Abstract
Pleiotropic genes are genes that affect more than one trait. For example, many genes required for pigmentation in the fruit fly Drosophila melanogaster also affect traits such as circadian rhythms, vision, and mating behavior. Here, we present evidence that two pigmentation genes, ebony and tan, which encode enzymes catalyzing reciprocal reactions in the melanin biosynthesis pathway, also affect cuticular hydrocarbon (CHC) composition in D. melanogaster females. More specifically, we report that ebony loss-of-function mutants have a CHC profile that is biased toward long (>25C) chain CHCs, whereas tan loss-of-function mutants have a CHC profile that is biased toward short (<25C) chain CHCs. Moreover, pharmacological inhibition of dopamine synthesis, a key step in the melanin synthesis pathway, reversed the changes in CHC composition seen in ebony mutants, making the CHC profiles similar to those seen in tan mutants. These observations suggest that genetic variation affecting ebony and/or tan activity might cause correlated changes in pigmentation and CHC composition in natural populations. We tested this possibility using the Drosophila Genetic Reference Panel (DGRP) and found that CHC composition covaried with pigmentation as well as levels of ebony and tan expression in newly eclosed adults in a manner consistent with the ebony and tan mutant phenotypes. These data suggest that the pleiotropic effects of ebony and tan might contribute to covariation of pigmentation and CHC profiles in Drosophila.
Keywords: Drosophila, pleiotropy, ebony, tan, pigmentation, cuticular hydrocarbons, trait covariation, dopamine
Introduction
When organisms adapt to novel environments, genetic changes often cause multiple traits to evolve. In some cases, organisms invading similar environments undergo similar shifts for suites of traits. In the threespine stickleback, for example, marine populations independently invading freshwater lake habitats have repeatedly evolved similar changes in defensive armor, behavior, and body shape (Walker and Bell, 2000; Schluter et al., 2004; Wark et al., 2011). Such correlated evolution might result from (i) selection favoring a particular suite of traits (i.e., selection targeting multiple unlinked loci), (ii) selection favoring a trait that is genetically linked to genes affecting other traits, or (iii) selection favoring a trait that varies due to genetic variation at a pleiotropic gene affecting multiple traits. In the case of the threespine stickleback, genetic variation linked to a single major gene, Eda, has been found to explain correlated differences in these traits among populations (Albert et al., 2008; Greenwood et al., 2016), suggesting that pleiotropy has played a role. Studies in various other plant and animal species also support the hypothesis that pleiotropy contributes to the coevolution of correlated traits (e.g., McKay et al., 2003; McLean et al., 2011; Duveau and Félix, 2012; Nagy et al., 2018).
In insects, genes determining body color are often pleiotropic. For example, in Drosophila, the yellow gene is required for the synthesis of black melanin and also affects mating behavior (Bastock, 1956; Drapeau et al., 2003, 2006). The genes pale and Dopa-decarboxylase, which encode enzymes that synthesize tyrosine-derived precursors for pigmentation, are also pleiotropic, affecting both body color and immunity (reviewed in Wittkopp and Beldade, 2009; Takahashi, 2013). In addition, prior work suggests that pigmentation genes might also affect cuticular hydrocarbon (CHC) profiles, which can affect desiccation (Gibbs et al., 1997; Gibbs, 1998; Foley and Telonis-Scott, 2011) and mate choice (reviewed in Yew and Chung, 2015). Specifically, a receptor for the tanning hormone bursicon and levels of the biogenic amine dopamine, which both affect cuticle pigmentation in Drosophila melanogaster, have been shown to influence CHC composition (Marican et al., 2004; Wicker-Thomas and Hamann, 2008; Flaven-Pouchon et al., 2016).
Here, we test whether the ebony and tan genes of D. melanogaster, which are required for the synthesis of dark melanins and yellow sclerotins from dopamine, respectively, also affect CHC composition. The ebony gene encodes a protein that converts dopamine into N-β-alanyl dopamine (NBAD), and the tan gene encodes a protein that catalyzes the reverse reaction, converting NBAD back into dopamine (Figure 1A). We report that loss-of-function mutations in both ebony and tan altered CHC length composition relative to wild-type flies in opposing directions. These opposing effects on CHC length composition are consistent with ebony and tan’s opposing biochemical functions in dopamine metabolism (Figure 1A). Indeed, pharmacological inhibition of dopamine synthesis in ebony mutants caused a tan-like CHC length profile. To examine the possibility that variation in ebony and/or tan activity might cause correlated changes in pigmentation and CHC composition in a natural population, we used lines from the Drosophila Genetic Reference Panel (DGRP) to test for covariation between pigmentation and CHC composition. We found that CHC length composition covaried not only with pigmentation but also with levels of ebony and tan expression in a manner consistent with the mutant analyses. In the discussion, we compare our data to studies of clinal variation in CHC composition and pigmentation to determine whether the pleiotropic effects we see might have contributed to correlated evolution of these traits.
FIGURE 1.
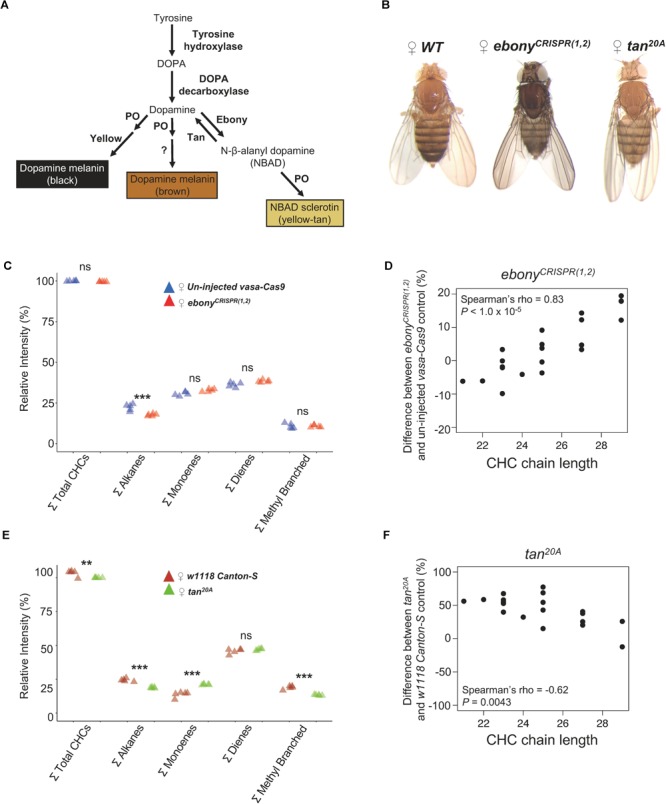
ebony and tan affect pigmentation and CHC composition in female Drosophila melanogaster. (A) Insect sclerotization and pigmentation synthesis pathway. Ebony converts dopamine into N-β-alanyl dopamine (NBAD) which is oxidized into yellow-colored NBAD sclerotin. Tan catalyzes the reverse reaction, converting NBAD back into dopamine that can be oxidized into black and brown melanins. (B) Photographs highlighting the effects of ebonyCRISPR(1,2) (darker) and tan20A (lighter) on body pigmentation compared to the un-injected vasa-Cas9 control line (WT). (C) Summary of ebonyCRISPR(1,2) effects on total summed CHC classes relative to un-injected vasa-Cas9 control females. (D) Difference in log-contrast of relative CHC intensity between ebonyCRISPR(1,2) and un-injected vasa-Cas9 control flies. (E) Summary of tan20A effects on total summed CHC classes relative to w1118 Canton-S control females. For (D,E), each triangle represents a single replicate of CHCs extracted from five pooled individuals (N = 5 replicates per genotype). (F) Difference in log-contrast of relative CHC intensity between tan20A and w1118 Canton-S control flies. Results of Tukey HSD post hoc tests following one-way ANOVA are shown: ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Materials and Methods
Fly Stocks and Maintenance
The following lines were used: P excision line tan20A (True et al., 2005) (courtesy of John True, Stony Brook University); the UAS-ebony-RNAi effector line was obtained from the Vienna Drosophila Resource Centre (Dietzl et al., 2007, KK106278); dsxGAL4 (Rideout et al., 2010) (courtesy of Stephen Goodwin, Oxford University); OK72-GAL4 (Ferveur et al., 1997) (courtesy of Scott Pletcher, University of Michigan); pannier-GAL4 (Calleja et al., 2000) was obtained from the Bloomington Drosophila Stock Center (BDSC 3039); vasa-Cas9 (Gratz et al., 2014, BDSC 51324) (courtesy of Rainbow Transgenics Inc.). All flies were grown at 23°C with a 12 h light-dark cycle on standard corn-meal fly medium.
