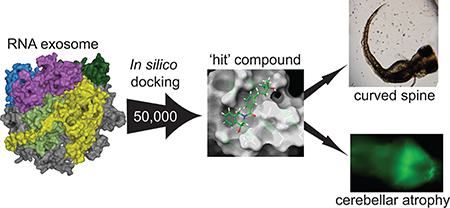
Abstract
Mutations of EXOSC3 have been linked to the rare neurological disorder known as Pontocerebellar Hypoplasia type 1B (PCH1B). EXOSC3 is one of three putative RNA-binding structural cap proteins that guide RNA into the RNA exosome, the cellular machinery that degrades RNA. Using RNAcompete, we identified a G-rich RNA motif binding to EXOSC3. Surface plasmon resonance (SPR) and microscale thermophoresis (MST) indicated an affinity in the low micromolar range of EXOSC3 for long and short G-rich RNA sequences. Although several PCH1B-causing mutations in EXOSC3 did not engage a specific RNA motif as shown by RNAcompete, they exhibited lower binding affinity to G-rich RNA as demonstrated by MST. To test the hypothesis that modification of the RNA–protein interface in EXOSC3 mutants may be phenocopied by small molecules, we performed an in-silico screen of 50 000 small molecules and used enzyme-linked immunosorbant assays (ELISAs) and MST to assess the ability of the molecules to inhibit RNA-binding by EXOSC3. We identified a small molecule, EXOSC3-RNA disrupting (ERD) compound 3 (ERD03), which (i) bound specifically to EXOSC3 in saturation transfer difference nuclear magnetic resonance (STD-NMR), (ii) disrupted the EXOSC3–RNA interaction in a concentration-dependent manner, and (iii) produced a PCH1B-like phenotype with a 50% reduction in the cerebellum and an abnormally curved spine in zebrafish embryos. This compound also induced modification of zebrafish RNA expression levels similar to that observed with a morpholino against EXOSC3. To our knowledge, this is the first example of a small molecule obtained by rational design that models the abnormal developmental effects of a neurodegenerative disease in a whole organism.
Graphical Abstract
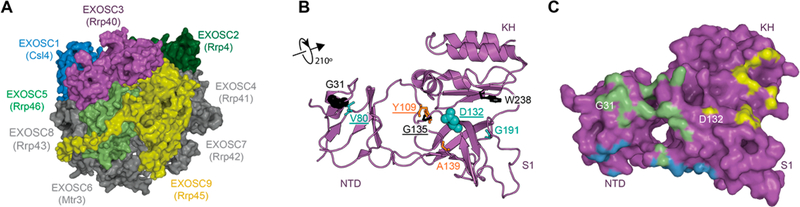
The RNA exosome is ubiquitous cellular machinery that degrades and processes RNA.1–3 The RNA exosome is involved in the processing of small nucleolar RNAs (snoRNAs), small nuclear RNAs (snRNAs), and rRNAs (rRNAs).4 The overall structure of the RNA exosome core (EXOSC1–9) is conserved from prokaryotes to higher eukaryotes and contains a ring-like structure composed of six RNase Pleckstrin homology (PH) domains that form a central channel able to accommodate single-stranded RNA and three additional proteins that form a “cap-like” structure5–8 (Figure 1A). The “cap proteins”—the entry point of RNAs threading through the exosome—are EXOSC1, −2, and −3. EXOSC2 and −3 contain two putative RNA-binding domains, the KH (heterogeneous nuclear ribonucleoprotein K homology) and S1 (ribosomal protein S1) domains, while EXOSC1 only contains an S1 domain; all three proteins have a similar N-terminal domain (NTD; Figures 1B and C). The S1 domains are positioned to bind RNA as it threads through the cap toward the core proteins and central channel of the RNA exosome,9 although it is not known if the KH domains also contribute to RNA binding.
Figure 1.
RNA exosome structure highlighting PCH1B-associated mutations in human EXOSC3. (A) The RNA exosome complex (PDB code: 2nn620) is composed of nine subunits. (B) Cartoon model of EXOSC3. PCH1B-associated amino acids shown as sticks (buried residues) and spheres (surface residues). Residues in black are associated with very poor survival,10,13 orange for those wherein survival is reduced when associated with D132A,10 and cyan for those associated with survival into adulthood.10,11,14 Mutations targeted in this study are underlined. NTD, N-terminal domain; S1, RNA binding S1; and KH, K homology domain. (C) Surface representation of EXOSC3 showing predicted contacts with other subunits. Colors refer to the EXOSC3 contacts with EXOSC5 (light green), EXOSC1 (blue), and EXOSC9 (yellow).
Pontocerebellar Hypoplasia 1B (PCH1B) is a recessive disorder characterized by cerebellar hypoplasia, variable pontine atrophy, and progressive microcephaly with global developmental delay.10 Whole genome sequencing of patients with PCH1B reported several mutations clustered in EXOSC3.10–14 Survival of the patients is correlated with the type of mutation and its association with a nonsense allele. Thus far, eight different point mutations have been reported, as either homozygous or heterozygous morphism associated with PCH1B (G31A, V80F, Y109N, D132A, G135E, A139P, and W238R;10–14 Figure 1C). The majority of these mutations cluster in the S1 domain, one mutation in the KH domain, and two in the NTD of EXOSC3.
Knockdown of EXOSC3 in zebrafish resulted in a PCH1B-like phenotype associated with common alterations in the expression of genes implicated in development including ataxin1b and homeobox gene HOXC.16 Another recent study demonstrated the lack of stability of a yeast analog of EXOSC3 W238R, and its failure to associate efficiently with the exosome in the presence of the wild-type yeast EXOSC3, Rrp40.18 Yet another study reported sequestration of EXOSC3-D132A in the cytosol of patient fibroblasts and an accumulation of mRNA in muscles of these patients; both were linked to mitochondrial dysfunction.19 Thus, accumulating evidence points to a link between mutations in EXOSC3 and PCH1B development.
How mutations in EXOSC3 lead to PCH1B is unknown. In this study, we interrogated how EXOSC3 mutations perturb interactions with other RNA exosome proteins through peptide array analysis and determined the RNA-binding preferences of EXOSC3 and several disease-causing mutations using RNAcompete and microscale thermophoresis (MST). We then used in-silico docking to target the RNA-binding interface of EXOSC3 and identified a small molecule, ERD03, able to induce a PCH1B-like phenotype in zebrafish. Using RNaseq and qPCR, we showed that RNA expression patterns were similar between EXOSC3-knockdown and ERD03-treated zebrafish. Finally, incubation of this compound with zebrafish embryos induced an atrophy of the cerebellum. To our knowledge, this is the first example of a small molecule obtained through targeted structure-based design that models a neurodegenerative disease.
RESULTS AND DISCUSSION
Assessing PCH1B Mutation’s Effect on Exosome Integrity
First, we compared the effect of the mutations on protein–protein contacts within the RNA exosome complex. Guided by the three-dimensional structure of the RNA exosome (PDB ID: 2nn6),20 we identified proteins that directly contact EXOSC3 (Figure 1A, Supporting Information Figure S1): EXOSC1 (another cap protein), EXOSC5, and EXOSC9. Among the PCH1B mutations,10,12–14 only two out of eight mutations, G31A and D132A, are located near protein–protein interfaces in the structure (Figures 1B,C).
