This work provides direct evidence that heterotrimeric G proteins can be activated in vivo by a cytoplasmic factor instead of by a GPCR. Specifically, DAPLE, a nonreceptor protein bearing an evolutionarily conserved Gα-binding and -activating (GBA) motif, triggers apical cell constriction during neurulation in Xenopus laevis embryos via G protein–dependent signaling.
Abstract
Heterotrimeric G proteins are signaling switches that control organismal morphogenesis across metazoans. In invertebrates, specific GPCRs instruct G proteins to promote collective apical cell constriction in the context of epithelial tissue morphogenesis. In contrast, tissue-specific factors that instruct G proteins during analogous processes in vertebrates are largely unknown. Here, we show that DAPLE, a non-GPCR protein linked to human neurodevelopmental disorders, is expressed specifically in the neural plate of Xenopus laevis embryos to trigger a G protein signaling pathway that promotes apical cell constriction during neurulation. DAPLE localizes to apical cell–cell junctions in the neuroepithelium, where it activates G protein signaling to drive actomyosin-dependent apical constriction and subsequent bending of the neural plate. This function is mediated by a Gα-binding-and-activating (GBA) motif that was acquired by DAPLE in vertebrates during evolution. These findings reveal that regulation of tissue remodeling during vertebrate development can be driven by an unconventional mechanism of heterotrimeric G protein activation that operates in lieu of GPCRs.
Introduction
Heterotrimeric G proteins (Gαβγ) are ubiquitous signaling switches involved in a vast array of physiological and pathological processes. Genetic deletion of heterotrimeric G proteins in mice causes embryonic malformations and lethality (Offermanns et al., 1998; Yu et al., 1998; Wettschureck et al., 2004; Lin et al., 2005; Plummer et al., 2012), highlighting their importance in morphogenesis. For example, deletion of Gβ1 causes neural tube (NT) defects and abnormal organization of the actin cytoskeleton in the embryonic brain (Okae and Iwakura, 2010). Because G proteins are ubiquitous, a critical question is how they are activated at the precise time and location to modulate the effector machinery driving morphogenesis, such as regulators of the actin cytoskeleton. For this, G protein activation must be achieved by factors that operate in a spatiotemporally restricted manner under morphogen-directed transcriptional programs (Gilmour et al., 2017). Logical candidates for this role are G protein–coupled receptors (GPCRs), a large family of proteins with guanine nucleotide exchange factor (GEF) activity toward G proteins (Gilman, 1987). Such GEF activity leads to the exchange of GDP for GTP on Gα subunits and subsequent dissociation of Gαβγ heterotrimers into Gα-GTP and “free” Gβγ subunits, both of which activate downstream effectors.
Studies in flies have established that GPCRs can work as tissue-specific G protein activators during morphogenesis. Formation of the ventral furrow in Drosophila melanogaster is a paradigm of tissue morphogenesis (Martin and Goldstein, 2014; Gilmour et al., 2017; Heer and Martin, 2017) that resembles some aspects of NT formation in vertebrates (Nikolopoulou et al., 2017). Both processes rely on mechanical deformation of an epithelial sheet by apical cell constriction, one of the most important cell shape changes that drive morphogenesis. In the ventral furrow, both Gα and Gβγ (Parks and Wieschaus, 1991; Kanesaki et al., 2013; Kerridge et al., 2016) contribute to the activation of a RhoGEF–Rho–Rok cascade that promotes contractility of an actomyosin meshwork enriched at the apical cortex and anchored at apical cell–cell junctions (Barrett et al., 1997; Kölsch et al., 2007). These forces translate into collective apical cell constriction that causes bending of the epithelial tissue. It has only recently been elucidated that tissue-specific activation of this G protein–actomyosin effector axis is achieved by the GPCR called Mist (mesoderm-invagination signal transducer; Manning et al., 2013), which is the long-sought receptor for the extracellular ligand Fog (Costa et al., 1994). Nevertheless, neither the receptors nor the ligand of this pathway is conserved out of insects (Manning and Rogers, 2014), leaving unanswered the question of how tissue-specific G protein activation drives apical cell constriction in the context of vertebrate morphogenesis (Gilmour et al., 2017).
The long-held tenet that activation of heterotrimeric G proteins is a function performed exclusively by GPCRs has been challenged recently. For example, some cytoplasmic factors bearing the so-called Gα-binding-and-activating (GBA) motif (de Opakua et al., 2017; DiGiacomo et al., 2018) possess GEF activity in vitro for Gα proteins of the Gi subfamily (Gαi1, Gαi2, or Gαi3; Garcia-Marcos et al., 2009, 2011; Aznar et al., 2015) and promote G protein signaling in cells, as determined by readouts for the generation of either GTP-bound Gαi (Lin et al., 2014; Lopez-Sanchez et al., 2014; Aznar et al., 2015; Midde et al., 2015) or free Gβγ (Garcia-Marcos et al., 2009; Aznar et al., 2015; Leyme et al., 2015, 2017; Midde et al., 2015; Parag-Sharma et al., 2016; Maziarz et al., 2018). Despite abundant information on the biochemical and signaling mechanisms of this growing family of regulators, direct in vivo evidence for a physiological process controlled by them is still lacking. We reasoned that DAPLE, a recently identified member of this group of GEFs, might serve as a G protein activator during embryonic morphogenesis based on the observation that congenital mutations in this gene cause neurodevelopmental disorders in humans (Ekici et al., 2010; Drielsma et al., 2012; Ruggeri et al., 2018; Wallis et al., 2018; Zwaveling-Soonawala et al., 2018; e.g., nonsyndromic hydrocephalus). Here, we show that DAPLE is a tissue-specific activator of G proteins that triggers a signaling cascade culminating in the apical constriction of neuroepithelial cells during neurulation. These findings shed light onto the mechanisms by which heterotrimeric G proteins control apical cell constriction in the context of vertebrate embryogenesis and establish that unconventional G protein activation by non-GPCR proteins controls a physiologically relevant process in vivo.
Results
DAPLE localizes to apical cell junctions and induces apical cell constriction
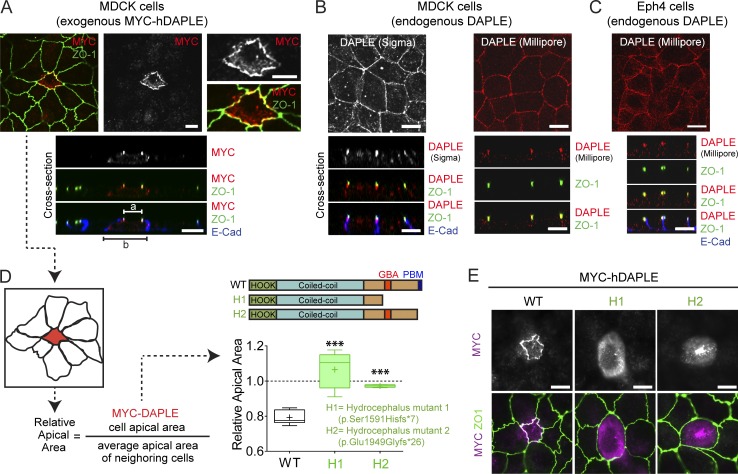
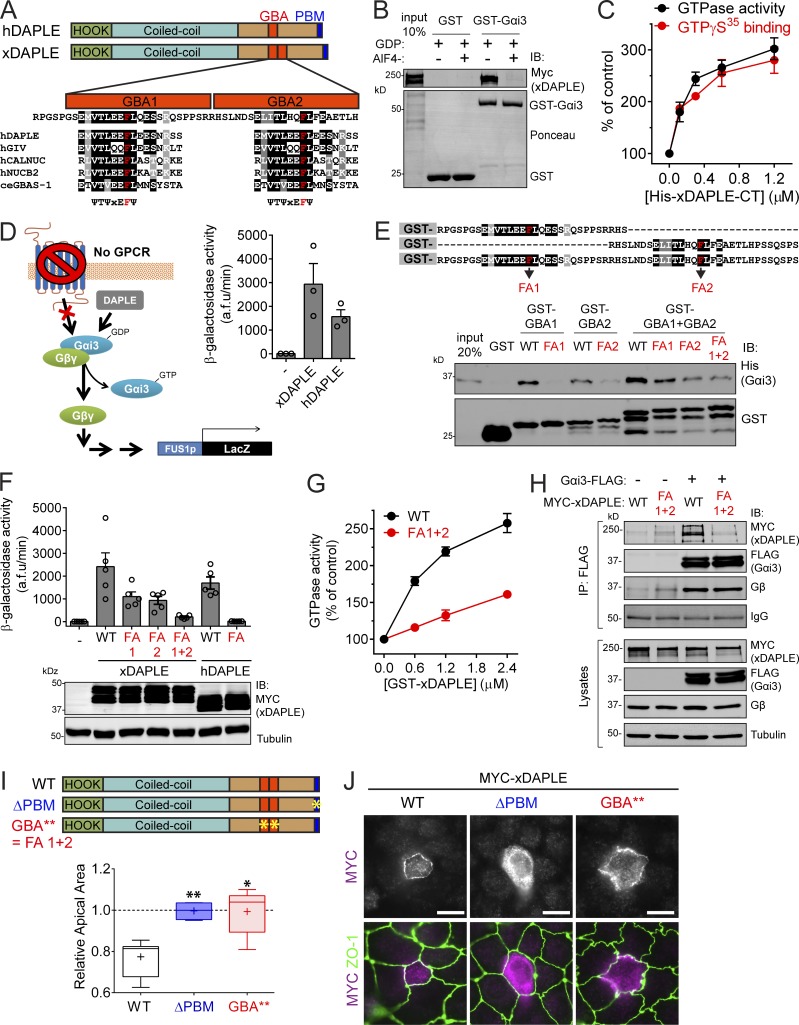
Expression of human DAPLE (hDAPLE) in MDCK cells, a polarized epithelial cell model, revealed two important points. One is that DAPLE is enriched at apical cell junctions, as determined by its partial colocalization with the apical junction marker ZO-1 (Fig. 1 A). This subcellular localization was also observed for endogenous DAPLE in two different epithelial cell lines (MDCK and EpH4) and using two different antibodies (Fig. 1, B and C). The second important observation is that expression of hDAPLE in MDCK cells induces apical constriction, as evidenced by the decreased apical area of DAPLE-expressing cells compared with their own basal area or the apical area of surrounding cells (Fig. 1, A and D).
Figure 1.
DAPLE localizes to apical cell junctions and promotes apical cell constriction. (A) Confocal fluorescence microscopy pictures of MDCK cells sparsely expressing ectopic MYC-hDAPLE costained for MYC (red), ZO-1 (green), and E-cadherin (blue). Top panels correspond to a view on the monolayer from the top, and panels on the bottom are optical cross sections of the monolayer. a and b segments correspond to the width of the apical and basal cell membrane domains. (B and C) Confocal fluorescence microscopy pictures of MDCK (B) or EpH4 (C) cell monolayers stained for DAPLE (Sigma-Aldrich or Millipore antibody, red), ZO-1 (green), and E-cadherin (blue) as indicated. The top panels correspond to views on the cell monolayers from the top, and panels on the bottom are optical cross sections of the monolayers. (D) Quantification of the relative apical area of DAPLE-transfected cells compared with neighboring, untransfected cells shows that hDAPLE WT, but not hydrocephalus-associated mutants (H1 and H2), cause apical constriction in MDCK cells. Results are presented as box-and-whiskers plots (minimum to maximum) of n = 5–9 independent experiments per condition. ***, P < 0.001 using the Mann–Whitney U test. (E) Fluorescence microscopy pictures of MDCK cells sparsely expressing the indicated MYC-hDAPLE constructs and costained for MYC (magenta) and ZO-1 (green) show that H1 and H2 mutants are not enriched at apical cell junctions like WT. Scale bars, 10 µm.
Recessive hDAPLE mutations identified in patients as causative of congenital nonsyndromic hydrocephalus are assumed to cause a loss of function. We reasoned that using some of these natural mutations that result in truncated proteins (H1 and H2; Ekici et al., 2010; Drielsma et al., 2012) as tools could provide clues about which domains of DAPLE are important for its function in apical constriction (Fig. 1 D). More specifically, one mutant (H2) lacks the last ∼80 aa in the C terminus of DAPLE, which include its PDZ-binding motif (PBM; Oshita et al., 2003), whereas the other mutant (H1) lacks a larger region that encompasses both the PBM and the GBA motif. When expressed in MDCK cells, these two hDAPLE mutants failed to induce apical constriction (Fig. 1 D), indicating that the missing protein domains are crucial for this function. In addition, we found that both mutants failed to concentrate at apical cell junctions (Fig. 1 E). Taken together, these findings suggest that the localization of DAPLE at apical cell junctions is important for its function of promoting constriction, which is consistent with the idea of apical junctions as one of the subcellular loci where actomyosin contractile forces are generated to drive apical constriction (Martin and Goldstein, 2014; Heer and Martin, 2017).
Both the PBM and GBA motif of DAPLE are required for apical cell constriction
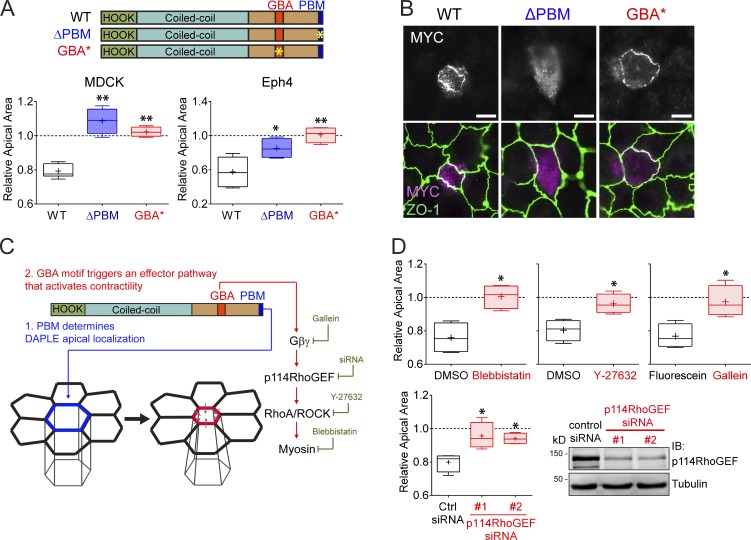
Next, we used synthetic mutants to gain deeper mechanistic insight into which domains of hDAPLE are required for apical cell constriction. For this, we used a PDZ-binding–deficient mutant (ΔPBM, last 3 aa deleted; Oshita et al., 2003) and a GEF-deficient mutant (GBA*; F1675 in the GBA motif mutated to alanine; Aznar et al., 2015; Fig. 2 A). Both mutants failed to induce apical constriction when expressed in two different cell lines, MDCK and EpH4 (Fig. 2 A).
Figure 2.
Both the PBM and GBA motif of DAPLE are required to promote apical cell constriction. (A) Quantification of the relative apical area of DAPLE-transfected cells compared with neighboring, untransfected cells shows that hDAPLE ΔPBM and hDAPLE GBA* mutants fail to promote apical constriction in MDCK (left) or EpH4 (right) cells. Results are presented as box-and-whiskers plots (error bars indicate minimum to maximum range) of n = 4–9 independent experiments per condition. *, P < 0.05; **, P < 0.01 using the Mann–Whitney U test. (B) Confocal fluorescence microscopy pictures of MDCK cells sparsely expressing the indicated MYC-hDAPLE constructs and costained for MYC (magenta) and ZO-1 (green) show that hDAPLE GBA* mutant is enriched at apical junctions like WT, while hDAPLE ΔPBM mutant is not. Scale bars, 10 µm. (C) Diagram depicting the different functions of the PBM and the GBA motif in DAPLE-induced apical cell constriction. A possible effector pathway to promote apical cell constriction though G protein activation is shown on the right, along with the treatments used in D to test it (green). (D) Box-and-whiskers plots (error bars indicate minimum to maximum range) for the quantification of relative apical area show that DAPLE-mediated apical constriction requires the activity of myosin (inhibited by blebbistatin), ROCK (inhibited by Y-27632), free Gβγ (inhibited by Gallein, but not its inactive analogue, fluorescein) and p114RhoGEF (inhibited by siRNA). n = 4 independent experiments per condition. *, P < 0.05 using the Mann–Whitney U test. The immunoblot (IB) on the bottom right shows the reduction of p114RhoGEF expression upon siRNA treatments.
