Abstract
As members of the basic helix-loop-helix (bHLH) family of transcription factors, E proteins function in the immune system by directing and maintaining a vast transcriptional network that regulates cell survival, proliferation, differentiation, and function. Proper activity of this network is essential to the functionality of the immune system. Aberrations in E protein expression or function can cause numerous defects, ranging from impaired lymphocyte development and immunodeficiency to aberrant function, cancer, and autoimmunity. Additionally, disruption of inhibitor of DNA-binding (Id) proteins, natural inhibitors of E proteins, can induce additional defects in development and function. Although E proteins have been investigated for several decades, their study continues to yield novel and exciting insights into the workings of the immune system. The goal of this chapter is to discuss the various classical roles of E proteins in lymphocyte development and highlight new and ongoing research into how these roles, if compromised, can lead to disease.
1. INTRODUCTION
The immune system maintains the health of the host by identifying, responding to, and subsequently eliminating harmful pathogens. These processes involve a multitude of cell types, which are loosely separated into two main branches of the immune system, the innate immune system and the adaptive immune system. A great deal of research has gone into elucidating the mechanisms and pathways involved in the development of the immune system. Of particular interest have been the developmental pathways of B and T cell development. As members of the adaptive immune system, B and T cells undergo an exceptionally complicated developmental process, including the acquisition of a diverse repertoire of antigen receptor specificities capable of recognizing virtually any antigen, previously encountered by the host or otherwise. Upon recognizing cognate antigen, B and T cells further adapt and evolve to better counter an identified threat. Following elimination of a pathogen, cells of the adaptive immune system form a pool of memory cells, capable of responding to a new challenge by the same pathogen with even greater rapidity and efficiency.
These processes supply the host with an effective, adaptive defense; however, the complex developmental and regulatory pathways that control the adaptive immune system can also be harmful if they are disrupted by genetic mutations. Production of B or T cells capable of responding to host proteins can initiate a destructive autoimmune response against critical tissues and organ systems in the body. Additionally, the high expression of particular lymphocyte-specific genes poses a potential problem as well. Translocation of various oncogenes to the transcriptional control of lymphocyte-specific regulatory elements, notably those of the antigen receptor genes, is a frequent event in tumorigenesis and is very common in leukemias and lymphomas. As such, it is critical that mechanisms exist to ensure that the immune system is kept in balance. These mechanisms have been and continue to be the subject of intense research.
One of the major regulatory mechanisms in directing lymphocyte development and function that has been frequently implicated in disease processes is the E protein transcriptional network. E proteins are members of the larger basic helix-loop-helix (bHLH) family and are widely expressed within the immune system. These proteins have been demonstrated to play critical roles at nearly every step of B and T cell development and function, from acquisition of a functional antigen receptor to cell survival and proliferation to maintaining proper functionality during an immune response. This chapter will focus primarily on the roles of E proteins in the development of B and T cells, their function within the immune system, and how these roles, when compromised, lead to severe consequences for the host.
2. E PROTEINS
E proteins are a family of transcription factors comprising a subgroup of the much larger basic bHLH family (Ephrussi, Church, Tonegawa, & Gilbert, 1985). The bHLH protein family comprises a group of widely expressed transcription factors involved in the development and maintenance of numerous cell types. bHLH proteins have been categorized into several classes. Most notable are the Class I bHLH proteins, which are widely expressed within the immune system and on which the majority of this chapter will be focused (Henthorn, Kiledjian, & Kadesch, 1990). These proteins recognize a canonical CANNTG DNA sequence, termed an E box. As such, Class I bHLH proteins are referred to as E proteins.
The E protein family is defined by the presence of several main protein domains: a C-terminal basic DNA-binding domain (the b in bHLH) and a helix-loop-helix domain (the HLH) comprising a pair of closely spaced alpha helices (Murre, McCaw, & Baltimore, 1989). These HLH domains facilitate the dimerization of bHLH proteins, an event that is required for their transcriptional activity (Murre & Baltimore, 1993). The bHLH domain has also been shown to interact with p300, a major component of the cell’s ubiquitous transcriptional machinery (Eckner, Yao, Oldread, & Livingston, 1996). bHLH proteins also contain two transcriptional activation domains, AD1 and AD2 (Aronheim, Shiran, Rosen, & Walker, 1993). AD2 is located within the central portion of the protein and is capable of driving expression of reporter constructs containing bHLH-regulated genes. AD1 is located at the N-terminus and has been shown to recruit the SAGA chromatin-remodeling complex (Massari et al., 1999).
Class I bHLH proteins (E proteins) include the E2A (also referred to as TCF-3), HEB (also referred to as TCF-12), and E2-2 (also referred to as TCF-4). The E2A and HEB genes encode several proteins by way of alternative splicing. The E2A gene encodes the proteins E12 and E47, while the HEB gene encodes the canonical HEB protein (HEBcan) as well as a shorter alternative variant (HEBalt) (Wang et al., 2006). While E47 is capable of readily binding DNA as a homodimer, E12 contains a different basic region, allowing it to only function efficiently as a heterodimer with other bHLH proteins (Sun & Baltimore, 1991). Mutations within these dimerization and DNA-binding regions can be disastrous, as shown by the recent discovery of a dominant-negative mutation in the E47 protein, which led to agammaglobulinemia and severe immunodeficiency (Boisson et al., 2013). Within developing B cells, dimers of E2A gene products are the predominant E protein transcriptional regulators, whereas developing T cells utilize primarily heterodimers of E2A and HEB gene products (Sawada & Littman, 1993). E protein dimers function by regulating a large array of genes. Dimerization between particular E proteins subtly alters the complex’s preferred DNA-binding sequence, suggesting a similar alteration in the set of genes being regulated (Hsu et al., 1994; Hu, Olson, & Kingston, 1992). E proteins have been shown to function as both transcriptional activators as well as transcriptional repressors, maintaining a vast transcriptional network (Greenbaum, Lazorchak, & Zhuang, 2004).
E protein activity is regulated by the inhibitor of differentiation (Id) gene family (Benezra, Davis, Lockshon, Turner, & Weintraub, 1990). Id proteins are similar to E proteins in that they also contain a conserved bHLH domain capable of dimerization with E proteins. However, Id proteins lack the basic DNA-binding domain. This lack of a DNA-binding domain effectively prevents the protein dimer from binding DNA and directing transcription. As such, Id proteins essentially inhibit E protein activity by out-competing functional E protein—E protein dimer formation in favor of nonfunctional E protein—Id protein dimer formation. This process effectively reverses the E protein transcriptional network, shutting down transcription of genes promoted by E proteins and removing repression of genes kept silent. In this way, E proteins can be thought of as a transcriptional “switch,” maintaining a network of gene expression until “switched off’ by upregulation of Id proteins.
Within the immune system, the primary active Id protein family members are Id3 and Id2 (Rivera, Johns, Quan, Johnson, & Murre, 2000; Sun, Copeland, Jenkins, & Baltimore, 1991). In lymphocytes, Id proteins are upregulated upon receipt of an activating signal. Id3 is rapidly upregulated following lymphocyte activation, while Id2 is upregulated more slowly (Bain et al., 2001). This suggests that Id3 and Id2 function in a semiredundant manner after activation to modulate the E protein transcriptional network, although their individual unique roles are not yet well understood.
2.1. E proteins and cell-cycle control
One of the major global transcriptional programs controlled by E proteins is the control of survival and cell-cycle progression. E2A-deficient cells develop an aggressive T cell leukemia (discussed in more detail below) characterized by rapid proliferation of developing thymocytes (Bain et al., 1997). Interestingly, restoration of E2A by ectopic expression in these cells did not arrest proliferation of these tumor cells, but rather resulted in cell death, indicating a role for E2A in cell survival as well (Engel & Murre, 1999). Subsequent experiments have demonstrated that E proteins do indeed regulate cell survival of developing lymphocytes, as conditional deletion of E2A was sufficient to drive cell death in otherwise healthy cells (Lazorchak, Wojciechowski, Dai, & Zhuang, 2006). Additional work has further implicated E proteins in regulating cell-cycle progression. Removal of E2A in some cell types was found to result in reduced levels of cyclin D2 and cyclin D3 and impaired entry into cell cycle (Zhao, Vilardi, Neely, & Choi, 2001). Interestingly, other work indicated that loss of E2A resulted in increased proliferation in lymphocytes (Park, Nolan, & Sun, 1999). Additionally, restoration of E2A in these cells halted growth. Thus, it appears that E proteins are capable of differentially regulating cell-cycle progression in a cell type-specific manner.
3. E PROTEINS IN LYMPHOCYTE DEVELOPMENT
As mentioned above, E proteins play many critical roles in lymphocyte development, particularly in the development of B and T cells. Developing B and T lymphocytes progress through a series of developmental stages in a highly regulated manner. E proteins have been shown to play a critical role in these processes.
3.1. Antigen receptor recombination
One of the primary characteristics of B and T cells is the acquisition of a highly diverse repertoire of antigen receptors (reviewed in detail here; Krangel, 2003). These antigen receptors are used to recognize and respond to antigens either directly, in the case of B cells, or after uptake, processing and subsequent presentation in the context of major histocompatibility complex (MHC) molecules in the case of T cells. These antigen receptor genes are the immunoglobulin heavy chain (IgH) and light chain (IgL) in B cells and the T cell receptor (TCR) genes in T cells. These antigen receptor genes share a unique structure, containing numerous sets of similar gene segments upstream of a conserved domain, termed the constant (C) region. These gene segments are divided into three subsets, termed variable (V), diversity (D), and joining (J) units. Upon successful recombination, the IgH and IgL chains combine to form the B cell receptor (BCR), also termed antibody. T cells contain two separate pairs of antigen receptor genes, giving rise to two distinct subsets of T cells. The TCR alpha and beta genes are capable of dimerization, producing an αβ TCR, while the TCR gamma and delta genes pair to produce a γδ TCR. Cells bearing these TCRs are referred to as αβ and γδ T cells, respectively. Interestingly, the TCRδ locus is housed within the TCRα locus, such that recombination of the TCRα locus removes a large part of the TCRδ locus, preventing further development toward the γδ lineage. While the mechanisms of recombination are similar between B cells, αβ T cells, and γδ T cells, the process and timing of recombination varies in each cell type. The regulation of these processes will be discussed in greater, cell type-specific detail later.