DGRP Stocks
The following inbred D. melanogaster lines from the DGRP (Ayroles et al., 2009; Mackay et al., 2012; Huang et al., 2014) were used in this study: RAL-208, RAL-303, RAL-324, RAL-335, RAL-357, RAL-358, RAL-360, RAL-365, RAL-380, RAL-399, RAL-517, RAL-555, RAL-705, RAL-707, RAL-732, RAL-774, RAL-786, RAL-799, RAL-820, RAL-852, RAL-714, RAL-437, RAL-861, and RAL-892. These lines consist of the set of 20 lines used in Miyagi et al. (2015) and additional three dark lines (RAL-714, RAL-437, and RAL-861), which were added to avoid line specific effects from a limited number of dark lines. All flies were grown at 25°C with a 12 h light-dark cycle on standard corn-meal fly medium.
Generation of ebony CRISPR Lines
New loss-of-function ebony mutants were constructed by synthesizing two single guide RNAs (gRNA), using a MEGAscript T7 Transcription Kit (Invitrogen), following the PCR-based protocol from Bassett et al. (2013), that target ebony’s first coding exon and co-injecting these at a total concentration of 100 ng/μL into embryos of a D. melanogaster vasa-Cas9 line (Gratz et al., 2014; BDSC 51324) (Supplementary Figure S1). These gRNAs were previously found to generate a high level of heritable germline transformants (Ren et al., 2014) (Supplementary Figure S1). We screened for germline transformants based on body pigmentation and confirmed via Sanger sequencing three unique ebony loss-of-function alleles, ebonyCRISPR(1,2) containing a 55 bp deletion, and ebonyCRISPR(3) and ebonyCRISPR(4), each containing an in-frame 3 bp deletion (Supplementary Figure S1). Each deletion caused flies to develop dark body pigmentation, indicating loss of Ebony activity (Figure 1B and Supplementary Figure S2A).
CHC Extraction and Measurements
For Figure 1, 2 and Supplementary Figures S2–S5, CHCs were extracted and analyzed as described below (CHC names and formulas are summarized in Supplementary Table S1). For the analyses using the DGRP (Figure 3, 4 and Supplementary Figure S6), all CHC data for females were obtained from Dembeck et al. (2015b); however, in the case of GC/MS peaks composed of more than two combined CHC components that differed in CHC chain length, the non-branched CHC chain length was used. Also, CHCs that were not detected in all strains were removed from the analyses.
FIGURE 2.
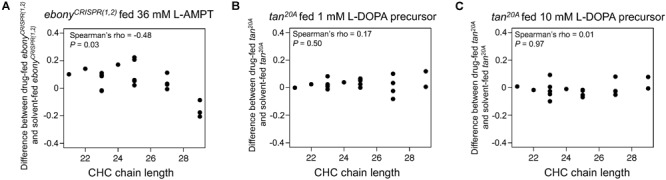
Effects of pharmacological treatments on CHC lengthening in ebonyCRISPR(1,2) and tan20A mutants. (A) Difference in log-contrast of relative CHC intensity between ebonyCRISPR(1,2) females fed 36 mM alpha methyl tyrosine (L-AMPT) and ebonyCRISPR(1,2) females fed a solvent control. (B) Difference in log-contrast relative of CHC intensity between tan20A females fed 1 mM methyl L-DOPA hydrochloride (L-DOPA precursor) and tan20A females fed a solvent control. (C) Difference in log-contrast of relative CHC intensity between tan20A females fed 10 mM L-DOPA precursor and tan20A females fed a solvent control.
FIGURE 3.
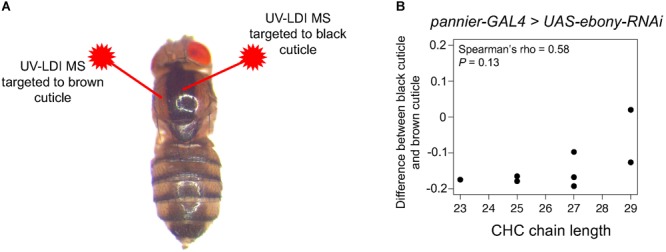
UV laser desorption/ionization mass spectrometry (UV-LDI MS) did not detect differences in short versus long CHCs between lightly and darkly pigmented cuticle. Female pannier-GAL4 flies were crossed to UAS-ebony-RNAi males to generate flies with a dark, heavily melanized stripe down the dorsal midline. (A) The UV-LDI MS lasers were targeted to light brown or dark black cuticle within the same fly (N = 3 biological replicates). (B) Difference in relative CHC intensity between black and brown cuticle.
FIGURE 4.
Abdominal pigmentation co-varies with CHC length profiles in the Drosophila Genetic Reference Panel (DGRP). Pigmentation scores and CHC data were obtained from Dembeck et al. (2015a,b). (A) Difference in log-contrast of relative CHC intensity between DGRP females with darkly pigmented fifth abdominal tergites (A5) (1.5 < score, N = 53) and the 155 line average. (B) Difference in log-contrast of relative CHC intensity between DGRP females with intermediately pigmented A5 (1 < score ≤ 1.5, N = 56) and the 155 line average. (C) Difference in log-contrast of relative CHC intensity between DGRP females with lightly pigmented A5 (score ≤ 1, N = 49) and the 155 line average.
Extraction
For each experiment, five replicate CHC samples of virgin female flies were prepared for each genotype or pharmacological treatment group. All ebony and tan mutant CHC extractions were performed on 3- to 4-day-old virgin females. We restricted our analysis to virgin females, because previous evidence studies suggested that a link between dopamine and CHC composition occurs in females but not males (Marican et al., 2004; Wicker-Thomas and Hamann, 2008). For pharmacological experiments, 1- to 2-day-old virgin females were treated for 4 days prior to CHC extraction. For GAL4/UAS experiments (Brand and Perrimon, 1993), virgin females were tested at 10–12 days. For each sample, five flies were placed in a single glass vial (Wheaton 224740 E–C Clear Glass Sample Vials) on ice. 120 μL of hexane (Sigma-Aldrich, St. Louis, MO, United States) spiked with 10 μg/mL of hexacosane (Sigma-Aldrich) was added to each vial and sealed with a cap. Vials were incubated at room temperature for 20 min. One hundred microliters of the cuticular extract was removed, transferred into a clean vial (Wheaton 0.25 mL with low volume insert), and stored at -20°C.
GC/MS Analysis
Gas chromatography mass spectrometry (GC/MS) analysis was performed on a 7820A GC system equipped with a 5975 Mass Selective Detector (Agilent Technologies, Inc., Santa Clara, CA, United States) and a HP-5 ms column [(5%-phenyl)-methylpolysiloxane, 30 m length, 250 μm ID, 0.25 μm film thickness; Agilent Technologies, Inc.]. Electron ionization (EI) energy was set at 70 eV. One microliter of the sample was injected in splitless mode and analyzed with helium flow at 1 mL/min. The following parameters were used: column was set at 40°C for 3 min, increased to 200°C at a rate of 35°C/min, then increased to 280°C at a rate of 20°C/min for 15 min. The MS was set to detect from m/z 33 to 500. Chromatograms and spectra were analyzed using MSD ChemStation (Agilent Technologies, Inc.). CHCs were identified on the basis of retention time and EI fragmentation pattern. The relative abundance for each CHC signal was calculated by normalizing the area under each CHC peak to the area of the hexacosane signal. To eliminate multicollinearity among sample peak amounts, a log-contrast transformation was applied to the resulting proportional values, using nC27 as the denominator (Blows and Allen, 1998; Yew et al., 2011):
| (1) |
To determine the relative change in CHC length between two genotypes, experimental groups, or groups of DGRP strains, the difference in relative intensity of individual CHC intensities of each group was calculated:
| (2) |
These values were then plotted against CHC chain length.
Ultraviolet Laser Desorption Ionization Mass Spectrometry (UV-LDI MS)
For intact fly analysis, individual animals were attached to a glass cover slip using adhesive pads (G304, Plano, Wetzlar, Germany). The cover slips were mounted on a custom-milled sample holder containing a rectangular, 1.8 mm deep well. Sample height was adjusted by choosing a stack of 0.2 mm-thick adhesive pads (G3347, Plano). Mass spectra were generated using a prototype orthogonal-extracting mass spectrometer (oTOF-MS) as described previously (Yew et al., 2011). The oTOF-MS was equipped with a modified oMALDI2 ion source (AB Sciex, Concord, ON, Canada) and an N2 laser (λ = 337 nm) operated at a pulse repetition rate of 30 Hz. N2 was used as buffer gas at p = 2 mbar. This elevated pressure is critical to achieve an efficient collisional cooling environment for generation of weakly bound [M + K]+ ions that constituted the major molecular ion species. Before starting the actual measurements, external mass calibration was achieved with red phosphorus, resulting in a mass accuracy of approximately 25 ppm. Approximately 900 laser shots were placed at one position to achieve a mass spectrum (30 s @30 Hz). All spectra were acquired in positive ion mode and processed using MS Analyst software (Analyst QS 2.0, AB Sciex, Concord, ON, Canada).
