Abstract
Purpose
The role of genetic polymorphisms in the pathogenesis of recurrent pregnancy loss (RPL) has been studied intensively. Complex diseases, including miscarriage, are believed to have a polygenic basis, and gene–gene interactions can play a significant role in the etiology of the disease. This study was conducted to investigate the association of gene–gene interactions with angiogenesis, endothelial dysfunction-related gene polymorphisms, and RPL.
Methods
A case–control study was conducted with 253 unrelated RPL patients with 2 or more spontaneous pregnancy losses and 339 healthy women with no history of pregnancy complications. Genotyping of single-nucleotide polymorphisms (SNPs) was performed using real-time polymerase chain reaction (real-time PCR), restriction fragment length polymorphism (RFLP), or allele-specific polymerase chain reaction methods.
Results
The genotypes 677TT of the MTHFR gene, 936TT, 936CT, and 634CC, 634GC of the VEGF gene, and allele 894T of the NOS3 gene were associated with a predisposition to RPL in the Russian population. A significant role of additive and epistatic effects in the gene–gene interactions of the SNPs of SERPINE-1, ACE, NOS3, MTHFR, and VEGF genes in RPL was demonstrated.
Conclusions
The results showed that gene–gene interactions are important for RPL susceptibility. Additionally, analysis of the genotype combinations of several allelic variants provides more information on RPL risk than analysis of independent polymorphic markers.
Keywords: Recurrent miscarriage, Single-nucleotide polymorphism, Gene–gene interactions, Endothelial dysfunction
Introduction
One of the most important problems in obstetrics is recurrent pregnancy loss (RPL). The prevalence of this pregnancy type is 1 to 5% [1–3]. RPL is one of the most troublesome areas in reproductive medicine because the etiology of this disease is often unknown, and modern diagnostics and known evidence-based treatment strategies are not sufficiently effective [2, 4].
Recurrent pregnancy loss is a complex disease that is often triggered by anatomical, infectious, immunological, endocrine, and genetic factors in various combinations [3]. According to current knowledge, maternal and paternal inheritance plays an important role in the development of RPL in the early stages of pregnancy [3, 5]. In conformity with the HuGE Navigator database, more than 230 candidate genes associated with RPL are known. They are related to thrombophilia and hypofibrinolysis, metabolism of folic acid and vitamin B12, the functioning of endothelial cells, metabolism of hormones, and the immune response [6]. However, the results of many studies have been either negative or non-replicable [1, 2, 5, 7]. Additionally, the findings of these studies are often contradictory, even when studying the same ethnic group, and are rarely replicable in other populations [8, 9]. Nevertheless, a generalized analysis of the data showed that several genetic markers have a significant association with RPL in several meta-analyses: C677T of the MTHFR gene (rs1801133), 4G/5G of the SERPINE-1 gene (rs1799889), G894T (rs2070744) of the NOS3 gene, the I/D polymorphism of the ACE gene (rs4646994), G215C (rs1042522) of the TP53 gene, and G634C (rs2010963) and C936T (rs3025039) of the VEGF-A gene. Therefore, these allelic variants undoubtedly are of interest in the study of the predisposition to the development of RPL [9–24].
The focus of genetic association studies for complex diseases has been gradually shifting from assessing independent genes to estimation of the interaction effects of genes [25]. Complex diseases, including miscarriage, are believed to have a polygenic basis, and gene–gene interactions may play a significant role in the etiology of the disease. Understanding the gene–gene interactions may also help to explain missing heritability of complex phenotypes and conflicting results from the analysis association study [26, 27].
Materials and methods
Study sample
This study was conducted in women of Russian descent who were referred to The Genetic Clinic at the Research Institute for Medical Genetics and Clinic of Scientific Research Institute of Obstetrics, Gynecology and Perinatology of Tomsk City during 2010–2014. The patient group consisted of 253 women who were diagnosed with RPL. The patients had at least two pregnancy losses up to 20 weeks, and they had no risk factors for RPL, such as anatomical abnormalities, chromosomal aberrations of partners and embryos, chronic infections, thrombophilia, metabolic disorders, and a positive lupus anticoagulant. The control group included 339 healthy women with at least two previous live births and no history of pregnancy loss or infertility. Additionally, the women enrolled in the control group had no pregnancy-associated complications. Patients with a history of pre-eclampsia, abruptio placentae, gestational diabetes mellitus, gestational hypertension, premature birth, or SGA (small-for-gestational age) birth also were excluded. The case and control groups were matched for age. Characteristics of the RPL and control group are shown in Table 1. Patients with RPL and healthy controls were all Caucasians of Russian ancestry. Written informed consent was obtained from all participating individuals. The study was conducted in accordance with the code of ethics of the Declaration of Helsinki, and approval was obtained from the local Ethical Committee of the Research Institute for Medical Genetics.
