The membrane dynamics underlying the biogenesis of autophagosomes, including the origin of the autophagosomal membrane, are still elusive. Shima et al. use a recently developed COPII vesicle–labeling system to show that COPII vesicles are a membrane source in autophagosome formation.
Abstract
A hallmark of autophagy is the de novo formation of double-membrane vesicles called autophagosomes, which sequester various cellular constituents for degradation in lysosomes or vacuoles. The membrane dynamics underlying the biogenesis of autophagosomes, including the origin of the autophagosomal membrane, are still elusive. Although previous studies suggested that COPII vesicles are closely associated with autophagosome biogenesis, it remains unclear whether these vesicles serve as a source of the autophagosomal membrane. Using a recently developed COPII vesicle–labeling system in fluorescence and immunoelectron microscopy in the budding yeast Saccharomyces cerevisiae, we show that the transmembrane cargo Axl2 is loaded into COPII vesicles in the ER. Axl2 is then transferred to autophagosome intermediates, ultimately becoming part of autophagosomal membranes. This study provides a definitive answer to a long-standing, fundamental question regarding the mechanisms of autophagosome formation by implicating COPII vesicles as a membrane source for autophagosomes.
Introduction
Macroautophagy (hereafter referred to as autophagy), an evolutionarily conserved degradation system in eukaryotes, is involved in various cellular activities and the pathogenesis of multiple human diseases (Yang and Klionsky, 2010; Ohsumi, 2014; Bento et al., 2016; Dikic and Elazar, 2018). In autophagy, cytoplasmic components, including proteins, RNAs, and organelles, are sequestered within double-membrane vesicles called autophagosomes and then transported into lysosomes or vacuoles. In Saccharomyces cerevisiae, the core autophagy-related (Atg) proteins assemble to organize the pre-autophagosomal structure (PAS) in the vicinity of the vacuole (Suzuki et al., 2001). Subsequently, a membranous component of the PAS (probably Golgi-derived vesicles containing Atg9; Mari et al., 2010; Yamamoto et al., 2012) expands into a cup-shaped membrane called the isolation membrane (or phagophore). This membrane subsequently becomes spherical and ultimately seals to complete the autophagosome. Previous studies have elucidated the molecular functions of the Atg proteins and their concerted actions, which are highly conserved in yeast and mammals (Nakatogawa et al., 2009; Mizushima et al., 2011). However, the molecular mechanisms underlying autophagosome formation, including the origin of the autophagosomal membrane, remain elusive.
Recent studies suggested several organelles as a source for the autophagosomal membrane, including the ER (Axe et al., 2008; Hayashi-Nishino et al., 2009; Ylä-Anttila et al., 2009), the ER–Golgi intermediate compartment (Ge et al., 2013), mitochondria (Hailey et al., 2010), the plasma membrane (Ravikumar et al., 2010), and the recycling endosome (Puri et al., 2013). In mammals, ER subdomains called omegasomes are formed upon induction of autophagy and are likely to act as a platform for formation of the autophagosome (Axe et al., 2008). Electron microcopy also revealed that the isolation membrane is connected with the ER via thin tubules (Hayashi-Nishino et al., 2009; Ylä-Anttila et al., 2009; Uemura et al., 2014). In yeast, previous studies suggested the involvement of coat protein complex II (COPII) vesicles, which mediate ER-to-Golgi transport (Barlowe et al., 1994; Jensen and Schekman, 2011). Mutants deficient for COPII vesicle formation are unable to form the autophagosome (Ishihara et al., 2001). In addition, the isolation membrane is associated with ER exit sites, where COPII vesicles are formed (Graef et al., 2013; Suzuki et al., 2013). Moreover, Atg9 interacts with the COPII protein Sec24, depending on the phosphorylation of Sec24 under autophagy-inducing conditions (Davis et al., 2016). In mammals, lipidation of the autophagosomal protein LC3 was shown to occur on special COPII vesicles generated from the ER–Golgi intermediate compartment under starvation conditions (Ge et al., 2013, 2014, 2017). These results allowed us to speculate that COPII vesicles serve to form the autophagosomal membrane. However, no direct evidence that COPII vesicles become part of the autophagosomal membrane has been provided.
In this study, we took advantage of a recently developed technique to intensely label COPII vesicles with the transmembrane cargo protein Axl2. We found that this protein is transferred from the ER to forming autophagosomal membranes, dependent upon its loading into COPII vesicles.