Pharmacology Experiments
For pharmacological treatments, standard corn-meal fly medium was liquefied and cooled to ca. 60°C before the addition of each respective drug or solvent control. Ten 1- to 2-day-old virgin females were placed in the vials for 4 days. To inhibit tyrosine hydroxylase activity, we prepared a 36 mM alpha methyl tyrosine (L-AMPT) (Sigma-Aldrich) diet. The pH of the solution was adjusted with concentrated HCl until the drug dissolved. A solvent control diet solution was prepared using identical procedures. For the dopamine treatments, 1 mM and 10 mM L-dopa precursor (Methyl L-DOPA hydrochloride) (Sigma-Aldrich) were dissolved in water before adding to liquefied fly media.
RNA Extraction
Female virgin flies were collected within 1 h of eclosion, and the heads were removed in RNAlater (Ambion) to separate the effect from transcripts in non-epidermal head tissues. The remaining head-less body samples were stored in RNAlater at -80°C until use. Three body samples from each line were placed in a 2 mL microtube with 400 μL TRIzol Reagent (Thermo Fisher Scientific, Tokyo, Japan) and an equivalent volume of 1.2 mm zirconia silica beads (Bio Medical Science). After shaking the tube at 3,200 rpm for 2 min using a Beads Crusher μT-12 (TAITEC, Koshigaya, Japan), 160 μl chloroform was added and mixed thoroughly. Total RNA in the aqueous phase was subsequently purified using silica-gel (Wakocil 5SIL, Wako, Osaka, Japan) based on the method of Boom et al. (1990) and was quantified using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific).
Quantitative Real-Time PCR (qRT-PCR)
First strand cDNA was synthesized from 1 μg total RNA by using a PrimeScript RT Reagent Kit with gDNA Eraser (Takara Bio, Kusatsu, Japan). qRT-PCR was performed in a 25 μL reaction volume with SYBR Premix Ex Taq II Tli RNaseH Plus (Takara Bio) on a Thermal Cycler Dice TP800 (Takara Bio). Primer pairs used for RT-qPCR were ebony: 5′-CTTAGTGTGAAACGGCCACAG-3′ and 5′-GCAGCGAACCCATCTTGAA-3′; tan: 5′-GTTGAGGGGCTTCGATAAGA-3′ and 5′-GTCCTCCGGAAAGATCCTG-3′;Act57B: 5′-CGTGTCATCCTTGGTTCGAGA-3′ and 5′-ACCGCGAGCGATTAACAAGTG-3′; Rp49: 5′-TCGGATCGATATGCTAAGCTG-3′ and 5′-TCGATCCGTAACCGATGTTG-3′. Act57B and Rp49 were used as internal control. Two replicate PCR reactions were performed for each cDNA sample and three biological replicates were obtained for each line.
Grouping DGRP Lines Based on Pigmentation Scores and ebony/tan Expression Levels
The DGRP lines (N = 155) with both pigmentation scores in Dembeck et al. (2015a) and CHC profiles in Dembeck et al. (2015b) were grouped into dark, intermediate, and light pigmentation lines using the pigmentation scores of the abdominal tergites from Dembeck et al. (2015a). The scores ranged from 0 for no dark pigmentation to 4 for 100% dark pigmentation in increments of 0.5, and were averaged across 10 individuals per line. Pigmentation grouping was done based on the score delimitations that split the lines most evenly into three groups. For the fifth tergite (A5), lines were categorized into following groups: dark (1.5 < score, N = 53), intermediate (1 < score ≤ 1.5, N = 56), and light (score ≤ 1, N = 49). For the sixth tergite (A6), lines were categorized into following groups: dark (3 < score, N = 51), intermediate (2 < score ≤ 3, N = 55), light (score ≤ 2, N = 49).
The 23 DGRP lines with varying ebony and tan expression levels were grouped into low, intermediate, and high expression lines using the qRT-PCR data. Since the normalized quantities are continuous values, grouping was done based on standard deviations (SD). For the ebony expression, lines were categorized into following groups: low (expression < mean – 0.5 SD, N = 6), intermediate (mean – 0.5 SD ≤ expression ≤ mean + 0.5 SD, N = 9), and high (mean + 0.5 SD < expression, N = 8). For the tan expression, lines were categorized into following groups: low (expression < mean - 0.5 SD, N = 10), intermediate (mean - 0.5 SD ≤ expression ≤ mean + 0.5 SD, N = 7), and high (mean + 0.5 SD < expression, N = 6).
Statistics
All statistical tests were performed in R for Mac version 3.3.3 (R Core Team, 2013) using one-way ANOVAs to test for statistically significant effects between more than two groups and post hoc Tukey HSD tests for multiple pairwise comparisons. We used Spearman’s rank correlation coefficient ρ to test for the significance of the association. All pairwise tests were two-tailed, and the level of significance was set as α = 0.05.
Results
Loss-of-Function Mutations in ebony and tan Have Reciprocal Effects on CHC Length Profiles
To determine whether the ebony gene affects CHCs, we created three new ebony mutant alleles via CRISPR/Cas9 gene editing. One allele, ebonyCRISPR(1,2), contained a 55 bp deletion that caused a frame-shift in ebony’s coding sequence (Supplementary Figure S1C). Flies homozygous for this ebonyCRISPR(1,2) allele showed dark body pigmentation similar to that described previously for loss-of-function ebony mutants (Bridges and Morgan, 1923) (Figure 1B). We measured CHC profiles in 3- to 4-day-old ebonyCRISPR(1,2) virgin females using gas chromatography (GC/MS) and found that ebonyCRISPR(1,2) flies showed lower levels of total alkanes relative to 3- to 4-day-old virgin females from the strain the guide RNAs were injected into (i.e., un-injected vasa-Cas9) (Figure 1C, one-way ANOVA: F9,40 = 4494, P < 2.0 × 10-16; post hoc Tukey HSD was significant for alkanes: P < 1.0 × 10-5).
We then tested whether ebonyCRISPR(1,2) females had different proportions of individual CHCs. We calculated the average difference in individual log-contrast transformed CHC relative intensities (see section “Materials and Methods”) between ebonyCRISPR(1,2) flies and un-injected vasa-Cas9 control flies and plotted these values against CHC chain length [varying from 21 carbons (C) to 29C] (Figure 1D and Supplementary Table S1). We found that ebonyCRISPR(1,2) flies tended to show lower levels of short chain CHCs (<25C) and higher levels of long chain CHCs (>25C), suggesting that disrupting the function of ebony causes a CHC lengthening effect (Figure 1D, Spearman’s ρ = 0.83, P < 1.0 × 10-5).
The two other ebony alleles generated using CRISPR/Cas9 gene editing [ebonyCRISPR(3) and ebonyCRISPR(4)] each had a single 3 bp in-frame deletion in the first coding exon (Supplementary Figures S1D,E), suggesting that they might have less severe effects on Ebony activity than the ebonyCRISPR(1,2) allele containing a 55 bp deletion causing a frame-shift. Consistent with this prediction, these ebony mutants also showed darker body pigmentation than wild-type flies (Supplementary Figure S2A), but did not show any bias toward longer CHCs (Supplementary Figures S2B,C, ebonyCRISPR(3): Spearman’s ρ = 0.22, P = 0.34; ebonyCRISPR(4): Spearman’s ρ = 0.07, P = 0.78).
To better understand the effects of reduced ebony expression on CHCs, we knocked down ebony expression in specific cell types using ebony-RNAi (Dietzl et al., 2007). First, we drove expression of ebony-RNAi with the dsxGAL4 driver (Rideout et al., 2010), which causes RNAi expression in the cuticle, fat body, CNS, and oenocytes among other tissues. We observed darker pigmentation in dsxGAL4 > UAS-ebony-RNAi flies than control flies (data not shown), suggesting that the ebony-RNAi effectively targeted and knocked down ebony expression. These dsxGAL4 > UAS-ebony-RNAi flies also showed a pattern of CHC lengthening similar to the ebonyCRISPR(1,2) mutants when compared to to dsxGAL4/+ control flies but not when compared to UAS-ebony-RNAi/+ control flies. This result might be due to leaky UAS-ebony-RNAi expression in the latter control flies that makes their profiles more similar to those of dsxGAL4 > UAS-ebony-RNAi flies (Supplementary Figures S3A,B, relative to dsxGAL4/+ control: Spearman’s ρ = 0.58, P < 0.007; relative to UAS-ebony-RNAi/+ control: Spearman’s ρ = 0.19, P = 0.42).