As a disease-causing mutant (EXOSC3-W238R) failed to associate efficiently with the exosome in the presence of the wild-type yeast EXOSC3, Rrp40,18 without being close to protein–protein interfaces, we sought to determine if other mutations of EXOSC3 (V80F, Y109N, D132A, and G135E) could alter its binding to EXOSC1, EXOSC5, and EXOSC9, either directly or through an allosteric affect. These mutations were selected based on (i) their location—V80F is in the NTD while Y109N, D132A, and G135E are clustered in the S1 domain (Figure 1B)—and (ii) their varying spectrum of patient survival10,13,14 (Supporting Information Figure S2A). While homozygous D132A mutation, the most common mutation, is linked to a mild phenotype, heterozygosity of D132A with V80F was found to cause a milder PCH1 phenotype.10,13,14 The homozygous G135E and heterozygous D132A/Y109N are found to cause severe phenotypes and are fatal within a few months of infancy.10,13
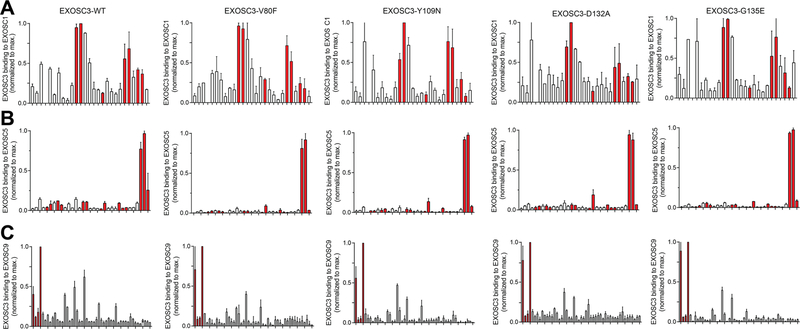
For all subsequent experiments, we purified EXOSC3-wildtype (EXOSC3-WT) and EXOSC3-V80F, EXOSC3-Y109N, EXOSC3-D132A, and EXOSC3-G135E (Supporting Information Figure S2B). To assess potential effects on protein—protein contacts, we constructed peptide arrays that encompass the three proteins interfacing with EXOSC3: EXOSC1, EXOSC5, and EXOSC9. The data were normalized to a maximum binding peptide that was consistently found in each array (Figure 2); the points of contacts based on the crystal structure are highlighted in red (Figure 2). There appears to be partial overlap of predictions and array data for EXOSC1 and −9 and very little overlap for EXOSC5 (Figure 2, Supporting Information Figure S3); the predicted contacts may be affected by crystal packing. On the basis of the normalized data, there are no differences in the ability of EXOSC3 mutants when compared to wild-type to bind to points of contacts of EXOSC1, −5, and −9 (Figure 2). These results suggest that mutations are unlikely to impact exosome integrity through loss of EXOSC3 contact.
Figure 2.
Effect of PCH1B mutations on EXOSC3 binding to adjacent subunits through a peptide array. Binding of EXOSC3-WT and four PCH1B mutants (V80F, Y109N, D132A, G135E) on immobilized 12-mer EXOSC1 (A), EXOSC5 (B), and EXOSC9 (C) peptides, in overlapping eight amino acid steps. The blots were scanned, and spot intensities were quantified and represented as a normalized signal. Predicted contacts with EXOSC3 are shown in red. Data: means ± SD (n = 2).
RNA-Binding by EXOSC3 Is Modified by PCH1B Mutations
As the mutations did not affect EXOSC3’s ability to interact with RNA exosome subunits and are clustered in the RNA-binding domains, we analyzed their impact on RNA binding using RNAcompete21,22 (see Supporting Information Methods) and microscale thermophoresis (MST).
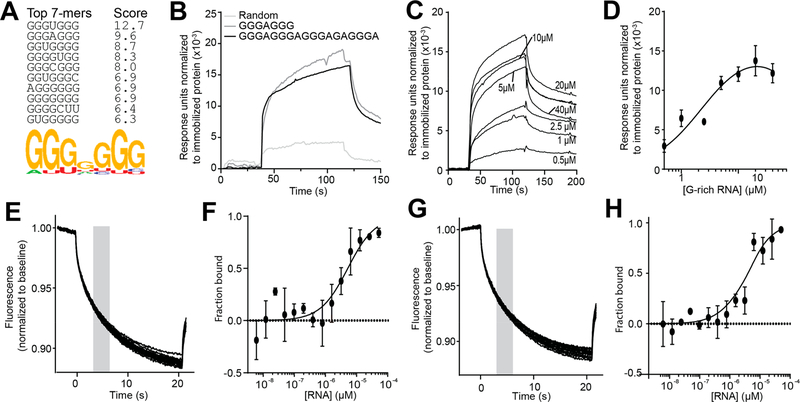
The top 10 7-mers that emerged from EXOSC3-WT converged on a G-rich sequence (top sequences: GGGUGGG and GGGAGGG; Figure 3A, Supporting Information Table 1, Supporting Information Figure S4A). We thus tested the ability of EXOSC3 to bind a short G-rich RNA (GGGAGGG), a random RNA (AGUCAUUC), and a long G-rich sequence (GGGAGGGAGGGAGAGGGA) using surface plasmon resonance (SPR). After normalizing the signal to the molecular weight of the corresponding RNA, we noted no difference in binding of EXOSC3 to the short and long G-rich sequences, while we observed little binding of the random RNA to EXOSC3 (Figure 3B). Further SPR experiments revealed binding of the long G-rich RNA to EXOSC3 with a dissociation constant (Kd) of 2.4 ± 1 μM (Figure 3C,D). MST showed an apparent Kd of 5.8 ± 1.3 μM of NT647-GST-EXOSC3 with long G-rich RNA (Figure 3E,F) and an apparent Kd of 3.6 ± 2.5 μM of NT647-GST-EXOSC3 with short G-rich RNA (Figure 3G,H).
Figure 3.
Profiling RNA binding landscape of EXOSC3. (A) RNA binding specificities of EXOSC3 were determined with RNAcompete. The top 10 binding 7-mer sequences and their Z-scores are shown. Below, the consensus RNA motif is shown as a position weight matrix. (B) SPR sensorgrams showing binding of 200 μm long G-rich RNA (GGGAGGGAGGGAGAGGGA), short G-rich RNA (GGGAGGG), and random RNA (AGUCAUUC) on immobilized EXOSC3. The sensorgrams were normalized to the molecular weight of the respective RNAs. (C) SPR sensorgrams showing binding of increasing concentrations of long G-rich RNA to immobilized EXOSC3 (~0.5 ng). (D) SPR data were fit to a one-site specific binding equation (r2 = 0.91) yielding an apparent Kd of 2.4 ± 1 μM. Data: means ± SD (n = 3). Some error bars are smaller than the symbols. (E) Thermographs of NT-647-GST-EXOSC3 with 0.0015–50 μM of long G-rich RNA. The gray bar represents the time point at which the MST measurements were used in F. (F) MST values yielded an apparent Kd of 5.8 ± 1.3 μM (see Methods for details). (G) Thermographs of NT-647-GST-EXOSC3 with 0.0015–50 μM of short G-rich RNA. The gray bar represents the time point at which the MST measurements were used in (H). (H) MST values yielded an apparent Kd of 3.58 ± 2.5 μM (see Methods for details).