The PBM, but not the GBA motif, of DAPLE is required for apical junction localization
We found that DAPLE ΔPBM failed to localize to apical junctions in MDCK cells (Fig. 2 B), whereas DAPLE GBA* was still enriched at apical cell junctions (Fig. 2 B), much like DAPLE WT, despite being defective in promoting apical cell constriction (Fig. 2 A). One conclusion drawn from these results is that PBM-dependent localization at cell junctions is necessary for DAPLE-mediated apical constriction. However, the PBM and junctional localization of DAPLE are not sufficient to drive cell-shape remodeling, because the GEF-deficient GBA* mutant, which contains an intact PBM, is enriched at apical cell junctions, similar to WT, yet fails to induce apical constriction. We conclude that the PBM and the GBA motif have two distinct functions required for apical cell constriction. (1) The PBM determines the apical junction localization of DAPLE, which serves as a positional cue for (2) the G protein regulatory activity mediated by the GBA motif, which serves as a signaling effector function (Fig. 2 C).
DAPLE promotes apical constriction via Gβγ and p114RhoGEF
Next, we set out to delineate the G protein–dependent signaling cascade involved in DAPLE-induced apical constriction (Fig. 2 C). For this, we quantified MDCK apical cell constriction upon expression of hDAPLE concurrently with different manipulations to interfere with downstream signaling events (Fig. 2, C and D). First, we reasoned that DAPLE-induced apical cell constriction might rely on myosin function downstream of Rho-associated protein kinase (ROCK), as this is a ubiquitous mechanism that directly promotes contractility. We found that treatment with blebbistatin, a myosin inhibitor, or Y-27632, a ROCK inhibitor, blocked DAPLE-induced apical cell constriction (Fig. 2 D). Which are the G proteins and G protein–dependent intermediaries by which DAPLE’s GBA motif impacts this ubiquitous contractile machinery? The GBA motif of DAPLE activates specifically Gα subunits of the Gi subfamily (Aznar et al., 2015), which are not known to directly modulate effectors involved in actin remodeling, such as RhoGEFs. However, DAPLE’s action on Gαi also causes release of free Gβγ from Gi heterotrimers and activation of Gβγ-dependent signaling (Aznar et al., 2015), which includes the regulation of several RhoGEFs (Aittaleb et al., 2010). We found that DAPLE-induced apical constriction is Gβγ dependent, as it is completely blunted by the Gβγ inhibitor Gallein (Smrcka, 2013; Fig. 2 D). Among the different RhoGEFs regulated by Gβγ, we reasoned that the best candidate to mediate the effect of DAPLE on apical cell constriction was p114RhoGEF. This is because p114RhoGEF is the only GEF for RhoA that, on one hand, is activated by Gβγ (Niu et al., 2003) and, on the other, operates at apical cell junctions to cause apical constriction of epithelial cells (Nakajima and Tanoue, 2011; Terry et al., 2011). Consistent with this hypothesis, we found that depletion of p114RhoGEF from MDCK cells with two independent siRNAs inhibited DAPLE-induced apical constriction (Fig. 2 D). None of the chemical or genetic manipulations described above disrupted the localization of hDAPLE at apical cell junctions (Fig. S1), which rules out that the observed effects are secondary to its mislocalization. Instead, this observation is consistent with the idea that the experimental manipulations used impact the G protein–dependent effector function of DAPLE, which is not required for its subcellular localization (Fig. 2 B). Taken together, our findings suggest that DAPLE triggers the following signaling cascade to promote apical cell constriction: GBA motif → Gβγ → p114RhoGEF → RhoA/ROCK → actomyosin contractility (Fig. 2 C).
DAPLE is required for neural plate bending during neurulation in Xenopus laevis
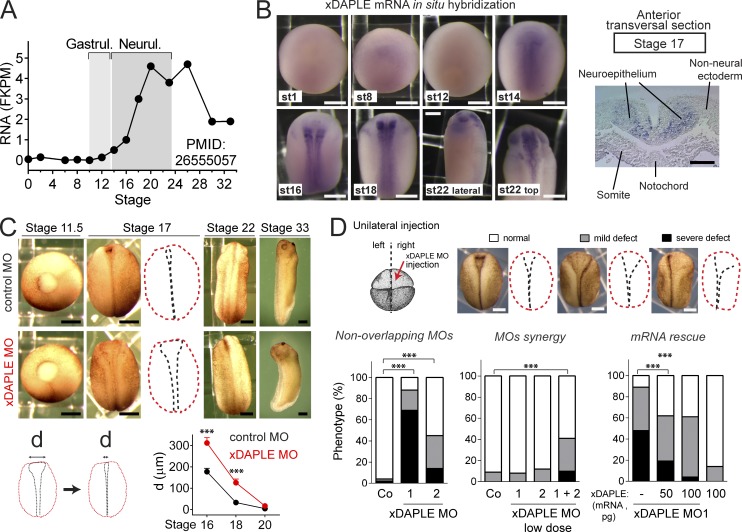
Apical cell constriction is important for epithelial remodeling during embryogenesis. We set out to investigate a putative function for DAPLE during embryonic development using Xenopus as a model organism because it has been extensively used to study the principles of morphogenesis and has conserved features with other vertebrates (Keller, 2002; Lowery and Sive, 2004, 2009). Xenopus DAPLE (xDAPLE or xDal; Kobayashi et al., 2005) mRNA is nearly absent from fertilization through gastrulation and then sharply induced during neurulation (Peshkin et al., 2015; Fig. 3 A). RNA in situ hybridization revealed that xDAPLE expression is spatially restricted to the neural plate (Fig. 3 B). Consistent with xDAPLE pattern of expression, we observed that morpholino (MO)-mediated depletion of xDAPLE (Fig. S2 A) does not affect embryo morphology up to gastrulation (stage 11.5) but causes a delay in the closure of the NT (stage 17; Fig. 3 C). Although the NT eventually closes (stage 22; Fig. 3 C), xDAPLE morphant embryos display mild axis curvature and head defects at later stages (stage 33; Fig. 3 C). Malformations were also evident in stage 46 embryonic brains (Fig. S2 B), which formed a closed structure but displayed a relative enlargement of ventricles. Taken together, these results indicate that xDAPLE is induced in the neural plate and is required for normal neurulation. More specifically, loss of DAPLE impairs neural plate bending without causing open NT defects.
Figure 3.
Loss of DAPLE delays NT closure in Xenopus. (A) Quantification of DAPLE mRNA abundance in whole Xenopus embryos at different stages by RNA sequencing showing induction during neurulation. Extracted from Peshkin et al. (2015). (B) Left: Whole-mount RNA in situ Hybridization for xDAPLE showing restricted expression in neural tissues from the onset of neurulation, but not at earlier stages (st). Right: anterior transversal section on the right shows specific expression in neuroepithelial cells. (C) xDAPLE morphants at different stages of development showing a delay in the closure of the neural plate compared with controls. Graph at the bottom shows a quantification of the distance between neural folds from 10 embryos at the indicated stages (average ± SEM). ***, P < 0.001 using the t test (two-tailed, unpaired). (D) Quantification of neural plate bending defects in embryos unilaterally injected with xDAPLE MOs and/or xDAPLE mRNA. n = 50–100 embryos per condition analyzed at stage 17. ***, P < 0.001 using the χ2 test. Validation of xDAPLE MOs is shown in Fig. S2. All scale bars represent 250 µm, except the one in the transversal section in B, which represents 50 µm.
Neural plate bending in Xenopus relies heavily on apical cell constriction and coincides spatiotemporally with the induction of xDAPLE expression. For these reasons, we focused our next efforts in validating the specificity (Blum et al., 2015) of the neural plate bending phenotypes as a consequence of loss of xDAPLE function. First, we found that bending defects were independently reproduced by two nonoverlapping xDAPLE MOs when injected unilaterally in embryos (Figs. 3 D and S2 A). Moreover, both MOs had synergistic effects (i.e., low MO doses that do not cause neural plate bending defects when injected individually do so when coinjected), suggesting specific action on a common target (Fig. 3 D). Finally, the MO-induced defects could be rescued by coinjection of xDAPLE mRNA (Fig. 3 D). These results confirm that loss of xDAPLE specifically impairs the bending of the neural plate.
Loss of DAPLE causes neural abnormalities in zebrafish embryos
To further substantiate the relatively mild neural phenotypes observed in Xenopus upon loss of DAPLE, we turned to zebrafish as a complementary model that is experimentally tractable for genetic editing by CRISPR-Cas9. Although formation of the NT in zebrafish differs from Xenopus (and mammals) in that it occurs from an intermediate closed structure (neural keel) rather than from a flat tissue (neural plate), some key tissue remodeling events are shared among these species (Lowery and Sive, 2004, 2009). In essence, formation of the hollow structure of the anterior NT relies on the stereotypical appearance of hinge points upon constriction of an epithelial tissue in zebrafish, as in other vertebrates like Xenopus (Lowery and Sive, 2004). mRNA in situ hybridization with two independent probes revealed that zebrafish DAPLE (zDAPLE) expression is undetectable at the gastrula stage (7.5 h postfertilization [hpf]) but readily observable during neurulation (16 and 22 hpf; Fig. S3, A and B). zDAPLE expression is spatially restricted to the neural rod (16 hpf) and the NT (22 and 28 hpf; Fig. S3, A and B). Thus, the spatiotemporal pattern of expression of DAPLE during zebrafish development is similar to Xenopus.
Next, we generated two independent DAPLE loss-of-function lines by targeting either exon 8 or exon 15 using CRISPR-Cas9. For each target region, we used a pair of sgRNAs separated ∼70–100 nt and selected two alleles with frameshift-inducing deletions (Fig. S3 C). Loss of DAPLE impaired hinge point formation in the forebrain and midbrain cavities at 22 hpf with a frequency of ∼95% (Fig. S3 D). Because this precedes the hydraulic inflation of ventricles, the observed abnormalities are directly related to the stereotypical formation of hinge points that occurs at this stage (Lowery and Sive, 2009). No overall developmental delay was observed based on somite count at 22 hpf (Fig. S3 E), and the only other difference noticed in these DAPLE loss-of-function zebrafish lines was a moderate curvature of the dorsal axis in ∼35% of the embryos at 28 hpf (Fig. S3 F), which disappeared at later stages. The impact of DAPLE loss of function on neural development was further substantiated by the analysis of embryos at later stages (56 hpf), which revealed mild abnormalities in the midbrain and forebrain ventricular cavities (Fig. S3 D). We conclude that expression of DAPLE during early development is required for proper neurodevelopment in two different vertebrate model animals, zebrafish and Xenopus.
Loss of DAPLE impairs apical cell constriction during neurulation
To characterize the tissue remodeling events that underlie the neurodevelopmental defects observed upon loss of DAPLE, we turned back to Xenopus, as these events have been extensively characterized in this experimental system. Because ectopic expression of DAPLE in cultured epithelial cells induces apical cell constriction (Figs. 1 and 2), we reasoned that the induction of xDAPLE expression during neurulation (Fig. 3 A) might promote apical constriction of neuroepithelial cells. Initial experiments ruled out other possible causes for the observed defects, such as neural differentiation defects or gross morphological abnormalities (Fig. S4). More specifically, xDAPLE MO-injected neural plates displayed normal expression of the neuroepithelial marker Sox3 (Fig. S4 A), and neural genes (N-tubulin, NCAM, and Zic3) were normally induced in xDAPLE morphants (Fig. S4 B). In Xenopus, stereotypical neural plate bending occurs simultaneously with anteroposterior elongation of the embryo, although both processes are not necessarily interdependent (Haigo et al., 2003). While neural plate bending was defective in xDAPLE morphants (Fig. 3), elongation remained normal during neurulation, and only a mild axis elongation defect was observed at later stages (Fig. S4 C). Thus, no major differentiation or elongation defects during neurulation seem to explain the impaired neural plate bending observed in DAPLE morphants.
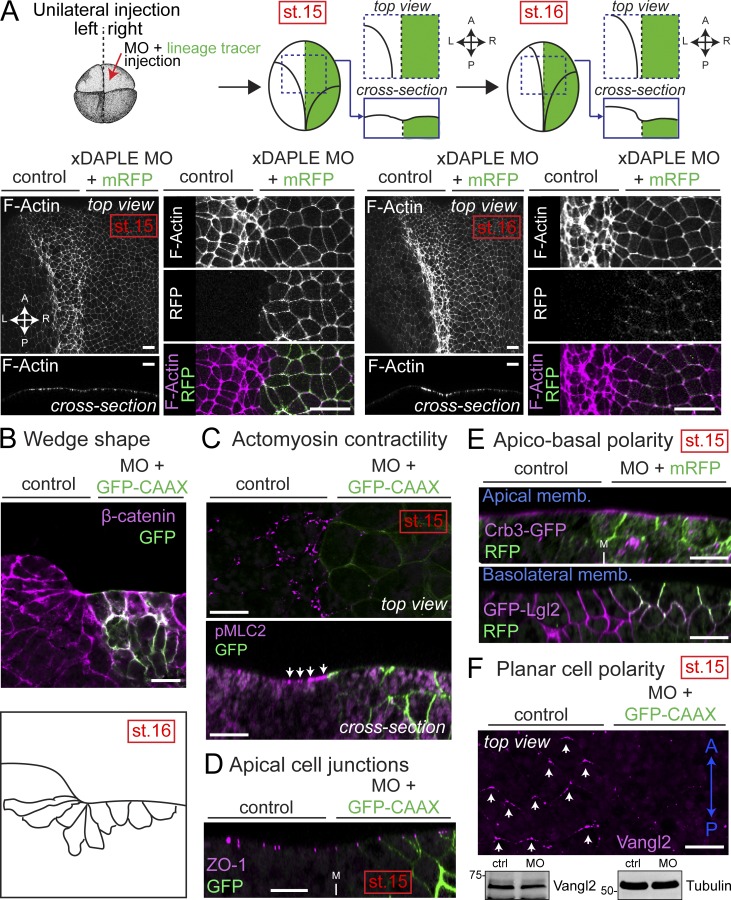
Next, we set out to characterize the cytoarchitectural features underlying the abnormal bending of the neuroepithelium (Fig. 4). We stained F-actin in embryos unilaterally injected with xDAPLE MO at stage 15 (the onset of the neural plate bending) and at stage 16 (when the neural plate bending is more pronounced; Fig. 4 A). xDAPLE MO-injected sides (identified by coexpression of a lineage tracer) displayed neuroepithelial cells with enlarged apical surfaces compared with control sides at both stages (Fig. 4 A), suggesting an apical constriction defect. Analysis of transversal sections of the neural plate revealed that cells of the outer layer of the neuroepithelium failed to acquire the characteristic wedge shape that accompanies apical cell constriction (Fig. 4 B). Staining with phospho(p)MLC2, a marker of actomyosin contractility, was lost in the apical cortex of DAPLE-depleted neuroepithelial cells (Fig. 4 C), thereby confirming that loss of DAPLE causes an apical constriction defect. Markers of apical cell junctions (ZO-1; Fig. 4 D), apical membranes (Crb3; Fig. 4 E), or basolateral membranes (Lgl2; Fig. 4 E) were unaltered in xDAPLE MO-injected sides, suggesting that apical constriction defects upon loss of DAPLE are not secondary to overt disruption of epithelial junction integrity and/or apicobasal polarity. On the other hand, we observed that DAPLE depletion disrupted the localization of the core planar cell polarity (PCP) protein Vangl2 (Fig. 4 F). As previously reported (Ossipova et al., 2015), we found that endogenous Vangl2 localizes preferentially to the anterior edge of the neuroepithelial cell cortex in controls, while xDAPLE depletion disrupts this localization without affecting total Vangl2 expression in the neural plate (Fig. 4 F). This is consistent with the observed apical constriction defects, as it has been reported that localization of PCP proteins correlates tightly with actomyosin-driven contraction (Butler and Wallingford, 2018) and that disruption of actomyosin contractility in the neural plate causes a loss of Vangl2 planar polarization (Ossipova et al., 2015). Taken together, these results indicate that loss of xDAPLE impairs the apical constriction of neuroepithelial cells during neurulation.
Figure 4.