We hypothesized that the effect on CHCs might be due to reducing ebony expression specifically in oenocytes because these cells synthesize many CHC precursor compounds (Wigglesworth, 1970). Therefore, we drove expression of ebony-RNAi using the OK72-GAL4 driver that is also expressed in oenocytes (Ferveur et al., 1997). These flies showed no significant difference in CHC length profiles (Supplementary Figure S3C, Spearman’s ρ = -0.01, P = 0.96), suggesting that ebony expression in non-oenocyte tissues expressing doublesex affects the overall length proportion of CHCs.
Next, we asked whether loss-of-function mutations in the tan gene also affect CHC composition. Specifically, we examined CHC composition in 3- to 4-day-old virgin females carrying a tan20A null allele, which contains an imprecise P-element excision that results in a 953 bp deletion that includes the presumptive promoter region (True et al., 2005). Because tan encodes a protein that catalyzes the reverse of the reaction catalyzed by Ebony (Figure 1A), we predicted that tan mutants might show the opposite effects on CHC composition. Similar to the ebonyCRISPR(1,2) mutants, tan20A females showed differences in the overall abundance of alkanes, but also total CHCs, monoenes, and methyl branched CHCs (Figure 1E, one-way ANOVA: F9,40 = 3586, P < 2.0 × 10-16; post hoc Tukey HSD was significant for total summed CHCs: P < 0.01, total summed alkanes: P < 0.001, total summed monoenes: P < 0.001, and total summed methyl branched: P < 0.001). More importantly, tan20A (w1118 tan20A) females tended to show higher levels of short chain CHCs relative to long chain CHCs when compared to w1118 Canton-S (CS) control flies, as predicted (Figure 1F, Spearman’s ρ = -0.62, P = 0.0043). Together, these results suggest that ebony and tan have reciprocal effects on both pigmentation synthesis (reviewed in True, 2003; True et al., 2005) and CHC length profiles. We note that this conclusion contradicts Wicker-Thomas and Hamann (2008)’s report that CHC profiles were similar in ebony or tan loss-of-function mutants and wild-type flies; however, the ebony and tan alleles used in this prior work might not have been nulls.
Pharmacological Inhibition of Tyrosine Hydroxylase Activity Reverses the CHC Lengthening Effect in ebonyCRISPR(1,2) Flies
We hypothesized that ebony and tan might have reciprocal effects on CHC length profiles because of their effects on dopamine metabolism. For example, because ebony encodes a protein that converts dopamine into NBAD (Figure 1A), we hypothesized that loss-of-function ebony mutants might accumulate dopamine (as reported in Hodgetts and Konopka, 1973) and that this dopamine might be shunted into other pathways, possibly affecting CHC lengthening. To explore this hypothesis, we fed 1- to 2-day-old adult female ebonyCRISPR(1,2) flies a tyrosine hydroxylase inhibitor, alpha methyl tyrosine (L-AMPT), for 4 days to determine whether inhibiting dopamine synthesis would reverse the CHC lengthening pattern we observed in ebonyCRISPR(1,2) flies. Relative to ebonyCRISPR(1,2) solvent-fed control flies, ebonyCRISPR(1,2) flies fed 36 mM L-AMPT did indeed reverse the CHC lengthening pattern we observed in ebonyCRISPR(1,2) flies, resulting in a shortening of CHCs similar to that observed in tan20A flies (Figure 2A, Spearman’s ρ = -0.48, P= 0.03). Feeding 1- to 2-day-old adult flies L-AMPT did not, however, affect body pigmentation (data not shown), consistent with body pigmentation being determined prior to and soon after eclosion (Hovemann et al., 1998). We also fed ebonyCRISPR(4) flies a 36 mM dose of L-AMPT to see if we could induce CHC shortening in an ebony mutant with unchanged CHC length composition. Similar to ebonyCRISPR(1,2) fed flies, we detected a significant negative correlation when comparing ebonyCRISPR(4) fed flies to an ebonyCRISPR(4) solvent-fed control (Supplementary Figure S4, Spearman’s ρ = -0.57, P= 0.009).
We next hypothesized that tan20A flies might have lower levels of circulating dopamine, because tan encodes a protein that converts NBAD back into dopamine (Figure 1A). To determine whether elevating dopamine levels in tan mutants would affect CHCs, we fed tan20A females a dopamine precursor, methyl L-DOPA hydrochloride (L-DOPA precursor), to see if elevating dopamine levels could reverse the CHC shortening pattern we observed in tan20A flies; however, neither the 1 mM nor 10 mM L-DOPA precursor treatments seemed to affect CHC length profiles when compared to tan20A solvent-fed control flies (Figure 2B,C, Spearman’s ρ = 0.17, P= 0.50; Spearman’s ρ = 0.01, P= 0.97, respectively). We also fed tan20A flies a higher 100 mM dose of the L-DOPA precursor, but all of these flies died before CHC extraction; these flies also showed darker cuticle pigmentation consistent with elevated dopamine. Finally, we fed 1 mM and 10 mM doses of L-DOPA precursor to wild-type (w1118 CS) females to see if we could induce CHC lengthening in a wild-type genetic background; instead, we observed a slight CHC shortening effect for the 1 mM dose and no effect for the 10 mM dose (Supplementary Figure S5, Spearman’s ρ = -0.52, P= 0.02; Spearman’s ρ = -0.36, P= 0.12, respectively). Together, these results indicate that inhibiting tyrosine hydroxylase activity in ebony mutants causes a CHC shortening effect like that observed in tan20A flies; however, increasing dopamine levels through feeding does not cause a CHC lengthening effect.
UV-LDI MS Data Suggests That ebony’s Effects on Pigmentation and CHC Length Profiles Are Not Linked at the Level of the Cuticle
Pigmentation synthesis in insect cuticles involves the secretion of biogenic amines (such as dopamine) by epidermal cells into the developing cuticle where they are oxidized into quinones that can form melanins or sclerotins that crosslink proteins (Figure 1A; reviewed in True, 2003; Riedel et al., 2011). To determine whether ebony’s effects on CHC length profiles depend on their function in pigmentation and sclerotization of the fly cuticle, we measured the relative abundance of individual CHCs in virgin females with different levels of pigmentation across the body. We crossed pannier-GAL4 (Calleja et al., 2000) females with males from the UAS-ebony-RNAi effector line to generate flies with a dark, heavily melanized stripe down the dorsal midline (Figure 3A). We then used UV laser desorption/ionization mass spectrometry (UV-LDI MS) to take repeated measurements of CHCs along the thorax of females, targeting inside and outside the dark stripe (Figure 3A). Although we observed an upward trend in abundance from short to long CHCs, we did not detect a significant CHC lengthening effect like that observed between ebonyCRISPR(1,2) flies and un-injected vasa-Cas9 females (Figure 3B, Spearman’s ρ = 0.58, P= 0.13). Within the black cuticle, most CHCs detected by UV-LDI MS showed a decrease in abundance relative to brown cuticle (Figure 3B). This result suggests that ebony does not affect CHC length profiles through the pigmentation/sclerotization synthesis pathway, at least at the level of CHC/pigment deposition in the cuticle.