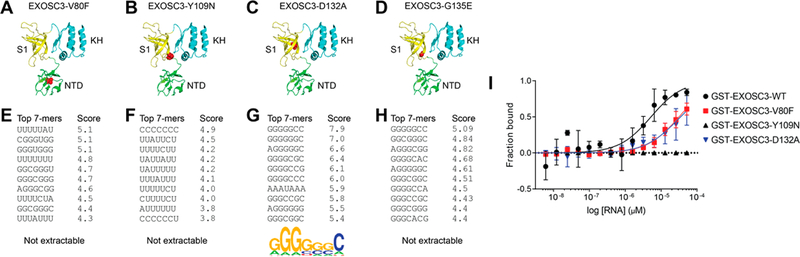
Together, our results demonstrate that EXOSC3 prefers G-rich RNA sequences. However, it is not clear whether the aforementioned EXOSC3 mutations associated with PCH1B also impact RNA binding. We thus used RNAcompete to analyze the RNA-binding preferences of EXOSC3-V80F, Y109N, D132A, and G135E (Figure 4B–D). Except EXOSC3-D132A, all mutants yielded low confidence RNA-compete data (Figure 4E–H, Supporting Information Table 1, Supporting Information Figure S4B–E). For all EXOSC3 mutants, the relative RNA-bindings, approximated by Z-score values, are reduced compared to EXOSC3-WT (Figure 4E–H and Supporting Information Figure S4). EXOSC3-D132A was the only mutant that yielded a motif—GGGGGGC—similar, but not identical, to the GGGAGGG motif of EXOSC3-WT (Figure 4G).
Figure 4.
PCH1B mutations on EXOSC3 altering the RNA binding landscape. Cartoon models of EXOSC3 mutants (in red ball–stick representation): V80F (A), Y109N (B), D132A (C), and G135E (D). The top 10 7-mers with their corresponding Z-scores show the RNA binding landscape for EXOSC3 mutants V80F (e), Y109N (F), D132A (G), and G135E (H); an RNA motif as a position weight matrix was extractable for D132A. (I) MST experiments yielded apparent Kd values of 31.8 ± 11 μM(V80F) and 46.8 ± 19 μM(D132A), which were higher compared to 5.8 ± 1.3 μM (EXOSC3-WT). No association could be detected between long G-rich RNA (GGGAGGGAGGGAGAGGGA) and GST-EXOSC3-Y109N; no binding curve was extractable. Means ± SD (n = 3). Some error bars are smaller than the symbols.
MST was next used to assess the effect of disease-causing mutations on EXOSC3’s binding to long G-rich RNA (Figure 4I). Robust thermodiffusion of labeled EXOSC3-WT, EXOSC3-V80F, EXOSC3-Y109N, and EXOSC3-D132A was observed; NT647-EXOSC3-G135E had a high propensity to adsorb to the capillaries and therefore was not used further. Compared to a Kd of 5.8 ± 1.3 μM of EXOSC3-WT (Figure 3F), EXOSC3-V80F and D132A decreased the Kd values for RNA to 31.8 ± 11 μM and 46.8 ± 19 μM, respectively (Figure 4I). No binding was detected for EXOSC3-Y109N under our conditions (Figure 4I).
Combined, our results demonstrate that EXOSC3 recognizes G-rich sequences and that mutations V80F, Y109N, D132A, and G135E have impaired and/or modified RNA binding. The defects in RNA binding and enrichment of RNA motifs of mutant EXOSC3 proteins shows some correlation with PCH1B disease severity. For example, mutant EXOSC3-V80F has reduced interaction with EXOSC1 and EXOSC9 (peptide array), only slightly decreases RNA binding (MST), and is linked to a good survival rate; mutant EXOSC3-Y109N, linked with a poor survival,13 showed no binding to G-rich RNA (MST). Mutant EXOSC3-G135E, also associated with poor survival,13 has no enriched binding motif (RNAcompete), and mutant EXOSC3-D132A, which has a similar motif as EXOSC3-WT, is linked to the mildest form of PCH1B.10,13
Rationale for Targeting EXOSC3/RNA Interface
Modeling neurodegenerative diseases is challenging given the complexity arising from development, identification of a vulnerable window of onset, and the multifactorial etiology of these diseases.26,27 Compounding the problem is the fact that often neurodegenerative diseases arise in patients due to diverse mutations within protein families, thus making it difficult to create models that capture the full spectrum of the disease. A differentiated approach is to understand the structural and mechanistic basis of diseases. We demonstrated that RNA binding is altered by mutations in the S1 domain of EXOSC3. Next, we asked if targeting RNA/EXOSC3 interfaces with small molecules could mimic the altered RNA-binding landscape of EXOSC3 to recapitulate cerebellar atrophy, the hallmark of PCH1B.
Our rationale was that successful identification of such molecules could enrich our understanding on the importance of the RNA-binding domain for EXOSC3’s function and lead to induction of a PCH1B-like phenotype in zebrafish embryos, thus creating a new model of this disease. Zebrafish have emerged as a useful model organism for developmental biology, because of their physiologic similarities to humans,28 their fast-developing rate, and their translucent skin during larval stages permitting brain imaging. Previous studies demonstrated that morpholino knockdown of EXOSC3 in zebrafish embryos caused the phenotype of a short, curved spine and underdeveloped brain, associated with poor motility and death by 3 days post-fertilization (dpf).10,16 Imaging of the Purkinje cells in EXOSC3-knockdown zebrafish further demonstrated cerebellar defects. Those observations modeled critical elements of the clinical disease of PCH1B and established EXOSC3-knockdown in the zebrafish as a facile model of this disease.
