Brukman et al. review cell–cell fusion mechanisms, focusing on the identity of the fusogens that mediate these processes and the regulation of their activities.
Abstract
Cell–cell fusion remains the least understood type of membrane fusion process. However, the last few years have brought about major advances in understanding fusion between gametes, myoblasts, macrophages, trophoblasts, epithelial, cancer, and other cells in normal development and in diseases. While different cell fusion processes appear to proceed via similar membrane rearrangements, proteins that have been identified as necessary and sufficient for cell fusion (fusogens) use diverse mechanisms. Some fusions are controlled by a single fusogen; other fusions depend on several proteins that either work together throughout the fusion pathway or drive distinct stages. Furthermore, some fusions require fusogens to be present on both fusing membranes, and in other fusions, fusogens have to be on only one of the membranes. Remarkably, some of the proteins that fuse cells also sculpt single cells, repair neurons, promote scission of endocytic vesicles, and seal phagosomes. In this review, we discuss the properties and diversity of the known proteins mediating cell–cell fusion and highlight their different working mechanisms in various contexts.
Introduction
The dynamic organization of cells depends on protein-controlled membrane remodeling processes that divide and fuse membranes. Fusion of intracellular membranes is a key stage in secretion, protein and lipid trafficking, and in the maintenance of ER and mitochondrial networks, and defects in these fusion processes have been linked to mitochondrial, lysosomal storage (Ballabio and Gieselmann, 2009), and degenerative disorders (Ranieri et al., 2013). Diverse enveloped viruses, including many human pathogens, infect cells by envelope–cell membrane fusion. Fusion between cells (referred to here as “cell fusion”), the focus of this review, is essential in fertilization and in development of tissues and organs such as skeletal muscles and placenta.
Fusion processes differ widely in the composition of the fusing membranes, biological context, and regulatory mechanisms. In some fusions, the proteins that mediate fusion (referred to as “fusion proteins” or “fusogens”) have to be present on only one of the fusing membranes (unilateral mechanism). Other fusions require the same or different fusogens to be present on both membranes (bilateral homotypic vs. bilateral heterotypic mechanisms). However, in all fusion processes, the function of the fusion protein machinery is to drive the transition from the pre-fusion to post-fusion state by bringing lipid bilayers into immediate contact, catalyzing the formation of energy-intensive fusion intermediates, and opening a fusion pore (Sapir et al., 2008). Fusion itself involves local rupture of the continuity of each of the lipid bilayers and their rejoining. The mechanisms and pathways underlying cell fusion have been studied in both biological and protein-free lipid bilayers using different theoretical and experimental approaches yielding several important concepts (Chernomordik and Kozlov, 2008; Markvoort and Marrink, 2011). Before fusion, characteristic distances between opposing plasma membranes are controlled by specific cell–cell adhesion proteins and vary in range from 10 to a few tens of nanometers (Leikina et al., 2004; Dhanyasi et al., 2015). Bringing membrane bilayers closer to each other requires displacement of membrane proteins toward the periphery of the fusion site and, at very close distances comparable with the thickness of the lipid monolayer (∼2 nm), overcoming very strong repulsive interactions related to hydration forces or thermal fluctuations (Chernomordik and Kozlov, 2003).
A strong bending of one or both membrane bilayers brings them into immediate contact (within a few nanometers) and facilitates a local disruption and rearrangement of the lipid monolayers (Chernomordik and Kozlov, 2003). The pathway of many fusion processes starts with hemifusion, a merger between contacting monolayers of the fusing bilayers that allows lipid mixing between the membranes (Chernomordik et al., 1987; Chernomordik and Kozlov, 2003). A subsequent merger of the distal monolayers generates a nascent fusion pore and allows content mixing (Fig. 1). While this fusion-through-hemifusion pathway was first described for fusion of protein-free bilayers formed from lipids that facilitate monolayer curvatures characteristic for either hemifusion intermediates or lipidic pores (Chernomordik et al., 1987), during biological fusion, proteins may serve as critical structural components of the early fusion intermediates. For instance, some studies suggest that Ca2+-triggered exocytosis involves formation of a proteinaceous fusion pore, the rim of which is entirely or partially lined by amino acid residues of transmembrane domains of SNARE proteins (Chang et al., 2017). It has been also suggested that under some conditions, influenza virus hemagglutinin initiates fusion by puncturing one of the contacting membranes to form a leaky “rupture-insertion” structure, and this structure by a yet-unexplained mechanism facilitates hemifusion and opening of a fusion pore (Haldar et al., 2018). The place of these intermediates in the productive fusion pathway that yields expanding fusion pores remains to be clarified. The hypothesis that fusion starts with a channel-like proteinaceous pore can be substantiated by finding the mechanisms that drive its transition to a larger lipidic pore and ways to specifically block this transition. To verify that the rupture-insertion structure is not a branch-off the normal fusion pathway, leakage measurements will have to be accompanied by content mixing assays. In our opinion, striking similarities between lipid dependences and properties of the key intermediates in diverse biological membrane fusion processes and in fusion of protein-free lipid bilayers (Chernomordik and Kozlov, 2003, 2008) argue for similar pathways and suggest that proteins catalyze a fusion-through-hemifusion fusion pathway (Fig. 1 B) that is intrinsic for membrane bilayers and driven by membrane bilayer stresses.
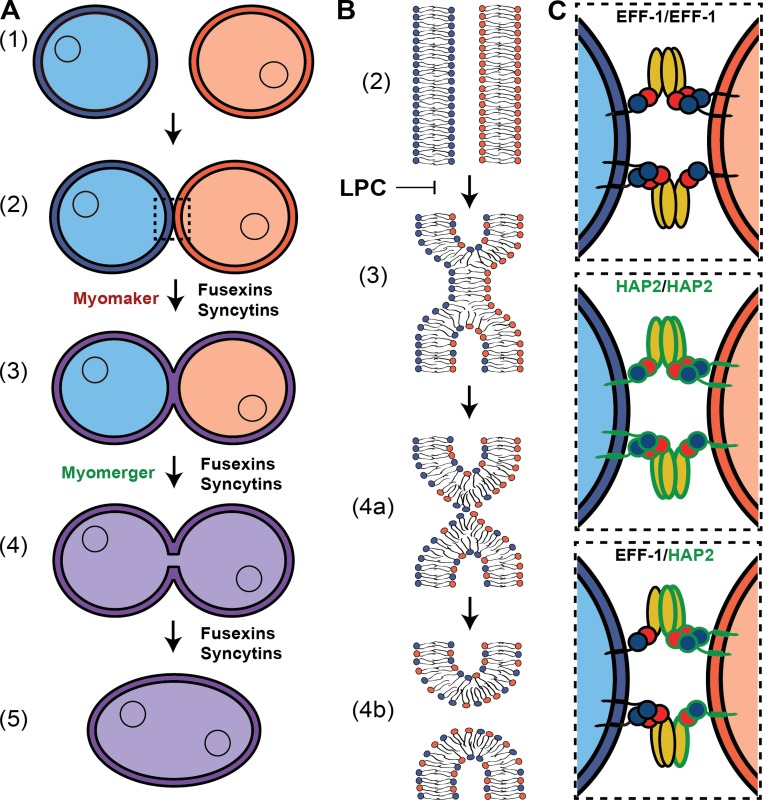
Figure 1.
Mechanisms of cell–cell fusion. (A) The pathway of cell–cell fusion. Ready-to-fuse cells (1) recognize and closely appose each other (2) and undergo hemifusion (3), i.e., the merger of the outer monolayers of two membrane bilayers, allowing redistribution of the lipid markers between the cells (note that both distal monolayers of the membranes and cell contents remain distinct). Opening of a fusion pore in the hemifusion structure allows the mixing of the cytoplasmic contents (4), and pore expansion completes joining of two cells into one (5). While Myomaker/Myomerger, syncytins, and fusexins seem to be for now the only proteins necessary for specific fusion processes, they are most likely working with other players, some of which, especially for myoblasts, are already identified. Fusexins and syncytins mediate all the stages of the fusion process; in contrast, Myomaker is required for an early stage involving the transition to hemifusion, while Myomerger is required for a later stage between hemifusion and opening of fusion pores (see the main text). (B) Schematic representation of the lipid rearrangements during the events explained in A. LPC blocks hemifusion by inhibiting the bending of the contacting monolayers (Chernomordik and Kozlov, 2003). (C) Inset from A 2: Protein fusogens are necessary to overcome the energetic barriers of hemifusion and opening and expansion of the fusion pore. Examples display bilateral and homotypic fusions mediated by C. elegans EFF-1 (upper panel) and Arabidopsis HAP2 (middle panel) as well as a bilateral and heterotypic fusion between them (lower panel; Valansi et al., 2017).
Up to a third of the cell nuclei in animals, from Caenorhabditis elegans to humans, are found in multinucleated cells formed by cell fusion (in C. elegans mostly in epithelia and in humans in skeletal muscle; Podbilewicz and White, 1994). However, most cells remain mononucleated, emphasizing that cell fusion is tightly regulated. In this review we discuss different cell fusion processes and proteins suggested to mediate them, intercalating videos illustrating the dynamics of cell fusion in different systems.
Challenges in identifying and exploring cell–cell fusion
Fusion processes mediated by viral and intracellular proteins are often triggered by calcium (exocytosis), acidification of endosomal compartments containing internalized virions (influenza virus), and/or virus interactions with receptors and fusion cofactors in host cells (human immunodeficiency virus [HIV] and Dengue virus). Such events are relatively fast (milliseconds to hours; reviewed in Jahn and Scheller, 2006; Podbilewicz, 2014). In contrast, complex and multistep differentiation processes that prepare cells for fusion in their biological context may take days, and the environmental cues that trigger the actual fusion events (seconds to minutes) are yet to be established.
How do we distinguish proteins involved in the actual fusion stage from proteins that only function at pre- and post-fusion stages? Identifying proteins as fusion proteins (fusogens) depends on diverse experimental approaches characterizing their fusogenic activities and structural characteristics. The gold standard is that a fusogen (or fusogenic complex) has to be (1) necessary for fusion, (2) present on the fusing membranes at the right time and place, and (3) sufficient to fuse membranes that normally do not fuse. Additionally, a tertiary structure similar to well-characterized fusogens is commonly used by many researchers for validation of new fusogens. Proteins that meet all these requirements are considered bona fide fusogens, and we suggest a scoring system based on gold standards for fusogens (Table 1). A decade ago, when a similar table was assembled, the best-characterized fusogens were viral and intracellular; the only well-characterized cell–cell fusogens were identified in the placenta of mammals (Syncytins [Syns]) and epithelia of nematodes (Fusion Family [FF]; Oren-Suissa and Podbilewicz, 2007). Recent studies have suggested new candidate proteins in fusion of gametes (Hapless 2 [HAP2]) and of muscle cells (Myomaker/Myomerger; Hernández and Podbilewicz, 2017; Sampath et al., 2018). Our discussion of different fusion processes and machineries will focus on recent mechanistic discoveries at the molecular, structural, and biochemical levels.
Table 1. Examples of candidate fusogens.
| Protein | Family | Organism | Suggested fusion event | Fusogenic scorea | Essential for fusion (0–2) | Structural similarity to fusogens (0–2) | Expressed at the time and place of fusion (0–2) | Sufficiency (0–4)b | References | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | |||||||||
| HA | Class I viral fusogens | Influenza virus | Viral infection | 10 | + | + | + | + | + | + | + | Reviewed in Kielian and Rey, 2006 |
| p14 | FAST proteins (Class IV viral fusogens) | Reptilian orthoreoviruses | Infected cells | 10 | + | + | + | + | + | + | + | Reviewed in Key and Duncan, 2014 |
| E1 | Fusexins (Class II viral fusogens) | Semliki Forest virus | Viral infection | 9.5 | + | + | + | + | + | + | ± | Reviewed in Kielian and Rey, 2006 |
| vSNARES, tSNARES | SNAREs (endoplasmic fusogens) | Eukaryotes and Archaea | Intracellular vesicles | 9 | + | + | + | + | + | ND | + | Reviewed in Jahn and Scheller, 2006 |
| EFF-1 | Fusexins (FF) | C. elegans; Nematodes | Epithelia, vulva, pharynx, axons | 9 | + | + | + | + | + | + | ND | Mohler et al., 2002; Shemer et al., 2004; Podbilewicz et al., 2006; Avinoam et al., 2011 |
| Syncytins | Class I viral fusogens | Placental mammals | Placenta, osteoclasts, myoblasts, cancer cells | 9 | + | + | + | + | + | + | ND | Blond et al., 2000; Mi et al., 2000; Bjerregaard et al., 2006; Antony et al., 2007; Dupressoir et al., 2011; Søe et al., 2011 |
| Atlastins | Dynamin (endoplasmic fusogen) | Eukaryotes | Endoplasmic reticulum | 8 | + | + | + | + | ND | ND | + | Reviewed in Hu and Rapoport, 2016 |
| HAP2/GCS1 | Fusexins (HAP2) | Arabidopsis; plants | Fertilization (sperm) | 8 | + | + | + | ND | + | + | ND | Mori et al., 2006; von Besser et al., 2006; Valansi et al., 2017 |
| AFF-1 | Fusexins (FF) | C. elegans; Nematodes | Epithelia, vulva, pharynx, dendrites | 8 | + | ± | + | + | + | + | ND | Sapir et al., 2007; Avinoam et al., 2011 |
| Myomaker | Multi-pass transmembrane protein | Mus musculus; Human | Myoblasts | 6 | + | ND | + | + | + | ND | ND | Millay et al., 2013; Bi et al., 2017; Gamage et al., 2017; Quinn et al., 2017; Zhang et al., 2017a; Leikina et al., 2018 |
| Myomerger | Single-pass transmembrane protein | |||||||||||
| HAP2/GCS1 | Fusexins (HAP2) | T. thermophila; Trypanosomes | Mating | 6 | + | + | + | ND | ND | ND | ND | Pinello et al., 2017 |
| HAP2/GCS1 | Fusexins (HAP2) | Chlamydomonas; Algae | Fertilization (mt− cell) | 6 | + | + | + | ND | ND | ND | ND | Liu et al., 2008; Fédry et al., 2017 |
| Bouncer | Ly6/uPAR | Danio reri and Oryzias latipes | Fertilization (egg) | 5 | + | − | + | + | ND | ND | ND | Herberg et al., 2018 |
| Izumo1 | IgSF | M. musculus; Mammals | Fertilization (sperm) | 4 | + | − | + | − | − | ND | ND | Inoue et al., 2005, 2013, 2015; Chalbi et al., 2014 |
| Juno | Folate receptors | M. musculus; Mammals | Fertilization (oocyte) | 4 | + | − | + | − | − | ND | ND | Bianchi et al., 2014; Kato et al., 2016 |
| SPE-9 | EGF repeats | C. elegans; Nematodes | Fertilization (sperm) | 4 | + | ND | + | ND | ND | ND | ND | Singson et al., 1998 |
| Sns | IgSF | Drosophila | FCM | 4 | + | ND | + | − | − | ND | ND | Bour et al., 2000; Shilagardi et al., 2013 |
| Duf/Kirre | IgSF | Drosophila | FC | 4 | + | ND | + | − | − | ND | ND | Ruiz-Gómez et al., 2000; Shilagardi et al., 2013 |
| Bindin | Bindins | S. purpuratus; Sea urchins | Fertilization (sperm) | 3 | ND | ND | + | ND | ND | ND | + | Vacquier and Moy, 1977; Glabe, 1985; Vacquier, 2012 |
| PRM1 | Tetraspanin integral protein | S. cerevisiae; Yeast | Mating | 3 | ± | ND | + | − | ND | ND | ND | Heiman and Walter, 2000; Olmo and Grote, 2010 |
The fusogenic score (0–10) was calculated using the following scoring system: + (requirement fulfilled) = max score, − (requirement not fulfilled) = 0 points, ND (not determined) = 0 points, and ± (requirement partially fulfilled) = half of max score.
