Abstract
Nucleic acid amplification tests (NAATs) provide high diagnostic accuracy for infectious diseases and quantitative results for monitoring viral infections. The majority of NAATs require complex equipment, cold chain dependent reagents, and skilled technicians to perform tests. This largely confines NAATs to centralized laboratories and can significantly delay appropriate patient care. Low-cost, point-of-care (POC) NAATs are especially needed in low-resource settings to provide patients with diagnosis and treatment planning in a single visit to improve patient care. In this work, we present a rapid POC NAAT with integrated sample preparation and amplification using electrokinetics and paper substrates. We use simultaneous isotachophoresis (ITP) and recombinase polymerase amplification (RPA) to rapidly extract, amplify, and detect target nucleic acids from serum and whole blood in a paper-based format. We demonstrate simultaneous ITP and RPA can consistently detect 5 copies per reaction in buffer and 10,000 copies per milliliter of human serum with no intermediate user steps. We also show preliminary extraction and amplification of DNA from whole blood samples. Our test is rapid (results in less than 20 minutes) and made from low-cost materials, indicating its potential for detecting infectious diseases and monitoring viral infections at the POC in low resource settings.
Graphical Abstract
INTRODUCTION
Nucleic acid amplification tests (NAATs) have replaced immunoassays, cell culture, microscopy, and other techniques to become the gold-standard for the accurate diagnosis of many infectious diseases.1–3 NAATs have been developed for numerous diseases due to their high diagnostic sensitivity and specificity, rapid time to result, and multiplexing strategies.4,5 NAATs are also employed by clinicians to quantify the viral load for several infectious diseases (e.g. HIV-1, Hepatitis B, Hepatitis C), providing crucial information for evaluating patient treatment plans.6–8 The majority of NAATs are performed in upper or mid-tier laboratories because they require complex protocols, cold chain dependent reagents, delicate instrumentation, reliable electrical power, qualified laboratory staff, and appropriate infrastructure to host equipment and materials. Laboratory-based NAATs necessitate significant logistics around specimen collection, transport, batched testing, and the return of results to clinicians and patients. In low resource settings this often results in delayed diagnoses that can negate patient management benefits.9 The challenges associated with laboratory-based NAATs have prompted the development of simple, affordable, and rapid point-of-care (POC) NAATs that can facilitate diagnosis and treatment in a single clinic visit.10,11 POC NAATs are especially needed in low resource settings where laboratory testing is limited and rapid testing is crucial for improved treatment (e.g. mitigating loss to follow-up which can lead to treatment failure and spread of disease).12,13
The World Health Organization developed the ASSURED design criteria (Affordable, Sensitive, Specific, User-friendly, Rapid and robust, Equipment-free, and Delivered) for POC diagnostics that describes the required qualities that adequately address the needs of patients in developing countries where infectious disease prevalence remains high.14 Significant challenges remain in the development of POC NAATs that perform sample-to-answer analysis with clinical specimens while meeting the ASSURED criteria. Towards this goal, several POC NAAT platforms are now commercially available including the Alere i, Alere q (Abbott Diagnostics), cobas Liat (Roche Diagnostics), and GeneXpert (Cepheid).15 These platforms offer a range of diagnostic tests for infectious diseases of global significance including HIV, influenza, and tuberculosis and many have been CLIA-waived.16–18 The tests provide rapid, single-step diagnostic results through automating the three operational steps of NAATs; namely, sample preparation, nucleic acid amplification, and detection of amplification. These commercial platforms employ mechanical systems for fluidic manipulation and precision heating that increase platform costs and electric power demands. Further innovation is still needed to lower the cost of capital equipment, perform quantitative tests, and reduce the complexity and costs of test cartridges for sustained use in low and middle income countries.
The integration of sample preparation, amplification, and detection into a robust, low-cost product is a formidable challenge. Significant research has been focused on individual strategies for each of these steps. NAATs can be performed on clinical specimens (e.g. blood, urine, sputum) that contain an array of complex biomolecules, and cellular debris that may inhibit downstream amplification and detection of nucleic acids from the target pathogen.13 Sample preparation is required to lyse the cell wall or virus envelope of pathogens and to then separate and concentrate target nucleic acids from other potent inhibitors in the sample. Lysis is commonly performed using mechanical, chemical, thermal, and enzymatic methods, or a combination therof.19,20 A widely-used method for nucleic acid purification in diagnostic tests is solid phase extraction (SPE), which requires multiple buffer exchanges to create purified target nucleic acids for amplification.21 Several studies have translated SPE to paper-based formats for the POC; however, they still require user steps for physical manipulation and buffer exchanges.22,23 Highly efficient nucleic acid extraction with no intermediate user steps remains a challenge in developing low cost diagnostics appropriate for the POC. An electrokinetic extraction technique called isotachophoresis (ITP) separates nucleic acids from complex samples using an electric field and a discontinuous buffer system of the leading electrolyte (LE) and trailing electrolyte (TE).24,25 ITP is a powerful separation and concentration technique that focuses charged species with electrophoretic mobilities less than the LE and greater than the TE into a plug at the interface of the two buffers. ITP in microchannels is capable of highly efficient purification of nucleic acids from complex samples.26–28 Recently, ITP in paper substrates was utilized to extract and preconcentrate fluorescent dyes and DNA in pure buffer.29–31 Paper-based ITP formats are well suited to POC diagnostic applications due to their reduced operational complexity, low material costs, ease of buffer control, and capacity for large sample volumes.29 No studies have demonstrated paper-based ITP for nucleic acid extraction from complex samples.
Following extraction of target nucleic acids, laboratory NAATs often use polymerase chain reaction (PCR) to amplify nucleic acids. This technique can be difficult to implement at the point-of-care due to the need for highly purified nucleic acids and energy-intensive thermocycling equipment that typically cannot tolerate the environmental extremes in many resource limited settings (e.g. dust, high humidity, high temperature).32 In the last two decades, several isothermal nucleic acid amplification methods (e.g. iSDA, HDA, LAMP, RPA) have been developed to amplify nucleic acids at a single reaction temperature and are more tolerant to inhibitors than traditional PCR.33–37 Isothermal amplification has been explored for POC NAATs using simple resistive heaters, water baths, or chemical heaters.38,39 Recombinase Polymerase Amplification (RPA) is a promising isothermal amplification strategy that is ideal for use in POC NAATs due to its sensitivity (<10 copies/rxn), specificity, speed (<20 minutes), low constant incubation temperature (25–43 °C), tolerance to inhibitors, and reagent stability at ambient temperatures.40,41 RPA amplicon detection can be performed using either endpoint techniques, such as gel electrophoresis, or by measuring fluorescence in real-time. Fluorescence detection of RPA involves a sequence-specific probe which creates a signal only after hybridizing to the target nucleic acid sequence and being cleaved by an exonuxclease.36 Real-time fluorescence monitoring of RPA requires moderately complex instrumentation, yet it can provide semi-quantitative information to assess severity of disease infection.42
Diagnostics researchers have investigated varied approaches for integrating sample preparation, amplification, and detection to create POC NAAT devices.43–47 Paper-based devices are common in the field due to their wicking properties, reagent storage, low cost, and ease of fabrication using minimal equipment. Rodriguez et al. developed a paper-based diagnostic for diagnosis of human papillomavirus (HPV) 16 DNA from clinical cervical samples.48 Sample preparation, amplification, and detection all occurs within the porous network, yet manual buffer exchanges are required. Lafleur et al. demonstrated an integrated paper-based device to provide sample-to-result analysis of nasal swab specimens.49 Their battery-powered approach performs sample lysis, dilution, isothermal amplification, and qualitative colorimetric detection in approximately one hour. Nucleic acid purification was not included in this work, though the presence of inhibiting materials make this a necessary step for sensitive detection when the target pathogen burden is low. These studies present promising platforms for integrating NAAT operations yet still require further development to process more complex samples and eliminate intermediate user steps.