Loss of DAPLE in Xenopus causes apical constriction defects during neurulation. (A) Whole-mount F-actin staining (magenta) of Xenopus embryos unilaterally coinjected with xDAPLE MO and a lineage tracer (mRFP or GFP-CAAX, green) showing enlarged apical surface of DAPLE-depleted neuroepithelial cells compared with uninjected control sides at stages 15 and 16. (B) Transversal cryosection stained for β-catenin (magenta) of the anterior neural plate of an embryo at stage 16 unilaterally coinjected with xDAPLE MO and a lineage tracer (GFP-CAAX, green). Outlines of cell borders are depicted in the bottom to show the lack of wedge shape morphology in the outer layer of neuroepithelial cells depleted of DAPLE. (C–F) Whole-mount pMLC2 (C), ZO-1 (D), GFP (E), and Vangl2 (F) staining (magenta) of Xenopus embryos unilaterally coinjected with xDAPLE MO and a lineage tracer (GFP-CAAX or mRFP; green). In E, embryos were bilaterally injected with Crb3-GFP (top) or GFP-Lgl2 (bottom). In F, staining for the lineage tracer (mRFP) is not shown for clarity, and an immunoblot from dissected neural plates is shown in the bottom. xDAPLE depleted sides show defective staining for actomyosin contractility and PCP markers at stage 15, while markers of apical cell junctions or apicobasal polarity are not changed. All images presented in this figure are representative results of n ≥ 3 experiments. All scale bars represent 25 µm, except those in A, which represent 50 µm.
Xenopus DAPLE contains two putative GBA motifs
Since hDAPLE promotes apical constriction cell autonomously via its G protein regulatory activity (Figs. 1 and 2), our next goal was to determine if the G protein regulatory activity of DAPLE is also required for apical cell constriction and neural plate bending in Xenopus embryos. However, the G protein regulatory activity of xDAPLE has not been established, and close examination of its protein sequence revealed that the presence of a functional GBA motif was not self-evident (Fig. 5 A). More specifically, we found that xDAPLE presents an imperfect sequence duplication of its GBA motif, resulting in two GBA-like sequences in tandem (GBA1 and GBA2; Fig. 5 A). Although both GBA-like sequences have high similarity to the consensus sequence ψ-T-ψ-x-E-F-ψ found in previously described GBA motifs (Fig. 5 A), the possible interference of flanking sequences with G protein coupling raised a concern about which one, if any, is a functional G protein regulatory motif.
Figure 5.
xDAPLE promotes G protein signaling and apical cell constriction via two GBA motifs in tandem. (A) Comparison of domain composition of hDAPLE and xDAPLE, and alignment of xDAPLE GBA1 and GBA2 with other GBA motifs. ce, C. elegans; h, Homo sapiens. (B) Full-length xDAPLE from HEK293T cell lysates binds to immobilized GST-Gαi3 when the G protein is loaded with GDP (inactive), but not when it is loaded with GDP-AlF4−. (C) Steady-state GTPase (black) and GTPγS binding (red) experiments showing that purified His-xDAPLE-CT accelerates nucleotide exchange of purified His-Gαi3. Results are the average ± SEM of n = 3 experiments. (D) xDAPLE-CT and hDAPLE-CT activate G protein signaling in a yeast-based β-galactosidase reporter assay. Diagram on the left depicts the pathway activated by DAPLE in yeast lacking the cognate GPCR and with the endogenous G protein replaced by human Gαi3. Results on the right are the average ± SEM of n = 3 experiments. (E) Protein–protein binding experiment showing that purified His-Gαi3 binds to both GBA1 and GBA2 of xDAPLE. Diagram on the top shows a detail of the sequence of the purified GST-fused xDAPLE constructs used in the experiment and the position of FA point mutations (in red). (F) G protein activity assays in yeast (as in D) show that both GBA1 and GBA2 have to be mutated simultaneously to abolish xDAPLE-mediated activation. Results are the average ± SEM of n = 5 experiments. (G) Steady-state GTPase experiments showing that activation of purified His-Gαi3 by GST-xDAPLE FA1+2 (red) is impaired compared GST-xDAPLE WT (black). Results are the average ± SEM of n = 3 experiments. Basal activity = 0.16 mol Pi/mol Gαi3/15 min. (H) Coimmunoprecipitation (IP) experiments showing that xDAPLE WT, but not xDAPLE FA1+2 mutant, binds to Gαi3-FLAG when expressed in HEK293T cells. Immunoblots (IB) of the FLAG IPs are shown on the top and equal aliquots of the starting lysates used for it are shown on the bottom. (I) Box-and-whiskers plots (minimum to maximum) for the quantification of relative apical area show xDAPLE ΔPBM and xDAPLE GBA** (FA1+2) mutants fail to promote apical constriction in MDCK cells compared with xDAPLE WT. Results are from n = 4–9 independent experiments. *, P < 0.05; **, P < 0.01 using the Mann–Whitney U test. (J) Fluorescence microscopy pictures of MDCK cells sparsely expressing the indicated MYC-xDAPLE constructs and costained for MYC (magenta) and ZO-1 (green) show that xDAPLE GBA** mutant is enriched at apical junctions like WT, while xDAPLE ΔPBM mutant is not. Scale bars, 10 µm.
xDAPLE is a bona fide GEF for Gαi
First, we examined if xDAPLE had the biochemical properties previously described for other GEFs of the GBA family. The binding site for the GBA motif is fully conserved in Gαi1, Gαi2, and Gαi3 in both mammals and Xenopus (Garcia-Marcos et al., 2010; Fuentealba et al., 2016; de Opakua et al., 2017). We found that full-length xDAPLE binds to inactive, GDP-loaded rat Gαi3, but not to active, GDP-AlF4−(a GTP mimetic)–loaded Gαi3 (Fig. 5 B), which is a property of all previously described GBA proteins (DiGiacomo et al., 2018; Maziarz et al., 2018) as well as other GEFs (Tall et al., 2003). To determine if xDAPLE has GEF activity, we performed two well-established enzymatic assays: the steady-state GTPase assay, which monitors the rate of nucleotide exchange indirectly, and the GTPγS-binding assay, which measures directly nucleotide exchange rate. We found that submicromolar to micromolar concentrations of purified xDAPLE-CT (C-terminal aa 1,638–1,932, which contains the GBA motifs) increased the steady-state GTPase activity of purified Gαi3 in a dose-dependent manner by approximately threefold compared with basal activity (Fig. 5 C). Very similar results were obtained with the GTPγS-binding assay (Fig. 5 C), which confirms that xDAPLE accelerates the rate of nucleotide exchange. Thus, xDAPLE has GEF activity in vitro.
To assess if xDAPLE also activates G protein signaling in a cellular context, we used a yeast-based assay previously implemented with the same purpose for assessing the activity of the endogenous yeast nonreceptor GEF Arr4/Get3 (Lee and Dohlman, 2008) or of ectopically expressed hDAPLE and other GBA proteins (Coleman et al., 2016; de Opakua et al., 2017; Maziarz et al., 2018). Briefly, we used a “humanized” Saccharomyces cerevisiae strain (Cismowski et al., 1999) that is genetically engineered to lack pheromone-sensitive GPCRs and replace the endogenous yeast Gα protein Gpa1 by human Gαi3 (Fig. 5 D). Thus, in this strain only an exogenous G protein activator can trigger a signaling pathway that is normally activated as a pheromone response and leads to activation of the FUS1 promoter. Expression of xDAPLE-CT led to G protein activation in cells, as determined by the activity of a β-galactosidase reporter of the FUS1 promoter (Fig. 5 D). The level of activation was at least as high as that observed for an analogous construct derived from hDAPLE (Fig. 5 D). In summary, xDAPLE has GEF activity in vitro and activates G protein signaling in cells.
Both the GBA1 and GBA2 motifs of xDAPLE bind and activate Gi proteins
Next, we set out to dissect whether only one or both of the putative GBA motifs (Fig. 5 A) were functional G protein regulatory sequences. First, we generated GST-fused constructs consisting of the GBA1 motif, the GBA2 motif, or both GBA motifs together to test their direct binding to G proteins (Fig. 5 E). We found that purified Gαi3 binds to GBA1 and to GBA2 separately, as well as to the construct containing both GBA motifs together (Fig. 5 E). Mutation of a conserved phenylalanine (F), previously reported to be essential for binding of other GBA motifs to G proteins, to alanine (FA) also impaired binding of both GBA motifs of xDAPLE to Gαi3 (Fig. 5 E). Next, we investigated if the loss of binding caused by these mutants translated into diminished activation of G protein signaling using the yeast assay described above. We found that single FA mutations (FA1 and FA2, corresponding to GBA1 and GBA2, respectively) lead to a partial reduction of xDAPLE-mediated signaling activation, whereas the double mutant (FA1+2) almost completely suppressed the response (Fig. 5 F). These findings with xDAPLE closely resembled those obtained with hDAPLE upon introduction of the FA mutation in its single GBA motif, which also caused an almost complete loss of function (Fig. 5 E). Similarly, steady-state GTPase assays with purified proteins confirmed that the double FA1+2 mutation also disrupts the GEF activity of xDAPLE (Fig. 5 G), which is analogous to previous results comparing hDAPLE WT and FA mutant (Aznar et al., 2015). We conclude that both GBA1 and GBA2 are required for xDAPLE-mediated G protein signaling and that simultaneous disruption of both motifs is required to abolish the G protein regulatory activity of xDAPLE.
xDAPLE promotes apical cell constriction via its G protein regulatory function
To validate the double FA1+2 mutation as a tool to disrupt the G protein regulatory activity of xDAPLE, we investigated if it also impaired G protein binding when introduced in the context of the full-length protein. We found that this is the case because xDAPLE FA1+2 was undetectable in coimmunoprecipitation experiments with Gαi3 in mammalian cells (Fig. 5 H). As an additional benchmark for this mutant, we tested whether it impaired xDAPLE-induced apical constriction. We found that, much like hDAPLE, expression of full-length xDAPLE WT in MDCK cells caused apical cell constriction and that this effect was not recapitulated by xDAPLE FA1+2 (which we henceforth renamed GBA**; Fig. 5 I). Also similar to the observations with hDAPLE, xDAPLE GBA** was enriched at apical cell junctions (Fig. 5 J), whereas xDAPLE ΔPBM failed to induce apical cell constriction and localized diffusely throughout the cytoplasm (Fig. 5, I and J). Taken together, these results show that the G protein regulatory function of xDAPLE is required for apical constriction in cultured epithelial cells.
xDAPLE-mediated G protein regulation is required for apical cell constriction during neurulation
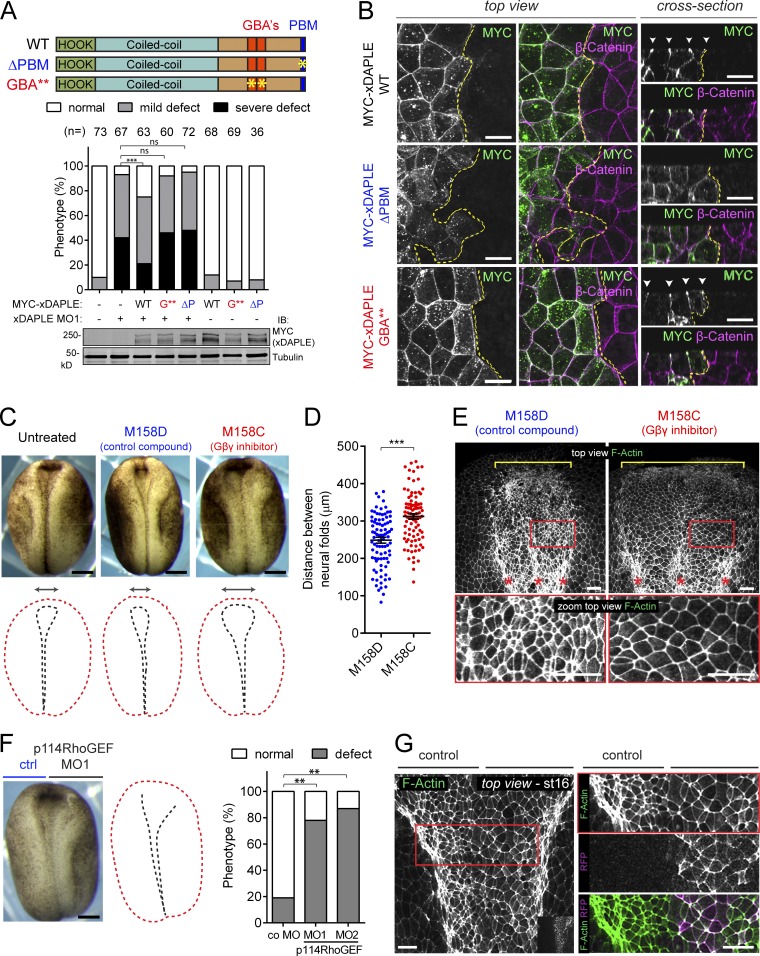
The results above confirm that the GBA** mutant disrupts the functions of xDAPLE that rely on its G protein regulatory activity, including apical cell constriction. Next, we implemented this mutant to investigate the impact of G protein activation by xDAPLE in vivo during neurulation in Xenopus. For this, we compared the ability of xDAPLE WT or GBA** to rescue neural plate bending defects in xDAPLE morphants, which are related to impaired apical constriction (Figs. 3 and 4). In parallel, we also investigated the effect of xDAPLE ΔPBM to probe further into the mechanism by which DAPLE regulates apical cell constriction. We found that in contrast to xDAPLE WT, both xDAPLE ΔPBM and xDAPLE GBA** failed to rescue MO-induced neural plate bending defects (Fig. 6 A). On the other hand, the two mutants behaved differently in terms of subcellular localization in neuroepithelial cells. While xDAPLE GBA** was enriched at apical cell junctions like xDAPLE WT, the ΔPBM mutant was not (Fig. 6 B). These findings are reminiscent of the results with analogous DAPLE mutants in apical constriction experiments with cultured epithelial cells (Figs. 2 and 5), suggesting that the PBM and GBA motifs of DAPLE have equivalent functions in apical constriction in vitro and in vivo. More specifically, while the PBM is required for proper neural plate bending by ensuring adequate subcellular localization, the GBA motifs are dispensable for localization but facilitate neural plate bending by activating a G protein–dependent effector pathway.
Figure 6.
Activation of G protein signaling by xDAPLE is required for apical cell constriction during neurulation. (A) Quantification of neural plate bending defects in embryos unilaterally injected with xDAPLE MO and/or xDAPLE mRNAs as in Fig. 3 D shows that xDAPLE WT, but neither ΔPBM (ΔP) nor GBA** (G**), rescues neural plate bending defects upon xDAPLE depletion. The number (n) of embryos per condition in indicated above the bars. ***, P < 0.001 or not significant (ns) using the χ2 test. (B) Confocal fluorescence microscopy pictures of the neural plate of Xenopus embryos unilaterally injected with MYC-xDAPLE mRNA and costained for MYC (green) and β-catenin (magenta). The left and middle panels correspond to a view on the neuroepithelium from the top, and panels on the right are optical cross sections. The yellow dotted line separates the injected from the uninjected side of the neuroepithelium. Scale bars, 20 µm. (C) Morphology of Xenopus embryos at stage 16 after treatment with the Gβγ inhibitor M158C or its inactive analogue, M158D. Scale bars, 250 µm. (D) Scatterplot for the quantification of the distance between neural folds from 90 embryos treated with M158D or M158C (average ± SEM). ***, P < 0.001 using the t test (two-tailed, unpaired). (E) Whole-mount F-actin staining of Xenopus embryos at stage 17 after treatment with the Gβγ inhibitor M158C or its inactive analogue, M158D. Red boxes in the top panels are magnified in the bottom panels to show the enlarged area of the neuroepithelial cells treated with M158D compared with M158C. Scale bars, 50 µm. (F) Quantification of neural plate bending defects in embryos unilaterally injected with p114RhoGEF MO1 (splicing-interfering, validated in Fig. S5) and p114RhoGEF MO2 (translation blocking). n = 80 embryos analyzed at stage 17. ***, P < 0.001 using the Fisher exact test. Scale bars, 250 µm. (G) Whole-mount F-actin staining (green) of Xenopus embryos unilaterally coinjected with p114RhoGEF MO1 and a lineage tracer (GFP-CAAX, magenta). The red box in the left panel is magnified in the panels on the right to show the enlarged area of the neuroepithelial cells depleted on p114RhoGEF compared with the control sides. Scale bars, 50 µm. All images presented in this figure are representative results of n ≥ 3 experiments.