Results and discussion
The transmembrane COPII cargo Axl2 localizes to autophagy-related membranes
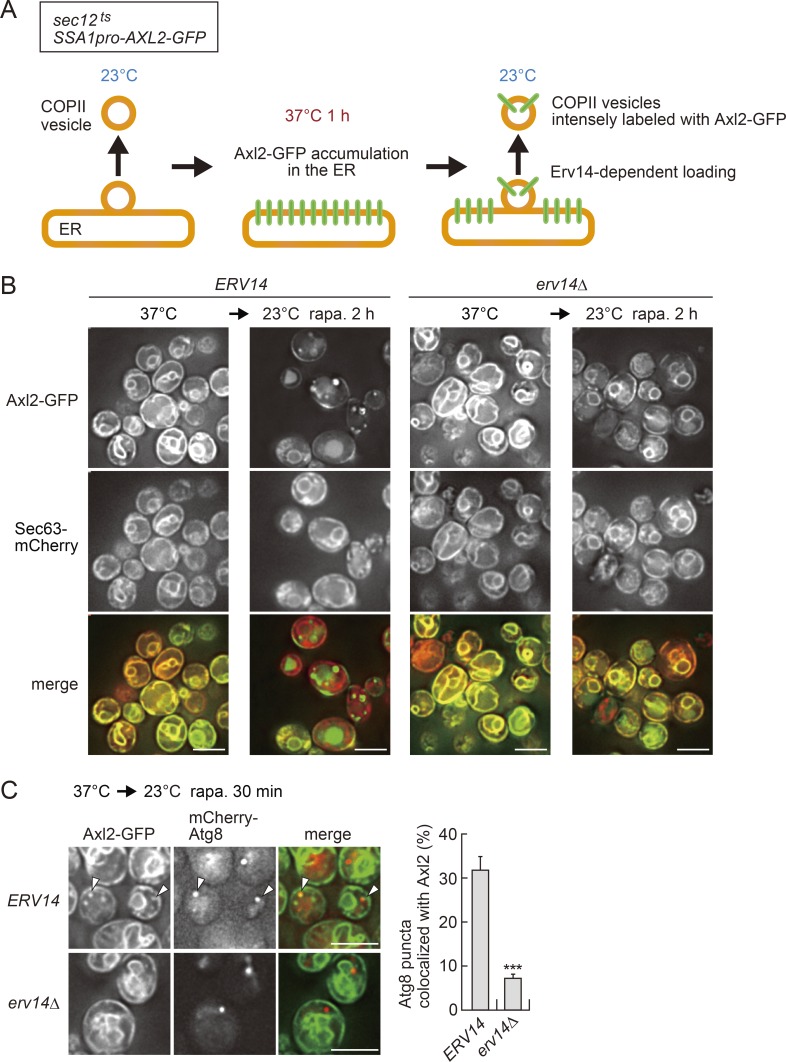
One straightforward approach to assessing whether COPII vesicles are used to form the autophagosome would be to microscopically observe whether the autophagosomal membrane is labeled with a COPII cargo protein. However, even when fluorescent proteins have been fused to COPII cargos, their localization to the autophagosomal membrane has not been observed. Although live imaging of COPII vesicles using a fluorescent protein–fused cargo is intrinsically difficult (probably due to the low copy numbers of cargo proteins and the high-speed movement of the vesicles), a recent study succeeded in circumventing this difficulty (Kurokawa et al., 2014). Axl2 is a transmembrane protein that is sorted into COPII vesicles in the ER and transported, via the Golgi apparatus, to the plasma membrane, where it is involved in budding of daughter cells (Roemer et al., 1996). In that study, Axl2 was fused to GFP and expressed under the control of the SSA1 heat shock promoter in a temperature-sensitive mutant of SEC12 (sec12ts), which encodes a guanine nucleotide-exchange factor required for COPII vesicle formation (Fig. 1 A; Kaiser and Schekman, 1990; Kurokawa et al., 2014). At a low temperature (23°C), these cells can form COPII vesicles, but the expression of AXL2-GFP is suppressed (Fig. 1 A, left). When the cells are shifted to a high temperature (37°C), synthesis of Axl2-GFP is strongly induced, whereas formation of COPII vesicles is blocked due to the inactivation of Sec12, resulting in accumulation of Axl2-GFP in the ER membrane (Fig. 1 A, middle). When the temperature is shifted back to 23°C, Axl2-GFP synthesis is shut off, and COPII vesicles heavily loaded for Axl2-GFP begin to exit from the ER (Fig. 1 A, right). Loading of Axl2 into COPII vesicles depends on the cargo receptor Erv14 (Powers and Barlowe, 1998). We sought to use this system to determine whether COPII vesicles are a source for the autophagosomal membrane.
Figure 1.
The COPII transmembrane cargo Axl2 is transferred from the ER to Atg8-positive structures. (A) Schematic diagram of the method for intense labeling of COPII vesicles. (B) sec12ts SSA1pro-AXL2-GFP cells were grown at 23°C, incubated at 37°C for 1 h, and then incubated at 23°C in the presence of rapamycin. After a 2-h incubation, the cells were observed under a fluorescence microscope. Scale bars, 5 µm. (C) sec12ts SSA1pro-AXL2-GFP cells were incubated at 37°C for 1 h and then treated with rapamycin at 23°C for 30 min, followed by fluorescence microscopy. Arrowheads show mCherry-Atg8 puncta positive for Axl2-GFP. Scale bars, 5 µm. The graph shows the proportion of mCherry-Atg8 puncta that were positive for Axl2-GFP. Values are means ± SD (n = 3). ***, P < 0.001 (unpaired two-tailed t test).
In yeast cells, the ER is mainly distributed in the cell periphery and the perinuclear region, as visualized with Sec63-mCherry (Fig. 1 B; Prinz et al., 2000). As reported previously (Kurokawa et al., 2014), when sec12ts cells were incubated at 37°C for 1 h, Axl2-GFP accumulated in the ER (Fig. 1 B). When the cells were shifted to 23°C in the presence of rapamycin, which inhibits Tor kinase complex 1 and thereby induces autophagy (Noda and Ohsumi, 1998), Axl2-GFP was immediately transported to the plasma membrane, as in the absence of rapamycin (Fig. 1 B; Kurokawa et al., 2014). This ER exit of Axl2-GFP was almost completely blocked in cells lacking Erv14 (Fig. 1 B). Thus, we confirmed that this COPII vesicle–labeling system properly operates under autophagy-inducing conditions.