One of the key challenges facing integrated NAAT diagnostics is the automated exchange of lysis, wash, elution buffers, and/or amplification reagents necessary for sensitive detection of pathogens. Two recent NAAT studies have leveraged ITP for integration of nucleic acid extraction and purification with nucleic acid amplification. Borysiak et al. developed an integrated microfluidic diagnostic that employed ITP to purify E. coli nucleic acids present in milk, along with heat activated pumps and capillary valves to drive the nucleic acids into a LAMP amplification reaction chamber.50 In this work, the only user steps were the application of the electric field and initiating heating for amplification. Eid et al. reported a similar assay leveraging ITP and RPA for L. monocytogenes detection from whole blood.51 They extracted nucleic acids into a reservoir using ITP, and then either pipetted target nucleic acids out of the chip into a separate tube or manually added necessary reagents into the chip reservoir for subsequent screening via RPA. Both studies utilized ITP for extraction followed by isothermal amplification in a separate well or chamber. However, there have been no studies to date that use either microchannels or porous substrates to concurrently perform both ITP and amplification—an important development for eliminating intermediate user steps and moving towards developing low cost components for scaled manufacturing.
In this study, we present a paper-based NAAT that integrates ITP and RPA to simultaneously extract and amplify target nucleic acids from serum or whole blood in less than 20 minutes. By applying an electric field, nucleic acids are isolated from the complex sample and focused with RPA reagents within an ITP plug where amplification occurs in a few minutes. A sequence-specific fluorescent probe enables real-time detection and provides sensitive and semi-quantitative results. We first determine the minimum number of copies per reaction required for RPA amplification within an ITP plug in a clean buffer sample. We then report on the limit of detection (LoD) and semi-quantitative results for simultaneous ITP-RPA using human serum. Finally, we present preliminary data detecting nucleic acids from whole blood via the use of a passive filter membrane that separates plasma from the whole blood.52 Simultaneous ITP-RPA in our low-cost paper-based device provides rapid results from relatively large sample volumes (20 μL of serum or 50 μL of whole blood), requiring no user steps in between sample preparation and amplification/detection. We propose that this device can decrease the complexity of traditional laboratory NAATs at a lower cost than POC NAATs that have recently entered the market.
EXPERIMENTAL SECTION
Simultaneous ITP-RPA overview and operation
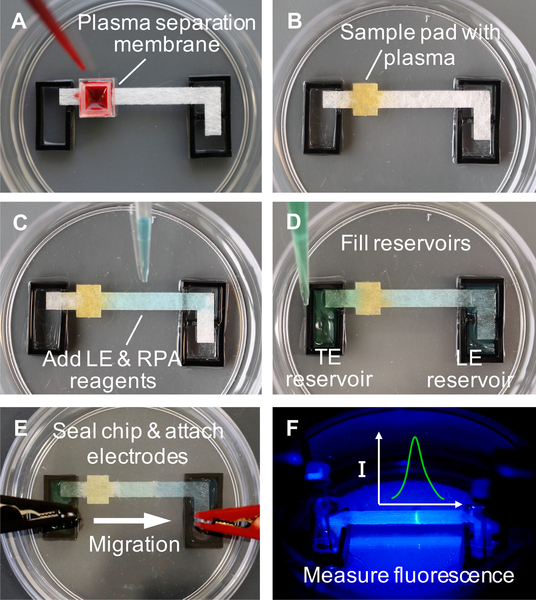
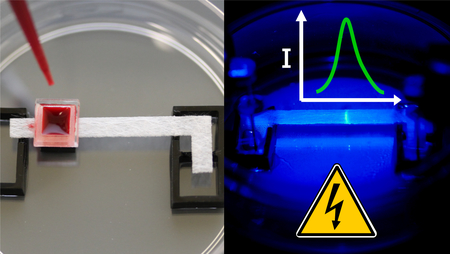
We conducted experiments with disposable devices that consist of a 30 by 3.5 mm porous glass fiber strip (GFCP203000, EMD Millipore, USA) placed between two liquid buffer reservoirs and housed within a sealed 60 mm petri dish (Fisher Scientific, USA), as shown in Figure 1A. The glass fiber strip and 3 mm thick acrylic reservoirs were fabricated with a CO2 laser cutter (Universal Laser Systems, USA). 22 gauge titanium wire electrodes (Unkamen Supplies, USA) are placed in the electrolyte reservoir and connected to a high voltage source meter. We place the device on top of a simple resistive heater plate (Mr. Coffee, USA), maintained at 36 °C using a temperature controller, to aid in heating the reaction and ensure consistency between experiments. In whole blood experiments, a 5 by 5 mm plasma separation membrane (Vivid GR, Pall Corporation, USA is placed over the sample pad area and used to separate plasma from whole blood.
Figure 1.
Operational steps of simultaneous ITP-RPA from whole blood. (A) Add 50 μL whole blood sample spiked with target DNA to plasma separation membrane which removes red and white blood cells. (B) Filtered plasma (~20 μL) wicks into the square sample pad region on the left side of the glass fiber strip, which is pretreated with proteinase K. The glass fiber strip is 3.5 mm wide and the square sample region is 4 mm long by 5 mm wide in order to accommodate at least 20 μL of plasma. Protein digestion proceeds for 3 minutes within the sample pad. Yellow dye is added to aid visualization of the plasma in these images. (C) Add a mixture of LE and RPA reagents to the center of the glass fiber strip, wetting the region from the sample pad to the LE reservoir. (D) Reservoirs are filled respectively with LE and TE solutions. (E) Seal the chip with a plastic lid embedded with electrodes. The positive electrode is submerged in the LE reservoir and the negative in the TE reservoir. The applied electric field initiates ITP-RPA and extracts nucleic acids from the filtered plasma. (F) A microscope records the fluorescence intensity emanating from the amplification reaction zone in the ITP plug.
Figure 1A shows whole blood (Bloodworks Northwest, USA) pipetted onto a plasma separation membrane where capillary action fills the sample pad with plasma while the red and white blood cells are removed via filtration. Experiments that used sterile-filtered human serum (Sigma-Aldrich, USA) did not require the use of the plasma separation membrane. To digest proteins that may interfere with the nucleic acid purification or amplification, the sample pad region of the glass fiber strip is pretreated by spotting with 5 μL of 0.05 μg/μL proteinase K (P8107S, New England Biolabs, USA) and 0.1% Triton X-100 (9002–93-1, Sigma-Aldrich, USA) and dried in a desiccator for 20 minutes. We allow 3 minutes for plasma protein digestion (Figure 1B).