The fusogenic score (0–10).
(A) Sufficient in situ (0–1). (B) Fuse heterologous cells (0–1). (C) Fuse pseudo-typed virus (0–1). (D) In vitro liposome fusion (0–1).
Gamete fusion
Sexual reproduction is widely distributed in the tree of life. In sexually reproducing organisms, permanent or transient cell–cell fusion is essential for genetic transfer (Video 1). Here we will focus on mechanisms of gamete fusion in eukaryotes.
Gamete fusion mediated by HAP2/Germ cell–specific 1 (GCS1)
Recent years have brought about a breakthrough in the identification of proteins that fuse gametes. HAP2, also named GCS1, is a type I transmembrane protein that functions in late stages of gamete fusion in different species including protists (Liu et al., 2008), flowering plants (Mori et al., 2006; von Besser et al., 2006), and invertebrates (reviewed in Hernández and Podbilewicz, 2017). It is localized in at least one of the fusing membranes at the moment of fertilization and is required for fusion (i.e., necessary and present; Mori et al., 2006; von Besser et al., 2006; Liu et al., 2008). Recent evidence in the algae Chlamydomonas reinhardtii (Fédry et al., 2017; Feng et al., 2018), the flowering plant Arabidopsis thaliana (Valansi et al., 2017; Fédry et al., 2018), and the ciliate Tetrahymena thermophila (Pinello et al., 2017) revealed that HAP2 has similarities with the eukaryotic somatic fusogen epithelial fusion failure-1 (EFF-1) and class II viral fusogens. Arabidopsis HAP2 expression in heterologous mammalian cells results in their hemifusion and cytoplasmic content mixing (Valansi et al., 2017). Moreover, vesicular stomatitis virus with a deletion of G glycoprotein–HAP2 virus expressing HAP2 instead of the viral G glycoprotein effectively enters cells. These studies of the HAP2 sufficiency for fusion indicate that HAP2 is indeed a bona fide fusogen (Valansi et al., 2017; Table 1). Yet it remains unclear whether HAP2 from different species use unilateral, bilateral, or hybrid mechanisms in vivo and in cell-free systems (Table 2). Arabidopsis HAP2 induces fusion only when it is present in both of the opposing membranes, suggesting a bilateral mechanism of action, similar to the related somatic fusogen EFF-1 (Valansi et al., 2017; Fig. 1 C). On the other hand, Chlamydomonas HAP2 is required only in the minus gamete (Liu et al., 2008), and although some HAP2 expression was detected in Arabidopsis ovules (Borges et al., 2008), the deletion of this gene produces male-specific sterility (Johnson et al., 2004; von Besser et al., 2006). This implies that in vivo, sperm HAP2 acts in trans with other unknown egg proteins or uses a unilateral mechanism (Valansi et al., 2017). Perhaps, unilateral fusion requires HAP2 and another sperm protein, and expression of HAP2 alone in a heterologous system is insufficient for its unilateral action. In the slime mold Dictyostelium discoideum, there are three mating types and at least two different genes encoding HAP2/GCS1 proteins. Genetic analyses of the mating-type specific gamete fusion in D. discoideum suggest that the fusogens form complexes in trans supporting a bilateral mechanism between Type I and II gametes (Okamoto et al., 2016). The structural similarities between HAP2 proteins and the class II viral fusogens suggest that they share common functional features. Class II viral fusogens possess an amphiphilic loop at the tip of domain DII that inserts into and destabilizes the host cell membrane (reviewed in Podbilewicz, 2014). Structural and biochemical analyses suggest that HAP2 proteins of Chlamydomonas, Arabidopsis, and trypanosomes interact with membranes through similar regions (composed of loops and/or α-helixes) containing hydrophobic amino acids (Fédry et al., 2017, 2018; Feng et al., 2018). However, this ability to interact with membranes does not imply unilateral action: other cellular fusogens that use bilateral mechanisms have domains that directly interact with membranes, yet still join trans-complexes which mediate fusion (e.g., atlastins and synaptotagmins; Table 2; Chapman, 2008; Faust et al., 2015; Liu et al., 2015b).
Table 2. Mechanism of action for some of the best candidate fusogens.
| Protein | Organism | Homotypic/Heterotypic | Bilateral/Unilateral | Dependent on regulators | Triggers | References |
|---|---|---|---|---|---|---|
| HA | Influenza virus | Heterotypic | Unilateral | Receptors | Low pH | Reviewed in Kielian and Rey, 2006 |
| p14 | Reptilian orthoreoviruses | Heterotypic | Unilateral | Cholesterol, calcium | ND | Reviewed in Key and Duncan, 2014 |
| E1 | Semliki Forest virus | Heterotypic | Unilateral | Cholesterol | Low pH | Reviewed in Kielian and Rey, 2006 |
| vSNARES, tSNARES | Eukaryotes and Archaea | Heterotypic | Bilateral | Synaptotagmin, complexin, and others | Docking | Reviewed in Jahn and Scheller, 2006 |
| EFF-1 | C. elegans; Nematodes | Hetero/Homo | Bilateral | Dynamin, vATPase, PS | ND | Mohler et al., 2002; Shemer et al., 2004; Podbilewicz et al., 2006; Avinoam et al., 2011; Neumann et al., 2015 |
| Atlastins | Eukaryotes | Homotypic | Bilateral | Dimerization | GTP hydrolysis (?) | Reviewed in Hu and Rapoport, 2016 |
| Syncytins | Placental mammals | Heterotypic | Unilateral | Receptor, Anxs, PS | Externalization of PS (?) | Blond et al., 2000; Mi et al., 2000; Bjerregaard et al., 2006; Dupressoir et al., 2011; Søe et al., 2011 |
| HAP2/GCS1 | Arabidopsis; plants | Heterotypic | Uni/Bi | ND | ND | Valansi et al., 2017 |
| AFF-1 | C. elegans; Nematodes | Hetero/Homo | Bilateral | ND | ND | Sapir et al., 2007; Avinoam et al., 2011 |
| Myomaker | M. musculus; Human | Homotypic | Bilateral | Anxs, PS binding proteins | PS exposure (?) | Millay et al., 2013; Bi et al., 2017; Gamage et al., 2017; Quinn et al., 2017; Zhang et al., 2017a; Leikina et al., 2018 |
| Myomerger | Heterotypic | Unilateral | ||||
| HAP2/GCS1 | T. thermophila; Trypanosomes | Hetero/Homo | Uni/Bi | ND | ND | Pinello et al., 2017 |
| HAP2/GCS1 | Chlamydomonas; Algae | Heterotypic | Unilateral | ND | ND | Fédry et al., 2017 |
ND, not determined; ?, not confirmed.
Fertilization in organisms lacking HAP2/GCS1
Despite the wide distribution of HAP2 among eukaryotes, some lineages like nematodes, fungi, and vertebrates lack any close HAP2 orthologue (Speijer et al., 2015; Valansi et al., 2017; Fédry et al., 2018). Although the fusogens involved in gamete fusion in these HAP2-lacking species are still unknown, several proteins have been shown to be relevant to this process, especially those related to the early recognition between gametes (Table 1). Examples include PRM1 from yeast (Heiman and Walter, 2000), the SPE-9 class from C. elegans (reviewed in Nishimura and L’Hernault, 2010; Video 1), and Bindin from sea urchins (Vacquier and Moy, 1977).
In mammals, certain proteins in both male and female gametes are necessary for gamete fusion, but none have been determined sufficient for membrane fusion (Wright and Bianchi, 2016). Izumo1, an immunoglobulin superfamily member, localizes to the sperm acrosomal membrane (Inoue et al., 2005). Following acrosome exocytosis, Izumo1 migrates to the equatorial segment of the sperm (Satouh et al., 2012), the site of fusion with the egg. Sperm of Izumo1 knockout mice fails to fuse, leading to male infertility (Inoue et al., 2005). Females lacking the GPI-anchored Izumo1 Receptor (Juno) are also infertile due to defects in gamete fusion (Bianchi et al., 2014). Shortly after sperm–egg fusion, Juno is shed from the egg membrane, preventing further sperm binding, thus contributing to the block of polyspermy (Bianchi et al., 2014). Juno and Izumo1 are conserved among mammals, including humans, where antibodies against Izumo1 and mutations in Juno are associated with female infertility (Clark and Naz, 2013; Yu et al., 2018). The interaction between these adhesion partners is bilateral in humans (Aydin et al., 2016; Ohto et al., 2016) and appears to be species specific (Bianchi and Wright, 2015). Heterologous cells expressing Juno or Izumo1 are able to adhere to sperm or eggs, respectively; however, this interaction is insufficient to mediate cell–cell fusion, suggesting the existence of additional molecular players (Inoue et al., 2013, 2015; Chalbi et al., 2014; Kato et al., 2016). In addition to Izumo1-Juno interactions, the egg-specific tetraspanin CD9 is necessary for gamete fusion (Kaji et al., 2000; Le Naour et al., 2000; Miyado et al., 2000) but its role seems to be related to the organization of microvilli (Runge et al., 2007). In sperm, the immunoglobulin-like protein SPACA6, is essential for gamete fusion (Lorenzetti et al., 2014). Recently, the egg GPI-anchored protein Bouncer was shown to be necessary during Zebrafish gamete fusion (Herberg et al., 2018). Bouncer mediates fertilization in a species-specific manner, and its heterologous expression in eggs of a different fish (Medaka) is sufficient to induce fusion with Zebrafish sperm (Herberg et al., 2018). Bouncer has a mammalian orthologue (SPACA4) that is sperm specific; more studies regarding the fusogenic role of both Bouncer and SPACA4 are required (Table 1).
At present, gamete fusogens in organisms lacking HAP2-like proteins remain to be identified. During evolution, HAP2 orthologues may have diverged so rapidly that bioinformatic tools are unable to detect distant phylogenetic relationships. In this sense, these hypothetical divergent proteins may conserve structural similarities to HAP2 or to other fusexins (e.g., EFF-1 and anchor cell fusion failure-1 [AFF-1] from nematodes; Table 1; Fédry et al., 2017; Pinello et al., 2017; Valansi et al., 2017). Another possibility is that HAP2 genes were replaced by different fusogens of viral origin acquired by horizontal gene transfer during evolution. Alternatively, these phylogenetic groups may depend on new eukaryotic fusogens unrelated to HAP2 or viral fusogens (Doms, 2017).
Somatic cell fusion and organ formation
In the next sections, we discuss cell–cell fusion in different tissues and organs and focus on fusion processes during the development of muscles, placenta, bones, stem cells, and synergid-endoplasm. We also review cell fusion in disease (cancer and viral infections) and then analyze the detailed map of cell–cell fusions in C. elegans, describing how fusion proteins sculpt cells in the epidermal, digestive, reproductive, and nervous systems.
Myoblast fusion
Skeletal muscles are composed of bundles of elongated multinucleated myofibers that form by fusion of mononucleated myoblasts. Myoblast fusion is necessary for myofiber maintenance, growth, and regeneration (Sampath et al., 2018).
Genetic work in Drosophila melanogaster embryos led to a model of myoblast fusion in which two distinct populations of myoblasts differentiate and cooperate during fiber formation. The founder cells (FCs) seed the formation of specific muscle fibers by fusion with fusion-competent myoblasts (FCMs; Bate, 1990; Video 2). More recent reports revealed a ring of FCM/FC adhesion molecules encircling the F-actin–enriched focus (Kesper et al., 2007). These podosome-like structures formed by plasma membranes of FCMs insert into FCs (Sens et al., 2010). The FCs mount a Myosin II– and spectrin-mediated response that controls the diameter and shape of the protrusions from the FCMs to promote fusion (Kim et al., 2015; Duan et al., 2018). In contrast to the mechanism of myoblast fusion at the tip of the protrusion suggested for Drosophila embryo, later in the development of indirect flight muscles, multiple fusion pores are formed in extended (∼4 µm) and tight (inter-cellular distance of ∼20 nm) membrane contacts (Dhanyasi et al., 2015). Studies in different organisms have identified molecular components involved in the generation of multinucleated muscle cells including actin, adhesion, and endocytic machineries; tetraspanin CD9, ferlins, JAM-B, and JAM-C receptors; and immunoglobulin domain–containing membrane receptors such as Kirrel in Zebrafish embryo (Tachibana and Hemler, 1999; Srinivas et al., 2007; Posey et al., 2011; Powell and Wright, 2011; Leikina et al., 2013; Kim et al., 2015; Schejter, 2016; Sampath et al., 2018).