We also checked whether autophagy occurs normally in the absence of Erv14. In the presence of rapamycin, the amino peptidase Ape1 is transported via autophagy to the vacuole, where it is processed into the mature form (Lynch-Day and Klionsky, 2010). We found that deletion of ERV14 (erv14Δ) did not affect the maturation of Ape1 (Fig. S1 A). Autophagic activity can also be assessed by monitoring GFP fragments generated by vacuolar degradation of the cytoplasmic protein Pgk1 fused to GFP (Welter et al., 2010). We found that Pgk1-GFP was degraded via autophagy in erv14Δ cells as efficiently as in WT cells (Fig. S1 B). We also confirmed that maturation of the vacuolar protease Pep4 proceeded normally in erv14Δ cells (Fig. S1 A). Thus, disruption of ERV14 does not affect autophagy.
The ubiquitin-like protein Atg8 is conjugated to phosphatidylethanolamine and thereby anchored to autophagy-related membranes, including the PAS, isolation membrane, and autophagosome. Consequently, these membranes can be observed by fluorescence microscopy as puncta of fluorescently labeled Atg8 (Suzuki et al., 2001). When sec12ts cells expressing AXL2-GFP and mCherry-ATG8 from the SSA1 and ATG8 promoters, respectively, were incubated at 37°C for 1 h and then treated with rapamycin at 23°C for 30 min, a significant proportion of mCherry-Atg8 puncta were positive for Axl2-GFP (Fig. 1 C). This Axl2 localization to Atg8 puncta was abolished by disruption of ERV14, although Atg8 puncta formed normally in these cells (Fig. 1 C). These results suggested that Axl2 localizes to autophagy-related membranes in a manner dependent on its exit from the ER via COPII vesicles. Colocalization of Axl2 with Atg8 was not observed in SEC12 (WT) cells, in which Axl2 continuously exits from the ER at 37°C (Fig. S2 A), suggesting that the intense, high-frequency loading of Axl2 into COPII vesicles, achieved by its ER accumulation followed by rapid release in a temperature-sensitive COPII mutant, would allow us to observe Axl2 localization to autophagy-related membranes.
Next, we incubated SEC12 cells at 37°C to accumulate Axl2-GFP at the plasma membrane and then shifted them to 23°C to shut off Axl2-GFP expression. When these cells were treated with rapamycin, Axl2-GFP did not localize to mCherry-Atg8 puncta, suggesting that Axl2 was not transported from the plasma membrane to autophagy-related membranes (Fig. S2 B). To accumulate Axl2-GFP at the Golgi apparatus, we performed similar experiments using a temperature-sensitive mutant of SEC7 (sec7ts), which encodes a guanine nucleotide-exchange factor that is responsible for vesicle formation at the Golgi (Franzusoff and Schekman, 1989) and also important for autophagosome formation (van der Vaart et al., 2010). Although these cells were treated with rapamycin at 23°C after incubation at 37°C, we did not observe Axl2 localization to Atg8 puncta, consistent with the idea that Axl2 is transferred from the ER to autophagy-related membranes rather than through the Golgi apparatus (Fig. S2 C).
Axl2 localizes to isolation membranes and autophagosomes
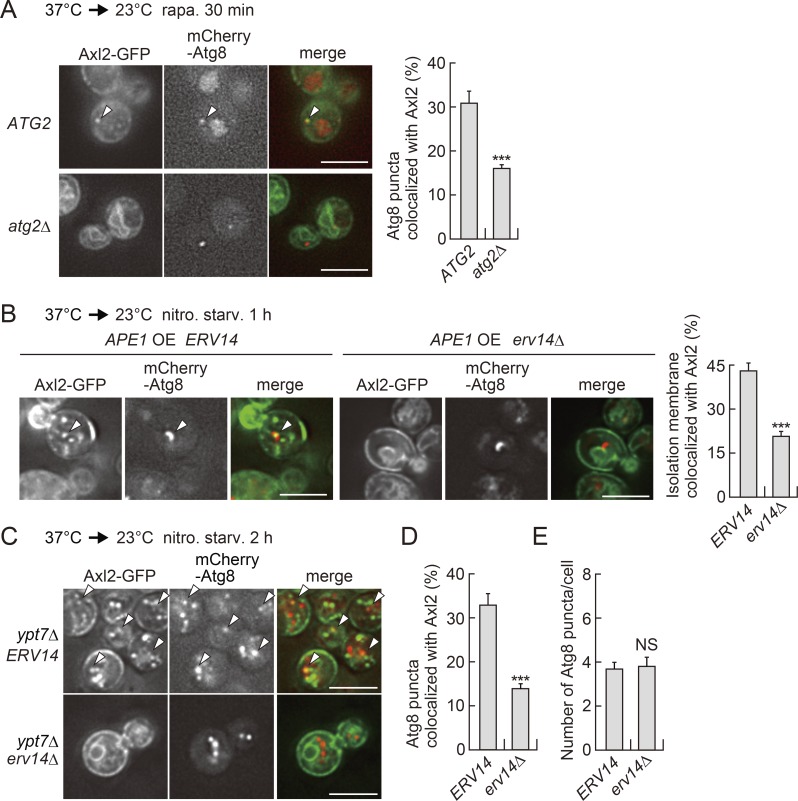
At the autophagosome formation site, the PAS is first organized, and then the isolation membrane expands to form the autophagosome. The Atg2–Atg18 complex is required for autophagosome formation (Shintani et al., 2001; Wang et al., 2001; Obara et al., 2008). Previous studies reported that the isolation membrane hardly expands in cells lacking the Atg2–Atg18 complex, although the other core Atg proteins, including Atg8 and Atg9 vesicles, are assembled (Suzuki et al., 2007, 2013; Gómez-Sánchez et al., 2018). Thus, the Atg8 puncta observed in atg2Δ cells represent the PAS or early isolation membranes. The colocalization of Axl2 with Atg8 was significantly diminished by deletion of ATG2, but it was still observed in the cells (Fig. 2 A). These results suggest that COPII vesicles are also recruited to the autophagosome formation site before the expansion of the isolation membrane initiates, although to a lesser extent.