The ITP buffers were developed to separate nucleic acids and RPA reagents from digested plasma that is rich with salts, polypeptides, and other biomolecules that inhibit nucleic acid amplification. ITP also removes the proteinase K (pI~8.9) as it is positively charged in our buffers. We employ a finite injection ITP configuration where sample spiked with target synthetic HIV-1 DNA is added directly to the sample pad of the membrane, separating the two ITP buffers: high mobility leading electrolyte (LE) and low mobility trailing electrolyte (TE). Finite injection provides superior nucleic acid extraction efficiency while still allowing adequate separation from amplification inhibitors in the sample.25
Commercially-available lyophilized RPA reagents (TwistAmp Exo, TwistDx Ltd., UK) are rehydrated in a solution of LE, RPA primers, and RPA probe. This reaction solution is added to the center of the glass fiber strip, wetting the region between the sample pad and LE reservoir through capillary action as shown in Figure 1C. The reservoirs are filled with LE and TE solutions, respectively, and form a fluidic connection through the wetted glass fiber strip (Figure 1D). Figure 2A shows a schematic depicting the location and concentration of the sample, target, and electrolytes before the experiment is initiated. Note that the RPA reagents are initially spatially separated from the target nucleic acids, which electromigrate to the center of the strip to mix with the RPA reagents and subsequently amplify. The chip is sealed with a plastic lid embedded with titanium electrodes which are positioned into the reservoirs, as shown in Figure 1E.
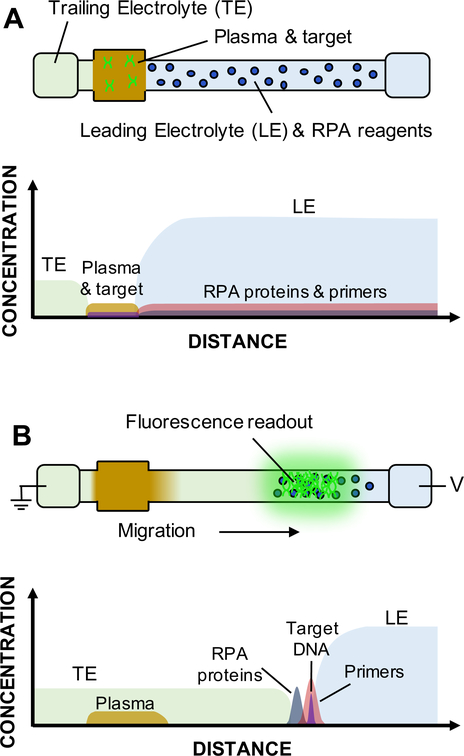
Figure 2.
Schematic of simultaneous ITP-RPA operation with filtered plasma. We show drawings of the glass fiber strip denoting locations of buffers, reagents, and sample, for the initial (A) and final (B) experimental time points. Approximate concentrations of each constituent are plotted with respect to distance. (A) Following passive filtration of the whole blood, plasma containing target DNA initially wets the sample pad on the left side of strip. LE and RPA reagents are disposed within the glass fiber membrane between the sample region and the LE reservoir. Pure LE and TE solutions reside in respective reservoirs. Initial spatial separation of reaction constituents prevents inhibitors in the sample from interacting with the amplification reagents. (B) Applying an electric field extracts DNA from the sample and focuses it with RPA reagents in the ITP plug. All constituents in the porous membrane electromigrate based on their respective charge and electrophoretic mobility. Nucleic acids and RPA proteins/enzymes speed past components of the plasma to focus at the interface of the LE and TE. The amplification reaction takes place within the concentrated ITP plug to create amplicons and detectable fluorescence.
Simultaneous ITP-RPA is initiated by the application of a voltage bias to the electrodes. ITP migrates the nucleic acids from the digested plasma or serum and focuses them with RPA reagents into the ITP plug at the interface of the TE and LE on the porous substrate. The RPA reaction initiates within the plug and target DNA is amplified. RPA-specific amplicons produced in the glass fiber membrane are detected using a sequence-specific fluorescent probe. The signal emanating from the plug is measured using epifluorescence microscopy, as shown in Figure 1F. In Figure 2B, we show concentration profiles of the RPA components and amplified target DNA developed within the ITP plug.
ITP and RPA conditions
The same TE and LE buffers were used for all the experiments. The TE buffer consists of β-alanine (107–95-9, Sigma), Tris (77–86-1, Sigma), polyvinylpyrrolidone (PVP) (9003–39-8, Sigma) and Triton X-100 at pH 8.9–9.1. The LE buffer consists of HCl (7647–01-0, Acros Organics, Belgium), Tris, MgCl2 (7786–30-3, Sigma), polyethylene glycol (PEG) average molecular weight 1,450 (25322–68-3, Sigma), PVP, Triton X-100, and tetramethylammonium chloride (75–57-0, Sigma) at pH 8.1. Reservoirs are filled respectively with 220 μL of LE and 125 μL of TE.
We developed a custom RPA reaction chemistry that is compatible with amplification in glass fiber membranes and the unique chemical composition required for ITP. The reaction solution consists of an RPA pellet rehydrated with LE solution, 1 μM forward primer, 1 μM reverse primer (Integrated DNA Technologies, USA), and 250 nM of sequence specific fluorescent probe (Biosearch Technologies, USA). In order to simplify proof-of-concept experiments, we use a synthetic DNA target (200 base pairs) synthesized using the gBlock gene sequence technology (Integrated DNA Technologies, USA). The sequence is adopted from the proviral HIV-1 DNA pol gene and contains the complementary sequence for the primers and probe adopted from Boyle et al.53 Full primer, probe, and target sequences are listed in the Supporting Information (SI).
Simultaneous ITP-RPA is performed by applying 150 V with a current compliance of 3.5 mA using a source meter (Model 2410, Keithley, USA). This resulted in 3.5 mA constant current conditions for the first 5 minutes of ITP before transitioning to a constant 150 V with current reducing exponentially to less than 1.0 mA after 15 minutes of electromigration (see Figure S-3 of the SI). These relatively high electric currents result from the selected ITP buffers and glass fiber strip’s large cross sectional area. Due to the low conductivity of the TE zone, a high electric field develops in this area, resulting in significant Joule heating. We leverage this effect to heat the ITP plug to the optimal temperature for RPA (35–40°C). Different temperature ranges can be achieved depending on the applied voltage, the composition of the ITP buffers, and the dimensions of the glass fiber strip (see the SI for thermography images of the strip). While this work utilized a resistive heater plate to aid heat control and provide consistency, we observed simultaneous ITP-RPA can be run using solely Joule heating to achieve an appropriate temperature for RPA within the ITP plug.
Data collection and analysis
We perform quantitative epifluorescence imaging (AZ100, Nikon, Japan) with a 0.5X (N=0.05) objective to visualize the fluorescent signal generated by the exonuclease-mediated digestion of the probe bound to RPA amplicons. Using an epifluorescence excitation (488 nm) and emission (518 nm) filter cube set (Omega Optics, USA) and a 16-bit, cooled CCD camera (Cascade 512B, Photometrics, USA), we captured grayscale images of fluorescence at one second intervals over a 15-minute period. The generated data was processed via a custom algorithm (MATLAB, MathWorks, USA) to determine the spatial and temporal fluorescence intensity. The algorithm subtracts the initial background from all images and computes the y-average intensity along the length of the strip (i.e. x-direction). This results in one-dimensional intensity profiles with respect to the length of the strip for distinct time points. The profiles are integrated over the x-direction to calculate a bulk fluorescence intensity for each time point. We employ a thresholding technique to eliminate some non-specific signal due to probe and enzyme interactions. For plotting amplification curves, fluorescence intensity values less than 114% of the initial average signal from the strip are neglected in calculating bulk fluorescence. The LoD calculation used a fluorescence threshold of 50% of the average initial signal in order to account for variations in non-specific signal from the no template controls. The full signal processing procedure is provided in the SI.