In-Silico Screen of EXOSC3 RNA-Protein Interfaces
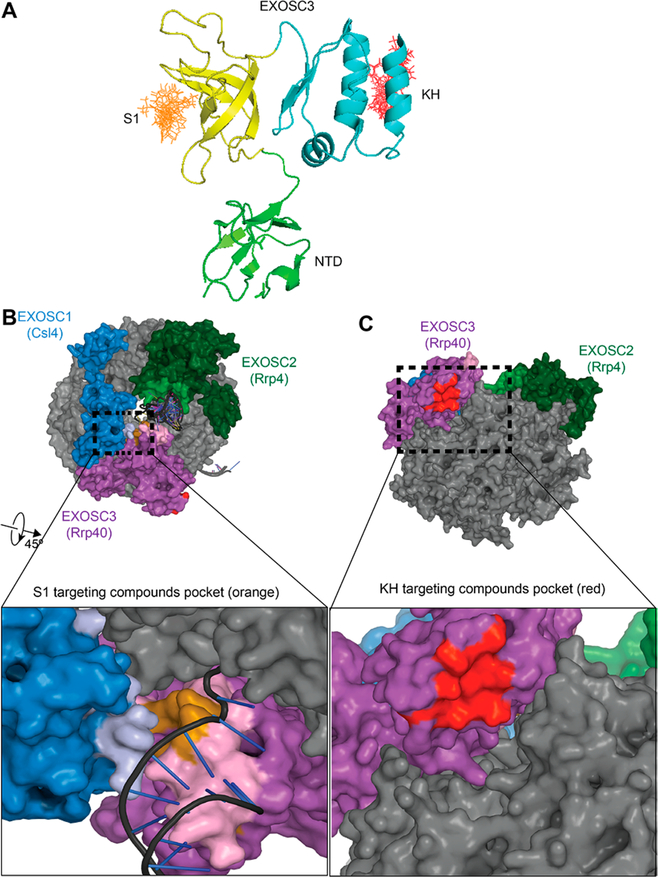
Using the Schrödinger Suite, we performed an in-silico docking experiment targeting residues in the two RNA binding domains—Gln244, Asn251, Ile252, Leu271, and Ser274 in the KH domain and Phe146, Glu147, Arg152, Asn188, and Met190 in the S1 domain. A 50 000-compound library was docked on a 10 Å grid surrounding these residues (Figure 5) using Glide. On the basis of this docking, two pockets emerged in the KH and S1 domains of EXOSC3 (Figure 5A). The S1-targeting compounds appeared to cluster in a region where RNA threads into the exosome (Figure 5B). The predicted RNA binding interface and the molecule binding site show a modest overlap (Figure 5B, inset). While the KH domain compounds are not positioned directly in the path of RNA threading into the exosome, the KH domain RNA-binding interface is accessible and may be necessary for RNA binding (Figure 5C).
Figure 5.
Docking of small molecules onto EXOSC3. (A) The top 20 molecules (stick representation) were docked on the S1 (yellow) or the KH (blue) domain of EXOSC3 (carton representation). (B) RNA exosome with core components colored in gray. Cap proteins EXOSC1, EXOSC2, and EXOSC3 are shown in blue, dark green, and purple, respectively. Close up of S1 domain binding site (orange) showing significant overlap with a predicted RNA interface on EXOSC3 (pink). Cartoon representation of RNA from yeast structure 5jea (for reference). EXOSC2 is omitted in the inset. (C) Rotated view of the RNA exosome showing the docking pocket in the KH domain (red). Close-up of KH targeting compounds docking site showing pocket accessibility.
Biophysical Triage of Putative EXOSC3-RNA-Disrupting (ERD) Compounds
The top 20 selected compounds from in-silico docking (Supporting Figure S5, Supporting Table 2) were divided into molecules that target the KH domain (EXOSC3 RNA Disruptors ERD01-ERD12) and molecules that target the S1 interface (ERD13-ERD20).
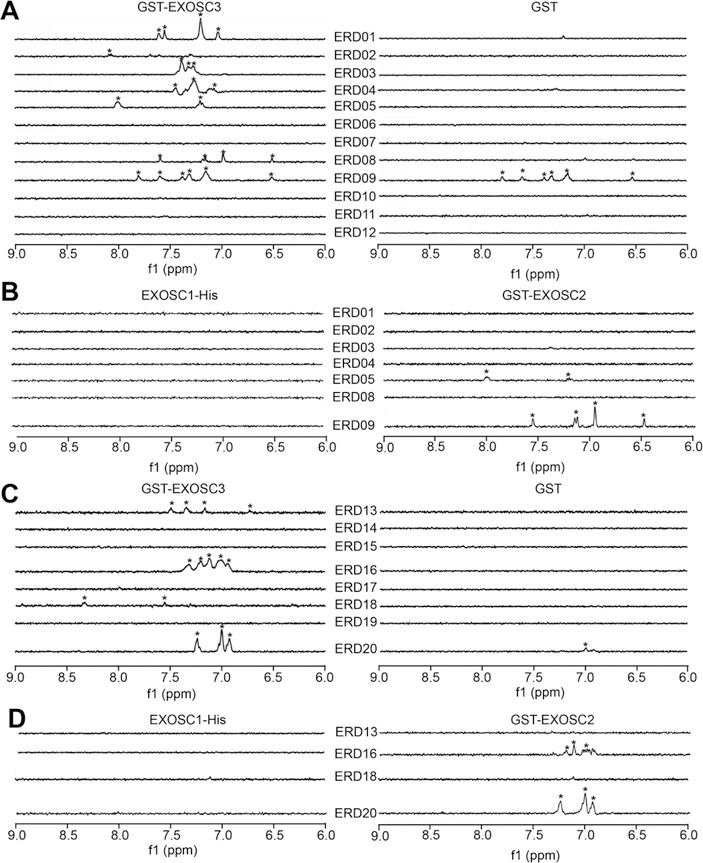
Following docking, saturation transfer difference NMR (STD-NMR) was used to test the ability of compounds to bind to EXOSC3. GST was used as a control for off-target binding. STD-NMR demonstrated that seven compounds targeting KH (ERD 01, 02, 03, 04, 05, 08, 09) and four compounds targeting S1 (ERD13, 16, 18, 20) bound EXOSC3 (Figure 6A,C). Compounds ERD09 and ERD20 also bound GST, typifying pan assay interference compounds (PAINS)30,31 (Figure 6A and C, right panels). Compounds ERD16, ERD20, and ERD05 also bound to GST-EXOSC2, but not to EXOSC1-His (Figure 6B,D).
Figure 6.
STD-NMR analysis of binding of small molecules to exosome subunits. 1D 1H STD-NMR showing on-resonance difference spectrum of 500 μM (EXOSC3/RNA disrupting (ERD)) compounds with 5 μM proteins. The aromatic region of the spectrum (6–9 ppm) is shown. (A) 1D 1H STD-NMR spectra of KH domain-targeting ERD01–ERD12 compounds binding to GST-EXOSC3 (left) or GST (right). (B) 1D 1H STD-NMR of ERD compounds that bound to GST-EXOSC3, but not GST, were tested against EXOSC1-His (left) and GST-EXOSC2 (right). (C, D) Same as A and B except with compounds ERD13–ERD20 (targeting S1 domain). (*protons reproducibly found in independent experiments (n = 3).)
With the above biophysical triage steps, we identified several molecules that met key criteria: (i) binding to EXOSC3 but not EXOSC1 or EXOSC2; (ii) no binding to GST; and (iii) did not fit the structure of PAINs compounds (“false positive” in high throughput screen). Selected compounds were ERD01, ERD02, ERD03, ERD04, and ERD08 (for the KH domain) and ERD13 and ERD18 (for the S1 domain).
