Abstract
We conduct a cartography of rhodopsin-like non-olfactory G protein-coupled receptors in the Ensembl database. The most recent genomic data (releases 90–92, 90 vertebrate genomes) are analyzed through the online interface and receptors mapped on phylogenetic guide trees that were constructed based on a set of ~14.000 amino acid sequences. This snapshot of genomic data suggest vertebrate genomes to harbour 142 clades of GPCRs without human orthologues. Among those, 69 have not to our knowledge been mentioned or studied previously in the literature, of which 28 are distant from existing receptors and likely new orphans. These newly identified receptors are candidates for more focused evolutionary studies such as chromosomal mapping as well for in-depth pharmacological characterization. Interestingly, we also show that 37 of the 72 human orphan (or recently deorphanized) receptors included in this study cluster into nineteen closely related groups, which implies that there are less ligands to be identified than previously anticipated. Altogether, this work has significant implications when discussing nomenclature issues for GPCRs.
Subject terms: G protein-coupled receptors, Data mining
Introduction
G protein-coupled receptors (GPCRs) are signalling proteins activated by for example neurotransmitters and neuromodulators and as such are involved in many physiological processes. Their location as a cellular gateway makes them key targets for drug discovery and chemical biology1–3, and 30–50% of drugs on the market have been reported to target GPCRs directly or indirectly4.
GPCRs are well known for sharing a three-dimensional architecture characterized by seven transmembrane α-helical segments5,6 connected by loops. In the rhodopsin family, a well conserved disulphide bridge often connects the second extracellular loop and the third transmembrane segment. This architecture is reflected at the sequence level by conserved sets of amino acids that serve structurally or as determinants of signal transduction, e.g. to name a few the E/DRY, CWxP, NPxxY motifs in the transmembrane segments 3, 6 and 7 of the rhodopsin family7,8. Additional family-specific motifs have been identified for examples in connecting loops9. Within each transmembrane segment, the most conserved amino acid will be referred to here as pivots (pivot amino acids are not always fully conserved: N1.50, 98% conservation; D2.50, 90%; R3.50, 95%; W4.50, 97%; P5.50, 78%; P6.50, 99%; P7.50, 88% see7). These amino acids are used as a basis of the widely used Ballesteros-Weinstein numbering10, where equivalent positions are reported by transmembrane segment number followed by relative distance to pivot, itself assigned the index 50.
The International Union of Basic and Clinical Pharmacology (IUPHAR) is responsible for the international classification of GPCRs and issues regular recommendations. GPCRs have been divided into five main families based on phylogenetic analyses11–13. The largest, Rhodopsin, in human ~700 members, subdivides into four branches (α, β, γ and δ) and 13 sub-branches11 and has originated through local duplications about 1400–1100 million years ago14. Families are further divided into subtypes15 that arouse, for many, 350–500 million years ago from two rounds of whole genome duplication (2R)16–22. In ray-finned fish (not in lobe-finned or jawless fishes), a third specific genome duplication took place about ~250 million years ago, leading to fish-specific duplicates23–25. In tunicates, GPCRs originated before the 2R and after the separation of invertebrates from chordates24–26. In lamprey, it remains unclear whether the cyclostome-gnathostome split at the origin of jawed vertebrates happened before or after the second round of genome duplication27–29.
As consequence of the origin of vertebrate GPCRs through the 2R/3R, in an ideal scenario, for each ancestral receptor we expect four/eight paralogues (that can be referred to as “ohnologues”16, many ohnologues are simply equivalent to subtypes). Nonetheless, local duplications and deletions (pseudogenes)30 may make the current day picture complex and, consequently, the expect four/eight ohnologues are most often not seen18,30. Furthermore, ligand binding preferences do not necessarily indicates the closest possible evolutionary relationships; some receptors have acquired the same ligand specificity several times, for example α1-, α2-, and β- adrenoceptors that are parts of the amine family31,32, or e.g. the cannabinoid receptors CB1 and CB2, and the recently deorphanized GPR55 that are activated by the same endogenous ligand 2-Arachidonoylglyserol33.
GPCRs have been identified and characterized in the 70’s and cloned in the 80’s, e.g. the well-studied rhodopsin34 and β2-adrenergic receptor35. The apparition of genomics in the 2000’s has brought an explosion in the amount of discovered GPCRs, yet GPCRs are in most cases named according to their human orthologues26,36–38. This leads to complex nomenclature issues especially when new clades without human orthologues are identified39–41. In addition, the repertoires in non-human vertebrate species are much less explored, and new subtypes or orphan receptors are often not annotated at all. The only way to date to tackle the issue of GPCRs in non-vertebrate species is the tedious manual annotation based on phylogenetic information.
Here, we conducted this manual annotation based on the online Ensembl database. The transcripts predicted in Ensembl are based on automated alignments matched to curated homologues sequences or ESTs. Ensembl gene trees are based on a consensus of five tree reconstruction methods: a maximum likelihood based on two types of distances and a neighbor-joining tree based on three types of distances. This annotation is a first step towards an experimental classification that would include cloning, pharmacological characterization, as well as a chromosomal analysis.
Results and Discussion
Overview
All data used for this study are available online and can be easily accessed using the codes provided (see the Experimental section).
This study was conducted based on the Ensembl genomic data42,43 twice with a five-year interval: 2012–2013, Ensembl release 67 (referred to as Ensembl.R67); and 2017–2018, Ensembl.R90-Ensembl.R.92. The text is organized according to the Ensembl.R91 trees (December 2017, available as an archive, see Experimental section). Some of the groupings may slightly differ in future releases. Ensembl.R92 (April.2018) became available during the final preparation of this manuscript and was used to solve a few ambiguities, as mentioned in text. The proposed new gene symbols were submitted to HUGO (Human Genome Nomenclature Committee) for initial consideration; for this purpose the suggested genes symbols were reevaluated against the Ensembl.R.94 (December 2018) and Ensembl.R.95 (February 2019) releases. They will be submitted to the other relevant committees on publication (MGNC, Mouse Genomic Nomenclature Committee; RGNC, Rat Genome and Nomenclature Committee, ZNC, Zebrafish Nomenclature Committee; XGC, Xenopus Gene Nomenclature Committee; CGNC, Chicken Gene Nomenclature Consortium). Therefore all gene symbols presented in this manuscript should be regraded as tentative until approved by the relevant committees.
Altogether, in Ensembl.R92 we identified 142 clusters of genes corresponding to non-olfactory rhodopsin-like GPCRs without human orthologues (Tables 1, 2), of which 69 have not been to our best knowledge previously described. Twenty-eight are distant from any group of receptors and likely orphan, and the others probable subtypes of existing receptors (Table 2). Not surprisingly, most of the new receptors are present in ray-finned fishes and only a few in placental mammals (Table 1). Nonetheless, most (23) of the new orphan receptors are found in two or more species clades (considering separately ray-finned and lobe-finned fishes, amphibians, birds, reptiles, monotreme, marsupial and placental mammalians). In addition, families that are evolutionary more recent and faster evolving such as purine (PUR) and chemokine (CHEM) contains a larger number of previously unidentified receptors (Table 2). We furthermore identify groups of ambiguous branching that need to wait for more and/or increased quality sequence data to be characterized (not annotated, see text below).
Table 1.
Species-specific counts of non-olfactory GPCRs clades without human orthologues in Ensembl.R91.
| Representative species | Lamprey | Fishes (Actinopterygii) | Fishes (Sarcopterygii) | Amphibian | Reptiles | Birds | Mammals | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Petromyzon marinus | Cypriniformes (Danio rerio); Characiformes (Astyanax mexicanus) | Lepisosteus oculatus + Takifugu rubripes and 7 others | Latimeria chalumnae | Xenopus tropicalis | Anolis carolinensis; Pelodiscus sinensis | Gallus gallus +4 others | Monotreme | Marsupial | Placental | |
| Whole genome duplication | 1R/2R? | 3R | 3R, except Lepisosteus oculatus 2R | 2R | 2R | 2R | 2R | 2R | 2R | 2R |
| Pictogram |
|
|

|

|

|

|

|

|

|

|
| Number of genomes | 1 | 2 | 9 | 1 | 1 | 2 | 5 | 1 | 3 | 65 |
| Paralogous GPCR clades identified | 22 | 118 | 134 | 95 | 69 | 74 | 52 | 16 | 19 | 11 |
Table 2.
Family-specific counts of non-olfactory GPCRs clades without human orthologues in Ensembl.R91.
| AMIN | MECA | PTGER | MLT | OPN | PEP | CHEM | SOG & MCH | LGR | MRG | PURIN | Others | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subtypes | 13 | 5 | 1 | 20 | 3 | 24 | 12 | 13 | 2 | 3 | 20 | 7 | 123 |
| Orphans | 2 | 1 | 1 | 0 | 0 | 1 | 5 | 2 | 0 | 0 | 10 | 2 | 24 |
| Total orphan + subtypes | 15 | 7 | 2 | 20 | 3 | 25 | 17 | 15 | 2 | 3 | 30 | 9 | 147 |
| Including, novel | 6 | 3 | 1 | 1 | 0 | 9 | 7 | 3 | 0 | 0 | 27 | 8 | 66 |
There are few aspects to keep in mind regarding manual annotations. First, we do not have enough manuscript space to detail the annotation of each of the 142 new receptor clades. Two examples are given as Supporting information. The annotation process is in our opinion straightforward enough to be reproducible by the reader using the online data. Secondly, this is not a full characterization of the novel receptors, which would require pharmacological studies and chromosomal organization studies. Thirdly, sometimes the branching alone do not allow a distinction between subtypes or new orphan receptor clades. Fourthly, small discrepancies may exist between the guide trees and the text of this article, essentially because the guide trees are older (R.67). We encourage the readers to look at the most recent data and build an informed opinion.