Acquisition of a functional antigen receptor occurs by splicing of the antigen receptor genes in a highly regulated manner. This splicing process is dependent on the recombination activating gene (RAG) family, known targets of E proteins (Hsu et al., 2003; Yu et al., 1999). RAG-mediated splicing begins upon transcriptional activation of the antigen receptor genes, a process that has been shown to be regulated in part by E proteins. RAG proteins recognize splice sites located between the various V, D, and J gene segments and facilitate the joining of these various gene segments into a functional antigen receptor gene. These events proceed in a highly regulated manner. Prior to RAG-mediated recombination, transcription of the germline IgH gene (in developing B cells) or the TCRβ, TCRγ, and TCRδ genes (in developing T cells) is initiated, opening up the chromatin environment surrounding the genes and making them more readily accessible to the recombination machinery. Animals lacking the E2A gene display an inability to initiate germline transcription of the IgH gene, leading to a block in B cell development, suggesting that E proteins play a role in initiating germline antigen receptor gene transcription (Bain et al., 1994; Zhuang, Soriano, & Weintraub, 1994). Further research showed that E proteins indeed play a direct role in activating germline transcription of antigen receptor loci and that ectopic expression of E47 alone is capable of initiating germline IgH transcripts in non-B cell lines (Choi, Shen, Radomska, Eckhardt, & Kadesch, 1996; Schlissel, Voronova, & Baltimore, 1991). Upon successful recombination of IgH (in B cells) the functional protein pairs with a conserved binding partner, the surrogate light chain to form a primordial antigen receptor. Successful recombination of the TCRβ gene is analogous to this process in T cells, pairing with the pre-Tα gene. This event allows the developing lymphocyte to receive a signal, leading to upregulation of Id proteins and subsequent reversal of E protein activity. This leads to repression of RAG genes as well as promotion of cell-cycle progression, proliferation, metabolic activity and expression of antiapoptotic genes, notably Bcl-2 (Maraskovsky et al., 1997). Additionally, successful recombination of both the TCRγ and TCRδ genes will result in the formation of a functional γδ TCR and yields a functional γδ T cell. Unsuccessful recombination resulting in an inability to pair with the surrogate light chain (in the case of B cells) or pre-Tα (in the case of αβT cells) will result in cell death, as failure to reverse E protein transcription will result in a lack of nutrient uptake, proliferation, and poor expression of antiapoptotic genes.
Following this proliferative burst, Id protein expression subsides, allowing E protein-mediated transcriptional control to resume (Bain et al., 2001). Proliferation ceases and RAG expression resumes, allowing recombination of the IgL genes in B cells and the TCRα gene in T cells (Yu et al., 1999). Successful recombination of the IgL or TCRα genes results in the development of a complete antigen receptor. The highly variable nature of this process results in the generation of a lymphocyte pool with a wide array of antigen specificities, although not all of these specificities are useful or desirable, notably in the case of autoreactive antigen receptors.
3.2. Lymphocyte selection
Following completion of V(D)J recombination, developing B and T cells must undergo a process of selection. Developing B or T cells capable of recognizing some form of antigen receive a signal through the antigen receptor, resulting in upregulation of Id proteins and a process similar, but not identical, to that described above will take place (these processes will be discussed in more detail below). RAG expression ceases, preventing further recombination of antigen receptor genes. Antiapoptotic genes begin to be expressed, promoting the survival of the cell. Any developing B or T cell that is incapable of recognizing some form of antigen will fail to receive this signal, resulting in cell death. While a B cell must simply express a functional antigen receptor, T cells must express a receptor capable of recognizing antigen peptide presented by MHC molecules (Anderson, Jenkinson, Moore, & Owen, 1993). Successful recognition of a peptide-MHC complex transmits a signal to the T cell that allows the cell to fully mature. This process is termed positive selection (Hogquist et al., 1994). Additionally, any B or T cells capable of recognizing host-derived antigens will be deleted via apoptosis in a process termed negative selection (Buch, Rieux-Laucat, Förster, & Rajewsky, 2002). Should a B or T cell develop bearing a TCR capable of recognizing self-antigens (termed an autoreactive TCR), it will receive a signal that is inherently different than one that yields positive selection. Receipt of a negative selection signal will result in cell death. This process ensures that the immune system will not recognize the host as a foreign pathogen and will not initiate immune responses against it. This lymphocyte-host nonaggression pact is referred to as self-tolerance and ensures that no lymphocytes capable of initiating an immune response against the host are permitted to develop. Negative selection, however, occasionally fails due to genetic defects or environmental perturbations. When this occurs, autoreactive B and/or T cells initiate an immune response against the host, resulting in a condition generally referred to as autoimmune disease. As mentioned above, autoimmune diseases destroy critical body tissues, resulting in dramatically decreased quality of life or even death.
Thus, proper recombination of the antigen receptor loci, and by extension, the role of E proteins in these processes, is critical to the survival of the health and survival of the host. An inability to produce an antigen receptor will result in severe immunodeficiency, rendering the host susceptible to numerous opportunistic pathogens. On the other hand, a failure to properly dispose of autoreactive cells can result in inappropriate lymphocyte-mediated destruction of body tissues, leading to reduced quality of life or even death.
4. E PROTEINS IN B CELL DEVELOPMENT
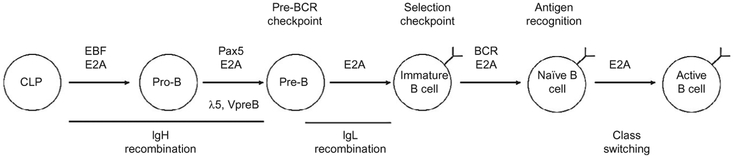
E proteins also play unique roles specific to B or T cells. Development of B cells from hematopoietic stem cells occurs in a highly regulated fashion, with E proteins, most significantly E2A, playing critical roles from a very early stage (Bain et al., 1994). During this process, developing B lymphocyte progenitors gradually lose the properties of hematopoietic stem cells while gaining B cell characteristics, becoming progressively more committed to the B cell lineage (see Fig. 4.1). A progenitor cell begins the journey to becoming a B cell when E protein activity is initiated in a common lymphoid progenitor cell (Dias, Månsson, Gurbuxani, Sigvardsson, & Kee, 2008). Activation of the E protein transcriptional network initiates expression of a series of additional transcription factors that cooperatively direct B cell development. The first of these is the early B cell factor (EBF) gene, which has been shown to be critically important in B cell development, permitting B cell progenitors to develop into pro-B cells (Lin & Grosschedl, 1995). Prior to its activation, the EBF gene is located at the transcriptionally repressive nuclear periphery (Lin, Benner, et al., 2012). Upon activation by E2A, the EBF gene relocates to a centralized, transcriptionally permissive environment and transcription begins (Lin, Benner, et al., 2012). Together, E2A and EBF begin to cooperatively initiate antigen receptor recombination (Romanow et al., 2000). Similar to the case of the EBF gene, the IgH locus also repositions itself away from the repressive nuclear periphery and DJ joining proceeds. It is likely that this repositioning facilitates not only germline transcription but also facilitates DNA accessibility for the RAG complex, as mice lacking EBF fail to generate B cells, as development is blocked at the pro-B cell stage prior to DJ rearrangement (Lin & Grosschedl, 1995). EBF has also recently been shown to promote B cell development by repressing the Id2 and Id3 genes, effectively ensuring that E2A activity is allowed to continue (Thal et al., 2009).
Figure 4.1.
Diagram of B cell development. Major developmental events involving E proteins and major E protein targets are indicated.
The coordinate activities of E2A and EBF also function to promote the transcription of the Pax5 (also known as BSAP) gene (Nutt, Heavey, Rolink, & Busslinger, 1999; Urbánek, Wang, Fetka, Wagner, & Busslinger, 1994). As with mice lacking E2A or EBF, mice lacking Pax5 also show a block in B cell development at the pro-B cell stage (Nutt et al., 1999). Unlike E2A or EBF-null B cells however, Pax5-deficient B cells have been shown to be capable of abandoning the B cell developmental pathway and adopting different cell fates, including T cells and macrophages (Nutt et al., 1999). To this end, Pax5-deficient B cells are capable of expressing several genes normally expressed in other cell types, including pre-Tα and macrophage colony-stimulating factor, while these genes are silenced in WT B cells, suggesting that Pax5 plays a role in silencing gene sets utilized by other lymphocyte lineages. Furthermore, experiments using conditional deletion of E2A in developing B cells have indicated that Pax5 alone is capable of driving B cell development beyond the pre-B cell stage (Kwon et al., 2008). However, these E2A-deficient mature B cells display impaired functionality, indicating that E proteins play additional distinct roles in B cell development and function apart from their roles in conjunction with Pax5 activity (Kwon et al., 2008). As such E2A and Pax5 function to promote commitment to the B cell lineage.
In addition to promoting commitment to the B cell lineage, Pax5 has been shown to play a role in IgH recombination. While EBF-deficient pro-B cells contain unrearranged IgH loci, Pax5-deficient pro-B cells complete DJ recombination, but fail to complete V-DJ recombination (Fuxa et al., 2004). There are several possible explanations for this phenomenon. First, Pax5-binding sites have been located near the V gene segments of the IgH locus. Additionally, Pax5 has been shown to regulate the conformation and location of the IgH locus. Prior to Pax5 expression, the IgH locus is located at the nuclear periphery; upon Pax5 expression, the locus relocates to the interior of the nucleus, suggesting that the location of the locus may play a role in the recombination process (Fuxa et al., 2004). Additional research into gene expression and RAG-mediated recombination suggests that localization at the nuclear periphery results in impaired V-DJ recombination. It is likely that these alterations in gene localization function to modulate the ability of RAG complexes to access the DNA (Chan et al., 2013). It is possible that E proteins or their cooperative activity with transcription factors such as Pax5 and/or EBF mediate this repositioning, as binding sites for these factors exist in several regulatory regions flanking the locus (Henthorn et al., 1990). Furthermore, Pax5 has been shown to promote chromatin interactions between recombined DJ segments and the distant V gene segments, bringing the two into close contact (Fuxa et al., 2004). In-depth analysis into these chromatin interactions has indicated that the distant V gene segments form large-scale looping structures, providing visual confirmation of earlier biochemical data (Sayegh, Jhunjhunwala, Riblet, & Murre, 2005). Thus, in addition to driving commitment to the B cell lineage, E2A and Pax5 also promote further development along the B cell developmental pathway by promoting completion of IgH recombination.
Following completion of IgH recombination, the newly rearranged IgH protein pairs with the surrogate light chain (composed of λ5 and V-pre-B, both E2A targets) and Igα and Igβ signaling chains, forming the pre-B cell receptor (Loffert et al., 1994). Developing B cells that fail to produce a functional, in-frame IgH locus are destined for apoptosis, while those cells that successfully express a functional pre-B cell receptor become pre-B cells and undergo several subsequent, simultaneous events. First, further IgH recombination is blocked in a process referred to as “allelic exclusion,” preventing simultaneous expression of multiple BCR specificities in a single B cell (Loffert et al., 1994). This prevents B cells from reacting to multiple, potentially unrelated, antigens. Second, a short burst of proliferation is initiated, allowing amplification of successfully recombined IgH chains (Yankee & Clark, 1999). During this time, E protein expression transiently declines, along with the E2A targets RAG1 and RAG2, preventing further recombination (Yu et al., 1999). Following this period of proliferation, E protein expression resumes and recombination of the Igκ and Igλ light chain (IgL) loci begins (Hardy & Hayakawa, 2001).
Similar to IgH recombination, E proteins also play roles in IgL recombination. Ectopic expression of E2A proteins in addition to the RAG genes has been shown to activate IgL transcription as well as V-J recombination in transformed cells (Romanow et al., 2000). E2A has been demonstrated to promote IgL recombination by inducing chromatin remodeling in conjunction with IRF-4 (Lazorchak, Schlissel, & Zhuang, 2006). Interestingly, these genes are also present during the pro-B cell stage, although IgL recombination does not occur. It is possible that EBF provides a mechanism for preventing IgL expression, as coexpression of E2A and EBF has been shown to drive DJ recombination on the IgH locus, while recombination of the IgL loci does not occur (Goebel et al., 2001).