Disruption of EXOSC3/RNA Interactions with ELISA
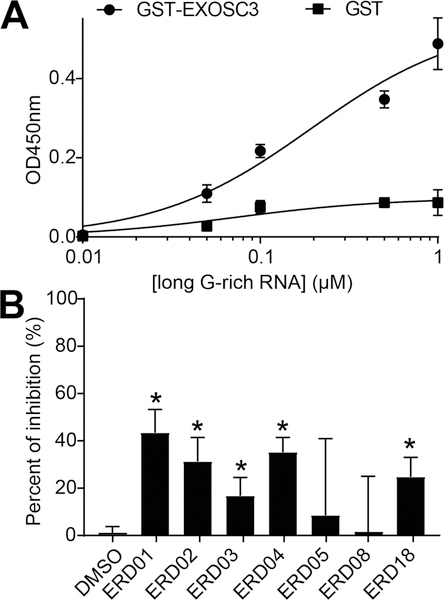
We next asked if these compounds, used at 50 μM, could disrupt EXOSC3–RNA interactions using an enzyme-linked immunosorbant assay (ELISA). As expected, binding was observed between G-rich RNA and EXOSC3 (Figure 7A). Among compounds that target the KH domain, ERD01 was the most effective with ~45% inhibition, followed by ERD04 and ERD02 (~38% and 35% inhibition, respectively), and ERD03 with ~18% inhibition. Among compounds that target the S1 domain, ERD18 inhibited binding by ~30% (Figure 7B).
Figure 7.
RNA/EXOSC3 disruption by small molecules. (A) 300 nM GST-EXOSC3 was incubated with increasing concentrations of long G-rich RNA. The data were fit to a one-site specific binding equation (r2 = 0.94). Long G-rich RNA bound to EXOSC3 with a half-saturation concentration of ~100 nM. Binding of GST alone demonstrated minimal background binding to RNA. Data: means ± SD (n = 6). Some error bars are smaller than the symbols. (B) ERD compounds specifically binding to EXOSC3 were tested for their ability to disrupt EXOSC3/RNA interaction. 50 μM ERDs were incubated with 0.2 μM RNA and GST-EXOSC3. Data: mean ± SD (*one-way ANOVA, p < 0.05, n = 6–12).
An ERD Compound Recapitulates PCH1B Phenotype in Development in Zebrafish Embryos
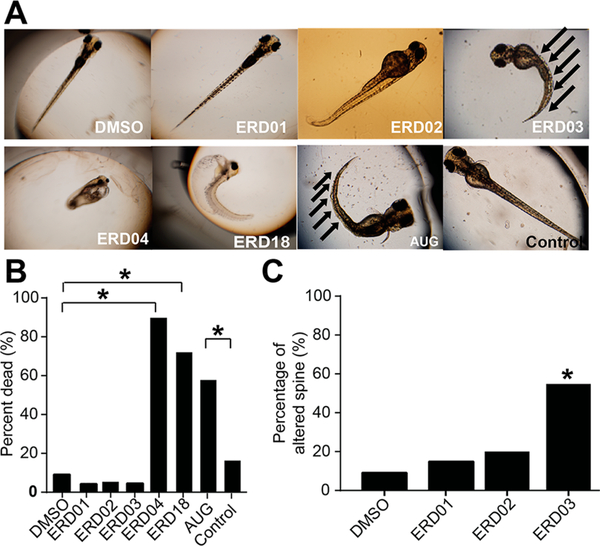
To examine possible functional consequences of ERD compounds, we incubated zebrafish embryos with selected ERDs and monitored their phenotype. No major differences are expected between the structures of human and zebrafish EXOSC3 proteins in terms of the sites that were targeted by the small molecules (Supporting Information Figure S6). A morpholino that targets the AUG translational initiation site (AUG morpholino) resulted in a pronounced phenotype with most zebrafish having a curved spine and a high incidence of death, similar to the phenotype previously reported;10 control morpholinos did not affect zebrafish development (Figure 8A,B). Of five ERD compounds tested in zebrafish, ERD18 and ERD04 led to ~75% and 90% death, respectively, by 3–5 days postfertilization (dpf), whereas ERD01, ERD02, and ERD03 induced death equivalent to DMSO. Only ERD03 caused significant spinal curvature (Figure 8A–C) and was thus selected as the lead compound that might phenocopy the PCH1B disease in zebrafish.
Figure 8.
ERD compounds induce a PCH1B-like phenotype in zebrafish. (A) Zebrafish embryos were incubated with 50 μM ERDs, DMSO (0.5%), or injected with morpholinos (AUG vs control) for 5 days before imaging. (B) The percent of dead zebrafish, assessed as having no heartbeat, is shown for each condition. (C) Bar graphs of percentage of zebrafish with short, curved spine (*Χ2 test, p < 0.001, n ≥ 50).
Characterization of Binding Affinity and Inhibition of RNA-EXOSC3 Interaction by ERD03
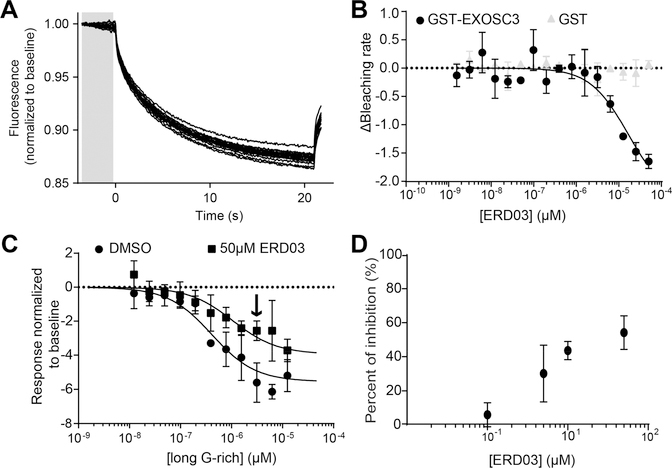
We showed that ERD03 binds specifically EXOSC3 (Figure 6), disrupts the EXOSC3/RNA interface (Figure 7), and causes a spinal curvature in zebrafish (Figure 8). To further characterize ERD03, we measured ERD03’s binding affinity for EXOSC3 using MST. Photobleaching experiments revealed that ERD03 bound specifically to EXOSC3 with an apparent Kd of 17 ± 7 μM (Figure 9A and B); no binding to GST was observed.
Figure 9.
Biophysical characterization of ERD03. (A) Thermographs of NT-647-GST-EXOSC3 with ranging concentrations of ERD03 (0.00153–50 μM). The gray bar represents the time point at which the MST measurements were used in B. (B) Photobleaching rates of EXOSC3’s initial fluorescence in the presence of 0.00153–50 μM ERD03 were used to determine ERD03 affinity for EXOSC3. The data were fitted (see Methods) with an apparent Kd =17 ± 7 μM (r2 = 0.96). GST protein was used as a negative control. Data: mean ± SD (n = 3). (C) MST values of EXOSC3 with 0.012–12.5 μM of long G-rich RNA and 1% (v/v) DMSO or 50 μM ERD03. Arrow denotes 3.12 μM RNA, which caused 54 ± 10% inhibition of EXOSC3-RNA by ERD03. (D) Concentration-dependent curves were obtained for ERD03’s disruption of RNA-EXOSC3 interaction at 3.12 μM of RNA. Data are represented as mean ± SD (n = 3).
Next, we measured inhibition of RNA/EXOSC3 by ERD03 using MST. We first incubated EXOSC3 with several concentrations of long G-rich RNA with DMSO or 50 μM ERD03 (Figure 9C). ERD03 decreased EXOSC3 binding to long G-rich RNA at several concentrations of RNA. At a concentration of 3.1 μM of RNA, ERD03 inhibited binding by 54% (Figure 9C, arrow). This 3.1 μM concentration of RNA was then used against several concentrations of ERD03. Data indicated a concentration-dependent inhibition for ERD03 (Figure 9D). Inhibition was also measured using ELISA but yielded less inhibition, albeit still with a concentration dependency (Supporting Information Figure S7A). We also tested the effect of ERD03 on EXOSC3 interaction with short G-rich RNA using MST, and we observed an inhibition similar to long G-rich RNA at 50 μM of ERD03 (Supporting Information Figure S7B).
