fast-scanning single-quadrupole mass spectrometer (Thermo-Finnigan), which was tuned and calibrated for mass resolution and mass accuracy on a daily basis. The samples analyzed via gas chromatography/MS were vacuum dried for 24 hours before derivatization, under dried nitrogen, using bistrimethyl-silyl-triflouroacetamide. Accurate metabolite identification was accomplished by comparing the MS spectra with Metabolon's library of spectra for >1000 purified standard compounds.
The samples were processed by Metabolon in two separate batches. To eliminate potential batch effects, the metabolite abundance tables were processed individually before merging them together into one table. This approach included removal of metabolites with >50% missing values across all samples; imputation of the remaining missing values with 10% of the minimum for each metabolite; determination of the median and the median of absolute deviations from the median (MAD) for every metabolite; and normalization using the median and MAD. After normalization, the median value for each metabolite was 0 and the variation was expressed in MAD units. The normalized tables (ie, one from each batch) were merged, keeping only the metabolites that were present in both tables.
Sequence-Based Community Comparisons
WGS-based taxonomic and functional profiles were evaluated for differential abundances using multtest.33 Features present in <10% of subjects were excluded, and Wilcoxon rank-sum tests with Benjamini-Hochberg corrections were performed on relative abundance data. Taxonomic profiles were analyzed at all taxonomic levels from phylum to species, and functional data were analyzed in the context of MetaCyc pathways and enzymatic reactions.
Metabolite-Based Community Comparisons
Differential abundance analysis of individual median and MAD normalized metabolites and their higher-order classes was performed using Wilcoxon rank-sum tests with Benjamini-Hochberg corrections in RStudio34 using R version 3.3.0. Higher-order classes were composed of member metabolites with fold changes ≥ |1.5|. Sulfated sterols/steroids and secondary bile acids were collapsed into additional separate groups.
Microbiome-Metabolite Data Integration
Pairwise Spearman correlations between clinical variables, metabolite profiles, and microbial community profiles were calculated with R. Also with R, Benjamini-Hochberg multiple testing corrections were calculated.
Using median/MAD-normalized metabolites, species, and functional pathway abundance data, a subject-species-metabolite pathway network was constructed using Cytoscape version 3.5.1.35 Analyte values were systematically filtered for those that displayed analysis of variance F values >7 in the comparison of IBS cases versus HCs, and the edge-weighted, spring-embedded layout was used to visualize potential separation of IBS cases and HCs on the basis of these multi-omics features.
Multivariate classifiers incorporating the top differentially abundant metagenomic and metabolite features were assembled and evaluated in Orange version 3.3.12.36 Features were prefiltered on the basis of differential abundance-based analysis of variance F scores, and the top 10 features (ie, metabolites, bacterial species, and/or functional pathway) were selected for inclusion in a series of classifier models. Specifically, classifier models, including random forest (RF), logistic regression [least absolute shrinkage selection operator (LASSO)], support vector machine, and naïve Bayes, were built and tested using fivefold cross validation.
Model performance was assessed as a function of classifier accuracy, precision, recall, area under the receiver operating characteristic curve, and calibration curves. All classifiers were developed under default settings in Orange, but specific details associated with each included the following: RF: 2500 trees were generated. Least absolute shrinkage selection operator (logistic regression): cost strength was set to 5 and L1 regularization was used. Support vector machine: the polynomial kernel was defined with γ constant equal to 0.01, the constant equal to 1.45, and the degree equal to 3. The cost was set to 1, regression loss ε was set at 0.10, and the numerical tolerance was set to 0.10. Naïve Bayes: no additional settings were available for potential modification. The ability of the features selected for classifier model construction to separate subjects in an ordination analysis was also evaluated. The 10 features were evaluated using principal components analysis in Orange under default settings.
After initial evaluation of all classifier models, the RF model was used with fivefold cross validation for the final classification. RF is a variant of decision tree approaches. Many decision trees are generated (2500 in this case) using different subsets of available features for each tree. The majority vote classification determined by all decision trees is more accurate than the classification determined by a single decision tree that uses all features at once. The f-fold cross validation (f = 5 in this case) is the method of choice in cases when the data set is small. The available data set is randomly divided into equal f-folds. The classification model is trained on f-1 folds and tested on the remaining single fold. This process is repeated until every fold is used once as a test fold.
Results
Subject Demographics and Clinical Variables
Seventy children (n = 36 with IBS and 34 HCs) completed the study, and 45 children (n = 23 with IBS and 22 HCs) provided sufficient material for all analyses (Figure 1). The distribution of IBS subtypes among the 23 IBS participants was as follows: IBS constipation predominant, 11; IBS unsubtyped, 10; and IBS diarrhea predominant, 2. IBS cases and HCs did not differ significantly with respect to age or sex distribution (Table 1). No differences were detected with respect to bowel movement frequency or mean stool form (Table 1), as has been reported previously.25 IBS children reported significantly more abdominal pain episodes and greater pain severity than HCs (Table 1). Pain frequency and severity were highly correlated with one another (Spearman r = 0.92, P < 0.05). No significant differences were detected between groups with respect to caloric consumption or relative intake of fat, carbohydrates, or protein based on 24-hour food diaries (Supplemental Table S1).
Table 1.
Demographic and Gastrointestinal Tract Characteristics in Children with IBS versus HCs
| Subject variable | Children with IBS (n = 23) | HCs (n = 22) | P value |
|---|---|---|---|
| Female sex, n (%) | 9 (39.1) | 9 (40.9) | 0.67 |
| Age, years | 9.7 ± 1.6 | 9.6 ± 1.5 | 0.72 |
| Abdominal pain frequency∗ | 9.7 ± 7.4 | 0.00 ± 0.0 | <0.001 |
| Abdominal pain severity† | 3.3 ± 1.4 | 0.00 ± 0.0 | <0.001 |
| Bowel movements‡ | 11.9 ± 4.8 | 10.6 ± 4.1 | 0.33 |
| Mean stool type§ | 3.3 ± 0.8 | 3.3 ± 0.7 | 0.88 |
Data are expressed as means ± SD unless otherwise indicated. Differences were evaluated using χ2 tests, t-tests, or U-tests, as appropriate.
Number of pain episodes reported in the 2-week pain and stooling diary.
Based on a rating scale of 0 to 10, with 10 being the most severe abdominal pain.
Number of bowel movements recorded in the 2-week pain and stooling diary.
Based on the Bristol Stool Form scale (range, 1 to 7).
Stool Microbial Community and Metabolic Capacity–Based Differences between IBS Children and HCs
An average of 116.8 million paired-end reads were produced per WGS library. Taxonomic profiling and differential abundance analysis identified the significant enrichment of Gammaproteobacteria in IBS (Wilcoxon rank-sum test, q = 0.03) (Table 2). Unclassified Clostridiales, the family-level designation for Flavonifractor, was enriched in the stool communities of IBS children; and at the genus and species levels, Flavonifractor, Flavonifractor plautii, and Lachnospiraceae bacterium 7_1_58FAA were similarly enriched (Wilcoxon rank-sum test, q < 0.05) (Table 2).
Table 2.