The limit of detection (LoD) is the lowest level of analyte that can reliably be measured and is determined according to the Clinical and Laboratory Standards Institute’s (CLSI) recommendations for in vitro diagnostic tests.54,55 The LoD is first determined as a signal intensity and then converted into corresponding analyte concentration (i.e. copies/mL).55 The sample intensity LoD is determined as LoD = μB + 1.645 σb + 1.645 σs, where μB is the mean intensity of the negative control samples, σb is the intensity standard deviation of the negative control samples, σS is the intensity sample standard deviation of the low concentration positives. Using these definitions, we have a 5% probability of committing either type I (false positive) or type II (false negative) error at the LoD.
RESULTS AND DISCUSSION
Simultaneous ITP-RPA
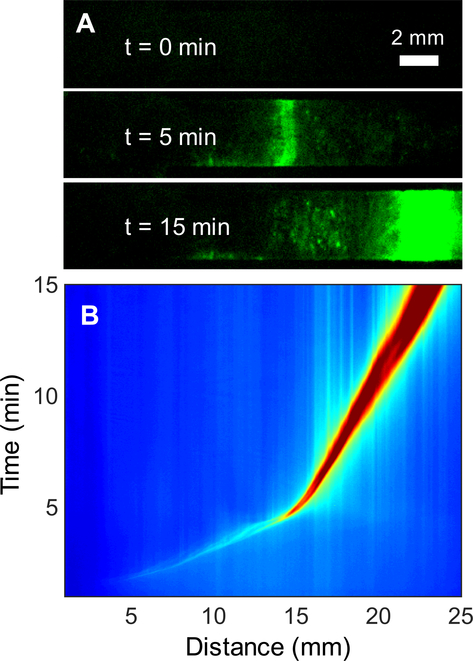
We present a method of amplifying DNA using an RPA reaction that occurs within an ITP plug on a paper substrate to determine the minimum number of copies needed for detectable amplification. These experiments were performed using only buffer as sample and therefore did not require sample preparation. We pipetted HIV-1 DNA suspended in 5 μL of LE onto the middle of the glass fiber strip pre-wetted with LE solution and RPA reagents. Figure 3A presents fluorescence images of a positive RPA reaction (10 cp/rxn) as it develops within the plug of the ITP system. When the experiment is initiated (t=0), there is no fluorescence emanating from the strip. Within 5 minutes the DNA, primers, probe, RPA proteins and enzymes focus halfway along the length of the strip, initiating RPA and generating a fluorescence signal. The signal intensity as well as the width of the ITP plug increase as the assay progresses. After 15 minutes the signal reaches its maximum, and the ITP plug nears the LE reservoir, eluting amplified nucleic acids into the reservoir. Trace fluorescence spots remain on the glass fiber in the wake of the reaction plug, possibly due to adsorption or entanglement of RPA amplicons within the fibrous network structure.
Figure 3.
Low-copy simultaneous ITP-RPA and real-time detection of target DNA in buffer. DNA is added to the center of the glass fiber strip to demonstrate successful amplification on glass fiber without extraction. (A) Fluorescence images at t=0, 5, 15 minutes of ITP-RPA over the 15-minute experiment. RPA reagents and target DNA initially applied to the glass fiber strip accumulate in the ITP plug. Amplification within the plug initiates after approximately 5 minutes. (B) Spatiotemporal map of a positive (10 cp/rxn) RPA experiment. Red and blue colors denote high and low fluorescence intensity, respectively. The fluorescence intensity of the plug rapidly increases after five minutes and migrating 15 mm down the strip.
Figure 3B shows a spatiotemporal intensity map of the positive RPA reaction depicted in Figure 3A. This figure represents the y-averaged intensity as a function of space and time, providing insight on the ITP migration dynamics. The map shows that fluorescent products from non-specific enzyme and probe interactions accumulate in the ITP plug and migrate with a constant velocity halfway along the strip (15 mm) in approximately 5 minutes. At this point, the fluorescence intensity within the plug significantly increases due to the amplification of the DNA, while the migration velocity decreases. The reduction in velocity aids the amplification as it increases the accumulation time for necessary RPA reactants to migrate into the plug and ensures that the plug does not reach the LE reservoir for at least 15 minutes. The ITP plug velocity is a function of the target electrophoretic mobility, local electric field strength, and electroosmotic counter flow.56,57 After 5 minutes the low conductivity trailing electrolyte has migrated sufficiently far down the strip such that electrical resistance results in the power supply switching from constant current to constant voltage. The net ITP plug migration velocity decreases, due to a combination of electrophoretic migration and electroosmotic flow. Electrophoretic velocity is a linear function of current, so after 5 minutes the current exponentially decreases below the 3.5 mA compliance, reducing the net plug velocity. Previous work on ITP has observed plug trajectories similar to Figure 3B.29,58
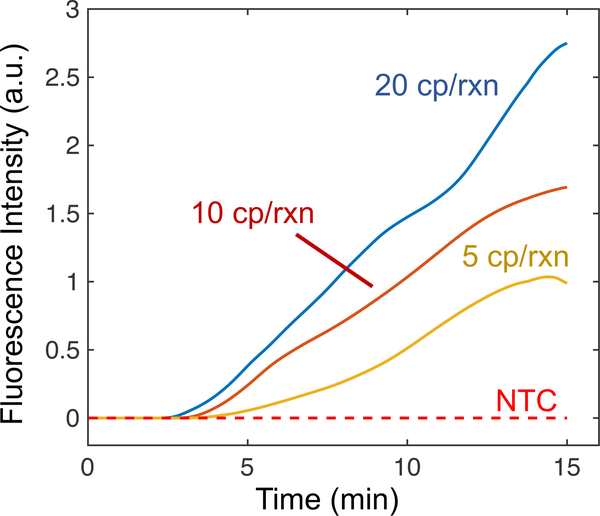
We present the normalized integrated fluorescence intensity of the RPA reporter probe as a function of time for low copy numbers of target DNA in Figure 4. The no template control (NTC) is shown as a constant zero intensity over the duration of the experiment (i.e. its signal is always below the threshold). When 5 copies of target are introduced, the intensity remains at zero for roughly four minutes where the amplification becomes visible and the intensity increases linearly until 15 minutes where it plateaus. The plateaus may be due to a limiting reagent (e.g. ATP required for recombinase functions) or deteriorating reaction conditions in the ITP plug over time (e.g. changes in ionic strength, temperature, and pH). Simultaneous ITP and RPA reactions have similar time to detection as compared to tube-based RPA assays.42,59 While RPA reactions are often exponential when amplifying high initial copies of target DNA, other studies have also reported linear amplification reactions when amplifying low copies of target DNA.42 The data indicates that simultaneous ITP-RPA can consistently detect 5 to 10 copies per reaction in pure buffer. This suggests that, with ideal nucleic acid purification and extraction, our simultaneous ITP-RPA assay should have similar sensitivity to standard RPA tube assays.36
Figure 4.
Simultaneous ITP-RPA experiments from pure buffer were analyzed to generate fluorescence curves with respect to time for low input DNA copy numbers. We present averaged data for 20 cp/rxn (2 replicates), 10 cp/rxn (3 replicates), 5 cp/rxn (3 replicates), and no template control (NTC) (3 replicates). The assay detected all replicates (8/8) with at least 5 cp/rxn of target DNA. NTC trials with 10 pg of sheared salmon sperm DNA did not amplify or alter the behavior of standard NTC experiments (shown in Figure S-9).
