Abstract
Background: Autism Spectrum Disorder (ASD) is a neurological disorder characterized by massive damage in various fields of development. Impaired social interaction and communication skills, unusual behavior or interests, and repetitive activities are considerably disabling in these patients. There are several challenges in diagnosis of ASD patients such as co-existing epilepsy, difference in clinician attitudes and possibly multifactorial etiology of autistic behavior among children and adults. Research in recent years has emphasized a possible connection between mutations in PTEN and macrocephaly (head circumference > 97th centile).
Methods: Articles in English Language were searched from international databases including Medline (PubMed), Google Scholar, Scopus, and CINHAL from January 1998 to January 2016.
Results: The results showed that among 2940 patients with behavioral disorders, 2755 individuals had ASD, and 35 cases with macrocephaly had mutations in PTEN. About 77% of the articles (7/9) analyzed mutations in PTEN in patients with head circumference more than 2SD away from the mean, but did not check mutations in this gene in other ASD patients without macrocephaly. To the best of our knowledge, this study is the first systematic review on human PTEN mutations and classical autistic behavior.
Conclusion: We conclude that the presence of macrocephaly may not be sufficient to examine the PTEN mutation in this group; however, surveying this gene in all cases of macrocephaly seems to be necessary.
Keywords: PTEN, Autism Spectrum Disorder, Macrocephaly, Systematic Review
↑ What is “already known” in this topic:
Autism spectrum disorder (ASD) is a heterogeneous group of complex neurodevelopment disorders with common features of intellectual disabilities and neurologic deficits. Previous studies have shown that specific genetic factors are associated with an increased risk of ASD.
→ What this article adds:
Here, we review molecular studies on PTEN in children with classic autism. Screening of PTEN with other genes related to brain development is important in patients with autism and macrocephaly.
Introduction
Autism Spectrum Disorders (ASD) comprise a heterogeneous group of neurodevelopmental disorders that are characterized by significant dysfunction in various fields of development such as social interaction skills, communication skills, behavior, and interests with a tendency towards repetitive activities. The term 'spectrum' is used to cover the broad range of behavioral difficulties that may be common among autistic patients. The main subgroups within the spectrum are classic autism, Pervasive Developmental Disorder– Not Otherwise Specified (PDD-NOS) and Asperger syndrome. PDD-NOS and Asperger syndrome have milder symptoms than classic autism (1, 2).
People with autism are careless and indifferent to feelings of others; in addition, they show unpredictable reactions in sensitive situations. These symptoms are not of the same intensity and can be weak or strong (3). Due to differences in the quality of social and communication skills in ASD, these patients are not diagnosed through clinical examination; instead, the diagnostic and statistical manual for mental disorders is usually used (4, 5). ASD prevalence has tripled since 1970 and research suggests that nowadays ASD has affected one out of 150 children (aged up to eight years) in the United States (6, 7).
ASD usually occurs early in life, often before the age of three, and is more common in boys than in girls with a ratio of four to five times (6). From a different view, ASD is a complex developmental and behavioral disorder that affects brain development and prevents progress in social and communicative skills (4, 5). Many reviews conducted in Iran and other areas have shown that genetic susceptibilities, various immunological and environmental factors are both involved in autistic behaviors. Figure 1 shows some causes of autistic behaviors (8-12). Besides, each subtype of ASD with or without epilepsy may be associated with different risk factors, suggesting a multifactorial etiology (8, 13-15).
Fig. 1.
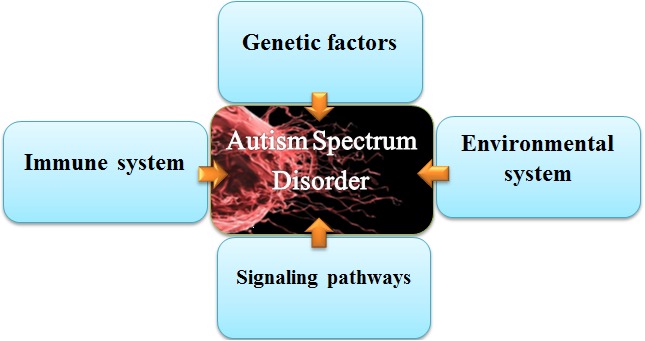
Etiologies of Autism spectrum disorders
Investigations in recent years have identified many environmental (such as exposure to heavy metals) and genetic (involvement of different genes and signaling pathways in brain development) factors that cause autism (16-24) (Table 1). A study by Atladóttir and colleagues in 2009 on 3325 children with autism showed a higher risk of having a child with autism when the family has the history of type 1 diabetes, rheumatoid diseases, and maternal celiac disease. Such familial autoimmune disorders in autism pedigree could well imply a combined etiology of a common genetic predisposition with exposure to antibodies, or with a change in prenatal fetal environment (25).