Differential Abundance of Bacterial Taxa in the WGS-Based Profiles of Gut Microbial Communities in Children with IBS (n = 23) and HCs (n = 22)
| Bacterial taxa | FC | P value | q Value |
|---|---|---|---|
| Class | |||
| Gammaproteobacteria | 9.57 | 0.003 | 0.03 |
| Verrucomicrobiae | 4.95 | 0.043 | 0.26 |
| Family | |||
| Unclassified Clostridiales | 3.11 | <0.001 | 0.01 |
| Oscillospiraceae | 1.85 | 0.007 | 0.13 |
| Enterobacteriaceae | 17.51 | 0.010 | 0.13 |
| Verrucomicrobiaceae | 4.95 | 0.043 | 0.41 |
| Genus | |||
| Flavonifractor | 7.01 | <0.001 | 0.02 |
| Oscillibacter | 1.84 | 0.007 | 0.19 |
| Eggerthella | 3.04 | 0.009 | 0.19 |
| Escherichia | 11.67 | 0.009 | 0.19 |
| Unclassified Clostridiaceae | 6.90 | 0.014 | 0.23 |
| Pseudoflavonifractor | ∗ | 0.022 | 0.30 |
| Holdemania | 2.22 | 0.028 | 0.31 |
| Akkermansia | 4.95 | 0.043 | 0.43 |
| Species | |||
| Flavonifractor plautii | 7.01 | <0.001 | 0.02 |
| Lachnospiraceae bacterium 7_1_58FAA | 3.25 | <0.001 | 0.02 |
| Lachnospiraceae bacterium 1_4_56FAA | ∗ | 0.004 | 0.22 |
| Oscillibacter species | 3.89 | 0.004 | 0.22 |
| Lachnospiraceae bacterium 5_1_57FAA | ∗ | 0.007 | 0.25 |
| Eggerthella species | 3.26 | 0.007 | 0.25 |
| Escherichia coli | 12.71 | 0.010 | 0.30 |
| Eubacterium 3_1_31 | ∗ | 0.014 | 0.33 |
| Clostridiaceae bacterium JC118 | 6.90 | 0.014 | 0.47 |
| Pseudoflavonifractor capillosus | ∗ | 0.022 | 0.57 |
| Clostridium symbiosum | 4.49 | 0.031 | 0.57 |
| Ruminococcus bromii | 0.02 | 0.033 | 0.57 |
| Subdoligranulum 4_3_54A2FAA | 3.40 | 0.043 | 0.57 |
| Akkermansia muciniphila | 4.95 | 0.043 | 0.57 |
Differences were evaluated using Wilcoxon rank-sum tests with Benjamini-Hochberg false-discovery rate (FDR) corrections. FC values >1 indicate enrichment in IBS. Taxa that remain significant (q < 0.05) after FDR correction are in bold.
FC, fold change; WGS, whole-genome sequencing.
A median value of 0 in HCs, precluding an FC calculation.
A total of 632 metabolic pathways and 6163 enzymatic reactions were identified among the functional metagenomic profiles. Differential abundance analyses yielded multiple differences with borderline significance between IBS and HCs after false-discovery rate correction (Wilcoxon rank-sum test, q < 0.10). Metabolic pathways (n = 58) (Supplemental Table S2) and enzymatic reactions (n = 182) (Supplemental Table S3) related to amino acid metabolism, phospholipid biosynthesis, and vitamin biosynthesis were relatively enriched in IBS, whereas enzymatic reactions related to carbohydrate metabolism were relatively depleted.
Higher-Order Metabolite Classes Differ in IBS Children versus HCs
Grouped into higher-order metabolite classes, steroids/sterols, bile acids, and products of phenylalanine and tyrosine degradation were significantly enriched in the stool of IBS children (Wilcoxon rank-sum test, q ≤ 0.05) (Table 3). Individual metabolites are listed in Supplemental Table S4, with false-discovery rate–corrected trends indicating the enrichment of multiple sterols, steroids, bile acids, and products of phenylalanine and tyrosine metabolism in IBS, as reflected by the higher-order metabolite findings (Table 3).
Table 3.
Differences in Metabolite Abundances, Grouped by Higher-Order Classes, Were Evaluated Using Wilcoxon Rank-Sum Tests, Followed by FDR Correction in Children with IBS (n = 23) and HCs (n = 22)
| Metabolite group | FC | P value | q Value |
|---|---|---|---|
| Steroid/sterol | 1.7 | 0.0002 | 0.016 |
| Deoxy-litho-ursodeoxycholate secondary bile acids | 2.6 | 0.0027 | 0.115 |
| Secondary bile acids | 1.7 | 0.0058 | 0.124 |
| Dihydroxy fatty acids | 1.3 | 0.0156 | 0.151 |
| Endocannabinoid | 3.2 | 0.0188 | 0.151 |
| Glycerolipid metabolism | 0.7 | 0.0188 | 0.151 |
| Guanidino and acetamido metabolism | 2.1 | 0.0193 | 0.151 |
| Phenylalanine and tyrosine metabolism∗ | 4.3 | 0.0328 | 0.226 |
| Hemoglobin and porphyrin metabolism | 0.6 | 0.0357 | 0.226 |
| Dipeptides | 1.5 | 0.0410 | 0.226 |
| Food component/plant∗ | 2.8 | 0.0410 | 0.226 |
| Monoacylglycerol∗ | 1.7 | 0.0421 | 0.226 |
Positive FC values indicate enrichment in IBS.
FC, fold change; FDR, false-discovery rate.
Based on aggregation of individual metabolites within each class that exhibited FC ≥ |1.5| in the comparison of IBS versus HC.
Abdominal Pain in IBS Is Associated with Key Microbes, Microbial Genes, and Microbial Metabolites
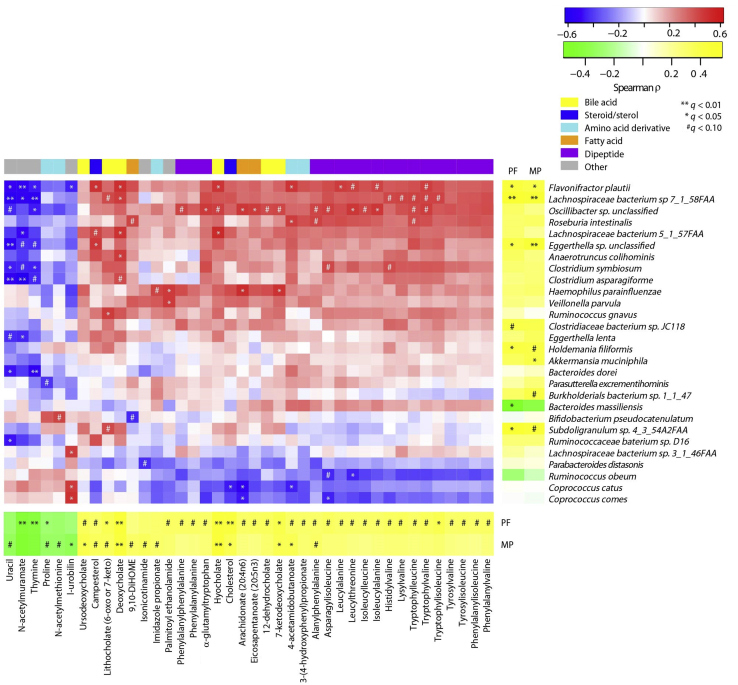
Significant correlations were identified between abdominal pain and the relative abundances of various microbial taxa, metagenomic functions, and metabolites. These associations included positive correlations between pain frequency and the relative abundances of several bacterial species, including F. plautii, L. bacterium 7_1_58FAA, and an unclassified Eggerthella (Figure 2). Mean pain was negatively correlated with the relative abundance of Bacteroides massiliensis (Figure 2).
Figure 2.