Limit of detection of simultaneous ITP-RPA
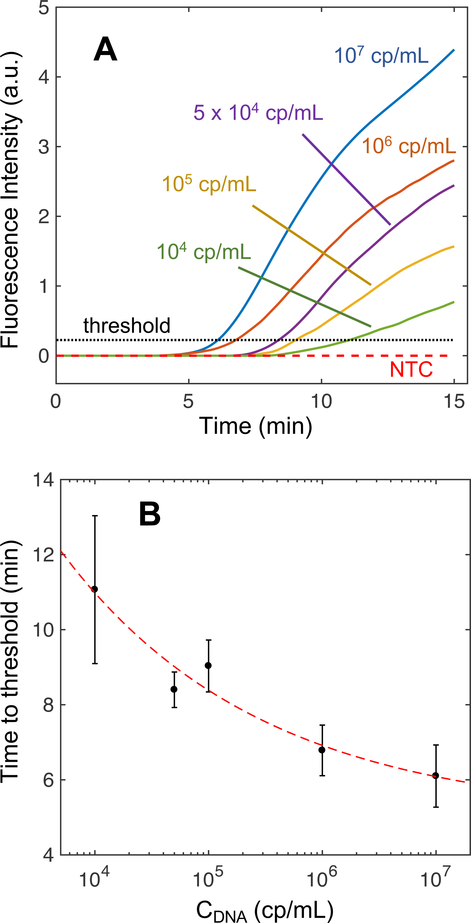
We conducted experiments to determine the LoD of simultaneous ITP-RPA from sterile-filtered human serum spiked with target DNA. We initially added 20 μL of serum to the sample pad for protein degradation using proteinase K then RPA reagents and ITP buffers were added to the paper and reservoirs. Application of an electric field initiated ITP to simultaneously extract DNA from the serum and focus it with RPA reagents, leading to RPA amplification within the ITP plug. Representative fluorescence images and spatiotemporal maps (Figure S-6 of the SI) were similar to those generated with pure DNA (Figure 3). We observe an increase in fluorescence trailing the plug that is hypothesized to be a result of proteins found in the serum clogging the fibrous glass fiber network or forming complexes with DNA and impacting the recovery of DNA.27 DNA bound to undigested serum proteins have reduced electrophoretic mobility which inhibits its electromigration to the ITP plug. The spatiotemporal map of a typical serum experiment (see Figure S-6B) shows a similar change in migration speed as Figure 3B. Figure 5A shows real-time, normalized integrated fluorescence for NTC and serum samples with target DNA spiked with concentrations ranging from 104 copies per mL (cp/mL) (i.e. 200 cp/rxn with a 20 μL sample) to 107 cp/mL (i.e. 2×105 cp/rxn). In the SI we provide integrated fluorescence signals for all replicates to give further insight into the qualitative reproducibility of the assay (Figure S-7). The simultaneous ITP-RPA reactions begin at zero normalized intensity and proceed to amplify in as soon as 5 minutes depending on the input target concentration in the serum.
Figure 5.
Fluorescence intensities from experiments during simultaneous DNA extraction and RPA amplification in an ITP plug from human serum spiked with target DNA. (A) Integrated RPA fluorescence intensities plotted with respect to time for a dilution series of DNA concentrations. Each respective intensity profile is an average of its replicates (N=3). Serum with DNA concentrations less than 104 cp/mL did not amplify, so the data is not shown here. (B) Average times to the fluorescence intensity threshold (A) for a dilution series of DNA concentrations in serum. Error bars represent one standard deviation above or below the mean time for the triplicates. The red dashed line (R2 = 0.95) indicates a power law that relates the time to threshold (t) to the initial concentration of the sample (Cdna) as t = 58.4 Cdna−025 + 5.01.
High-copy experiments amplify in less time, more rapidly (greater slope), and with higher endpoint fluorescence than low-copy experiments, generally plateauing at higher fluorescence values. We hypothesize that endpoint fluorescence is reduced in low-copy samples due to non-specific amplification from primer interactions consuming necessary reagents, such as nucleotides and ATP. Figure 5B plots the mean time to the fluorescence threshold inferred from Figure 5A with respect to the initial target concentration. The error bars represent the standard deviation of the time to threshold for the triplicates at each concentration. Time to threshold relates to copy number via a power law for concentrations from 104 to 107 copies/mL. These results are similar to those of Rohrman et al. which showed semi-quantitative HIV-1 DNA detection using a tube-based realtime RPA assay.42 Improvements in extraction efficiency and mitigating adsorption are expected to reduce the LoD and improve the consistency of the test. The observed trend between time to threshold and concentration cannot adequately quantitate input DNA, but it distinguishes high, low, and undetectable levels of target DNA. Therefore we refer to simultaneous ITP-RPA as semi-quantitative, a desirable diagnostic feature in applications such HIV viral load monitoring.60,61 Using the CLSI definition, we determined the LoD of simultaneous ITP-RPA to be 104 cp/mL, equivalent to 200 cp/rxn using a 20 μL sample volume.54 The LoD calculation accounted for variations in non-specific fluorescence signal between NTC experiments (illustrated in Figure S-8). This performance is comparable to similar prototypic POC integrated diagnostic platforms.48,49
Simultaneous ITP-RPA using whole blood
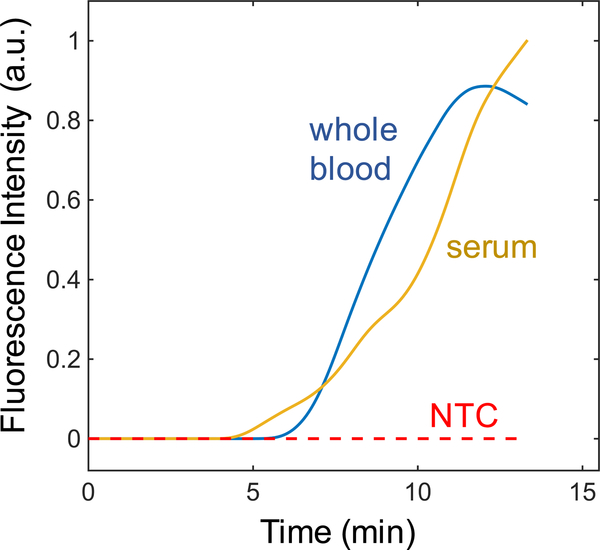
Here we present simultaneous ITP-RPA of target DNA spiked into whole blood samples. Whole blood requires filtration to remove red blood cells and peripheral blood mononuclear cells (PBMCs), which are known to inhibit RPA amplification and contain elevated levels of genomic DNA which can impede RPA.41,62 Whole blood spiked with synthetic HIV-1 DNA was applied directly to the blood separation filter and passively fractionated for 3 minutes to isolate plasma in the glass fiber membrane. Following protein digestion, simultaneous ITP-RPA was performed directly from this sample without any further processing. Figure 6 shows ITP-RPA averaged fluorescence intensities from target spiked into serum and whole blood under identical experimental conditions. The similarity of the amplification curves for serum and fractionated whole blood demonstrate that the ITP-RPA system can amplify target nucleic acid targets in unprocessed whole blood in under 20 minutes (5 minutes filtering and digesting + 13 minutes separation and amplification) using a blood cell filter.