Efficient neural plate bending requires Gβγ signaling and p114RhoGEF
We reasoned that if DAPLE-induced apical constriction occurs via Gβγ and the Gβγ-activated RhoGEF p114RhoGEF in cultured cells (Fig. 2), neural plate bending in vivo would rely on a similar mechanism. To investigate this, we first tested the effect of the Gβγ inhibitor M158C, as well as its inactive analogue M158D (Bonacci et al., 2006), on Xenopus neurulation. Embryos treated with M158C, but not those treated with M158D, phenocopied loss of xDAPLE (Fig. 6 C), i.e., they displayed neural plate bending defects, as manifested by lager distances between the anterior neural folds (Fig. 6, C and D) and enlarged neuroepithelial apical surfaces (Fig. 6 E) compared with controls. Unilateral depletion of p114RhoGEF using a splicing-interfering MO (MO1; validated in Fig. S5), also resulted in neural plate bending defects (Fig. 6, F and G), which were reproduced by a second MO (MO2, translation blocking; Fig. 6 F). Neuroepithelial cells injected with p114RhoGEF MO also displayed enlarged apical surfaces (Fig. 6 G). In conclusion, disruption of Gβγ-dependent signaling or depletion of p114RhoGEF mimics the apical constriction defects observed in the neural plate upon loss of DAPLE.
Discussion
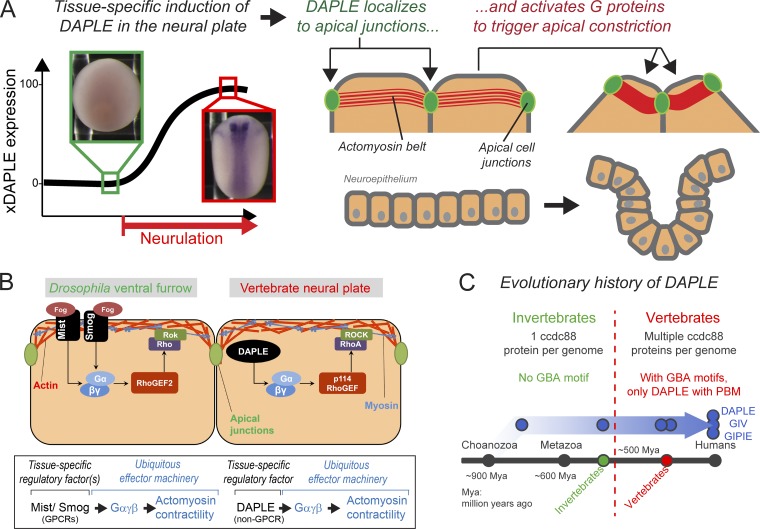
The main finding of this work is the identification of DAPLE as a tissue-specific factor that activates G protein signaling to control apical cell constriction in vivo during embryonic development (Fig. 7 A). There are two significant implications for this finding on our understanding of the fundamental mechanisms that govern tissue remodeling during development. The first one is that although examples of tissue-specific G protein activators that regulate apical cell constriction have been reported in morphogenetic processes of invertebrates, their corresponding counterparts in vertebrates had remained elusive until now (Fig. 7 B). The second important implication relates to the nature of the tissue-specific G protein regulator per se, which is a cytoplasmic factor rather than a receptor of the GPCR superfamily.
Figure 7.
Mechanism of G protein–mediated regulation of apical cell constriction during neurulation by the non-GPCR protein DAPLE. (A) Expression of DAPLE is specifically induced during neurulation. Upon expression, DAPLE localizes to apical cell junctions of neuroepithelial cells, where it triggers G protein activation that leads to apical cell constriction and the subsequent bending of the neural plate. (B) Theme and variations of G protein–regulated apical cell constriction during epithelial tissue morphogenesis in vertebrates versus invertebrates. Heterotrimeric G proteins are part of a conserved ubiquitous machinery that controls actomyosin contractility, but they are regulated differently across species. In vertebrates, DAPLE fulfills the role performed by GPCRs in invertebrates as tissue-specific activators of signaling that drives apical cell constriction. (C) The G protein regulatory function of DAPLE (i.e., its GBA motif) was acquired during evolution in the transition from invertebrates to vertebrates, suggesting that the unconventional mechanism of G protein activation described here is an evolutionary innovation for epithelial remodeling in vertebrates.
GPCRs have been the focus of extensive research for decades, but much less is known about cytoplasmic factors that work as alternative activation inputs for heterotrimeric G proteins. One challenge in establishing the physiological relevance of such factors is the difficulty in relating in vitro biochemical features, like having GEF activity, to specific functions in vivo. However, DAPLE belongs to a group of cytoplasmic activators of heterotrimeric G proteins that is more tractable in this regard, as they rely on a structurally characterized (de Opakua et al., 2017; DiGiacomo et al., 2018) stretch of ∼30 aa, the GBA motif, to activate G protein signaling. By leveraging this feature, here we provide for the first time direct evidence for the physiological relevance of an unconventional mechanism of G protein activation. Specific disruption of the GBA motif of DAPLE reveals that apical constriction of neuroepithelial cells relies on non–GPCR-mediated G protein activation in vivo for proper neural plate bending during neurulation.
Heterotrimeric G proteins are ubiquitously expressed during embryonic development in animal species, including Gαi subunits in Xenopus (Peshkin et al., 2015; Fuentealba et al., 2016; Zhang et al., 2018). Thus, their functional consequences in development are greatly determined by factors that control their activity in a temporally and spatially defined manner. Our findings suggest a variation on a common theme in the G protein signaling pathways that control apical constriction during embryogenesis across evolutionarily distant organisms (Fig. 7 B). Formation of the ventral furrow in Drosophila and formation of the NT in vertebrates are two paradigms of tissue morphogenesis that rely on apical cell constriction. In both systems, there is an analogous effector machinery composed of ubiquitous proteins that directly regulate the actin cytoskeleton; i.e., in flies, Gαβγ signaling directly activates Rho/Rok-mediated actomyosin contractility via RhoGEF2 (Manning and Rogers, 2014; Gilmour et al., 2017), whereas in frogs, Gαβγ signaling directly activates RhoA/ROCK-mediated actomyosin contractility via p114RhoGEF. A significant variation on the theme is the nature of the upstream tissue-specific factors that ensure precise spatiotemporal control of the morphogenetic process. In flies, this is performed by the GPCR Mist (Manning et al., 2013), in coordination with another ubiquitous GPCR, Smog (Kerridge et al., 2016), whereas in vertebrates, it is performed by DAPLE, a non-GPCR protein (Fig. 7 B). Interestingly, the acquisition of a GBA motif by DAPLE seems to be an evolutionary innovation of vertebrates (Fig. 7 C). DAPLE has two paralogs in most vertebrate species (GIV [Gα-Interacting Vesicle-associated protein] and GIPIE [GRP78-Interacting Protein Induced by ER stress]), which together form the ccdc88 family. In contrast, there is only one ccdc88 orthologue in invertebrates, which lacks the GBA motif (Coleman et al., 2016). It was only upon the expansion of the ccdc88 family in vertebrates that two of the members (GIV and DAPLE) acquired the ability to regulate G proteins via GBA motif (Coleman et al., 2016). Of this two, only DAPLE has been reported to possess a PBM, the other domain required for its function in apical cell constriction. It is tempting to speculate that the acquisition of the GBA motif in vertebrates might have occurred as an evolutionary adaptation to make cytoplasmic factors like DAPLE operate in lieu of certain morphogenetic roles of GPCRs found in invertebrates. However, it is likely that GPCRs work as tissue-specific factors in other morphogenetic processes of vertebrates. For example, it has been shown that loss of PAR1 and PAR2 causes NT closure defects in mice (Camerer et al., 2010), which appears to relate to a specific role in neural fold fusion. Also, we cannot rule out that some GPCRs might be involved, maybe by cooperation with DAPLE, in apical cell constriction during neurulation, but they still remain to be identified.
The G protein regulatory activity of DAPLE seems to be primarily controlled by its level of expression and subcellular localization. This is consistent with previous findings showing that physical proximity between GBA GEFs and their G protein substrates is a major driver for signaling activation. Gi family proteins are constitutively anchored to cell membranes, and the relocalization of DAPLE and other GBA proteins to cell membranes, as observed in response to different stimuli (Ghosh et al., 2010; Aznar et al., 2015; Leyme et al., 2015), is sufficient to trigger G protein activation similar in magnitude to that exerted by GPCRs (Parag-Sharma et al., 2016; Maziarz et al., 2018). Although the localization of DAPLE at cell–cell junctions via its PBM motif might be sufficient to constitutively activate G proteins in that location, it is possible that its activity can be further modulated by other mechanisms such as posttranslational modifications or in response to external cues. For example, it has been shown that DAPLE translocates to the cell cortex and mediates G protein activation in response to Wnt ligands in cultured cells (Aznar et al., 2015). Another important point that requires further investigation is the identification of the factors responsible for targeting DAPLE to apical cell junctions via PBM binding.
Understanding the formation of the NT is of great biomedical interest due to its direct linkage to congenital malformations in humans (Nikolopoulou et al., 2017). Our findings show that loss of DAPLE causes mild overall defects in neurulation, which do not result in open NT defects. This suggests that DAPLE may cooperate with other redundant and/or compensatory mechanisms during NT morphogenesis. Components of the core PCP machinery are critical drivers of NT closure (Nishimura et al., 2012; Sokol, 2015). While global disruption of PCP causes open NT defects (Wallingford and Harland, 2002), loss of individual PCP components in Xenopus, such as Prickle2, results in mild neurulation defects strikingly similar to those observed upon loss of DAPLE (Butler and Wallingford, 2018). Here, we show that loss of DAPLE disrupts the planar polarized distribution of the PCP protein Vangl2 in the neural plate, suggesting a functional interaction between PCP and DAPLE in this context. Moreover, recent evidence indicates that core PCP proteins also require DAPLE for proper planar polarized distribution in other tissues (Siletti et al., 2017; Takagishi et al., 2017). Interestingly, DAPLE was originally discovered based on its ability to bind Disheveled (Dvl), a core component of the PCP machinery, via its PBM (Oshita et al., 2003). However, contrary to Dvl, DAPLE localization is not planar polarized in the neural plate, and a dominant-negative Dvl (xdd1) that disrupts PCP and NT closure (Wallingford and Harland, 2002) does not affect the subcellular localization of DAPLE in the neural plate of Xenopus (not shown). It is possible that the impact of DAPLE on Vangl2 localization is not due to a direct physical interaction with PCP components and is a secondary consequence of defects in apical cell constriction upon loss of DAPLE, as it has been recently shown that localization of PCP core proteins during neural plate closure might rely on contractility (Ossipova et al., 2015; Sokol, 2016; Butler and Wallingford, 2018). Further investigation will be needed to clarify the possible relationship between DAPLE and PCP during neurulation.
Materials and methods
Plasmids and MOs
The plasmid used for the expression of full-length MYC-xDAPLE and for the generation of xDAPLE mRNA in situ hybridization probes (pCS2-6XMYC-xDAPLE), as well the plasmid for expression of xDAPLE (aa 1–1,293) mRNA preceded by its 5′ UTR (pCS2-5′UTR-1293-MYC) was kindly provided by T. Michiue (University of Tokyo, Tokyo, Japan; Kobayashi et al., 2005). The plasmid used to generate Cas9 mRNA for zebrafish embryo injections (pT3TS-nCas9n) was from Addgene (46757; Jao et al., 2013). Plasmids for the expression of GFP-CAAX (pCS2-eGFP-CAAX) and membrane RFP (mRFP; pCS2-mRFP) in Xenopus were a gift from S. Sokol (Icahn School of Medicine at Mount Sinai, New York, NY; Ossipova et al., 2015), and those for the expression of Crb3-GFP (pCS2-Crb3-GFP) and GFP-Lgl2 (pCS2-GFP-Lgl2) were kindly provided by C. Wylie and S.W. Cha (Cincinnati Children’s Hospital Medical Center, Cincinnati, OH; Wang et al., 2013). The plasmid for expression of MYC-hDAPLE (pCS2-6XMYC-hDAPLE) was generated by inserting the hDAPLE sequence amplified by PCR into the EcoRI site of pCS2-6XMYC. The following mutants of DAPLE were generated by site directed mutagenesis (QuickChange II; Agilent) in pCS2-6XMYC-hDAPLE or pCS2-6XMYC-xDAPLE. All the constructs were confirmed by Sanger DNA sequencing. hDAPLE H1 (Hydrocephalus mutant 1) was created by introducing a sequence (G1590-HSQLPG*) that recapitulates that generated by the natural mutation p.S1591HfsX7 mutation reported by Ekici et al. (2010), and hDAPLE H2 (Hydrocephalus mutant 2) was created by introducing a sequence (G1948-GGHHHPCPGRAQPLRGRRGPGAGLQ*) that recapitulates that generated by the natural mutation p.E1949GfsX26 reported by Drielsma et al. (2012). ΔPBM mutants of both hDAPLE and xDAPLE were generated by mutating the tyrosine codon located right before the PBM (i.e., deleting the last three amino acids, GCV) into a stop codon. GBA motif mutants were created by mutating F1675 to alanine in hDAPLE (FA or GBA* mutant) or by mutating F1662 to alanine (FA1) or F1689 to alanine (FA2) in the GBA1 or GBA2 motif of xDAPLE, respectively (FA1+2 or GBA** denotes a double mutant bearing both mutations simultaneously). Plasmids for expression of GST-Gαi3 (pGEX-KG-Gαi3) or His-Gαi3 in bacteria (pET28b-Gαi3) and for expression of Gαi3-FLAG in mammalian cells (p3xFLAG-CMV14-Gαi3) have been previously described (Garcia-Marcos et al., 2010). Plasmids for the expression of His-xDAPLE-CT (pLIC-His-xDAPLE-CT) in bacteria or xDAPLE in yeast (pLIC-YES2-xDAPLE-CT) were generated by ligation independent cloning (Stols et al., 2002) of a PCR amplified fragment of xDAPLE-CT (aa 1,638–1,932) into pLIC-His (also known as pMCSG7; Stols et al., 2002) or pLIC-YES2 (Coleman et al., 2016), respectively. Plasmids for bacterial production of GST-GBA1 (xDAPLE aa 1,648–1,677), GST-GBA2 (xDAPLE aa 1,675–1,704), or GST-GBA1+GBA2 (xDAPLE aa 1,648–1,704) were generated by ligation independent cloning of PCR amplified fragments into a previously described plasmid (pLIC-GST; Cabrita et al., 2006; kindly provided by J. Sondek, University of North Carolina at Chapel Hill, Chapel Hill, NC). The pLIC-YES2-hDAPLE (aa 1,650–1,880) plasmid used for expressing hDAPLE in yeast was described previously (de Opakua et al., 2017).
The following MOs were purchased from Gene Tools: xDAPLE MO (also known as xDAPLE MO1) targeting xDAPLE (transcription start site [TSS] −1/+24 nt: 5′-CTGTTGGGAAATGGTAGTATCCATG-3′; Kobayashi et al., 2005), xDAPLE MO2 (TSS −87/−63 nt: 5′-AGACCCTGAATGGGCTTTAGAAGCT-3′), p114RhoGEF MO1 targeting the p114RhoGEF exon 1–intron 1 boundary (5′-AATTATATTACTTACCTAGCTCTGC-3′), p114RhoGEF MO2 (TSS −2/+19 nt: 5′-CTCTATTGGTGACAGTCATTC-3′), and control MO (5′-CCTCTTACCTCAGTTACAATTTATA-3′).
