Abstractsss
Papillomavirus replication is tightly linked to squamous epithelial differentiation which in turn is governed to a large extent by epigenetic remodeling of genomes within the differentiating squamous epithelial cells. Over the past years it became evident that epigenetic and in particular differential methylation events substantially contribute to the regulation of the papillomavirus life cycle. Moreover, there is now good evidence that the initial trigger for HPV-mediated transformation of squamous epithelial cells is mediated by methylation of distinct CpG dinucleotides within E2-binding sites of the papillomavirus upstream regulatory region (URR). These findings have important implications for novel diagnostic markers but also for novel and indeed targeted therapy strategies for HPV linked neoplastic lesions.
1. Introduction
Papillomaviruses are a large and diverse group of epitheliotropic double-stranded DNA viruses that accompany the evolution of vertebrates since at least 350 million years [1]. They became well adapted to their host tissue, the squamous epithelia of the skin and mucosal surfaces in the oropharynx and the genital tract. The vast majority of papillomaviruses multiply their genomes and produce progeny viruses in their hosts without damage or any detectable pathologies. Only few papillomavirus types cause clinically relevant lesions that reach from minor and benign proliferative epithelial lesions like small warts to extensive giant condylomas and even pre-malignant lesions that may in some circumstances progress to invasive carcinomas. There is apparently a host cell tropism as some of the β-papillomavirus genotypes cause predominantly warts on feet and hands [1], whereas the α-papillomaviruses cause warts of the genital tract. A subgroup of about 15 of the α-types (high risk (HR)-HPV types) cause initially minor lesions of mucosal surfaces, which under defined circumstances may progress to invasive carcinomas (reviewed in Ref. [2]). This tissue restriction could in part be mediated by limited infection routes of the virus to distinct squamous epithelial sites. However, it is also conceivable that the restriction of distinct papillomavirus-induced lesions to specific squamous epithelial sites is linked to intrinsic factors within the epithelial host cells.
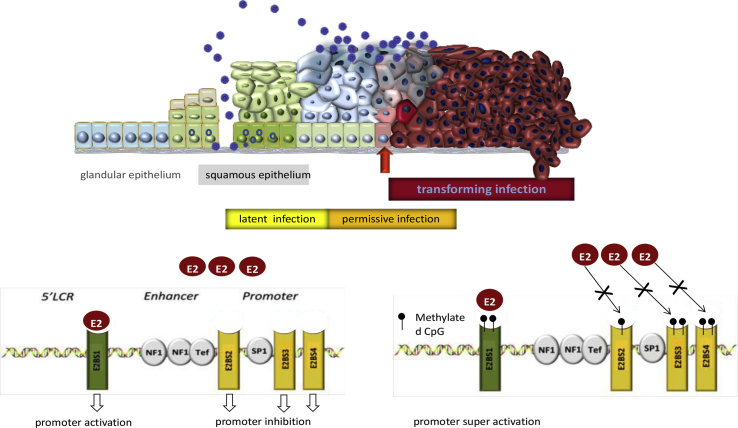
Although the viral life cycle of papillomaviruses starts essentially with the infection of basal epithelial stem cells at the bottom of the squamous epithelium (Fig. 1), they replicate their viral genomes in the intermediate epithelial layers and release progeny viral particles only in superficial squamous epithelial cells of stratum spinosum and corneum at the very surface of the squamous epithelium. Restricting the expression of the more abundant viral proteins to these superficial, post-mitotic differentiated squamous cells reduces exposure of viral antigens to the immune system and thus the chance to trigger a robust immune response.
Fig. 1.
Schematic representation of the infection mode of human papillomaviruses: Upper panel: The viral genome is released from its capsid upon entry into basal cells of the epithelium. In most instances it appears to be silenced for example by extensive methylation. Only under yet not defined conditions and only in individual cells gene expression may trigger proliferation of infected stem cells and replication in the terminally differentiated post-mitotic cells of the intermediate layers of the squamo epithelium resulting in a permissive infection. For oncogenic HPV types and in squamous columnar junction cells within the transformation zone of the uterine cervix active expression of the viral oncogenes (E6 and E7) may trigger genomic instability and subsequent neoplastic transformation (transforming infection) that can be highlighted by overexpression of p16INK4a. Lower panel: The activity of the E6-E7 genes is in part regulated by the HPV E2 protein. (Left) low levels of E2 bind to and activate the E2-binding site 1 resulting in increased levels of E6-E7 and also E2. (Right) higher levels of E2 bind in addition to the low affinity E2BS 3 and 4 resulting in inhibition of transcription. Methylation of the E2BS 1 results in super-activation of the E2BS 1 whereas methylation of the E2BS 3 and 4 prevents binding of E2 and thus blocks the transcriptional repression of E6-E7 which in turn triggers transformation. The figure was adapted to figures published in Ref. [4].