Formation of muscle fibers is inhibited by the hemifusion-inhibiting lipid lysophosphatidylcholine (LPC), promoted by the phosphoinositide PI(4,5)P2, and associated with cell surface exposure of phosphatidylserine (PS; van den Eijnde et al., 2001; Hochreiter-Hufford et al., 2013; Leikina et al., 2013; Bothe et al., 2014; Whitlock et al., 2018). Myoblast fusion is also dependent on PS-binding proteins annexins (Anxs), stabilins, and the PS-exposure–mediating protein TMEM16E (Leikina et al., 2013; Kim et al., 2016; Hamoud et al., 2018; Whitlock et al., 2018). Two muscle-specific proteins, Myomaker (Tmem8c) and Myomerger (Gm7325/Myomixer/Minion), have been shown to be essential for vertebrate myoblast fusion (Millay et al., 2013; Bi et al., 2017; Gamage et al., 2017; Quinn et al., 2017; Zhang et al., 2017a; Leikina et al., 2018). Myomaker has seven transmembrane domains, and Myomerger is an 84–amino acid integral membrane protein with a C-terminal ectodomain (Millay et al., 2013; Leikina et al., 2018). Expressing both Myomaker and Myomerger in fibroblasts, but not separately, is sufficient to induce cell–cell fusion. In contrast to Myomaker, which is able to support fusion only if expressed in both fusing cells, Myomerger is required in only one of the cells (Table 2).
To identify the proteins and lipids directly involved in myoblast fusion, ready-to-fuse murine myoblasts were accumulated in the presence of LPC without blocking pre-fusion differentiation, followed by LPC removal to observe robust and synchronized fusion (Leikina et al., 2013). Specific treatments such as antibodies and inhibitors applied during LPC removal focused the analysis of the contributions of candidate proteins and lipids on the cell fusion stage of myogenesis (Leikina et al., 2013, 2018). This approach combined with application of three complementary fusion assays (syncytium formation, lipid mixing, and content mixing) allowed researchers to distinguish proteins involved only in pre-fusion stages of myogenesis from proteins involved in hemifusion or in pore formation and expansion (Fig. 1). It was shown that cell-surface AnxA1 and AnxA5 (Leikina et al., 2013) and Myomaker (Gamage et al., 2017; Leikina et al., 2018) function at the hemifusion stage of myoblast fusion. The transition from myoblast hemifusion to syncytium formation (multinucleation) depends on cell metabolism, dynamin 2 activity, and phosphoinositide PI(4,5)P2 concentration (Leikina et al., 2013), as well as the cell-surface Myomerger (Leikina et al., 2018). While fusion of Myomerger-deficient myoblasts stall at hemifusion, Myomaker-deficient myoblasts do not form even early hemifusion intermediates, confirming that Myomaker functions at or upstream of hemifusion and that Myomerger probably drives pore formation (Leikina et al., 2018). Myomaker does not need Myomerger to mediate hemifusion, and Myomerger does not need Myomaker to complete the fusion once hemifusion is reached, demonstrating independent and distinct functions of these proteins at different fusion steps (Fig. 1 A). The specific roles of Myomaker in hemifusion and Myomerger in fusion pore opening, as well as the mechanisms by which muscle microenvironment trigger and coordinate fusion-related proteins and lipids in distinct stages of the fusion pathway, remain to be clarified (Schejter, 2016; Sampath et al., 2018; Table 2).
Placental trophoblast fusion
Fusion of villous cytotrophoblasts throughout pregnancy generates syncytiotrophoblast, a giant cell with more than 10 billion nuclei and an ∼10-m2 surface area that serves as the main fetomaternal barrier (Pötgens et al., 2004; Bolze et al., 2017). This cell fusion depends on the endogenous retroviral proteins, namely, Syn1 and Syn2 in primates, and SynA and SynB in rodents. Interactions of Syn1 and Syn2 with their receptors, ubiquitous neutral amino acid transporters ASCT1 and ASCT2, trigger this fusogenic restructuring (Pötgens et al., 2004; reviewed in Aguilar et al., 2013). Syncytin-mediated trophoblast fusion is accompanied by formation of podosome-like plasma membrane protrusions (Wang et al., 2014) and PS exposure (Pötgens et al., 2004), and involves AnxA5 (Degrelle et al., 2017). Specific contributions of different syncytins and the mechanisms that ensure the appropriate timing and specificity of this fusion process (trophoblasts fuse only with each other and the syncytiotrophoblast) remain to be determined (see Movie S 3 in Wang et al., 2014).
Osteoclast precursor fusion
Bone remodeling during development and in bone maintenance depends on the balance between bone formation by osteoblasts and bone resorption by osteoclasts. The latter are generated by fusion of osteoclast precursors (preosteoclasts) derived from monocyte/macrophage lineage after macrophage colony stimulating factor and RANKL stimulation (Levaot et al., 2015). Larger osteoclasts with multiple nuclei resorb bones better than mononucleated osteoclasts (Lees and Heersche, 1999), and thus changes in fusion efficiency are expected to disrupt normal bone remodeling. As with myoblasts and trophoblasts, fusion of preosteoclasts involves actin-enriched podosome-like protrusions (Oikawa and Matsuo, 2012) and distinct populations of precursor cells that differ in protein expression and in fusion competence (Levaot et al., 2015; and see Movie 1 therein). Several proteins, including CD9 (Ishii et al., 2018), a dendritic cell specific transmembrane protein (Yagi et al., 2005), the endocytotic machinery including dynamin 2, clathrin, and AP-2 (Shin et al., 2014; Verma et al., 2014), and Syn1 (Søe et al., 2011), are involved in osteoclast formation and may function in fusion itself (Table 1).
Recent analysis using a fusion synchronization approach has established that dendritic cell specific transmembrane protein and Syn1 are involved in the human preosteoclast hemifusion (Verma et al., 2018). In addition, this fusion stage depends on TMEM16F-mediated exposure of PS, AnxA5 (PS-binding protein), and S100A4 (Anx-binding protein; Table 2). Again bearing similarity to myoblasts, generation of multinucleated osteoclasts depends on cell metabolism and dynamin 2 (Shin et al., 2014; Verma et al., 2014) at the stage of the expansion of local membrane connections (Verma et al., 2014). These findings indicate that preosteoclast fusion is controlled by a complex multiprotein fusion machinery, and future studies will likely expand the list of the protein components involved and clarify their role.
Stem cell fusion and fusion in liver and eye
While the role of cell fusion in myogenesis, osteoclastogenesis, placentogenesis, and fertilization is well established, there are additional biological processes that have been hypothesized to depend on fusion. In analogy to fusion of differentiating skeletal muscle stem cells (satellite cells) that plays a key role in muscle regeneration, fusion has been suggested to be involved in the repair and regeneration of other tissues mediated by adult and embryonic stem cells (Pesaresi et al., 2018). Bone marrow adult stem cells (BMSCs) differentiate into many lineages including hepatocytes, neurons, and cardiomyocytes (Terada et al., 2002; Guo et al., 2018). Through bloodstream circulation, BMSCs come into contact with different tissues, allowing these cells to be involved in the regeneration of different types of tissues. Do BMSCs facilitate the regeneration by fusion to the resident cells? This hypothesis has been substantiated by reports that bone marrow–derived cells can fuse with certain types of cells including Purkinje neurons, cardiomyocytes, and hepatocytes (reviewed in Pesaresi et al., 2018). In addition to a possible role of BMSC fusion, tissue regeneration can also involve fusion between mesenchymal and embryonic stem cells (Sottile et al., 2016). In contrast to the very efficient fusion of the satellite cells, BMSC fusion and mesenchymal stem cell fusion events are very rare (generally under 2% of cells in the population and as low as ∼1 in 500,000 cells [Terada et al., 2002]). This hinders unambiguous analysis of their potential role and mechanism in tissue regeneration, and the significance of their in vivo regeneration of tissues other than skeletal muscle is still lacking solid evidence (Kajstura et al., 2005; Lizier et al., 2018; Pesaresi et al., 2018).
Cell fusion has been also discussed as a possible mechanism of formation of polyploid liver cells, but while hepatocytes can fuse in vitro, the physiological relevance in vivo remains controversial (Lizier et al., 2018). In another poorly understood example of a potentially important cell fusion processes, terminal differentiation of fiber cells in the vertebrate lens results in their partial fusion that generates the lens syncytia in which fusion pores connecting constituent cells facilitate intercellular diffusion but do not expand (Shi et al., 2009).
Fertilization-independent cell fusion between the persistent synergid and endosperm
In flowering plants, a somatic cell–cell fusion that occurs after double fertilization has been shown to eliminate the persistent synergid signaling cell in a process alternative to apoptosis that is mechanistically independent of sexual cell fusions (Maruyama et al., 2015; reviewed in Maruyama et al., 2016). The persistent synergid is necessary for pollen attraction. During plant double fertilization, the pollen tube releases two sperm into the ovule. One sperm fuses with the egg while the other fuses to the central cell to form the endosperm. A few hours later, the endosperm fuses with the persistent synergid cell (see Movie S1 in Maruyama et al., 2015). After the fusion, the nucleus of the persistent synergid cell becomes disorganized while the endosperm nucleus divides. While the gamete fusion events are HAP2-mediated, and require actin polymerization and protein secretion, the somatic synergid–endosperm fusion is independent of HAP2 and filamentous actin, and dependent on cyclin-dependent kinases (Motomura et al., 2018).
Cell fusion in disease
Disrupted or unbalanced cell fusion in developmental processes is linked to human diseases. Defects in sperm–egg fusion lead to male infertility (Mou and Xie, 2017). Impediments in myoblast fusion can be perinatally lethal in mice and have been associated with some human myopathies (Di Gioia et al., 2017; Sampath et al., 2018). Syncytin-mediated trophoblast fusion is critical for normal pregnancy, and defects in this fusion have been linked to preeclampsia (Bolze et al., 2017). Unbalanced bone remodeling due to excessive and insufficient osteoclast fusion can lead to osteoporosis and osteopetrosis (Yagi et al., 2005). Fusion between cells that do not normally fuse has been also linked to diseases including cancer and viral infections, discussed in the following sections.
Cell fusion in cancer
In the early 1900s, Otto Aichel suggested that leukocyte-like characteristics of metastatic cancer cells that facilitate their migration through the blood are acquired by their fusion with white blood cells (Aichel, 1911). Since then, many studies have substantiated the hypothesis that fusion among cancer cells and between cancer cells and nonmalignant cells can contribute to initiation and progression of cancer and, specifically, aneuploidy, drug resistance, and metastatic potential characteristic of malignant cells. Indeed, cancer cells do fuse with each other (Noubissi and Ogle, 2016; Uygur et al., 2019). Different cancer cells also spontaneously fuse with nonmalignant cells. For instance, prostate cancer cells fuse with stromal and skeletal muscle cells, and breast cancer cells fuse with normal mammary gland cells and with endothelial cells (Kerbel et al., 1983). Hybrid cells generated by cancer cell/dendritic cell fusion can actually induce an anti-tumor immune response and can potentially be used as a treatment for colorectal and renal cancer (Koido, 2016). However, in most cases, such hybrid cells have cancer stem cell properties with elevated metastatic potential, proliferation rate, and drug resistance (reviewed in Bastida-Ruiz et al., 2016; Noubissi and Ogle, 2016; Gast et al., 2018; Wang et al., 2018). Most recently, it has been shown that fusion between cancer cells and leukocytes increases tumor heterogeneity, and the number of the hybrid cells in blood of human patients correlates with cancer stage (Gast et al., 2018; Video 3). The hypothesis that drastic changes in the properties of the cells upon their fusion can initiate and promote malignancy is further supported by the elegant demonstration that fusion between nontransformed, cytogenetically stable epithelial cells brought about by a chemical fusogen initiates chromosomal instability, cell transformation, and malignancy (Zhou et al., 2015).
In a recent study aimed at modeling the effects of muscle cells surrounding the prostate gland on prostate cancer cells, coculturing prostate cancer cells with primary skeletal or smooth muscle cells resulted in cancer cell fusion (Uygur et al., 2019). Fusion between cancer cells was found to expand the subpopulations of the cells with cancer stem cell features, suggesting that this fusion reaction promotes cancer progression. This novel system obtains a relatively high efficiency of cancer cell fusion with 10–20% of cell nuclei located in fusion-generated multinucleated cells, facilitating the analysis of the underlying mechanisms (Uygur et al., 2019). Cancer cell fusion in prostate cancer/muscle cell cocultures involves a placental fusogen Syn1 (implicated in cancer cell fusion [Noubissi and Ogle, 2016]) and AnxA5 (Uygur et al., 2019). Fusion is associated with up-regulation of these proteins and is inhibited by blocking their expression. Human prostate cancer cells have higher levels of Syn1 and AnxA5 expression than nonmalignant tissues. The case for the direct involvement in cancer cell fusion is especially strong for Syn1 as blocking fusogenic refolding of Syn1 with a peptide inhibitor abolishes fusion (Uygur et al., 2019). Interestingly, ASCT2, a Syn1 receptor, also has a role in cancer cell fusion, as evidenced by the finding that knocking down ASCT2 or Syn1 inhibits fusion between breast cancer cells and endothelial cells (Bjerregaard et al., 2006). Knocking down ASCT2 also inhibits cell proliferation and growth of different tumors (Wang et al., 2015).