Cell culture, treatments, and transfections
MDCK (CRL2936; ATCC), HEK293T (CRL3216; ATCC), and EpH4 (from X. Varelas, Boston University, Boston, MA) were grown at 37°C, 5%CO2 in DMEM supplemented with 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, and 1% L-glutamine. MDCK or EpH4 cells were seeded on glass coverslips placed in 24-well plates at a density of 250,000 cells per well. Next day, cells were transfected with 1 µg of plasmid DNA (encoding MYC-hDAPLE or MYC-xDAPLE) with Turbofect (R0531; Thermo Fisher Scientific) for MDCK or Lipofectamine LTX (15338100; Life Technologies) for EpH4 following the manufacturer’s instructions. 30 h after transfection, confluent cell monolayers were processed for immunofluorescence staining as described in Immunofluorescence analysis of cultured cells. For pharmacological treatments (Fig. 2 D), MDCK cells were transfected with MYC-hDAPLE WT exactly as described above and treated with compounds as described for the next 30 h after transfection. Cells were treated for 90 min with 50 µM Blebbistatin (sc-204253; Santa Cruz) or an equivalent volume of DMSO (control), for 30 min with 10 µM Y-27632 (1254; Tocris) or an equivalent volume of DMSO (control), and for 30 min with 10 µM Gallein (A0601; Tokyo Chemical Industry) or 10 µM Fluorescein (control, F0095; Tokyo Chemical Industry).
For the experiments testing the effect of p114RhoGEF depletion on DAPLE-mediated apical constriction (Fig. 2 D), cells were first transfected with siRNA for silencing p114RhoGEF expression and then transfected with MYC-hDAPLE as follows. On day 1, 500,000 MDCK cells were reverse transfected in a six-well plate with 20 nM of the custom-made stealth RNA (Life Technologies) using RNAiMAX reagent (13778030; Life Technologies) according to the manufacturer’s protocol. On day 3, 400,000 RNAi-transfected cells were reseeded on glass coverslips in 24-well plates and transfected 6 h later with Turbofect exactly as described above. On day 5, confluent cell monolayers were processed for immunofluorescence staining as described in Immunofluorescence analysis of cultured cells and quantification of apical cell constriction. Previously validated (Nakajima and Tanoue, 2011) stealth RNAi oligo sequences for p114RhoGEF (canine) were p114RhoGEF siRNA#1 5′-UGGCCACAAUGAGGCGGUCAAUCAU-3′ and p114RhoGEF siRNA#2 5′-GGCCAACGAGGAGAAAGCCAUGUUU-3′. The stealth RNAi negative control, medium GC, was also purchased from Life Technologies (12935300).
Immunofluorescence analysis of cultured cells and quantification of apical cell constriction
Cells grown on glass coverslips were fixed in 100% methanol at −20°C for 10 min, washed three times with PBS, and incubated in blocking solution (10% [vol/vol] normal goat serum and 0.1% [vol/vol] Triton X-100 in PBS) for 1 h. Cells were incubated overnight at 4°C in blocking solution with the following primary antibodies: rabbit polyclonal MYC (1:300; C3956; Sigma-Aldrich), rabbit polyclonal DAPLE (1:100; ABS515; Millipore), rabbit polyclonal DAPLE (1:100; HPA005832; Sigma-Aldrich), mouse monoclonal E-Cadherin (1:100; rr1; Developmental Studies Hybridoma Bank [DSHB]), mouse monoclonal MYC (1:200; 2276 [9B11]; Cell Signaling Technology), and rat monoclonal ZO-1 (1:100; R26.4c; DSHB). Coverslips were washed three times in PBS and incubated in secondary antibodies for 1 h at room temperature in blocking solution. The secondary antibodies used were goat anti-mouse Alexa Fluor 488 (1:400; A11017; Invitrogen) and goat anti-rabbit Alexa Fluor 594 (1:400; A11072; Invitrogen) and goat anti-rat Alexa Fluor 647 (1:400; A21247; Invitrogen) diluted in blocking solution. Coverslips were mounted in ProLong Diamond Antifade (P36965; Invitrogen) and imaged at room temperature by fluorescence microscopy.
For experiments to determine the colocalization of DAPLE with markers in the xyz planes (Fig. 1, A–C), a Zeiss LSM 700 microscope was used. Stacks of confocal pictures of 0.321 µm thickness along the z axis were taken with a 63× oil-immersion objective (NA 1.4, working distance 0.19 mm) using ZEN software. Images were processed for presentation using ImageJ software (National Institutes of Health), and individual images were assembled in Photoshop and Illustrator software (Adobe).
For experiments to assess apical constriction of cells (Figs. 1 E, 2 B, 5 J, and S1), a Zeiss Axio Observer Z1 fluorescence microscope equipped with a digital camera (C10600/ORCA-R2; Hamamatsu Photonics) was used. Images were taken with a 63× oil-immersion objective (NA 1.4, working distance 0.19 mm) using ZEN software. Cells were costained for MYC (MYC-DAPLE) and ZO-1 as described above. Random fields containing MYC-positive cells were selected and images of the apical cell domain acquired by bringing into focus the ZO-1 signals. Image analysis was done with ImageJ and the apical areas of MYC-DAPLE–expressing and neighboring cells (as determined by shared cell-cell apical boundaries) were determined by delineating the cell boundaries manually or by using the “magic wand” plugin when possible. The relative apical area was calculated by dividing the apical area of the MYC-positive cell by the average of the apical area of its neighboring cells (as represented in Fig. 1 D). MYC-positive cells in direct contact with another MYC-positive cell or overexpressing high levels of MYC-DAPLE in aggregate-like structures were excluded from the analysis. Results are presented as box-and-whiskers plots (minimum to maximum) of the averaged relative apical area across individual experiments. Statistical P values were calculated with the Mann–Whitney U test in Prism (GraphPad).
Xenopus embryo culture, microinjections, and morphological analysis
Frog studies were performed with WT Xenopus animals (Nasco) according to Boston University Institutional Animal Care and Use Committee–approved protocol AN14092, in compliance with the Guide for the Care and Use of Laboratory Animals. Egg laying was induced by dorsal lymph injection of 600 U of human chorionic gonadotrophin (028938; Merck). In vitro fertilization and embryo culture were performed in 0.1× Marc’s modified Ringer’s medium as previously described (Newport and Kirschner, 1982). Dejellied embryos from at least three independent frog fertilizations were used in all of the experiments described below.
mRNAs for injections in frog embryos were prepared using the SP6 mMessage mMachine Kit (AM1340; Ambion). MOs and mRNAs were injected dorsally the two- to four-cell stage to target the presumptive neuroectoderm in a single (unilateral injections) or both blastomeres (bilateral injections) as indicated in the figure legends and cultured at 16°C. For each mRNA and MO, an initial round of titration experiments was performed to determine the optimal dose to be injected. Quantities of MOs injected per blastomere were as follows: xDAPLE MO1, 20 ng (except in the rescue experiments in Figs. 3 D and 6 A, in which it was 10 ng); xDAPLE MO2, 40 ng; p114RhoGEF MO1, 20 ng; and p114RhoGEF MO2, 20 ng. The experiment to test the combinatorial effect of xDAPLE MOs at low dose in Fig. 3 D was performed with 0.8 ng xDAPLE MO1 and 10 ng xDAPLE MO2. Control MO was always injected at a dose that matched the highest amount of any other MO used in the same experiment. Unless otherwise indicated in the figures, the quantities of mRNA injected per blastomere were as follows: xDAPLE, 100 pg; mRFP, 50 pg; GFP-CAAX, 50 pg; GFP-Lgl2, 50 pg; and Crb3-GFP, 50 pg. For the experiments with pharmacological inhibitors of Gβγ (M158C and M158D, obtained from the National Cancer Institute Developmental Therapeutics Program, compound numbers 158110 and 158112, respectively), compounds (30 µM) were added to the media at stage 12 and maintained until fixation at stage 16.
Morphological analyses were performed on embryos manually devitellinized with blunt-ended fine forceps with a Leica MZ6 dissection microscope. Morphometric quantifications (distance between neural folds in Figs. 3 C and 6 D and length-to-width ratio in Fig. S4 C) were performed with ImageJ on pictures of fixed whole embryos positioned with the dorsal side toward the lens of the microscope and taken at the indicated stages. Quantification of neural plate bending defects (Fig. 3 D and Fig. 6, A and F) was performed at stage 17 on embryos fixed with MEMFA (100 mM MOPS, pH 7.4, 2 mM EGTA, 1 mM MgSO4, and 3.7% [vol/vol] formaldehyde). Embryos were scored as normal or defective by comparing MO-injected sides to uninjected control sides. All the pictures of whole Xenopus embryos were taken in PBS at room temperature with a Canon XSi camera connected to a Leica MZ6 dissection microscope.
For the analysis of protein expression in whole embryos by immunoblotting (Figs. S2 and 6 A), two embryos were resuspended in 60 µl lysis buffer (20 mM Hepes, pH 7.2, 5 mM Mg(CH3COO)2, 125 mM K(CH3COO), 0.4% Triton X-100, and 1 mM DTT supplemented with a protease inhibitor cocktail [S8830]; Sigma-Aldrich) and homogenized by pipetting. After centrifugation for 10 min at 14,000 ×g at 4°C, supernatants were supplemented with Laemmli sample buffer and boiled 5 min. Preparation of samples for Vangl2 immunoblotting Fig. 4 F was performed the same way except that the starting material was neural plates dissected from 20 embryos.
mRNA in situ hybridization of Xenopus embryos
mRNA in situ hybridization of Xenopus embryos was performed essentially as described previously (Sive et al., 2000). Antisense and sense digoxigenin-11-UTP (DIG-UTP)–labeled RNA probes of ∼1,250 nt were generated using pCS2-6XMYC-xDAPLE linearized with EcoRI or XhoI, respectively, as templates with T7 (11175025910; Roche) or SP6 (11175025910; Roche) labeling kits. Embryos were fixed at the stages indicated in the figures by incubation in MEMFA for 1 h at room temperature, followed by dehydration in methanol at −20°C. For hybridization, embryos of different stages underwent all subsequent procedures in parallel. Embryos were rehydrated gradually in PBS, washed three times with PBS supplemented with 0.1% (vol/vol) Tween-20 (Ptw), and treated with proteinase K (P8107S; New England Biolabs) at 2 µg/ml for 20 min. Embryos were immediately acetylated by two sequential incubations of 5 min in 0.1 M Triethanolamine, pH 7.5 (T1502; Sigma-Aldrich), followed two sequential incubations in 0.25% (vol/vol) acetic anhydride (A6404; Sigma-Aldrich). The embryos were washed twice in Ptw and fixed in Ptw + 4% paraformaldehyde for 20 min. After five washes in Ptw, the embryos were preincubated 6 h at 60°C in hybridization buffer (50% [vol/vol] deionized formamide, yeast RNA 1 mg/ml, heparin 100 µg/ml, 0.02% [wt/vol] BSA, 0.02% [wt/vol] polyvinylpyrrolidone [PVP-40], 0.02% [wt/vol] Ficoll-400, 5× SSC, 150 mM NaCl, 15 mM sodium citrate, pH 7, 0.1% [vol/vol] Tween-20, 0.1% [wt/vol] CHAPS, and EDTA 10 mM, pH 8). The solution was then replaced with fresh hybridization buffer and the DIG-labeled RNA probe at 0.5 µg/ml and incubated overnight at 60°C. The embryos were washed at 60°C once with hybridization buffer for 10 min, three times with 2× SSC for 20 min, and then twice in 0.2× SSC for 30 min. Embryos were then washed twice with PBS supplemented with 0.1% (vol/vol) Triton X-100 (Ptx) at room temperature, blocked in Ptx supplemented with 10% (vol/vol) normal goat serum for 1 h and incubated overnight at 4°C with anti-DIG antibodies conjugated to alkaline phosphatase (1:2,000 dilution; 1093274; Roche) in Ptx supplemented with 10% (vol/vol) normal goat serum. Embryos were washed 10 times for 20 min in Ptx and twice with alkaline phosphatase buffer (100 mM Tris-HCl, pH 9.5, 50 mM MgCl2, 100 mM NaCl, 0.1% [vol/vol] Tween-20, and 5 mM Levamisole). Alkaline phosphatase–based staining of the embryos was performed by incubation in substrate solution 1 tablet of 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (1697471; Roche) in 100 mM NaCl, 0.1% [vol/vol] Tween-20, and 2 mM Levamisole) at room temperature. After optimal signal development (6–8 h), the staining reaction was simultaneously stopped for all specimens analyzed in a given experiment by transferring the embryos to Bouin’s fixative (10% [vol/vol] formaldehyde and 5% [vol/vol] glacial acetic acid) followed by overnight incubation. Bouin’s fixative was washed in 30% Ptw/70% ethanol (vol/vol; 10 times 10 min), bleached in a solution containing 1% (vol/vol) H2O2, 5% (vol/vol) formamide, and 0.5× SSC, and photographed in PBS at room temperature with a Canon XSi camera connected to a Leica MZ6 microscope. Transversal cross sections of prestained embryos in Fig. 3 B were performed using a standard xylene/paraffin procedure (Sive et al., 2000).
RT-PCR of Xenopus embryos
mRNA from five Xenopus embryos was isolated with TRIzol (Thermo Fisher Scientific), and cDNA was generated using Superscript II First-Strand synthesis (11904018; Thermo Fisher Scientific). PCRs to evaluate the induction of neural genes were performed with Taq polymerase (M0273S; New England Biolabs) using the previously validated primers (Nakata et al., 1997) NCAM (5′-GCGGGTACCTTCTAATAGTCAC-3′ and 5′-GGCTTGGCTGTGGTTCTGAAGG-3′), N-Tubulin (5′-ATGCTGATCTACGCAAAC-3′ and 5′-AGATAGCAGCTACTGTGAG-3′), Zic3 (5′-CAACAGTGAGGAACCTTCCA-3′ and 5′-GGGCTTTGTTAGTCTGTAGC-3′), and ODC (5′-CAGCTAGCTGTGGTGTGG-3′ and 5′-CAACATGGAAACTCACACC-3′). To validate on-target activity of p114RhoGEF MO1, cDNA from stage 16 embryos bilaterally injected with p114RhoGEF MO1 were prepared exactly as described above. PCRs to assess the retention of intron 1 were performed with Q5 High-Fidelity DNA polymerase (M0491S; New England Biolabs) with the primers complementary to exon 1 (5′-CTCGGAATGACTGTCACCAA-3′) and exon 2 (5′-CAACAGCACAACTCCACGAT-3′).
Fluorescence microscopy and histological analysis of Xenopus embryos
For whole-mount F-actin staining (Fig. 4 A and Fig. 6, E and G), embryos were fixed in MEMFA for 2 h and washed three times in PBT (PBS supplemented with 0.1% [vol/vol] Triton X-100). Neural plates were dissected and incubated overnight with 0.28 µM Alexa Fluor 488–conjugated Phalloidin (PHDG1-A; Cytoskeleton Inc.) in PBT (methanol present in the stock of Phalloidin was evaporated at 37°C before diluting in PBT). Embryos were washed three times for 20 min with PBT and imaged in PBS at room temperature in homemade chambers built as previously described (Wallingford, 2010) with a Zeiss LSM 700 laser scanning inverted confocal microscope controlled by the manufacturer’s software. Pictures of 2 µm thickness were taken with a 20× objective (NA 0.8, working distance 0.55 mm) with a digital 0.5× zoom. Maximum intensity projections were generated with ImageJ, and individual images were assembled for presentation using Photoshop and Illustrator.
For whole-mount immunostaining (Fig. 4, C–F; and Fig. 6 B), embryos were fixed in 2% (vol/vol) TCA solution for 30 min at room temperature and washed three times with 0.3% (vol/vol) Triton X-100 in PBS for 30 min (Ossipova et al., 2015). Neural plates were dissected and incubated in blocking solution (PBS supplemented with 0.1% [vol/vol] Triton X-100 and 10% [vol/vol] normal goat serum). Neural plate explants were incubated with the following primary antibodies overnight in blocking solution: pMLC2 (rabbit polyclonal, 1:100; ab2480; Abcam); ZO-1 (rabbit polyclonal, 1:100; 61–7300; Zymed); β-Catenin (rabbit polyclonal, 1:400; ab16051; Abcam), Vangl2 (rabbit polyclonal; gift from S. Sokol), MYC (mouse monoclonal; 9E10; DSHB), GFP (mouse monoclonal, 1:100; sc-9996; Santa Cruz), and RFP (rabbit polyclonal, 1:200; ab62341; Abcam). Neural plate explants were washed five times for 1 h in PBT and incubated overnight with secondary antibodies goat anti-mouse Alexa Fluor 488 (1:400; A11017; Invitrogen) and goat anti-rabbit Alexa Fluor 594 (1:400; A11072; Invitrogen) diluted in blocking solution. Explants were washed five times for 1 h in PBT, cleared in Murray’s clear (Benzyl benzoate/Benzyl alcohol, 2:1) before imaging at room temperature in homemade chambers with a Zeiss LSM 700 laser scanning inverted confocal microscope controlled by the manufacturer’s software. Stacks of confocal pictures of 1 µm thickness along the z axis were taken with a 40× objective (NA 1.3, working distance 0.21 mm) using ZEN software. Images were processed for presentation using Photoshop and Illustrator.