All in all, these biological features of papillomavirus infections argue for a precise control of viral gene expression in the infected squamous epithelial host cells, which is tightly coupled to the squamous epithelial differentiation program. There is now increasing evidence that regulation of the differential gene expression programs in squamous epithelial cells is mediated by epigenetic control that in particular involves DNA methylation (reviewed in Ref. [3]). DNA methylation is a key factor to suppress the expression of genes involved in the induction of cell cycle arrest and terminal differentiation in epithelial stem cells. Therefore, the overall methylation levels in de-differentiated squamous epithelial stem cells tend to be substantially higher if compared to the differentiated post-mitotic cells of the intermediate and superficial layers of the squamous epithelium.
2. DNA methylation of viral genomes in the PV life cycle
Expression of the HPV early genes is controlled by transcriptional control elements located in the upstream regulatory region (URR) (see Fig. 1, lower panel). Due to its overall far reaching medical impact, most of the insights in the regulation of HPV genomes have been gathered by studying HPV16. The HPV 16 URR encompasses a complex pattern of transcription factor binding sites for cellular and viral transcription factors. Among the various cellular transcription factors in particular the SP1 site located in the promoter region activates transcription upon binding of the SP1 protein. The key factor regulating the transcriptional activity of the HPV genome, however, is the HPV E2 protein, that controls its activity by an autoregulated feedback mechanism (reviewed in Ref. [4]. Early data from Kim et al. showed already that the activity of the viral promoter can be modulated by the methylation status of the E2 binding sites (E2BS) within the upstream regulatory region (URR) of the virus [5]. Moreover, they showed that the viral URR becomes hypo-methylated upon squamous cell differentiation in cells capable of replicating the viral genomes and producing progeny viral particles. These data were corroborated by further studies showing that in cells capable of producing papillomavirus particles, the URR tends to become hypomethylated in more differentiated cells [6]. However, the E2BSs and in particular the SP1 binding site seem to be highly methylated in the very superficial cell layers consistent with the downregulation of the early genes and the activation of the late genes due to the SP1 site methylation in these fully differentiated cells (early late shift) [7]. Interestingly, the latter report also provided evidence that in squamous epithelial cells in histopathological normal tissue adjacent to HPV induced lesions, all CpG dinucleotides in the URR were methylated in all layers of the epithelium. This observation points to a “latent” type of HPV infection in cells that remain unaffected from the presence of PV genomes, presumably due to a complete silencing of the PV genomes in all layers of the differentiating cells [7]. These data support the assumption that shifts of the methylation patterns of the PV genomes in the differentiating epithelium are linked to their respective gene expression activities (reviewed in Ref. [8]).
A further notion substantiating this concept is the fact that the PV life cycle starts with the entrance of a PV particle into a basal cell of the squamous epithelium (Fig. 1). The genome enclosed in the infecting particle is therefore derived from superficial cells expressing the late PV genes but not the early genes following the early late switch in superficial cells. If direct epigenetic modifications of the viral genomes may be involved in their regulation, it is reasonable to assume that they would be preserved on the viral genomes packaged into the infectious PV capsids in the superficial cells. Upon infection of other basal cells, however, no expression of the late genes L1 and L2 has ever been observed. This suggests that, upon infection, the incoming viral genomes are re-programmed by epigenetic factors within the infected epithelial stem cells. The detailed molecular mechanisms involved in HPV genome methylation alterations within basal epithelial cells, however, remain to be clarified.
3. DNA methylation and transformation of squamous epithelial cells
HPV-induced transformation of epithelial cells reveals a remarkable predilection for few squamocolumnar junction cells (SCJCs) displaying distinct differentiation patterns [9]. A similar tropism of HPV-mediated transformation is also observed for the dentate line in the anal canal and distinct epithelial cells in the tonsillar crypts of the oropharyngeal Waldeyer epithelium. These observations suggest that only few epithelial cells with distinct characteristics are predominantly vulnerable to transformation by oncogenic human papillomaviruses.