To examine whether the compounds interfered with protein–protein interactions in the RNA exosome proteins, we tested EXOSC3’s interactions with EXOSC1, EXOSC5, and EXOSC9 using peptide arrays in the presence of ERD03 and ERD01: no major differences were found between the peptide–protein interactions of EXOSC3 with the compounds (Supporting Information Figure S8).
Optimization of ERD03 to gain better potency would be advantageous in order to more finely tune EXOSC3/RNA inhibition. The docking model of ERD03 (Supporting Information Figure S5) with EXOSC3 provides us a starting point for lead optimization. The structure of ERD03 consists of an isoquinoline ring with a butylbenzoyl group attached to a nitrogen atom (Supporting Information Table 2), and an initial lead optimization program could focus on the butylbenzoyl group with an aim to gain additional hydrogen bonding and other interactions in the active site.
ERD03 Recapitulates EXOSC3 Knockdown Effects on RNA Transcriptomic Profiles in Zebrafish
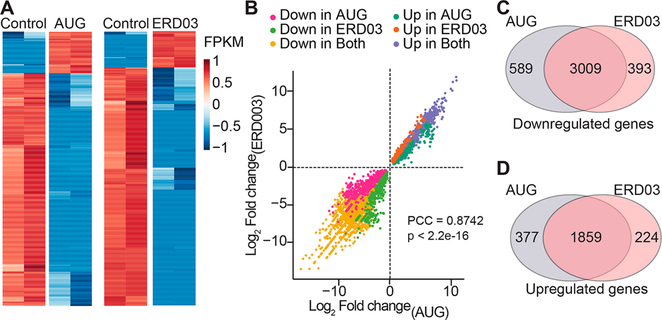
Knockout of the EXOSC3 in mouse and in zebrafish has been previously shown to induce accumulation of certain types of RNA including long noncoding RNA (lncRNA).16,32,33 To address if ERD03 does the same, RNaseq analysis was performed on 3 dpf zebrafish incubated with ERD03 or injected with AUG morpholino (Figure 10, Supporting Information Table 3). Most transcripts appear to be affected in a similar fashion by AUG injection and ERD03 incubation compared to the control (Figure 10A). Change in relative expression of each transcript, expressed as log2 fold change, correlated well between the two conditions (Figure 10B) as demonstrated by a Pearson correlation factor of 0.87 (p < 2.2e-16). A total of 3009 RNAs were downregulated and 1859 RNAs were upregulated in both AUG and ERD03 treatments (Figure 10C,D), leading to an ~80% overlap of differentially expressed RNAs (75%, p < 0.01; 83%, p < 0.05). As previously described, lncRNAs constituted a greater percentage of the upregulated vs downregulated genes (13.5% vs 6%, p < 0.001 by Fisher’s exact test). There is substantial similarity and overlap in the effect on lncRNAs between EXOSC3 knockdown and ERD03 treatment. The observed strong positive correlation in gene expression suggests that pathways downstream of EXOSC3 are being impacted in a similar manner and degree between the ERD03 and EXOSC3 knockdown backgrounds. These data further support our assertion that ERD03 can be used as a pharmacological tool to modulate EXOSC3 activity and mimic a PCH1B phenotype.
Figure 10.
Transcriptomics in zebrafish after ERD03 incubation or EXOSC3-knockdown. (A) Heatmap of log2 FPKM (fragments per kilobase of transcript per million mapped reads) values for the top 1000 differentially expressed genes in morpholino knockdown samples (AUG) or samples treated with ERD03 compared to control. FPKM represents the absolute value of a transcript expression. (B) Distribution of log2 fold-change values for all differentially expressed genes (relative to control with an adjusted p < 0.01) in the ERD03-treated and AUG-knockdown samples. Values are color-coded based on the treatment in which they were significantly differentially expressed (relative to controls). The Pearson correlation coefficient (PCC) was used to assess similarity between AUG and ERD03 treatments (p < 2.2e-16). Venn diagram depicting distribution of overlapping downregulated (C) or upregulated (D) RNAs in the treatments. Full data: https://www.ncbi.nlm.nih.gov/sra-bioproject, ID PRJNA470927.
ERD03 Induces an Atrophy of the Zebrafish Cerebellum, a Clinical Feature of PCH1B
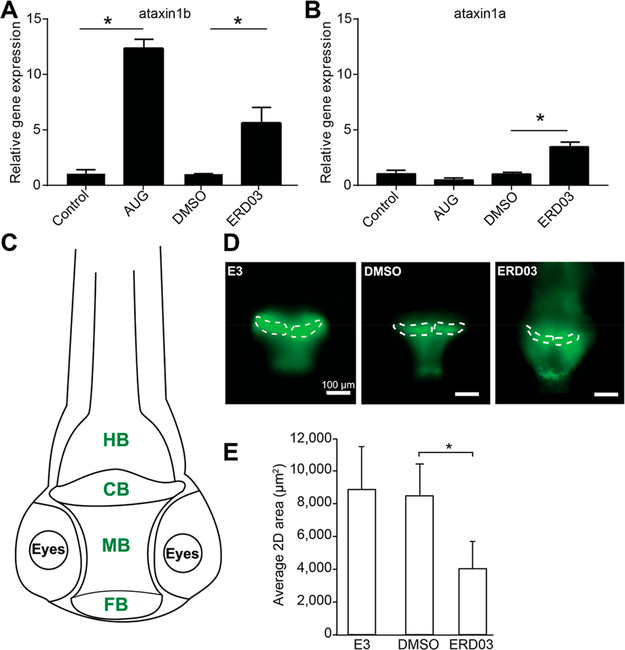
PCH1B patients exhibit cerebellar atrophy with dysmorphic Purkinje cells,10 and EXOSC3 knockdown in zebrafish previously showed significant cerebellum defects.16 Ataxin, a DNA-binding protein encoded by the ATXN1 gene, is important for differentiation of Purkinje cells in the mouse,34 and defects in ATXN1 were linked to spinocerebellar ataxia type 1,35 a disorder similar to PCH1B, characterized by progressive movement problems due to Purkinje cell degeneration and subsequent cerebellar atrophy.36 Furthermore, ataxin1b mRNA was previously reported to be significantly upregulated with EXOSC3-knockdown in zebrafish embryos and was linked to a toxic effect on the cerebellum development.16 We performed quantitative RT-PCR (qPCR) on RNA extracted from zebrafish incubated with ERD03, DMSO control, or injected with AUG morpholino (Figure 11A). ERD03 induced a ~6-fold upregulation of ataxin1b mRNA (Figure 11A). Although ataxin1a mRNA was not modified by AUG morpholino, there is a minor accumulation of ataxin1a mRNA in embryos with ERD03 (Figure 11B).
Figure 11.