Following successful recombination of an Igλ or Igκ chain, the recombined IgL protein pairs with the previously recombined IgH protein to produce the BCR, also termed antibody. In the event that the resulting BCR recognizes a host antigen, it is deleted through apoptosis in a process called “negative selection” (Nossal, 1983). Those B cells that do not recognize host antigen are then released from the bone marrow to colonize the body’s peripheral lymphoid organs. By preventing B cells capable of responding to host antigens from completing the developmental process, the B cell pool becomes tolerant to its host. Disruption of this process often leads to an autoimmune response against the host, with B cells often producing antibodies against antigens such as DNA and ribosomal proteins as well as other host proteins particular to individual autoimmune diseases.
As discussed above, the E protein network plays critical roles in B cell development, ranging from transcriptional regulation of key developmental genes to antigen receptor recombination, but the mechanisms underpinning this regulatory network have largely remained elusive. However, recent research has begun to bring these mechanisms to light. With the development of large-scale biochemistry and DNA sequencing, it has become possible to examine transcription factor activity on a global scale. These experiments have revealed intriguing roles for E2A beyond simply turning genes on or off. E2A has been shown to regulate gene transcription on several levels. Recent work has demonstrated that global chromatin accessibility is regulated, both positively and negatively, in part by E proteins (Lin, Jhunjhunwala, et al., 2010). E proteins have been shown to be capable of recruiting chromatin-remodeling complexes to the various genes they regulate (Lazorchak, Schlissel, et al., 2006; Sakamoto et al., 2012). E protein-mediated chromatin remodeling has been shown to be increasingly finely regulated by many of the transcription factors described above. In concert with EBF, E proteins have been shown to fine-tune the E protein network by initiating chromatin remodeling at loci jointly regulated by E2A and EBF prior to transcription initiation, making these loci effectively “poised” for transcription (Treiber et al., 2010). In this manner, E protein activity initiates a complex global transcriptional program that develops in concert with the cells themselves, evolving as additional developmental coregulators such as EBF and Pax5 are progressively activated (Lin, Jhunjhunwala, et al., 2010). These events fine-tune the E protein network and facilitate the completion of upcoming developmental processes, while shutting down activities occurring in previously completed developmental stages.
5. E PROTEIN ROLES IN MATURE B CELLS
Even after B cells complete development, the E protein network remains an integral part of B cell activity. B cells that pass the selection checkpoint are released into the periphery where they patrol the body and protect it from pathogens. Upon recognizing a pathogen via the BCR, the B cell is activated and undergoes a number of changes, many of them involving E proteins. In resting B cells, E2A levels are low, while E2A is highly expressed in activated B cells (Quong, Harris, Swain, & Murre, 1999). Shortly after BCR stimulation, the Id3 gene is rapidly upregulated (Deed, Hara, Atherton, Peters, & Norton, 1997). B cells lacking the Id3 gene display impaired proliferation, consistent with the role of E proteins in regulating cell cycle (Pan, Sato, Frederick, Sun, & Zhuang, 1999). Additionally, activated B cells undergo class switching, altering their BCR to better respond to the activating pathogen (Goldfarb, Flores, & Lewandowska, 1996). Notably, B cells lacking the E2A gene fail to initiate class switching, although most other events in B cell activation proceed normally, including expression of the activation markers CD69 and CD44 (Quong et al., 1999). This is largely due to the role of E proteins in regulating the expression of AID, the molecule responsible for class switching (Sayegh, Quong, Agata, & Murre, 2003). Further supporting the role of E proteins in class switching, ectopic expression of the Id1 gene in B cells yields an impairment similar to the one observed in E2A-deficient cells (Goldfarb et al., 1996).
While the roles of E2A in B cell development have been studied in depth, the HEB and E2-2 genes have also been shown to play roles in B cell development. Unlike mice lacking E2A, mice lacking either E2-2 or HEB are able to successfully produce mature B cells; they display a dramatic reduction in numbers of pro-B cells, suggesting roles for these genes in promoting survival at the pro-B cell stage (Zhuang, Cheng, & Weintraub, 1996).
6. E PROTEIN ROLES IN T CELL DEVELOPMENT
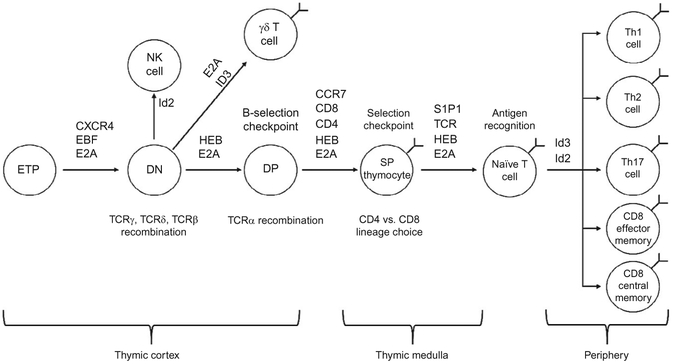
Similar to developing B cells, E proteins have been shown to play critical roles in T cell development as well. Whereas homodimers of E2A gene products predominate in developing B cells, heterodimers of E2A and HEB gene products predominate in thymocytes (Sawada & Littman, 1993). T cell development begins when early thymic progenitor cells migrate from the bone marrow to the thymus. The thymus is a complex organ, containing two major regions, the outer cortex and the inner medulla (see Fig. 4.2). Similar to developing B cells, thymocytes must undergo recombination of their antigen receptor genes. During this process, they must also migrate to particular regions of the thymus for development to proceed properly. Interestingly, E proteins have been shown to play important roles in the completion of this migratory process, while disruption of the E protein network can produce dramatic impairments in thymocyte development.
Figure 4.2.
Diagram of T cell development. Major developmental events involving E proteins and major E protein targets are indicated.
Developing thymocytes are broadly classified by their expression of the CD4 and CD8 coreceptor molecules as well as expression of an αβ or γδ TCR. Upon entry into the thymus, thymocytes express neither CD4 nor CD8 and are referred to as DN cells. These cells migrate into the thymic cortex and initiate recombination of the TCRβ, TCRγ, and TCRδ genes. This process is dependent on CXCR4, a chemokine receptor (Plotkin, Prockop, Lepique, & Petrie, 2003). Expression of CXCR4 has been shown to be regulated in part by E proteins, as cells lacking E2A and HEB fail to upregulate this marker (Jones & Zhuang, 2007). As in developing B cells, expression of E proteins is required for activation of germline antigen receptor transcription, although full activation of the TCRβ, TCRγ, and TCR-δ loci has been shown to require expression of both E2A as well as HEB (Ghosh, Romanow, & Murre, 2001; Jia, Dai, & Zhuang, 2008; Langerak, Wolvers-Tettero, Van Gastel-Mol, Oud, & Van Dongen, 2001). Again, germline transcription of the antigen receptor genes is coupled with subnuclear repositioning of the DNA, paving the way for RAG-mediated recombination (Schlimgen, Reddy, Singh, & Krangel, 2008). Successful recombination of the TCRγ and TCRδ genes yields a γδ T cell, while successful recombination of the TCRβ gene allows the TCRβ protein to pair with the pre-T cell receptor alpha (pre-Tα) chain, analogous to the surrogate light chain of B cells (Kruisbeek et al., 2000). This process, called β-selection, initiates a burst of proliferation as well as initiation of CD8 and CD4 expression, becoming double-positive (DP) thymocytes (Barndt, Dai, & Zhuang, 2000). Again, E proteins play key roles in these processes. Upon pre-Tα expression, Id3 expression is upregulated, inhibiting E protein activity (Engel, Johns, Bain, Rivera, & Murre, 2001). It is likely that E proteins play a role in restricting passage of DN thymocytes to the DP stage, as elimination of E2A is sufficient to permit DN thymocytes to transition to the DP stage without a rearranged TCRβ allele (Engel et al., 2001). Additionally, the HEB gene has been shown to be required for proper expression of pre-Tα, leading to a block in T cell development at the β-selection checkpoint (Herblot, Steff Hugo, Aplan, & Hoang, 2000). Furthermore, overexpression of Id3 recapitulates the phenotype observed in HEB-deficient thymocytes, generating a block in thymocyte development, partly by blocking pre-Tα expression (Blom et al., 1999).
Following β-selection, a developing αβ T cell downregulates CXCR4 and migrates to the thymic medulla, a process requiring upregulation of the CCR7 chemokine receptor. As with CXCR4 expression, CCR7 expression is also regulated by E proteins, albeit in an opposite (repressive) manner (Jones & Zhuang, 2007). Recombination of the TCRβ genes ceases, with the unrearranged allele relocating to the nuclear periphery so as to prevent further recombination (Chan et al., 2013). Removal of E proteins allows for spontaneous upregulation of CCR7 along with continued development of thymocytes in the absence of β-selection. This defect will be discussed in more detail below. Upon migration into the thymic medulla, recombination of the TCR-β genes ceases and TCRα recombination begins (Von Boehmer, 1994). Following successful TCRα recombination, a functional αβ TCR can be expressed on the cell surface. Cells expressing an αβ TCR then undergo the processes of positive and negative selection. As mentioned above, in order to successfully pass the positive selection checkpoint, a given TCR must be capable of recognizing antigenic peptide presented by either MHC class I or MHC class II (reviewed in detail here; Robey & Fowlkes, 1994). Thymocytes-recognizing antigen presented by MHC class I will lose CD4 expression, becoming CD8 single-positive (SP) cells, while those recognizing antigen presented by MHC class II will lose CD8 expression and become CD4 SP cells. Again, E proteins play key roles in positive selection. Upon receiving a signal through the TCR, Id3 is rapidly upregulated (Bain et al., 2001). Id3 upregulation is coupled with upregulation of S1P1, a molecule required for exit from the thymus (Matloubian et al., 2004). When E2A and HEB are removed in DP thymocytes, it appears that Id protein upregulation is no longer required for continued development, permitting developing T cells to bypass the need for any signal through the TCR (Jones & Zhuang, 2007), removing the need for an antigen receptor signal results in the development of T cells lacking a functional TCR. Interestingly, removal of E2A and HEB in developing T cells results in a complete block in the development of CD4 T cells, suggesting that the CD8 lineage is a “default” developmental pathway.