Table 1. Several risk factors in ASD .
| Several Environment factors | References |
| Exposure to high levels of mercury, styrene and chromium ,trichloroethylene, and vinyl chloride during pregnancy | (18, 20) |
| Factors associated with high levels of vitamin D deficiency | (19) |
| Air pollution | (18) |
| Maternal lifestyle , maternal and paternal age | (22, 23) |
| Several Genetic Factors | References |
| Receptor genes | (17) |
| Ig-CAM genes | (24) |
| Scaffolding genes | (24) |
| PTEN | (12) |
Although the exact cause of autism is not yet clear, studies in recent years have shown that signaling pathways and several genes are involved in both human and mouse brain development (17, 26, 27).
Autism and PTEN Mutations
Homologous Phosphatase and Tensin Gene (PTEN), known as a tumor suppressor gene, is located on chromosome 10 (12, 28). Deletion or mutation in this gene is associated with a large number of human cancers and neurological disorders. Mutations in highly conserved exons of this gene (PTEN ENSG00000171862) can cause a great variety of problems (12, 28-30). Studies show that exons 1 and 9 of this gene have a few coding regions, and the product of this gene is the PTEN protein with 403 amino acids and 5 domains (12, 28, 31). This protein regulates the PI3K signaling pathway through the phosphorylation of PIP3 (12). In addition to cancer, mutation in PTEN can cause disorders such as Cowden syndrome (30), Bannayan-Riley-Ruvalcaba syndrome (BRRS) (15) and autism (12). On the other hand, PTEN mutation can cause cancer or dysfunction in myelin formation, cell polarity, axon/dendrite structure, cell migration, maintenance of neuronal flows and neural stem cell differentiation (32-35). Studies have shown that PTEN with its phosphatase activity and use of extracellular signal transduction can lead to cell polarity (33, 35).
One of the ways of assessing brain development in ASD is by measuring the head circumference which is done in both children and infants. It was suggested that all people with high head circumference (more than 97 centile or 2 standard deviations above the mean) be analyzed for mutations in PTEN (3). Many studies have shown that patients with autism have a larger head circumference than their peers (36, 37); this feature is more evident in boys with autism in their first year of life (38, 39). In 2003, Shi et al. showed that over-expression of PTEN leads to changes in axon structure and neuronal and glial cell population (35). Studies in the past few years have shown different PTEN mutations can cause different phenotypes; for example, PTEN deletion leads to seizure in Cowden syndrome and increases brain size in mouse models (27, 40). Other mutations, depending on the location, influencing PTEN protein show different phenotypes (34, 41, 42).
In general, these studies show that heterogeneity in the etiology of autism is a major reason for classification problems of this disease spectrum. Because of PTEN's role in brain development and macrocephaly in children with autistic behavior, we decided to comprehensively and systematically study all articles written on mutations of human PTEN and their relationship with classic autism.
Methods
Search strategy
The main protocol for this systematic review was designed a priori, and search terms and the inclusion and exclusion criteria were selected in an attempt to access all relevant articles. All original articles were searched from international databases including Medline (PubMed), Google Scholar, Scopus, and CINHAL. We arranged a systematic review of relevant papers published in English between January 1998 to January 2016. In this systematic review in accordance to Figure 2 a carefully designed search strategy was performed to identify relevant papers evaluating the PTEN mutation spectrum in Patients with Macrocephaly and Classic Autism. We searched relevant keywords indexed in the Medical Subject Heading (MeSH) or Embase, and the following search phrases were used: ‘classic autism’, ‘autism spectrum disorders’, ‘human autistic behavior’, ‘PTEN mutation’, ‘Macrocephaly Autism Syndrome’ and ‘macrocephaly.
Fig. 2.
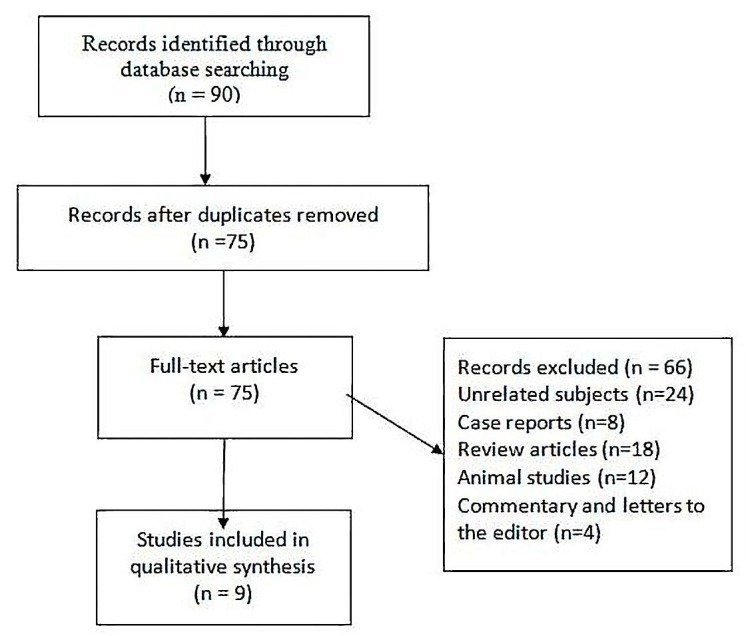
Flow chart of the study selection
Articles were selected in three phases:
First: Three authors independently analyzed the titles and abstracts of all articles.