Amine and trace amine receptors (AMIN)
In the AMIN family (Fig. 1a,b) we suggest one new orphan receptor and four new subtypes that to our best knowledge have not been previously reported. This family was among the first discovered and its evolution has been comprehensively studied (see e.g.44), in particular, for dopamine receptors40,45.
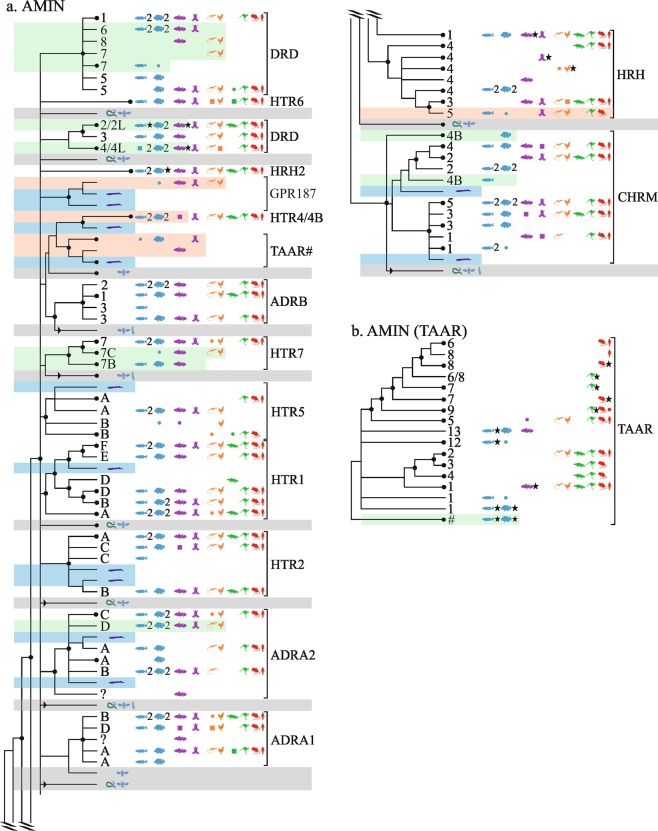
Figure 1.
Overview of vertebrate GPCR clades mapped onto neighbor-joining trees of the α-branch (1/2). AMIN (a,b). Some branches are collapsed together. Species symbol, see Table 1. Squares, genes mistakenly removed during the curation process; circles, sequences of a representative species added in Ensembl.R90-R91 releases. Internal duplications (stars), presence of 3R duplicates (number “2”). Isolated non-assigned sequences (“?”) or genes in Ensembl.R67 that are not found in further releases (“Δ”). Bootstrap values above 65% (black circles). Highlights: new receptors that have (green) or have not (light red) been to our knowledge previously reported; presence of putative ancestral sequences from lamprey (light blue) or early vertebrates or invertebrates (light grey). Some branches have been collated together.
Gene tree ADRA2A,2B, 2C; ADRB1,2,3; DRD1,5; DRD2,3,4; HTR2A,2B,2C; HTR6; HRH2
In the dopamine D2,3,4 receptor family, there are two new fish-specific clades found in coelacanth and ray-finned fishes: see ENSDARP00000127653, named here D2L (gene: DRD2L)(not to be confused with the long splice variant of D2); and see ENSAMXP00000016690, named D4L. Previous work characterized the expression in zebrafish of a single D3 and three D2 receptors46, whereas this study suggest seven zebrafish receptors in the D2,3,4 family: D2a, D2b, D2L, D3, D4a, D4b, and D4L. A previously cloned and pharmacologically characterized α2-adrenoceptor D subtype (ADRA2D) subtype was also found9,47. Near the histamine receptor H2 (HRH2), rooted by two lampreys, a gene cluster from gar/coelacanth/amphibian/sauropsids(7), see ENSPMAP00000006842, was named GPR187.
ADRA1A,1B,1C; HTR1A,1B,1C,1D,1E; HTR5,7
We identified at least two unannotated 5-hydroxytryptamine subtypes: 5-HT7B (HTR7B) found in gar/fishes(10)/coelacanth, see ENSLACP00000008875, that has likely been cloned in zebrafish48; and a set of genes in gar/coelacanth/sauropsids(7), see ENSLACP00000011078, that we name 5-HT7C (HTR7C). A cluster of five genes from gar/coelacanth/birds(3), see ENSLOCP00000003684, may be orthologues of the mammalian 5-HT5B. The 5-HT5B is pseudogenic in humans, but well characterized in mice49.
HRH1,3,4; CHRM1,2,3,4,5
In the histamine H1,3,4 subtree, a set of genes from gar/fishes(2)/amphibian/sauropsids(7)/mammals(4, 2 marsupials), see ENSLOCP00000006664, was named H5 (HRH5). Near the muscarinic cholinergic receptors 4 and 2, a group of ray-finned fish genes (8), see ENSDARP00000128513, was named CHRM4B; these muscarinic receptors have been cloned from zebrafish50.
HTR4; TAAR
Equally distant from 5-HT4 and trace amine (TAAR) receptors, a monophyletic group containing fishes(9)/coelacanth/gar/amphibian, see ENSLACP00000014144, was left unnanotated. Near the 5-HT4 subtree, a set containing gar/fishes(10)/coelacanth genes, see ENSLACP00000004821, was named 5-HT4B (gene HTR4B; not orthologous to human 5-HT4 since this later has spotted gar). The TAAR have been studied in mouse, rat, human and chimpanzee51 and fishes, where their repertoire is substantially larger than in human52,53. A set of ray-finned fish genes, see ENSLOCP00000022119, may belong to a previously unannotated subtype.
Melanocortin/EDG/Cannabinoid/Adenosine (MECA) receptors
In the MECA family (Fig. 2a) we identified one new orphans, a new subtype, and a fish-specific receptor clade.
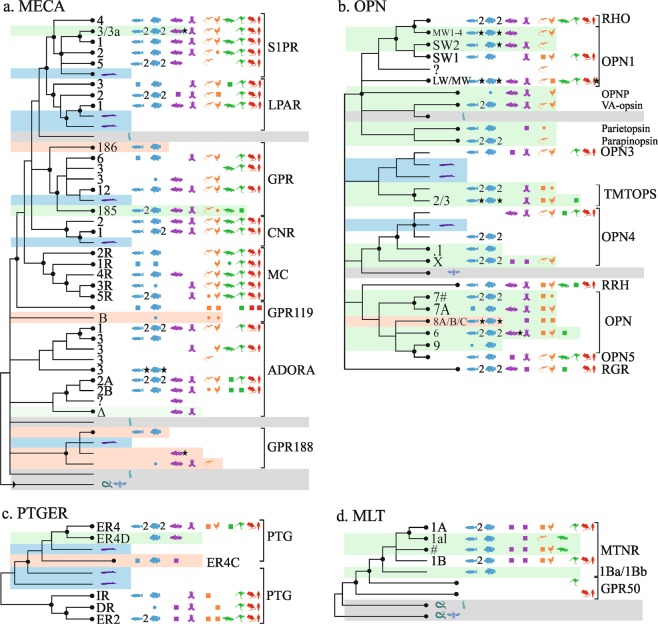
Figure 2.
Overview of vertebrate GPCR clades mapped onto neighbor-joining trees of the α-branch (2/2). MECA (a); OPN (b); PTGER (c); MLT (d). Caption otherwise similar to Fig. 1.
ADORA1,2,3
Two sets of genes are likely subtypes of a new orphan receptor, GPR188, rooted by lamprey and ascidian sequences, see ENSCINP00000025465. The first set contains gar/coelacanth(duplicated)/fishes(9), see ENSLOCP00000022091; and the second includes fishes(2)/coelacanth/amphibian/reptiles(2), see ENSGMOP00000016757. The duplicated coelacanth sequences open the possibility of a third subtype, but not conclusively.
MC1,2,3,4,5R; GPR119
The orphan receptor GPR119 has a potential subtype named GPR119B, found in gar/sauropsids(5), see ENSLOCP00000002170.
S1PR1,2,3,4; LPAR1,2,3; GPR3,6,12; CNR1,2
A cluster of ray-finned fishes (9) genes suggest a new receptor close to GPR3, see ENSAMXP00000025994. Nearby, another set has genes of gar/fishes(9)/coelacanth/amphibian/sauropsids(3)/marsupial(1)/monotreme(1), see ENSLOCP00000021577, and corresponds to GPR185; this receptor has been cloned in Xenopus laevis (named also GPRx54). Two novel sphingosine-1-phosphate receptor subtypes have been previously reported in zebrafish55, which should correspond here to a set of fishes(17)/coelacanth/reptile(1) and to S1PR3a in fishes(10)/coelacanth.
Opsin (OPN) receptors
In the OPN family (Fig. 2b), one new fish-specific opsin was identified. OPN has been extensively studied, in particular cone visual pigments56, UV-sensitive photoreceptors57, and melanopsins58. Zebrafish has 10 classical visual photo pigments and 32 non-visual opsins57,59.
OPN1MW/LW/SW; OPN3/4/5; RHO; RRH
Near OPN8a, receptors from a set found in gar/fishes(7), see ENSLOCP00000020544, were named OPN8b. They come in addition to the OPN4–9 in zebrafish59.
Prostaglandin (PTGER) receptors
In the PTGER family, one new subtype was identified (Fig. 2c).
PTGER1,2,3,4; PTGDR; PTGIR; PTGFR; TBXA2R
Near PTGER4, a group of fishes(7)/coelacanth/spotted gar, see ENSLACP00000020254, is a probable new subtype, PTGER4D that has been previously characterized in zebrafish60. In addition, a group of gar/fishes(11)/coelacanth genes, rooted by lamprey, see ENSAMXP00000018684, suggests a new subtype PTGER4C.
Melatonin (MLT) receptors
In the melatonin tree, no new receptors could be identified (Fig. 2d).