Disruption of Id proteins in developing T cells also results in numerous developmental defects. Elimination of the Id3 gene results in several major phenotypes. The first is the preferential development and/or expansion of a unique subset of γδ T cells (Ueda-Hayakawa, Mahlios, & Zhuang, 2009). These cells all bear a TCR using the Vγ1.1 and Vδ6.3 gene segments and share many characteristics with innate immune cells, including cytokine production, expression of PLZF and dependence on the signaling adaptor protein SAP (Verykokakis et al., 2010). This population is developed very early in life and is maintained through self-renewal. Deletion of Id3 in these Vγ1.1/Vδ6.3+ cells was found to cause a dramatic expansion early in life, although Vγ1.1/Vδ6.3+ cells developed later in life did not share this property (Zhang, Dai, Li, & Zhuang, 2013). Interestingly, this phenotype shows a peculiar strain-dependence, as Id3 deficiency on the C57BL/6 background results in a strong phenotype, while the same deletion on the 129/sv background does not (Azuara, Grigoriadou, Lembezat, Nagler Anderson, & Pereira, 2001). This difference in γδ T cell development was traced to a strain-specific mutation in the Id2 gene resulting in weaker Id2 expression in C57BL/6 mice (Zhang, Lin, Dai, & Zhuang, 2014). Id3-deficient mice have also been shown to have defects in thymocyte selection (Rivera et al., 2000). A disproportionately small number of DP thymocytes are capable of making the leap to mature SP cells, suggesting that the early upregulation of Id3 plays a critical role in the development of a large number of thymocytes. These defects in T cell development are coupled with the initiation of an autoimmune disease reminiscent of human Sjogren’s Syndrome (SS) (Li, Dai, & Zhuang, 2004). These data also suggest that Id2 is capable of compensating for the loss of Id3, at least partially. While Id2 can compensate for some roles of Id3 in thymocyte development, it also plays unique roles as well. Disruption of the Id2 gene has been shown to result in a block in natural killer (NK) and natural killer-T (NKT) cell development, while αβ and γδ development proceeds seemingly normally (Boos, Yokota, Eberl, & Kee, 2007). Further investigation into the roles of E proteins in NK cell development indicated that combined deletion of Id2 and Id3 in DN thymocytes was sufficient to drive a dramatic expansion of invariant NKT (iNKT) cells (Li, Wu, Jiang, & Zhuang, 2013). E proteins were also shown to have a role in regulating the proliferation of these iNKT cells (Hu et al., 2013; Li et al., 2013; Verykokakis et al., 2013). Intriguingly, partial restoration of Id proteins resulted in a switch from iNKT development to γδ T cell development, suggesting that fine regulation of E protein activity is required in the development of various T cell subsets (Li et al., 2013).
While the removal of Id3 or Id2 alone permits relatively normal αβ T cell development, removal of both Id2 and Id3 in DP thymocytes results in profound developmental aberrations. While removal of the E2A and HEB genes results in the development of large numbers of CD8 T cells, many of which lack a functional TCR, deletion of Id2 and Id3 results in a complete lack of CD8+ T cells (Jones-Mason et al., 2012). Additionally, development of CD4+ T cells is severely restricted. Taken together with the results derived from E2A/HEB double-knockout T cells, these data indicate that E proteins are required for the development of CD4+ T cells.
7. ROLES OF E PROTEINS IN MATURE T CELLS
Following maturation within the thymus, naïve T cells migrate to the periphery. There, they protect the host from invading pathogens. Upon recognition of a pathogen via the TCR, a naïve T cell becomes activated and begins to adapt its response to the current pathogenic challenge. During this process, a naïve T cell response will further develop into one of a number of effector responses, each with its own signature array of inflammatory mediators. For example, an intracellular pathogen, such as a virus, induces a T helper type 1 (Th1) response, including production of IFN-γ. This response features significant activity by CD8 cytotoxic cells, which eliminate infected host cells. By contrast, extracellular pathogens give rise to other responses. A Th2 response is characterized by production of IL-4, IL-5, and IL-13 as well as marked recruitment of B cell activity, which aids in removal of blood-borne pathogens.
As ever, E proteins play important roles in regulating these processes. Id3 expression has been shown to be relatively high in naïve T cells (Miyazaki et al., 2011). Elimination of the Id3 gene results in spontaneous upregulation of surface markers characteristic of differentiated, effector-memory cells (Miyazaki et al., 2011). These results indicate that Id3-mediated suppression of E proteins in naïve T cells is required to prevent spontaneous maturation into effector cells. These results were further supported by experiments investigating the various roles of Id2 and Id3 in naïve T cells (Yang et al., 2011). Id3 expression was found to correlate with long-lived memory cell formation, while Id2 was associated with the development of short-lived, effector-memory cells (Yang et al., 2011). Additionally, deletion of either Id2 or Id3 resulted in a failure to generate effector-memory or long-lived memory cells, respectively. The role of Id3 in supporting memory cell formation is likely regulated in part by Blimp-1, as high Blimp-1 expression downregulated Id3 and limited the generation of long-lived memory cells (Ji et al., 2011). Id proteins have also been implicated in the adoption of particular T helper responses. A large-scale gene-association study found that Id2 and Id3 are differentially regulated in particular T helper populations (Lund et al., 2007). This evidence is further supported by the finding that mice lacking Id2 display disproportionately Th2-skewed T cell responses (Kusunoki et al., 2003). The role of Id2 in this developmental skewing is in fact profound enough to affect autoimmune disease progression. An analysis of Id2-deficient T cells demonstrated that removal of Id2 in T cells was sufficient to protect mice from experimentally induced autoimmune encephalomyelitis, a Th17-mediated disease (Lin, Jones-Mason, et al., 2012). Although pathogenic cells were developed, they did so at a greatly reduced rate, as proapoptotic proteins were found to be upregulated in Id2-deficient T cells, leading to increased cell death (Lin, Jones-Mason, et al., 2012). These results further highlight the role of the E protein system in maintaining and regulating the T cell response to antigen.
8. E PROTEINS IN LYMPHOID DISEASES
As demonstrated above, E proteins play critical roles in the highly complex developmental pathways and subsequent functions of B and T cells. These roles include controlling cell proliferation, ensuring developmental checkpoints remain enforced, maintaining self-tolerance and regulating the effector functions of mature cells. In general, E proteins tend to function as gatekeepers, ensuring a cell does not proceed to another developmental stage before it has completed the steps required to do so. By enforcing these checkpoints, E proteins keep the immune system operating normally. Because E proteins play so many roles in these sensitive processes, it is critical that their function remain undisturbed, as serious consequences could result from defects in these processes. Failures in selection or antigen receptor generation could lead to severe immunodeficiency. Failures in selection could lead to autoimmune reactions. Failures in cell-cycle regulation could lead to tumor formation. The remainder of this chapter will focus on the ways in which these developmental mechanisms can be co-opted to induce diseases, particularly autoimmunity and cancer.
8.1. E proteins in autoimmunity
SS is an autoimmune disease characterized by progressive destruction of the salivary and lachrymal glands, resulting in impaired saliva and tear production (Bloch, Buchanan, Wohl, & Bunim, 1965). This gland destruction is mediated by lymphocytic infiltration into the gland tissue, provoking an inflammatory response within the gland (Greenspan, Daniels, Talal, & Sylvester, 1974). This inflammatory response results in development of fibrotic scar tissue within the gland, leading to increasingly poor secretory function (Tarpley, Anderson, & White, 1974). In humans, SS primarily affects women, typically becoming apparent in the fourth or fifth decade of life. Among available mouse models, the Id3 knockout mouse has been established as a unique model for human primary SS (Lee, Gauna, Pauley, Park, & Cha, 2012).
The Id3-deficient mouse develops many of the same characteristics of human SS (Li et al., 2004). Significantly impaired gland function is readily apparent in Id3 knockout mice within 2 months of age. By 4 months, pronounced lymphocytic infiltration begins to occur. The initial infiltrating lymphocytes are primarily αβ T cells, with B cells arriving shortly thereafter. Beyond 6 months of age, mice begin to show outward signs of disease: increased water consumption and lesions around the face due to repeated scratching of dry eyes. Autoantibody production, notably anti-Ro and anti-La, typically begins around 1 year of age (Alexander, Hirsch, Arnett, Provost, & Stevens, 1981; Alexander, Arnett, Provost, & Stevens, 1983). Historically, SS was thought of as a Th1-mediated disease, as human patients frequently displayed elevated levels of IFN-γ, although animal models did not typically share this phenotype (Dalavanga, Drosos, & Moutsopoulos, 1985; Mahlios & Zhuang, 2011; Szodoray, Alex, Brun, Centola, & Jonsson, 2004). Additionally, it has been demonstrated that removal of Id3 in T cells alone was sufficient to initiate the disease process (Guo et al., 2011). Adoptive transfer of Id3-deficient T cells into sublethally irradiated WT hosts was also sufficient to transfer disease (Li et al., 2004). However, additional cell types have proved to be important disease mediators as well, as depletion of B cells was capable of improving disease symptoms in mice (Hayakawa, Tedder, & Zhuang, 2007). In addition to B cell involvement, mice suffering from SS were found to contain markedly elevated numbers of mast cells within the gland tissue (Mahlios & Zhuang, 2011). Together, the immune cells drive extensive fibrotic remodeling of the gland tissue, leading to degradation of gland function. Interestingly, elevated E protein activity has been implicated in driving fibrotic remodeling in other tissues, suggesting an additional potential nonlymphoid role in gland impairment (Slattery, Mcmorrow, & Ryan, 2006). Furthermore, with later discoveries of additional effector T cell subtypes and the cytokines they produce, notably IL-13, it became clear that the disease process was in fact far more complicated than originally thought (McKenzie et al., 1993).
Identification of IL-13 as a part of the Th2 effector response led to reevaluation of several immune processes, such as allergic processes and epithelial inflammation (De Vries, 1998; Grünig et al., 1998). This paradigm shift was not limited to typical foreign-antigen responses, as elevated levels of IL-13 were soon identified in patients with SS symptoms (Mitsias et al., 2002). This discovery was also observed in the Id3-deficient mouse model of SS (Mahlios & Zhuang, 2011). Additional research into the role of IL-13 in SS pathogenesis has been enlightening. Neutralization of circulating IL-13 was sufficient to improve gland function in Id3-deficient mice and caused a mild reduction in the numbers of mast cells residing within the gland tissue, although the source of IL-13 remained elusive (Mahlios & Zhuang, 2011). As IL-13 is known to be produced by T cells, elimination of αβ T cells in Id3-deficient mice was sufficient to reduce IL-13 concentrations in serum to near-WT levels, though IL-13 was still slightly elevated. Elimination of αβ T cells was also capable of impeding the disease process; disease symptoms still appeared in Id3/TCRβ double-deficient mice, albeit much later than in Id3 knockout mice (Mahlios & Zhuang, 2011).
The above results indicate that SS is a highly complex disorder with numerous cell types involved in its pathogenesis. Many of the cell types known to be involved (CD4+ T cells, γδ T cells, B cells, mast cells, etc.) likely contribute to gland impairment in multiple ways, ranging from gland infiltration to cytokine production to tissue remodeling. Recent research in both humans and animal models has highlighted the importance of IL-13, particularly T cell-derived IL-13, in disease progression. These findings raise the possibility of IL-13 as a potential diagnostic tool or therapeutic target in the treatment of patients with SS.
8.2. E proteins in cancer
The prevalent roles of E proteins in regulating gene expression and developmental progression ensure that lymphocytes develop and function in a way that is beneficial to the host. We have already discussed how defects in lymphocyte development and selection can turn the immune system against the host. The role of E proteins in regulating gene expression can also be dysregulated, oftentimes resulting in tumorigenesis.
8.2.1. Burkitt Lymphoma
Many B cell cancers arise from a translocation event between the IgH promoter, which is constitutively active in B cells, and the c-Myc oncogene, a prosurvival transcription factor. Upon placing c-Myc under the control of the IgH promoter, the resultant B cell is highly resistant to apoptosis. This event can lead to several different varieties of B cell cancers, including both Burkitt’s Lymphoma (BL) and Diffuse Large B Cell Lymphoma (DLBCL). Intriguingly, recent work has shown that, upon B cell activation, the c-Myc locus becomes transcriptionally active and frequently repositions itself adjacent to the IgH locus, greatly facilitating this translocation event (Osborne et al., 2007). While both BL and DLBCL often share certain underlying mutations, recent research has shown them to be quite different. For example, low-dose cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP) therapy has been successfully used in the treatment of DLBCL, however, much larger doses have been shown to be required for effective treatment of BL (Butler & Hainsworth, 1993). Additionally, use of treatments capable of passing the blood-brain barrier have been shown to be required for proper treatment of BL, while these measures have proven unnecessary for DLBCL (Bishop, Rao, & Wilson, 2000). These remarkable differences between cancers sharing such similar mutations have led to a great deal of research into the genetic differences underlying these disparities.