Figure 2.
COPII vesicles are recruited to forming autophagosomes. (A) Axl2 localization to Atg8 puncta in atg2Δ cells was examined as described in Fig. 1 C. Scale bars, 5 µm. Values in the graph are means ± SD (n = 3). ***, P < 0.001 (unpaired two-tailed t test). (B) sec12ts SSA1pro-AXL2-GFP cells overexpressing Ape1 were incubated at 37°C for 1 h, transferred to nitrogen-starvation medium at 23°C for 1 h, and then observed by fluorescence microscopy. Arrowheads show an Axl2-GFP punctum on an isolation membrane visualized with mCherry-Atg8. Scale bars, 5 µm. The graph shows the proportion of Atg8-positive isolation membranes that colocalized with Axl2-GFP. Values are means ± SD (n = 3). ***, P < 0.001 (unpaired two-tailed t test). (C–E) sec12ts SSA1pro-AXL2-GFP ypt7Δ cells were incubated at 37°C for 1 h, transferred to nitrogen-starvation medium at 23°C for 2 h, and then observed by fluorescence microscopy. Arrowheads show Axl2-positive Atg8 puncta. Scale bars, 5 µm. The graphs show the proportion of mCherry-Atg8 puncta that colocalized with Axl2-GFP (D) and the number of Atg8 puncta per cell (E). Values represent means ± SD (n = 3). ***, P < 0.001 (unpaired two-tailed t test).
When the autophagosome cargo Ape1 is overexpressed in yeast cells, it self-assembles into a giant complex. When autophagy is induced in these cells, isolation membranes expanding along the surface of the giant Ape1 complex can be visualized by fluorescence microscopy as extended Atg8-positive structures (Suzuki et al., 2013). Axl2-GFP localized to these structures in an Erv14-dependent manner (Fig. 2 B). These results suggest that COPII vesicles are employed during expansion of the isolation membrane. However, Axl2-GFP did not stain the entire isolation membrane (see Discussion).
We also investigated whether Axl2-GFP resides on complete autophagosomes. In cells lacking Ypt7, a GTPase required for autophagosome–vacuole fusion, autophagosomes accumulate in the cytoplasm under starvation conditions (Kirisako et al., 1999). Our fluorescence microscopy analysis revealed that a significant proportion of autophagosomes, observed as mCherry-Atg8 puncta in ypt7Δ cells, were labeled with Axl2-GFP (Fig. 2, C and D). Deletion of ERV14 substantially diminished the autophagosomal localization of Axl2-GFP (Fig. 2, C and D), although it did not affect the number of autophagosomes per se (Fig. 2, C and E). These results suggest that Axl2-GFP in the ER membrane is transferred to the isolation membrane via COPII vesicles and ultimately becomes part of the autophagosomal membrane. Residual colocalization of Axl2-GFP with mCherry-Atg8 in erv14Δ cells would at least partly represent ER fragments sequestered within autophagosomes (see the next section).
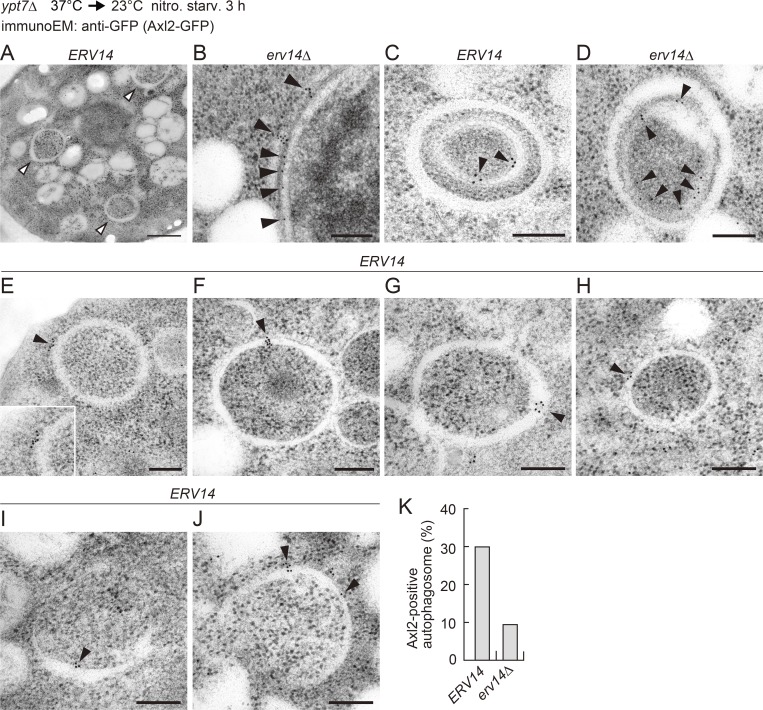
We recently reported that fragments of the ER are selectively sequestered into the autophagosome for degradation in the vacuole under nitrogen-starvation conditions (Mochida et al., 2015). This observation raised the possibility that our fluorescence microscopy experiments were detecting Axl2-GFP signals in ER fragments within autophagosomes rather than in the autophagosomal membrane per se. To address this possibility, we performed immunoelectron microscopy using anti-GFP antibody in order to closely examine the autophagosomal localization of Axl2-GFP (Fig. 3). In yeast cells, autophagosomes are easily identifiable based on their morphological characteristics: double membrane–bound vesicles containing cytoplasmic components, including ribosomes, which appear as electron-dense dots (Fig. 3 A; Takeshige et al., 1992). Immunogold particles representing Axl2-GFP were observed in the ER membrane with higher frequency in erv14Δ cells, in which exit of Axl2-GFP from the ER was blocked (Fig. 3 B). In addition, as expected, double-membrane vesicles derived from the perinuclear ER (or nuclear envelope; Mochida et al., 2015) within autophagosomes were decorated with gold particles (Fig. 3, C and D). Nonetheless, signals for Axl2-GFP were also detected precisely on autophagosomal membranes (Fig. 3, E–H and K). By contrast, autophagosomal membranes were rarely labeled with gold particles in erv14Δ cells. Although at low frequency, Axl2-GFP signals were also found on isolation membranes (unclosed, cup-shaped membranes; Fig. 3, I and J). Taken together, these results suggest that COPII vesicles become part of the autophagosomal membrane during its formation.