Spearman correlations between abdominal pain–associated species and metabolites in 45 pediatric subjects. Relationships between whole-genome sequencing–based species abundances and metabolites (middle), metabolites and abdominal pain (bottom), and species and abdominal pain (right) are depicted. Each metabolite depicted herein correlates with abdominal pain frequency (PF) and/or mean abdominal pain (MP). All of the included species are significantly correlated with abdominal pain and/or abdominal pain–associated metabolites. False-discovery rate–corrected statistical significance is denoted as follows: *q < 0.05, **q < 0.01, and #q < 0.10. n = 23 pediatric subjects with IBS; n = 22 HCs. DiHome, (12Z)-9,10-dihydroxyoctadec-12-enoic acid.
Positive correlations were observed (across all subjects) between abdominal pain frequency and severity and the relative abundances of multiple functional pathways. Examples of these include fucose and rhamnose degradation, the tricarboxylic acid cycle, phospholipid biosynthesis, and assimilatory sulfate reduction (Supplemental Table S5). In contrast, pain frequency and severity were negatively correlated with pathways related to fatty acid salvage and nucleotide biosynthesis (Supplemental Table S5).
Positive correlations were also observed between pain frequency and severity and concentrations of steroids and sterols, multiple bile acids, and multiple protein-degradation products (Table 4 and Figure 2). Negative correlations were observed between pain and the concentrations of the peptidoglycan component N-acetylmuramate.
Table 4.
Correlations between Metabolites and MP or PF in 45 Pediatric Subjects (n = 23 IBS Cases and 22 HCs)
| Pain metric | Metabolite | Metabolic category | ρ Value | q Value |
|---|---|---|---|---|
| MP | Hyocholate | Bile acids | 0.52 | 0.003 |
| MP | Deoxycholate | Bile acids | 0.49 | 0.008 |
| MP | Ursodeoxycholate | Bile acids | 0.43 | 0.027 |
| MP | 7-Ketodeoxycholate | Bile acids | 0.41 | 0.036 |
| MP | N-acetylmuramate | Amino sugars | −0.51 | 0.004 |
| MP | 4-Acetamidobutanoate | Guanidino and acetamido metabolism | 0.40 | 0.039 |
| MP | l-Urobilin | Hemoglobin and porphyrin metabolism | −0.39 | 0.049 |
| MP | Thymine | Pyrimidine metabolism | −0.47 | 0.013 |
| MP | Cholesterol | Steroids/sterols | 0.41 | 0.034 |
| PF | Deoxycholate | Bile acids | 0.52 | 0.004 |
| PF | Hyocholate | Bile acids | 0.51 | 0.005 |
| PF | 7-Ketodeoxycholate | Bile acids | 0.43 | 0.027 |
| PF | Lithocholate (6-oxo or 7-keto) | Bile acids | 0.41 | 0.034 |
| PF | N-acetylmuramate | Amino sugars | −0.48 | 0.009 |
| PF | Tryptophylisoleucine | Dipeptide | 0.43 | 0.027 |
| PF | Thymine | Pyrimidine metabolism | −0.45 | 0.017 |
| PF | Cholesterol | Steroids/sterols | 0.49 | 0.007 |
Relationships were evaluated using Spearman correlations and Benjamini-Hochberg false-discovery rate corrections.
MP, mean pain; PF, pain frequency.
Notably, many of the pain frequency– and/or severity-related metabolites and bacterial species highlighted above also shared significant correlations with one another. The relative abundances of F. plautii, L. bacterium 7_1_58FAA, and an unclassified Oscillibacter were correlated with deoxycholate, hyocholate, and other bile acids, as well as multiple pain frequency–associated dipeptides (Figure 2).
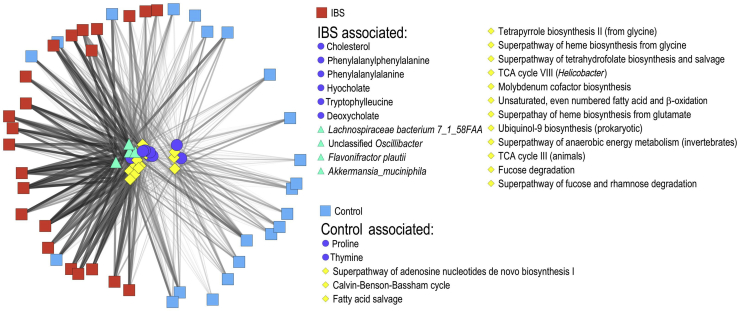
A Combination of Microbial and Metabolite Features Correctly Predict IBS Status
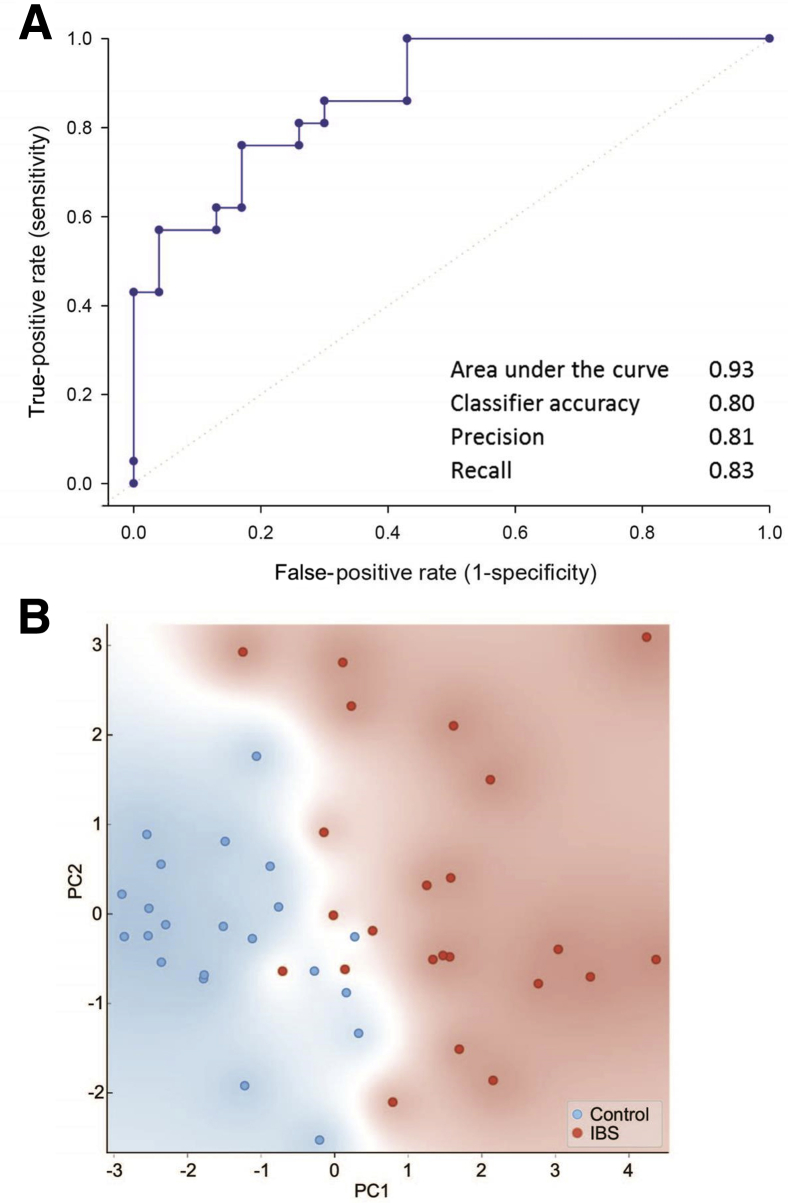
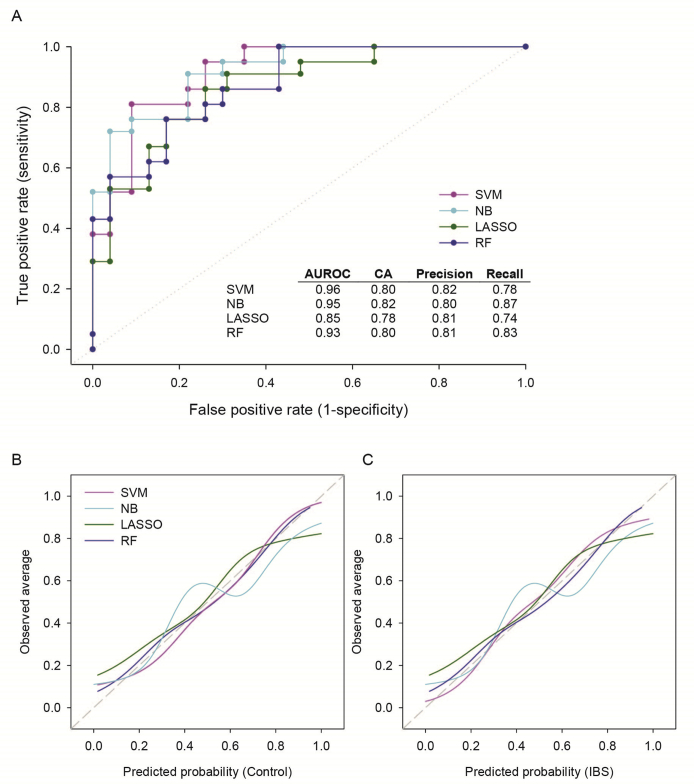
On the basis of 27 metabolite, pathway, and species features, each with analysis of variance F scores >7, separation of IBS cases from HCs was achieved via network analysis (Figure 3). Further limiting these features to the 10 most differentially abundant metabolites, bacterial species, and functional pathways, an RF classifier was developed, which distinguished IBS cases from HCs with an area under the receiver operating characteristic curve of 0.93 and a classification accuracy of 0.80 (Figure 4A). In developing the RF model, multiple classification algorithms were evaluated. Each classifier model was trained using fivefold cross validation. Although most classifier models performed comparably (Supplemental Figure S1A), the RF model was consistently better fitting than the others with respect to predicted versus observed class probabilities via calibration curve analysis (Supplemental Figure S1, B and C). The classification accuracies of the five individual RF folds were similar, with an average classification accuracy of 0.80 and an SD of 0.15.