Several groups have recently performed extensive comparative analysis of BL and DLBCL (Love et al., 2012; Richter et al., 2012; Schmitz et al., 2012). These groups discovered that, while BL and DLBCL may share an initial mutation, many secondary mutations are unique to each tumor. Indeed, more than two thirds of BL tumors have been found to contain mutations within the Id3 gene (Love et al., 2012; Richter et al., 2012). Interestingly, the majority of these mutations were determined to be within the bHLH region of the protein and were frequently nonsense or frameshift mutations (Love et al., 2012; Richter et al., 2012). These results suggest that the secondary mutations within the Id3 locus are loss-of-function mutations, resulting in dysregulation of the E protein transcriptional network. Intriguingly, these mutations were almost entirely absent from DLBCL tumors. Additional work by Staudt and colleagues corroborated this finding and additionally identified frequent mutations within the E2A gene (Schmitz et al., 2012). While mutations within the Id3 gene were biallelic and predominantly resulted in a loss-of-function, the mutations within the E2A gene typically monoallelic and resulted in elevated transcription. Strikingly, in BL tumors bearing E2A mutations, nearly all mutations were restricted to the bHLH or DNA-binding domains of the E47 splice-variant, while the E12 variant remained unchanged. In many E2A alleles containing mutations within the DNA-binding domain, these changes resulted in alterations in the canonical “CANNTG” binding sequence preferred by E protein dimers (Schmitz et al., 2012). Additionally, E47 was expressed at a higher level than E12 in these tumors, suggesting a nonredundant role for these proteins in BL pathogenesis.
While BL tumors have been shown to contain secondary mutations in Id3 and/or E2A, the effects of these mutations are not well understood. As mentioned above, mutations in Id3 and E2A are typically found within the bHLH region. In addition to increased expression of E47 (in the case of E2A mutations), one of the primary effects of these mutations has been shown to be an inhibition of dimerization between E proteins and Id proteins (Schmitz et al., 2012). In the case of Id3 and E2A, this inhibition results in an inability of Id3 to reverse the E protein transcriptional network. In BL tumors bearing Id3 mutations, this has been shown to result in increased progression through the cell cycle. Expression of several genes involved in cell-cycle initiation, including CDK7, E2F1, and MCM10 were found to be elevated in BL cells bearing Id3 mutations. Additionally, introduction of individual BL-derived Id3 mutant alleles into BL lines containing wild-type Id3 genes resulted in a significantly reduced proportion of cells in G1 as well as an increased proportion of cells entering S phase (Love et al., 2012). Furthermore, reintroduction of wild-type Id3 into BL cells expressing Id3 mutants was sufficient to slow the rate at which cells entered the cell cycle (Love et al., 2012).
In addition to altering progression into the cell cycle, mutations in Id3 and E2A have been shown to have notable effects on cell survival. Reintroduction of E2A expression has been shown to result in significantly increased cell death in several tumor lines, while alteration of E2A activity is a major oncogenic factor in primary BL cells (Engel & Murre, 1999; Schmitz et al., 2012). Recent work has shown that E protein regulation of PI3K signaling may be responsible for this increased survival (Schmitz et al., 2012). PI3Ks regulate a wide range of biological processes by generating lipid-based second-messenger molecules (Okkenhaug & Vanhaesebroeck, 2003). In lymphocytes, PI3K plays an important role in antigen receptor and coreceptor signal transduction. Notably, PI3K signaling is known to activate AKT, which in turn regulates cell growth, survival, and metabolism (Stocker et al., 2002; Vanhaesebroeck & Alessi, 2000). PI3K is activated within seconds of tyrosine phosphorylation of antigen receptor molecules, notably Igα and Igβ (also known as CD79a and CD79b, respectively) (Okada, Maeda, Iwamatsu, Gotoh, & Kurosaki, 2000; Tuveson, Carter, Soltoff & Fearon, 1993). Following PI3K activation, AKT is phosphorylated and activates survival, proliferation, and metabolic pathways, largely through activation of the mTOR pathway (Mendoza, Er, & Blenis, 2011). This pathway is negatively regulated by tyrosine phosphatase proteins, notably SHP-1 (Sathish et al., 2001). SHP-1 dephosphorylates tyrosine residues on antigen receptor and coreceptor molecules, inhibiting activation of AKT. In BL tumors bearing mutations in E2A or Id3, levels of SHP-1 were found to be lower than in WT cells; additionally, knockdown of E2A was found to increase SHP-1 levels while simultaneously decreasing phospho-AKT levels (Schmitz et al., 2012). These results suggest that, in addition to regulating entry into the cell cycle, disruptions in the E protein system can also alter antigen receptor signaling to further promote growth, survival, and nutrient uptake.
Taken together, the results discussed above suggest that mutations targeting the E protein transcriptional network are capable of supporting the oncogenic nature of c-Myc translocation in a manner distinct from other c-Myc-driven cancers. It has been suggested that these mutations, as well as their downstream effects, may be used as diagnostic criteria, allowing clinicians to better identify cases of BL and separate them from other, similar cancers, thereby allowing improved, more specialized treatment. It is also possible that a more complete understanding of the mechanisms and cellular pathways involved in and unique to BL may allow for the development of new therapeutic options, yielding improved prognoses for patients.
8.2.2. E2A–PBX1 translocation in B cell acute lymphocytic leukemia
As mentioned previously, E proteins control a large transcriptional network, regulating events such as lymphocyte development, cell survival and proliferation. Loss of E or Id proteins can cause significant defects in these processes. Alterations in the gene networks regulated by E proteins can have similarly deleterious effects. E proteins have also been found to play a role in other types of B cell cancers, notably pre-B cell Acute Lymphocytic Leukemia (pre-B ALL). The t(1;19)(q23;p13.3) translocation is found in approximately 25% of patients with pre-B ALL. In the vast majority of these patients, the translocation event joins the promoter and transcriptional activation domain of the E2A gene with the DNA-binding region of the Pre-B Cell Homeobox-1 (PBX1) gene. While E2A is widely expressed throughout the body, PBX1 is not normally expressed within the immune system. PBX1 mediates its transcriptional activity by forming part of a molecular complex that includes Class 1 Homeobox (HOX) proteins as well as Meis1 and pKnox1 (Knoepfler, Calvo, Chen, Antonarakis, & Kamps, 1997). In addition to disrupting one copy of the E2A gene, which has been shown to lead to an increased incidence of lymphoid tumors, this fusion event results in constitutive activation of many of the genes regulated by PBX1 in cells expressing E2A. This transactivation has been shown to require both the DNA-binding region of PBX1 as well as the transcriptional activation domain of E2A, as disruption of either of these sequences eliminates the tumorigenicity of the E2A—PBX1 fusion protein (Monica et al., 1994). Monica and colleagues further characterized the effects of mutations within the E2A portion of E2A—PBX1, noting that disruption of the AD1 domain resulted in a loss of transcriptional activation in both lymphoid and fibroblast lines, while disruption of AD2 only impaired expression in fibroblast lines, indicating potential separate roles for the AD1 and AD2 domains in tumorigenesis (Monica et al., 1994).
While both E2A and E2A—PBX1 are potent transcriptional activators in lymphoid cells, PBX1 alone is not (Lu & Kamps, 1996). In fact, expression of PBX1 has been shown to inhibit some of the activity of other HOX proteins (Lu & Kamps, 1996). Intriguingly, the E2A—PBX1 fusion protein produces further aberrations in transcriptional activity beyond simple abnormal expression of PBX1. While normal activity of PBX1 requires interactions with Meis1 and pKnox1, the E2A—PBX1 fusion protein is incapable of interacting with these proteins (Knoepfler et al., 1997). This is likely due to the fact that the residues required for interaction with Meis1 and pKnox1 are contained in the portion of PBX1 replaced by the E2A gene (Knoepfler et al., 1997). Thus, the transcriptional activity of the E2A—PBX1 protein results in expression of a different set of genes than does native PBX1. Later research into the tumorigenicity of the E2A—PBX1 fusion protein yielded several intriguing gene targets, in particular, Wnt16 and EB-1. Wnt16 is a member of the Wnt family of proteins, which are involved in promoting cell growth. The Wnt family of proteins is expressed in numerous cell types and has been shown to be frequently mutated in several types of cancers (Polakis, 2000). Interestingly, Wnt16 is not normally expressed in pre-B cells (McWhirter et al., 1999). However, a role for the Wnt signaling pathway in early hematopoiesis and proliferation of developing B cells has been described (Austin et al., 1997; Reya et al., 2000; Van Den Berg et al., 1998). These results indicate that the E2A—PBX1 fusion protein may exert some of its negative effects by aberrantly activating the Wnt signaling pathway, leading to inappropriate proliferation of pre-B cells.
EB-1 was identified by Kamps and colleagues while investigating genes activated by E2A-PBX1 (Fu et al., 1999). The EB-1 gene encodes protein containing a phosphotyrosine-binding element and two SAM domains, similar to other genes playing roles in tyrosine kinase signal transduction and cell proliferation. EB-1 expression is normally restricted to the brain and testes, however, upon introduction of E2A—PBX1, EB-1 expression in developing B cells increased nearly 100-fold. Such a result suggests that E2A—PBX1-driven EB1 expression may alter one or more of the signaling pathways responsible for regulating cell growth.
Taken together, the above observations indicate that the E2A—PBX1 translocation leads to pre-B ALL in part by disrupting the sensitive E protein transcriptional network as well as initiating the aberrant expression of several tissue-specific genes via the unique transcriptional activity of E2A—PBX1. Several of these genes have been shown to regulate cell growth and proliferation, further amplifying the oncogenic effects of the initial translocation event. The Wnt pathway has been well studied in the context of neurobiology, particularly in Alzheimer’s disease (Caricasole et al., 2003). The association of Wnt pathway activity in E2A—PBX1 tumorigenesis may provide opportunities for the development of new therapeutic strategies (Barker & Clevers, 2006).
8.2.3. E proteins in T cell cancers
E proteins have also been shown to play significant roles in T cell leukemias and lymphomas as well (Yan et al., 1997). While mice deficient in E2A often die perinatally, those that survive their infancy often develop an aggressive T cell lymphoma comprised of immature thymic progenitors. It is likely that the loss of E2A results in enhanced proliferation, as reintroduction of E2A proteins is able to induce cell death in E2A-deficient tumors (Engel & Murre, 1999). Furthermore, as mentioned above, DN thymocytes lacking E proteins are capable of spontaneously bypassing the β-selection checkpoint. Given that one of the consequences of β-selection is rapid proliferation during the period of Id3-mediated E protein suppression, disruption of E protein-mediated cell-cycle control seems a likely culprit in the development of these tumors. In fact, recent work has demonstrated that loss of E2A is indeed a major player in Sézary Syndrome, a subtype of human T cell lymphoma (Steininger et al., 2011). Restoration of E2A in these tumor cells resulted in a reduction in proliferation and increased cell death, corroborating previous data implicating E proteins in cell-cycle control. E2A-deficient tumors are characterized by a greatly enlarged thymus with an abnormal architecture, lymphadenopathy and frequent metastasis to other lymphoid and nonlymphoid organs. These tumorigenic T cells expressed little to no surface TCR, highlighting their immaturity. The lack of surface TCR on these cells is likely due to the removal of the requirement for β-selection. Interestingly, particular tumor incidences were found to be made up of either CD4+/CD8+ cells, CD4low/CD8+, or CD4−/CD8+ cells, lending support the role of E proteins in regulating T cell lineage choice.