With regard to the tumor microenvironment, interactions with muscle apparently trigger fusion of prostate cancer cells by raising concentrations of anti-inflammatory interleukins 4 and 13 in the medium (Uygur et al., 2019). Cancer cell fusion and fusion-dependent disease progression can be also triggered by inflammation, hypoxia, and oxidative stress, and have been associated with apoptotic pathways (Mohr et al., 2015). All these fusion-triggering processes have been linked to PS externalization, and many different cancer cells have an unusually high cell surface concentration of PS (Sharma and Kanwar, 2018). It remains to be clarified whether cancer cell fusion also depends on cell surface PS and at what stage. Better understanding of the mechanisms and steps of cancer cell fusion their role in cancer initiation and progression will hopefully help in development of new diagnostic tools and treatment options for the disease.
Cell fusion in viral infection
In addition to virus–cell membrane fusion, a key stage in enveloped virus entry, some viruses are thought to use cell–cell fusion to spread infection between contacting cells. Cells infected with nonenveloped viruses such as baboon reovirus express fusion-associated small-transmembrane (FAST) proteins. FAST proteins facilitate virus spread between the cells by inducing cell fusion between infected and noninfected cells (Ciechonska et al., 2014; Table 1). T lymphocytes infected with HIV also express viral fusogen (HIV Env) and have been reported to form syncytia in lymph nodes of HIV patients and HIV infected humanized mice as well as in cell culture systems (reviewed in Symeonides et al., 2015; Compton and Schwartz, 2017). While these Env-mediated syncytia have been suggested to significantly contribute to HIV spread (Symeonides et al., 2015), their role in the replication and pathogenesis of HIV-1 in vivo, and, more generally, the role of cell–cell fusion in different viral infections, still await additional analysis (Compton and Schwartz, 2017).
Cell fusion sculpts tissues in C. elegans
The complete anatomy of C. elegans is known at EM resolution (White, 1988), and this is the only known organism with an invariant cell lineage (Sulston et al., 1983), revealing that one third of all the somatic cells that are born as mononucleated cells fuse during development to become multinucleated (Podbilewicz and White, 1994). The timing and locations of the somatic cell–cell fusions during morphogenesis of the embryonic epidermis are fully described (Video 4; Podbilewicz and White, 1994; Mohler et al., 1998; del Campo et al., 2005; Gattegno et al., 2007). During postembryonic (larval) development, additional cells merge with the major embryonic syncytium hyp7 forming the largest worm cell, containing 139 nuclei in the adult hermaphrodite (Podbilewicz and White, 1994; Yochem et al., 1998). Additional multinucleate cells form in the epidermis and during organogenesis of the vulva, uterus, and hymen in the reproductive system (Sharma-Kishore et al., 1999; Kolotuev and Podbilewicz, 2008; Weinstein and Podbilewicz, 2016). In the digestive system, fusions occur in epithelial and myoepithelial cells of the pharynx and in different glands (Shemer et al., 2004). All these cell fusions are highly regulated at the transcriptional, translational, and posttranslational levels to ensure only correct partners fuse in spatial and temporal settings (Shemer and Podbilewicz, 2002; Margalit et al., 2007; Sapir et al., 2007; Alper and Podbilewicz, 2008; Brabin et al., 2011). Precise combinations of signaling pathways including Notch, Wnt, and growth factors control each cell fusion event (Cassata et al., 2005; Rasmussen et al., 2008) in unison with microRNAs and heterochronic genes that temporally regulate the merger of cells and intracellular trafficking of fusogens and actin cytoskeleton which tightly control the correct localization and activity of the fusion machinery itself (Shinn-Thomas et al., 2016; Smurova and Podbilewicz, 2016; Zhang et al., 2017b).
EFF-1 and AFF-1 merge cells to sculpt epithelia and tubular organs
Genetic screens in C. elegans identified two genes essential for developmental cell fusion events (reviewed in Hernández and Podbilewicz, 2017). Mutations in eff-1 result in failure in most fusion events in the epidermis, reproductive, and digestive systems (Mohler et al., 2002). It is essential to define the shape of the epidermis, vulva, pharynx, and uterus. Loss of function of EFF-1 also results in abnormal cell fates and defective migration of unfused cells, and lower fertility (reviewed in Podbilewicz and Chernomordik, 2006; Shinn-Thomas and Mohler, 2011). EFF-1 is a type I membrane glycoprotein with primary sequence similarity to proteins in arthropods, ctenophores, some protists, and chordates (reviewed in Avinoam and Podbilewicz, 2011), and its crystal structure reveals similarity to viral class II glycoproteins (Pérez-Vargas et al., 2014). The second gene identified was named aff-1. It is a paralog of eff-1 and is essential for cell fusions in the formation of the hymen, vulva, epidermis, and pharyngeal muscles (Sapir et al., 2007).
Genetic mosaic analysis at single-cell resolution in C. elegans, ectopic expression in insect cells, and analyses of cell fusion dynamics showed that EFF-1 is required in both fusing cells (Shemer et al., 2004; del Campo et al., 2005; Podbilewicz et al., 2006; Gattegno et al., 2007; Yang et al., 2017). AFF-1 is also required in both fusing cells in worms, insect, and mammalian cells (Sapir et al., 2007; Avinoam et al., 2011). EFF-1::GFP is rapidly endocytosed via dynamin-dependent receptor-mediated endocytosis and mostly localized in RAB-5–positive early endosomes. Failure to endocytose EFF-1 results in excess fusion and lethality (Smurova and Podbilewicz, 2017; Video 4). Independent support for the bilateral mechanism for EFF-1 and AFF-1 comes from fusion between pseudotyped vesicular stomatitis virus with a deletion of G glycoprotein–AFF-1 to cells (Avinoam et al., 2011; Fridman, 2012). Even more remarkable, EFF-1 and HAP-2 can similarly interact in trans in such heterologous systems (Valansi et al., 2017; Fig. 1 C).
Although EFF-1 and AFF-1 are required in both fusing membranes in vivo (C. elegans) and in simpler systems (reviewed in Podbilewicz, 2014), these fusogens may possess some supplementary unilateral activity via partial insertion of amphipathic domains, as noted for HAP2, only presumably much weaker (Liu et al., 2008; Fédry et al., 2017, 2018). Moreover, EFF-1/AFF-1–mediated fusions apparently involve lateral cooperation in cis between fusogens (Avinoam et al., 2011). This has also been shown for intracellular fusogenic complexes such as synaptotagmins, SNAREs, and atlastins (Earles et al., 2001; Hernandez et al., 2014; Liu et al., 2015a). The solved structures of EFF-1 and HAP2 provide a framework to help determine the mechanisms of action of these exoplasmic fusogens (fusexins; Table 1), and their ability to act in a heterotypic way demonstrates mechanistic conservation of action even between plants and animals (Fig. 1 C and Table 2).
Neuronal fusion
Neuronal fusion was documented in invertebrates more than 50 yr ago (Hoy et al., 1967), and in the vertebrate peripheral nervous system (reviewed in Giordano-Santini et al., 2016). A system to cut axons using laser microsurgery (axotomy) was implemented in C. elegans (Yanik et al., 2004) and was used in a breakthrough experiment which determined that EFF-1 fuses cut axons in the PLM mechanosensory neuron (Ghosh-Roy and Chisholm, 2010). EFF-1 directly mediates the reconnection process, and RAB-5–mediated endocytosis of EFF-1 controls the axonal fusion remodeling (Linton et al., 2018). Based on rescue experiments, EFF-1 acts cell-autonomously in the PLM neuron during the axonal auto-fusion process (Fig. 2), and EFF-1::GFP localizes to the regenerating growth cone (Neumann et al., 2015). The reconnection by fusion of the severed PLM axons enables a recovery of the neuronal function of touch sensitivity, and intra-axonal vesicular transport is also restored (Video 5). PS exposure on the axonal outer membrane correlates with axonal fusion and increases with age, although auto-fusion capability itself declines with age. The microRNA let-7 also inhibits the ability to recover functionality by EFF-1–mediated auto-fusion (Abay et al., 2017; Basu et al., 2017). Similar to its role in muscle fusion, PS is exposed and binds to secreted transthyretin (TTR-52) following axotomy. Axonal regeneration depends on, among others, NRF-5 (secreted lipid binding protein) and various components of the phagocytosis pathway (e.g., the engulfment receptor CED-1, CED-6 [Engulfment Adaptor PTB Domain, GULP1], and CED-7 [ABC transporter]; Neumann et al., 2015). The mechanisms underlying axonal repair by self-fusion following experimental injury in C. elegans have potential future applications in neurodegeneration and repair of neuronal injuries in vertebrates (Ghosh-Roy and Chisholm, 2010; Neumann et al., 2011). In summary, EFF-1–mediated auto-fusion is highly regulated and plays a vital role in recovery of injured axons both structurally and functionally.
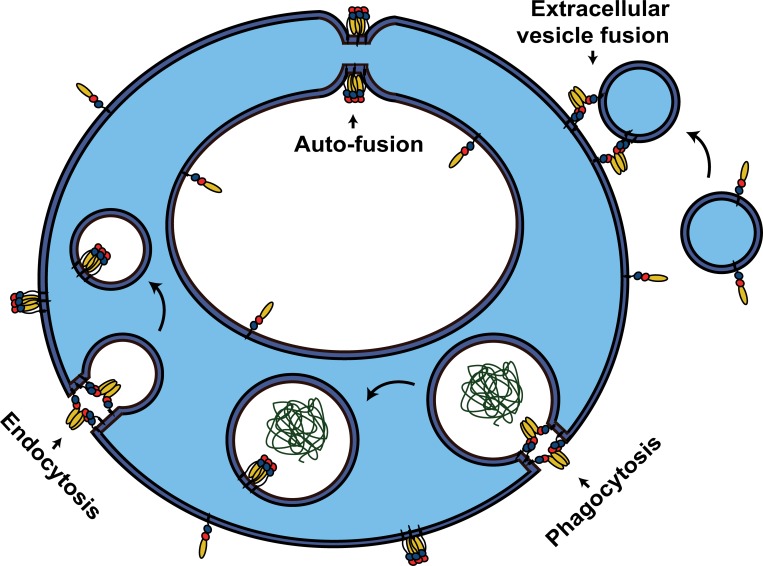
Figure 2.
Alternative functions for cell–cell fusogens. Membrane remodeling activity of EFF-1 and AFF-1 proteins is not limited to mediating cell–cell fusion events. Auto-fusion: a single cell fuses with itself to form donut-shaped cells that can stack and elongate to form tubes, or alternatively join a severed process, as in neuronal regeneration. Extracellular vesicle fusion: AFF-1 proteins can mediate the fusion between a vesicular carrier and the cell. Phagocytosis (EFF-1–mediated) and endocytosis (AFF-1–mediated): Fission events occur to seal the fission pore of the forming intracellular vesicle. Note that while endoplasmic fusogens (e.g., SNAREs and atlastins) act from the cytoplasmic space (light blue areas), EFF-1 and AFF-1 cell–cell fusogens induce fusion from the extracellular space (exoplasmic fusogens in white areas).
EFF-1 has also been shown to control morphogenesis and maintenance of the stereotypic and complex dendritic trees of the PVD neuron (Oren-Suissa et al., 2010). EFF-1 can fuse and “prune” dendrites to model them during larval development, through adulthood and in response to laser microsurgery (dendrotomy; reviewed in Soulavie and Sundaram, 2016). In contrast with the PLM axon, cut dendrites can auto-fuse in a process that requires AFF-1 noncell autonomously (Oren-Suissa et al., 2010, 2017). Dendrotomy induces production of AFF-1–containing extracellular vesicles derived from epidermal seam cells. A model was proposed in which AFF-1 proteins on extracellular vesicles fuse the dendrites and repair the lesion remodeling the dendritic trees from the outside (Oren-Suissa et al., 2017; Fig. 2). Another extrinsic function for EFF-1 in the sculpting of PVD dendritic trees is based on the epidermal localization of SAX-7(L1CAM) Ig domain protein (Zhu et al., 2017) and was recently reviewed (Inberg et al., 2019). Reduction in EFF-1 activity resulted in sprouting of dendrites, increasing the probability of repair by fusion (Oren-Suissa et al., 2017). Similar to axonal repair, dendritic regeneration is impaired in older adults, but can be rescued by ectopic expression of AFF-1 or through mutations in DAF-2 (Insulin growth factor-1 receptor), which is related to increased lifespan (Kravtsov et al., 2017). In summary, age-dependent remodeling of arborized dendrites is dependent on fusogens through auto-fusion and extracellular vesicles.
Other functions of cell–cell fusogens in auto-fusion, endocytosis, and phagocytosis
Some cell–cell fusogens also seem to play roles in other fusion processes. In different organisms, there are examples of a single cell fusing different parts of its own membrane to generate single-cell donuts (reviewed in Sundaram and Cohen, 2017). In C. elegans, EFF-1 and AFF-1 independently mediate auto-fusion of cells that wrap and form donuts that connect to construct tubes in the excretory and digestive systems (Rasmussen et al., 2008; Stone et al., 2009; Fig. 2). The formation of small capillaries in vertebrates also uses auto-fusion strategies during vascular pruning (Lenard et al., 2015). Thus, auto-fusion can be a universal strategy to sculpt seamless donuts and small-scale tube structures (Soulavie and Sundaram, 2016).
Another novel function for AFF-1 during excretory duct elongation in C. elegans was recently uncovered (Soulavie et al., 2018). Using inducible AFF-1 degradation, it was shown how AFF-1 is necessary for auto-fusion of a seamless donut as previously discussed (Stone et al., 2009), but its subsequent elongation to form a tube is independent of its early function (Soulavie et al., 2018). During auto-fusion, AFF-1::mCherry localizes to apical junctions, while during subsequent tube cell elongation, it localizes mostly to basal membranes. During endocytosis, AFF-1 localizes at the necks of endocytic invaginations (Fig. 2). aff-1 mutants in C. elegans have a shortened excretory duct cell, with accumulated membrane inclusions and vesicles suggesting a block in endocytosis from the basal membrane (Soulavie et al., 2018). This work suggests that AFF-1 mediates scission of basal membrane endosomes and facilitates polarized apical exocytosis to elongate seamless tubes. These results uncover a novel and exciting function for the exoplasmic fusogen AFF-1 in endocytic fission and seamless tube elongation by membrane scission. It is conceivable that other exoplasmic fusogens such as syncytins, HAP2, and Myomaker/Myomerger have similar endocytic and tube elongation functions in placenta, gametes, and muscles, respectively.