MLT, GPR50
Ambiguities in this tree were lifted in Ensembl.R92. A complete set containing gar/fishes(10)/coelacanth/amphibian(duplicated)/sauropsids(7)/platypus, see ENSLOCP00000018152, clusters with GPR50 (data from R.95). Another set includes lamprey/gar/fishes(9)/amphibian/sauropsids(2)/platypus, see ENSLOCP00000014362, was named MTNR1al. A third set of fishes (4), see ENSDARP00000070419, was named MTNR1Ba/1Bb. Three MLT subtypes have been previously cloned in zebrafish61.
Peptide (PEP) receptors
In the PEP family (Fig. 3a–c), we identified ten vertebrate receptors without human orthologues, one of which is likely a new orphan. PEP is generally conserved, retaining many lamprey sequences at the root of receptor clades.
Figure 3.
Overview of vertebrate GPCR clades mapped onto neighbor-joining trees of the β-branch. PEP receptors. Caption otherwise similar to Fig. 1.
NTSR, GPR39, MLNR, GHRS, NMUR, TRHR
Near GPR39 and neurotensin receptors (gene: NTSR), rooted by lamprey, a set of gar/fishes(3)/coelacanth/amphibian genes, see ENSXETP00000007421, suggest a new orphan, GPR189.
The motilin receptor (gene: MLNR) has one novel subtype, MLNRB that contains coelacanth/amphibian/reptiles(2), see ENSLACP00000009190. A closely branched set of gar/fishes(9) genes, see ENSLOCP00000011352, may additionally belong to MLNRB.
The growth hormone secretagogue receptor (GHSR) tree includes three groups of fishes: one containing gar/zebrafish, see ENSLOCP00000009072, likely orthologous to human GHSR; one containing gar/fishes(10)/coelacanth, see ENSLOCP00000000036, that was named GHSR2; and a set of fishes (11, duplicates in cave fish), see ENSAMXP00000003803, also orthologous to human GHSR. Four goldfish receptors (GHS-R1a type 1/type 2, GHS-R2a type 1/type 2) have been previously suggested62,63.
The thyrotropin-releasing hormone receptors 1–3 (TRHR) are resolved in Ensembl.R92 as one orthologue and two paralogues of the human TRHR receptor. Four receptors (THR1a, THR1b, THR2, THR3) have been previously characterized in medaka fish64 and three cloned in frog65.
The neuromedin U receptors (NMUR) are divided into NMUR1 and NMUR2, both with lamprey orthologues (ENSPMAP00000009319, ENSPMAG00000001504). Two sets of fish genes were assigned to NMUR1 (based on Ensembl.R92). A set of sequences found in lamprey (ENSPMAP00000007962) and gar/fishes(10)/coelacanth, see ENSLOCP00000004052, was named NMUR3.
NPFF; QFRPR; HCTR; NPY4,8,6,1; NPY5; NPY7; NPY2R; PRLHR
Two groups of genes are likely ohnologues of NPFFR1: a gar/fishes(11) set, see ENSLOCP00000019807, named here NPFFR1L2/3; and gar/fishes(3), see ENSLOCP00000018374, named NPFFR1L1. Two reptile genes branch with either of these receptors.
Two groups of two novel prolactin releasing hormone receptors (PRLHR) are found: the first group is rooted by lamprey (ENSPMAP00000011235) and comprised of one set comprising gar/fishes(16, duplicated)/coelacanth/amphibian/birds(3), see ENSLOCP00000018414, named here PRLHR2; and another set with gar/amphibian/sauropsids(7)/marsupials(2), see ENSLOCP00000022432, named PRLHR3. The other group is also rooted by lamprey (ENSPMAP00000008908) and comprises a gar/fishes(7)/coelacanth/amphibian/sauropsids(6) set, named PRLHR4; and another gar/fishes(9)/coelacanth set, see ENSLACP00000014694 (removed from Ensembl subsequent releases). Three homologues of the mammalian receptors have been cloned in chicken66, presumably PRLHR2–4.
In the neuropeptide Y receptor family, three unannotated receptors NPY6, NPY7, and NPY8 have been described previously67–70.
AVPR; OXTR; NPSR; GNHR
Three clusters of vasopressin (AVPR) receptor genes are found: one with gar/amphibian/sauropsids(6), see ENSLOCP00000001209, named AVPR2B; one comprised of fishes(4) genes, see ENSDARP00000119491, named AVPR2l; and one with gar/fishes(14)/coelacanth, see ENSLOCP00000013628, named AVPR2C. Presumably AVPR2l and AVPR2C have been previously reported and named respectively V2-like and V2B71.
We found several interesting gonadotropin-releasing hormone receptors (GNRHR) groups, most including lamprey sequences at their root, whose annotation will require further study since Ensembl.R92 has clarified some of the branchings, but not all. The first contains gar/fishes(27)/coelacanth/amphibian, see ENSLOCP00000017682. The second includes fishes(16)/coelacanth/amphibian/sauropsids(6), see ENSLOCP00000010658. The remaining groups are coelacanth/amphibian/reptile(1), reptiles(5), and placental(31)/marsupial(3) mammals(see ENSMEUP00000014678). The evolution of GNRH receptors has been previously studied72 and four zebrafish GNRHR previously cloned73.
GRPR; BRS3; NMBR; EDNRA,B; GPR37,37L
A group constituted of gar/fishes(8)/coelacanth/amphibian/sauropsids(7)/platypus, see ENSLOCP00000018439, likely corresponds to a previously reported endothelin receptor, EDNRB2 74.
TACR1,2,3; PROKR1,2; GPR83; GPR165
Ensembl.R92 resolves Pgr15l as a new orphan present in many species: gar/fishes(8)/coelacanth/amphibian/sauropsids(6)/mammals(14 genes from rodents/rabbit), see ENSLACP00000018091. Ensembl.R95 reveals a new subtype of GPR83, annotated GPR83L, and Pgr15lto be a mammalian duplicate of GPR165.
Chemokine (CHEM) receptors
In CHEM, we report eight novel receptors, of which four are likely orphans, one divided into two subtypes (Fig. 4a). The assignment of receptors in CHEMS is difficult due to a fast evolution rate and numerous internal duplications, and additional subtypes will certainly be defined when more data becomes available. Evolution of the chemokine system has been comprehensively studied75.
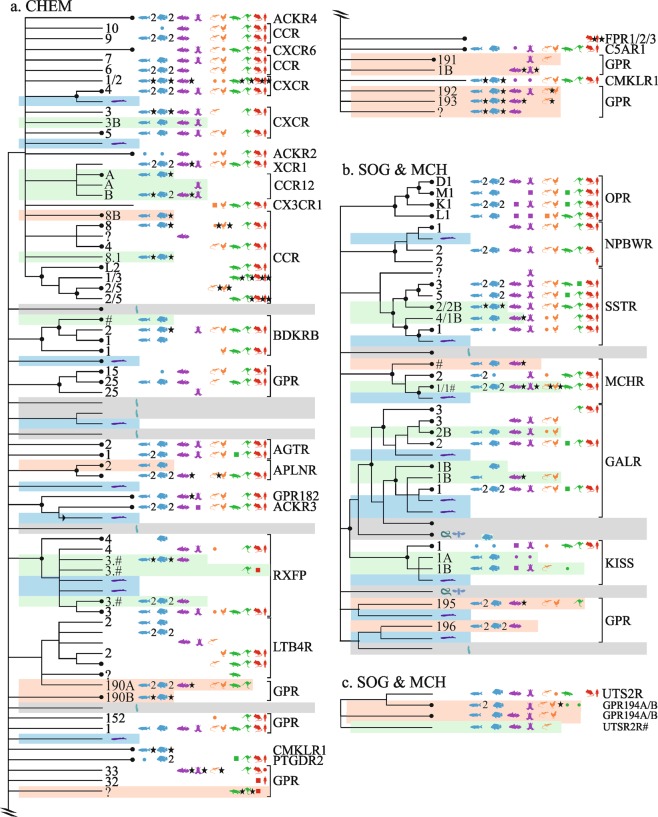
Figure 4.
Overview of vertebrate GPCR clades mapped onto neighbor-joining trees of the γ-branch. (a) CHEM (b,c) SOG and MCH receptors. Caption otherwise similar to Fig. 1.
LTB4R, FPR1,2,3, GPR32, GPR33, CMKLR1, C5AR1, C3AR1, PTGDR2, GPR1, GPR152
Near the leukotriene-4 receptors (LTB4Rs) we suggest a new orphan receptor GPR190, composed of two (or three) subtypes. A set of gar(duplicated)/fishes(16)/coelacanth(duplicated)/sauropsids(7) genes, see ENSLOCP00000021707, was named GPR190A. The second subtype is composed of gar(duplicated)/fishes(32,internal duplications)/marsupials(2), see ENSLOCP00000021705, and named GPR190B. A lonely platypus sequence (ENSOANP00000021820), complete with respect to its GPCR signature, may represent a third subtype. Two sets of genes near LTB4R1/2 (in Ensembl.R92) may be orthologues of existing human receptors (sets removed in E.95). Three LTB4R have been identified in zebrafish76.
The branching near the formyl peptide receptors FRP1,2,3 and GPR32 is especially blurred, and Ensembl.R92 helps understand the recent history. FRP1,2,3 have arose through local duplications, and marsupial(3) sequences, see ENSSHAP00000001972, are likely orthologous to the FRP1,2,3 set. GPR32 is likely rooted by seven monotreme and marsupial genes, see ENSOANP00000022386; and the FRP1,2,3-GPR32 group is likely rooted by a sequence from turtle, see ENSPSIP00000009727.
Near GPR1 (in Ensembl.R92), a complex cluster contains four sets of coelacanth genes (some duplicated) branching with two sets of fishes and one set (9 duplicates) of amphibian genes, see ENSLACG00000005198. This group was named GPR1B.