Figure 6.
Simultaneous ITP-RPA of target spiked into whole blood. The test was conducted on human plasma that results from microfiltration of whole blood through an integrated plasma separation membrane. We plot normalized fluorescence amplification curves for simultaneous ITP-RPA experiments using whole blood with target DNA and serum with target DNA. We show these different samples amplify similarly using our paper-based device, suggesting feasibility of diagnostic testing from whole blood samples.
CONCLUSION
We present a paper-based NAAT that employs ITP and RPA to simultaneously extract and amplify target nucleic acids from serum or whole blood in under 20 minutes. By applying an electric field, nucleic acids are isolated from the complex sample and focused with RPA reagents within an ITP plug where amplification occurs in a few minutes and does not require any intermediate user steps or complex mechanical fluidic control for buffer exchanges. Our device processes undiluted whole blood or serum, eliminating a common dilution user step for diagnostics. To our knowledge, this work is the first nucleic acid amplification reaction performed within an ITP plug. While this technique may be possible in a microchannel, our paper-based ITP system has several favorable features for POC diagnostic use. Porous membranes easily integrate plasma separation membranes via capillary action, and wicking properties ease sample loading and buffer addition. We show paper-based ITP allows for large undiluted sample volumes (20 μL of serum or 50 μL of whole blood). There are also significant challenges in paper-based devices including evaporation, potential for contamination, and drying or burning of paper substrates due to relatively high electrical currents during ITP.
The use of viscous crowding agents and mixing of RPA reactions after 5 minutes of initiation is key to efficient RPA amplification in tubes when amplifying a low copy number of target molecules.63 The mixing is typically performed manually or in an automated system using steel beads and a magnet. By employing ITP for nucleic acid migration through an aqueous solution of RPA reagents, we perform this key mixing step leveraging electroosmotic flow (EOF) to disperse the reactants in the ITP plug. Optimal concentrations of the polymer PVP and surfactant Triton X-100 prevented excessive EOF and allowed for adequate mixing. Polymers (PEG and PVP) and surfactants in our ITP buffers did not hinder DNA transport through the porous membrane or disturb the separation process.
The simultaneous ITP-RPA assay has an LoD of 104 cp/mL of human serum (i.e. equivalent of 200 cp/rxn using a 20 μL serum sample). We demonstrate detection of initial synthetic HIV-1 DNA concentrations of 104 cp/mL to 107 cp/mL, well within the clinical range for HIV-1 viral loads.62 We use HIV-1 DNA in this work as a first step towards our goal of viral RNA detection and monitoring. Simultaneous ITP-RPA is a versatile technique that may be applied to detect a wide range of DNA or RNA targets from whole blood or serum at concentrations greater than 104 cp/mL, such as Zika virus, Dengue virus, and malaria.64–66 Our LoD is comparable to several commercial nucleic acid-based tests for viral hepatitis B DNA and hepatitis C RNA, both infections where physicians seek quantitative diagnostic information.67,68 Monitoring these infectious diseases may require several other technical challenges, including lysis, primer design, and reverse transcription. We observed 5 copy sensitivity for simultaneous ITP-RPA from buffer which suggests high efficiency extraction and purification of DNA from serum will dramatically improve our LoD. We hypothesize that DNA adsorption to the porous membrane and insufficient serum protein degradation prevent target DNA from electromigrating to the ITP plug for amplification. Future work will focus on improving the efficiency of ITP nucleic acid extraction.
Simultaneous ITP-RPA is a novel approach for integrating nucleic acid extraction and amplification, an operational step for which there are few elegant solutions requiring no valves, pumps, robotics, or user intervention. Our paper-based device has the potential to reduce the cost and improve the speed and ease of use of POC nucleic-acid based testing. While our test uses low-cost materials and buffers, this study required a fluorescence microscopy station and benchtop power supply. A compact fluorescence imaging unit and a power supply may be used to develop a low-cost, rapid NAAT for use in primary care settings with limited resources.29,50
Supplementary Material
ACKNOWLEDGEMENTS
Research reported in this publication was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under Award Number R01EB022630. Mr. Bender was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number TL1TR000422 as well as a National Science Foundation Graduate Research Fellowship. Mr. Borysiak was supported by a Graduate Assistance in Areas of National Need (GAANN) fellowship. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the National Science Foundation.
Footnotes
SUPPORTING INFORMATION
Figures and tables providing further details on the assay conditions, experimental approach, data analysis algorithm, and supplemental experimental data. Video showing fluorescence microscopy of simultaneous ITP-RPA.
REFERENCES
- (1).Caliendo AM; Gilbert DN; Ginocchio CC; Hanson KE; May L; Quinn TC; Tenover FC; Alland D; Blaschke AJ; Bonomo RA; et al. Better Tests, Better Care: Improved Diagnostics for Infectious Diseases. Clin. Infect. Dis 2013, 57 (suppl 3), S139–S170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Tietz Fundamentals of Clinical Chemistry and Molecular Diagnostics, Seventh edition.; Burtis CA, Bruns DE, Sawyer BG, Tietz NW, Eds.; Elsevier/Saunders: St. Louis, Missouri, 2015. [Google Scholar]
- (3).Olano JP; Walker DH Diagnosing Emerging and Reemerging Infectious Diseases: The Pivotal Role of the Pathologist. Arch. Pathol. Lab. Med 2011, 135 (1), 83–91. [DOI] [PubMed] [Google Scholar]
- (4).Mothershed EA; Whitney AM Nucleic Acid-Based Methods for the Detection of Bacterial Pathogens: Present and Future Considerations for the Clinical Laboratory. Clin. Chim. Acta 2006, 363 (1–2), 206–220. [DOI] [PubMed] [Google Scholar]
- (5).Mackay IM Real-Time PCR in the Microbiology Laboratory. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis 2004, 10 (3), 190–212. [DOI] [PubMed] [Google Scholar]
- (6).Saag MS; Holodniy M; Kuritzkes DR; O’Brien WA; Coombs R; Poscher ME; Jacobsen DM; Shaw GM; Richman DD; Volberding PA HIV Viral Load Markers in Clinical Practice. Nat. Med 1996, 2 (6), 625–629. [DOI] [PubMed] [Google Scholar]
- (7).Abe A; Inoue K; Tanaka T; Kato J; Kajiyama N; Kawaguchi R; Tanaka S; Yoshiba M; Kohara M Quantitation of Hepatitis B Virus Genomic DNA by Real-Time Detection PCR. J. Clin. Microbiol 1999, 37 (9), 2899–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Gretch DR Diagnostic Tests for Hepatitis C. Hepatology 1997, 26 (S3). [DOI] [PubMed] [Google Scholar]
- (9).Price CP Regular Review: Point of Care Testing. BMJ 2001, 322 (7297), 1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Peeling RW; Mabey D Point-of-Care Tests for Diagnosing Infections in the Developing World. Clin. Microbiol. Infect 2010, 16 (8), 1062–1069. [DOI] [PubMed] [Google Scholar]
- (11).Yager P; Domingo GJ; Gerdes J Point-of-Care Diagnostics for Global Health. Annu. Rev. Biomed. Eng 2008, 10 (1), 107–144. [DOI] [PubMed] [Google Scholar]
- (12).Drain PK; Hyle EP; Noubary F; Freedberg KA; Wilson D; Bishai WR; Rodriguez W; Bassett IV Diagnostic Point-of-Care Tests in Resource-Limited Settings. Lancet Infect. Dis. 2014, 14 (3), 239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Chin CD; Linder V; Sia SK Lab-on-a-Chip Devices for Global Health: Past Studies and Future Opportunities. Lab Chip 2007, 7 (1), 41–57. [DOI] [PubMed] [Google Scholar]
- (14).Kettler H; White K; Hawkes SJ; others. Mapping the Landscape of Diagnostics for Sexually Transmitted Infections: Key Findings and Recommendations. 2004.