For immunostaining of tissue sections (Figs. 4 B and S4 A), embryos were fixed in 2% TCA solution for 30 min at room temperature and washed three times with 0.3% Triton X-100 in PBS for 30 min (Ossipova et al., 2015). Embryos were embedded in a solution of 15% (vol/vol) fish gelatin and 15% (wt/vol) sucrose, oriented under the dissection microscope in embedding molds (4565; Sakura), and quickly flash frozen. Blocks were sectioned within 4 h using a Cryostat (HM 550; Microm) at 10 µm thickness as previously described (Lee et al., 2012). Sections were transferred to poly-L-lysine–coated slides and stored at −80°C. Slides were taken out of the freezer, dried at room temperature in a ventilated area for 30 min, and incubated for 1 min in acetone. After three 10-min washes in PBS, slides were incubated in blocking solution (PBS supplemented with 0.1% [vol/vol] Triton X-100 and 10% [vol/vol] normal goat serum) for 30 min. The following primary antibodies diluted in blocking solution were incubated overnight at 4°C: Sox3 (rabbit polyclonal, 1:200, gift from M. Klymkowsky, University of Colorado, Boulder, CO), GFP (mouse monoclonal, 1:100; sc-9996; Santa Cruz), or β-Catenin (rabbit polyclonal, 1:400; ab16051; Abcam). After three 10-min washes in PBS, slides were incubated for 90 min in secondary antibodies goat anti-mouse Alexa Fluor 488 (1:400; A11017; Invitrogen) and goat anti-rabbit Alexa Fluor 594 (1:400; A11072; Invitrogen) diluted in blocking solution. Sections were washed three times in PBT for 10 min and mounted with ProLong Diamond Antifade (P36965; Invitrogen). Pictures were taken at room temperature with a Zeiss Axio Observer Z1 microscope equipped with a digital camera (C10600/ORCA-R2; Hamamatsu Photonics) and controlled by the manufacturer’s software. Pictures were taken with a 40× objective (NA 1.4, working distance 0.4 mm).
Histological transversal sections of stage 46 embryos (Fig. S2) were prepared following a standard xylene/paraffin procedure. Briefly, embryos were fixed in MEMFA for 2 h at room temperature and subsequently dehydrated in ethanol at 4°C for at least 1 d. Embryos were gradually transferred to xylene, embedded in paraplast (39501006; McCormick) in embedding molds (22363555; Thermo Fisher Scientific) at 57°C. Embryos were oriented under the dissection microscope and quickly transferred to 4°C to solidify the block in the desired orientation. Sections of 5 µm were prepared with a microtome (820; American Optical Company), transferred to slides, dried at room temperature, and stored at 4°C. For hematoxylin and eosin staining, slides were deparaffined, gradually rehydrated, and stained with hematoxylin (GHS332; Sigma-Aldrich) and eosin (HT110232; Sigma-Aldrich) and mounted in Permount (SP15; Thermo Fisher Scientific). Pictures were taken at room temperature with a Nikon Eclipse TE2000-E microscope equipped with a digital camera (DS-Fi2; Nikon) and controlled by NIS Elements. Pictures were taken with a 10× objective (NA 0.45, working distance 4 mm).
Generation of zebrafish mutant lines and morphological analysis
Zebrafish (Danio rerio) experiments were performed according to Boston University Institutional Animal Care and Use Committee–approved protocol AN-15558, in compliance with the Guide for the Care and Use of Laboratory Animals. Two independent zDAPLE mutant lines were generated using CRISPR-Cas9, one by targeting exon 8 and another targeting exon 15. For each locus, two sgRNAs targeting sequences separated by ∼70–100 nt were used (sgRNA.1 and sgRNA.2 for exon 8 and sgRNA.3 and sgRNA.4 for exon 15). sgRNA target sites were selected using CRISPRscan (Moreno-Mateos et al., 2015), yielding the following sequences: sgRNA.1, 5′-GGGAGCTCACGGCCTGGGTCAGG-3′; sgRNA.2, 5′-CGAGGCCCGAGTGAGGGAGGTGG-3′; sgRNA.3, 5′-GAGAAAGGGACGTGCGCTCAGGG-3′; and sgRNA.4, 5′-AGGCCAAACTGCGGCGCTCCCGG-3′, where the PAM sequence (NGG) is indicated in bold. sgRNA templates were generated by annealing and polymerase-mediated extension of a forward oligo containing the T7 promoter sequence, the 20-nt sgRNA target sequence (without the PAM sequence), and a 15-nt sequence complementary to the reverse oligo containing the invariable Cas9-binding scaffold. PCR reactions with Q5 high-fidelity polymerase (M0491; New England Biolabs) were performed as follows: 1 cycle at 95°C for 3 min, 35 cycles (at 95°C for 3 min, 45°C for 30 s, and 72°C for 20 s), and 1 cycle at 72°C for 5 min. Reactions were purified with a PCR purification kit (T1030; New England Biolabs). Approximately 120–150 ng DNA was used as a template for a T7 in vitro transcription reaction with the AmpliScribe-T7-Flash transcription kit (ASF3507; Epicentre). In vitro transcription sgRNA products were purified (74104; Qiagen) and quantified. Zebrafish embryos were injected at one-cell stage with Cas9 mRNA (100 pg) together with one of the two sgRNA pairs (30 pg of each sgRNA) to generate the two independent lines. Injected embryos were raised to adulthood and potential founders (F0) were backcrossed with WT animals. Their progenies were screened for the presence of ∼70- to 100-nt deletions in exon 8 or exon 15 by PCR genotyping (see below). Deletion fragments were gel extracted (K0691; Thermo Fisher Scientific) and analyzed by Sanger DNA sequencing to identify frameshift deletions in each locus (zDAPLEex8 and zDAPLEex15). Heterozygote mutants were intercrossed to obtain zygotic homozygous mutants and homozygous WT littermates, which in turn were incrossed to yield the maternal-and-zygotic (MZ) zDAPLEex8 and MZ zDAPLEex15, and their corresponding WT controls used for phenotypic analyses.
PCR genotyping was performed from genomic DNA extracted of adult fins as previously described (Meeker et al., 2007). Each fin was collected in PCR tubes containing 100 µl of 100 mM NaOH and boiled for 15 min at 95°C. Samples were neutralized with Tris-HCl, pH 7.4, and diluted with 100 µl water. 2 µl of each genomic DNA preparation was subsequently used in a PCR reaction using Taq DNA polymerase (M0273; New England Biolabs) following manufacturer protocol and run in a 2% agarose gel. The following primers were used: 5′-TGAGCGAGATGAGTCTGCTG-3′ and 5′-CATTGTTGGTTCACTCTGGGTA-3′ for exon 8 and 5′-CCAGTGTCTTTTTGTAGGTCCTTT-3′ and 5′-GACGAGGAACTTCCCTTTCC-3′ for exon 15.
For phenotype analysis, embryos were imaged at room temperature with a Zeiss Discovery.V12 equipped with an AxioCam MRc. For somite count at 22 hpf and quantification of axis curvature at 28 hpf (Fig. S3 F), embryos were dechorionated with 150 µg/ml pronase (P5147; Sigma-Aldrich) 1 h before the time of analysis. Somite counting was done on fixed specimens. For the quantification of NT defects, animals were imaged 22 hpf and scored as having a defect when ventricular hinge points were not discernible. Some of these animals were manually dechorionated, anesthetized with 150 µg/ml Tricaine (A-5040; Sigma-Aldrich), and microinjected with one nanoliter of a solution of Texas Red Dextran beads (D1830; Invitrogen) at 2.5 mg/ml in the brain ventricles to facilitate the visualization of phenotypes for presentation (as in Fig. S3 D). The same procedure was followed for the images taken at 56 hpf (Fig. S3 D), except that the embryos were treated with 1-phenyl 2-thiourea at 200 µM (P7629; Sigma-Aldrich) from 22 hpf to prevent pigment formation. The transversal view of 22-hpf embryonic brains shown in Fig. S3 D was generated from reconstructed confocal pictures of whole-mount stained embryos. For this, embryos were fixed overnight at 4°C in Dent’s fixative (80% methanol/20% DMSO [vol/vol]). Embryos were gradually rehydrated in PBS. After three 15-min washes with PBT, embryos were then deyolked and their anterior part explanted. Explants were incubated in blocking solution (PBT supplemented with 10% [vol/vol] normal goat serum) for 1 h, followed by an overnight incubation at 4°C with anti-β-catenin primary antibody (rabbit polyclonal, 1:400; ab16051; Abcam). After five washes with PBT, embryos were incubated overnight at 4°C with goat anti-rabbit Alexa Fluor 488 secondary antibodies (1:400; A11070; Invitrogen). After five washes in PBT, embryos were transferred to glycerol for 1 h and mounted for confocal fluorescence microscopy analysis using a Zeiss LSM 700 microscope. Stacks of confocal pictures of 1 µm thickness along the z axis were acquired at room temperature with a 40× objective (NA 1.3, working distance 0.21 mm) with a zoom of 0.5× using ZEN software. Images were processed for presentation using ZEN software, and individual images were assembled in Photoshop and Illustrator.
mRNA in situ hybridization in zebrafish embryos
mRNA in situ hybridization of zebrafish embryos was performed as described by Thisse and Thisse (2008). DIG-UTP–labeled RNA probes complementary to either zDAPLE exon 15 (coding sequence 1,666–2,166) or C terminus (coding sequence 5,494–5,994) were synthesized using the SP6 DIG-UTP labeling kit (11175025910; Roche). For this, PCR products of either the complementary sequence (antisense probe) or the sense sequence (sense probe) fused to the SP6 promoter sequence were used as templates. Embryos were dechorionated with 150 µg/ml pronase (P5147; Sigma-Aldrich), fixed at the desired developmental stage by overnight incubation at 4°C with 4% (wt/vol) paraformaldehyde in PBS, and subsequently dehydrated at −20°C with methanol. Embryos were gradually rehydrated in PBS, washed four times with PBS supplemented with 0.1% (wt/vol) Tween-20 (Ptw), and treated with 10 µg/ml proteinase K (P8107S; New England Biolabs). Proteinase K digestion was stopped by replacing the solution with 4% (wt/vol) paraformaldehyde in PBS and incubation for 20 min. After four washes with Ptw (5 min each), embryos were incubated in hybridization mix (50% (vol/vol) deionized formamide, 5× SSC, 0.1% [vol/vol] Tween-20, 50 µg/ml heparin, and yeast RNA 0.5 mg/ml) for 4 h at 70°C, followed by an overnight incubation at 70°C with 50 ng DIG-labeled RNA probe in 200 µl hybridization mix. Embryos were gradually exchanged into 2× SSC at 70°C and subsequently washed twice with 0.2× SSC at 70°C. After incubation for 4 h at room temperature in blocking solution (Ptw supplemented with 10% normal goat serum), embryos were incubated overnight at 4°C with anti-DIG antibodies conjugated to alkaline phosphatase (1:10,000 in blocking solution; 1093274; Roche). Embryos were washed six times with Ptw for 15 min, three times for 5 min with alkaline phosphatase buffer (100 mM Tris-HCl, pH 9.5, 50 mM MgCl2, 100 mM NaCl, 0.1% [vol/vol] Tween-20, and 5 mM Levamisole) and then incubated in staining solution prepared by dissolving one tablet of 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium in alkaline phosphatase buffer. After optimal staining was observed, reactions were simultaneously stopped for all stages analyzed by transferring embryos stop solution (PBS, pH 5.5, 1 mM EDTA, and 0.1% Tween 20). Embryos were photographed at room temperature in PBS as whole-mount specimens or in glycerol after removal of the yolk as flat-mounted specimens.
Protein expression and purification
His-tagged and GST-tagged proteins were expressed in BL21(DE3) Escherichia coli (Life Technologies) transformed with the corresponding plasmids by overnight induction at 23°C with 1 mM IPTG. Protein purification was performed following previously described protocols (Garcia-Marcos et al., 2009, 2010). Briefly, bacteria pelleted from 1 liter of culture were resuspended in 25 ml buffer (50 mM NaH2PO4, pH 7.4, 300 mM NaCl, 10 mM imidazole, and 1% [vol/vol] Triton X-100 supplemented with protease inhibitor cocktail [Leupeptin 1 µM, Pepstatin 2.5 µM, Aprotinin 0.2 µM, and PMSF 1 mM]). For Gαi3, this buffer was supplemented with 25 µM GDP and 5 mM MgCl2. After sonication (four cycles, with pulses lasting 20 s/cycle, with a 1-min interval between cycles to prevent heating), lysates were centrifuged at 12,000 ×g for 20 min at 4°C. The soluble fraction (supernatant) of the lysate was used for affinity purification on HisPur Cobalt or glutathione-agarose resins (Pierce) and eluted with lysis buffer supplemented with 250 mM imidazole or with 50 mM Tris-HCl, pH 8, 100 mM NaCl, and 30 mM reduced glutathione, respectively. Proteins were dialyzed overnight at 4°C against PBS, except for Gαi3 proteins, which were buffer exchanged into 20 mM Tris-HCl, pH 7.4, 20 mM NaCl, 1 mM MgCl2, 1 mM DTT, 10 µM GDP, and 5% (vol/vol) glycerol using a HiTrap Desalting column (GE Healthcare) or cycles of dilution/concentration with 10,000-D cutoff centrifugal filters. All protein samples were aliquoted and stored at −80°C.
In vitro protein-binding assays with GST-fused proteins
GST-GBA fusions of xDAPLE (5 µg) were immobilized on glutathione-agarose beads in PBS for 90 min at room temperature. Beads were washed twice with PBS, resuspended in 300 µl of binding buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 0.4% [vol/vol] NP-40, 10 mM MgCl2, 5 mM EDTA, 2 mM DTT, and 30 µM GDP), and incubated 4 h at 4°C with constant tumbling in the presence of purified His-Gαi3 (0.1 µg). Beads were washed four times with 1 ml of wash buffer (4.3 mM Na2HPO4, 1.4 mM KH2PO4, pH 7.4, 137 mM NaCl, 2.7 mM KCl, 0.1% [vol/vol] Tween-20, 10 mM MgCl2, 5 mM EDTA, 1 mM DTT, and 30 µM GDP) and resin-bound proteins eluted with Laemmli sample buffer by incubation at 37°C for 10 min. Proteins were separated by SDS-PAGE, transferred to polyvinylidene fluoride membranes, and stained with Ponceau S or immunoblotted as indicated in the Immunoblotting section. The experiments with immobilized GST-Gαi3 (20 µg) were performed the same way, except that immobilization was done in binding buffer, and for some conditions (AlF4−), the binding and wash buffers were supplemented with 30 µM AlCl3 and 10 mM NaF. Lysates of HEK293T cells expressing MYC-xDAPLE WT (approximately one fourth of a p10 dish transfected with 6 µg DNA by the calcium phosphate method) were used as the source of the soluble ligand. Briefly, cells were washed with PBS, scrapped in PBS, and pelleted before resuspension in cold lysis buffer (20 mM Hepes, pH 7.2, 5 mM Mg(CH3COO)2, 125 mM K(CH3COO), 0.4% [vol/vol] Triton X-100, 1 mM DTT, 10 mM β-glycerophosphate, and 0.5 mM Na3VO4 supplemented with a protease inhibitor cocktail [S8830]; Sigma-Aldrich) and clearing (14,000 ×g, 10 min).