Near GPR33 and the chemerin chemokine-like receptor 1 (CMKLR1), taking shape in Ensembl.R92, there is a set comprised of fishes(2)/amphibian/reptile(lizard) genes, see ENSXETP00000062533, that was named GPR191. Nearby is another orphan, with several subtypes, named GPR193 and composed of: coelacanth/reptile(lizard), see eg ENSLACP00000014297; gar/fishes(20, internally duplicated), see ENSLOCP00000021780; isolated genes from lizard (ENSACAP00000004236) and coelacanth (ENSLACP00000021116). There is also a large set of gar/fishes (31 genes, see si:dkey-117a8.4) of ambiguous positioning. Nearby, a set of gar/fishes(26, internal duplications) genes, see ENSLOCP00000022270, was named GPR192; GPR192 may be divided into subtypes since it is branched to coelacanth/sauropsid (5) and amphibian/reptiles(5) sets, indicating an internal duplication in tetrapods.
CCR1,2,3,4,5,6,7,8,9,10; ACKR2,4; CXCR1,2,3,4,5,6; CX3CR1; XCR1
Three sets of fish genes were difficult to assign. Two of these sets are resolved in Ensembl.R92 and tentatively assigned to the fishes orthologous to CCR8, see ENSLOCP00000022371, as well as to a fish-specific clade containing gar/fishes(8), see ENSLOCP00000000990, named CCR8B. A third group comprising gar(duplicated)/fishes(14)coelacanth, see ENSLOCP00000022422, is located at the root of the mammalian CCR1,2,3,4,5 and CX3CR1 and probably correspond to orthologues of CCR2 and CCR5. We also identified already reported fish-specific receptors, CXCR3B characterized in common carp77 (ENSLACP00000016977) and CCR12A,B identified in orange-spotted grouper (see ENSLOCP00000022265)78.
AGTR1,2; BDKRB1,2; GPR15,25,182; APLNR; ACKR3; RXFP3,4
The bradykinin receptor B2 (BDKRB2) has an additional set gar/fishes(6) genes, probable result of a local duplication, see ENSLOCP00000021801. A set of gar/fishes(20)/coelacanth (also a fragmental wallaby sequence), see ENSLACP00000020198, may be paralogous to the apelin receptor (APLNR). A second group of fishes(11) nearby, see ENSAMXP00000021714, was assigned as APLNR2.
Human relaxin/insulin-like family peptide receptor 3 (RXFP3) has orthologues in fish (ENSLOCP00000021654); a paralogous group comprising gar/fishes(18)/coelacanth, see ENSLACP00000009554. Near RXFP4 (that has fish orthologues too) there is evidence of a new subtype comprised of gar/fishes(22, duplicated)/coelacanth/mammal(2, including one marsupial) sequences, see ENSLOCP00000022049), as well as a fish-specific set of duplicates (9, annotated rxfp3.2b). These new receptors have been named here RXFP3.# pending further studies and probably have been previously described as RXFP3–2 and RXFP3–379,80.
Somatostatin-opioid-galanin (SOG) and melanin-concentrating hormone (MCH) receptors
In the SOG and MCH families we suggest five previously unreported receptor types (Fig. 4b,c), including three orphans. For a review of neuropeptide signaling systems see81.
OPRD1,-M1,-K1,-L1; NPBWR1,2; SSTR1,2,3,4,5; MCHR1,2
In the somatostatin (SSTR) receptor group, near SSTR2 a gar/fishes(5) group named SSTR2B, probable result of an internal duplication, see ENSLOCP00000021860, has been described previously as SSTR682. Nearby, a group of fishes and coelacanth represent a new SSTR subtype, SSTR1B, since SSTR4 has lost its fishes with the exception of coelacanth82,83.
Branching close to MCHR2, there is a gar/fish(10)/coelacanth(duplicated) set, see ENSLACP00000004413. Near MCHR1, two new subtypes are suggested: a coelacanth/amphibian/reptile(1) set, see ENSLACP00000017963; and a larger set comprising gar/fishes(14,duplicated)/coelacanth/sauropsids(5), see ENSLACP00000009737. Mammals have five melanocortin receptors and one or two MCH receptors, zebrafish six melanocortin (extra MC5RB) and three MCH receptors; fugu four MCR (MC3R missing) and two MCHR; Two MCHR receptors have been cloned in goldfish and four MCHR receptors have been cloned in Xenopus tropicalis84. No annotations are proposed due to the complexity of the Ensembl trees for this subfamily.
CCKAR; CCKBR; GPR19; KISS1R; GALR1,2,3
Near the Kisspeptin receptor 1 (KISS1), three unassigned sets of genes were detected. The first contains gar/amphibian, see ENSLOCP00000002856, and was named KISS1RA. The second set contains gar/fishes(10)/coelacanth/amphibian, see ENSLOCP00000003036, and was named KISS1RB; this receptor has likely been studied in ray-finned fishes85. A related set contains gar/coelacanth/reptiles(2), see ENSLOCP00000008391, and the tree furthermore includes fragmental sequences at the root of KISS1 (not discussed here).
In Ensembl.R92, the human galanin receptor 1 (GALR1) has two sets of paralogous fish genes (human GALR1 having its own fishes), rooted by a lamprey sequence (ENSPMAP00000000353). A set of gar/fishes(7)/coelacanth(2)/sauropsids(7) genes, see ENSLACP00000010695, was named GALR1B. Near GALR3, a paralogous branch with gar/fishes(10)/coelacanth/amphibian/sauropsids(4), see ENSLOCP00000001326, was named GALR2B. Avians have been reported to have two new types in addition to Galr1 and Galr2: GalR1, GalR1-like, GalR2-like; four galanin receptors have been isolated from European sea bass (named GalR1a, 1b, 2a, 2b)86,87.
Two new orphans were identified near the cholecystokinin (CCK) A and B receptors (grouped with PEP in Ensembl.R92). One is composed of fishes(8)/coelacanth(2)/sauropsids(6), see ENSLOCP00000021858, and was named GPR195; this receptor may have two subtypes because of coelacanth duplicates. The other is fish-specific and in Ensembl.R92 occupies its own tree, see ENSLOCP00000007402. It contains lamprey, 3R-duplicated ray-finned fishes(16), and coelacanth, and was named as GPR196.
UTS2R
Previously four urotensin 2 receptor (UTS2R) subtypes have been named as UTS2R2–582. A set of gar/fishes(6)/coelacanth/amphibian/sauropsids(6), see ENSLACP00000012826, is orthologous to human UTS2R; A set from gar/fishes(9)/coelacanth/amphibian, see ENSLACP00000003013 is a probable new UTS2R subtype.The Ensembl.R95 gene tree suggest two subtypes of a new orphan receptor, GPR194: gar/fishes(2)/sauropsids/marsupials(2)/monotreme, see ENSLOCP00000022204, named GPR194A; and gar/fishes(10)/amphibian/birds(3), see ENSLOCP00000021948, named GPR194B.
Leucine-rich repeat (LGR) and Mas-related (MRG) receptors
In the MRG and LGR families, no new vertebrate receptors were found (Fig. 5a,b).
Figure 5.
Overview of vertebrate GPCR clades mapped onto neighbor-joining trees of the δ-branch. (a) LGR (b) MGR (c) PUR. Caption similar to that of Fig. 1.
LGR4,5,6; LHCGR; FSHR; TSHR; RXFP1,2
The LGRs are rooted by a set of fish genes (6, see ENSTRUP00000003361) likely orthologous to human LGR5. A set of gar/fishes(2)/coelacanth/amphibian/sauropsids(5)/marsupials(2) genes, see ENSLOCP00000018098, corresponds to the previously described RXFP1B79, annotated here RXFP2L (marsupial and bird genes not found in R.95). A fish-specific luteinizing hormone receptor (LH), see ENSLOCP00000020088, has also been reported88.
MRGPRD; MRGPRF; MRGPRG; MRGPRE; MRGPRX1,2,3,4; MRGPRa,b; MAS1;1L
The evolution of the MAS1, MAS1 oncogene-like, and MAS-related GPR (MRG) families is complexified by numerous internal duplications. A large group of sauropsid sequences, locally duplicated, could be orthologous to the mammalian MRGPR, see ENSGALP00000047269, named here MRGPR1; A group of placental and marsupial mammals, see ENSSHAP00000018195, named MRGPRH. Nearby is a cluster of reptile genes, see ENSACAP00000019338, which may be orthologous or may represent a new subtype. MAS receptors have been reported to arise in amphibians89, and the MAS family tree is rooted by a group of amphibians, MAS1L, see ENSXETP00000063951. Three clusters of fish genes were positioned in Ensembl.R91 with MAS/MRGPRD but in the newer Ensembl.R92 are aggregated with GPR1B (see ENSLACP00000001563, described above in the CHEM/CMKRL1 tree).
Purine (PUR) receptors
The PUR family is the most uncharted and we identified 27 new receptors, including 12 orphans (Fig. 5c).
LPAR4,5,6; PTAFR; F2LR1/2/3; P2RY8; P2YR10; F2R; GPR4,18,35,55,65,68,132,174
Near LPAR5, a set of gar(duplicated)/fishes(2)/coelacanth(3, internally duplicated) was assigned to a new orphan probably with two subtypes, see ENSLOCP00000021495, named LPAR5A/B. Near PTAFR, a set of genes comprising gar/coelacanth(duplicated)/sauropsids(2)/mammalian (2, one marsupial), see ENSLOCP00000002041, was named GPR199.
Near LPAR4/6, a set with gar/fishes(4)/coelacanth/reptile(1), see ENSLOCP00000021661, was named GPR202. Another orphan is composed two amphibian and two lamprey sequences, see ENSXETP00000036876, named GPR200. Near GPR55, a full set of genes from gar/fishes(2)/coelacanth(duplicated)/sauropsids(5)/monotreme(1), see ENSLOCP00000021497, was named GPR55B.