Second: Unrelated papers were excluded that were not in agreement with the inclusion criteria.
Third: All potentially related studies (based on the inclusion and exclusion criteria) were identified and analyzed in details by 3 researchers separately.
Eligibility criteria
Inclusion and exclusion criteria were as follow:
Inclusion criteria are researching PTEN mutations in classic autism, experimental and non-experimental study designs, quasi-experimental, cohort, case-control, longitudinal, and cross-sectional observational studies.
Exclusion criteria are all those that do not match inclusion criteria, unrelated subjects, case reports, and animal studies. Furthermore, duplicate articles, commentary, and letters to the editor were discarded.
Data Extraction
Three authors reviewed all full-text articles. Some disagreements were resolved by means of discussion. According to Table 2, a structured checklist was designed for the extraction of data on the 1) author, 2) year of published, 3) Ratio of macrocephaly 4) PTEN mutations, 5) and number of patients diagnosed with autism in selected articles. Furthermore, all patients’ information available in the literature including clinical features, mutation type, head circumference, and presence or absence of macrocephaly, family history, mental retardation, speech delay, developmental delay and inheritance of PTEN mutation was analyzed in our article. In our study, all related papers to PTEN mutations and autism were listed by publication date.
Table 2. Ratio of macrocephaly, PTEN mutations and ASD behaviors in previous articles .
| Authors | Ratio of macrocephaly in individuals with PTEN mutations | Descriptions |
Ratio of mutation/ autism |
Ratio of macrocephaly/ autism | References |
|
Butler et al 2005 |
(3/3) | Only ASD patients who had macrocephaly, were analyzed for PTEN mutations | (3/18) |
(18/18) *1 |
(44) |
| Buxbaum et al 2007 | (6/6) | Only ASD patients who had macrocephaly, were analyzed for PTEN mutations | (6/35) | (35/35) | (47) |
|
Gail E Herman et al 2007 |
(2/2) | Only ASD patients who had macrocephaly, were analyzed for PTEN mutations |
(2/16) *3 |
(16/40) | (48) |
|
Orrico A 2008 |
(2/2) | Only ASD patients who had macrocephaly, were analyzed for PTEN mutations | (2/19) | (19/19) | (29) |
|
Varga MS 2009 |
(5/5) | First checked PTEN mutations in all autistic children with or without macrocephaly then head circumferences were analyzed | (5/60) | (27/60) | (49) |
|
O'Roak et al 2012 |
(3/3) | First checked PTEN mutations in all autistic children with or without macrocephaly then head circumferences were analyzed | (3/2446) |
*2 ? |
(43) |
| Vanderver et al 2012 | (7/7) | Only ASD patients who had macrocephaly, were analyzed for PTEN mutations | (7/7) | (7/7) | (26) |
| Marchese et al 2014 | (1/1) | Only ASD patients who had macrocephaly, were analyzed for PTEN mutations | (1/31) | (31/81) | (14) |
|
Hobert et al 2014 |
(6/6) | Only ASD patients who had macrocephaly, were analyzed for PTEN mutations | (6/22) | (22/49) | (52) |
| Results | 35/35 | 77% (7 out of 9) of the articles have assessed PTEN mutations in autistic children with high head circumference (more than 2SD away from the mean), but PTEN mutations in other ASD patients without macrocephaly, have not been analyzed. | 35/2654 |
?/2755 *2 |
*1- All patients had macrocephaly
*2- There was no information about head circumference in other patients without mutations.
*3- In this article PTEN was sequenced in 16 individuals with macrocephaly.
Results
Nine papers were included for final analysis. Table 2 shows the ratio of macrocephaly in individuals with PTEN mutations, the ratio of mutation/autism, the ratio of macrocephaly/autism, sample size and reference and conceptual approach.
In general, 2940 patients were reported with PTEN mutation and behaviors resembling ASD of which 2755 were diagnosed with ASD (classical autism), and 35 cases had mutations in PTEN (Table 2). Eight articles had small sample sizes, and major samples were from O'Roak's study (43), who examined 44 genes in 2446 individuals with autism (Table 2).
Overview of the studies
Butler study in 2005 showed that only 3 of 18 children with autism had mutations in PTEN (H93R (exon 4), D252G (exon 7), and F241S (exon 7)). The average head circumference (HC) of these three children were 5SD away from the mean, and additionally, they had developmental delay but did not show signs of seizure, except one who probably had suspected seizure symptoms. One of the three children had a de novo mutation of H93R (exon4 codon 93 CAT> CGT) without signs of Cowden syndrome and had inherited macrocephaly (+4SD away from the mean) from his father. He had a brother with Down syndrome.(44) According to the article, H93Y mutation had previously been reported in a 46-year-old Japanese man with CS by Kohno et al (44, 45). This alternative study indicated that missense mutations at the same residue could cause different symptoms because different mutations affect PTEN lipid phosphatase activity to certain different degrees. For example, in-vitro studies investigated G129E and G129Rand showed that these mutations could have different impacts on protein and lipid phosphatase activity of PTEN (44).