Abdominal Pigmentation Covaries With CHC Length Profiles in the Drosophila Genetic Reference Panel (DGRP)
The effects of ebony and tan mutants on CHC profiles described above suggest that variation in these genes might contribute to variation in both pigmentation and CHC profiles. Recently, Dembeck et al. (2015a,b) analyzed the genetic architecture of abdominal pigmentation and CHC composition in female D. melanogaster lines from the DGRP: Dembeck et al. (2015a) quantified abdominal pigmentation intensity in the fifth and sixth abdominal tergites (A5 and A6), and Dembeck et al. (2015b) investigated CHC profiles from the majority of the panel, but the relationship between the two traits was not examined. Using data from the 155 DGRP lines for which both pigmentation scores and CHC profiles were published, we tested the hypothesis that natural variation in pigmentation covaries with natural variation in CHC length profiles. In order to investigate CHC composition in a way that was comparable to the experiments described above, we divided the 155 DGRP lines into dark (N = 53), intermediate (N = 56), and light (N = 46) pigmentation groups using the fifth abdominal tergite (A5) pigmentation scores (0–4) from Dembeck et al. (2015a). Next, we tested whether females from dark, intermediate, or light pigmentation groups showed differences in their abundance of CHCs with different chain lengths relative to the 155 DGRP line average. We found that the group with the darkest A5 pigmentation showed lower levels of short chain CHCs and higher levels of long chain CHCs relative to the 155 line average (Figure 4A, Spearman’s ρ = 0.44, P< 0.01); the group with intermediate A5 pigmentation showed no relationship with CHC chain length (Figure 4B, Spearman’s ρ = 0.002, P= 0.98); and the group with lightest A5 pigmentation showed the opposite pattern as the dark group (Figure 4C, Spearman’s ρ = -0.57, P= 1.0 × 10-3). We also compared CHC profiles in dark (N = 51), intermediate (N = 55), and light (N = 49) groups based on pigmentation of the sixth abdominal tergite (A6), and found that, unexpectedly, the dark group did not show a significant CHC lengthening effect (Supplementary Figure S6A, Spearman’s ρ = 0.19, P= 0.25), and the intermediate group showed a CHC lengthening effect (Supplementary Figure S6B, Spearman’s ρ = 0.44, P< 0.01). However, the light group showed a significant CHC shortening effect as expected (Supplementary Figure S6C, Spearman’s ρ = -0.68, P< 1.0 × 10-5). These data suggest that darkly pigmented DGRP females show a pattern of CHC lengthening similar to the darkly pigmented loss-of-function ebonyCRISPR(1,2) flies, and lightly pigmented DGRP females show a pattern of CHC shortening similar to lightly pigmented loss-of-function tan20A flies.
ebony and tan Expression Covaries With CHC Length Profiles in the DGRP
The DGRP genome-wide association (GWAS) study from Dembeck et al. (2015a) revealed that top variants associated with pigmentation are in ebony, tan, and bab1, consistent with variation in ebony expression level observed in the DGRP lines (Miyagi et al., 2015) and associations between pigmentation and these genes in studies of other D. melanogaster populations (Rebeiz et al., 2009a,b; Takahashi and Takano-Shimizu, 2011; Telonis-Scott et al., 2011; Bastide et al., 2013; Endler et al., 2016, 2018). We therefore hypothesized that the differences in CHC length profiles seen in darkly and lightly pigmented DGRP females might be a consequence of expression variation at ebony and/or tan.
Using qRT-PCR, we quantified ebony and tan expression within 1 h after eclosion, which is when pigments determining adult body color are actively produced, in a sample of 23 DGRP lines that showed variable pigmentation. We then tested whether variation in ebony and tan expression covaried with CHC length profiles by categorizing the 23 DGRP lines into groups of low, intermediate, and high ebony or tan expression levels based on the qRT-PCR results, examining the average difference in individual CHC abundances between each expression group relative to the 23 line average, and plotting these values against CHC chain length (Figure 5).
FIGURE 5.
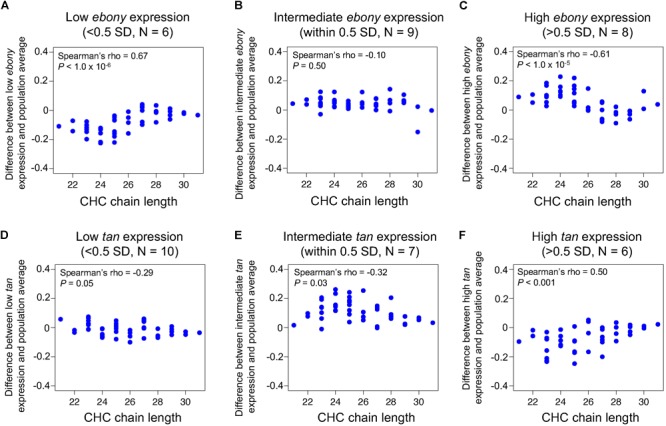
Variation in ebony and tan expression co-varies with CHC length profiles in the DGRP. CHC data was obtained from Dembeck et al. (2015b), and ebony and tan expression was quantified via qRT-PCR for 23 DGRP lines. (A) Difference in log-contrast of relative CHC intensity between DGRP females with low ebony expression and the 23 line average. (B) Difference in log-contrast of relative CHC intensity between DGRP females with intermediate ebony expression and the 23 line average. (C) Difference in log-contrast of relative CHC intensity between DGRP females with low ebony expression and the 23 line average. (D) Difference in log-contrast of relative CHC intensity between DGRP females with low tan expression and the 23 line average. (E) Difference in log-contrast of relative CHC intensity between DGRP females with intermediate tan expression and the 23 line average. (F) Difference in log-contrast of relative CHC intensity between DGRP females with high tan expression and the 23 line average.
Consistent with our hypothesis, the DGRP lines with low ebony expression showed lower levels of short chain CHCs, lines with high ebony expression showed higher levels of short chain CHCs, and lines with intermediate expression showed no change in CHC profiles (Figure 5A–C, Spearman’s ρ = 0.67, P < 1.0 × 10-6, Spearman’s ρ = -0.61, P< 1.0 × 10-5, Spearman’s ρ = -0.10, P = 0.50, respectively). Reciprocally, the DGRP lines with low or intermediate tan expression showed a slight increase in short chain CHCs, and lines with high tan expression showed a significant decrease in short chain CHCs (Figure 5D–F, Spearman’s ρ = -0.29, P = 0.05, Spearman’s ρ = -0.32, P = 0.03, Spearman’s ρ = 0.50, P < 0.001, respectively). Taken together, our results suggest that differences in ebony and tan gene expression have pleiotropic effects on both pigmentation and CHC length profiles that might cause these traits to covary in natural D. melanogaster populations.
Discussion
Pigmentation genes are often pleiotropic, with effects on vision, circadian rhythms, immunity, and mating behavior (reviewed in Wittkopp and Beldade, 2009; Takahashi, 2013). Here, we show that ebony and tan also affect CHC production, with the two genes altering CHC length profiles in opposing directions: ebonyCRISPR(1,2) mutants had significantly higher levels of long chain CHCs, and tan20 mutants had significantly higher levels of short chain CHCs. Our results suggest (1) that ebony and tan have a previously undescribed role in CHC synthesis and/or deposition and (2) that pleiotropy of both genes might influence the covariation of pigmentation and CHC composition.
Considering the Pleiotropic Effects of ebony and tan Through Changes in Dopamine Metabolism
Previous work has shown that changes in dopamine metabolism influence CHC composition in Drosophila melanogaster. Specifically, females homozygous for loss-of-function Dopa-decarboxylase (Ddc) temperature-sensitive alleles showed changes in CHC composition that could be reversed with dopamine feeding (Marican et al., 2004; Wicker-Thomas and Hamann, 2008). Additionally, inhibiting dopamine synthesis by feeding wild-type females the tyrosine hydroxylase inhibitor L-AMPT altered CHC composition in a similar direction as the loss-of-function alleles (Marican et al., 2004; Wicker-Thomas and Hamann, 2008). We found that feeding with L-AMPT affects CHC length composition, causing ebonyCRISPR(1,2) and ebonyCRISPR(3) mutants to have a more tan20-like CHC length profile (Figure 2A and Supplementary Figure S4). This result suggests that ebony and tan may affect CHC length composition through dopamine metabolism, but feeding tan20 and wild-type females dopamine did not lead to CHC lengthening (Figure 2B,C and Supplementary Figure S5). Why did L-AMPT feeding affect CHC length composition while dopamine feeding did not? One possible reason is that L-AMPT is a potent inhibitor of tyrosine hydroxylase activity (Spector et al., 1965), which processes tyrosine that flies ingest, whereas dopamine feeding might not cause significant changes in dopamine abundance in tissues relevant to CHC synthesis.
Another gene suggesting a possible link between CHC composition and dopamine is the D. melanogaster apterous gene. Loss of apterous gene function causes an increase in the proportion of long chain CHCs (Wicker and Jallon, 1995), and apterous mutants also show high levels of dopamine (Gruntenko et al., 2003, 2005, 2012). These mutants also show low levels of juvenile hormone (JH) (Altaratz et al., 1991), and treating decapitated females with methoprene to increase JH synthesis caused a decrease in long chain CHCs (Wicker and Jallon, 1995). The CHC lengthening and increased dopamine levels seen in apterous mutants resemble ebony mutants, but it is unknown whether ebony mutants show altered JH profiles. Further evidence supporting a role of JH and other ecdysteroids in determining CHC chain length comes from houseflies (Blomquist et al., 1987). In D. melanogaster, ecdysteroid signaling was found to be required not only for CHC synthesis but also survival of the oenocyte cells that synthesize CHCs (Chiang et al., 2016). An interesting future direction would be to test whether changes in dopamine metabolism in ebony or tan mutants influence CHC length composition through JH signaling. More broadly, a thorough genetic analysis focused on tissue-specific manipulation of dopamine is needed to deepen our understanding about its role in CHC synthesis.