Steady-state GTPase assay
This assay was performed as described previously (Garcia-Marcos et al., 2010). Briefly, His-Gαi3 (100 nM) was preincubated with different concentrations of His-xDAPLE-CT (aa 1,638–1,932) or GST-xDAPLE (aa 1,648–1,704) for 60 min at 4°C or 15 min at 30°C in assay buffer (20 mM Na-Hepes, pH 8, 100 mM NaCl, 1 mM EDTA, 25 mM MgCl2, 1 mM DTT, and 0.05% [wt/vol] C12E10). GTPase reactions were initiated at 30°C by adding an equal volume of assay buffer containing 1 µM [γ-32P]GTP (∼50 cpm/fmol). Duplicate aliquots (25 µl) were removed at 15 min and reactions stopped with 975 µl of ice-cold 5% (wt/vol) activated charcoal in 20 mM H3PO4, pH 3. Initial time-course experiments confirmed that the GTPase activity of Gαi3 in the presence or absence of xDAPLE is linear for at least 15 min (Fig. S6). Samples were then centrifuged for 10 min at 10,000 ×g, and 500 µl of the resultant supernatant was scintillation counted to quantify [32P]Pi released. Background [32P]Pi detected at 15 min in the absence of G protein was subtracted from each reaction and data expressed as percentage of the Pi produced by His-Gαi3 in the absence of xDAPLE. Background counts were <5% of the counts detected in the presence of G proteins.
GTPγS-binding assay
His-Gαi3 (100 nM) was preincubated with different concentrations of His-xDAPLE-CT (aa 1,638–1,932) for 15 min at 30°C in assay buffer (20 mM Na-Hepes, pH 8, 100 mM NaCl, 1 mM EDTA, 25 mM MgCl2, 1 mM DTT, and 0.05% [wt/vol] C12E10]. Reactions were initiated at 30°C by adding an equal volume of assay buffer containing 1 µM [35S] GTPγS (∼50 cpm/fmol). Duplicate aliquots (25 µl) were removed at different time points, and binding of radioactive nucleotide was stopped by addition of 3 ml ice-cold wash buffer (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, and 25 mm MgCl2). The quenched reactions were rapidly passed through BA-85 nitrocellulose filters (GE Healthcare) and washed with 4 ml cold wash buffer. Filters were dried and subjected to liquid scintillation counting. Background [35S]GTPγS detected at 15 min in the absence of G protein was subtracted from each reaction and data expressed as percentage of the [35S]GTPγS produced by His-Gαi3 in the absence of xDAPLE. Background counts were <5% of the counts detected in the presence of G proteins.
Yeast-based G protein activation reporter assay
The previously described (Cismowski et al., 1999) S. cerevisiae strain CY7967 (MATα GPA1(1–41)-Gαi3 far1Δ fus1p-HIS3 can1 ste14:trp1:LYS2 ste3Δ lys2 ura3 leu2 trp1 his3; kindly provided by J. Broach, Penn State University, Hershey, PA) was used for all yeast experiments. The main features of this strain are that the only pheromone responsive GPCR (Ste3) is deleted, the endogenous Gα-subunit Gpa1 is replaced by a chimeric Gpa1(1–41)-human Gαi3 (36–354), and the cell cycle arrest–inducing protein far1 is deleted. In this strain, the pheromone response pathway can be up-regulated by the ectopic expression of activators of human Gαi3 and does not result in the cell cycle arrest that occurs in the native pheromone response (Cismowski et al., 1999). Plasmid transformations were performed using the lithium acetate method. CY7967 was first transformed with a centromeric plasmid (CEN TRP) encoding the LacZ gene under the control of the FUS1 promoter, which is activated by the pheromone response pathway. The PFUS1::LacZ-expressing strain was transformed with pLIC-YES2 plasmids (2 µm, URA) encoding xDAPLE WT, FA1, FA2, FA1+2, and hDAPLE WT and FA as described in Plasmids and MOs. Double transformants were selected in synthetic defined–TRP-URA media. Individual colonies were inoculated into 3 ml of synthetic defined–galactose-TRP-URA and incubated overnight at 30°C to induce the expression of the proteins of interest under the control of the galactose-inducible promoter of pLIC-YES2. This starting culture was used to inoculate 20 ml of synthetic defined–galactose-TRP-URA at 0.3 OD600. Exponentially growing cells (∼0.7–0.8 OD600, 4–5 h) were pelleted to prepare samples for subsequent assays. β-Galactosidase activity assays were performed as described previously (Coleman et al., 2016). Pellets corresponding to 0.5 OD600 (in duplicates) were washed once with PBS + 0.1% (wt/vol) BSA and resuspended in 200 µl assay buffer (60 mM Na2PO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgCl2, 0.25% [vol/vol] β-mercaptoethanol, 0.01% [wt/vol] SDS, and 10% [vol/vol] chloroform) and vortexed. 100 µl was transferred to 96-well plates in duplicates and reactions started by the addition of 50 µl of the fluorogenic β-galactosidase substrate fluorescein di-β-D-galactopyranoside (100 µM final). Fluorescence (excitation 485 ± 10 nm, emission 528 ± 10 nm) was measured every 2 min for 90 min at 30°C in a Biotek H1 synergy plate reader. Enzymatic activity was expressed as arbitrary fluorescence units per minute. Samples for immunoblotting were prepared exactly as described previously (Coleman et al., 2016) using a TCA precipitation method.
Immunoprecipitation
Approximately 2 million HEK293T cells were seeded in 10-cm dishes and transfected the day after using calcium phosphate with plasmids encoding FLAG-Gαi3 (3 µg) and/or MYC-xDAPLE WT or FA1+2 (also known as GBA**) mutant (6 µg). 24 h after transfection, cells were lysed on ice with 500 µl lysis buffer (20 mM Hepes, pH 7.2, 5 mM Mg(CH3COO)2, 125 mM K(CH3COO), 0.4% [vol/vol] Triton X-100, 1 mM DTT, 10 mM β-glycerophosphate, 0.5 mM Na3VO4, and 30 µM GDP supplemented with a protease inhibitor cocktail [S8830]; Sigma-Aldrich) and cleared (14,000 ×g, 10 min). Cleared lysates were incubated with 2 µg FLAG antibodies (F1804; Sigma-Aldrich) for 4 h at 4°C with constant tumbling. 35 μl Protein G agarose beads preblocked with 5% (wt/vol) BSA (2 h at room temperature) was added to the tubes and incubated for 90 min at 4°C. Beads were washed three times in wash buffer (4.3 mM Na2HPO4, 1.4 mM KH2PO4, pH 7.4, 137 mM NaCl, 2.7 mMKCl, 0.1% [vol/vol] Tween-20, 10 mM MgCl2, 5 mM EDTA, 1 mM DTT, and 30 µM GDP) and proteins eluted by adding Laemmli sample buffer and boiling for 5 min. Proteins were separated by SDS-PAGE and immunoblotted with antibodies as indicated in the Immunoblotting section.
Immunoblotting
Proteins were separated by SDS-PAGE and transferred to polyvinylidene fluoride membranes, which were sequentially incubated with primary and secondary antibodies The primary antibodies were used at the following dilutions: MYC (1:1,000; 2276 (9B11); Cell Signaling Technology), His (1:12,500; H1029; Sigma-Aldrich), FLAG (1:2,000; F1804; Sigma-Aldrich), GST (1:1,000; sc-459; Santa Cruz) α-tubulin (1:2,500; T6074; Sigma-Aldrich), Gβ (1:1,000; sc-261; Santa Cruz), Vangl2 (1:500; from S. Sokol; Ossipova et al., 2015), and p114RhoGEF (1:1,000; PA5-21429; Thermo Fisher Scientific). The secondary antibodies were goat anti-rabbit Alexa Fluor 680 (1:10,000; A21077; Invitrogen) and goat anti-mouse IRDye 800 (1:10,000; 926-32210; Li-Cor). Infrared imaging of immunoblots was performed using an Odyssey Infrared Imaging System (Li-Cor Biosciences). Images were processed using the ImageJ software and assembled for presentation using Photoshop and Illustrator.
Statistical analysis
Each experiment was performed at least three times. For experiments displaying pooled data, results are expressed as average ± SEM, frequency distributions, scatterplots, or box-and-whisker plots, as indicated in the figure legends. For other experiments, one representative result is presented. Statistical significance between various conditions was assessed by determining P values using a t test (two-tailed, unpaired), χ2, Fisher exact test, or Mann–Whitney U test, as indicated in the figure legends, using Prism (GraphPad). The parametric t test was calculated with data that passed the normality test (D’Agostino and Pearson omnibus normality test) calculated with GraphPad.
Online supplemental material
Fig. S1 shows that apical cell junction localization of DAPLE is not disrupted by different treatments that inhibit DAPLE-mediated apical cell constriction. Fig. S2 shows validation of two DAPLE translation-blocking MOs and embryonic brain defects in xDAPLE morphants. Fig. S3 shows that loss of DAPLE causes mild neurodevelopmental defects in zebrafish. Fig. S4 shows that loss of xDAPLE does not affect neural cell differentiation or axis elongation during neurulation. Fig. S5 shows validation of a splicing-interfering MO against p114RhoGEF in Xenopus. Fig. S6 shows that basal and DAPLE-stimulated steady-state GTPase activity of Gαi3 are linear for at least 15 min.
Supplementary Material
Acknowledgments
We thank Emily Mann, Covadonga Lopez Ramos, and Stefan Broselid for their technical help. We thank V. Trinkaus-Randall for giving access to the Confocal Microscopy Facility at Boston University School of Medicine. We thank S. Sokol for plasmids and for the Vangl2 antibody, M. Klymkowsky for the Sox3 antibody, Pradipta Ghosh (University of California, San Diego, La Jolla, CA) for the full-length hDAPLE construct, T. Michiue for the xDAPLE plasmids, and X. Varelas for sharing reagents and for helpful discussions.
This work was supported by National Institutes of Health grants R01GM108733 and R01GM130120 (to M. Garcia-Marcos), R01GM098367 (to I. Dominguez), and R00HD071968 (to D. Cifuentes).
The authors declare no competing financial interests.
Author contributions: A. Marivin, V. Morozova, I. Walawalkar, A. Leyme, D.A. Kretov, D. Cifuentes, and M. Garcia-Marcos conducted experiments. A. Marivin, D. Cifuentes, I. Dominguez, and M. Garcia-Marcos designed experiments. A. Marivin, V. Morozova, I. Walawalkar, D. Cifuentes, I. Dominguez, and M. Garcia-Marcos analyzed data. A. Marivin and M. Garcia-Marcos wrote the manuscript. M. Garcia-Marcos conceived and supervised the project.
References
- Aittaleb M., Boguth C.A., and Tesmer J.J.. 2010. Structure and function of heterotrimeric G protein-regulated Rho guanine nucleotide exchange factors. Mol. Pharmacol. 77:111–125. 10.1124/mol.109.061234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aznar N., Midde K.K., Dunkel Y., Lopez-Sanchez I., Pavlova Y., Marivin A., Barbazán J., Murray F., Nitsche U., Janssen K.P., et al. 2015. Daple is a novel non-receptor GEF required for trimeric G protein activation in Wnt signaling. eLife. 4:e07091 10.7554/eLife.07091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett K., Leptin M., and Settleman J.. 1997. The Rho GTPase and a putative RhoGEF mediate a signaling pathway for the cell shape changes in Drosophila gastrulation. Cell. 91:905–915. 10.1016/S0092-8674(00)80482-1 [DOI] [PubMed] [Google Scholar]
- Blum M., De Robertis E.M., Wallingford J.B., and Niehrs C.. 2015. Morpholinos: Antisense and Sensibility. Dev. Cell. 35:145–149. 10.1016/j.devcel.2015.09.017 [DOI] [PubMed] [Google Scholar]
- Bonacci T.M., Mathews J.L., Yuan C., Lehmann D.M., Malik S., Wu D., Font J.L., Bidlack J.M., and Smrcka A.V.. 2006. Differential targeting of Gbetagamma-subunit signaling with small molecules. Science. 312:443–446. 10.1126/science.1120378 [DOI] [PubMed] [Google Scholar]
- Butler M.T., and Wallingford J.B.. 2018. Spatial and temporal analysis of PCP protein dynamics during neural tube closure. eLife. 7:e36456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrita L.D., Dai W., and Bottomley S.P.. 2006. A family of E. coli expression vectors for laboratory scale and high throughput soluble protein production. BMC Biotechnol. 6:12 10.1186/1472-6750-6-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerer E., Barker A., Duong D.N., Ganesan R., Kataoka H., Cornelissen I., Darragh M.R., Hussain A., Zheng Y.W., Srinivasan Y., et al. 2010. Local protease signaling contributes to neural tube closure in the mouse embryo. Dev. Cell. 18:25–38. 10.1016/j.devcel.2009.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cismowski M.J., Takesono A., Ma C., Lizano J.S., Xie X., Fuernkranz H., Lanier S.M., and Duzic E.. 1999. Genetic screens in yeast to identify mammalian nonreceptor modulators of G-protein signaling. Nat. Biotechnol. 17:878–883. 10.1038/12867 [DOI] [PubMed] [Google Scholar]
- Coleman B.D., Marivin A., Parag-Sharma K., DiGiacomo V., Kim S., Pepper J.S., Casler J., Nguyen L.T., Koelle M.R., and Garcia-Marcos M.. 2016. Evolutionary Conservation of a GPCR-Independent Mechanism of Trimeric G Protein Activation. Mol. Biol. Evol. 33:820–837. 10.1093/molbev/msv336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M., Wilson E.T., and Wieschaus E.. 1994. A putative cell signal encoded by the folded gastrulation gene coordinates cell shape changes during Drosophila gastrulation. Cell. 76:1075–1089. 10.1016/0092-8674(94)90384-0 [DOI] [PubMed] [Google Scholar]
- de Opakua A.I., Parag-Sharma K., DiGiacomo V., Merino N., Leyme A., Marivin A., Villate M., Nguyen L.T., de la Cruz-Morcillo M.A., Blanco-Canosa J.B., et al. 2017. Molecular mechanism of Gαi activation by non-GPCR proteins with a Gα-Binding and Activating motif. Nat. Commun. 8:15163 10.1038/ncomms15163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiacomo V., Marivin A., and Garcia-Marcos M.. 2018. When Heterotrimeric G Proteins Are Not Activated by G Protein-Coupled Receptors: Structural Insights and Evolutionary Conservation. Biochemistry. 57:255–257. 10.1021/acs.biochem.7b00845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drielsma A., Jalas C., Simonis N., Désir J., Simanovsky N., Pirson I., Elpeleg O., Abramowicz M., and Edvardson S.. 2012. Two novel CCDC88C mutations confirm the role of DAPLE in autosomal recessive congenital hydrocephalus. J. Med. Genet. 49:708–712. 10.1136/jmedgenet-2012-101190 [DOI] [PubMed] [Google Scholar]
- Ekici A.B., Hilfinger D., Jatzwauk M., Thiel C.T., Wenzel D., Lorenz I., Boltshauser E., Goecke T.W., Staatz G., Morris-Rosendahl D.J., et al. 2010. Disturbed Wnt Signalling due to a Mutation in CCDC88C Causes an Autosomal Recessive Non-Syndromic Hydrocephalus with Medial Diverticulum. Mol. Syndromol. 1:99–112. 10.1159/000319859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentealba J., Toro-Tapia G., Rodriguez M., Arriagada C., Maureira A., Beyer A., Villaseca S., Leal J.I., Hinrichs M.V., Olate J., et al. 2016. Expression profiles of the Gα subunits during Xenopus tropicalis embryonic development. Gene Expr. Patterns. 22:15–25. 10.1016/j.gep.2016.09.001 [DOI] [PubMed] [Google Scholar]
- Garcia-Marcos M., Ghosh P., and Farquhar M.G.. 2009. GIV is a nonreceptor GEF for G alpha i with a unique motif that regulates Akt signaling. Proc. Natl. Acad. Sci. USA. 106:3178–3183. 10.1073/pnas.0900294106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Marcos M., Ghosh P., Ear J., and Farquhar M.G.. 2010. A structural determinant that renders G alpha(i) sensitive to activation by GIV/girdin is required to promote cell migration. J. Biol. Chem. 285:12765–12777. 10.1074/jbc.M109.045161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Marcos M., Kietrsunthorn P.S., Wang H., Ghosh P., and Farquhar M.G.. 2011. G Protein binding sites on Calnuc (nucleobindin 1) and NUCB2 (nucleobindin 2) define a new class of G(alpha)i-regulatory motifs. J. Biol. Chem. 286:28138–28149. 10.1074/jbc.M110.204099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P., Beas A.O., Bornheimer S.J., Garcia-Marcos M., Forry E.P., Johannson C., Ear J., Jung B.H., Cabrera B., Carethers J.M., and Farquhar M.G.. 2010. A Galphai-GIV molecular complex binds epidermal growth factor receptor and determines whether cells migrate or proliferate. Mol. Biol. Cell. 21:2338–2354. 10.1091/mbc.e10-01-0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman A.G. 1987. G proteins: transducers of receptor-generated signals. Annu. Rev. Biochem. 56:615–649. 10.1146/annurev.bi.56.070187.003151 [DOI] [PubMed] [Google Scholar]