Buxbaum et al. (2007) surveyed 88 children with ASD and macrocephaly including 35 classic autism cases, but only 45 had met the DSM-IV criteria (Diagnostic and Statistical Manual of Mental Disorders, 4th edition), and 7 had mutations in PTEN; however, mutations in this gene were not detected in other children with macrocephaly (but without classic autism). Authors also examined rare variants in their control sample with different ethnicity groups (Caucasian, African American, Caucasian, Chinese, and Japanese). They found two similar variants (1026C → A / 903G → A) in both patient and control subjects, which Zhou et al. (2003) (15) and Sarquis et al. (2006) (46) had previously reported as polymorphisms. They also found PTEN mutations in all ASD patients including exon 8 (c.976G→ A; D326N), exon 1 (c.66C→G; D22E) and intron 3 (IVS3-9T→C) and four variants in promoter (1088C→T/1084C→T/ 1026C→A/ 903G→A) (47).
In 2007 Herman GE and his colleagues showed that among 71 patients with primary diagnosis of autism based on DSM-IV criteria,(4) 57 had autistic behavior of whom 40 cases of classical autism, 3 Rett syndrome, 10 PDD-NOS, and four Asperger syndrome; however, only 16 patients had been analyzed for PTEN. Of 16 patients, two macrocephaly cases had heterozygous mutations (R130X, 530insT) in PTEN and two had chromosomal abnormalities. In total, macrocephaly was detected in 19 patients (48).
Varga et al. (2009) showed that in 114 patients with ASD, developmental delay/mental retardation and/or macrocephaly, 11 mutations were found, but only 5 of which (R130X,E157G,L139X,520insT, IVS6-3 C>G) were identified in sixty patients with autism symptoms, 27 of whom had macrocephaly. This study showed that all individuals with mutations in PTEN had macrocephaly (=5/5), but the reverse was not true (49).
In contrast, in a short report by Orrico at the same time, it was shown that among 40 patients with neurodevelopmental disorders and macrocephaly, 19 cases (on the basis of diagnostic criteria of autism(50)) had autistic behavior. Two PTEN mutations (p.Y176C, p.N276S) were found among these 40 patients with neurodevelopmental disorders, of whom two had both severe mental retardation (MR) and autism (29).
The study of Schaaf conducted in 2011 as oligogenic heterozygosity in individuals with high functioning autism spectrum disorders was excluded from our study because of inadequate information (51). The following year, O'Roak and colleagues assessed 44 genes in 2446 ASD patients and found 27 new mutations in 16 genes. They believed that CHD8, DYRK1A, GRIN2B, TBR1,PTEN, and TBL1XR1 are involved in etiology of only 1% of ASD sporadic patients. In this study all 3 patients with PTEN mutations (p. T131I/ p. C136MfsX44/ p. T167N) were macrocephalic, and their non-verbal intellectual quotients (NVIQ) were 50, 33 and 77, respectively. This study showed that macrocephaly was not specific for PTEN in ASD; patients with mutation in the CHD8 gene showed a higher range of head circumference than those with PTEN mutations. Microcephaly also was observed in 3 patients with classic autism with mutations in DYRK1A gene in this article (43).
Some of the data from the publication by Genomic Medicine Institute Group in America revealed that among 69 patients with autistic behavior (49 autistics, 13 PDD-NOS, and 7 Asperger’s), 22 had macrocephaly. Moreover, several biochemical analyses such as Asp, Gly, Tau, Cys and Met were measured in urine or plasma in autistic children with or without PTEN mutation. Only 18 children with ASD had macrocephaly and just 6 of them had both mutations in PTEN (p.H141TFS*39, P. L70V, P.M1I, P. R335*, C.209+5G>A, P.M1I) and macrocephaly. Biochemical studies of these children showed that 72% of all children had a reduction in the amount of plasma Cys, and 87 percent showed increasing amounts of urine aspartic acid (52).
In 2014, Marchese and colleagues at the Clinical Neurophysiology Laboratory in Italy analyzed 81 ASD patients (50 autism-epilepsy phenotypes (AEP) and 31 classical ASD)) with or without epilepsy, of which 31 patients had macrocephaly, while only 6 out of 31 had severe macrocephaly. After a survey of PTEN in patients with macrocephaly and ASD, a de novo mutation (c.43delA) was found in one child with severe macrocephaly. This child also had an intellectual disability and seizures (14).
The result of another study conducted by Vanderver et al. (2014) on a survey of 23 patients with unclassified white matter disorders with or without ASD, revealed that 7 individuals with macrocephaly and ASD had 7 mutations in PTEN (26).