Figure 3.
A multi-omics network of bacterial species (green triangles), metagenomic pathways (yellow diamonds), and metabolite abundances (blue spheres) separates pediatric IBS cases (red squares) from HCs (cyan squares). Features (ie, species, pathways, and metabolites) were included if they had F values >7 in the comparison of IBS cases versus HCs. The edge-weighted, spring-embedded layout was used to visualize network structure. n = 23 pediatric IBS cases; n = 22 HCs. TCA, tricarboxylic acid.
Figure 4.
Multivariate classification based on a lean set of multi-omics features correctly distinguished IBS cases from HCs with a high degree of accuracy. A: Receiver operating characteristic curve of the random forest (RF) classifier and its associated accuracy and precision rates. Classifier metrics were generated using fivefold cross validation. Random classification is represented by the dotted line. B: Principal component (PC) analysis of subjects based on the set of species, pathways, and metabolites used to train the RF classifier. Background shading indicates point density of IBS cases versus HCs.
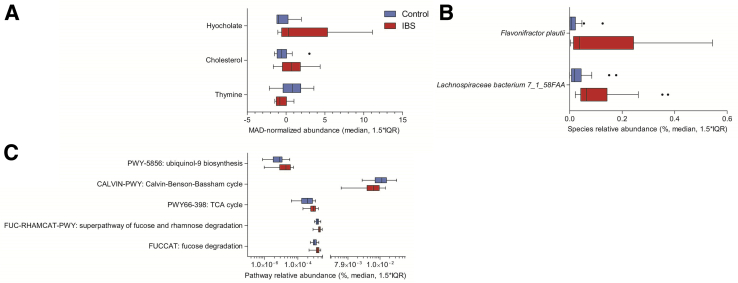
Principal components analysis, based on these same 10 features, resulted in separation of IBS cases and HCs along principal component 1 (Figure 4B). Metabolites contributing to classifier success were hyocholate, cholesterol, and thymine (Figure 5A). Bacterial species leveraged by the classifier were F. plautii and L. bacterium 7_1_58FAA (Figure 5B), and the functional pathways included those related to ubiquinol biosynthesis (PWY-5856) and carbohydrate metabolism (tricarboxylic acid cycle, fucose and rhamnose degradation, and Calvin cycle) (Figure 5C).
Figure 5.
Abundances and distributions of the metabolites (A), bacterial species (B), and functional pathways (C) on which the classifiers were trained. Boxplots depict median and first and third quartile values, whereas whiskers indicate 1.5 times the interquartile range (IQR). Species, pathway, and metabolite values were assessed in IBS cases and HCs. B: Only the bacterial species are significantly differentially abundant (false-discovery rate–corrected q = 0.02). n = 23 IBS cases (A–C); n = 22 HCs (A–C). MAD, median of the absolute deviations from the median; TCA, tricarboxylic acid.
Discussion
Consensus among IBS microbiome studies has been challenged for multiple reasons, including the use of recall-based classification and symptom-based definitions of IBS, which have changed over time and are open to interpretation.37, 38 At the same time, effective nutritional interventions [eg, low-fermentable oligo-, di-, mono-saccharides and polyols (FODMAP) diets] in subsets of patients with IBS have stressed the importance of accurate disease stratification.6, 19, 39, 40 To advance the stratification and management of IBS patients, rigorous disease phenotyping is essential. In this study, participants were first screened using the validated pediatric Rome III questionnaire; then, expert recommendations were followed by phenotyping IBS patients on the basis of a 2-week diary20 and applying an established algorithm evaluating stool form and frequency to ensure that subjects met IBS criteria.22 With this approach, deeper insights were gained into the pathobiology and disease stratification of pediatric IBS. The molecular and biochemical findings point the way forward in beginning to apply microbiome-based molecular diagnostics to chronic human disease phenotypes.
Using this well-vetted cohort, these studies confirmed some previously reported findings from our own work9, 25 and that of others, in children. This extends knowledge in the field with the identification of metagenomic and metabolomic features that may serve as diagnostic biomarkers and contribute to our understanding of the biology of IBS. By leveraging the greater depth and precision of WGS compared with 16S rRNA–based sequencing, a species-resolved, multi-omics classifier, capable of distinguishing IBS cases from controls with 80% accuracy, was developed and significant new associations were identified between abdominal pain frequency and/or severity and key bacterial taxa, microbial functions, and fecal metabolites.