The receptors GPR4, GPR68, GPR65 and GPR132 branch together with a new orphan, named GPR201, divided into two subtypes. One of these subtypes contains gar(7 duplicates)/fishes(2)/reptile(1)/marsupials(2), see ENSLACP00000013584; the other contains gar/fishes(10)/coelacanth, see ENSLACP00000020885.
Near GPR65 (GPR68 in E.95) there is a set of fishes(5)/coelacanth, see ENSLACP00000000626, was named GPR68B. Near GPR132 a set of gar/fishes(10)/coelacanth genes, see ENSLACP00000009633, was assigned to GPR184. Near P2RY10 and GPR174, a group of gar/fishes(3)/coelacanth genes, see ENSLACP00000004667, was named GPR203.
The coagulation factor II receptor F2RL3 is prone to local duplications, for example in spotted gar (see ENSLOCP00000014196) or coelacanth (see ENSLACP00000003039). Near F2RL3, a group of amphibian/birds(5), see ENSXETP00000005770, was named F2RL4. A set of fishes(18) paralogous to human F2R, see ENSAMXP00000020201, need further consideration for annotation.
Near P2RY8, a group constituted of gar/fish/coelacanth/amphibian/reptiles(2), see ENSLOCP00000021876, was named P2RY8B. Nearby a coelacanth duplicate (ENSLACP00000004454) could not be assigned. In P2RY10, a set of receptors resulting from a mammalian-specific internal duplication, see ENSMUSP00000133122, were assigned to P2RY10B.
GPR20,31; HCAR1,2,3; OXER1
Near GPR20 we suggest a new orphan, divided into two subtypes: a group of constituted of gar/fishes(6)/reptiles(3)/monotreme(1), see ENSLOCP00000001967, named here GPR207A; and gar/coelacanth(duplicated), see ENSLOCP00000021450, named GPR207B. A set of fishes(21), see ENSLOCP00000021469, is probably orthologous to the mammalian oxoeicosanoid receptor 1 (OXER1); in such case a set of fish/amphibian(2) genes, see ENSLOCP00000020331, is paralogous. The subtypes 1–3 of the hydroxycarboxylic acid receptors HCA, found only in mammals, are closely related and thus the likely result of recent duplications. Two sets of fishes are branched to the mammalian subtypes, of which gar/fishes(8)/coelacanth, see ENSLOCP00000022252, was named HCAR1L.
GPR34,82,87,171; P2RY12,13,14
P2RY13 has an extra set of fishes(3), see ENSLOCP00000002272, indicating an internal duplication. Near GPR34 there are two groups of sequences: one gar/fishes(41, many duplicates), see ENSLOCP00000021478, named here GPR206; another set with gar/fish/amphibian/sauropsids(5), see ENSLOCP00000016952, was named GPR34B.
SUCNR1; OXGR1; P2RY1,2,4,6; GPR17; GPR183; CYSTLR1,2
Branched distantly from others is a new orphan divided into three sets: two with genes from gar(2)/coelacanth/reptile, see ENSACAP00000008615, named GPR205C and GPR205B; the second gar/fishes(9)/coelacanth/amphibian/sauropsids(4), see ENSACAP00000013831, named GPR205A. Near P2RY1 (NPSR1 in R.95) there is a cluster of genes from gar/fishes(23)/coelacanth/amphibian(triplicates)/sauropsids(7), see ENSLOCP00000022457, that we named NPSR1L. Near P2RY6 there are two sets of unannotated genes: a complete group with gar/fishes(3)/coelacanth/reptile(1)/mammals(21), see ENSLOCP00000021492, named here P2RY16; and, distant and rooted by duplicated lampreys (see ENSPMAP00000011278), a cluster comprised of gar/fishes(18, duplicates)/coelacanth/amphibian, see ENSLOCP00000021662, that we named GPR204. A set of genes from ray-finned fishes(10)/coelacanth(duplicated)/sauropsids(5)/mammals(5), including one monotreme and one marsupial), see ENSLOCP00000021877, was assigned as GPR197. A gar/fishes(8) specific group of genes, see ENSLOCP00000014291, is branched near GPR17, and we named it GPR17B. Another group rooted by lamprey (triplicate, ENSPMAP00000011192) comprised of gar/fishes(4)/coelacanth, see ENSPMAP00000011192, was named CYSLTR3A. Two other fishes (gar, cave fish) nearby open the possibility of another subtype CYSLTR3B.
FFAR1,2,3
Free fatty acid receptors (FFAR) have generally undergone many internal duplications. A small gene cluster from coelacanth/amphibian/reptile, see ENSLACP00000002775, is likely a new new subtype FFAR2#(annotation in process). A set of 32 genes from fishes, see ENSLOCP00000006360, is likely belonging to FFAR2# (Ensembl.R92).
GPR141; GPR101,161,176
These two clusters of orphan receptors were initially aggregated with PURIN receptors but form independent groups in Ensembl.R92. A set containing gar/fishes(10)/coelacanth, see ENSLOCP00000014127, was named GPR141C. A set with coelacanth/sauropsids(4)/mammals(15, including two monotremes), see ENSLACP00000005080, was named GPR141B. A set of fishes(10)/amphibian, see ENSXETP00000035434, was named GPR198.
Other families
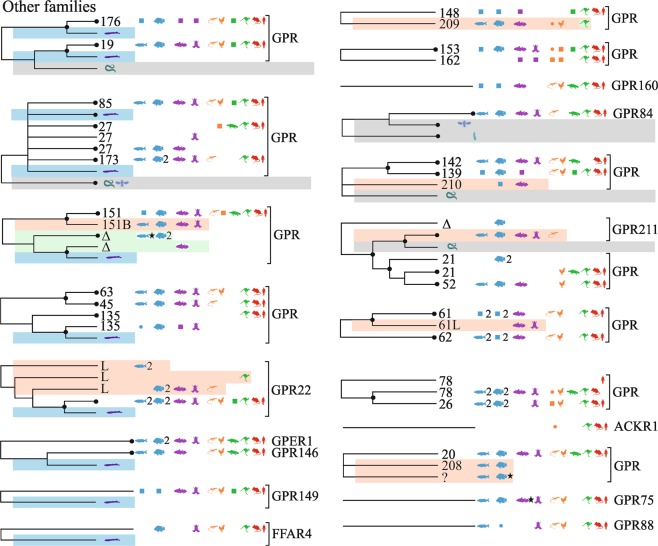
We grouped together receptors that we could not link to the main families, and in this group we identify seven novel receptors (Fig. 6). Interestingly, nine of the orphan groups are closely associated with a lamprey or tunicate sequence, showing an ancient origin; and there are clear grouping for some of the orphans into subtype groups.
Figure 6.
Overview of vertebrate GPCR clades mapped onto neighbor-joining trees for orphans without clear families. Caption otherwise similar to that of Fig. 1.
GPR22
A set from gar/fishes(17,duplicated)/coelacanth/amphibian/reptile/marsupial, see ENSLOCP00000017943, is a new subtype GPR22L, which may divide further into different subtypes.
GPR139; GPR142
Three sets of genes indicate an orphan divided into three subtypes: coelacanth/fishes(4), see ENSLACP00000014196, named here GPR210A; gar/coelacanth, see ENSLACP00000019189 (not in R.95); gar/fishes(2)/coelacanth, see ENSLACP00000015589, named GPR210B.
GPER1
A new subtype in ray-fin fishes (8) has been previously reported, which may be a fish-specific duplicate (3R)90.
GPR20
An independent group of fishes(8), see ENSAMXP00000025847, was named GPR208; a set of fishes(10)/coelacanth/amphibian/sauropsids(7)/marsupial/monotreme genes, see ENSAMXP00000019172, is distant from GPR20’s placental mammals and may represent a new subtype. A group of gar/fishes(15) is also nearby in Ensembl.R92 (annotated lpar5b), see ENSLOCP00000021496.
GPR151
A set of fishes(9)/coelacanth/amphibian, see ENSLACP00000009338, was named GPR151B.
GPR148
A set of fish(2)/sauropsids(5)/marsupial(1), see ENSLOCP00000021506, was named GPR209. A second spotted gar, see ENSLOCP00000021505, may indicate another subtype.
GPR61
A set coelacanth/ambiphian, see ENSLACP00000015922, is likely paralogous to GPR62.
GPR21,52
A set of gar/fishes(10)/coelacanth/amphibian/lizard, see ENSLACP00000013616, named GPR211.
Conservation across families
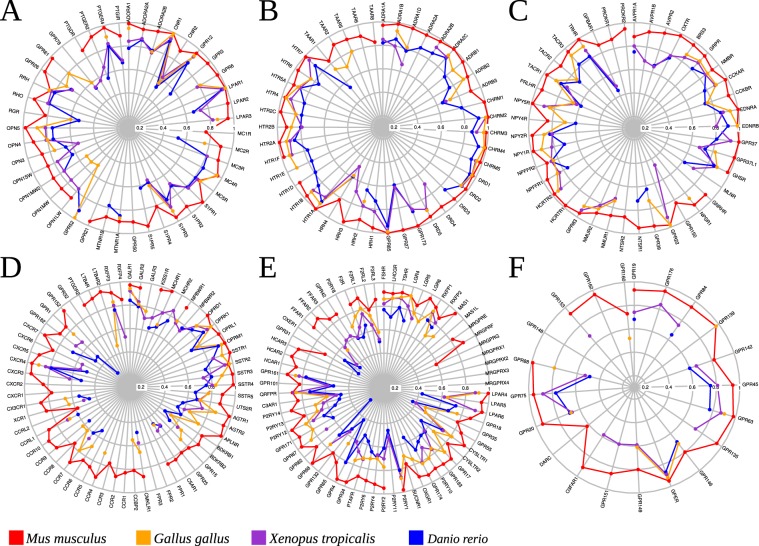
We used the set of sequences aligned across families to study the relative conservation of the subtypes across four representatives species (human, mouse, bird, amphibian and fish) (see Supporting information; 189 aligned positions). Generally, as expected the more “ancient” branches such as α-branch and β-branch are relatively conserved compared to more recent branches (χ-branch and δ-branch). In the α-branch (Fig. 7A,B), the most conserved are LPAR1 and CNR1 in MECA that share sequence identity above 90% towards their human orthologue (human-zebrafish LPAR1, 96%; human-mouse LPAR1, 99%; human-zebrafish CNR1 92% and human-mouse CNR1 99%). Conservation is also high in the AMIN family (Fig. 7A) where the DRD1/2, ADRA2C and CHRM3/4/5 have percent sequence identities above 90% towards the human receptors. In the β-branch, in PEP GPR22 and HCRTR2 maintain sequence identity above 90% from amphibian to mouse (Fig. 7C). Few receptors of the γ-branch maintain sequence identity above 80% through species (Fig. 7D), but this is the case of OPRD1, OPRK1, GALR1 and SSTR1 (most conserved OPRM1, human-zebrafish 93%; human-mouse 99%). In the δ-branch conservation is less (Fig. 7E), and the most conserved are TSHR (80–93%) and P2RY1 (81–98%). In the unassigned receptors (Fig. 7F), GPER1 is the most conserved (human-zebrafish 87%; human-mouse 95%). Not surprisingly, GPR85 (human-zebrafish, 94%; human-mouse, 100%), an orphan receptor closely related to the amine family and named "Super Conserved Receptor" shows a very high conservation too (initially grouped with AMIN, Fig. 7A).