CHC Lengthening in ebony Mutants Does Not Seem to Depend on Changes at the Level of the Cuticle
Data from our tyrosine hydroxylase inhibition experiments supported the hypothesis that elevated dopamine levels in ebony mutants (as reported in Hodgetts and Konopka, 1973) affect CHC lengthening; however, it remains unclear which cells require ebony expression (and possibly dopamine metabolism) to influence CHC synthesis. We hypothesized that ebony-dependent changes of the fly cuticle itself might affect CHC deposition during fly development or CHC extraction in the laboratory, and found that all but one detected CHC showed an overall decrease in abundance in dark cuticle relative to light cuticle. We note that these differences might be due to changes in the physical properties of dark versus light cuticle as they interact with the laser of the UV-LDI instrument. We also note that ebonyCRISPR(3) and ebonyCRISPR(4) mutants had darkly pigmented cuticle like ebonyCRISPR(1,2) mutants but CHC length profiles similar to wild-type flies, suggesting that ebony and tan’s effects on CHC length composition can be separated from their role in pigmentation synthesis. For example, ebony expression in glia is necessary for normal circadian rhythms in D. melanogaster but not pigmentation (Suh and Jackson, 2007). It is also possible that ebony actually affects CHC composition through changes in pigmentation precursors within epidermal cells underneath the cuticle, which might not have been detected by our UV-LDI MS analysis in the thorax. We tested whether knocking down ebony in oenocytes in the abdomen affected CHC length composition and found that it did not, thus the specific cells required for ebony and tan’s effects on CHC synthesis remain unknown.
Patterns of CHC Composition and Pigmentation Along Clines in Natural Populations
Identifying the pleiotropic effects of ebony and tan on pigmentation and CHCs is important because it suggests that these genes might contribute to the covariation of both traits in natural populations. For example, selection for ebony- or tan-dependent pigmentation variation might also cause variation in CHC length composition without selection acting directly on this trait. Alternatively, selection for long chain CHCs with higher melting temperatures (Gibbs and Pomonis, 1995; Gibbs, 1998) in drier climates might cause a correlated increase in pigmentation intensity. Indeed, we found that variation in abdominal pigmentation covaries with both ebony and tan gene expression as well as CHC length profiles in directions predicted by ebony and tan mutants among the DGRP lines, which were derived from flies isolated from a single, natural population (Ayroles et al., 2009; Mackay et al., 2012; Huang et al., 2014). However, this finding does not necessarily imply variation in both traits is caused by the same gene(s) nor that these traits will always co-evolve; for example, individuals with dark pigmentation may coincidentally possess alleles that are in linkage disequilibrium that cause a CHC lengthening phenotype. Comparing the phenotypic frequency of pigmentation and CHC length composition phenotypes within and between the same populations that are undergoing adaptation to common environments will help answer this question. In Africa, for example, D. melanogaster populations repeatedly show a strong positive correlation between elevation and dark pigmentation, suggesting that environments at high altitudes might select for darkly pigmented flies (or some other trait that correlates with pigmentation) (Pool and Aquadro, 2007; Bastide et al., 2014). It will be interesting to know whether these populations also show an increase in abundance of long chain CHCs.
Both pigmentation and CHC length profiles vary along altitudinal and latitudinal clines in natural Drosophila populations, suggesting that ecological factors such as humidity or temperature play a role in shaping variation in at least one of these traits. At higher altitudes or latitudes, populations often showed darker pigmentation profiles in Europe, India, and Australia (Heed and Krishnamurthy, 1959; David et al., 1985; Capy et al., 1988; Das, 1995; Munjal et al., 1997; Parkash and Munjal, 1999; Pool and Aquadro, 2007; Parkash et al., 2008a,b; Telonis-Scott et al., 2011; Matute and Harris, 2013). In Africa, however, latitude and pigmentation intensity showed a negative correlation, so this relationship is not universal (Bastide et al., 2014). For CHCs, Rajpurohit et al. (2017) reported that D. melanogaster populations at higher latitudes showed more short chain CHCs, whereas populations at lower latitudes showed more long chain CHCs in the United States. Frentiu and Chenoweth (2010) similarly found that populations at high latitudes along a cline in Australia showed more short chain CHCs and fewer long chain CHCs. These patterns do not match predictions based on the pleiotropy we observed: flies at higher latitudes tend to have darker pigmentation and higher levels of short chain CHCs whereas ebonyCRISPR(1,2) mutants, for example, have darker pigmentation and lower levels of short chain CHCs. To the best of our knowledge, pigmentation (nor ebony or tan expression) and CHC length composition have not been simultaneously measured in flies from the same cline, making it difficult to discern whether pigmentation and CHC composition covary in the wild in ways predicted by the mutant data. For example, Frentiu and Chenoweth (2010) measured CHCs from populations along the east coast of Australia, but they did not include populations from higher latitude coastal regions with darker pigmentation and lower ebony expression in newly eclosed adults (Telonis-Scott et al., 2011). Comparing variation in both traits within and between populations along latitudinal and/or altitudinal clines will make it clearer if and to what extent pigmentation and CHC composition covary and whether variation in these features is accompanied by changes in ebony and tan expression.
Author Contributions
JM, NA, PW, JY, and AT conceived the project. JM, NA, TB, KD, and JY collected the data. JM, NA, TB, KD, JY, and AT analyzed the data. JM, PW, JY, and AT wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank members of the Takahashi, Wittkopp, Yew Labs, and Aki Ejima for helpful discussions; John True, Stephen Goodwin, Scott Pletcher, Rainbow Transgenics Inc., the Bloomington Drosophila Stock Center, and the Vienna Drosophila RNAi Center for fly stocks; and Rainbow Transgenics Inc., for fly injections.
Footnotes
Funding. This work was supported by a Department of Ecology and Evolutionary Biology, University of Michigan, Nancy W. Walls Research Award, National Institutes of Health training (Grant No. T32GM007544), and Howard Hughes Medical Institute Janelia Graduate Research Fellowship awarded to JM; the German Research Foundation (Grant No. DR 416/10-1) awarded to KD; National Institutes of Health (Grant No. 1R35GM118073) awarded to PW; Department of Defense, United States Army Research Office (Grant No. W911NF1610216) and National Institutes of Health (Grant No. 1P20GM125508) awarded to JY; The Sumitomo Foundation Grant for Basic Science Research Projects (Grant No. 160999) and JSPS KAKENHI (Grant No. JP19H03276) to AT.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.00518/full#supplementary-material
References
- Albert A. Y., Sawaya S., Vines T. H., Knecht A. K., Miller C. T., Summers B. R., et al. (2008). The genetics of adaptive shape shift in stickleback: pleiotropy and effect size. Evolution 62 76–85. [DOI] [PubMed] [Google Scholar]
- Altaratz M., Applebaum S. W., Richard D. S., Gilbert L. I., Segal D. (1991). Regulation of juvenile hormone synthesis in wild-type and apterous mutant Drosophila. Mol. Cell. Endocrinol. 81 205–216. 10.1016/0303-7207(91)90219-i [DOI] [PubMed] [Google Scholar]