Although previous studies have demonstrated that microbial community features may be useful in distinguishing or identifying individual IBS subtypes9, 21 or IBS severity,12 they have generally relied on relatively large numbers of operational taxonomic unit–based features, have lacked the taxonomic resolution and functional information afforded by WGS, and/or have been informed by microbial features alone. By leveraging both metagenomic and metabolomic information, a lean (ie, 10 features), species-resolved classifier was identified, which is capable of distinguishing pediatric IBS cases from controls with an area under the curve of 0.93 and ≥80% accuracy. In comparison, a recently published classifier for IBS disease diagnosis using 16S rRNA sequencing based on 157 fecal microbial genera reported an area under the curve of 0.81.41 In our estimation, a disease classifier at or around 80% would represent a significant advance in the diagnosis of these functional gastrointestinal tract disorders. In contrast to disorders such as celiac disease or inflammatory bowel disease, current phenotypic assessments of IBS lack biochemical, microbial, molecular, or pathologic features, as part of a routine diagnostic workup.
A classifier with improved accuracy may be clinically impactful. The classifier's ability to identify patients with IBS supports the hypothesis that the microbiome and microbiome-associated metabolites are clinically relevant features of IBS. In addition, a classifier may help identify the subpopulation of children with IBS who are more likely to benefit from nutritional interventions, such as low-FODMAP diets.6 In contrast, subjects with IBS without the presence of the classifier features may require other therapies (eg, guided imagery). Ultimately, such multi-omics–based disease classifiers may be improved in the future and offer refined diagnostic approaches for coupling patients with functional gastrointestinal tract disorders and proper nutritional or medical interventions.
The relative abundances of bacterial taxa highlighted in this report may yield different contributions to disease classifier success, and bacterial taxa may share significant correlations with pain and/or pain-associated metabolites. Although additional studies are needed to address the potential pathobiological relevance of these microbial taxa to childhood IBS, evidence from the literature suggests a plausible role for each taxon. Flavonifractor plautii was significantly enriched in IBS cases and was correlated with recurrent abdominal pain. Flavonifractor plautii proteins can elicit enhanced IgG responses in postinfectious IBS patients,11 and the enrichment of F. plautii has been described in nonpostinfectious IBS and other functional GI tract disorders, too.42, 43 Enrichment of the genus Flavonifractor was described in adults with comorbid IBS diarrhea predominant and depression,42 as was the enrichment of F. plautii in children with autism spectrum disorder, abdominal pain, and functional constipation.43 Flavonifractor plautii is a flavonoid degrader, and polyphenolic compounds, including flavonoids and their derivatives, have the potential to inhibit carbohydrate metabolism.44 Extending this notion, potentially impaired carbohydrate metabolism, by F. plautii or other polyphenol degraders, may help to explain why low fermentable carbohydrate diets provide relief for some patients experiencing IBS.6, 45
Similarly, L. bacterium 7_1_58FAA, an isolate originally obtained from inflamed colonic tissue (https://www.beiresources.org/Catalog/bacteria/HM-153.aspx, last accessed August 22, 2018) was significantly enriched in IBS children and correlated with both pain and pain-associated metabolites. Although L. bacterium 7_1_58FAA and F. plautii share a somewhat disparate relationship taxonomically, the two are closely related on a genomic level. Lachnospiraceae bacterium 7_1_58FAA and F. plautii share approximately 75% of their gene content with a pairwise average nucleotide identity exceeding 98% [average nucleotide identity (ANI)-clique, 20; https://img.jgi.doe.gov, last accessed August 22, 2018]. Such nucleotide identity values exceed minimum definitions for intraspecies pairs,46 suggest that these may represent the same species, indicate that one or both may be misclassified, and highlight the challenges and potential pitfalls of relying on 16S rRNA gene–based classifications alone. Various unclassified Lachnospiraceae have been described in association with IBS on the basis of 16S rRNA gene survey data.7, 13 Lachnospiraceae-produced flagellins can stimulate immune responses in some IBS patients,47 and unclassified Lachnospiraceae operational taxonomic units were identified as indicator species for IBS and associated with innate immune activation, altered motility, intestinal barrier dysfunction, and anxiety-like behaviors in a humanized mouse model of IBS.13
Secondary bile acids, including our classification-relevant hyocholate, were also enriched in the stool of IBS children and significantly correlated with abdominal pain. Reports have linked primary bile acids in stool and serum with changes in stool form,16 colonic transit rates,15 perceived pain,16 and gut microbiota16 in some adults with IBS; and rectal or sigmoid infusion of deoxycholic acid, a secondary bile acid, induces discomfort in healthy adults48 and pain in adult IBS patients.49, 50 It has been suggested that secondary bile acids may heighten pain perception by altering intracellular Ca2+ ion and/or cholinergic neurotransmitter concentrations.49 Taken together, our findings and those of others suggest that both primary and secondary bile acids may influence IBS symptoms, but their relative importance may vary as a function of age, microbiome composition,26 or predominant stool type. For example, primary bile acid concentrations tend to be greater in adults with IBS diarrhea predominant than IBS constipation predominant,16 but most IBS children studied had IBS constipation predominant, the most common pediatric IBS subtype.25
Significant correlations were also identified between fecal cholesterol concentrations and pain frequency, a novel finding in the context of IBS. Steroids and related compounds can influence microbial growth and function, as many bacterial species encode steroid catabolism genes.51 Microbially modified steroids may exert physiological effects on the host, acting, for example, as neurosteroids and modulating neuronal excitability through γ-aminobutyric acid receptors.52 γ-Aminobutyric acid receptors are involved in visceral sensation processing and, as such, may be relevant to visceral hypersensitivity in IBS.8 Similarly, microbially modified steroids may alter central brain processing and plasticity, affecting both central and peripheral visceral nociception.53 Two steroid metabolites were found to be significantly enriched (each >25-fold increase) in children with IBS versus HCs (pregnenolone steroid monosulfate and 5-α-pregnan-3-β,20 α-diol disulfate). They were shown to modulate the γ-aminobutyric acid A receptors.54 Also, steroid hormones enriched in woman were shown to dramatically potentiate visceral pain responses to stimuli in rat models.55 These two metabolites represent sulfated steroids, and related steroids in this group may be differentially abundant in other chronic pain conditions.56 A potential confounding factor is that exogenous steroid therapy and nonsteroidal anti-inflammatory drugs may alter steroid sulfation profiles.57 Microbial and human cells may both modify steroid compounds by different mechanisms, including sulfation, and these modified steroids, including neurosteroids, may alter nociceptive thresholds.
Recent reports in the literature also support the plausible role of other classifier features in contributing to IBS-related dysbiosis and pain. For example, enhanced pyrimidine metabolism, including that of thymine, may be indicative of dysbiosis.58 Greater abundance of tricarboxylic acid cycle genes and increased potential for the biosynthesis of ubiquinol, an antioxidant compound, may reflect increased exposure to or capacity to handle oxidative stress.59 Enhanced metagenomic potential for fucose degradation has been reported in adults with comorbid chronic fatigue syndrome and IBS (versus controls).60
Despite the frequency of recurrent abdominal pain in school-aged children and the clinical need for better diagnostic and management strategies, the identification of eligible participants and rigorous phenotyping we applied resulted in the recruitment of a relatively small cohort. Although this may limit the generalizability of the results, our classifiers rely on features that differentiated cases from controls in our own study and are supported by a variety of previously published studies, as discussed earlier in this section. Additional work is needed to validate these findings in a larger cohort and determine the degree to which they are capable of distinguishing IBS from other functional GI tract disorders.
Conclusions
The identification of microbial and metabolic features that distinguish IBS cases from controls may improve IBS diagnosis, and the identification of gut microbes and metabolites potentially contributing to or serving as indicators of visceral pain will likely contribute to impactful disease management. The bacterial features and metabolites described in this report appear to be closely linked with abdominal pain and emphasize the importance of the microbiome-gut-brain axis to human health. Additional studies evaluating the ability of these bacterial features and metabolites in distinguishing IBS from other functional GI tract disorders, as well as their roles in pathophysiology of childhood IBS, are needed and may lead to improved diagnostics and therapeutics for IBS.