Figure 3.
COPII vesicles become part of autophagosomal membranes. (A–J) sec12ts SSA1pro-AXL2-GFP ypt7Δ cells were incubated at 37°C for 1 h, and then autophagy was induced at 23°C by nitrogen starvation for 3 h, followed by immunoelectron microscopy using anti-GFP antibodies. Open arrowheads indicate autophagosomes, and closed arrowheads indicate GFP signals (gold particles). Scale bars: 500 nm (A), 200 nm (B–J). (K) At least 30 autophagosomes in ERV14 and erv14Δ cells were examined, and the proportion of autophagosomes that contained gold particles on their membranes is shown in the graph.
Observation of the transfer of lipids or transmembrane proteins constitutes direct evidence for the contribution of one organelle as a membrane source for the biogenesis of another organelle. In this study, we showed that the transmembrane protein Axl2 is transferred from the ER, via COPII vesicles, to forming autophagosomal membranes. In our analysis, ∼30% of autophagy-related membranes were positive for Axl2 (Fig. 1 C; Fig. 2, A and D; and Fig. 3 K), implying that COPII vesicles are not employed to form all autophagosomes. However, given that almost all autophagosomes are formed in the close vicinity of ER exit sites and that the formation of COPII vesicles is required for autophagosome formation (Ishihara et al., 2001; Graef et al., 2013; Suzuki et al., 2013), COPII vesicles should be an essential source of membrane for autophagosome formation. Therefore, it is more likely that the partial labeling of autophagy-related membranes with Axl2-GFP is due to the nature of the experimental system, in which a limited proportion of COPII vesicles were intensely loaded with Axl2-GFP. Moreover, cells might contain autophagosomes that were formed after most Axl2-GFP had exited the ER (Axl2-GFP expression was shut off upon the temperature downshift).
Although Axl2 localization to Atg8 puncta was significantly diminished by ATG2 deletion, colocalization of Axl2 and Atg8 was still observed (Fig. 2 A), suggesting that COPII vesicles are also recruited at an early stage in autophagosome formation. This idea is consistent with the previous report that defects in COPII vesicle formation impair the PAS localization of Atg proteins (Graef et al., 2013). Nonetheless, the fact that Axl2 colocalized with Atg8 puncta to a greater extent in WT cells than in atg2Δ cells suggests that more COPII vesicles are employed during the expansion of the isolation membrane.
Whereas our results provide the evidence that COPII vesicles (the ER) are a source for the autophagosomal membrane, previous studies have proposed that other organelles also supply membranes to generate autophagosomes (Lamb et al., 2013; Ge et al., 2015; Pavel and Rubinsztein, 2017). As discussed, these different conclusions are not mutually exclusive; in a redundant or nonredundant manner, some organelles may supply membranes as building blocks, whereas others supply proteins or lipids that play specific roles for autophagosome formation. Our observation that Axl2-GFP resided in part of the isolation membrane (Fig. 2 B) may represent the contribution of COPII vesicles to autophagosome formation in this manner.
Recent studies reported that puncta of LC3 (an Atg8 homologue) are closely associated with ER exit sites in mammalian cells (Graef et al., 2013; Zhao et al., 2017). In addition, FIP200, a component of the ULK1 kinase complex that initiates autophagosome formation, interacts with SEC12 (Ge et al., 2017). These results suggest that COPII vesicles are used to form autophagosomal membranes in mammalian cells as well. On the other hand, in mammals, omegasomes and isolation membrane–associated tubules are also involved in autophagosome formation (Axe et al., 2008; Hayashi-Nishino et al., 2009; Ylä-Anttila et al., 2009; Uemura et al., 2014). Hence, it is important to clarify how ER exit sites and COPII vesicles cooperate with these autophagy-related ER subdomains to form autophagosomes.
An inherent, essential role for COPII vesicles is to transport proteins and membranes from the ER to the Golgi apparatus. How are COPII vesicles redirected from transport to the Golgi to the PAS or isolation membrane? First, physical proximity to ER exit sites would facilitate the capture of COPII vesicles by these autophagy-related membranes. In addition, the Rab GTPase Ypt1, which localizes to COPII vesicles, interacts with components of the Atg1 kinase complex in yeast (Wang et al., 2013). A recent study also reported that the COPII coat protein Sec24 is phosphorylated by Hrr25 and interacts with Atg9 (Davis et al., 2016). These protein–protein interactions can target COPII vesicles to the autophagy-related membranes. Subsequently, membrane fusion should occur between these membranes, allowing COPII vesicles to become part of the nascent autophagosomal membranes. It is also noteworthy that autophagy occurs normally in a mutant of Uso1 (Tan et al., 2013), which binds Ypt1 to tether COPII vesicles to the Golgi, suggesting that COPII vesicle fusion to the Golgi is not necessary for autophagosome formation.