EFF-1 has another unexpected role in a novel engulfment pathway clearing the distal process of the C. elegans tail spike cell (TSC) and the CEM sex-specific neuron. Mutants in eff-1 fail to clear the distal TSC segment, and this phenotype can be rescued by expressing EFF-1 in the epidermal hyp10 cell (Ghose et al., 2018). This clearance pathway is independent of the classic engulfment pathways that eliminate the soma through CED-5/DOCK180 and CED-1. Thus, EFF-1 acts as a fission-inducing component necessary for sealing of the phagosome containing the distal process of the TSC. In eff-1 mutants, the phagosome containing the distal process is unsealed, as evidenced by FRAP of muscle-secreted GFP surrounding the distal segment, proving continuity of extracellular GFP into the phagosome area. Moreover, EFF-1 localizes to the phagosome arm tips at the putative sealing region (Ghose et al., 2018). Previous investigations have proposed that the fission machinery responsible for endocytic and phagocytic scission acts from the endoplasmic (cytoplasmic) domain of the cellular membranes in eukaryotes. However, EFF-1 mediates sealing of the phagosome by a scission-inducing activity from the exoplasmic domain of the plasma membrane (Fig. 2). This surprising discovery may solve the mystery of the identity of the fission machinery that seals the phagosome. This mechanism for sealing of phagosomes may even have evolutionary implications in the origin of eukaryotes.
Concluding remarks
Fusion of plasma membranes is an essential and dynamic stage in fertilization and organ development, and in pathological processes like viral infections, cancer, and neuronal injury (Videos 1, 2, 3, 4, and 5). The identification of fusogens requires a complex analysis, and we suggest a scoring system based on the gold standards suggested above (Table 1). Ongoing work on identification of proteins that mediate cell fusion has to consider that in this multistep pathway fusion-initiation and fusion-completion may be performed by different proteins, neither of which can mediate fusion on its own. Moreover, each of the fusion stages can depend on the concerted activity of several proteins (Fig. 1). The identity of fusogens involved in gamete fusion in vertebrates, nematodes, and fungi is still missing, and even characterized cell fusion mechanisms are still not completely elucidated (Table 2). Cell–cell fusogens also function in diverse and unexpected cellular processes such as endocytic scission, sealing of phagosomes, auto-fusion during tube formation, remodeling of injured neurons, and fusion of extracellular vesicles to cells (Fig. 2). These recently discovered functions of eukaryotic fusogens promise to reveal universal mechanisms essential to every aspect of life. In the last decade it has become widely accepted that while many proteins control different aspects of cell fusion, only the fusogens are both necessary and sufficient to merge cells together. Recent structural data indicate eukaryotic fusogens have striking structural and functional similarities with better-characterized intracellular and viral fusogens, which opens exciting avenues for new candidate discoveries. Another crucial aspect involves the transcriptional, translational, and posttranslational regulation of the cell fusion process. In addition, membrane lipid composition, intracellular trafficking, and the cytoskeleton alter the activity, localization, and expression of fusogens so that they fuse the right cells at the right place and time.
As discussed above, some mechanistic motifs, including fusion dependence on actin-enriched protrusions, PS, AnxA5, and dynamin, are apparently shared by several different cell fusion processes, suggesting that some of the regulatory mechanisms can be conserved. Future research will determine how fusogens fuse cells, how cellular machineries regulate their activity, and the identity of yet unidentified fusogens.
Online supplemental material
Video 1 shows sperm–oocyte fusion in C. elegans. Video 2 shows myoblast fusion in Drosophila embryos. Video 3 shows mouse-derived macrophage-cancer cell fusion in vitro. Video 4 shows syncytia formation in the dorsal epidermis in C. elegans embryos. Video 5 shows axonal fusion: neuronal repair mechanism.
Supplementary Material
Acknowledgments
We thank members of our laboratories for their intellectual support. We are also grateful to Yael Iosilevskii, Ori Avinoam, and Dan Cassel for critically reading and commenting on the manuscript.
Work in our laboratories was supported by grants from the United States-Israel Binational Science Foundation (grant 2013151 to L.V. Chernomordik and B. Podbilewicz) and the Israel Science Foundation (grants 442/12, 257/17, and 2462/18 to B. Podbilewicz). The research in the L.V. Chernomordik laboratory was also supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
The authors declare no competing financial interests.
Author contributions: N.G. Brukman, B. Uygur, B. Podbilewicz, and L.V. Chernomordik wrote and edited the manuscript.
References
- Abay Z.C., Wong M.Y.-Y., Teoh J.-S., Vijayaraghavan T., Hilliard M.A., and Neumann B.. 2017. Phosphatidylserine save-me signals drive functional recovery of severed axons in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 114:E10196–E10205. 10.1073/pnas.1703807114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar P.S., Baylies M.K., Fleissner A., Helming L., Inoue N., Podbilewicz B., Wang H., and Wong M.. 2013. Genetic basis of cell-cell fusion mechanisms. Trends Genet. 29:427–437. 10.1016/j.tig.2013.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aichel O. 1911. About cell fusion with qualitatively abnormal chromosome distribution as cause for tumor formation / Über zellverschmelzung mit qualitativ abnormer chromosomenverteilung als ursache der geschwulstbildung. In Vorträge und aufsätze über entvickelungsmechanik der organismen. Roux W., editor. Wilhelm Engelmann, Leipzig: 92–111. [Google Scholar]
- Alper S., and Podbilewicz B.. 2008. Cell Fusion in Caenorhabditis elegans. In Methods in molecular biology. Humana Press, Clifton, NJ. 53–74. [DOI] [PubMed] [Google Scholar]
- Antony J.M., Ellestad K.K., Hammond R., Imaizumi K., Mallet F., Warren K.G., and Power C.. 2007. The human endogenous retrovirus envelope glycoprotein, syncytin-1, regulates neuroinflammation and its receptor expression in multiple sclerosis: a role for endoplasmic reticulum chaperones in astrocytes. J. Immunol. 179:1210–1224. 10.4049/jimmunol.179.2.1210 [DOI] [PubMed] [Google Scholar]
- Avinoam O., and Podbilewicz B.. 2011. Eukaryotic Cell–Cell Fusion Families. Curr. Top. Membr. 68:209–234. 10.1016/B978-0-12-385891-7.00009-X [DOI] [PubMed] [Google Scholar]
- Avinoam O., Fridman K., Valansi C., Abutbul I., Zeev-Ben-Mordehai T., Maurer U.E., Sapir A., Danino D., Grünewald K., White J.M., and Podbilewicz B.. 2011. Conserved eukaryotic fusogens can fuse viral envelopes to cells. Science. 332:589–592. 10.1126/science.1202333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin H., Sultana A., Li S., Thavalingam A., and Lee J.E.. 2016. Molecular architecture of the human sperm IZUMO1 and egg JUNO fertilization complex. Nature. 534:562–565. 10.1038/nature18595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballabio A., and Gieselmann V.. 2009. Lysosomal disorders: from storage to cellular damage. Biochim. Biophys. Acta. 1793:684–696. 10.1016/j.bbamcr.2008.12.001 [DOI] [PubMed] [Google Scholar]
- Bastida-Ruiz D., Van Hoesen K., and Cohen M.. 2016. The Dark Side of Cell Fusion. Int. J. Mol. Sci. 17:638 10.3390/ijms17050638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A., Dey S., Puri D., Das Saha N., Sabharwal V., Thyagarajan P., Srivastava P., Koushika S.P., and Ghosh-Roy A.. 2017. let-7 miRNA controls CED-7 homotypic adhesion and EFF-1-mediated axonal self-fusion to restore touch sensation following injury. Proc. Natl. Acad. Sci. USA. 114:E10206–E10215. 10.1073/pnas.1704372114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate M. 1990. The embryonic development of larval muscles in Drosophila. Development. 110:791–804. [DOI] [PubMed] [Google Scholar]
- Bi P., Ramirez-Martinez A., Li H., Cannavino J., McAnally J.R., Shelton J.M., Sánchez-Ortiz E., Bassel-Duby R., and Olson E.N.. 2017. Control of muscle formation by the fusogenic micropeptide myomixer. Science. 356:323–327. 10.1126/science.aam9361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi E., and Wright G.J.. 2015. Cross-species fertilization: The hamster egg receptor, Juno, binds the human sperm ligand, Izumo1. Philos. Trans. R. Soc. B Biol. Sci. 370:20140101 10.1098/rstb.2014.0101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi E., Doe B., Goulding D., and Wright G.J.. 2014. Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature. 508:483–487. 10.1038/nature13203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerregaard B., Holck S., Christensen I.J., and Larsson L.-I.. 2006. Syncytin is involved in breast cancer-endothelial cell fusions. Cell. Mol. Life Sci. 63:1906–1911. 10.1007/s00018-006-6201-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blond J.L., Lavillette D., Cheynet V., Bouton O., Oriol G., Chapel-Fernandes S., Mandrand B., Mallet F., and Cosset F.L.. 2000. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 74:3321–3329. 10.1128/JVI.74.7.3321-3329.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolze P.-A., Mommert M., and Mallet F.. 2017. Contribution of Syncytins and Other Endogenous Retroviral Envelopes to Human Placenta Pathologies. Prog. Mol. Biol. Transl. Sci. 145:111–162. 10.1016/bs.pmbts.2016.12.005 [DOI] [PubMed] [Google Scholar]
- Borges F., Gomes G., Gardner R., Moreno N., McCormick S., Feijó J.A., and Becker J.D.. 2008. Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol. 148:1168–1181. 10.1104/pp.108.125229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothe I., Deng S., and Baylies M.. 2014. PI(4,5)P2 regulates myoblast fusion through Arp2/3 regulator localization at the fusion site. Development. 141:2289–2301. 10.1242/dev.100743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bour B.A., Chakravarti M., West J.M., and Abmayr S.M.. 2000. Drosophila SNS, a member of the immunoglobulin superfamily that is essential for myoblast fusion. Genes Dev. 14:1498–1511. 10.1101/gad.14.12.1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabin C., Appleford P.J., and Woollard A.. 2011. The Caenorhabditis elegans GATA factor ELT-1 works through the cell proliferation regulator BRO-1 and the Fusogen EFF-1 to maintain the seam stem-like fate. PLoS Genet. 7:e1002200 10.1371/journal.pgen.1002200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassata G., Shemer G., Morandi P., Donhauser R., Podbilewicz B., and Baumeister R.. 2005. ceh-16/engrailed patterns the embryonic epidermis of Caenorhabditis elegans. Development. 132:739–749. 10.1242/dev.01638 [DOI] [PubMed] [Google Scholar]
- Chalbi M., Barraud-Lange V., Ravaux B., Howan K., Rodriguez N., Soule P., Ndzoudi A., Boucheix C., Rubinstein E., Wolf J.P., et al. 2014. Binding of sperm protein Izumo1 and its egg receptor Juno drives Cd9 accumulation in the intercellular contact area prior to fusion during mammalian fertilization. Development. 141:3732–3739. 10.1242/dev.111534 [DOI] [PubMed] [Google Scholar]
- Chang C.W., Chiang C.W., and Jackson M.B.. 2017. Fusion pores and their control of neurotransmitter and hormone release. J. Gen. Physiol. 149:301–322. 10.1085/jgp.201611724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E.R. 2008. How does synaptotagmin trigger neurotransmitter release? Annu. Rev. Biochem. 77:615–641. 10.1146/annurev.biochem.77.062005.101135 [DOI] [PubMed] [Google Scholar]
- Chernomordik L.V., and Kozlov M.M.. 2003. Protein-lipid interplay in fusion and fission of biological membranes. Annu. Rev. Biochem. 72:175–207. 10.1146/annurev.biochem.72.121801.161504 [DOI] [PubMed] [Google Scholar]
- Chernomordik L.V., and Kozlov M.M.. 2008. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 15:675–683. 10.1038/nsmb.1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik L.V., Melikyan G.B., and Chizmadzhev Y.A.. 1987. Biomembrane fusion: a new concept derived from model studies using two interacting planar lipid bilayers. Biochim. Biophys. Acta. 906:309–352. 10.1016/0304-4157(87)90016-5 [DOI] [PubMed] [Google Scholar]
- Ciechonska M., Key T., and Duncan R.. 2014. Efficient reovirus- and measles virus-mediated pore expansion during syncytium formation is dependent on annexin A1 and intracellular calcium. J. Virol. 88:6137–6147. 10.1128/JVI.00121-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S., and Naz R.K.. 2013. Presence and incidence of izumo antibodies in sera of immunoinfertile women and men. Am. J. Reprod. Immunol. 69:256–263. 10.1111/aji.12060 [DOI] [PubMed] [Google Scholar]