Acknowledgments
We thank Rebecca Cappello and Elizabeth Menard for contributions to subject screening, recruitment, and sample processing; Wen Y. Chong for contributions to subject screening and sample processing; Yue Shang, Delphine Saulnier, and Tulin Ayvaz for contributions to specimen extraction and sequence library preparation; Nadim Ajami for contributions to specimen management and sequencing, as well as critical feedback on the manuscript; Kevin Riehle and Aleks Milosavljevic for data analysis support; and Karen Prince for contributions to the development and preparation of figures.
R.J.S., M.H., and J.V. designed the study; R.J.S., J.V., T.C.S., J.F.P., and R.A.G. managed the project; E.M.W. and M.R.-G. recruited subjects and collected subject data; R.A.L., M.R.-G., S.R., G.A.M., and D.M.M. performed sample processing and sequencing; E.B.H., N.O., B.P.C., E.M.W., M.D., J.L.C., and T.-A.M. analyzed the data; E.B.H., B.P.C., N.O., R.A.L., T.C.S., R.J.S., and J.V. wrote and edited the manuscript; all authors commented on the manuscript and approved the final draft.
Footnotes
Supported by the National Institute of Diabetes and Digestive and Kidney Diseases grants UH2DK093990 (J.V.) and UH3DK083990 (J.V.), R21DK096323-01 (T.C.S.), K23DK101688 (B.P.C.) and R03DK117219 (B.P.C.); the National Center for Complementary and Integrative Medicine grant RO1AT004326 (J.V.); the National Cancer Institute grant U01CA170930 (J.V.); the National Human Genome Research Institute grant U54HG004973 (R.A.G.); the National Institute of Nursing Research grants R01NR05337 (R.J.S.), R01NR013497 (R.J.S.), and RC2NR011959 (R.J.S. and M.H.); the National Institute of Allergy and Infectious Diseases grants U011AI24290-01 (T.C.S.) and R01AI10091401 (T.C.S.); the Autism Speaks Gastrointestinal and Neurobehavioral Processes grant 9455 (R.A.L.); the Baylor College of Medicine Caroline Weiss Law Fund for Research in Molecular Medicine (B.P.C.); and Daffy's Foundation (R.J.S.). Shared resources that helped advance this project were also supported, in part, by NIH grant P30 DK56338 (Texas Medical Center Digestive Diseases Center).
E.B.H. and N.O. contributed equally to this work.
Disclosures: E.B.H. received research support from Cargill, Inc., and is employed by and holds stock in Diversigen, Inc.; B.P.C. provided consultancy for Mead Johnson Nutrition; J.L.C. is employed by Diversigen, Inc., and holds stock in the company; T.C.S. received research support from and/or provided consultancy for Merck, Cubist, Nivalis, Rebiotix, Assembly BioSciences, and Mead Johnson Nutrition; R.J.S. provided consultancy for Nutrinia, IMHealth, and Biogaia AB, and received restricted research support from Mead-Johnson; J.V. received unrestricted research support from Biogaia AB (Stockholm, Sweden) and serves on the Scientific Advisory Boards of Biomica, Plexus Worldwide, and Seed Health; no study sponsors were involved in the design of the study, collection, analysis, or interpretation of the data, or the writing of the manuscript.
Current address of E.B.H and J.L.C., Diversigen, Inc., Houston, TX.
Supplemental material for this article can be found at https://doi.org/10.1016/j.jmoldx.2019.01.006.
Supplemental Data
Supplemental Figure S1.
Receiver operating characteristic (ROC) and calibration curves indicating the classification success and quality of the class-based (ie, IBS case versus HC) probability predictions generated by each of the classifier models considered [random forest (RF), support vector machine (SVM), and naïve Bayes (NB)]. A: ROC curves and their associated accuracy and precision metrics. Random classification is represented by the dotted line. B and C: Calibration curves for each of the classifiers are depicted relative to their ability to correctly classify HCs (B) and IBS cases (C). The calibration curve for an ideal model would plot along the 45-degree reference dashed line, whereas deviations from this line indicate tendencies to overpredict or underpredict class probabilities. For both HCs (B) and IBS cases (C), the RF and SVM models tend to be the best calibrated. AUROC, area under the ROC curve; CA, classification accuracy; LASSO, least absolute shrinkage selection operator.
References
- 1.Rasquin A., Di Lorenzo C., Forbes D., Guiraldes E., Hyams J.S., Staiano A., Walker L.S. Childhood functional gastrointestinal disorders: child/adolescent. Gastroenterology. 2006;130:1527–1537. doi: 10.1053/j.gastro.2005.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quigley E.M., Abdel-Hamid H., Barbara G., Bhatia S.J., Boeckxstaens G., De Giorgio R., Delvaux M., Drossman D.A., Foxx-Orenstein A.E., Guarner F., Gwee K.A., Harris L.A., Hungin A.P., Hunt R.H., Kellow J.E., Khalif I.L., Kruis W., Lindberg G., Olano C., Moraes-Filho J.P., Schiller L.R., Schmulson M., Simren M., Tzeuton C. A global perspective on irritable bowel syndrome: a consensus statement of the World Gastroenterology Organisation Summit Task Force on irritable bowel syndrome. J Clin Gastroenterol. 2012;46:356–366. doi: 10.1097/MCG.0b013e318247157c. [DOI] [PubMed] [Google Scholar]
- 3.Cain K.C., Headstrom P., Jarrett M.E., Motzer S.A., Park H., Burr R.L., Surawicz C.M., Heitkemper M.M. Abdominal pain impacts quality of life in women with irritable bowel syndrome. Am J Gastroenterol. 2006;101:124–132. doi: 10.1111/j.1572-0241.2006.00404.x. [DOI] [PubMed] [Google Scholar]
- 4.Varni J.W., Shulman R.J., Self M.M., Nurko S., Saps M., Saeed S.A., Bendo C.B., Patel A.S., Dark C.V., Zacur G.M., Pohl J.F., Pediatric Quality of Life Inventory Gastrointestinal Symptoms Module Testing Study Consortium Symptom profiles in patients with irritable bowel syndrome or functional abdominal pain compared with healthy controls. J Pediatr Gastroenterol Nutr. 2015;61:323–329. doi: 10.1097/MPG.0000000000000795. [DOI] [PubMed] [Google Scholar]
- 5.Chumpitazi B.P., Shulman R.J. Underlying molecular and cellular mechanisms in childhood irritable bowel syndrome. Mol Cell Pediatr. 2016;3:11. doi: 10.1186/s40348-016-0036-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chumpitazi B.P., Cope J.L., Hollister E.B., Tsai C.M., McMeans A.R., Luna R.A., Versalovic J., Shulman R.J. Randomised clinical trial: gut microbiome biomarkers are associated with clinical response to a low FODMAP diet in children with the irritable bowel syndrome. Aliment Pharmacol Ther. 2015;42:418–427. doi: 10.1111/apt.13286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salonen A., de Vos W.M., Palva A. Gastrointestinal microbiota in irritable bowel syndrome: present state and perspectives. Microbiology. 2010;156:3205–3215. doi: 10.1099/mic.0.043257-0. [DOI] [PubMed] [Google Scholar]
- 8.Greenwood-Van Meerveld B., Moloney R.D., Johnson A.C., Vicario M. Mechanisms of stress-induced visceral pain: implications in irritable bowel syndrome. J Neuroendocrinol. 2016;28 doi: 10.1111/jne.12361. [DOI] [PubMed] [Google Scholar]