- Gilmour D., Rembold M., and Leptin M.. 2017. From morphogen to morphogenesis and back. Nature. 541:311–320. 10.1038/nature21348 [DOI] [PubMed] [Google Scholar]
- Haigo S.L., Hildebrand J.D., Harland R.M., and Wallingford J.B.. 2003. Shroom induces apical constriction and is required for hingepoint formation during neural tube closure. Curr. Biol. 13:2125–2137. 10.1016/j.cub.2003.11.054 [DOI] [PubMed] [Google Scholar]
- Heer N.C., and Martin A.C.. 2017. Tension, contraction and tissue morphogenesis. Development. 144:4249–4260. 10.1242/dev.151282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao L.E., Wente S.R., and Chen W.. 2013. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA. 110:13904–13909. 10.1073/pnas.1308335110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanesaki T., Hirose S., Grosshans J., and Fuse N.. 2013. Heterotrimeric G protein signaling governs the cortical stability during apical constriction in Drosophila gastrulation. Mech. Dev. 130:132–142. 10.1016/j.mod.2012.10.001 [DOI] [PubMed] [Google Scholar]
- Keller R. 2002. Shaping the vertebrate body plan by polarized embryonic cell movements. Science. 298:1950–1954. 10.1126/science.1079478 [DOI] [PubMed] [Google Scholar]
- Kerridge S., Munjal A., Philippe J.M., Jha A., de las Bayonas A.G., Saurin A.J., and Lecuit T.. 2016. Modular activation of Rho1 by GPCR signalling imparts polarized myosin II activation during morphogenesis. Nat. Cell Biol. 18:261–270. 10.1038/ncb3302 [DOI] [PubMed] [Google Scholar]
- Kobayashi H., Michiue T., Yukita A., Danno H., Sakurai K., Fukui A., Kikuchi A., and Asashima M.. 2005. Novel Daple-like protein positively regulates both the Wnt/beta-catenin pathway and the Wnt/JNK pathway in Xenopus. Mech. Dev. 122:1138–1153. 10.1016/j.mod.2005.05.003 [DOI] [PubMed] [Google Scholar]
- Kölsch V., Seher T., Fernandez-Ballester G.J., Serrano L., and Leptin M.. 2007. Control of Drosophila gastrulation by apical localization of adherens junctions and RhoGEF2. Science. 315:384–386. 10.1126/science.1134833 [DOI] [PubMed] [Google Scholar]
- Lee M.J., and Dohlman H.G.. 2008. Coactivation of G protein signaling by cell-surface receptors and an intracellular exchange factor. Curr. Biol. 18:211–215. 10.1016/j.cub.2008.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.S., Sokol S.Y., Moody S.A., and Daar I.O.. 2012. Using 32-cell stage Xenopus embryos to probe PCP signaling. Methods Mol. Biol. 839:91–104. 10.1007/978-1-61779-510-7_8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyme A., Marivin A., Perez-Gutierrez L., Nguyen L.T., and Garcia-Marcos M.. 2015. Integrins activate trimeric G proteins via the nonreceptor protein GIV/Girdin. J. Cell Biol. 210:1165–1184. 10.1083/jcb.201506041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyme A., Marivin A., Maziarz M., DiGiacomo V., Papakonstantinou M.P., Patel P.P., Blanco-Canosa J.B., Walawalkar I.A., Rodriguez-Davila G., Dominguez I., and Garcia-Marcos M.. 2017. Specific inhibition of GPCR-independent G protein signaling by a rationally engineered protein. Proc. Natl. Acad. Sci. USA. 114:E10319–E10328. 10.1073/pnas.1707992114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Ear J., Midde K., Lopez-Sanchez I., Aznar N., Garcia-Marcos M., Kufareva I., Abagyan R., and Ghosh P.. 2014. Structural basis for activation of trimeric Gi proteins by multiple growth factor receptors via GIV/Girdin. Mol. Biol. Cell. 25:3654–3671. 10.1091/mbc.e14-05-0978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F., Sepich D.S., Chen S., Topczewski J., Yin C., Solnica-Krezel L., and Hamm H.. 2005. Essential roles of Galpha12/13 signaling in distinct cell behaviors driving zebrafish convergence and extension gastrulation movements. J. Cell Biol. 169:777–787. 10.1083/jcb.200501104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Sanchez I., Dunkel Y., Roh Y.S., Mittal Y., De Minicis S., Muranyi A., Singh S., Shanmugam K., Aroonsakool N., Murray F., et al. 2014. GIV/Girdin is a central hub for profibrogenic signalling networks during liver fibrosis. Nat. Commun. 5:4451 10.1038/ncomms5451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery L.A., and Sive H.. 2004. Strategies of vertebrate neurulation and a re-evaluation of teleost neural tube formation. Mech. Dev. 121:1189–1197. 10.1016/j.mod.2004.04.022 [DOI] [PubMed] [Google Scholar]
- Lowery L.A., and Sive H.. 2009. Totally tubular: the mystery behind function and origin of the brain ventricular system. BioEssays. 31:446–458. 10.1002/bies.200800207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning A.J., and Rogers S.L.. 2014. The Fog signaling pathway: insights into signaling in morphogenesis. Dev. Biol. 394:6–14. 10.1016/j.ydbio.2014.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning A.J., Peters K.A., Peifer M., and Rogers S.L.. 2013. Regulation of epithelial morphogenesis by the G protein-coupled receptor mist and its ligand fog. Sci. Signal. 6:ra98 10.1126/scisignal.2004427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A.C., and Goldstein B.. 2014. Apical constriction: themes and variations on a cellular mechanism driving morphogenesis. Development. 141:1987–1998. 10.1242/dev.102228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maziarz M., Broselid S., DiGiacomo V., Park J.C., Luebbers A., Garcia-Navarrete L., Blanco-Canosa J.B., Baillie G.S., and Garcia-Marcos M.. 2018. A biochemical and genetic discovery pipeline identifies PLCδ4b as a nonreceptor activator of heterotrimeric G-proteins. J. Biol. Chem. 293:16964–16983. 10.1074/jbc.RA118.003580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeker N.D., Hutchinson S.A., Ho L., and Trede N.S.. 2007. Method for isolation of PCR-ready genomic DNA from zebrafish tissues. Biotechniques. 43:610–614. 10.2144/000112619 [DOI] [PubMed] [Google Scholar]
- Midde K.K., Aznar N., Laederich M.B., Ma G.S., Kunkel M.T., Newton A.C., and Ghosh P.. 2015. Multimodular biosensors reveal a novel platform for activation of G proteins by growth factor receptors. Proc. Natl. Acad. Sci. USA. 112:E937–E946. 10.1073/pnas.1420140112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Mateos M.A., Vejnar C.E., Beaudoin J.D., Fernandez J.P., Mis E.K., Khokha M.K., and Giraldez A.J.. 2015. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods. 12:982–988. 10.1038/nmeth.3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima H., and Tanoue T.. 2011. Lulu2 regulates the circumferential actomyosin tensile system in epithelial cells through p114RhoGEF. J. Cell Biol. 195:245–261. 10.1083/jcb.201104118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata K., Nagai T., Aruga J., and Mikoshiba K.. 1997. Xenopus Zic3, a primary regulator both in neural and neural crest development. Proc. Natl. Acad. Sci. USA. 94:11980–11985. 10.1073/pnas.94.22.11980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J., and Kirschner M.. 1982. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 30:675–686. 10.1016/0092-8674(82)90272-0 [DOI] [PubMed] [Google Scholar]
- Nikolopoulou E., Galea G.L., Rolo A., Greene N.D., and Copp A.J.. 2017. Neural tube closure: cellular, molecular and biomechanical mechanisms. Development. 144:552–566. 10.1242/dev.145904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T., Honda H., and Takeichi M.. 2012. Planar cell polarity links axes of spatial dynamics in neural-tube closure. Cell. 149:1084–1097. 10.1016/j.cell.2012.04.021 [DOI] [PubMed] [Google Scholar]
- Niu J., Profirovic J., Pan H., Vaiskunaite R., and Voyno-Yasenetskaya T.. 2003. G Protein betagamma subunits stimulate p114RhoGEF, a guanine nucleotide exchange factor for RhoA and Rac1: regulation of cell shape and reactive oxygen species production. Circ. Res. 93:848–856. 10.1161/01.RES.0000097607.14733.0C [DOI] [PubMed] [Google Scholar]
- Offermanns S., Zhao L.P., Gohla A., Sarosi I., Simon M.I., and Wilkie T.M.. 1998. Embryonic cardiomyocyte hypoplasia and craniofacial defects in G alpha q/G alpha 11-mutant mice. EMBO J. 17:4304–4312. 10.1093/emboj/17.15.4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okae H., and Iwakura Y.. 2010. Neural tube defects and impaired neural progenitor cell proliferation in Gbeta1-deficient mice. Dev. Dyn. 239:1089–1101. 10.1002/dvdy.22256 [DOI] [PubMed] [Google Scholar]
- Oshita A., Kishida S., Kobayashi H., Michiue T., Asahara T., Asashima M., and Kikuchi A.. 2003. Identification and characterization of a novel Dvl-binding protein that suppresses Wnt signalling pathway. Genes Cells. 8:1005–1017. 10.1111/j.1365-2443.2003.00692.x [DOI] [PubMed] [Google Scholar]
- Ossipova O., Kim K., and Sokol S.Y.. 2015. Planar polarization of Vangl2 in the vertebrate neural plate is controlled by Wnt and Myosin II signaling. Biol. Open. 4:722–730. 10.1242/bio.201511676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parag-Sharma K., Leyme A., DiGiacomo V., Marivin A., Broselid S., and Garcia-Marcos M.. 2016. Membrane Recruitment of the Non-receptor Protein GIV/Girdin (Gα-interacting, Vesicle-associated Protein/Girdin) Is Sufficient for Activating Heterotrimeric G Protein Signaling. J. Biol. Chem. 291:27098–27111. 10.1074/jbc.M116.764431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks S., and Wieschaus E.. 1991. The Drosophila gastrulation gene concertina encodes a G alpha-like protein. Cell. 64:447–458. 10.1016/0092-8674(91)90652-F [DOI] [PubMed] [Google Scholar]
- Peshkin L., Wühr M., Pearl E., Haas W., Freeman R.M. Jr., Gerhart J.C., Klein A.M., Horb M., Gygi S.P., and Kirschner M.W.. 2015. On the Relationship of Protein and mRNA Dynamics in Vertebrate Embryonic Development. Dev. Cell. 35:383–394. 10.1016/j.devcel.2015.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer N.W., Spicher K., Malphurs J., Akiyama H., Abramowitz J., Nürnberg B., and Birnbaumer L.. 2012. Development of the mammalian axial skeleton requires signaling through the Gα(i) subfamily of heterotrimeric G proteins. Proc. Natl. Acad. Sci. USA. 109:21366–21371. 10.1073/pnas.1219810110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggeri G., Timms A.E., Cheng C., Weiss A., Kollros P., Chapman T., Tully H., and Mirzaa G.M.. 2018. Bi-allelic mutations of CCDC88C are a rare cause of severe congenital hydrocephalus. Am. J. Med. Genet. A. 176:676–681. 10.1002/ajmg.a.38592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siletti K., Tarchini B., and Hudspeth A.J.. 2017. Daple coordinates organ-wide and cell-intrinsic polarity to pattern inner-ear hair bundles. Proc. Natl. Acad. Sci. USA. 114:E11170–E11179. 10.1073/pnas.1716522115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive H.L., Grainger R.M., and Harland R.M.. 2000. Early Development of Xenopus laevis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Smrcka A.V. 2013. Molecular targeting of Gα and Gβγ subunits: a potential approach for cancer therapeutics. Trends Pharmacol. Sci. 34:290–298. 10.1016/j.tips.2013.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol S.Y. 2015. Spatial and temporal aspects of Wnt signaling and planar cell polarity during vertebrate embryonic development. Semin. Cell Dev. Biol. 42:78–85. 10.1016/j.semcdb.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol S.Y. 2016. Mechanotransduction During Vertebrate Neurulation. Curr. Top. Dev. Biol. 117:359–376. 10.1016/bs.ctdb.2015.11.036 [DOI] [PubMed] [Google Scholar]
- Stols L., Gu M., Dieckman L., Raffen R., Collart F.R., and Donnelly M.I.. 2002. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr. Purif. 25:8–15. 10.1006/prep.2001.1603 [DOI] [PubMed] [Google Scholar]
- Takagishi M., Sawada M., Ohata S., Asai N., Enomoto A., Takahashi K., Weng L., Ushida K., Ara H., Matsui S., et al. 2017. Daple Coordinates Planar Polarized Microtubule Dynamics in Ependymal Cells and Contributes to Hydrocephalus. Cell Reports. 20:960–972. 10.1016/j.celrep.2017.06.089 [DOI] [PubMed] [Google Scholar]
- Tall G.G., Krumins A.M., and Gilman A.G.. 2003. Mammalian Ric-8A (synembryn) is a heterotrimeric Galpha protein guanine nucleotide exchange factor. J. Biol. Chem. 278:8356–8362. 10.1074/jbc.M211862200 [DOI] [PubMed] [Google Scholar]
- Terry S.J., Zihni C., Elbediwy A., Vitiello E., Leefa Chong San I.V., Balda M.S., and Matter K.. 2011. Spatially restricted activation of RhoA signalling at epithelial junctions by p114RhoGEF drives junction formation and morphogenesis. Nat. Cell Biol. 13:159–166. 10.1038/ncb2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C., and Thisse B.. 2008. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3:59–69. 10.1038/nprot.2007.514 [DOI] [PubMed] [Google Scholar]
- Wallingford J.B.2010. Low-magnification live imaging of Xenopus embryos for cell and developmental biology. Cold Spring Harb Protoc. 2010:pdb.prot5425. [DOI] [PubMed]
- Wallingford J.B., and Harland R.M.. 2002. Neural tube closure requires Dishevelled-dependent convergent extension of the midline. Development. 129:5815–5825. 10.1242/dev.00123 [DOI] [PubMed] [Google Scholar]
- Wallis M., Baumer A., Smaili W., Jaouad I.C., Sefiani A., Jacobson E., Bowyer L., Mowat D., and Rauch A.. 2018. Surprisingly good outcome in antenatal diagnosis of severe hydrocephalus related to CCDC88C deficiency. Eur. J. Med. Genet. 61:189–196. 10.1016/j.ejmg.2017.12.002 [DOI] [PubMed] [Google Scholar]
- Wang S., Cha S.W., Zorn A.M., and Wylie C.. 2013. Par6b regulates the dynamics of apicobasal polarity during development of the stratified Xenopus epidermis. PLoS One. 8:e76854 10.1371/journal.pone.0076854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettschureck N., Moers A., and Offermanns S.. 2004. Mouse models to study G-protein-mediated signaling. Pharmacol. Ther. 101:75–89. 10.1016/j.pharmthera.2003.10.005 [DOI] [PubMed] [Google Scholar]
- Yu S., Yu D., Lee E., Eckhaus M., Lee R., Corria Z., Accili D., Westphal H., and Weinstein L.S.. 1998. Variable and tissue-specific hormone resistance in heterotrimeric Gs protein alpha-subunit (Gsalpha) knockout mice is due to tissue-specific imprinting of the gsalpha gene. Proc. Natl. Acad. Sci. USA. 95:8715–8720. 10.1073/pnas.95.15.8715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Tran U., and Wessely O.. 2018. Polycystin 1 loss of function is directly linked to an imbalance in G-protein signaling in the kidney. Development. 145:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaveling-Soonawala N., Alders M., Jongejan A., Kovacic L., Duijkers F.A., Maas S.M., Fliers E., van Trotsenburg A.S.P., and Hennekam R.C.. 2018. Clues for Polygenic Inheritance of Pituitary Stalk Interruption Syndrome From Exome Sequencing in 20 Patients. J. Clin. Endocrinol. Metab. 103:415–428. 10.1210/jc.2017-01660 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.