Activation of the viral E6-E7 oncogenes and subsequent cellular transformation induces epigenetic re-programming of the infected host cells. Among others, these epigenetic alterations promote overexpression of the cellular protein p16INK4a [10], which is therefore applied as a highly sensitive and specific biomarker for HPV-triggered transformation in epithelial cells at the uterine cervix (reviewed in Ref. [11]). In non-transforming, virus-producing low grade squamous intraepithelial lesions (LSIL)s (also referred to as permissive infections; Fig. 1) the expression of the E6 and E7 oncogenes is tightly controlled by the viral E2 protein (Fig. 1). Low levels of E2 activate the early promoter in the HPV URR by binding to the high-affinity E2-binding site (E2BS) 1. Promoter activation subsequently results in increasing levels of E6-E7 as well as E2 itself. At higher levels, E2 may now also bind to the low-affinity E2BSs 3 and 4 promoting the down-regulation of URR promoter activity and decreasing the expression of the E6-E7 oncogenes (reviewed in Ref. [4]). These findings suggested that loss of E2 function is a critical initial event allowing for uncontrolled E6-E7 expression and subsequent cellular transformation inducing further epigenetic remodeling [4,12]. In many HPV-induced invasive cancers, the HPV genomes become integrated into host cell chromosomes. The integration event frequently results in the structural disruption of the E2 open reading frame (orf) of the episomal viral genome. These observations led to the hypothesis that integration of the PV genome is essentially required to disrupt the E2 orf and thereby inactivate the transformation repressing function of the E2 protein (reviewed in Ref. [12]). This concept was questioned, however, when it became clear, that the activation of the E6-E7 oncogenes may substantially precede the integration of the viral genomes and thus disruption of the E2 orf (reviewed in Refs. [4,13]). This suggests that the mere inactivation of the E2 orf via integration into host cell chromosomes cannot be the major reason for the loss of E2 function in HPV-transformed cells [4].
We therefore hypothesized that differential methylation of transcription factor binding sites may contribute to the regulation of the expression of the HPV genome in the respective epithelial host cells. To test this hypothesis, we analyzed the methylation pattern of the HPV 16 URR in micro-dissected basal-, intermediate-, and superficial epithelial cells in productive parts of the epithelium [7]. The data revealed complex differences of CpG-dinucleotides pending on the differentiation status of the host cells. This observation is in line with the different HPV gene expression patterns in these lesions. In addition, differential methylation levels of CpGs within HPV 16 URR and in particular the E2BSs were observed in transforming infections (see Fig. 1) characterized by overexpression of E6 E7 and thus p16INK4a [7,14]. Functional studies indicate that methylation of the E2BS1 leads to the recruitment of an – up to now unspecified – transcription complex to this binding site, thereby increasing E2-dependent promoter activity and thus E6-E7 oncogene expression [7]. Binding of the HPV E2 protein to the unmethylated E2BSs 3 and 4 inhibits promoter activity by displacing the promoter-activating transcription factor SP1 from the SP1 binding site that lies in close proximity to the E2BSs 3 and 4 [14]. If the E2BSs 3 and 4 become methylated the E2 protein cannot bind any longer to these sites, thereby abrogating the E2-mediated promoter-inhibition. Taking those observations together, methylation of the E2BSs within the viral URR results in a loss of E2 function. This phenomenon can explain the uncontrolled HPV E6-E7 oncogene expression in HPV-transformed cells without evidence of viral integration and with intact E2 gene copies. This concept is further supported by the observation that HPV integration and aberrant methylation of the E2BSs tend to be negatively correlated in various HPV-driven cancers [15,16].
We therefore propose a model whereby aberrant methylation of the HPV E2BSs represents the initial trigger for aberrant HPV oncogene expression and can be regarded as functionally equivalent to the “break through” mutations in other cancers [17]. Subsequent E6-E7-mediated chromosomal instability might consequently favor integration of HPV genomes in a proportion of HPV-transformed cells as a secondary event and thereby promote sub-clonal disease progression in a subset of HPV-transformed lesions. Once the viral genomes become integrated into host cell chromosomes, the increasing degree of genomic instability may induce secondary epigenetic re-programming that potentially renders the expression of the HPV oncogenes independent from their direct control by the HPV E2 protein in some of the respective clones. Strikingly, while E6-E7 oncogene overexpression can be triggered by aberrant viral genome methylation, E6-E7 themselves in turn increase the methylation levels within the infected cell by stimulating expression and activity of the DNA methyltransferase (DNMTI) 1 [18,19] suggesting that all HPV-driven cancers may depend on ongoing E6-E7 driven DNA-methyltransferase activities that may keep them in a squamous epithelial stemness-like differentiation status [3].