- 9.Saulnier D.M., Riehle K., Mistretta T.A., Diaz M.A., Mandal D., Raza S., Weidler E.M., Qin X., Coarfa C., Milosavljevic A., Petrosino J.F., Highlander S., Gibbs R., Lynch S.V., Shulman R.J., Versalovic J. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology. 2011;141:1782–1791. doi: 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simrén M., Barbara G., Flint H.J., Spiegel B.M.R., Spiller R.C., Vanner S., Verdu E.F., Whorwell P.J., Zoetendal E.G. Intestinal microbiota in functional bowel disorders: a Rome foundation report. Gut. 2013;62:159–176. doi: 10.1136/gutjnl-2012-302167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pike B.L., Paden K.A., Alcala A.N., Jaep K.M., Gormley R.P., Maue A.C., Christmann B.S., Elson C.O., Riddle M.S., Porter C.K. Immunological biomarkers in postinfectious irritable bowel syndrome. J Travel Med. 2015;22:242–250. doi: 10.1111/jtm.12218. [DOI] [PubMed] [Google Scholar]
- 12.Tap J., Derrien M., Tornblom H., Brazeilles R., Cools-Portier S., Dore J., Storsrud S., Le Neve B., Ohman L., Simren M. Identification of an intestinal microbiota signature associated with severity of irritable bowel syndrome. Gastroenterology. 2017;152:111–123.e118. doi: 10.1053/j.gastro.2016.09.049. [DOI] [PubMed] [Google Scholar]
- 13.De Palma G., Lynch M.D., Lu J., Dang V.T., Deng Y., Jury J., Umeh G., Miranda P.M., Pigrau Pastor M., Sidani S., Pinto-Sanchez M.I., Philip V., McLean P.G., Hagelsieb M.G., Surette M.G., Bergonzelli G.E., Verdu E.F., Britz-McKibbin P., Neufeld J.D., Collins S.M., Bercik P. Transplantation of fecal microbiota from patients with irritable bowel syndrome alters gut function and behavior in recipient mice. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aaf6397. eaaf6397. [DOI] [PubMed] [Google Scholar]
- 14.Rajilic-Stojanovic M., Biagi E., Heilig H.G., Kajander K., Kekkonen R.A., Tims S., de Vos W.M. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792–1801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 15.Bajor A., Tornblom H., Rudling M., Ung K.A., Simren M. Increased colonic bile acid exposure: a relevant factor for symptoms and treatment in IBS. Gut. 2015;64:84–92. doi: 10.1136/gutjnl-2013-305965. [DOI] [PubMed] [Google Scholar]
- 16.Dior M., Delagreverie H., Duboc H., Jouet P., Coffin B., Brot L., Humbert L., Trugnan G., Seksik P., Sokol H., Rainteau D., Sabate J.M. Interplay between bile acid metabolism and microbiota in irritable bowel syndrome. Neurogastroenterol Motil. 2016;28:1330–1340. doi: 10.1111/nmo.12829. [DOI] [PubMed] [Google Scholar]
- 17.Ponnusamy K., Choi J.N., Kim J., Lee S.Y., Lee C.H. Microbial community and metabolomic comparison of irritable bowel syndrome faeces. J Med Microbiol. 2011;60:817–827. doi: 10.1099/jmm.0.028126-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lackner J.M., Jaccard J., Keefer L., Firth R., Carosella A.M., Sitrin M., Brenner D. Representing the IRG: the accuracy of patient-reported measures for GI symptoms: a comparison of real time and retrospective reports. Neurogastroenterol Motil. 2014;26:1802–1811. doi: 10.1111/nmo.12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Self M.M., Williams A.E., Czyzewski D.I., Weidler E.M., Shulman R.J. Agreement between prospective diary data and retrospective questionnaire report of abdominal pain and stooling symptoms in children with irritable bowel syndrome. Neurogastroenterol Motil. 2015;27:1110–1119. doi: 10.1111/nmo.12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lacy B.E., Mearin F., Chang L., Chey W.D., Lembo A.J., Simren M., Spiller R. Bowel disorders. Gastroenterology. 2016;150:1393–1407.e5. doi: 10.1053/j.gastro.2016.02.031. [DOI] [PubMed] [Google Scholar]
- 21.Shankar V., Reo N.V., Paliy O. Simultaneous fecal microbial and metabolite profiling enables accurate classification of pediatric irritable bowel syndrome. Microbiome. 2015;3:73. doi: 10.1186/s40168-015-0139-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Czyzewski D.I., Lane M.M., Weidler E.M., Williams A.E., Swank P.R., Shulman R.J. The interpretation of Rome III criteria and method of assessment affect the irritable bowel syndrome classification of children. Aliment Pharmacol Ther. 2011;33:403–411. doi: 10.1111/j.1365-2036.2010.04535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shulman R.J., Eakin M.N., Jarrett M., Czyzewski D.I., Zeltzer L.K. Characteristics of pain and stooling in children with recurrent abdominal pain. J Pediatr Gastroenterol Nutr. 2007;44:203–208. doi: 10.1097/01.mpg.0000243437.39710.c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Baeyer C.L., Spagrud L.J., McCormick J.C., Choo E., Neville K., Connelly M.A. Three new datasets supporting use of the Numerical Rating Scale (NRS-11) for children's self-reports of pain intensity. Pain. 2009;143:223–227. doi: 10.1016/j.pain.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Weidler E.M., Self M.M., Czyzewski D.I., Shulman R.J., Chumpitazi B.P. Stooling characteristics in children with irritable bowel syndrome. Clin Gastroenterol Hepatol. 2017;15:140–141. doi: 10.1016/j.cgh.2016.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollister E.B., Riehle K., Luna R.A., Weidler E.M., Rubio-Gonzales M., Mistretta T.A., Raza S., Doddapaneni H.V., Metcalf G.A., Muzny D.M., Gibbs R.A., Petrosino J.F., Shulman R.J., Versalovic J. Structure and function of the healthy pre-adolescent pediatric gut microbiome. Microbiome. 2015;3:36. doi: 10.1186/s40168-015-0101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abubucker S., Segata N., Goll J., Schubert A.M., Izard J., Cantarel B.L., Rodriguez-Mueller B., Zucker J., Thiagarajan M., Henrissat B., White O., Kelley S.T., Methé B., Schloss P.D., Gevers D., Mitreva M., Huttenhower C. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Truong D.T., Franzosa E.A., Tickle T.L., Scholz M., Weingart G., Pasolli E., Tett A., Huttenhower C., Segata N. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 30.Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 31.Suzek B.E., Huang H., McGarvey P., Mazumder R., Wu C.H. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 32.Caspi R., Altman T., Dreher K., Fulcher C.A., Subhraveti P., Keseler I.M., Kothari A., Krummenacker M., Latendresse M., Mueller L.A., Ong Q., Paley S., Pujar A., Shearer A.G., Travers M., Weerasinghe D., Zhang P., Karp P.D. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2012;40:D742–D753. doi: 10.1093/nar/gkr1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollard K., Dudoit S., van der Laan M. Multiple testing procedures: the multtest package and applications to genomics. In: Gentleman R., Carey V.J., Huber W., Irizarry R.A., Dudoit S., editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor Statistics for Biology and Health. Springer; New York, NY: 2005. pp. 249–271. [Google Scholar]