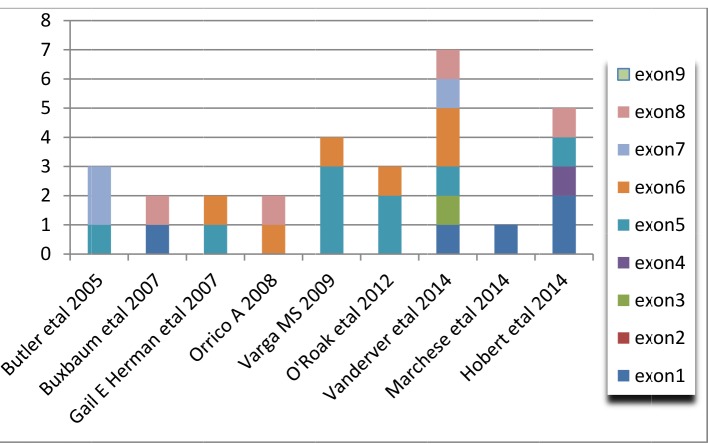
Frequencies of mutations in each exon was found in 9 articles. The studies reviewed in this paper reveal that the exon 5 mutation rate is higher than other PTEN exons. Figure 3 shows the mutation rate of each exon in each study separately. This figure shows that in children with classic autistic behavior, about 28 mutations were found in all studies, ten mutations were related to exon 5, and 6 cases were related to exon 6, respectively; these are the highest percentage of mutations in these studies. Figure 3 shows only coding mutations of patients who are in the classic autistic group, and the rest of the mutations were not included. The information in this figure is derived from databases such as dbSNP, HGMD, and Ensembl. Of course, some of them has been obtained through predictive software and needs to be further investigated.
Fig. 3.
Prevalence of PTEN exons mutation in 9 studies
Selection bias in some articles
In addition, about 77% (7 out of 9) of the articles have assessed PTEN mutations in autistic children with high head circumference (more than 2SD away from the mean), but PTEN mutations in other ASD patients without macrocephaly, have not been analyzed. For example, Marchese et al. (2014) studied 81 ASD patients, 31 of whom with macrocephaly were selected for PTEN mutation analysis (14).
Forty-nine ASD patients were checked by Hobert et al. (2014) for the presence of macrocephaly, then all individuals with head circumferences of more than 2SD away from the mean were analyzed for PTEN mutations (52). However, articles by Varga et al. and O'Roak et al., first checked PTEN mutations in all autistic children with or without macrocephaly; then, head circumferences were analyzed .(43, 49).
O'Roak et al. examined 44 genes, and showed that in addition to PTEN, other genes (CHD8 and DYRK1A) could be involved in changing head circumference; the impact of increasing head circumference maybe even more than PTEN (43).
Discussion
We conducted a Systematic review of the literature on the PTEN mutations in patients with macrocephaly and classic autism. To the best of our knowledge, this is the first review of its type to cover all PTEN mutations in coding regions in classic autism.
Autism is a highly heterogeneous disease with environmental and genetic factors involved in its etiology (9, 13, 17, 53, 54). Studies in the past few decades and the discovery of genes in autistic behaviors have created this belief that genetic factors play an important role in ASD, as well as brain development and neural pathways. Loss of PTEN function in the brain has important effects on neuronal morphogenesis and synaptic connections. The PTEN pathway can interact with other signaling pathways (major PI3K/PTEN/AKT pathway) to control social interaction and communication skills (17, 29, 55).
Unfortunately, due to lack of diagnostic markers and inappropriate classification of ASD, there is no definitive diagnosis and appropriate treatment for these children at birth or in utero.
Various studies have shown that genetics and related factors play an important role in the pathogenesis of autism (12, 29, 54). Thus, genetic screening can be a useful way of diagnosing diseases that are directly related to genotype-phenotype correlations. However, for heterogeneous diseases like autism, it is a very time-consuming and costly task to survey all genes and environmental and immunological factors. Therefore, studies to identify important genes and their effects on the pathogenesis of autism are valuable and useful. We surveyed one of the most important human genes involved in the development of brain in this study. Our study shows that most articles have some traits of bias in their own way (selection bias in choosing patients to check mutations in PTEN) and the presence of macrocephaly in their patients was a reason for PTEN mutation testing. This bias is clearly shown in Table 2.
A few papers with advanced genetic techniques have recently examined PTEN in all classic autism patients with or without severe macrocephaly (53, 55). Detection of both genetic and clinical factors related to ASD with or without macrocephaly can enhance early diagnosis of such children and timely planning of interventions to address their psychological and medical needs.
Conclusion
We conclude that presence of macrocephaly may not be sufficient to examine the PTEN mutation in this group; however, surveying this gene in all cases of macrocephaly seems to be necessary (due to the important role of PTEN in brain development and cancer). In order to achieve effective results, it is necessary to examine the genes involved in cerebral development.
Acknowledgments
We thank all professors and other participants who sincerely helped us during this study.