ERD03 effect on cerebellum development. Gene expression analysis of atxn1b (A) and atxn1a (B) in zebrafish following ERD03 incubation or EXOSC3-knockdown. Data: mean ± SD (*one-way ANOVA, p < 0.001, n ≥ 50). (C) Schematic depicting the localization of the zebrafish brain. The TDL6 zebrafish line expresses GFP in neurons of the central nervous system. Green text represents tissues expressing GFP. HB, hindbrain; CB, cerebellum; MB, midbrain; FB, forebrain. (D) Fluorescent images of 3dpf zebrafish embryos’ brains incubated with E3 buffer, DMSO (0.5%, v/v), or 50 μM ERD03. The area used to calculate cerebellum average size is represented in white within each image. (E) Average cerebellum size per 2D cross-section is shown. Data: mean ± SD (*one-way ANOVA with Tukey HSD posthoc test, p < 0.05, n = 3).
Because of the ability of ERD03 to induce ataxin1b mRNA accumulation in zebrafish, linked to an underdeveloped cerebellum, we imaged zebrafish brains in the presence of ERD03. We used the Tübingen driver line (TDL6) zebrafish line where galactose (Gal4)-driven green-fluorescent protein (GFP) expression marks neurons of the central nervous system (Figure 11C).37 We incubated TDL6 zebrafish embryos with either buffer E3, DMSO, or 50 μM ERD03 and imaged the brains at 3 dpf (Figure 11D). Incubation with ERD03 resulted in zebrafish cerebella half the size of control brains in DMSO (Figure 11D and E). Taken together, these results demonstrate that disrupting the EXOSC3–RNA interaction in zebrafish embryos recapitulates several hallmarks of PCH1B: (i) modification of RNA transcription profiles, (ii) spine curvature, and (iii) accumulation of atxn1b mRNA combined with an atrophy of the cerebellum. On the basis of these observations, we propose that ERD03 could be used as a chemical tool to model the progression of PCH1B neurological disease through disruption of the EXOSC3/RNA interactions. Further studies with ERD03 will shed light on mechanisms of RNA processing and development of the cerebellum. We may uncover additional mechanisms to the RNA displacement since ERD03 binding is 3-fold weaker than RNA binding to EXOSC3; alternatively, such a dramatic phenotype in the zebrafish may be due to cumulative RNA defects.
The translational significance of this work is the possibility of discovering small molecules to manage progression of PCH1B-phenotype in model organisms and could be applicable to other neurological disorders. This chemical approach to mimic neurological disorders caused by mutations may be an instructive example of how to model rare diseases and could be utilized widely in the drug discovery field to develop testable mechanistic hypotheses and drive therapeutic development. To our knowledge, this is the first example of a small molecule obtained by rational design that models the abnormal developmental effects of a neurodegenerative disease in a whole organism and could become an invaluable tool when genetic modifications are difficult to obtain in conventional model species.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the 2014 AHSC-CDA Arizona Health Sciences Career Development Award and AZ ABRC ADHS16–162407 Arizona grant (to M.K.); NSF grant IOS 1444490 (to E.L. and M.B.); NIH R01HG008613 (to T.R.H. and Q.D.M.) and NIH R01GM61721; and USAMRAA W81XWH-17–1-0333 (to C.K.).
Footnotes
The authors declare no competing financial interest.
EXPERIMENTAL PROCEDURES
See the Supporting Information Methods.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschem-bio.8b00745.
RNAcompete 7-mer sequences and Z-scores (XLSX)
RNaseq results (XLSX)
experimental procedures; detailed map of interactions between EXOSC3, EXOSC1, EXOSC5, and EXOSC9; stability of disease causing mutations on EXOSC3; raw data of peptide arrays between EXOSC3 and EXOSC1, EXOSC5, and EXOSC9; Z-score distribution of RNA 7-mers from RNAcompete on EXOSC3 proteins; top hits from in silico docking poses for each domain; comparison of human structures of EXOSC3 with homology model of zebrafish EXOSC3; ERD03 disruption of EXOSC3-RNA interaction by MST; effect of ERD03 and ERD18 binding to adjacent subunits assayed by peptide array; compound structures of top hits from in silico docking; primers used for protein cloning; primers used to generate EXOSC3 mutants; qPCR primers used to assess mRNA levels in zebrafish embryos (PDF)
REFERENCES
- (1).Mitchell P, Petfalski E, Shevchenko A, Mann M, and Tollervey D (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′ to 5′ exoribonucleases. Cell 91, 457–466. [DOI] [PubMed] [Google Scholar]
- (2).Decker CJ (1998) The exosome: a versatile RNA processing machine. Curr. Biol 8, R238–R240. [DOI] [PubMed] [Google Scholar]
- (3).Anderson JSJ, and Parker R (1998) The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SK12 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 17, 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, and Tollervey D (1999) Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18, 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Makino DL, Baumgartner M, and Conti E (2013) Crystal structure of an rna-bound 11-subunit eukaryotic exosome complex. Nature 495, 70–75. [DOI] [PubMed] [Google Scholar]
- (6).Kowalinski E, Kogel A, Ebert J, Reichelt P, Stegmann E, Habermann B, and Conti E (2016) Structure of a Cytoplasmic 11-Subunit RNA Exosome Complex. Mol. Cell 63, 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Makino DL, and Conti E (2013) Structure determination of an 11-subunit exosome in complex with RNA by molecular replacement. Acta Crystallogr., Sect. D: Biol. Crystallogr 69, 2226–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Wasmuth EV, Januszyk K, and Lima CD (2014) Structure of an Rrp6-RNA exosome complex bound to poly(A) RNA. Nature 511, 435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Januszyk K, and Lima CD (2010) Structural components and architectures of RNA exosomes. Adv. Exp. Med. Biol 702, 9–28. [PMC free article] [PubMed] [Google Scholar]
- (10).Wan J, Yourshaw M, Mamsa H, Rudnik-Schoneborn S, Menezes MP, Hong JE, Leong DW, Senderek J, Salman MS, Chitayat D, Seeman P, Von Moers A, Graul-Neumann L, Kornberg AJ, Castro-Gago M, Sobrido MJ, Sanefuji M, Shieh PB, Salamon N, Kim RC, Vinters HV, Chen Z, Zerres K, Ryan MM, Nelson SF, and Jen JC (2012) Mutations in the RNA exosome component gene EXOSC3 cause pontocerebellar hypoplasia and spinal motor neuron degeneration. Nat. Genet 44, 704–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Halevy A, Lerer I, Cohen R, Kornreich L, Shuper A, Gamliel M, Zimerman B. El Korabi I, Meiner V, Straussberg R, and Lossos A (2014) Novel EXOSC3 mutation causes complicated hereditary spastic paraplegia. J. Neurol 261, 2165–2169. [DOI] [PubMed] [Google Scholar]