- (15).Abel G Current Status and Future Prospects of Point-of-Care Testing around the Globe. Expert Rev. Mol. Diagn 2015, 15 (7), 853–855. [DOI] [PubMed] [Google Scholar]
- (16).Hsiao N; Dunning L; Kroon M; Myer L Laboratory Evaluation of the Alere q Point-of-Care System for Early Infant HIV Diagnosis. PloS One 2016, 11 (3), e0152672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Boehme CC; Nabeta P; Henostroza G; Raqib R; Rahim Z; Gerhardt M; Sanga E; Hoelscher M; Notomi T; Hase T; et al. Operational Feasibility of Using Loop-Mediated Isothermal Amplification for Diagnosis of Pulmonary Tuberculosis in Microscopy Centers of Developing Countries. J. Clin. Microbiol 2007, 45 (6), 1936–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Melchers WJG; Kuijpers J; Sickler JJ; Rahamat-Langendoen J Lab-in-a-Tube: Real-Time Molecular Point-of-Care Diagnostics for Influenza A and B Using the Cobas® Liat® System. J. Med. Virol 2017, 89 (8), 1382–1386. [DOI] [PubMed] [Google Scholar]
- (19).Mariella R Sample Preparation: The Weak Link in Microfluidics-Based Biodetection. Biomed. Microdevices 2008, 10 (6), 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Brehm-Stecher B; Young C; Jaykus L-A; Tortorello ML Sample Preparation: The Forgotten Beginning. J. Food Prot 2009, 72 (8), 1774–1789. [DOI] [PubMed] [Google Scholar]
- (21).Boom R; Sol CJ; Salimans MM; Jansen CL; Wertheim-van Dillen PM; van der Noordaa J Rapid and Simple Method for Purification of Nucleic Acids. J. Clin. Microbiol 1990, 28 (3), 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Linnes JC; Fan A; Rodriguez NM; Lemieux B; Kong H; Klapperich CM Paper-Based Molecular Diagnostic for Chlamydia Trachomatis. RSC Adv 2014, 4 (80), 42245–42251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Rodriguez NM; Linnes JC; Fan A; Ellenson CK; Pollock NR; Klapperich CM Paper-Based RNA Extraction, in Situ Isothermal Amplification, and Lateral Flow Detection for Low-Cost, Rapid Diagnosis of Influenza A (H1N1) from Clinical Specimens. Anal. Chem 2015, 87 (15), 7872–7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Everaerts FM; Beckers JL; Verheggen TP Some Theoretical and Practical Aspects of Isotachophoretical Analysis. Ann. N. Y. Acad. Sci 1973, 209 (1), 419–444. [DOI] [PubMed] [Google Scholar]
- (25).Rogacs A; Marshall LA; Santiago JG Purification of Nucleic Acids Using Isotachophoresis. J. Chromatogr. A 2014, 1335, 105–120. [DOI] [PubMed] [Google Scholar]
- (26).Kondratova V; Serd’uk O; Shelepov V; Lichtenstein A Concentration and Isolation of DNA from Biological Fluids by Agarose Gel Isotachophoresis. BioTechniques 2005, 39 (5), 695–699. [DOI] [PubMed] [Google Scholar]
- (27).Persat A; Marshall LA; Santiago JG Purification of Nucleic Acids from Whole Blood Using Isotachophoresis. Anal. Chem 2009, 81 (22), 9507–9511. [DOI] [PubMed] [Google Scholar]
- (28).Marshall LA; Rogacs A; Meinhart CD; Santiago JG An Injection Molded Microchip for Nucleic Acid Purification from 25 Microliter Samples Using Isotachophoresis. J. Chromatogr. A 2014, 1331, 139–142. [DOI] [PubMed] [Google Scholar]
- (29).Moghadam BY; Connelly KT; Posner JD Isotachophoretic Preconcenetration on Paper-Based Microfluidic Devices. Anal. Chem 2014, 86 (12), 5829–5837. [DOI] [PubMed] [Google Scholar]
- (30).Rosenfeld T; Bercovici M 1000-Fold Sample Focusing on Paper-Based Microfluidic Devices. Lab Chip 2014, 14 (23), 4465–4474. [DOI] [PubMed] [Google Scholar]
- (31).Li X; Luo L; Crooks RM Low-Voltage Paper Isotachophoresis Device for DNA Focusing. Lab Chip 2015, 15 (20), 4090–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Raizada N; Sachdeva KS; Sreenivas A; Vadera B; Gupta RS; Parmar M; Kulsange S; Babre A; Thakur R; Gray C; et al. Feasibility of Decentralised Deployment of Xpert MTB/RIF Test at Lower Level of Health System in India. PloS One 2014, 9 (2), e89301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Toley BJ; Covelli I; Belousov Y; Ramachandran S; Kline E; Scarr N; Vermeulen N; Mahoney W; Lutz BR; Yager P Isothermal Strand Displacement Amplification (ISDA): A Rapid and Sensitive Method of Nucleic Acid Amplification for Point-of-Care Diagnosis. The Analyst 2015, 140 (22), 7540–7549. [DOI] [PubMed] [Google Scholar]
- (34).Vincent M; Xu Y; Kong H Helicase-Dependent Isothermal DNA Amplification. EMBO Rep. 2004, 5 (8), 795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Notomi T; Mori Y; Tomita N; Kanda H Loop-Mediated Isothermal Amplification (LAMP): Principle, Features, and Future Prospects. J. Microbiol 2015, 53 (1), 1–5. [DOI] [PubMed] [Google Scholar]
- (36).Piepenburg O; Williams CH; Stemple DL; Armes NA DNA Detection Using Recombination Proteins. PLoS Biol. 2006, 4 (7), e204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Craw P; Balachandran W Isothermal Nucleic Acid Amplification Technologies for Point-of-Care Diagnostics: A Critical Review. Lab. Chip 2012, 12 (14), 2469. [DOI] [PubMed] [Google Scholar]
- (38).Lillis L; Lehman D; Singhal MC; Cantera J; Singleton J; Labarre P; Toyama A; Piepenburg O; Parker M; Wood R; et al. Non-Instrumented Incubation of a Recombinase Polymerase Amplification Assay for the Rapid and Sensitive Detection of Proviral HIV-1 DNA. PLoS ONE 2014, 9 (9), e108189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Niemz A; Ferguson TM; Boyle DS Point-of-Care Nucleic Acid Testing for Infectious Diseases. Trends Biotechnol. 2011, 29 (5), 240–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Euler M; Wang Y; Nentwich O; Piepenburg O; Hufert FT; Weidmann M Recombinase Polymerase Amplification Assay for Rapid Detection of Rift Valley Fever Virus. J. Clin. Virol 2012, 54 (4), 308–312. [DOI] [PubMed] [Google Scholar]
- (41).Rohrman B; Richards-Kortum R Inhibition of Recombinase Polymerase Amplification by Background DNA: A Lateral Flow-Based Method for Enriching Target DNA. Anal. Chem 2015, 87 (3), 1963–1967. [DOI] [PubMed] [Google Scholar]