- Ayroles J. F., Carbone M. A., Stone E. A., Jordan K. W., Lyman R. F., Magwire M. M., et al. (2009). Systems genetics of complex traits in Drosophila melanogaster. Nat. Genet. 41 299–307. 10.1038/ng.332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett A. R., Tibbit C., Ponting C. P., Liu J.-L. (2013). Highly efficient targeted mutagenesis of Drosophila with CRISPR/Cas9 system. Cell Rep. 4 220–228. 10.1016/j.celrep.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastide H., Betancourt A., Nolte V., Tobler R., Stöbe P., Futschik A., et al. (2013). A genome-wide, fine-scale map of natural pigmentation variation in Drosophila melanogaster. PLoS Genetics 9:e1003534. 10.1371/journal.pgen.1003534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastide H., Yassin A., Johanning E. J., Pool J. E. (2014). Pigmentation in Drosophila melanogaster reaches its maximum in Ethiopia and correlates most strongly with ultra-violet radiation in sub-Saharan Africa. BMC Evol. Biol. 14:179. 10.1186/s12862-014-0179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastock M. (1956). A gene mutation which changes a behavior pattern. Evolution 10 421–439. 10.1111/j.1558-5646.1956.tb02868.x [DOI] [Google Scholar]
- Blomquist G. J., Dilwith J. W., Adams T. S. (1987). “Biosynthesis and endocrine regulation of sex pheromone production in Diptera,” in Pheromone Biochemistry ed. Blomquist G. J. (Berlin: Elsevier; ) 217–250. 10.1016/b978-0-12-564485-3.50012-9 [DOI] [Google Scholar]
- Blows M. W., Allen R. A. (1998). Levels of mate recognition within and between two Drosophila species and their hybrids. Am. Nat. 152 826–837. 10.1086/286211 [DOI] [PubMed] [Google Scholar]
- Boom R., Sol C. J., Salimans M. M., Jansen C. L., Wertheim-van Dillen P. M., Van der Noordaa J. (1990). Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 401–415. [DOI] [PubMed] [Google Scholar]
- Bridges C. B., Morgan T. H. (1923). Third-Chromosome Group of Mutant Characters of Drosophila melanogaster. Washington, DC: Carnegie Institution Of Washington. [Google Scholar]
- Calleja M., Herranz H., Estella C., Casal J., Lawrence P., Simpson P., et al. (2000). Generation of medial and lateral dorsal body domains by the pannier gene of Drosophila. Development 127 3971–3980. [DOI] [PubMed] [Google Scholar]
- Capy P., David J. R., Robertson A. (1988). Thoracic trident pigmentation in natural populations of Drosophila simulans: a comparison with D. melanogaster. Heredity 61:263 10.1038/hdy.1988.114 [DOI] [Google Scholar]
- Chiang Y. N., Tan K. J., Chung H., Lavrynenko O., Shevchenko A., Yew J. Y. (2016). Steroid hormone signaling is essential for pheromone production and oenocyte survival. PLoS Genetics 12:e1006126. 10.1371/journal.pgen.1006126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A. (1995). Abdominal pigmentation in Drosophila melanogaster females from natural Indian populations. J. Zool. Syst. Evol. Res. 33 84–87. 10.1111/j.1439-0469.1995.tb00213.x [DOI] [Google Scholar]
- David J. R., Capy P., Payant V., Tsakas S. (1985). Thoracic trident pigmentation in Drosophila melanogaster: differentiation of geographical populations. Génét. Sélect. Évol. 17 211. 10.1051/gse [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembeck L. M., Böröczky K., Huang W., Schal C., Anholt R. R., Mackay T. F. (2015a). Genetic architecture of natural variation in cuticular hydrocarbon composition in Drosophila melanogaster. eLife 4:e09861. 10.7554/eLife.09861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembeck L. M., Huang W., Magwire M. M., Lawrence F., Lyman R. F., Mackay T. F. (2015b). Genetic architecture of abdominal pigmentation in Drosophila melanogaster. PLoS Genetics 11:e1005163. 10.1371/journal.pgen.1005163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., Fellner M., et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448 151–156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Drapeau M. D., Cyran S. A., Viering M. M., Geyer P. K., Long A. D. (2006). A cis-regulatory sequence within the yellow locus of Drosophila melanogaster required for normal male mating success. Genetics 172 1009–1030. 10.1534/genetics.105.045666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapeau M. D., Radovic A., Wittkopp P. J., Long A. D. (2003). A gene necessary for normal male courtship, yellow, acts downstream of fruitless in the Drosophila melanogaster larval brain. J. Neurobiol. 55 53–72. 10.1002/neu.10196 [DOI] [PubMed] [Google Scholar]
- Duveau F., Félix M. A. (2012). Role of pleiotropy in the evolution of a cryptic developmental variation in Caenorhabditis elegans. PLoS Biology 10:e1001230. 10.1371/journal.pbio.1001230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler L., Betancourt A. J., Nolte V., Schlötterer C. (2016). Reconciling differences in pool-GWAS between populations: a case study of female abdominal pigmentation in Drosophila melanogaster. Genetics 202 843–855. 10.1534/genetics.115.183376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler L., Gibert J. M., Nolte V., Schlötterer C. (2018). Pleiotropic effects of regulatory variation in tan result in correlation of two pigmentation traits in Drosophila melanogaster. Mol. Ecol. 27 3207–3218. 10.1111/mec.14781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferveur J. F., Savarit F., O’kane C. J., Sureau G., Greenspan R. J., Jallon J. M. (1997). Genetic feminization of pheromones and its behavioral consequences in Drosophila males. Science 276 1555–1558. 10.1126/science.276.5318.1555 [DOI] [PubMed] [Google Scholar]
- Flaven-Pouchon J., Farine J.-P., Ewer J., Ferveur J.-F. (2016). Regulation of cuticular hydrocarbon profile maturation by Drosophila tanning hormone, bursicon, and its interaction with desaturase activity. Insect Biochem. Mol. Biol. 79 87–96. 10.1016/j.ibmb.2016.10.007 [DOI] [PubMed] [Google Scholar]
- Foley B. R., Telonis-Scott M. (2011). Quantitative genetic analysis suggests causal association between cuticular hydrocarbon composition and desiccation survival in Drosophila melanogaster. Heredity 106 68. 10.1038/hdy.2010.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentiu F. D., Chenoweth S. F. (2010). Clines in cuticular hydrocarbons in two Drosophila species with independent population histories. Evolution 64 1784–1794 10.1111/j.1558-5646.2009.00936.x [DOI] [PubMed] [Google Scholar]
- Gibbs A., Pomonis J. G. (1995). Physical properties of insect cuticular hydrocarbons: the effects of chain length, methyl-branching and unsaturation. Comp. Biochem. Physiol. B 112 243–249. 10.1016/0305-0491(95)00081-x [DOI] [Google Scholar]
- Gibbs A. G. (1998). Water-proofing properties of cuticular lipids. Am. Zool. 38 471–482. 10.1371/journal.pgen.1005708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs A. G., Chippindale A. K., Rose M. R. (1997). Physiological mechanisms of evolved desiccation resistance in Drosophila melanogaster. J. Exp. Biol. 200 1821–1832. [DOI] [PubMed] [Google Scholar]
- Gratz S. J., Ukken F. P., Rubinstein C. D., Thiede G., Donohue L. K., Cummings A. M., et al. (2014). Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196 961–971. 10.1534/genetics.113.160713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood A. K., Mills M. G., Wark A. R., Archambeault S. L., Peichel C. L. (2016). Evolution of schooling behavior in threespine sticklebacks is shaped by the Eda gene. Genetics 203 677–681. 10.1534/genetics.116.188342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruntenko N. E., Chentsova N. A., Andreenkova E. V., Bownes M., Segal D., Adonyeva N. V., et al. (2003). Stress response in a juvenile hormone-deficient Drosophila melanogaster mutant apterous. Insect Mol. Biol. 12 353–363. 10.1046/j.1365-2583.2003.00419.x [DOI] [PubMed] [Google Scholar]
- Gruntenko N. E., Karpova E. K., Alekseev A. A., Chentsova N. A., Saprykina Z. V., Bownes M., et al. (2005). Effects of dopamine on juvenile hormone metabolism and fitness in Drosophila virilis. J. Insect Physiol. 51 959–968. 10.1016/j.jinsphys.2005.04.010 [DOI] [PubMed] [Google Scholar]
- Gruntenko N. Å, Laukhina O. V., Bogomolova E. V., Karpova E. K., Menshanov P. N., Romanova I. V.et al. (2012). Downregulation of the dopamine D2-like receptor in corpus allatum affects juvenile hormone synthesis in Drosophila melanogaster females. J. Insect Physiol. 58 348–355. 10.1016/j.jinsphys.2011.12.006 [DOI] [PubMed] [Google Scholar]
- Heed W. B., Krishnamurthy N. B. (1959). Genetic studies on the cardini group of Drosophila in the West Indies. Univ. Texas Publ. 5914 155–179. [Google Scholar]
- Hodgetts R. B., Konopka R. J. (1973). Tyrosine and catecholamine metabolism in wild-type Drosophila melanogaster and a mutant, ebony. J. Insect Physiol. 19 1211–1220. 10.1016/0022-1910(73)90205-9 [DOI] [PubMed] [Google Scholar]