- Compton A.A., and Schwartz O.. 2017. They Might Be Giants: Does Syncytium Formation Sink or Spread HIV Infection? PLoS Pathog. 13:e1006099 10.1371/journal.ppat.1006099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrelle S.A., Gerbaud P., Leconte L., Ferreira F., and Pidoux G.. 2017. Annexin-A5 organized in 2D-network at the plasmalemma eases human trophoblast fusion. Sci. Rep. 7:42173 10.1038/srep42173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Campo J.J., Opoku-Serebuoh E., Isaacson A.B., Scranton V.L., Tucker M., Han M., and Mohler W.A.. 2005. Fusogenic activity of EFF-1 is regulated via dynamic localization in fusing somatic cells of C. elegans. Curr. Biol. 15:413–423. 10.1016/j.cub.2005.01.054 [DOI] [PubMed] [Google Scholar]
- Dhanyasi N., Segal D., Shimoni E., Shinder V., Shilo B.-Z., VijayRaghavan K., and Schejter E.D.. 2015. Surface apposition and multiple cell contacts promote myoblast fusion in Drosophila flight muscles. J. Cell Biol. 211:191–203. 10.1083/jcb.201503005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Gioia S.A., Connors S., Matsunami N., Cannavino J., Rose M.F., Gilette N.M., Artoni P., de Macena Sobreira N.L., Chan W.-M., Webb B.D., et al. Moebius Syndrome Research Consortium . 2017. A defect in myoblast fusion underlies Carey-Fineman-Ziter syndrome. Nat. Commun. 8:16077 10.1038/ncomms16077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doms R.W. 2017. What Came First-the Virus or the Egg? Cell. 168:755–757. 10.1016/j.cell.2017.02.012 [DOI] [PubMed] [Google Scholar]
- Duan R., Kim J.H., Shilagardi K., Schiffhauer E.S., Lee D.M., Son S., Li S., Thomas C., Luo T., Fletcher D.A., et al. 2018. Spectrin is a mechanoresponsive protein shaping fusogenic synapse architecture during myoblast fusion. Nat. Cell Biol. 20:688–698. 10.1038/s41556-018-0106-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A., Vernochet C., Harper F., Guégan J., Dessen P., Pierron G., and Heidmann T.. 2011. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc. Natl. Acad. Sci. USA. 108:E1164–E1173. 10.1073/pnas.1112304108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earles C.A., Bai J., Wang P., and Chapman E.R.. 2001. The tandem C2 domains of synaptotagmin contain redundant Ca2+ binding sites that cooperate to engage t-SNAREs and trigger exocytosis. J. Cell Biol. 154:1117–1123. 10.1083/jcb.200105020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust J.E., Desai T., Verma A., Ulengin I., Sun T.-L., Moss T.J., Betancourt-Solis M.A., Huang H.W., Lee T., and McNew J.A.. 2015. The Atlastin C-terminal tail is an amphipathic helix that perturbs the bilayer structure during endoplasmic reticulum homotypic fusion. J. Biol. Chem. 290:4772–4783. 10.1074/jbc.M114.601823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fédry J., Liu Y., Péhau-Arnaudet G., Pei J., Li W., Tortorici M.A., Traincard F., Meola A., Bricogne G., Grishin N.V., et al. 2017. The Ancient Gamete Fusogen HAP2 Is a Eukaryotic Class II Fusion Protein. Cell. 168:904–915.e10. 10.1016/j.cell.2017.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fédry J., Forcina J., Legrand P., Péhau-Arnaudet G., Haouz A., Johnson M., Rey F.A., and Krey T.. 2018. Evolutionary diversification of the HAP2 membrane insertion motifs to drive gamete fusion across eukaryotes. PLoS Biol. 16:e2006357 10.1371/journal.pbio.2006357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Dong X., Pinello J., Zhang J., Lu C., Iacob R.E., Engen J.R., Snell W.J., and Springer T.A.. 2018. Fusion surface structure, function, and dynamics of gamete fusogen HAP2. eLife. 7:e39772 10.7554/eLife.39772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman K. 2012. Ultrastructure and function of AFF-1 and EFF-1 in membrane remodeling. Technion - Israel Institute of Technology, Thesis, Israel. [Google Scholar]
- Gamage D.G., Leikina E., Quinn M.E., Ratinov A., Chernomordik L.V., and Millay D.P.. 2017. Insights into the localization and function of myomaker during myoblast fusion. J. Biol. Chem. 292:17272–17289. 10.1074/jbc.M117.811372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gast C.E., Silk A.D., Zarour L., Riegler L., Burkhart J.G., Gustafson K.T., Parappilly M.S., Roh-Johnson M., Goodman J.R., Olson B., et al. 2018. Cell fusion potentiates tumor heterogeneity and reveals circulating hybrid cells that correlate with stage and survival. Sci. Adv. 4:eaat7828 10.1126/sciadv.aat7828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattegno T., Mittal A., Valansi C., Nguyen K.C.Q., Hall D.H., Chernomordik L.V., and Podbilewicz B.. 2007. Genetic control of fusion pore expansion in the epidermis of Caenorhabditis elegans. Mol. Biol. Cell. 18:1153–1166. 10.1091/mbc.e06-09-0855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghose P., Rashid A., Insley P., Trivedi M., Shah P., Singhal A., Lu Y., Bao Z., and Shaham S.. 2018. EFF-1 fusogen promotes phagosome sealing during cell process clearance in Caenorhabditis elegans. Nat. Cell Biol. 20:393–399. 10.1038/s41556-018-0068-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh-Roy A., and Chisholm A.D.. 2010. Caenorhabditis elegans: a new model organism for studies of axon regeneration. Dev. Dyn. 239:1460–1464. 10.1002/dvdy.22253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano-Santini R., Linton C., and Hilliard M.A.. 2016. Cell-cell fusion in the nervous system: Alternative mechanisms of development, injury, and repair. Semin. Cell Dev. Biol. 60:146–154. 10.1016/j.semcdb.2016.06.019 [DOI] [PubMed] [Google Scholar]
- Glabe C.G. 1985. Interaction of the sperm adhesive protein, bindin, with phospholipid vesicles. II. Bindin induces the fusion of mixed-phase vesicles that contain phosphatidylcholine and phosphatidylserine in vitro. J. Cell Biol. 100:800–806. 10.1083/jcb.100.3.800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Bai Y., Zhang L., Zhang B., Zagidullin N., Carvalho K., Du Z., and Cai B.. 2018. Cardiomyocyte differentiation of mesenchymal stem cells from bone marrow: new regulators and its implications. Stem Cell Res. Ther. 9:44 10.1186/s13287-018-0773-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar S., Mekhedov E., McCormick C.D., Blank P.S., and Zimmerberg J.. 2018. Lipid-dependence of target membrane stability during influenza viral fusion. J. Cell Sci. 132:jcs218321 10.1242/jcs.218321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamoud N., Tran V., Aimi T., Kakegawa W., Lahaie S., Thibault M.-P., Pelletier A., Wong G.W., Kim I.-S., Kania A., et al. 2018. Spatiotemporal regulation of the GPCR activity of BAI3 by C1qL4 and Stabilin-2 controls myoblast fusion. Nat. Commun. 9:4470 10.1038/s41467-018-06897-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M.G., and Walter P.. 2000. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J. Cell Biol. 151:719–730. 10.1083/jcb.151.3.719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herberg S., Gert K.R., Schleiffer A., and Pauli A.. 2018. The Ly6/uPAR protein Bouncer is necessary and sufficient for species-specific fertilization. Science. 361:1029–1033. 10.1126/science.aat7113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez J.M., Kreutzberger A.J.B., Kiessling V., Tamm L.K., and Jahn R.. 2014. Variable cooperativity in SNARE-mediated membrane fusion. Proc. Natl. Acad. Sci. USA. 111:12037–12042. 10.1073/pnas.1407435111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández J.M., and Podbilewicz B.. 2017. The hallmarks of cell-cell fusion. Development. 144:4481–4495. 10.1242/dev.155523 [DOI] [PubMed] [Google Scholar]
- Hochreiter-Hufford A.E., Lee C.S., Kinchen J.M., Sokolowski J.D., Arandjelovic S., Call J.A., Klibanov A.L., Yan Z., Mandell J.W., and Ravichandran K.S.. 2013. Phosphatidylserine receptor BAI1 and apoptotic cells as new promoters of myoblast fusion. Nature. 497:263–267. 10.1038/nature12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy R.R., Bittner G.D., and Kennedy D.. 1967. Regeneration in crustacean motoneurons: evidence for axonal fusion. Science. 156:251–252. 10.1126/science.156.3772.251 [DOI] [PubMed] [Google Scholar]
- Hu J., and Rapoport T.A.. 2016. Fusion of the endoplasmic reticulum by membrane-bound GTPases. Semin. Cell Dev. Biol. 60:105–111. 10.1016/j.semcdb.2016.06.001 [DOI] [PubMed] [Google Scholar]
- Inberg S., Meledin A., Kravtsov V., Iosilevskii Y., Oren-Suissa M., and Podbilewicz B.. 2019. Lessons from worm dendritic patterning. Annu. Rev. Neurosci. In press. [DOI] [PubMed] [Google Scholar]
- Inoue N., Ikawa M., Isotani A., and Okabe M.. 2005. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature. 434:234–238. 10.1038/nature03362 [DOI] [PubMed] [Google Scholar]
- Inoue N., Hamada D., Kamikubo H., Hirata K., Kataoka M., Yamamoto M., Ikawa M., Okabe M., and Hagihara Y.. 2013. Molecular dissection of IZUMO1, a sperm protein essential for sperm-egg fusion. Development. 140:3221–3229. 10.1242/dev.094854 [DOI] [PubMed] [Google Scholar]
- Inoue N., Hagihara Y., Wright D., Suzuki T., and Wada I.. 2015. Oocyte-triggered dimerization of sperm IZUMO1 promotes sperm-egg fusion in mice. Nat. Commun. 6:8858 10.1038/ncomms9858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T., Ruiz-Torruella M., Ikeda A., Shindo S., Movila A., Mawardi H., Albassam A., Kayal R.A., Al-Dharrab A.A., Egashira K., et al. 2018. OC-STAMP promotes osteoclast fusion for pathogenic bone resorption in periodontitis via up-regulation of permissive fusogen CD9. FASEB J. 32:4016–4030. 10.1096/fj.201701424R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R., and Scheller R.H.. 2006. SNAREs--engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7:631–643. 10.1038/nrm2002 [DOI] [PubMed] [Google Scholar]
- Johnson M.A., von Besser K., Zhou Q., Smith E., Aux G., Patton D., Levin J.Z., and Preuss D.. 2004. Arabidopsis hapless mutations define essential gametophytic functions. Genetics. 168:971–982. 10.1534/genetics.104.029447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K., Oda S., Shikano T., Ohnuki T., Uematsu Y., Sakagami J., Tada N., Miyazaki S., and Kudo A.. 2000. The gamete fusion process is defective in eggs of Cd9-deficient mice. Nat. Genet. 24:279–282. 10.1038/73502 [DOI] [PubMed] [Google Scholar]
- Kajstura J., Rota M., Whang B., Cascapera S., Hosoda T., Bearzi C., Nurzynska D., Kasahara H., Zias E., Bonafé M., et al. 2005. Bone marrow cells differentiate in cardiac cell lineages after infarction independently of cell fusion. Circ. Res. 96:127–137. 10.1161/01.RES.0000151843.79801.60 [DOI] [PubMed] [Google Scholar]
- Kato K., Satouh Y., Nishimasu H., Kurabayashi A., Morita J., Fujihara Y., Oji A., Ishitani R., Ikawa M., and Nureki O.. 2016. Structural and functional insights into IZUMO1 recognition by JUNO in mammalian fertilization. Nat. Commun. 7:12198 10.1038/ncomms12198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerbel R.S., Lagarde A.E., Dennis J.W., and Donaghue T.P.. 1983. Spontaneous fusion in vivo between normal host and tumor cells: possible contribution to tumor progression and metastasis studied with a lectin-resistant mutant tumor. Mol. Cell. Biol. 3:523–538. 10.1128/MCB.3.4.523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesper D.A., Stute C., Buttgereit D., Kreisköther N., Vishnu S., Fischbach K.-F., and Renkawitz-Pohl R.. 2007. Myoblast fusion in Drosophila melanogaster is mediated through a fusion-restricted myogenic-adhesive structure (FuRMAS). Dev. Dyn. 236:404–415. 10.1002/dvdy.21035 [DOI] [PubMed] [Google Scholar]
- Key T., and Duncan R.. 2014. A compact, multifunctional fusion module directs cholesterol-dependent homomultimerization and syncytiogenic efficiency of reovirus p10 FAST proteins. PLoS Pathog. 10:e1004023 10.1371/journal.ppat.1004023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielian M., and Rey F.A.. 2006. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 4:67–76. 10.1038/nrmicro1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G.W., Park S.Y., and Kim I.S.. 2016. Novel function of stabilin-2 in myoblast fusion: the recognition of extracellular phosphatidylserine as a “fuse-me” signal. BMB Rep. 49:303–304. 10.5483/BMBRep.2016.49.6.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Jin P., Duan R., and Chen E.H.. 2015. Mechanisms of myoblast fusion during muscle development. Curr. Opin. Genet. Dev. 32:162–170. 10.1016/j.gde.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koido S. 2016. Dendritic-Tumor Fusion Cell-Based Cancer Vaccines. Int. J. Mol. Sci. 17:828 10.3390/ijms17060828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolotuev I., and Podbilewicz B.. 2008. Changing of the cell division axes drives vulva evolution in nematodes. Dev. Biol. 313:142–154. 10.1016/j.ydbio.2007.10.010 [DOI] [PubMed] [Google Scholar]
- Kravtsov V., Oren-Suissa M., and Podbilewicz B.. 2017. The fusogen AFF-1 can rejuvenate the regenerative potential of adult dendritic trees by self-fusion. Development. 144:2364–2374. 10.1242/dev.150037 [DOI] [PubMed] [Google Scholar]
- Lees R.L., and Heersche J.N.. 1999. Macrophage colony stimulating factor increases bone resorption in dispersed osteoclast cultures by increasing osteoclast size. J. Bone Miner. Res. 14:937–945. 10.1359/jbmr.1999.14.6.937 [DOI] [PubMed] [Google Scholar]