- (42).Crannell ZA; Rohrman B; Richards-Kortum R Quantification of HIV-1 DNA Using Real-Time Recombinase Polymerase Amplification. Anal. Chem 2014, 86 (12), 5615–5619. [DOI] [PubMed] [Google Scholar]
- (43).Kloke A; Fiebach AR; Zhang S; Drechsel L; Niekrawietz S; Hoehl MM; Kneusel R; Panthel K; Steigert J; von Stetten F; et al. The LabTube - a Novel Microfluidic Platform for Assay Automation in Laboratory Centrifuges. Lab. Chip 2014, 14 (9), 1527. [DOI] [PubMed] [Google Scholar]
- (44).Connelly JT; Rolland JP; Whitesides GM “Paper Machine” for Molecular Diagnostics. Anal. Chem 2015, 87 (15), 7595–7601. [DOI] [PubMed] [Google Scholar]
- (45).Ferguson TM; Weigel KM; Lakey Becker A; Ontengco D; Narita M; Tolstorukov I; Doebler R; Cangelosi GA; Niemz A Pilot Study of a Rapid and Minimally Instrumented Sputum Sample Preparation Method for Molecular Diagnosis of Tuberculosis. Sci. Rep 2016, 6, 19541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Liu C; Mauk MG; Hart R; Qiu X; Bau HH A Self-Heating Cartridge for Molecular Diagnostics. Lab. Chip 2011, 11 (16), 2686–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Roskos K; Hickerson AI; Lu H-W; Ferguson TM; Shinde DN; Klaue Y; Niemz A Simple System for Isothermal DNA Amplification Coupled to Lateral Flow Detection. PLOS ONE 2013, 8 (7), e69355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Rodriguez NM; Wong WS; Liu L; Dewar R; Klapperich CM A Fully Integrated Paperfluidic Molecular Diagnostic Chip for the Extraction, Amplification, and Detection of Nucleic Acids from Clinical Samples. Lab Chip 2016, 16 (4), 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Lafleur LK; Bishop JD; Heiniger EK; Gallagher RP; Wheeler MD; Kauffman P; Zhang X; Kline EC; Buser JR; Kumar S; et al. A Rapid, Instrument-Free, Sample-to-Result Nucleic Acid Amplification Test. Lab Chip 2016, 16 (19), 3777–3787. [DOI] [PubMed] [Google Scholar]
- (50).Borysiak MD; Kimura KW; Posner JD NAIL: Nucleic Acid Detection Using Isotachophoresis and Loop-Mediated Isothermal Amplification. Lab Chip 2015, 15 (7), 1697–1707. [DOI] [PubMed] [Google Scholar]
- (51).Eid C; Santiago JG Assay for Listeria Monocytogenes Cells in Whole Blood Using Isotachophoresis and Recombinase Polymerase Amplification. The Analyst 2017, 142 (1), 48–54. [DOI] [PubMed] [Google Scholar]
- (52).Nabatiyan A; Parpia ZA; Elghanian R; Kelso DM Membrane-Based Plasma Collection Device for Point-of-Care Diagnosis of HIV. J. Virol. Methods 2011, 173 (1), 37–42. [DOI] [PubMed] [Google Scholar]
- (53).Boyle DS; Lehman DA; Lillis L; Peterson D; Singhal M; Armes N; Parker M; Piepenburg O; Overbaugh J Rapid Detection of HIV-1 Proviral DNA for Early Infant Diagnosis Using Recombinase Polymerase Amplification. mBio 2013, 4 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Tholen DW Protocols for Determination of Limits of Detection and Limits of Quantitation: Approved Guideline; NCCLS: Wayne, Pa, 2004. [Google Scholar]
- (55).Borysiak MD; Thompson MJ; Posner JD Translating Diagnostic Assays from the Laboratory to the Clinic: Analytical and Clinical Metrics for Device Development and Evaluation. Lab Chip 2016, 16 (8), 1293–1313. [DOI] [PubMed] [Google Scholar]
- (56).Bhattacharyya S; Gopmandal PP; Baier T; Hardt S Sample Dispersion in Isotachophoresis with Poiseuille Counterflow. Phys. Fluids 2013, 25 (2), 022001. [Google Scholar]
- (57).Garcia-Schwarz G; Bercovici M; Marshall LA; Santiago JG Sample Dispersion in Isotachophoresis. J. Fluid Mech 2011, 679, 455–475. [Google Scholar]
- (58).Khurana TK; Santiago JG Effects of Carbon Dioxide on Peak Mode Isotachophoresis: Simultaneous Preconcentration and Separation. Lab. Chip 2009, 9 (10), 1377. [DOI] [PubMed] [Google Scholar]
- (59).Lillis L; Lehman DA; Siverson JB; Weis J; Cantera J; Parker M; Piepenburg O; Overbaugh J; Boyle DS Cross-Subtype Detection of HIV-1 Using Reverse Transcription and Recombinase Polymerase Amplification. J. Virol. Methods 2016, 230, 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Luft LM; Gill MJ; Church DL HIV-1 Viral Diversity and Its Implications for Viral Load Testing: Review of Current Platforms. Int. J. Infect. Dis 2011, 15 (10), e661–e670. [DOI] [PubMed] [Google Scholar]
- (61).Ritchie AV; Ushiro-Lumb I; Edemaga D; Joshi HA; De Ruiter A; Szumilin E; Jendrulek I; McGuire M; Goel N; Sharma PI; et al. SAMBA HIV Semiquantitative Test, a New Point-of-Care Viral-Load-Monitoring Assay for Resource-Limited Settings. J. Clin. Microbiol 2014, 52 (9), 3377–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Kersting S; Rausch V; Bier FF; von Nickisch-Rosenegk M Rapid Detection of Plasmodium Falciparum with Isothermal Recombinase Polymerase Amplification and Lateral Flow Analysis. Malar. J 2014, 13 (1), 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Lillis L; Siverson J; Lee A; Cantera J; Parker M; Piepenburg O; Lehman DA; Boyle DS Factors Influencing Recombinase Polymerase Amplification (RPA) Assay Outcomes at Point of Care. Mol. Cell. Probes 2016, 30 (2), 74–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Pabbaraju K; Wong S; Gill K; Fonseca K; Tipples GA; Tellier R Simultaneous Detection of Zika, Chikungunya and Dengue Viruses by a Multiplex Real-Time RT-PCR Assay. J. Clin. Virol 2016, 83, 66–71. [DOI] [PubMed] [Google Scholar]
- (65).Ren P; Ortiz DA; Terzian ACB; Colombo TE; Nogueira ML; Vasilakis N; Loeffelholz MJ Evaluation of Aptima Zika Virus Assay. J. Clin. Microbiol 2017, 55 (7), 2198–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Moody A Rapid Diagnostic Tests for Malaria Parasites. Clin. Microbiol. Rev 2002, 15 (1), 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Pawlotsky J-M Molecular Diagnosis of Viral Hepatitis. Gastroenterology 2002, 122 (6), 1554–1568. [DOI] [PubMed] [Google Scholar]
- (68).Krajden M; McNabb G; Petric M The Laboratory Diagnosis of Hepatitis B Virus. Can. J. Infect. Dis. Med. Microbiol 2005, 16 (2), 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.