- (12).Schwabova J, Brozkova DS, Petrak B, Mojzisova M, Pavlickova K, Haberlova J, Mrazkova L, Hedvicakova P, Hornofova L, Kaluzova M, Fencl F, Krutova M, Zamecnik J, and Seeman P (2013) Homozygous EXOSC3 mutation c.92G→C, p.G31A is a founder mutation causing severe pontocerebellar hypoplasia type 1 among the Czech Roma. J. Neurogenet 27, 163–169. [DOI] [PubMed] [Google Scholar]
- (13).Eggens VRC, Barth PG, Niermeijer JMF, Berg JN, Darin N, Dixit A, Fluss J, Foulds N, Fowler D, Hortobagyi T, Jacques T, King MD, Makrythanasis P, Mate A, Nicoll JAR, O’Rourke D, Price S, Williams AN, Wilson L, Suri M, Sztriha L, Dijns-De Wissel MB, Van Meegen MT, Van Ruissen F, Aronica E, Troost D, Majoie CBLM, Marquering HA, Poll-Thé BT, and Baas F (2014) EXOSC3 mutations in pontocerebellar hypoplasia type 1: Novel mutations and genotype-phenotype correlations. Orphanet J. Rare Dis, 9, 23,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Zanni G, Scotton C, Passarelli C, Fang M, Barresi S, Dallapiccola B, Wu B, Gualandi F, Ferlini A, Bertini E, and Wei W (2013) Exome sequencing in a family with intellectual disability, early onset spasticity, and cerebellar atrophy detects a novel mutation in EXOSC3. Neurogenetics 14, 247–250. [DOI] [PubMed] [Google Scholar]
- (15).Boczonadi V, Müller JS, Pyle A, Munkley J, Dor T, Quartararo J, Ferrero I, Karcagi V, Giunta M, Polvikoski T, Birchall D, Princzinger A, Cinnamon Y, Lutzkendorf S, Piko H, Reza M, Florez L, Santibanez-Koref M, Griffin H, Schuelke M, Elpeleg O, Kalaydjieva L, Lochmuller H, Elliott DJ, Chinnery PF, Edvardson S, and Horvath R (2014) EXOSC8 mutations alter mRNA metabolism and cause hypomyelination with spinal muscular atrophy and cerebellar hypoplasia. Nat. Commun, 5, DOI: 10.1038/ncomms5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Giunta M, Edvardson S, Xu Y, Schuelke M, Gomez-Duran A, Boczonadi V, Elpeleg O, Müller JS, and Horvath R (2016) Altered RNA metabolism due to a homozygous RBM7 mutation in a patient with spinal motor neuropathy. Hum. Mol. Genet 25, 2985–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Gillespie A, Gabunilas J, Jen JC, and Chanfreau GF (2017) Mutations of EXOSC3/Rrp40p associated with neurological diseases impact ribosomal RNA processing functions of the exosome. RNA 23, 466–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Fasken MB, Losh JS, Leung SW, Brutus S, Avin B, Vaught JC, Potter-Birriel J, Craig T, Conn GL, Mills-Lujan K, Corbett AH, and van Hoof A (2017) Insight into the RNA exosome complex through modeling pontocerebellar hypoplasia type 1b disease mutations in yeast. Genetics 205, 221–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Schottmann G, Picker-Minh S, Schwarz JM, Gill E, Rodenburg RJT, Stenzel W, Kaindl AM, and Schuelke M (2017) Recessive mutation in EXOSC3 associates with mitochondrial dysfunction and pontocerebellar hypoplasia. Mitochondrion 37, 46–54. [DOI] [PubMed] [Google Scholar]
- (20).Liu Q, Greimann JC, and Lima CD (2006) Reconstitution, Activities, and Structure of the Eukaryotic RNA Exosome. Cell 127, 1223–1237. [DOI] [PubMed] [Google Scholar]
- (21).Ray D, Ha KCH, Nie K, Zheng H, Hughes TR, and Morris QD (2017) RNAcompete methodology and application to determine sequence preferences of unconventional RNA-binding proteins. Methods 118–119, 3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Ray D, Kazan H, Chan ET, Castillo LP, Chaudhry S, Talukder S, Blencowe BJ, Morris Q, and Hughes TR (2009) Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat. Biotechnol 27, 667–670. [DOI] [PubMed] [Google Scholar]
- (23).Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, Na H, Irimia M, Matzat LH, Dale RK, Smith SA, Yarosh CA, Kelly SM, Nabet B, Mecenas D, Li W, Laishram RS, Qiao M, Lipshitz HD, Piano F, Corbett AH, Carstens RP, Frey BJ, Anderson RA, Lynch KW, Penalva LOF, Lei EP, Fraser AG, Blencowe BJ, Morris QD, and Hughes TR (2013) A compendium of RNA-binding motifs for decoding gene regulation. Nature 499, 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Lunde BM, Moore C, and Varani G (2007) {RNA}-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol 8, 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Sauer E (2013) Structure and RNA-binding properties of the bacterial LSm protein Hfq. RNA Biol. 10, 610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Noble W, and Burns MP (2010) Challenges in neurodegeneration research. Front. Psychiatry, DOI: 10.3389/fpsyt.2010.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Young AB (2009) Four decades of neurodegenerative disease: How far we have come! J. Neurosci. 29, 12722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Cao Y, Jiang L, Zhao L, Zhou X, Wang N, Zhang P, Tang Y, and Zhou J (2015) Evaluation of the in vivo safety profiles of Rictor inhibition using a zebrafish model. Curr. Pharm. Des 21, 1645–1653. [DOI] [PubMed] [Google Scholar]
- (29).Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, and Mainz DT (2006) Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem 49, 6177–6196. [DOI] [PubMed] [Google Scholar]
- (30).Dahlin JL, Nissink JWM, Strasser JM, Francis S, Higgins L, Zhou H, Zhang Z, and Walters MA (2015) PAINS in the assay: Chemical mechanisms of assay interference and promiscuous enzymatic inhibition observed during a HTS. J. Med. Chem 58, 2091–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Capuzzi SJ, Muratov EN, and Tropsha A (2017) Phantom PAINS: Problems with the Utility of Alerts for P an- A ssay in terference Compound S. J. Chem. Inf. Model 57, 417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Pefanis E, Wang J, Rothschild G, Lim J, Chao J, Rabadan R, Economides AN, and Basu U (2014) Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature 514, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu ZP, Economides AN, Bradner JE, Rabadan R, and Basu U (2015) RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 161, 774–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Ebner B. a, Ingram MA, Barnes JA, Duvick LA, Frisch JL, Clark HB, Zoghbi HY, Ebner TJ, and Orr HT (2013) Purkinje cell ataxin-1 modulates climbing fiber synaptic input in developing and adult mouse cerebellum. J. Neurosci 33, 5806–5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Matilla-Dueñas A, Goold R, and Giunti P (2008) Clinical, genetic, molecular, and pathophysiological insights into spinocerebellar ataxia type 1. Cerebellum 7, 106–14. [DOI] [PubMed] [Google Scholar]
- (36).Huang M, and Verbeek DS (2018) Why do so many genetic insults lead to Purkinje Cell degeneration and spinocerebellar ataxia? Neurosci. Lett, DOI: 10.1016/j.neulet.2018.02.004. [DOI] [PubMed] [Google Scholar]
- (37).Levesque MP, Krauss J, Koehler C, Boden C, and Harris MP (2013) New Tools for the Identification of Developmentally Regulated Enhancer Regions in Embryonic and Adult Zebrafish. Zebrafish 10, 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.