Financial support and sponsorship
This study was approved and supported by grant no. 94-04-30-26878 by Research Deputy, Iran University of Medical Sciences, Tehran, Iran.
Conflicts of interest
The authors declare no competing financial interest.
Cite this article as: Zahedi Abghari F, Moradi Y, Akouchekian M. PTEN gene mutations in patients with macrocephaly and classic autism: A systematic review. Med J Islam Repub Iran. 2019 (25 Feb);33:10. https://doi.org/10.34171/mjiri.33.10
References
- 1.Benvenuto A, Moavero R, Alessandrelli R, Manzi B, Curatolo P. Syndromic autism: causes and pathogenetic pathways. World J Pediatr. 2009;5(3):169–76. doi: 10.1007/s12519-009-0033-2. [DOI] [PubMed] [Google Scholar]
- 2.Walker DR, Thompson A, Zwaigenbaum L, Goldberg J, Bryson SE, Mahoney WJ. et al. Specifying PDD-NOS: a comparison of PDD-NOS, Asperger syndrome, and autism. J Am Acad Child Psy. 2004;43(2):172–80. doi: 10.1097/00004583-200402000-00012. [DOI] [PubMed] [Google Scholar]
- 3.Carter M, Scherer S. Autism spectrum disorder in the genetics clinic: a review. Clin Genet. 2013;83(5):399–407. doi: 10.1111/cge.12101. [DOI] [PubMed] [Google Scholar]
- 4.King BH, Navot N, Bernier R, Webb SJ. Update on diagnostic classification in autism. Curr Opin Psychiatr. 2014;27(2):105–9. doi: 10.1097/YCO.0000000000000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Volkmar FR, State M, Klin A. Autism and autism spectrum disorders: diagnostic issues for the coming decade. J Child Psychol Psyc. 2009;50(1-2):108–15. doi: 10.1111/j.1469-7610.2008.02010.x. [DOI] [PubMed] [Google Scholar]
- 6.Johnson CP, Myers SM. Identification and evaluation of children with autism spectrum disorders. Pediatrics. 2007;120(5):1183–215. doi: 10.1542/peds.2007-2361. [DOI] [PubMed] [Google Scholar]
- 7.Van Naarden Braun K, Pettygrove S, Daniels J, Miller L, Nicholas J, Baio J. et al. Evaluation of a methodology for a collaborative multiple source surveillance network for autism spectrum disorders--Autism and Developmental Disabilities Monitoring Network, 14 sites, United States, 2002. MMWR Surveill Summ. 2007;56(1):29–40. [PubMed] [Google Scholar]
- 8.Lord C, Cook EH, Leventhal BL, Amaral DG. Autism spectrum disorders. Neuron. 2000;28(2):355–63. doi: 10.1016/s0896-6273(00)00115-x. [DOI] [PubMed] [Google Scholar]
- 9.Kinney DK, Barch DH, Chayka B, Napoleon S, Munir KM. Environmental risk factors for autism: do they help cause de novo genetic mutations that contribute to the disorder? Med Hypotheses. 2010;74(1):102–6. doi: 10.1016/j.mehy.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shelton JF, Hertz-Picciotto I, Pessah IN. Tipping the balance of autism risk: potential mechanisms linking pesticides and autism. Environ Health Persp. 2012;120(7):944. doi: 10.1289/ehp.1104553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Chauhan A, Sheikh AM, Patil S, Chauhan V, Li XM. et al. Elevated immune response in the brain of autistic patients. J Neuroimmunol. 2009;207(1):111–6. doi: 10.1016/j.jneuroim.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou J, Parada LF. PTEN signaling in autism spectrum disorders . Curr Opin Neurobiol. 2012;22(5):873–9. doi: 10.1016/j.conb.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Robinson EB, Neale BM, Hyman SE. Genetic research in autism spectrum disorders. Curr Opin Pediatr. 2015;27(6):685. doi: 10.1097/MOP.0000000000000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marchese M, Conti V, Valvo G, Moro F, Muratori F, Tancredi R. et al. Autism-epilepsy phenotype with macrocephaly suggests PTEN, but not GLIALCAM, genetic screening. BMC Med Genet. 2014;15(1):26. doi: 10.1186/1471-2350-15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou XP, Waite KA, Pilarski R, Hampel H, Fernandez MJ, Bos C. et al. Germline PTEN promoter mutations and deletions in Cowden/Bannayan-Riley-Ruvalcaba syndrome result in aberrant PTEN protein and dysregulation of the phosphoinositol-3-kinase/Akt pathway . Am J Hum Genet. 2003;73(2):404–11. doi: 10.1086/377109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yassa HA. Autism: A form of lead and mercury toxicity. Environ Toxicol Phar. 2014;38(3):1016–24. doi: 10.1016/j.etap.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Rosti RO, Sadek AA, Vaux KK, Gleeson JG. The genetic landscape of autism spectrum disorders. Dev Med Child Neurol. 2014;56(1):12–8. doi: 10.1111/dmcn.12278. [DOI] [PubMed] [Google Scholar]