Our results demonstrate that COPII vesicles provide material for formation of the autophagosomal membrane, emphasizing the importance of addressing the following key questions. First, how do COPII vesicles fuse with autophagy-related membranes? SNARE proteins are likely to mediate this reaction, as with other intracellular membrane fusion events (Jahn and Scheller, 2006). Although previous studies showed that yeast mutants for SNAREs, which are responsible for fusion between COPII vesicles and the Golgi, are defective in autophagy (Nair et al., 2011; Tan et al., 2013), it remains to be elucidated whether and how these SNAREs are involved in autophagosome formation. It is also important to reveal how COPII vesicles contribute to autophagosome formation. Are COPII vesicles a major source for expansion of the autophagosomal membrane? Do they supply proteins or lipids required for autophagosome formation? It would also be valuable to know the composition and morphology of COPII vesicles used to form autophagosomes, which may differ from those of conventional COPII vesicles. Clarifying these issues will further advance our understanding of the mechanism of autophagosome formation.
Materials and methods
Yeast strains and media
The yeast strains used in this study are listed in Table S1. Gene knockout and tagging of fluorescent proteins were performed by a standard PCR-based method (Janke et al., 2004). To construct the strains expressing Axl2-GFP, the plasmid pRS304-p2HSE-AXL2-GFP (gifted from Dr. Kazuo Kurokawa and Dr. Akihiko Nakano, RIKEN, Saitama, Japan) was linearized with SnaBI and introduced into the parental strains to allow integration of the plasmid into the chromosomal TRP1 locus. Yeast strains were cultured in YPD medium (1% yeast extract, 2% peptone, and 2% glucose) or SD+CA medium (0.17% yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate, 0.5% casamino acid, and 2% glucose) with appropriate supplements. To induce autophagy, cells were shifted to SD-N medium (0.17% yeast nitrogen base without amino acids and ammonium sulfate and 2% glucose) or treated with rapamycin (53123–88-9; LCL).
Fluorescence microscopy
Fluorescence microscopy was performed using a DeltaVision Elite imaging system (GE Healthcare) equipped with a scientific CMOS camera (pco.edge 5.5; PCO AG) and a 60× objective lens (PLAPON, NA/1.42; Olympus). Five z-stack images at 0.2-µm intervals were acquired and deconvolved using the SoftWoRx software. Images were processed using Fiji (ImageJ).
Electron microscopy
Electron microscopy was performed by Tokai Electron Microscopy, Inc. based on a rapid freezing and freeze-fixation method. Yeast cell pellets sandwiched with copper disks were quickly frozen in liquid propane at −175°C and then treated with 0.5% tannic acid in acetone and 2% distilled water at −80°C for 2 d. These samples were dehydrated using anhydrous acetone and anhydrous ethanol, infiltrated with a 50:50 mixture of ethanol and resin (LR white; London Resin Co. Ltd.) at 4°C for 2 h, and then infiltrated with 100% LR white at 4°C for 30 min, followed by polymerization at 50°C overnight. The polymerized resins were ultra-thin sectioned at a thickness of 80 nm using an ultramicrotome (Ultracut UCT; Leica). The sections were placed on nicked grids, which were subjected to immunostaining with anti-GFP antibodies (ab6556; Abcam) and secondary antibodies conjugated to 10-nm gold particles (ab27234; Abcam), followed by treatment with 2% glutaraldehyde. These samples were stained with 2% uranyl acetate and then with lead stain solution (Sigma-Aldrich) and observed on a transmission electron microscope (JEM-1400 Plus; JEOL). Images were acquired using a CCD camera (Veleta; Olympus).
Immunoblots
Cell pellets were suspended in 20% trichloroacetic acid, incubated on ice for 20 min, centrifuged at 15,000 g for 5 min, washed with cold acetone, suspended in SDS sample buffer (100 mM Tris-HCl, pH 7.5, 10% glycerol, 2% SDS, and 20 mM dithiothreitol), and incubated at 65°C for 10 min. The cells were then disrupted on a FastPrep-24 homogenizer (MP Biomedicals) with 0.5-mm YZB zirconia beads (Yasui Kikai). The samples were clarified by centrifugation, and the supernatants were analyzed by SDS-PAGE, followed by immunoblotting with antibodies against GFP (mouse monoclonal, JL-8; Clontech), Pgk1 (mouse monoclonal, 459250; Invitrogen), Ape1, and Pep4 (rabbit polyclonal, anti-API-2, and anti-PrA, respectively; gifted from Dr. Yoshinori Ohsumi, Tokyo Institute of Technology, Yokohama, Japan).
Statistical analysis
The significance of differences was evaluated by an unpaired two-tailed t test.
Online supplemental material
Fig. S1 shows autophagic activity in erv14Δ cells. Fig. S2 shows Axl2 localization in SEC12 or sec7ts cells. Table S1 lists yeast strains used in this study.
Supplementary Material
Acknowledgments
We thank the members of our laboratory for materials, discussions, and technical and secretarial support; Dr. Kazuo Kurokawa, Dr. Akihiko Nakano, and Dr. Hayashi Yamamoto for providing the plasmids; Dr. Yoshinori Ohsumi for providing the antibodies; and the Biomaterial Analysis Center, Technical Department at Tokyo Tech for DNA sequencing service.