Figure 7.
Percent sequence identity (189 aligned positions) to their human orthologues for four representative species: mouse (Mus musculus), chicken (Gallus gallus), western clawed frog (Xenopus tropicalis) and zebrafish (Danio rerio). (A,B), α- (C), β- (D), γ- (E) δ- branches and (F) other families.
Concluding remarks
The novel receptors are identified based on sequence data alone. The complete integrity of the sequences, validated by the conserved GPCR motifs, suggest that these are not pseudogenic. The groupings have been highly stable during the last five years, especially inside monophyletic groups, and most likely to remain so. Generally, the branchings in the fast-evolving PUR and CHEM families (see eg especially near FRP1,2,3 and GPR32,33), and for distant sequences such as lamprey, have been the most rearranged over the different Ensembl releases. The automated rearrangements have been nonetheless parsimonous, ie groups have been reassigned as orthologues rather than paralogues of known receptors.
This study raise numerous nomenclature issues, but in the same time sets a framework on the extent of yet-to-be studied vertebrate receptors. A difficult issue is that without establishing ligand binding preferences new sequences cannot be defined as “subtypes” or “orphan” receptors; we thus chose to keep the “subtype” denomination only for the likely cases. Importantly, the ligand binding preferences are not fully indicative of close evolutionary relationships; some receptors have acquired the same ligand specificity several times, for example α1-, α2-, and β- adrenoceptors31,32.
This study maps the extent of orthologues of human GPCRs to vertebrate species. Furthermore, it explores the pool of yet-to-be-studied GPCRs, suggesting at minima 69 sets of novel receptors in vertebrates not orthologous to human. The clustering data allows to group orphan receptors and thus probably brings down the number of endogenous ligands yet to be identified. Nineteen of these groups are maintained in Ensembl.R92 and thus worthy of consideration: In MECA, GPR3/6/12; in PEP, GPR83/165, EDNR/GPR37/37L1, GPR39/NTSR; in CHEM, GPR15/25, GPR182/ACK3; in PUR, GPR20/35/55, GPR101/161, P2RY10/GPR174, GPR17/183, GPR87/P2RY14, GPR31/HCAR; and in other families GPR85/27/173, GPR45/63/135, GPR153/162, GPR139/142, GPR21/52, GPR61/62, GPR26/78; for a few of these receptors an endogenous ligand has been suggested91. The data at the basis of this study are available online (see the Experimental section) and will continue to be improved following especially the sequencing of novel genomes and the increased coverage of existing genomes.
Experimental Section
Ensembl data
The Ensembl.R90 includes 89 vertebrate genomes: eleven ray-finned fishes (e.g. zebrafish, spotted gar), one lobe-finned fish, one amphibian, five birds, two reptilians, three marsupial, one monotreme and 65 eutherian mammals, as well as early vertebrates (lamprey, two tunicates) and invertebrates (one insect and one nematode). The most recent release, Ensembl.R92, includes 94 genomes.
New receptors were identified based on the branching presented on the Ensembl-generated trees. Three genomes are at the cornerstone for assessing a new receptors: coelacanth, frog, and spotted gar, and we separate them (“spotted gar” referred to simply as “gar”) from the other fishes. Spotted gar diverged from the teleost lineage before the 3R92). New orphan receptors were named in numerical order starting from 181, using the numbers left empty (for human genes) by the HUGO Gene Nomenclature Committee. New subtypes were identified either numerically or alphabetically.
Guide trees
The data used to build the guide trees are based on an automatically extracted and curated set of 14.000 sequences is described as Supporting Information. This earlier data are used in this manuscript only to build guide trees and compute sequence identities in transmembrane regions.
Electronic supplementary material
Acknowledgements
This work was financially supported by Waldemar von Frenckells and Oskar Öflunds foundations (M.R), by a grant from Higher Education Commission (HEC) of Pakistan under the indigenous Ph.D. program (074-1844-PS4-406) (ZUR. T.) and by the University of Helsinki internal funding (H.X.). Dr Elspeth Bruford from the HUGO nomenclature committee is thanked for the inital assement of the suggested gene symbols, an input that helped improve the manuscript. Dr Vigneshwari Subramanian is thanked for the R scripts used to create Figure 7. The Drug Discovery and Chemical Biology Consortium is thanked for organizing the computational infrastructure in the Xhaard group. CSC-IT is thanked for providing access to supercomputing resources. The European COST action (CA18133, ERNEST) is thanked for organizing a network of GPCR researchers.
Author Contributions
Sequence retrieval and bioinformatics Z.U.R.T., A.K., H.X.; Data analysis, M.R., H.X.; Prepared the Figures and Tables, M.R; Drawn the pictograms, M.R.; Wrote the manuscript (main contribution), M.R., H.X., Study design, H.X.
Data Accessibility
All genomic data from the Ensembl releases is freely accessible online in few simple steps: (1) log to http://www.ensembl.org or to the Ensembl archives https://www.ensembl.org/info/website/archives/index.html (2) query the name of the receptor of interest or the provided codes; (3) select (usually) the first Gene hit on the list; (4) click on “gene tree” in the left-hand panel; (5) browse the tree data: in particular, branches can be expanded and sequences visualized. The sequence data used to construct the guide trees is accessible upon request. Note added following acceptance: The genes identified in this manuscript have been submitted to the HGNC, and will be submitted to the MGNC, RGNC, XNC, and ZNC nomenclature committees on publication.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33120-8.
References
- 1.Ahonen TJ, et al. Synthesis of 7β-hydroxy-8-ketone opioid derivatives with antagonist activity at mu- and delta-opioid receptors. Eur. J. Med. Chem. 2018;151:495–507. doi: 10.1016/j.ejmech.2018.02.074. [DOI] [PubMed] [Google Scholar]
- 2.Leino TO, et al. Azulene-based compounds for targeting orexin receptors. Eur. J. Med. Chem. 2018;157:88–100. doi: 10.1016/j.ejmech.2018.07.040. [DOI] [PubMed] [Google Scholar]
- 3.Turku A, et al. Pharmacophore Model To Discover OX 1 and OX 2 Orexin Receptor Ligands. J. Med. Chem. 2016;59:8263–8275. doi: 10.1021/acs.jmedchem.6b00333. [DOI] [PubMed] [Google Scholar]
- 4.Hauser AS, Attwood MM, Rask-Andersen M, Schiöth HB, Gloriam DE. Trends in GPCR drug discovery: new agents, targets and indications. Nat. Rev. Drug Discov. 2017;16:829–842. doi: 10.1038/nrd.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–45. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 6.Wang C, et al. Structure of the human smoothened receptor bound to an antitumour agent. Nature. 2013;497:338–43. doi: 10.1038/nature12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isberg V, et al. GPCRDB: an information system for G protein-coupled receptors. Nucleic Acids Res. 2014;42:D422–D425. doi: 10.1093/nar/gkt1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isberg V, et al. Generic GPCR residue numbers – aligning topology maps while minding the gaps. Trends Pharmacol. Sci. 2015;36:22–31. doi: 10.1016/j.tips.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruuskanen JO, et al. Identification of duplicated fourth alpha2-adrenergic receptor subtype by cloning and mapping of five receptor genes in zebrafish. Mol. Biol. Evol. 2004;21:14–28. doi: 10.1093/molbev/msg224. [DOI] [PubMed] [Google Scholar]
- 10.Ballesteros JA, Weinstein H. [19] Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995;25:366–428. doi: 10.1016/S1043-9471(05)80049-7. [DOI] [Google Scholar]
- 11.Fredriksson R, Lagerström MC, Lundin L-G, Schiöth HB. The G-Protein-Coupled Receptors in the Human Genome Form Five Main Families. Phylogenetic Analysis, Paralogon Groups, and Fingerprints. Mol. Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 12.Fredriksson R, Schiöth HB. The Repertoire of G-Protein-Coupled Receptors in Fully Sequenced Genomes. Mol. Pharmacol. 2005;67:1414–1425. doi: 10.1124/mol.104.009001. [DOI] [PubMed] [Google Scholar]
- 13.Kolakowski LF. GCRDb: a G-protein-coupled receptor database. Receptors Channels. 1994;2:1–7. [PubMed] [Google Scholar]
- 14.Krishnan A, Almén MS, Fredriksson R, Schiöth HB. The origin of GPCRs: identification of mammalian like Rhodopsin, Adhesion, Glutamate and Frizzled GPCRs in fungi. PLoS One. 2012;7:e29817. doi: 10.1371/journal.pone.0029817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bylund DB, et al. Pharmacological characteristics of alpha 2-adrenergic receptors: comparison of pharmacologically defined subtypes with subtypes identified by molecular cloning. Mol. Pharmacol. 1992;42:1–5. [PubMed] [Google Scholar]
- 16.Ohno Susumu. Evolution by Gene Duplication. Berlin, Heidelberg: Springer Berlin Heidelberg; 1970. [Google Scholar]