Last, recent work investigating the Id3-deficient mouse model of SS revealed an intriguing finding. Mice lacking the Id3 gene were found to occasionally develop a lymphoma comprised of TCRγδ+ T cells, a condition which is very rare occurrence in humans (Lin, Jhunjhunwala, et al., 2010). These lymphomas presented themselves primarily with splenomegaly, hepatomegaly, and lymphadenopathy. Additionally, tumor cells were found within the bone marrow, kidneys, lungs, and thymus. Indeed, splenic involvement was such that the overall architecture was almost completely destroyed. Interestingly, the majority of these lymphomas were comprised of cells bearing TCRs featuring the Vγ1.1 gene segment, likely due to the preferential development of these cells in Id3-deficient mice. Some lymphomas consisted of cells using the Vγ3 gene segment, but none were found to use Vγ2. This suggests that Id3 may play some role in tumor suppression in addition to its role in suppressing the development of Vγ1.1+ cells. It is possible that the Id3-deficient mouse model may also be useful in the study of human γδ T cell lymphoma, as no additional model of this disease is known to exist.
9. CONCLUSION
Since their discovery, E proteins and their inhibitors, Id proteins, have proven to be major players in the immune system at virtually every stage. Even though they were first discovered decades ago, ongoing research continues to uncover new intricacies of the E proteins. Recent research has repeatedly demonstrated the roles of E and Id proteins in maintaining the state of a lymphocyte, preventing them from improper maturation, proliferation, and functional activity (Miyazaki et al., 2011). Breakdowns in the E protein system can cause serious defects in the immune system, causing aberrant activity and autoimmunity (Guo et al., 2011). Subtle changes in E protein function, even those that alter dimerization properties can also produce profound immunodeficiencies (Boisson et al., 2013). New research has also shown that E protein functionality can differentiate types of cancer formerly thought to be highly similar (Love et al., 2012; Richter et al., 2012). Because E proteins and Id proteins play such numerous and such varied roles in lymphocyte development and function, sometimes in a cell type-specific manner, and because their expression level must remain finely tuned, their use as potential therapeutic targets has been limited. For example, loss of E protein activity blocks B cell development, but can also lead to aberrant T cell development (Bain et al., 1994, 1997). Furthermore, simply restoring E protein activity (or removal of Id proteins) can also yield deleterious results in these models (Engel & Murre, 1999). As such, more research is required to better understand the mechanisms by which breakdowns in the E protein system contribute to developmental and functional abnormalities in particular cell types and particular stages of development.
Fortunately, recent research into the targets of the E protein network has been promising. The evolution of large-scale sequencing has identified numerous downstream gene targets of E proteins, as well as transcription factors, some of them cell type-specific, involved in fine-tuning the E protein network (Lin, Jhunjhunwala, et al., 2010). However, further research is necessary to more fully understand which E protein functions are ubiquitous and which are unique to particular cell types or cell stages. By better understanding how the E protein network is regulated in particular cell types, E protein activity can be better connected to the pathways that modulate its downstream effects. A more complete understanding of the ways in which E proteins function to regulate downstream events in the many cellular processes involved in development could yield therapeutic targets useful in the treatment of the many diseases that can result from abnormalities in the E protein system.
ACKNOWLEDGMENTS
The authors would like to thank Baojun Zhang, Yen-yu Lin, Elizabeth Chan, Jia Li, and Sumedha Roy for helpful comments during the preparation of this chapter.
REFERENCES
- Alexander EL, Arnett FC, Provost TT, & Stevens MB (1983). Sjögren’s syndrome: Association of anti-Ro (SS-A) antibodies with vasculitis, hematologic abnormalities, and serologic hyperreactivity. Annals of Internal Medicine, 98(2), 155–159. [DOI] [PubMed] [Google Scholar]
- Alexander E, Hirsch T, Arnett F, Provost T, & Stevens M (1981). Ro (SSA) and La (SSB) antibodies in the clinical spectrum of Sjogren’s syndrome. The Journal of Rheumatology, 9(2), 239–246. [PubMed] [Google Scholar]
- Anderson G, Jenkinson EJ, Moore NC, & Owen JJ (1993). MHC class II-positive epithelium and mesenchyme cells are both required for T-cell development in the thymus. Nature, 362, 70–73. [DOI] [PubMed] [Google Scholar]
- Aronheim A, Shiran R, Rosen A, & Walker MD (1993). The E2A gene product contains two separable and functionally distinct transcription activation domains. Proceedings of the National Academy of Sciences, 90(17), 8063–8067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin TW, Solar GP, Ziegler FC, Liem L, & Matthews W (1997). A role for the Wnt gene family in hematopoiesis: Expansion of multilineage progenitor cells. Blood, 89(10), 3624–3635. [PubMed] [Google Scholar]
- Azuara V, Grigoriadou K, Lembezat MP, Nagler Anderson C, & Pereira P (2001). Strain-specific TCR repertoire selection of IL-4-producing Thy-1dull γ δ thymocytes. European Journal of Immunology, 31(1), 205–214. [DOI] [PubMed] [Google Scholar]
- Bain G, Cravatt CB, Loomans C, Alberola-Ila J, Hedrick SM, & Murre C (2001). Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras-ERK MAPK cascade. Nature Immunology, 2(2), 165–171. [DOI] [PubMed] [Google Scholar]
- Bain G, Engel I, Maandag ER, Te Riele H, Voland JR, Sharp LL, et al. (1997). E2A deficiency leads to abnormalities in alphabeta T-cell development and to rapid development of T-cell lymphomas. Molecular and Cellular Biology, 17(8), 4782–4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G, Maandag ECR, Izon DJ, Amsen D, Kruisbeek AM, Weintraub BC, et al. (1994). E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell, 79(5), 885–892. [DOI] [PubMed] [Google Scholar]
- Barker N, & Clevers H (2006). Mining the Wnt pathway for cancer therapeutics. Nature Reviews Drug Discovery, 5(12), 997–1014. [DOI] [PubMed] [Google Scholar]
- Barndt RJ, Dai M, & Zhuang Y (2000). Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Molecular and Cellular Biology, 20(18), 6677–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, & Weintraub H (1990). The protein Id: A negative regulator of helix-loop-helix DNA binding proteins. Cell, 61(1), 49–59. [DOI] [PubMed] [Google Scholar]
- Bishop PC, Rao VK, & Wilson WH (2000). Burkitt’s lymphoma: Molecular pathogenesis and treatment. Cancer Investigation, 18(6), 574–583. [DOI] [PubMed] [Google Scholar]
- Bloch K, Buchanan W, Wohl M, & Bunim J (1965). Sjogren’s syndrome: A clinical, pathological, and serological study of sixty-two cases. Medicine, 44(3), 187–231. [PubMed] [Google Scholar]
- Blom B, Heemskerk MH, Verschuren MC, Van Dongen JJ, Stegmann AP, Bakker AQ, et al. (1999). Disruption of αβ but not of γδ T cell development by overexpression of the helix-loop-helix protein Id3 in committed T cell progenitors. The EMBO Journal, 18(10), 2793–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson B, Wang Y-D, Bosompem A, Ma CS, Lim A, Kochetkov T, et al. (2013). A recurrent dominant negative E47 mutation causes agammaglobulinemia and BCR-B cells. The Journal of Clinical Investigation, 123(11), 4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos MD, Yokota Y, Eberl G, & Kee BL (2007). Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. The Journal of Experimental Medicine, 204(5), 1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch T, Rieux-Laucat F, Förster I, & Rajewsky K (2002). Failure of HY-specific thymocytes to escape negative selection by receptor editing. Immunity, 16(5), 707–718. [DOI] [PubMed] [Google Scholar]
- Butler RD, & Hainsworth JD (1993). Optimal therapy for small noncleaved cell lymphoma In Dana WD (Ed.), Malignant lymphomas, including Hodgkin’s disease: diagnosis, management, and special problems (pp. 65–67). Boston, MA: Springer. [DOI] [PubMed] [Google Scholar]
- Caricasole A, Copani A, Caruso A, Caraci F, Iacovelli L, Sortino MA, et al. (2003). The Wnt pathway, cell-cycle activation and β-amyloid: Novel therapeutic strategies in Alzheimer’s disease? Trends in Pharmacological Sciences, 24(5), 233–238. [DOI] [PubMed] [Google Scholar]
- Chan EA, Teng G, Corbett E, Choudhury KR, Bassing CH, Schatz DG, et al. (2013). Peripheral subnuclear positioning suppresses Tcrb recombination and segregates Tcrb alleles from RAG2. Proceedings of the National Academy of Sciences, 110(48), E4628–E4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JK, Shen C-P, Radomska H, Eckhardt L, & Kadesch T (1996). E47 activates the Ig-heavy chain and TdT loci in non-B cells. The EMBO Journal, 15(18), 5014. [PMC free article] [PubMed] [Google Scholar]
- Dalavanga Y, Drosos A, & Moutsopoulos H (1985). Labial salivary gland immunopathology in Sjogren’s syndrome. Scandinavian Journal of Rheumatology. Supplement, 61, 67–70. [PubMed] [Google Scholar]
- De Vries JE (1998). The role of IL-13 and its receptor in allergy and inflammatory responses. Journal of Allergy and Clinical Immunology, 102(2), 165–169. [DOI] [PubMed] [Google Scholar]
- Deed RW, Hara E, Atherton GT, Peters G, & Norton JD (1997). Regulation of Id3 cell cycle function by Cdk-2-dependent phosphorylation. Molecular and Cellular Biology, 17(12), 6815–6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias S, Månsson R, Gurbuxani S, Sigvardsson M, & Kee BL (2008). E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity, 29(2), 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckner R, Yao T-P, Oldread E, & Livingston DM (1996). Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes & Development, 10(19), 2478–2490. [DOI] [PubMed] [Google Scholar]
- Engel I, Johns C, Bain G, Rivera RR, & Murre C (2001). Early thymocyte development is regulated by modulation of E2A protein activity. The Journal of Experimental Medicine, 194(6), 733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel I, & Murre C (1999). Ectopic expression of E47 or E12 promotes the death of E2A-deficient lymphomas. Proceedings of the National Academy of Sciences, 96(3), 996–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A, Church GM, Tonegawa S, & Gilbert W (1985). B lineage-specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science, 227(4683), 134–140. [DOI] [PubMed] [Google Scholar]
- Fu X, Roberts WG, Nobile V, Shapiro R, & Kamps MP (1999). mAngiogenin-3, a target gene of oncoprotein E2a-Pbx1, encodes a new angiogenic member of the angiogenin family. Growth Factors, 17(2), 125–137. [DOI] [PubMed] [Google Scholar]
- Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, & Busslinger M (2004). Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes & Development, 18(4), 411–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh JK, Romanow WJ, & Murre C (2001). Induction of a diverse T cell receptor γ/δ repertoire by the helix-loop-helix proteins E2A and HEB in nonlymphoid cells. The Journal of Experimental Medicine, 193(6), 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel P, Janney N, Valenzuela JR, Romanow WJ, Murre C, & Feeney AJ (2001). Localized gene-specific induction of accessibility to V (D) J recombination induced by E2A and early B cell factor in nonlymphoid cells. The Journal of Experimental Medicine, 194(5), 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb AN, Flores JP, & Lewandowska K (1996). Involvement of the E2A basic helix-loop-helix protein in immunoglobulin heavy chain class switching. Molecular Immunology, 33(11), 947–956. [DOI] [PubMed] [Google Scholar]
- Greenbaum S, Lazorchak AS, & Zhuang Y (2004). Differential functions for the transcription factor E2A in positive and negative gene regulation in pre-B lymphocytes. Journal of Biological Chemistry, 279(43), 45028–45035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan J, Daniels T, Talal N, & Sylvester R (1974). The histopathology of Sjögren’s syndrome in labial salivary gland biopsies. Oral Surgery, Oral Medicine, Oral Pathology, 37(2), 217–229. [DOI] [PubMed] [Google Scholar]
- Grünig G, Warnock M, Wakil AE, Venkayya R, Brombacher F, Rennick DM, et al. (1998). Requirement for IL-13 independently of IL-4 in experimental asthma. Science, 282(5397), 2261–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Li H, Han M, Xu T, Wu X, & Zhuang Y (2011). Modeling Sjögren’s syndrome with Id3 conditional knockout mice. Immunology Letters, 135(1), 34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy RR, & Hayakawa K (2001). B cell development pathways. Annual Review of Immunology, 19(1), 595–621. [DOI] [PubMed] [Google Scholar]
- Hayakawa I, Tedder TF, & Zhuang Y (2007). B-lymphocyte depletion ameliorates Sjögren’s syndrome in Id3 knockout mice. Immunology, 122(1), 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henthorn P, Kiledjian M, & Kadesch T (1990). Two distinct transcription factors that bind the immunoglobulin enhancer microE5/kappa 2 motif. Science, 247(4941), 467–470. [DOI] [PubMed] [Google Scholar]
- Herblot S, Steff A-M, Hugo P, Aplan PD, & Hoang T (2000). SCL and LMO1 alter thymocyte differentiation: Inhibition of E2A-HEB function and pre-Tα chain expression. Nature Immunology, 1(2), 138–144. [DOI] [PubMed] [Google Scholar]
- Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, & Carbone FR (1994). T cell receptor antagonist peptides induce positive selection. Cell, 76(1), 17–27. [DOI] [PubMed] [Google Scholar]
- Hsu H-L, Huang L, Tsan JT, Funk W, Wright WE, Hu J-S, et al. (1994). Preferred sequences for DNA recognition by the TAL1 helix-loop-helix proteins. Molecular and Cellular Biology, 14(2), 1256–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu L-Y, Lauring J, Liang H-E, Greenbaum S, Cado D, Zhuang Y, et al. (2003). A conserved transcriptional enhancer regulates RAG gene expression in developing B cells. Immunity, 19(1), 105–117. [DOI] [PubMed] [Google Scholar]
- Hu J-S, Olson E, & Kingston R (1992). HEB, a helix-loop-helix protein related to E2A and ITF2 that can modulate the DNA-binding ability of myogenic regulatory factors. Molecular and Cellular Biology, 12(3), 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T, Wang H, Simmons A, Bajaña S, Zhao Y, Kovats S, et al. (2013). Increased level of e protein activity during invariant NKT development promotes differentiation of invariant NKT2 and invariant NKT17 subsets. The Journal of Immunology, 191(10), 5065–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Pos Z, Rao M, Klebanoff CA, Yu Z, Sukumar M, et al. (2011). Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8 + T cells. Nature Immunology, 12(12), 1230–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Dai M, & Zhuang Y (2008). E proteins are required to activate germline transcription of the TCR Vβ8. 2 gene. European Journal of Immunology, 38(10), 2806–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ME, & Zhuang Y (2007). Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity, 27(6), 860–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Mason ME, Zhao X, Kappes D, Lasorella A, Iavarone A, & Zhuang Y (2012). E protein transcription factors are required for the development of CD4 + lineage T cells. Immunity, 36(3), 348–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Calvo KR, Chen H, Antonarakis SE, & Kamps MP (1997). Meis1 and pKnox1 bind DNA cooperatively with Pbx1 utilizing an interaction surface disrupted in oncoprotein E2a-Pbx1. Proceedings of the National Academy of Sciences, 94(26), 14553–14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krangel MS (2003). Gene segment selection in V (D) J recombination: Accessibility and beyond. Nature Immunology, 4(7), 624–630. [DOI] [PubMed] [Google Scholar]
- Kruisbeek AM, Haks MC, Carleton M, Wiest DL, Michie AM, & Zúñiga-Pflücker JC (2000). Branching out to gain control: How the pre-TCR is linked to multiple functions. Immunology Today, 21(12), 637–644. [DOI] [PubMed] [Google Scholar]
- Kusunoki T, Sugai M, Katakai T, Omatsu Y, Iyoda T, Inaba K, et al. (2003). TH2 dominance and defective development of a CD8 + dendritic cell subset in Id2-deficient mice. Journal of Allergy and Clinical Immunology, 111(1), 136–142. [DOI] [PubMed] [Google Scholar]
- Kwon K, Hutter C, Sun Q, Bilic I, Cobaleda C, Malin S, et al. (2008). Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity, 28(6), 751–762. [DOI] [PubMed] [Google Scholar]
- Langerak AW, Wolvers-Tettero IL, Van Gastel-Mol EJ, Oud ME, & Van Dongen JJ (2001). Basic helix-loop-helix proteins E2A and HEB induce immature T-cell receptor rearrangements in nonlymphoid cells. Blood, 98(8), 2456–2465. [DOI] [PubMed] [Google Scholar]
- Lazorchak AS, Schlissel MS, & Zhuang Y (2006). E2A and IRF-4/Pip promote chromatin modification and transcription of the immunoglobulin κ locus in pre-B cells. Molecular and Cellular Biology, 26(3), 810–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazorchak AS, Wojciechowski J, Dai M, & Zhuang Y (2006). E2A promotes the survival of precursor and mature B lymphocytes. The Journal of Immunology, 177(4), 2495–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Gauna AE, Pauley KM, Park Y-J, & Cha S (2012). Animal models in autoimmune diseases: Lessons learned from mouse models for Sjögren’s syndrome. Clinical Reviews in Allergy & Immunology, 42(1), 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Dai M, & Zhuang Y (2004). AT cell intrinsic role of Id3 in a mouse model for primary Sjögren’s syndrome. Immunity, 21(4), 551–560. [DOI] [PubMed] [Google Scholar]
- Li J, Wu D, Jiang N, & Zhuang Y (2013). Combined deletion of Id2 and Id3 genes reveals multiple roles for E proteins in invariant NKT cell development and expansion. The Journal of Immunology, 191(10), 5052–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Benner C, Mansson R, Heinz S, Miyazaki K, Miyazaki M, et al. (2012). Global changes in the nuclear positioning of genes and intra-and interdomain genomic interactions that orchestrate B cell fate. Nature Immunology, 13(12), 1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, & Grosschedl R (1995). Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature, 376(6537), 263–267. [DOI] [PubMed] [Google Scholar]
- Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, et al. (2010). A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nature Immunology, 11(7), 635–643. http://www.nature.com/ni/journal/v11/n7/suppinfo/ni.1891_S1.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, et al. (2010). Mutation of inhibitory helix-loop-helix protein Id3 causes γδ T-cell lymphoma in mice. Blood, 116(25), 5615–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-Y, Jones-Mason ME, Inoue M, Lasorella A, Iavarone A, Li Q-J, et al. (2012). Transcriptional regulator Id2 is required for the CD4 T cell immune response in the development of experimental autoimmune encephalomyelitis. The Journal of Immunology, 189(3), 1400–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffert D, Schaal S, Ehlich A, Hardy RR, Zou YR, Müller W, et al. (1994). Early B cell development in the mouse: Insights from mutations introduced by gene targeting. Immunological Reviews, 137(1), 135–153. [DOI] [PubMed] [Google Scholar]
- Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, et al. (2012). The genetic landscape of mutations in Burkitt lymphoma. Nature Genetics, 44(12), 1321–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, & Kamps MP (1996). Structural determinants within Pbx1 that mediate cooperative DNA binding with pentapeptide-containing Hox proteins: Proposal for a model of a Pbx1-Hox-DNA complex. Molecular and Cellular Biology, 16(4), 1632–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund RJ, Löytömäki M, Naumanen T, Dixon C, Chen Z, Ahlfors H, et al. (2007). Genome-wide identification of novel genes involved in early Th1 and Th2 cell differentiation. The Journal of Immunology, 178(6), 3648–3660. [DOI] [PubMed] [Google Scholar]
- Mahlios J, & Zhuang Y (2011). Contribution of IL-13 to early exocrinopathy in Id3 −/− mice. Molecular Immunology, 49(1), 227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraskovsky E, O’Reilly LA, Teepe M, Corcoran LM, Peschon JJ, & Strasser A (1997). Bcl-2 can rescue T lymphocyte development in interleukin-7 receptor-deficient mice but not in mutant rag-1 −/− Mice. Cell, 89(7), 1011–1019. [DOI] [PubMed] [Google Scholar]
- Massari ME, Grant PA, Pray-Grant MG, Berger SL, Workman JL, & Murre C (1999). A conserved motif present in a class of helix-loop-helix proteins activates transcription by direct recruitment of the SAGA complex. Molecular Cell, 4(1), 63–73. [DOI] [PubMed] [Google Scholar]
- Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, et al. (2004). Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature, 427(6972), 355–360. [DOI] [PubMed] [Google Scholar]
- McKenzie A, Culpepper J, De Waal Malefyt R, Briere F, Punnonen J, Aversa G, et al. (1993). Interleukin 13, a T-cell-derived cytokine that regulates human monocyte and B-cell function. Proceedings of the National Academy of Sciences, 90(8), 3735–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhirter JR, Neuteboom STC, Wancewicz EV, Monia BP, Downing JR, & Murre C (1999). Proceedings of the National Academy of Sciences, 96(20), 11464–11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza MC, Er EE, & Blenis J (2011). The Ras-ERK and PI3K-mTOR pathways: Cross-talk and compensation. Trends in Biochemical Sciences, 36(6), 320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsias D, Tzioufas A, Veiopoulou C, Zintzaras E, Tassios I, Kogopoulou O, et al. (2002). The Th1/Th2 cytokine balance changes with the progress of the immunopathological lesion of Sjogren’s syndrome. Clinical& Experimental Immunology, 128(3), 562–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki M, Rivera RR, Miyazaki K, Lin YC, Agata Y, & Murre C (2011). The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nature Immunology, 12(10), 992–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monica K, LeBrun DP, Dedera DA, Brown R, & Cleary ML (1994). Transformation properties of the E2a-Pbx1 chimeric oncoprotein: Fusion with E2a is essential, but the Pbx1 homeodomain is dispensable. Molecular and Cellular Biology, 14(12), 8304–8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C, & Baltimore D (1993). The helix-loop-helix motif: Structure and function. Cold Spring Harbor Monograph Series, 22, 861. [Google Scholar]