This work was supported in part by the Ministry of Education, Culture, Sports, Science and Technology of Japan (KAKENHI Grants-in-Aid for Scientific Research 25111003 and 25711005 to H. Nakatogawa), the Japan Science and Technology Agency (CREST grant JPMJCR13M7 to H. Nakatogawa), and the Tokyo Tech Fund (STAR Grant to H. Nakatogawa).
The authors declare no competing financial interests.
Author contributions: T. Shima and H. Nakatogawa designed the project. T. Shima performed most of the experiments with the help of H. Kirisako. T. Shima and H. Nakatogawa wrote the manuscript. All authors analyzed and discussed the results and commented on the manuscript.
References
- Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., and Ktistakis N.T.. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701. 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C., Orci L., Yeung T., Hosobuchi M., Hamamoto S., Salama N., Rexach M.F., Ravazzola M., Amherdt M., and Schekman R.. 1994. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 77:895–907. 10.1016/0092-8674(94)90138-4 [DOI] [PubMed] [Google Scholar]
- Bento C.F., Renna M., Ghislat G., Puri C., Ashkenazi A., Vicinanza M., Menzies F.M., and Rubinsztein D.C.. 2016. Mammalian Autophagy: How Does It Work? Annu. Rev. Biochem. 85:685–713. 10.1146/annurev-biochem-060815-014556 [DOI] [PubMed] [Google Scholar]
- Davis S., Wang J., Zhu M., Stahmer K., Lakshminarayan R., Ghassemian M., Jiang Y., Miller E.A., and Ferro-Novick S.. 2016. Sec24 phosphorylation regulates autophagosome abundance during nutrient deprivation. eLife. 5:1–22. 10.7554/eLife.21167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I., and Elazar Z.. 2018. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19:349–364. 10.1038/s41580-018-0003-4 [DOI] [PubMed] [Google Scholar]
- Franzusoff A., and Schekman R.. 1989. Functional compartments of the yeast Golgi apparatus are defined by the sec7 mutation. EMBO J. 8:2695–2702. 10.1002/j.1460-2075.1989.tb08410.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L., Melville D., Zhang M., and Schekman R.. 2013. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. eLife. 2:e00947 10.7554/eLife.00947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L., Zhang M., and Schekman R.. 2014. Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. eLife. 3:e04135 10.7554/eLife.04135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L., Wilz L., and Schekman R.. 2015. Biogenesis of autophagosomal precursors for LC3 lipidation from the ER-Golgi intermediate compartment. Autophagy. 11:2372–2374. 10.1080/15548627.2015.1105422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L., Zhang M., Kenny S.J., Liu D., Maeda M., Saito K., Mathur A., Xu K., and Schekman R.. 2017. Remodeling of ER-exit sites initiates a membrane supply pathway for autophagosome biogenesis. EMBO Rep. 18:1586–1603. 10.15252/embr.201744559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Sánchez R., Rose J., Guimarães R., Mari M., Papinski D., Rieter E., Geerts W.J., Hardenberg R., Kraft C., Ungermann C., and Reggiori F.. 2018. Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. J. Cell Biol. 217:2743–2763. 10.1083/jcb.201710116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef M., Friedman J.R., Graham C., Babu M., and Nunnari J.. 2013. ER exit sites are physical and functional core autophagosome biogenesis components. Mol. Biol. Cell. 24:2918–2931. 10.1091/mbc.e13-07-0381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey D.W., Rambold A.S., Satpute-Krishnan P., Mitra K., Sougrat R., Kim P.K., and Lippincott-Schwartz J.. 2010. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 141:656–667. 10.1016/j.cell.2010.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., and Yamamoto A.. 2009. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11:1433–1437. 10.1038/ncb1991 [DOI] [PubMed] [Google Scholar]
- Ishihara N., Hamasaki M., Yokota S., Suzuki K., Kamada Y., Kihara A., Yoshimori T., Noda T., and Ohsumi Y.. 2001. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell. 12:3690–3702. 10.1091/mbc.12.11.3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R., and Scheller R.H.. 2006. SNAREs—engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7:631–643. 10.1038/nrm2002 [DOI] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., and Knop M.. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962. 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Jensen D., and Schekman R.. 2011. COPII-mediated vesicle formation at a glance. J. Cell Sci. 124:1–4. 10.1242/jcs.069773 [DOI] [PubMed] [Google Scholar]
- Kaiser C.A., and Schekman R.. 1990. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 61:723–733. 10.1016/0092-8674(90)90483-U [DOI] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., and Ohsumi Y.. 1999. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147:435–446. 10.1083/jcb.147.2.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K., Okamoto M., and Nakano A.. 2014. Contact of cis-Golgi with ER exit sites executes cargo capture and delivery from the ER. Nat. Commun. 5:3653 10.1038/ncomms4653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C.A., Yoshimori T., and Tooze S.A.. 2013. The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14:759–774. 10.1038/nrm3696 [DOI] [PubMed] [Google Scholar]
- Lynch-Day M.A., and Klionsky D.J.. 2010. The Cvt pathway as a model for selective autophagy. FEBS Lett. 584:1359–1366. 10.1016/j.febslet.2010.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari M., Griffith J., Rieter E., Krishnappa L., Klionsky D.J., and Reggiori F.. 2010. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J. Cell Biol. 190:1005–1022. 10.1083/jcb.200912089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., and Ohsumi Y.. 2011. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27:107–132. 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- Mochida K., Oikawa Y., Kimura Y., Kirisako H., Hirano H., Ohsumi Y., and Nakatogawa H.. 2015. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 522:359–362. 10.1038/nature14506 [DOI] [PubMed] [Google Scholar]