- 34.Team: R . RStudio, Inc.; Boston, MA: 2015. RStudio: Integrated Development of R. [Google Scholar]
- 35.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demsar J., Curk T., Erjavec A. Orange: data mining toolbox in phyton. J Mach Learn Res. 2013;14:2349–2353. [Google Scholar]
- 37.Gwee K.A., Ghoshal U.C. The Rome criteria divides, distorts and dilutes the prevalence of irritable bowel syndrome. Saudi J Gastroenterol. 2010;16:143–144. doi: 10.4103/1319-3767.65178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moayyedi P., Ford A.C. Symptom-based diagnostic criteria for irritable bowel syndrome: the more things change, the more they stay the same. Gastroenterol Clin North Am. 2011;40:87–103. doi: 10.1016/j.gtc.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 39.Self M.M., Czyzewski D.I., Chumpitazi B.P., Weidler E.M., Shulman R.J. Subtypes of irritable bowel syndrome in children and adolescents. Clin Gastroenterol Hepatol. 2014;12:1468–1473. doi: 10.1016/j.cgh.2014.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chogle A., Sztainberg M., Bass L., Youssef N.N., Miranda A., Nurko S., Hyman P., Cocjin J., Di Lorenzo C., Saps M. Accuracy of pain recall in children. J Pediatr Gastroenterol Nutr. 2012;55:288–291. doi: 10.1097/MPG.0b013e31824cf08a. [DOI] [PubMed] [Google Scholar]
- 41.Gao X., Lin H., Dong Q. A Dirichlet-multinomial Bayes classifier for disease diagnosis with microbial compositions. mSphere. 2017;2 doi: 10.1128/mSphereDirect.00536-17. pii: e00536-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y., Zhang L., Wang X., Wang Z., Zhang J., Jiang R., Wang X., Wang K., Liu Z., Xia Z., Xu Z., Nie Y., Lv X., Wu X., Zhu H., Duan L. Similar fecal microbiota signatures in patients with diarrhea-predominant irritable bowel syndrome and patients with depression. Clin Gastroenterol Hepatol. 2016;14:1602–1611.e1605. doi: 10.1016/j.cgh.2016.05.033. [DOI] [PubMed] [Google Scholar]
- 43.Luna R.A., Oezguen N., Balderas M., Venkatachalam A., Runge J.K., Versalovic J., Veenstra-VanderWeele J., Anderson G.M., Savidge T., Williams K.C. Distinct microbiome-neuroimmune signatures correlate with functional abdominal pain in children with autism spectrum disorder. Cell Mol Gastroenterol Hepatol. 2017;3:218–230. doi: 10.1016/j.jcmgh.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moco S., Martin F.P., Rezzi S. Metabolomics view on gut microbiome modulation by polyphenol-rich foods. J Proteome Res. 2012;11:4781–4790. doi: 10.1021/pr300581s. [DOI] [PubMed] [Google Scholar]
- 45.McIntosh K., Reed D.E., Schneider T., Dang F., Keshteli A.H., De Palma G., Madsen K., Bercik P., Vanner S. FODMAPs alter symptoms and the metabolome of patients with IBS: a randomised controlled trial. Gut. 2017;66:1241–1251. doi: 10.1136/gutjnl-2015-311339. [DOI] [PubMed] [Google Scholar]
- 46.Varghese N.J., Mukherjee S., Ivanova N., Konstantinidis K.T., Mavrommatis K., Kyrpides N.C., Pati A. Microbial species delineation using whole genome sequences. Nucleic Acids Res. 2015;43:6761–6771. doi: 10.1093/nar/gkv657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schoepfer A.M., Schaffer T., Seibold-Schmid B., Muller S., Seibold F. Antibodies to flagellin indicate reactivity to bacterial antigens in IBS patients. Neurogastroenterol Motil. 2008;20:1110–1118. doi: 10.1111/j.1365-2982.2008.01166.x. [DOI] [PubMed] [Google Scholar]
- 48.Edwards C.A., Brown S., Baxter A.J., Bannister J.J., Read N.W. Effect of bile acid on anorectal function in man. Gut. 1989;30:383–386. doi: 10.1136/gut.30.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor I., Basu P., Hammond P., Darby C., Flynn M. Effect of bile acid perfusion on colonic motor function in patients with the irritable colon syndrome. Gut. 1980;21:843–847. doi: 10.1136/gut.21.10.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coremans G., Tack J., Vantrappen G., Janssens J., Annese V. Is the irritable bowel really irritable? Ital J Gastroenterol. 1991;23:39–40. [PubMed] [Google Scholar]
- 51.Bergstrand L.H., Cardenas E., Holert J., Van Hamme J.D., Mohn W.W. Delineation of steroid-degrading microorganisms through comparative genomic analysis. MBio. 2016;7:e00166. doi: 10.1128/mBio.00166-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carver C.M., Reddy D.S. Neurosteroid interactions with synaptic and extrasynaptic GABA(A) receptors: regulation of subunit plasticity, phasic and tonic inhibition, and neuronal network excitability. Psychopharmacology (Berl) 2013;230:151–188. doi: 10.1007/s00213-013-3276-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moloney R.D., Johnson A.C., O'Mahony S.M., Dinan T.G., Greenwood-Van Meerveld B., Cryan J.F. Stress and the microbiota-gut-brain axis in visceral pain: relevance to irritable bowel syndrome. CNS Neurosci Ther. 2016;22:102–117. doi: 10.1111/cns.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gibbs T.T., Russek S.J., Farb D.H. Sulfated steroids as endogenous neuromodulators. Pharmacol Biochem Behav. 2006;84:555–567. doi: 10.1016/j.pbb.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 55.Peroni R.N., Orliac M.L., Becu-Villalobos D., Huidobro-Toro J.P., Adler-Graschinsky E., Celuch S.M. Sex-linked differences in the vasorelaxant effects of anandamide in vascular mesenteric beds: role of oestrogens. Eur J Pharmacol. 2004;493:151–160. doi: 10.1016/j.ejphar.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 56.Parker K.S., Crowley J.R., Stephens-Shields A.J., van Bokhoven A., Lucia M.S., Lai H.H., Andriole G.L., Hooton T.M., Mullins C., Henderson J.P. Urinary metabolomics identifies a molecular correlate of interstitial cystitis/bladder pain syndrome in a Multidisciplinary Approach to the Study of Chronic Pelvic Pain (MAPP) Research Network cohort. EBioMedicine. 2016;7:167–174. doi: 10.1016/j.ebiom.2016.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cohen I.V., Cirulli E.T., Mitchell M.W., Jonsson T.J., Yu J., Shah N., Spector T.D., Guo L., Venter J.C., Telenti A. Acetaminophen (paracetamol) use modifies the sulfation of sex hormones. EBioMedicine. 2018;28:316–323. doi: 10.1016/j.ebiom.2018.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Larsen P.E., Dai Y. Metabolome of human gut microbiome is predictive of host dysbiosis. Gigascience. 2015;4:42. doi: 10.1186/s13742-015-0084-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 60.Nagy-Szakal D., Williams B.L., Mishra N., Che X., Lee B., Bateman L., Klimas N.G., Komaroff A.L., Levine S., Montoya J.G., Peterson D.L., Ramanan D., Jain K., Eddy M.L., Hornig M., Lipkin W.I. Fecal metagenomic profiles in subgroups of patients with myalgic encephalomyelitis/chronic fatigue syndrome. Microbiome. 2017;5:44. doi: 10.1186/s40168-017-0261-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.