- Murre C, McCaw PS, & Baltimore D (1989). A new DNA binding and dimerization motifin immunoglobulin enhancer binding, daughterless, MyoD and myc proteins. Cell, 56(5), 777–783. [DOI] [PubMed] [Google Scholar]
- Nossal G (1983). Cellular mechanisms of immunologic tolerance. Annual Review of Immunology, 1(1), 33–62. [DOI] [PubMed] [Google Scholar]
- Nutt SL, Heavey B, Rolink AG, & Busslinger M (1999). Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature, 401(6753), 556–562. [DOI] [PubMed] [Google Scholar]
- Okada T, Maeda A, Iwamatsu A, Gotoh K, & Kurosaki T (2000). BCAP: The tyrosine kinase substrate that connects B cell receptor to phosphoinositide 3-kinase activation. Immunity, 13(6), 817–827. [DOI] [PubMed] [Google Scholar]
- Okkenhaug K, & Vanhaesebroeck B (2003). PI3K in lymphocyte development, differentiation and activation. Nature Reviews Immunology, 3(4), 317–330. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, et al. (2007). Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biology, 5(8), e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Sato S, Frederick JP, Sun X-H, & Zhuang Y (1999). Impaired immune responses and B-cell proliferation in mice lacking the Id3 gene. Molecular and Cellular Biology, 19(9), 5969–5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park ST, Nolan GP, & Sun X-H (1999). Growth inhibition and apoptosis due to restoration of E2A activity in T cell acute lymphoblastic leukemia cells. The Journal of Experimental Medicine, 189(3), 501–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin J, Prockop SE, Lepique A, & Petrie HT (2003). Critical role for CXCR4 signaling in progenitor localization and T cell differentiation in the postnatal thymus. The Journal of Immunology, 171(9), 4521–4527. [DOI] [PubMed] [Google Scholar]
- Polakis P (2000). Wnt signaling and cancer. Genes and Development, 14(15), 1837–1851. [PubMed] [Google Scholar]
- Quong MW, Harris DP, Swain SL, & Murre C (1999). E2A activity is induced during B-cell activation to promote immunoglobulin class switch recombination. The EMBO Journal, 18(22), 6307–6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, O’Riordan M, Okamura R, Devaney E, Willert K, Nusse R, et al. (2000). Wnt signaling regulates B lymphocyte proliferation through a LEF-1 dependent mechanism. Immunity, 13(1), 15–24. [DOI] [PubMed] [Google Scholar]
- Richter J, Schlesner M, Hoffmann S, Kreuz M, Leich E, Burkhardt B, et al. (2012). Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nature Genetics, 44(12), 1316–1320. [DOI] [PubMed] [Google Scholar]
- Rivera RR, Johns CP, Quan J, Johnson RS, & Murre C (2000). Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity, 12(1), 17–26. [DOI] [PubMed] [Google Scholar]
- Robey E, & Fowlkes B (1994). Selective events in T cell development. Annual Review of Immunology, 12(1), 675–705. [DOI] [PubMed] [Google Scholar]
- Romanow WJ, Langerak AW, Goebel P, Wolvers-Tettero IL, Van Dongen JJ, Feeney AJ, et al. (2000). E2A and EBF act in synergy with the V (D) J recombinase to generate a diverse immunoglobulin repertoire in nonlymphoid cells. Molecular Cell, 5(2), 343–353. [DOI] [PubMed] [Google Scholar]
- Sakamoto S, Wakae K, Anzai Y, Murai K, Tamaki N, Miyazaki M, et al. (2012). E2A and CBP/p300 act in synergy to promote chromatin accessibility of the immunoglobulin κ locus. The Journal of Immunology, 188(11), 5547–5560. [DOI] [PubMed] [Google Scholar]
- Sathish JG, Johnson KG, Leroy FG, Fuller KJ, Hallett MB, Brennan P, et al. (2001). Requirement for CD28 costimulation is lower in SHP 1 deficient T cells. European Journal of Immunology, 31(12), 3649–3658. [DOI] [PubMed] [Google Scholar]
- Sawada S, & Littman D (1993). A heterodimer of HEB and an E12-related protein interacts with the CD4 enhancer and regulates its activity in T-cell lines. Molecular and Cellular Biology, 13(9), 5620–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayegh C, Jhunjhunwala S, Riblet R, & Murre C (2005). Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes & Development, 19(3), 322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayegh CE, Quong MW, Agata Y, & Murre C (2003). E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nature Immunology, 4(6), 586–593. [DOI] [PubMed] [Google Scholar]
- Schlimgen RJ, Reddy KL, Singh H, & Krangel MS (2008). Initiation of allelic exclusion by stochastic interaction of Tcrb alleles with repressive nuclear compartments. Nature Immunology, 9(7), 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlissel M, Voronova A, & Baltimore D (1991). Helix-loop-helix transcription factor E47 activates germ-line immunoglobulin heavy-chain gene transcription and rearrangement in a pre-T-cell line. Genes & Development, 5(8), 1367–1376. [DOI] [PubMed] [Google Scholar]
- Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, et al. (2012). Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature, 490(7418), 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery C, Mcmorrow T, & Ryan MP (2006). Overexpression of E2A proteins induces epithelial-mesenchymal transition in human renal proximal tubular epithelial cells suggesting a potential role in renal fibrosis. FEBS Letters, 580(17), 4021–4030. [DOI] [PubMed] [Google Scholar]
- Steininger A, Möbs M, Ullmann R, Köchert K, Kreher S, Lamprecht B, et al. (2011). Genomic loss of the putative tumor suppressor gene E2A in human lymphoma. The Journal of Experimental Medicine, 208(8), 1585–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker H, Andjelkovic M, Oldham S, Laffargue M, Wymann MP, Hemmings BA, et al. (2002). Living with lethal PIP3 levels: Viability of flies lacking PTEN restored by a PH domain mutation in Akt/PKB. Science, 295(5562), 2088–2091. [DOI] [PubMed] [Google Scholar]
- Sun X-H, & Baltimore D (1991). An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell, 64(2), 459–470. [DOI] [PubMed] [Google Scholar]
- Sun X-H, Copeland NG, Jenkins NA, & Baltimore D (1991). Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Molecular and Cellular Biology, 11(11), 5603–5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szodoray P, Alex P, Brun J, Centola M, & Jonsson R (2004). Circulating cytokines in primary Sjögren’s syndrome determined by a multiplex cytokine array system. Scandinavian Journal of Immunology, 59(6), 592–599. [DOI] [PubMed] [Google Scholar]
- Tarpley TM Jr., Anderson LG, & White CL (1974). Minor salivary gland involvement in Sjögren’s syndrome. Oral Surgery, Oral Medicine, Oral Pathology, 37(1), 64–74. [DOI] [PubMed] [Google Scholar]
- Thal MA, Carvalho TL, He T, Kim H-G, Gao H, Hagman J, et al. (2009). Ebf1-mediated down-regulation of Id2 and Id3 is essential for specification of the B cell lineage. Proceedings of the National Academy of Sciences, 106(2), 552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber T, Mandel EM, Pott S, Györy I, Firner S, Liu ET,et al. (2010). Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription-independent poising of chromatin. Immunity, 32(5), 714–725. [DOI] [PubMed] [Google Scholar]
- Tuveson DA, Carter RH, Soltoff SP, & Fearon DT (1993). CD19 of B cells as a surrogate kinase insert region to bind phosphatidylinositol 3-kinase. Science, 260(5110), 986–989. [DOI] [PubMed] [Google Scholar]
- Ueda-Hayakawa I, Mahlios J, & Zhuang Y (2009). Id3 restricts the developmental potential of γδ lineage during thymopoiesis. The Journal of Immunology, 182(9), 5306–5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbánek P, Wang Z-Q, Fetka I, Wagner EF, & Busslinger M (1994). Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5BSAP. Cell, 79(5), 901–912. [DOI] [PubMed] [Google Scholar]
- Van Den Berg DJ, Sharma AK, Bruno E, & Hoffman R (1998). Role of members of the Wnt gene family in human hematopoiesis. Blood, 92(9), 3189–3202. [PubMed] [Google Scholar]
- Vanhaesebroeck B, & Alessi D (2000). The PI3K-PDK1 connection: More than just a road to PKB. Biochemical Journal, 346, 561–576. [PMC free article] [PubMed] [Google Scholar]
- Verykokakis M, Boos MD, Bendelac A, Adams EJ, Pereira P, & Kee BL (2010). Inhibitor of DNA binding 3 limits development of murine Slam-associated adaptor protein-dependent “innate” γδ T cells. PLoS ONE, 5(2), e9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verykokakis M, Krishnamoorthy V, Iavarone A, Lasorella A, Sigvardsson M, & Kee BL (2013). Essential functions for ID proteins at multiple checkpoints in invariant NKT cell development. The Journal of Immunology, 191(12), 5973–5983. 10.4049/jimmunol.1301521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Boehmer H (1994). Positive selection of lymphocytes. Cell, 76(2), 219–228. [DOI] [PubMed] [Google Scholar]
- Wang D, Claus CL, Vaccarelli G, Braunstein M, Schmitt TM, Zúñiga-Pflücker JC, et al. (2006). The basic helix-loop-helix transcription factor HEBAlt is expressed in pro-T cells and enhances the generation of T cell precursors. The Journal of Immunology, 177(1), 109–119. [DOI] [PubMed] [Google Scholar]
- Yan W, Young AZ, Soares VC, Kelley R, Benezra R, & Zhuang Y (1997). Molecular and Cellular Biology, 17(12), 7317–7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, et al. (2011). The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nature Immunology, 12(12), 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankee T, & Clark E (1999). Signaling through the B cell antigen receptor in developing B cells. Reviews in Immunogenetics, 2(2), 185–203. [PubMed] [Google Scholar]
- Yu W, Misulovin Z, Suh H, Hardy RR, Jankovic M, Yannoutsos N, et al. (1999). Coordinate regulation of RAG1 and RAG2 by cell type-specific DNA elements 5’ of RAG2. Science, 285(5430), 1080–1084. [DOI] [PubMed] [Google Scholar]
- Zhang B, Dai M, Li Q-J, & Zhuang Y (2013). Tracking proliferative history in lymphocyte development with Cre-mediated sister chromatid recombination. PLoS Genetics, 9(10), e1003887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Lin Y-Y, Dai M, & Zhuang Y (2014). Id3 and Id2 act as a dual safety mechanism in regulating the development and population size of innate-like γδ T cells. The Journal of Immunology, 192, 1055–1063, 1302694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Vilardi A, Neely RJ, & Choi JK (2001). Promotion of cell cycle progression by basic helix-loop-helix E2A. Molecular and Cellular Biology, 21(18), 6346–6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Cheng P, & Weintraub H (1996). B-lymphocyte development is regulated by the combined dosage of three basic helix-loop-helix genes, E2A, E2-2, and HEB. Molecular and Cellular Biology, 16(6), 2898–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Soriano P, & Weintraub H (1994). The helix-loop-helix gene E2A is required for B cell formation. Cell, 79(5), 875–884. [DOI] [PubMed] [Google Scholar]