- Leikina E., Mittal A., Cho M.-S., Melikov K., Kozlov M.M., and Chernomordik L.V.. 2004. Influenza hemagglutinins outside of the contact zone are necessary for fusion pore expansion. J. Biol. Chem. 279:26526–26532. 10.1074/jbc.M401883200 [DOI] [PubMed] [Google Scholar]
- Leikina E., Melikov K., Sanyal S., Verma S.K., Eun B., Gebert C., Pfeifer K., Lizunov V.A., Kozlov M.M., and Chernomordik L.V.. 2013. Extracellular annexins and dynamin are important for sequential steps in myoblast fusion. J. Cell Biol. 200:109–123. 10.1083/jcb.201207012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leikina E., Gamage D.G., Prasad V., Goykhberg J., Crowe M., Diao J., Kozlov M.M., Chernomordik L.V., and Millay D.P.. 2018. Myomaker and Myomerger Work Independently to Control Distinct Steps of Membrane Remodeling during Myoblast Fusion. Dev. Cell. 46:767–780.e7. 10.1016/j.devcel.2018.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Naour F., Rubinstein E., Jasmin C., Prenant M., and Boucheix C.. 2000. Severely reduced female fertility in CD9-deficient mice. Science. 287:319–321. 10.1126/science.287.5451.319 [DOI] [PubMed] [Google Scholar]
- Lenard A., Daetwyler S., Betz C., Ellertsdottir E., Belting H.-G., Huisken J., and Affolter M.. 2015. Endothelial cell self-fusion during vascular pruning. PLoS Biol. 13:e1002126 10.1371/journal.pbio.1002126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levaot N., Ottolenghi A., Mann M., Guterman-Ram G., Kam Z., and Geiger B.. 2015. Osteoclast fusion is initiated by a small subset of RANKL-stimulated monocyte progenitors, which can fuse to RANKL-unstimulated progenitors. Bone. 79:21–28. 10.1016/j.bone.2015.05.021 [DOI] [PubMed] [Google Scholar]
- Linton C., Neumann B., Giordano-Santini R., and Hilliard M.A.. 2018. RAB-5 regulates regenerative axonal fusion by controlling EFF-1 endocytosis. bioRxiv. 339150 10.1101/339150 [DOI] [Google Scholar]
- Liu T.Y., Bian X., Romano F.B., Shemesh T., Rapoport T.A., and Hu J.. 2015a Cis and trans interactions between atlastin molecules during membrane fusion. Proc. Natl. Acad. Sci. USA. 112:E1851–E1860. 10.1073/pnas.1504368112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Tewari R., Ning J., Blagborough A.M., Garbom S., Pei J., Grishin N.V., Steele R.E., Sinden R.E., Snell W.J., and Billker O.. 2008. The conserved plant sterility gene HAP2 functions after attachment of fusogenic membranes in Chlamydomonas and Plasmodium gametes. Genes Dev. 22:1051–1068. 10.1101/gad.1656508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Pei J., Grishin N., and Snell W.J.. 2015b The cytoplasmic domain of the gamete membrane fusion protein HAP2 targets the protein to the fusion site in Chlamydomonas and regulates the fusion reaction. Development. 142:962–971. 10.1242/dev.118844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizier M., Castelli A., Montagna C., Lucchini F., Vezzoni P., and Faggioli F.. 2018. Cell fusion in the liver, revisited. World J. Hepatol. 10:213–221. 10.4254/wjh.v10.i2.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzetti D., Poirier C., Zhao M., Overbeek P.A., Harrison W., and Bishop C.E.. 2014. A transgenic insertion on mouse chromosome 17 inactivates a novel immunoglobulin superfamily gene potentially involved in sperm-egg fusion. Mamm. Genome. 25:141–148. 10.1007/s00335-013-9491-x [DOI] [PubMed] [Google Scholar]
- Margalit A., Neufeld E., Feinstein N., Wilson K.L., Podbilewicz B., and Gruenbaum Y.. 2007. Barrier to autointegration factor blocks premature cell fusion and maintains adult muscle integrity in C. elegans. J. Cell Biol. 178:661–673. 10.1083/jcb.200704049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markvoort A.J., and Marrink S.J.. 2011. Lipid acrobatics in the membrane fusion arena. Curr. Top. Membr. 68:259–294. 10.1016/B978-0-12-385891-7.00011-8 [DOI] [PubMed] [Google Scholar]
- Maruyama D., Völz R., Takeuchi H., Mori T., Igawa T., Kurihara D., Kawashima T., Ueda M., Ito M., Umeda M., et al. 2015. Rapid Elimination of the Persistent Synergid through a Cell Fusion Mechanism. Cell. 161:907–918. 10.1016/j.cell.2015.03.018 [DOI] [PubMed] [Google Scholar]
- Maruyama D., Ohtsu M., and Higashiyama T.. 2016. Cell fusion and nuclear fusion in plants. Semin. Cell Dev. Biol. 60:127–135. 10.1016/j.semcdb.2016.07.024 [DOI] [PubMed] [Google Scholar]
- Mi S., Lee X., Li X., Veldman G.M., Finnerty H., Racie L., LaVallie E., Tang X.-Y., Edouard P., Howes S., et al. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 403:785–789. 10.1038/35001608 [DOI] [PubMed] [Google Scholar]
- Millay D.P., O’Rourke J.R., Sutherland L.B., Bezprozvannaya S., Shelton J.M., Bassel-Duby R., and Olson E.N.. 2013. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature. 499:301–305. 10.1038/nature12343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyado K., Yamada G., Yamada S., Hasuwa H., Nakamura Y., Ryu F., Suzuki K., Kosai K., Inoue K., Ogura A., et al. 2000. Requirement of CD9 on the egg plasma membrane for fertilization. Science. 287:321–324. 10.1126/science.287.5451.321 [DOI] [PubMed] [Google Scholar]
- Mohler W.A., Simske J.S., Williams-Masson E.M., Hardin J.D., and White J.G.. 1998. Dynamics and ultrastructure of developmental cell fusions in the Caenorhabditis elegans hypodermis. Curr. Biol. 8:1087–1090. 10.1016/S0960-9822(98)70447-6 [DOI] [PubMed] [Google Scholar]
- Mohler W.A., Shemer G., del Campo J.J., Valansi C., Opoku-Serebuoh E., Scranton V., Assaf N., White J.G., and Podbilewicz B.. 2002. The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev. Cell. 2:355–362. 10.1016/S1534-5807(02)00129-6 [DOI] [PubMed] [Google Scholar]
- Mohr M., Zaenker K.S., and Dittmar T.. 2015. Fusion in cancer: an explanatory model for aneuploidy, metastasis formation, and drug resistance. Methods Mol. Biol. 1313:21–40. 10.1007/978-1-4939-2703-6_2 [DOI] [PubMed] [Google Scholar]
- Mori T., Kuroiwa H., Higashiyama T., and Kuroiwa T.. 2006. GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat. Cell Biol. 8:64–71. 10.1038/ncb1345 [DOI] [PubMed] [Google Scholar]
- Motomura K., Kawashima T., Berger F., Kinoshita T., Higashiyama T., and Maruyama D.. 2018. A pharmacological study of Arabidopsis cell fusion between the persistent synergid and endosperm. J. Cell Sci. 131:jcs204123 10.1242/jcs.204123 [DOI] [PubMed] [Google Scholar]
- Mou L., and Xie N.. 2017. Male infertility-related molecules involved in sperm-oocyte fusion. J. Reprod. Dev. 63:1–7. 10.1262/jrd.2016-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B., Nguyen K.C.Q., Hall D.H., Ben-Yakar A., and Hilliard M.A.. 2011. Axonal regeneration proceeds through specific axonal fusion in transected C. elegans neurons. Dev. Dyn. 240:1365–1372. 10.1002/dvdy.22606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B., Coakley S., Giordano-Santini R., Linton C., Lee E.S., Nakagawa A., Xue D., and Hilliard M.A.. 2015. EFF-1-mediated regenerative axonal fusion requires components of the apoptotic pathway. Nature. 517:219–222. 10.1038/nature14102 [DOI] [PubMed] [Google Scholar]
- Nishimura H., and L’Hernault S.W.. 2010. Spermatogenesis-defective (spe) mutants of the nematode Caenorhabditis elegans provide clues to solve the puzzle of male germline functions during reproduction. Dev. Dyn. 239:1502–1514. 10.1002/dvdy.22271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noubissi F.K., and Ogle B.M.. 2016. Cancer Cell Fusion: Mechanisms Slowly Unravel. Int. J. Mol. Sci. 17:1587 10.3390/ijms17091587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto U., Ishida H., Krayukhina E., Uchiyama S., Inoue N., and Shimizu T.. 2016. Structure of IZUMO1-JUNO reveals sperm-oocyte recognition during mammalian fertilization. Nature. 534:566–569. 10.1038/nature18596 [DOI] [PubMed] [Google Scholar]
- Oikawa T., and Matsuo K.. 2012. Possible role of IRTKS in Tks5-driven osteoclast fusion. Commun. Integr. Biol. 5:511–515. 10.4161/cib.21252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M., Yamada L., Fujisaki Y., Bloomfield G., Yoshida K., Kuwayama H., Sawada H., Mori T., and Urushihara H.. 2016. Two HAP2-GCS1 homologs responsible for gamete interactions in the cellular slime mold with multiple mating types: Implication for common mechanisms of sexual reproduction shared by plants and protozoa and for male-female differentiation. Dev. Biol. 415:6–13. 10.1016/j.ydbio.2016.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmo V.N., and Grote E.. 2010. Prm1 targeting to contact sites enhances fusion during mating in Saccharomyces cerevisiae. Eukaryot. Cell. 9:1538–1548. 10.1128/EC.00116-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren-Suissa M., and Podbilewicz B.. 2007. Cell fusion during development. Trends Cell Biol. 17:537–546. 10.1016/j.tcb.2007.09.004 [DOI] [PubMed] [Google Scholar]
- Oren-Suissa M., Hall D.H., Treinin M., Shemer G., and Podbilewicz B.. 2010. The fusogen EFF-1 controls sculpting of mechanosensory dendrites. Science. 328:1285–1288. 10.1126/science.1189095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren-Suissa M., Gattegno T., Kravtsov V., and Podbilewicz B.. 2017. Extrinsic Repair of Injured Dendrites as a Paradigm for Regeneration by Fusion in Caenorhabditis elegans. Genetics. 206:215–230. 10.1534/genetics.116.196386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Vargas J., Krey T., Valansi C., Avinoam O., Haouz A., Jamin M., Raveh-Barak H., Podbilewicz B., and Rey F.A.. 2014. Structural basis of eukaryotic cell-cell fusion. Cell. 157:407–419. 10.1016/j.cell.2014.02.020 [DOI] [PubMed] [Google Scholar]
- Pesaresi M., Sebastian-Perez R., and Cosma M.P.. 2018. Dedifferentiation, transdifferentiation and cell fusion: in vivo reprogramming strategies for regenerative medicine. FEBS J. 10.1111/febs.14633 [DOI] [PubMed] [Google Scholar]
- Pinello J.F., Lai A.L., Millet J.K., Cassidy-Hanley D., Freed J.H., and Clark T.G.. 2017. Structure-Function Studies Link Class II Viral Fusogens with the Ancestral Gamete Fusion Protein HAP2. Curr. Biol. 27:651–660. 10.1016/j.cub.2017.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podbilewicz B. 2014. Virus and cell fusion mechanisms. Annu. Rev. Cell Dev. Biol. 30:111–139. 10.1146/annurev-cellbio-101512-122422 [DOI] [PubMed] [Google Scholar]
- Podbilewicz B., and Chernomordik L.V.. 2006. Cell Fusion in Development and Disease. In Protein-Lipid Interactions. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany. 219–244. [Google Scholar]
- Podbilewicz B., and White J.G.. 1994. Cell fusions in the developing epithelial of C. elegans. Dev. Biol. 161:408–424. 10.1006/dbio.1994.1041 [DOI] [PubMed] [Google Scholar]
- Podbilewicz B., Leikina E., Sapir A., Valansi C., Suissa M., Shemer G., and Chernomordik L.V.. 2006. The C. elegans developmental fusogen EFF-1 mediates homotypic fusion in heterologous cells and in vivo. Dev. Cell. 11:471–481. 10.1016/j.devcel.2006.09.004 [DOI] [PubMed] [Google Scholar]
- Posey A.D. Jr., Demonbreun A., and McNally E.M.. 2011. Ferlin proteins in myoblast fusion and muscle growth. Curr. Top. Dev. Biol. 96:203–230. 10.1016/B978-0-12-385940-2.00008-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pötgens A.J.G., Drewlo S., Kokozidou M., and Kaufmann P.. 2004. Syncytin: the major regulator of trophoblast fusion? Recent developments and hypotheses on its action. Hum. Reprod. Update. 10:487–496. 10.1093/humupd/dmh039 [DOI] [PubMed] [Google Scholar]
- Powell G.T., and Wright G.J.. 2011. Jamb and jamc are essential for vertebrate myocyte fusion. PLoS Biol. 9:e1001216 10.1371/journal.pbio.1001216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn M.E., Goh Q., Kurosaka M., Gamage D.G., Petrany M.J., Prasad V., and Millay D.P.. 2017. Myomerger induces fusion of non-fusogenic cells and is required for skeletal muscle development. Nat. Commun. 8:15665 10.1038/ncomms15665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranieri M., Brajkovic S., Riboldi G., Ronchi D., Rizzo F., Bresolin N., Corti S., and Comi G.P.. 2013. Mitochondrial fusion proteins and human diseases. Neurol. Res. Int. 2013:293893 10.1155/2013/293893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen J.P., English K., Tenlen J.R., and Priess J.R.. 2008. Notch signaling and morphogenesis of single-cell tubes in the C. elegans digestive tract. Dev. Cell. 14:559–569. 10.1016/j.devcel.2008.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Gómez M., Coutts N., Price A., Taylor M.V., and Bate M.. 2000. Drosophila dumbfounded: a myoblast attractant essential for fusion. Cell. 102:189–198. 10.1016/S0092-8674(00)00024-6 [DOI] [PubMed] [Google Scholar]
- Runge K.E., Evans J.E., He Z.-Y., Gupta S., McDonald K.L., Stahlberg H., Primakoff P., and Myles D.G.. 2007. Oocyte CD9 is enriched on the microvillar membrane and required for normal microvillar shape and distribution. Dev. Biol. 304:317–325. 10.1016/j.ydbio.2006.12.041 [DOI] [PubMed] [Google Scholar]