- 17.Lundin LG. Evolution of the vertebrate genome as reflected in paralogous chromosomal regions in man and the house mouse. Genomics. 1993;16:1–19. doi: 10.1006/geno.1993.1133. [DOI] [PubMed] [Google Scholar]
- 18.Dehal P, Boore JL. Two Rounds of Whole Genome Duplication in the Ancestral Vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Fernàndez J. The genesis and evolution of homeobox gene clusters. Nat. Rev. Genet. 2005;6:881–892. doi: 10.1038/nrg1723. [DOI] [PubMed] [Google Scholar]
- 20.Lagman D, et al. The vertebrate ancestral repertoire of visual opsins, transducin alpha subunits and oxytocin/vasopressin receptors was established by duplication of their shared genomic region in the two rounds of early vertebrate genome duplications. BMC Evol. Biol. 2013;13:238. doi: 10.1186/1471-2148-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yun S, et al. Prevertebrate Local Gene Duplication Facilitated Expansion of the Neuropeptide GPCR Superfamily. Mol. Biol. Evol. 2015;32:2803–17. doi: 10.1093/molbev/msv179. [DOI] [PubMed] [Google Scholar]
- 22.Pasquier J, et al. Molecular evolution of GPCRs: Kisspeptin/kisspeptin receptors. J. Mol. Endocrinol. 2014;52:T101–17. doi: 10.1530/JME-13-0224. [DOI] [PubMed] [Google Scholar]
- 23.Amores A, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–4. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 24.Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD) Bioessays. 2005;27:937–45. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- 25.Steinke D, Hoegg S, Brinkmann H, Meyer A. Three rounds (1R/2R/3R) of genome duplications and the evolution of the glycolytic pathway in vertebrates. BMC Biol. 2006;4:16. doi: 10.1186/1741-7007-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamesh N, Aradhyam GK, Manoj N. The repertoire of G protein-coupled receptors in the sea squirt Ciona intestinalis. BMC Evol. Biol. 2008;8:129. doi: 10.1186/1471-2148-8-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Escriva H, Manzon L, Youson J, Laudet V. Analysis of lamprey and hagfish genes reveals a complex history of gene duplications during early vertebrate evolution. Mol. Biol. Evol. 2002;19:1440–50. doi: 10.1093/oxfordjournals.molbev.a004207. [DOI] [PubMed] [Google Scholar]
- 28.Kuraku S, Meyer A, Kuratani S. Timing of Genome Duplications Relative to the Origin of the Vertebrates: Did Cyclostomes Diverge before or after? Mol. Biol. Evol. 2008;26:47–59. doi: 10.1093/molbev/msn222. [DOI] [PubMed] [Google Scholar]
- 29.Hedges SB, Kumar S. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 30.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–5. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 31.Vincent, J. D., Cardinaud, B. & Vernier, P. Evolution of monoamine receptors and the origin of motivational and emotional systems in vertebrates. Bull. Acad. Natl. Med. 182, 1505-14; discussion 1515–6 (1998). [PubMed]
- 32.Xhaard H, Rantanen V-V, Nyrönen T, Johnson MS. Molecular evolution of adrenoceptors and dopamine receptors: implications for the binding of catecholamines. J. Med. Chem. 2006;49:1706–19. doi: 10.1021/jm0511031. [DOI] [PubMed] [Google Scholar]
- 33.Pertwee RG. GPR55: a new member of the cannabinoid receptor clan? Br. J. Pharmacol. 2007;152:984–6. doi: 10.1038/sj.bjp.0707464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nathans J, Hogness DS. Isolation, sequence analysis, and intron-exon arrangement of the gene encoding bovine rhodopsin. Cell. 1983;34:807–14. doi: 10.1016/0092-8674(83)90537-8. [DOI] [PubMed] [Google Scholar]
- 35.Dixon RAF, et al. Cloning of the gene and cDNA for mammalian β-adrenergic receptor and homology with rhodopsin. Nature. 1986;321:75–79. doi: 10.1038/321075a0. [DOI] [PubMed] [Google Scholar]
- 36.Gloriam DE, Fredriksson R, Schiöth HB. The G protein-coupled receptor subset of the rat genome. BMC Genomics. 2007;8:338. doi: 10.1186/1471-2164-8-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagerström MC, et al. The G protein-coupled receptor subset of the chicken genome. PLoS Comput. Biol. 2006;2:e54. doi: 10.1371/journal.pcbi.0020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarkar A, Kumar S, Sundar D. The G protein-coupled receptors in the pufferfish Takifugu rubripes. BMC Bioinformatics. 2011;12(Suppl 1):S3. doi: 10.1186/1471-2105-12-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harding SD, et al. The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Res. 2017;46:D1091–D1106. doi: 10.1093/nar/gkx1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamoto K, et al. Evolution of Dopamine Receptor Genes of the D1 Class in Vertebrates. Mol. Biol. Evol. 2013;30:833–843. doi: 10.1093/molbev/mss268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoyer D. 5-HT Receptor Nomenclature: Naming Names, Does It Matter? A Tribute to Maurice Rapport. ACS Chem. Neurosci. 2017;8:908–919. doi: 10.1021/acschemneuro.7b00011. [DOI] [PubMed] [Google Scholar]
- 42.Flicek P, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aken BL, et al. Ensembl 2017. Nucleic Acids Res. 2017;45:D635–D642. doi: 10.1093/nar/gkw1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vernier P, Cardinaud B, Valdenaire O, Philippe H, Vincent JD. An evolutionary view of drug-receptor interaction: the bioamine receptor family. Trends Pharmacol. Sci. 1995;16:375–81. doi: 10.1016/S0165-6147(00)89078-1. [DOI] [PubMed] [Google Scholar]
- 45.Haug-Baltzell A, Jarvis ED, McCarthy FM, Lyons E. Identification of dopamine receptors across the extant avian family tree and analysis with other clades uncovers a polyploid expansion among vertebrates. Front. Neurosci. 2015;9:361. doi: 10.3389/fnins.2015.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boehmler W, et al. Evolution and expression of D2 and D3 dopamine receptor genes in zebrafish. Dev. Dyn. 2004;230:481–93. doi: 10.1002/dvdy.20075. [DOI] [PubMed] [Google Scholar]
- 47.Ruuskanen JO, et al. Conserved structural, pharmacological and functional properties among the three human and five zebrafish α2-adrenoceptors. Br. J. Pharmacol. 2005;144:165–177. doi: 10.1038/sj.bjp.0706057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sourbron J, et al. Serotonergic Modulation as Effective Treatment for Dravet Syndrome in a Zebrafish Mutant Model. ACS Chem. Neurosci. 2016;7:588–98. doi: 10.1021/acschemneuro.5b00342. [DOI] [PubMed] [Google Scholar]
- 49.Matthes H, et al. Mouse 5-hydroxytryptamine5A and 5-hydroxytryptamine5B receptors define a new family of serotonin receptors: cloning, functional expression, and chromosomal localization. Mol. Pharmacol. 1993;43:313–9. [PubMed] [Google Scholar]
- 50.Nuckels RJ, Forstner MRJ, Capalbo-Pitts EL, García DM. Developmental expression of muscarinic receptors in the eyes of zebrafish. Brain Res. 2011;1405:85–94. doi: 10.1016/j.brainres.2011.06.016. [DOI] [PubMed] [Google Scholar]
- 51.Lindemann L, et al. Trace amine-associated receptors form structurally and functionally distinct subfamilies of novel G protein-coupled receptors. Genomics. 2005;85:372–85. doi: 10.1016/j.ygeno.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 52.Gloriam DEI, et al. The repertoire of trace amine G-protein-coupled receptors: large expansion in zebrafish. Mol. Phylogenet. Evol. 2005;35:470–82. doi: 10.1016/j.ympev.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 53.Hashiguchi Y, Nishida M. Evolution of trace amine associated receptor (TAAR) gene family in vertebrates: lineage-specific expansions and degradations of a second class of vertebrate chemosensory receptors expressed in the olfactory epithelium. Mol. Biol. Evol. 2007;24:2099–107. doi: 10.1093/molbev/msm140. [DOI] [PubMed] [Google Scholar]
- 54.Ríos-Cardona D, Ricardo-González RR, Chawla A, Ferrell JE. A role for GPRx, a novel GPR3/6/12-related G-protein coupled receptor, in the maintenance of meiotic arrest in Xenopus laevis oocytes. Dev. Biol. 2008;317:380–8. doi: 10.1016/j.ydbio.2008.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hisano Y, et al. Comprehensive analysis of sphingosine-1-phosphate receptor mutants during zebrafish embryogenesis. Genes Cells. 2015;20:647–58. doi: 10.1111/gtc.12259. [DOI] [PubMed] [Google Scholar]
- 56.Chinen A, Hamaoka T, Yamada Y, Kawamura S. Gene duplication and spectral diversification of cone visual pigments of zebrafish. Genetics. 2003;163:663–75. doi: 10.1093/genetics/163.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sato K, et al. Two UV-Sensitive Photoreceptor Proteins, Opn5m and Opn5m2 in Ray-Finned Fish with Distinct Molecular Properties and Broad Distribution in the Retina and Brain. PLoS One. 2016;11:e0155339. doi: 10.1371/journal.pone.0155339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bellingham J, et al. Evolution of melanopsin photoreceptors: discovery and characterization of a new melanopsin in nonmammalian vertebrates. PLoS Biol. 2006;4:e254. doi: 10.1371/journal.pbio.0040254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davies WIL, et al. An extended family of novel vertebrate photopigments is widely expressed and displays a diversity of function. Genome Res. 2015;25:1666–79. doi: 10.1101/gr.189886.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iwasaki R, et al. Molecular and pharmacological characterization of zebrafish ‘contractile’ and ‘inhibitory’ prostanoid receptors. Biochem. Biophys. Res. Commun. 2013;438:353–358. doi: 10.1016/j.bbrc.2013.07.075. [DOI] [PubMed] [Google Scholar]