- 18.Talbott EO, Marshall LP, Rager JR, Arena VC, Sharma RK, Stacy SL. Air toxics and the risk of autism spectrum disorder: the results of a population based case–control study in southwestern Pennsylvania. Environ Health. 2015;14(1):80. doi: 10.1186/s12940-015-0064-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kočovská E, Fernell E, Billstedt E, Minnis H, Gillberg C. Vitamin D and autism: clinical review. Res Dev Disabil. 2012;33(5):1541–50. doi: 10.1016/j.ridd.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 20.Angelidou A, Asadi S, Alysandratos K-D, Karagkouni A, Kourembanas S, Theoharides TC. Perinatal stress, brain inflammation and risk of autism-review and proposal. BMC Pediatr. 2012;12(1):89. doi: 10.1186/1471-2431-12-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Windham GC, Zhang L, Gunier R, Croen LA, Grether JK. Autism spectrum disorders in relation to distribution of hazardous air pollutants in the San Francisco Bay area. Environ Health Persp. 2006;114(9):1438. doi: 10.1289/ehp.9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grether JK, Anderson MC, Croen LA, Smith D, Windham GC. Risk of autism and increasing maternal and paternal age in a large north American population. Am J Epidemiol. 2009;170(9):1118–26. doi: 10.1093/aje/kwp247. [DOI] [PubMed] [Google Scholar]
- 23.Lyall K, Schmidt RJ, Hertz-Picciotto I. Maternal lifestyle and environmental risk factors for autism spectrum disorders. Int J Epidemiol. 2014;43(2):443–64. doi: 10.1093/ije/dyt282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bourgeron T. A synaptic trek to autism. Curr Opin Neurobiol. 2009;19(2):231–4. doi: 10.1016/j.conb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Atladóttir HÓ, Pedersen MG, Thorsen P, Mortensen PB, Deleuran B, Eaton WW. et al. Association of family history of autoimmune diseases and autism spectrum disorders. Pediatrics. 2009;124(2):687–94. doi: 10.1542/peds.2008-2445. [DOI] [PubMed] [Google Scholar]
- 26.Vanderver A, Tonduti D, Kahn I, Schmidt J, Medne L, Vento J. et al. Characteristic brain magnetic resonance imaging pattern in patients with macrocephaly and PTEN mutations . Am J Med Genet A. 2014;164A(3):627–33. doi: 10.1002/ajmg.a.36309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Backman SA, Stambolic V, Suzuki A, Haight J, Elia A, Pretorius J. et al. Deletion of PTEN in mouse brain causes seizures, ataxia and defects in soma size resembling Lhermitte-Duclos disease . Nat Genet. 2001;29(4):396–403. doi: 10.1038/ng782. [DOI] [PubMed] [Google Scholar]
- 28.Worby CA, Dixon JE. PTEN. Annu Rev Biochem. 2014;83:641–69. doi: 10.1146/annurev-biochem-082411-113907. [DOI] [PubMed] [Google Scholar]
- 29.Orrico A, Galli L, Buoni S, Orsi A, Vonella G, Sorrentino V. Novel PTEN mutations in neurodevelopmental disorders and macrocephaly . Clin Genet. 2009;75(2):195–8. doi: 10.1111/j.1399-0004.2008.01074.x. [DOI] [PubMed] [Google Scholar]
- 30.Goffin A, Hoefsloot LH, Bosgoed E, Swillen A, Fryns JP. PTEN mutation in a family with Cowden syndrome and autism . Am J Med Genet. 2001;105(6):521–4. doi: 10.1002/ajmg.1477. [DOI] [PubMed] [Google Scholar]
- 31.Bermudez Brito M, Goulielmaki E, Papakonstanti EA. Focus on PTEN Regulation . Front Oncol. 2015;5:166. doi: 10.3389/fonc.2015.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Diepen MT, Eickholt BJ. Function of PTEN during the formation and maintenance of neuronal circuits in the brain . Dev Neurosci. 2007;30(1-3):59–64. doi: 10.1159/000109852. [DOI] [PubMed] [Google Scholar]
- 33.Veleva-Rotse BO, Barnes AP. Brain patterning perturbations following PTEN loss . Front Mol Neurosci. 2014;7:35. doi: 10.3389/fnmol.2014.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyu J, Yu X, He L, Cheng T, Zhou J, Cheng C. et al. The protein phosphatase activity of PTEN is essential for regulating neural stem cell differentiation . Mol Brain. 2015;8:26. doi: 10.1186/s13041-015-0114-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi SH, Jan LY, Jan YN. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell. 2003;112(1):63–75. doi: 10.1016/s0092-8674(02)01249-7. [DOI] [PubMed] [Google Scholar]
- 36.Aylward EH, Minshew NJ, Field K, Sparks B, Singh N. Effects of age on brain volume and head circumference in autism. Neurology. 2002;59(2):175–83. doi: 10.1212/wnl.59.2.175. [DOI] [PubMed] [Google Scholar]
- 37.Courchesne E, Carper R, Akshoomoff N. Evidence of brain overgrowth in the first year of life in autism. Jama. 2003;290(3):337–44. doi: 10.1001/jama.290.3.337. [DOI] [PubMed] [Google Scholar]