- Sampath S.C., Sampath S.C., and Millay D.P.. 2018. Myoblast fusion confusion: the resolution begins. Skelet. Muscle. 8:3 10.1186/s13395-017-0149-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapir A., Choi J., Leikina E., Avinoam O., Valansi C., Chernomordik L.V., Newman A.P., and Podbilewicz B.. 2007. AFF-1, a FOS-1-regulated fusogen, mediates fusion of the anchor cell in C. elegans. Dev. Cell. 12:683–698. 10.1016/j.devcel.2007.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapir A., Avinoam O., Podbilewicz B., and Chernomordik L.V.. 2008. Viral and developmental cell fusion mechanisms: conservation and divergence. Dev. Cell. 14:11–21. 10.1016/j.devcel.2007.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satouh Y., Inoue N., Ikawa M., and Okabe M.. 2012. Visualization of the moment of mouse sperm-egg fusion and dynamic localization of IZUMO1. J. Cell Sci. 125:4985–4990. 10.1242/jcs.100867 [DOI] [PubMed] [Google Scholar]
- Schejter E.D. 2016. Myoblast fusion: Experimental systems and cellular mechanisms. Semin. Cell Dev. Biol. 60:112–120. 10.1016/j.semcdb.2016.07.016 [DOI] [PubMed] [Google Scholar]
- Sens K.L., Zhang S., Jin P., Duan R., Zhang G., Luo F., Parachini L., and Chen E.H.. 2010. An invasive podosome-like structure promotes fusion pore formation during myoblast fusion. J. Cell Biol. 191:1013–1027. 10.1083/jcb.201006006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma B., and Kanwar S.S.. 2018. Phosphatidylserine: A cancer cell targeting biomarker. Semin. Cancer Biol. 52:17–25. 10.1016/j.semcancer.2017.08.012 [DOI] [PubMed] [Google Scholar]
- Sharma-Kishore R., White J.G., Southgate E., and Podbilewicz B.. 1999. Formation of the vulva in Caenorhabditis elegans: a paradigm for organogenesis. Development. 126:691–699. [DOI] [PubMed] [Google Scholar]
- Shemer G., and Podbilewicz B.. 2002. LIN-39/Hox triggers cell division and represses EFF-1/fusogen-dependent vulval cell fusion. Genes Dev. 16:3136–3141. 10.1101/gad.251202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemer G., Suissa M., Kolotuev I., Nguyen K.C.Q., Hall D.H., and Podbilewicz B.. 2004. EFF-1 is sufficient to initiate and execute tissue-specific cell fusion in C. elegans. Curr. Biol. 14:1587–1591. 10.1016/j.cub.2004.07.059 [DOI] [PubMed] [Google Scholar]
- Shi Y., Barton K., De Maria A., Petrash J.M., Shiels A., and Bassnett S.. 2009. The stratified syncytium of the vertebrate lens. J. Cell Sci. 122:1607–1615. 10.1242/jcs.045203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilagardi K., Li S., Luo F., Marikar F., Duan R., Jin P., Kim J.H., Murnen K., and Chen E.H.. 2013. Actin-propelled invasive membrane protrusions promote fusogenic protein engagement during cell-cell fusion. Science. 340:359–363. 10.1126/science.1234781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin N.-Y., Choi H., Neff L., Wu Y., Saito H., Ferguson S.M., De Camilli P., and Baron R.. 2014. Dynamin and endocytosis are required for the fusion of osteoclasts and myoblasts. J. Cell Biol. 207:73–89. 10.1083/jcb.201401137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinn-Thomas J.H., and Mohler W.A.. 2011. New Insights into the Mechanisms and Roles of Cell–Cell Fusion. Int. Rev. Cell Mol. Biol. 289:149–289209. [DOI] [PubMed] [Google Scholar]
- Shinn-Thomas J.H., del Campo J.J., Wang J., and Mohler W.A.. 2016. The EFF-1A Cytoplasmic Domain Influences Hypodermal Cell Fusions in C. elegans But Is Not Dependent on 14-3-3 Proteins. PLoS One. 11:e0146874 10.1371/journal.pone.0146874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singson A., Mercer K.B., and L’Hernault S.W.. 1998. The C. elegans spe-9 gene encodes a sperm transmembrane protein that contains EGF-like repeats and is required for fertilization. Cell. 93:71–79. 10.1016/S0092-8674(00)81147-2 [DOI] [PubMed] [Google Scholar]
- Smurova K., and Podbilewicz B.. 2016. RAB-5- and DYNAMIN-1-Mediated Endocytosis of EFF-1 Fusogen Controls Cell-Cell Fusion. Cell Reports. 14:1517–1527. 10.1016/j.celrep.2016.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smurova K., and Podbilewicz B.. 2017. Endocytosis regulates membrane localization and function of the fusogen EFF-1. Small GTPases. 8:177–180. 10.1080/21541248.2016.1211399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søe K., Andersen T.L., Hobolt-Pedersen A.-S., Bjerregaard B., Larsson L.-I., and Delaissé J.-M.. 2011. Involvement of human endogenous retroviral syncytin-1 in human osteoclast fusion. Bone. 48:837–846. 10.1016/j.bone.2010.11.011 [DOI] [PubMed] [Google Scholar]
- Sottile F., Aulicino F., Theka I., and Cosma M.P.. 2016. Mesenchymal stem cells generate distinct functional hybrids in vitro via cell fusion or entosis. Sci. Rep. 6:36863 10.1038/srep36863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulavie F., and Sundaram M.V.. 2016. Auto-fusion and the shaping of neurons and tubes. Semin. Cell Dev. Biol. 60:136–145. 10.1016/j.semcdb.2016.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulavie F., Hall D.H., and Sundaram M.V.. 2018. The AFF-1 exoplasmic fusogen is required for endocytic scission and seamless tube elongation. Nat. Commun. 9:1741 10.1038/s41467-018-04091-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speijer D., Lukeš J., and Eliáš M.. 2015. Sex is a ubiquitous, ancient, and inherent attribute of eukaryotic life. Proc. Natl. Acad. Sci. USA. 112:8827–8834. 10.1073/pnas.1501725112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas B.P., Woo J., Leong W.Y., and Roy S.. 2007. A conserved molecular pathway mediates myoblast fusion in insects and vertebrates. Nat. Genet. 39:781–786. 10.1038/ng2055 [DOI] [PubMed] [Google Scholar]
- Stone C.E., Hall D.H., and Sundaram M.V.. 2009. Lipocalin signaling controls unicellular tube development in the Caenorhabditis elegans excretory system. Dev. Biol. 329:201–211. 10.1016/j.ydbio.2009.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J.E., Schierenberg E., White J.G., and Thomson J.N.. 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100:64–119. 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- Sundaram M.V., and Cohen J.D.. 2017. Time to make the doughnuts: Building and shaping seamless tubes. Semin. Cell Dev. Biol. 67:123–131. 10.1016/j.semcdb.2016.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symeonides M., Murooka T.T., Bellfy L.N., Roy N.H., Mempel T.R., and Thali M.. 2015. HIV-1-Induced Small T Cell Syncytia Can Transfer Virus Particles to Target Cells through Transient Contacts. Viruses. 7:6590–6603. 10.3390/v7122959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana I., and Hemler M.E.. 1999. Role of transmembrane 4 superfamily (TM4SF) proteins CD9 and CD81 in muscle cell fusion and myotube maintenance. J. Cell Biol. 146:893–904. 10.1083/jcb.146.4.893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada N., Hamazaki T., Oka M., Hoki M., Mastalerz D.M., Nakano Y., Meyer E.M., Morel L., Petersen B.E., and Scott E.W.. 2002. Bone marrow cells adopt the phenotype of other cells by spontaneous cell fusion. Nature. 416:542–545. 10.1038/nature730 [DOI] [PubMed] [Google Scholar]
- Uygur B., Leikina E., Melikov K., Villasmil R., Verma S.K., Vary C.P.H., and Chernomordik L.V.. 2019. Interactions with muscle cells boost fusion, stemness, and drug resistance of prostate cancer cells. Mol. Cancer Res. 17:806–820. 10.1158/1541-7786.MCR-18-0500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacquier V.D. 2012. The quest for the sea urchin egg receptor for sperm. Biochem. Biophys. Res. Commun. 425:583–587. 10.1016/j.bbrc.2012.07.132 [DOI] [PubMed] [Google Scholar]
- Vacquier V.D., and Moy G.W.. 1977. Isolation of bindin: the protein responsible for adhesion of sperm to sea urchin eggs. Proc. Natl. Acad. Sci. USA. 74:2456–2460. 10.1073/pnas.74.6.2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valansi C., Moi D., Leikina E., Matveev E., Graña M., Chernomordik L.V., Romero H., Aguilar P.S., and Podbilewicz B.. 2017. Arabidopsis HAP2/GCS1 is a gamete fusion protein homologous to somatic and viral fusogens. J. Cell Biol. 216:571–581. 10.1083/jcb.201610093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Eijnde S.M., van den Hoff M.J., Reutelingsperger C.P., van Heerde W.L., Henfling M.E., Vermeij-Keers C., Schutte B., Borgers M., and Ramaekers F.C.. 2001. Transient expression of phosphatidylserine at cell-cell contact areas is required for myotube formation. J. Cell Sci. 114:3631–3642. [DOI] [PubMed] [Google Scholar]
- Verma S.K., Leikina E., Melikov K., and Chernomordik L.V.. 2014. Late stages of the synchronized macrophage fusion in osteoclast formation depend on dynamin. Biochem. J. 464:293–300. 10.1042/BJ20141233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S.K., Leikina E., Melikov K., Gebert C., Kram V., Young M.F., Uygur B., and Chernomordik L.V.. 2018. Cell-surface phosphatidylserine regulates osteoclast precursor fusion. J. Biol. Chem. 293:254–270. 10.1074/jbc.M117.809681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Besser K., Frank A.C., Johnson M.A., and Preuss D.. 2006. Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development. 133:4761–4769. 10.1242/dev.02683 [DOI] [PubMed] [Google Scholar]
- Wang Q., Hardie R.-A., Hoy A.J., van Geldermalsen M., Gao D., Fazli L., Sadowski M.C., Balaban S., Schreuder M., Nagarajah R., et al. 2015. Targeting ASCT2-mediated glutamine uptake blocks prostate cancer growth and tumour development. J. Pathol. 236:278–289. 10.1002/path.4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Dang Y.L., Zheng R., Li Y., Li W., Lu X., Wang L.J., Zhu C., Lin H.Y., and Wang H.. 2014. Live cell imaging of in vitro human trophoblast syncytialization. Biol. Reprod. 90:117 10.1095/biolreprod.113.114892 [DOI] [PubMed] [Google Scholar]
- Wang Z., Yuan Y., Zhang L., Min Z., Zhou D., Yu S., Wang P., Ju S., Jun L., and Fu J.. 2018. Impact of cell fusion in myeloma marrow microenvironment on tumor progression. Oncotarget. 9:30997–31006. 10.18632/oncotarget.25742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein N., and Podbilewicz B.. 2016. Organogenesis of the C. elegans Vulva and Control of Cell Fusion. In Organogenetic Gene Networks. Springer International Publishing, Switzerland: 9–56. [Google Scholar]
- White J.G. 1988. The Anatomy. In The Nematode Caenorhabditis elegans. Wood W.B., editor. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: 81–122. [Google Scholar]
- Whitlock J.M., Yu K., Cui Y.Y., and Hartzell H.C.. 2018. Anoctamin 5/TMEM16E facilitates muscle precursor cell fusion. J. Gen. Physiol. 150:1498–1509. 10.1085/jgp.201812097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G.J., and Bianchi E.. 2016. The challenges involved in elucidating the molecular basis of sperm-egg recognition in mammals and approaches to overcome them. Cell Tissue Res. 363:227–235. 10.1007/s00441-015-2243-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi M., Miyamoto T., Sawatani Y., Iwamoto K., Hosogane N., Fujita N., Morita K., Ninomiya K., Suzuki T., Miyamoto K., et al. 2005. DC-STAMP is essential for cell-cell fusion in osteoclasts and foreign body giant cells. J. Exp. Med. 202:345–351. 10.1084/jem.20050645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Zhang Y., Li W.-J., Jiang Y., Zhu Z., Hu H., Li W., Wu J.-W., Wang Z.-X., Dong M.-Q., et al. 2017. Spectraplakin Induces Positive Feedback between Fusogens and the Actin Cytoskeleton to Promote Cell-Cell Fusion. Dev. Cell. 41:107–120.e4. 10.1016/j.devcel.2017.03.006 [DOI] [PubMed] [Google Scholar]
- Yanik M.F., Cinar H., Cinar H.N., Chisholm A.D., Jin Y., and Ben-Yakar A.. 2004. Neurosurgery: functional regeneration after laser axotomy. Nature. 432:822 10.1038/432822a [DOI] [PubMed] [Google Scholar]
- Yochem J., Gu T., and Han M.. 1998. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics. 149:1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Zhao H., Chen T., Tian Y., Li M., Wu K., Bian Y., Su S., Cao Y., Ning Y., et al. 2018. Mutational analysis of IZUMO1R in women with fertilization failure and polyspermy after in vitro fertilization. J. Assist. Reprod. Genet. 35:539–544. 10.1007/s10815-017-1101-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Vashisht A.A., O’Rourke J., Corbel S.Y., Moran R., Romero A., Miraglia L., Zhang J., Durrant E., Schmedt C., et al. 2017a The microprotein Minion controls cell fusion and muscle formation. Nat. Commun. 8:15664 10.1038/ncomms15664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yang Y., Zhu Z., and Ou G.. 2017b WASP-Arp2/3-dependent actin polymerization influences fusogen localization during cell-cell fusion in Caenorhabditiselegans embryos. Biol. Open. 6:1324–1328. 10.1242/bio.026807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Merchak K., Lee W., Grande J.P., Cascalho M., and Platt J.L.. 2015. Cell Fusion Connects Oncogenesis with Tumor Evolution. Am. J. Pathol. 185:2049–2060. 10.1016/j.ajpath.2015.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T., Liang X., Wang X.M., and Shen K.. 2017. Dynein and EFF-1 control dendrite morphology by regulating the localization pattern of SAX-7 in epidermal cells. J. Cell Sci. 130:4063–4071. 10.1242/jcs.201699 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.