4. Diagnostic implications
The impact of epigenetic modifications of oncogenic HPV types is further supported by many studies showing that less differentiated HPV-transformed squamous epithelial cells carry HPV genomes that are highly methylated throughout CpG dinucleotides within the L1 and L2 gene region [20,21] and in addition hyper-methylated cellular genes (reviewed in Ref. [22]). This finding is now increasingly being used as morphology-independent biomarker to detect transforming HPV infections and supports the impact of epigenetic re-programming in HPV-driven cancers and their pre-malignant lesions.
5. Therapeutic implications of the HPV epigenome
The emerging role of specific methylation events in the control of the viral life cycle and in particular its functional contribution to HPV-mediated neoplastic transformation let us hypothesize that treatment of HPV-transformed cells with demethylating drugs, such as the DNMT inhibitor 5-aza-2′-deoxycytidine, may reverse the aberrant methylation patterns of the E2BSs, down-regulate HPV E6-E7 oncogene expression and consequently lead to a regression of the HPV-transformed phenotype. This hypothesis was confirmed by treating a panel of HPV-transformed cell lines reacting with dose-dependent demethylation, down-regulation of HPV oncogene expression and pronounced inhibition of cellular proliferation upon demethylating treatment [23]. Moreover, the biologic effects of demethylating treatment might not only be mediated by demethylation of the E2BSs but also by global epigenetic re-programming of the affected cells, such as demethylation and subsequent re-expression of previously silenced tumor suppressor genes or miRNAs implicated in the suppression of HPV oncoprotein expression [23].
In conclusion, our growing understanding of these functionally important epigenetic alterations in HPV-infected cells now enables the design of novel treatment strategies targeting primarily epigenetic alterations in HPV-induced pre- and invasive cancers and may thus pave the way to an indeed targeted therapy for HPV-driven cancers.
References
- 1.Egawa N., Doorbar J. The low-risk papillomaviruses. Virus Res. 2017;231:119–127. doi: 10.1016/j.virusres.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 2.Doorbar J., Egawa N., Griffin H., Kranjec C., Murakami I. Human papillomavirus molecular biology and disease association. Rev. Med. Virol. 2015;25(Suppl 1):2–23. doi: 10.1002/rmv.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orioli D., Dellambra E. Epigenetic regulation of skin cells in natural aging and premature aging diseases. Cells. 2018;7 doi: 10.3390/cells7120268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doeberitz M., Vinokurova S. Host factors in HPV-related carcinogenesis: cellular mechanisms controlling HPV infections. Arch. Med. Res. 2009;40:435–442. doi: 10.1016/j.arcmed.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Kim K., Garner-Hamrick P.A., Fisher C., Lee D., Lambert P.F. Methylation patterns of papillomavirus DNA, its influence on E2 function, and implications in viral infection. J. Virol. 2003;77:12450–12459. doi: 10.1128/JVI.77.23.12450-12459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simanaviciene V., Popendikyte V., Gudleviciene Z., Zvirbliene A. Different DNA methylation pattern of HPV16, HPV18 and HPV51 genomes in asymptomatic HPV infection as compared to cervical neoplasia. Virology. 2015;484:227–233. doi: 10.1016/j.virol.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Vinokurova S., von Knebel Doeberitz M. Differential methylation of the HPV 16 upstream regulatory region during epithelial differentiation and neoplastic transformation. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johannsen E., Lambert P.F. Epigenetics of human papillomaviruses. Virology. 2013;445:205–212. doi: 10.1016/j.virol.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herfs M., Yamamoto Y., Laury A., Wang X., Nucci M.R., McLaughlin-Drubin M.E., Munger K., Feldman S., McKeon F.D., Xian W. A discrete population of squamocolumnar junction cells implicated in the pathogenesis of cervical cancer. Proc. Natl. Acad. Sci. U. S. A. 2012;109:10516–10521. doi: 10.1073/pnas.1202684109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McLaughlin-Drubin M.E., Crum C.P., Munger K. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc. Natl. Acad. Sci. U. S. A. 2011;108:2130–2135. doi: 10.1073/pnas.1009933108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergeron C., Ronco G., Reuschenbach M., Wentzensen N., Arbyn M., Stoler M., von Knebel Doeberitz M. The clinical impact of using p16(INK4a) immunochemistry in cervical histopathology and cytology: an update of recent developments. Int. J. Cancer. 2015;136:2741–2751. doi: 10.1002/ijc.28900. [DOI] [PubMed] [Google Scholar]
- 12.Woodman C.B., Collins S.I., Young L.S. The natural history of cervical HPV infection: unresolved issues. Nat. Rev. Canc. 2007;7:11–22. doi: 10.1038/nrc2050. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez A.F., Esteller M. Viral epigenomes in human tumorigenesis. Oncogene. 2010;29:1405–1420. doi: 10.1038/onc.2009.517. [DOI] [PubMed] [Google Scholar]
- 14.Leung T.W., Liu S.S., Leung R.C., Chu M.M., Cheung A.N., Ngan H.Y. HPV 16 E2 binding sites 1 and 2 become more methylated than E2 binding site 4 during cervical carcinogenesis. J. Med. Virol. 2015;87:1022–1033. doi: 10.1002/jmv.24129. [DOI] [PubMed] [Google Scholar]
- 15.Chaiwongkot A., Vinokurova S., Pientong C., Ekalaksananan T., Kongyingyoes B., Kleebkaow P., Chumworathayi B., Patarapadungkit N., Reuschenbach M., von Knebel Doeberitz M. Differential methylation of E2 binding sites in episomal and integrated HPV 16 genomes in preinvasive and invasive cervical lesions. Int. J. Cancer. 2013;132:2087–2094. doi: 10.1002/ijc.27906. [DOI] [PubMed] [Google Scholar]
- 16.Reuschenbach M., Huebbers C.U., Prigge E.S., Bermejo J.L., Kalteis M.S., Preuss S.F., Seuthe I.M., Kolligs J., Speel E.J., Olthof N. Methylation status of HPV16 E2-binding sites classifies subtypes of HPV-associated oropharyngeal cancers. Cancer. 2015;121:1966–1976. doi: 10.1002/cncr.29315. [DOI] [PubMed] [Google Scholar]
- 17.Vogelstein B., Kinzler K.W. The path to cancer --Three strikes and you're out. N. Engl. J. Med. 2015;373:1895–1898. doi: 10.1056/NEJMp1508811. [DOI] [PubMed] [Google Scholar]
- 18.Au Yeung C.L., Tsang W.P., Tsang T.Y., Co N.N., Yau P.L., Kwok T.T. HPV-16 E6 upregulation of DNMT1 through repression of tumor suppressor p53. Oncol. Rep. 2010;24:1599–1604. doi: 10.3892/or_00001023. [DOI] [PubMed] [Google Scholar]
- 19.Burgers W.A., Blanchon L., Pradhan S., de Launoit Y., Kouzarides T., Fuks F. Viral oncoproteins target the DNA methyltransferases. Oncogene. 2007;26:1650–1655. doi: 10.1038/sj.onc.1209950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clarke M.A., Gradissimo A., Schiffman M., Lam J., Sollecito C.C., Fetterman B., Lorey T., Poitras N., Raine-Bennett T.R., Castle P.E. Human papillomavirus DNA methylation as a biomarker for cervical precancer: consistency across 12 genotypes and potential impact on management of HPV-positive women. Clin. Cancer Res. 2018;24:2194–2202. doi: 10.1158/1078-0432.CCR-17-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cook D.A., Krajden M., Brentnall A.R., Gondara L., Chan T., Law J.H., Smith L.W., van Niekerk D.J., Ogilvie G.S., Coldman A.J. Evaluation of a validated methylation triage signature for human papillomavirus positive women in the HPV FOCAL cervical cancer screening trial. Int. J. Cancer. 2019 May 15;144(10):2587–2595. doi: 10.1002/ijc.31976. Epub 2018 Dec 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steenbergen R.D., Snijders P.J., Heideman D.A., Meijer C.J. Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions. Nat. Rev. Canc. 2014;14:395–405. doi: 10.1038/nrc3728. [DOI] [PubMed] [Google Scholar]
- 23.Stich M., Ganss L., Puschhof J., Prigge E.S., Reuschenbach M., Guiterrez A., Vinokurova S., von Knebel Doeberitz M. 5-aza-2'-deoxycytidine (DAC) treatment downregulates the HPV E6 and E7 oncogene expression and blocks neoplastic growth of HPV-associated cancer cells. Oncotarget. 2017;8:52104–52117. doi: 10.18632/oncotarget.10631. [DOI] [PMC free article] [PubMed] [Google Scholar]