- Nair U., Jotwani A., Geng J., Gammoh N., Richerson D., Yen W.L., Griffith J., Nag S., Wang K., Moss T., et al. . 2011. SNARE proteins are required for macroautophagy. Cell. 146:290–302. 10.1016/j.cell.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H., Suzuki K., Kamada Y., and Ohsumi Y.. 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10:458–467. 10.1038/nrm2708 [DOI] [PubMed] [Google Scholar]
- Noda T., and Ohsumi Y.. 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273:3963–3966. 10.1074/jbc.273.7.3963 [DOI] [PubMed] [Google Scholar]
- Obara K., Sekito T., Niimi K., and Ohsumi Y.. 2008. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J. Biol. Chem. 283:23972–23980. 10.1074/jbc.M803180200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y. 2014. Historical landmarks of autophagy research. Cell Res. 24:9–23. 10.1038/cr.2013.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavel M., and Rubinsztein D.C.. 2017. Mammalian autophagy and the plasma membrane. FEBS J. 284:672–679. 10.1111/febs.13931 [DOI] [PubMed] [Google Scholar]
- Powers J., and Barlowe C.. 1998. Transport of axl2p depends on erv14p, an ER-vesicle protein related to the Drosophila cornichon gene product. J. Cell Biol. 142:1209–1222. 10.1083/jcb.142.5.1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz W.A., Grzyb L., Veenhuis M., Kahana J.A., Silver P.A., and Rapoport T.A.. 2000. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 150:461–474. 10.1083/jcb.150.3.461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri C., Renna M., Bento C.F., Moreau K., and Rubinsztein D.C.. 2013. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 154:1285–1299. 10.1016/j.cell.2013.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B., Moreau K., Jahreiss L., Puri C., and Rubinsztein D.C.. 2010. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 12:747–757. 10.1038/ncb2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer T., Madden K., Chang J., and Snyder M.. 1996. Selection of axial growth sites in yeast requires Axl2p, a novel plasma membrane glycoprotein. Genes Dev. 10:777–793. 10.1101/gad.10.7.777 [DOI] [PubMed] [Google Scholar]
- Shintani T., Suzuki K., Kamada Y., Noda T., and Ohsumi Y.. 2001. Apg2p functions in autophagosome formation on the perivacuolar structure. J. Biol. Chem. 276:30452–30460. 10.1074/jbc.M102346200 [DOI] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., and Ohsumi Y.. 2001. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20:5971–5981. 10.1093/emboj/20.21.5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kubota Y., Sekito T., and Ohsumi Y.. 2007. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 12:209–218. 10.1111/j.1365-2443.2007.01050.x [DOI] [PubMed] [Google Scholar]
- Suzuki K., Akioka M., Kondo-Kakuta C., Yamamoto H., and Ohsumi Y.. 2013. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J. Cell Sci. 126:2534–2544. 10.1242/jcs.122960 [DOI] [PubMed] [Google Scholar]
- Takeshige K., Baba M., Tsuboi S., Noda T., and Ohsumi Y.. 1992. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119:301–311. 10.1083/jcb.119.2.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D., Cai Y., Wang J., Zhang J., Menon S., Chou H.-T., Ferro-Novick S., Reinisch K.M., and Walz T.. 2013. The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc. Natl. Acad. Sci. USA. 110:19432–19437. 10.1073/pnas.1316356110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T., Yamamoto M., Kametaka A., Sou Y.S., Yabashi A., Yamada A., Annoh H., Kametaka S., Komatsu M., and Waguri S.. 2014. A cluster of thin tubular structures mediates transformation of the endoplasmic reticulum to autophagic isolation membrane. Mol. Cell. Biol. 34:1695–1706. 10.1128/MCB.01327-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart A., Griffith J., and Reggiori F.. 2010. Exit from the Golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 21:2270–2284. 10.1091/mbc.e09-04-0345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.W., Kim J., Huang W.P., Abeliovich H., Stromhaug P.E., Dunn W.A. Jr., and Klionsky D.J.. 2001. Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. J. Biol. Chem. 276:30442–30451. 10.1074/jbc.M102342200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Menon S., Yamasaki A., Chou H.-T., Walz T., Jiang Y., and Ferro-Novick S.. 2013. Ypt1 recruits the Atg1 kinase to the preautophagosomal structure. Proc. Natl. Acad. Sci. USA. 110:9800–9805. 10.1073/pnas.1302337110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter E., Thumm M., and Krick R.. 2010. Quantification of nonselective bulk autophagy in S. cerevisiae using Pgk1-GFP. Autophagy. 6:794–797. 10.4161/auto.6.6.12348 [DOI] [PubMed] [Google Scholar]
- Yamamoto H., Kakuta S., Watanabe T.M., Kitamura A., Sekito T., Kondo-Kakuta C., Ichikawa R., Kinjo M., and Ohsumi Y.. 2012. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J. Cell Biol. 198:219–233. 10.1083/jcb.201202061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., and Klionsky D.J.. 2010. Eaten alive: a history of macroautophagy. Nat. Cell Biol. 12:814–822. 10.1038/ncb0910-814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylä-Anttila P., Vihinen H., Jokitalo E., and Eskelinen E.L.. 2009. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 5:1180–1185. 10.4161/auto.5.8.10274 [DOI] [PubMed] [Google Scholar]
- Zhao Y.G., Chen Y., Miao G., Zhao H., Qu W., Li D., Wang Z., Liu N., Li L., Chen S., et al. . 2017. The ER-Localized Transmembrane Protein EPG-3/VMP1 Regulates SERCA Activity to Control ER-Isolation Membrane Contacts for Autophagosome Formation. Mol. Cell. 67:974–989.e6. 10.1016/j.molcel.2017.08.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.