- 61.Reppert SM, Weaver DR, Cassone VM, Godson C, Kolakowski LF. Melatonin receptors are for the birds: molecular analysis of two receptor subtypes differentially expressed in chick brain. Neuron. 1995;15:1003–15. doi: 10.1016/0896-6273(95)90090-X. [DOI] [PubMed] [Google Scholar]
- 62.Kaiya H, Mori T, Miyazato M, Kangawa K. Ghrelin receptor (GHS-R)-like receptor and its genomic organisation in rainbow trout, Oncorhynchus mykiss. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 2009;153:438–50. doi: 10.1016/j.cbpa.2009.04.612. [DOI] [PubMed] [Google Scholar]
- 63.Kaiya H, Miura T, Matsuda K, Miyazato M, Kangawa K. Two functional growth hormone secretagogue receptor (ghrelin receptor) type 1a and 2a in goldfish, Carassius auratus. Mol. Cell. Endocrinol. 2010;327:25–39. doi: 10.1016/j.mce.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 64.Mekuchi M, et al. Molecular cloning, gene structure, molecular evolution and expression analyses of thyrotropin-releasing hormone receptors from medaka (Oryzias latipes) Gen. Comp. Endocrinol. 2011;170:374–80. doi: 10.1016/j.ygcen.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 65.Bidaud I, et al. Distribution of the mRNAs encoding the thyrotropin-releasing hormone (TRH) precursor and three TRH receptors in the brain and pituitary of Xenopus laevis: effect of background color adaptation on TRH and TRH receptor gene expression. J. Comp. Neurol. 2004;477:11–28. doi: 10.1002/cne.20235. [DOI] [PubMed] [Google Scholar]
- 66.Wang Y, et al. Identification of the receptors for prolactin-releasing peptide (PrRP) and Carassius RFamide peptide (C-RFa) in chickens. Endocrinology. 2012;153:1861–74. doi: 10.1210/en.2011-1719. [DOI] [PubMed] [Google Scholar]
- 67.Fredriksson R, Larson ET, Yan Y-L, Postlethwait J-H, Larhammar D. Novel neuropeptide Y Y2-like receptor subtype in zebrafish and frogs supports early vertebrate chromosome duplications. J. Mol. Evol. 2004;58:106–14. doi: 10.1007/s00239-003-2529-z. [DOI] [PubMed] [Google Scholar]
- 68.Fredriksson R, Sjödin P, Larson ET, Conlon JM, Larhammar D. Cloning and characterization of a zebrafish Y2 receptor. Regul. Pept. 2006;133:32–40. doi: 10.1016/j.regpep.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 69.Larsson TA, et al. Early vertebrate chromosome duplications and the evolution of the neuropeptide Y receptor gene regions. BMC Evol. Biol. 2008;8:184. doi: 10.1186/1471-2148-8-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Larhammar D, Bergqvist CA. Ancient Grandeur of the Vertebrate Neuropeptide Y System Shown by the Coelacanth Latimeria chalumnae. Front. Neurosci. 2013;7:27. doi: 10.3389/fnins.2013.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ocampo Daza D, Lewicka M, Larhammar D. The oxytocin/vasopressin receptor family has at least five members in the gnathostome lineage, inclucing two distinct V2 subtypes. Gen. Comp. Endocrinol. 2012;175:135–43. doi: 10.1016/j.ygcen.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 72.Kim D-K, et al. Revisiting the evolution of gonadotropin-releasing hormones and their receptors in vertebrates: secrets hidden in genomes. Gen. Comp. Endocrinol. 2011;170:68–78. doi: 10.1016/j.ygcen.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 73.Tello JA, Wu S, Rivier JE, Sherwood NM. Four functional GnRH receptors in zebrafish: analysis of structure, signaling, synteny and phylogeny. Integr. Comp. Biol. 2008;48:570–87. doi: 10.1093/icb/icn070. [DOI] [PubMed] [Google Scholar]
- 74.Hyndman KA, Miyamoto MM, Evans DH. Phylogeny, taxonomy, and evolution of the endothelin receptor gene family. Mol. Phylogenet. Evol. 2009;52:677–687. doi: 10.1016/j.ympev.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 75.DeVries ME, et al. Defining the origins and evolution of the chemokine/chemokine receptor system. J. Immunol. 2006;176:401–15. doi: 10.4049/jimmunol.176.1.401. [DOI] [PubMed] [Google Scholar]
- 76.Okuno T, Ishitani T, Yokomizo T. Biochemical characterization of three BLT receptors in zebrafish. PLoS One. 2015;10:e0117888. doi: 10.1371/journal.pone.0117888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chadzinska M, Golbach L, Pijanowski L, Scheer M, Verburg-van Kemenade BM. L. Characterization and expression analysis of an interferon-γ2 induced chemokine receptor CXCR3 in common carp (Cyprinus carpio L.) Dev. Comp. Immunol. 2014;47:68–76. doi: 10.1016/j.dci.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 78.Ni L-Y, et al. Identification and expression analysis of three XCR1-like receptors from Epinephelus coioides after Cryptocaryon irritans infection. Fish Shellfish Immunol. 2017;67:95–102. doi: 10.1016/j.fsi.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 79.Good S, Yegorov S, Martijn J, Franck J, Bogerd J. New Insights into Ligand-Receptor Pairing and Coevolution of Relaxin Family Peptides and Their Receptors in Teleosts. Int. J. Evol. Biol. 2012;2012:1–14. doi: 10.1155/2012/310278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yegorov S, Good S. Using paleogenomics to study the evolution of gene families: origin and duplication history of the relaxin family hormones and their receptors. PLoS One. 2012;7:e32923. doi: 10.1371/journal.pone.0032923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Elphick MR, Mirabeau O, Larhammar D. Evolution of neuropeptide signalling systems. J. Exp. Biol. 2018;221:jeb151092. doi: 10.1242/jeb.151092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tostivint H, et al. MOLECULAR EVOLUTION OF GPCRS: Somatostatin/urotensin II receptors. J. Mol. Endocrinol. 2014;52:T61–T86. doi: 10.1530/JME-13-0274. [DOI] [PubMed] [Google Scholar]
- 83.Mizusawa K, et al. Molecular cloning and expression of two melanin-concentrating hormone receptors in goldfish. Peptides. 2009;30:1990–6. doi: 10.1016/j.peptides.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 84.Kobayashi Y, Hamamoto A, Hirayama T, Saito Y. Molecular cloning, expression, and signaling pathway of four melanin-concentrating hormone receptors from Xenopus tropicalis. Gen. Comp. Endocrinol. 2015;212:114–123. doi: 10.1016/j.ygcen.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 85.Nocillado JN, et al. The Kiss2 receptor (Kiss2r) gene in Southern Bluefin Tuna, Thunnus maccoyii and in Yellowtail Kingfish, Seriola lalandi - functional analysis and isolation of transcript variants. Mol. Cell. Endocrinol. 2012;362:211–20. doi: 10.1016/j.mce.2012.06.024. [DOI] [PubMed] [Google Scholar]
- 86.Ho JCW, Jacobs T, Wang Y, Leung FC. Identification and characterization of the chicken galanin receptor GalR2 and a novel GalR2-like receptor (GalR2-L) Gen. Comp. Endocrinol. 2012;179:305–12. doi: 10.1016/j.ygcen.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 87.Martins RST, et al. Novel galanin receptors in teleost fish: Identification, expression and regulation by sex steroids. Gen. Comp. Endocrinol. 2014;205:109–120. doi: 10.1016/j.ygcen.2014.06.030. [DOI] [PubMed] [Google Scholar]
- 88.Maugars G, Dufour S. Demonstration of the Coexistence of Duplicated LH Receptors in Teleosts, and Their Origin in Ancestral Actinopterygians. PLoS One. 2015;10:e0135184. doi: 10.1371/journal.pone.0135184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bader M, Alenina N, Andrade-Navarro MA, Santos RA. Mas and Its Related G Protein-Coupled Receptors, Mrgprs. Pharmacol. Rev. 2014;66:1080–1105. doi: 10.1124/pr.113.008136. [DOI] [PubMed] [Google Scholar]
- 90.Pinto PIS, et al. Duplicated membrane estrogen receptors in the European sea bass (Dicentrarchus labrax): phylogeny, expression and regulation throughout the reproductive cycle. J. Steroid Biochem. Mol. Biol. 2017 doi: 10.1016/j.jsbmb.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 91.Alexander SP, et al. The concise Guide to Pharmacology 2017/18: G protein-coupled receptors. Br. J. Pharmacol. 2017;174(Suppl 1):S17–S129. doi: 10.1111/bph.13878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pasquier J, et al. Evolution of gene expression after whole-genome duplication: New insights from the spotted gar genome. J. Exp. Zool. Part B Mol. Dev. Evol. 2017;328:709–721. doi: 10.1002/jez.b.22770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genomic data from the Ensembl releases is freely accessible online in few simple steps: (1) log to http://www.ensembl.org or to the Ensembl archives https://www.ensembl.org/info/website/archives/index.html (2) query the name of the receptor of interest or the provided codes; (3) select (usually) the first Gene hit on the list; (4) click on “gene tree” in the left-hand panel; (5) browse the tree data: in particular, branches can be expanded and sequences visualized. The sequence data used to construct the guide trees is accessible upon request. Note added following acceptance: The genes identified in this manuscript have been submitted to the HGNC, and will be submitted to the MGNC, RGNC, XNC, and ZNC nomenclature committees on publication.