- 38.Campbell DJ, Chang J, Chawarska K. Early generalized overgrowth in autism spectrum disorder: prevalence rates, gender effects, and clinical outcomes. J Am Acad Child Psy. 2014;53(10):1063–73. doi: 10.1016/j.jaac.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chawarska K, Campbell D, Chen L, Shic F, Klin A, Chang J. Early generalized overgrowth in boys with autism. Arch Gen Psychiat. 2011;68(10):1021–31. doi: 10.1001/archgenpsychiatry.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwon C-H, Luikart BW, Powell CM, Zhou J, Matheny SA, Zhang W. et al. PTEN regulates neuronal arborization and social interaction in mice . Neuron. 2006;50(3):377–88. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marsh DJ, Dahia P, Caron S, Kum JB, Frayling IM, Tomlinson I. et al. Germline PTEN mutations in Cowden syndrome-like families . J Med Genet. 1998;35(11):881–5. doi: 10.1136/jmg.35.11.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eng C. PTEN: one gene, many syndromes. Hum Mutat. 2003;22(3):183–98. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- 43.O'Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG. et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338(6114):1619–22. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butler MG, Dasouki MJ, Zhou XP, Talebizadeh Z, Brown M, Takahashi TN. et al. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations . J Med Genet. 2005;42(4):318–21. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kohno T, Takahashi M, Fukutomi T, Ushio K, Yokota J. Germline mutations of the PTEN/MMAC1 gene in Japanese patients with Cowden disease. Cancer Sci. 1998;89(5):471–4. doi: 10.1111/j.1349-7006.1998.tb03285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarquis MS, Agrawal S, Shen L, Pilarski R, Zhou XP, Eng C. Distinct expression profiles for PTEN transcript and its splice variants in Cowden syndrome and Bannayan-Riley-Ruvalcaba syndrome . Am J Hum Genet. 2006;79(1):23–30. doi: 10.1086/504392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buxbaum JD, Cai G, Chaste P, Nygren G, Goldsmith J, Reichert J. et al. Mutation screening of the PTEN gene in patients with autism spectrum disorders and macrocephaly . Am J Med Genet B Neuropsychiatr Genet. 2007;144b(4):484–91. doi: 10.1002/ajmg.b.30493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herman GE, Henninger N, Ratliff-Schaub K, Pastore M, Fitzgerald S, McBride KL. Genetic testing in autism: how much is enough? Genet Med. 2007;9(5):268–74. doi: 10.1097/gim.0b013e31804d683b. [DOI] [PubMed] [Google Scholar]
- 49.Varga EA, Pastore M, Prior T, Herman GE, McBride KL. The prevalence of PTEN mutations in a clinical pediatric cohort with autism spectrum disorders, developmental delay, and macrocephaly . Genet Med. 2009;11(2):111–7. doi: 10.1097/GIM.0b013e31818fd762. [DOI] [PubMed] [Google Scholar]
- 50.Kulage KM, Smaldone AM, Cohn EG. How will DSM-5 affect autism diagnosis? A systematic literature review and meta-analysis. J Autism Dev Disord. 2014;44(8):1918–32. doi: 10.1007/s10803-014-2065-2. [DOI] [PubMed] [Google Scholar]
- 51.Schaaf CP, Sabo A, Sakai Y, Crosby J, Muzny D, Hawes A. et al. Oligogenic heterozygosity in individuals with high-functioning autism spectrum disorders. Hum Mol Genet. 2011;20(17):3366–75. doi: 10.1093/hmg/ddr243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hobert JA, Embacher R, Mester JL, Frazier TW, 2nd 2nd, Eng C. Biochemical screening and PTEN mutation analysis in individuals with autism spectrum disorders and macrocephaly . Eur J Hum Genet. 2014;22(2):273–6. doi: 10.1038/ejhg.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frazier TW, Embacher R, Tilot AK, Koenig K, Mester J, Eng C. Molecular and phenotypic abnormalities in individuals with germline heterozygous PTEN mutations and autism . Mol Psychiatry. 2015;20(9):1132–8. doi: 10.1038/mp.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chaste P, Leboyer M. Autism risk factors: genes, environment, and gene-environment interactions. Dialogues Clin Neurosci. 2012;14(3):281–92. doi: 10.31887/DCNS.2012.14.3/pchaste. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tammimies K, Marshall CR, Walker S, Kaur G, Thiruvahindrapuram B, Lionel AC. et al. Molecular diagnostic yield of chromosomal microarray analysis and whole-exome sequencing in children with autism spectrum disorder. Jama. 2015;314(9):895–903. doi: 10.1001/jama.2015.10078. [DOI] [PubMed] [Google Scholar]