- Hovemann B. T., Ryseck R. P., Walldorf U., Störtkuhl K. F., Dietzel I. D., Dessen E. (1998). The Drosophila ebony gene is closely related to microbial peptide synthetases and shows specific cuticle and nervous system expression. Gene 221 1–9. 10.1016/s0378-1119(98)00440-5 [DOI] [PubMed] [Google Scholar]
- Huang W., Massouras A., Inoue Y., Peiffer J., Ràmia M., Tarone A. M., et al. (2014). Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 24 1193–1208. 10.1101/gr.171546.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., Zhu D., et al. (2012). The Drosophila melanogaster genetic reference panel. Nature 482 173–178. 10.1038/nature10811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marican C., Duportets L., Birman S., Jallon J. M. (2004). Female-specific regulation of cuticular hydrocarbon biosynthesis by dopamine in Drosophila melanogaster. Insect Biochem. Mol. Biol. 34 823–830. 10.1016/j.ibmb.2004.05.002 [DOI] [PubMed] [Google Scholar]
- Matute D. R., Harris A. (2013). The influence of abdominal pigmentation on desiccation and ultraviolet resistance in two species of Drosophila. Evolution 67 2451–2460. 10.1111/evo.12122 [DOI] [PubMed] [Google Scholar]
- McKay J. K., Richards J. H., Mitchell-Olds T. (2003). Genetics of drought adaptation in Arabidopsis thaliana: I. Pleiotropy contributes to genetic correlations among ecological traits. Mol. Ecol. 12 1137–1151. 10.1046/j.1365-294x.2003.01833.x [DOI] [PubMed] [Google Scholar]
- McLean C. Y., Reno P. L., Pollen A. A., Bassan A. I., Capellini T. D., Guenther C., et al. (2011). Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature 471:216. 10.1038/nature09774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi R., Akiyama N., Osada N., Takahashi A. (2015). Complex patterns of cis-regulatory polymorphisms in ebony underlie standing pigmentation variation in Drosophila melanogaster. Mol. Ecol. 24 5829–5841. 10.1111/mec.13432 [DOI] [PubMed] [Google Scholar]
- Munjal A. K., Karan D., Gibert P., Moreteau B., Parkash R., David J. R. (1997). Thoracic trident pigmentation in Drosophila melanogaster: latitudinal and altitudinal clines in Indian populations. Genet. Select. Evol. 29:601 10.1186/1297-9686-29-5-601 [DOI] [Google Scholar]
- Nagy O., Nuez I., Savisaar R., Peluffo A. E., Yassin A., Lang M., et al. (2018). Correlated evolution of two copulatory organs via a single cis-regulatory nucleotide change. Curr. Biol. 28 3450–3457. 10.1016/j.cub.2018.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkash R., Munjal A. K. (1999). Phenotypic variability of thoracic pigmentation in Indian populations of Drosophila melanogaster. J. Zool. Syst. Evol. Res. 37 133–140. 10.1111/j.1439-0469.1999.00112.x [DOI] [Google Scholar]
- Parkash R., Rajpurohit S., Ramniwas S. (2008a). Changes in body melanisation and desiccation resistance in highland vs. lowland populations of D. melanogaster. J. Insect Physiol. 54 1050–1056. 10.1016/j.jinsphys.2008.04.008 [DOI] [PubMed] [Google Scholar]
- Parkash R., Sharma V., Kalra B. (2008b). Climatic adaptations of body melanisation in Drosophila melanogaster from Western Himalayas. Fly 2 111–117. 10.4161/fly.6351 [DOI] [PubMed] [Google Scholar]
- Pool J. E., Aquadro C. F. (2007). The genetic basis of adaptive pigmentation variation in Drosophila melanogaster. Mol. Ecol. 16 2844–2851. 10.1111/j.1365-294x.2007.03324.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2013). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Rajpurohit S., Hanus R., Vrkoslav V., Behrman E. L., Bergland A. O., Petrov D., et al. (2017). Adaptive dynamics of cuticular hydrocarbons in Drosophila. J. Evol. Biol. 30 66–80. 10.1111/jeb.12988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M., Pool J. E., Kassner V. A., Aquadro C. F., Carroll S. B. (2009a). Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science 326 1663–1667. 10.1126/science.1178357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M., Ramos-Womack M., Jeong S., Andolfatto P., Werner T., True J., et al. (2009b). Evolution of the tan locus contributed to pigment loss in Drosophila santomea: a response to Matute et al. Cell 139 1189–1196. 10.1016/j.cell.2009.11.004 [DOI] [PubMed] [Google Scholar]
- Ren X., Yang Z., Xu J., Sun J., Mao D., Hu Y., et al. (2014). Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep. 9 1151–1162. 10.1016/j.celrep.2014.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout E. J., Dornan A. J., Neville M. C., Eadie S., Goodwin S. F. (2010). Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat. Neurosci. 13:458. 10.1038/nn.2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel F., Vorkel D., Eaton S. (2011). Megalin-dependent yellow endocytosis restricts melanization in the Drosophila cuticle. Development 138 149–158. 10.1242/dev.056309 [DOI] [PubMed] [Google Scholar]
- Schluter D., Clifford E. A., Nemethy M., McKinnon J. S. (2004). Parallel evolution and inheritance of quantitative traits. Am. Nat. 163 809–822. 10.1086/383621 [DOI] [PubMed] [Google Scholar]
- Spector S., Sjoerdsma A., Udenfriend S. (1965). Blockade of endogenous norepinephrine synthesis by α-methyl-tyrosine, an inhibitor of tyrosine hydroxylase. J. Pharmacol. Exp. Ther. 147 86–95. [PubMed] [Google Scholar]
- Suh J., Jackson F. R. (2007). Drosophila ebony activity is required in glia for the circadian regulation of locomotor activity. Neuron 55 435–447. 10.1016/j.neuron.2007.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A. (2013). Pigmentation and behavior: potential association through pleiotropic genes in Drosophila. Genes Genet. Syst. 88 165–174. 10.1266/ggs.88.165 [DOI] [PubMed] [Google Scholar]
- Takahashi A., Takano-Shimizu T. (2011). Divergent enhancer haplotype of ebony on inversion In(3R)Payne associated with pigmentation variation in a tropical population of Drosophila melanogaster. Mol. Ecol. 20 4277–4287. 10.1111/j.1365-294X.2011.05260.x [DOI] [PubMed] [Google Scholar]
- Telonis-Scott M., Hoffmann A. A., Sgro C. M. (2011). The molecular genetics of clinal variation: a case study of ebony and thoracic trident pigmentation in Drosophila melanogaster from eastern Australia. Mol. Ecol. 20 2100–2110. 10.1111/j.1365-294X.2011.05089.x [DOI] [PubMed] [Google Scholar]
- True J. R. (2003). Insect melanism: the molecules matter. Trends Ecol. Evol. 18 640–647. 10.1016/j.tree.2003.09.006 [DOI] [Google Scholar]
- True J. R., Yeh S.-D., Hovemann B. T., Kemme T., Meinertzhagen I. A., Edwards T. N., et al. (2005). Drosophila tan encodes a novel hydrolase required in pigmentation and vision. PLoS Genetics 1:e63. 10.1371/journal.pgen.0010063.eor [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. A., Bell M. A. (2000). Net evolutionary trajectories of body shape evolution within a microgeographic radiation of threespine sticklebacks (Gasterosteus aculeatus). J. Zool. 252 293–302. 10.1017/s0952836900000030 [DOI] [Google Scholar]
- Wark A. R., Greenwood A. K., Taylor E. M., Yoshida K., Peichel C. L. (2011). Heritable differences in schooling behavior among threespine sticklebacks revealed by a novel assay. PLoS One 6:e18316. 10.1371/journal.pone.0018316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker C., Jallon J. M. (1995). Hormonal control of sex pheromone biosynthesis in Drosophila melanogaster. J. Insect Physiol. 41 65–70. 10.1016/0022-1910(94)00074-q [DOI] [Google Scholar]
- Wicker-Thomas C., Hamann M. (2008). Interaction of dopamine, female pheromones, locomotion and sex behavior in Drosophila melanogaster. J. Insect Physiol. 54 1423–1431. 10.1016/j.jinsphys.2008.08.005 [DOI] [PubMed] [Google Scholar]
- Wigglesworth V. B. (1970). Structural lipids in the insect cuticle and the function of the oenocytes. Tissue Cell 2 155–179. [DOI] [PubMed] [Google Scholar]
- Wittkopp P. J., Beldade P. (2009). Development and evolution of insect pigmentation: genetic mechanisms and the potential consequences of pleiotropy. Semin. Cell Dev. Biol. 20 65–71. 10.1016/j.semcdb.2008.10.002 [DOI] [PubMed] [Google Scholar]
- Yew J. Y., Chung H. (2015). Insect pheromones: An overview of function, form, and discovery. Prog. Lipid Res. 59 88–105. 10.1016/j.plipres.2015.06.001 [DOI] [PubMed] [Google Scholar]
- Yew J. Y., Dreisewerd K., De Oliveira C. C., Etges W. J. (2011). Male-specific transfer and fine scale spatial differences of newly identified cuticular hydrocarbons and triacylglycerides in a Drosophila species pair. PLoS One 6:e16898. 10.1371/journal.pone.0016898 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