Table 1.
Characteristics of the RPL and control group
| RPL group | Control group | p value* | |
|---|---|---|---|
| Age (year) (range) | 29.5 ± 4.5 (19–44) | 27.3 ± 4.6 (18–38) | 0.239 |
| BMI (kg/m2) (range) | 23.8 ± 4.7 (20–27) | 24.2 ± 4.1 (20–27) | 0.788 |
| Weeks of early fetal losses, median (range) | 8.7 ± 1.1 (3–20) | 0 | 0.001 |
| Number of RPL (range) | 2.6 ± 0.8 (2–6) | 0 | 0.001 |
| Number of pregnancies (range) | 2.8 ± 0.9 (2–6) | 2.2 ± 0.73 (2–4) | 0.001 |
p < 0.05 is considered statistically significant
*Student’s t test was used to compare the age in patients and controls, while χ2 analysis was used to compare the other parameters
DNA extraction and genotyping
Genomic DNA from the peripheral blood of patients was extracted by a modified phenol/chloroform method. Seven functional single-nucleotide polymorphisms (SNPs) were selected from angiotensin-converting enzyme (ACE), methylenetetrahydrofolate reductase (MTHFR), plasminogen activator inhibitor-1 (SERPINE-1/PAI-1), nitric oxide synthase 3 (NOS3), tumor protein 53 (TP53), and vascular endothelial growth factor (VEGF) genes (Table 2) based on our previous studies [9].
Table 2.
Genes and variants analyzed in this study
| Gene name | Chromosome | SNP (rs#) | Localization SNP in the gene | Alleles |
|---|---|---|---|---|
| ACE | 17q22-q24 | I/D (rs4646994) | 16 intron | I/D |
| MTHFR | 1p36.3 | C677T (rs1801133) | 5 exon | С/T |
| SERPINE1 | 7q21.3-q22 | 5G/4G (rs1799889) | 5′-UTR | 4G/5G |
| NOS3 | 7q36 | G894T (rs1799983) | 7 exon | G/T |
| VEGF | 6p21.3 | C936T (rs3025039) | 3′-UTR | C/T |
| G634C (rs2010963) | 5′-UTR | G/C | ||
| TP53 | 17p13.1 | G215C (rs1042522) | 4 exon | G/C |
Genotyping of rs4646994, rs1801133, rs1799889, and rs1799983 markers was carried out using restriction fragment length polymorphism (RFLP) analysis or allele-specific polymerase chain reaction according to the previously described protocols [28–31]. Genotyping of rs3025039, rs2010963, and rs1042522 polymorphic variants was performed in the real-time mode using PCR implemented by the TaqMan Genotyping Assay (Applied Biosystems Inc., Foster, USA).
Statistical analyses
Statistical processing of the results was performed using the statistical software package Statistica 10.0 and MDR. We used Pearson’s chi-squared test with Yates’ correction and Fisher’s exact test to compare allele and genotype frequencies. To estimate SNP association with the pathological phenotype, the odds ratio (OR) and confidence intervals (CI) for OR (95% CI) were calculated. The concordance of genotype frequencies with Hardy–Weinberg equilibrium was estimated by χ2 test. Linkage disequilibrium (LD) between SNP pairs was assessed using D’ and Pearson’s coefficients [32] in the HaploView 4.2 program. Gene–gene interaction analysis was performed using Multifactor Dimensionality Reduction software by reducing a multifaceted dimension with the help of an exhaustive search algorithm.
Results
Genotype and allele frequency distribution in the patient and control groups
Table 3 illustrates the genotype and allele frequency distribution of the polymorphic variants, compliance of the genotype frequency distribution with the Hardy–Weinberg equilibrium, and level of significance obtained while comparing the controls and patients with RPL. The distribution of genotype frequencies in all the controls corresponded to Hardy–Weinberg equilibrium. According to the database of single-nucleotide polymorphisms [33], the allele and genotype frequencies obtained in our population were consistent with the European population for all polymorphisms.
Table 3.
Distribution of allele and genotype frequencies in the study group
| SNPs (genes) | Genotypes/alleles | RPL, n = 253 | Controls, n = 339 | p 1 |
|---|---|---|---|---|
| Percentage | ||||
| C677T (MTHFR) | CC | 51 | 62 | 0.01* |
| CT | 39 | 33 | ||
| TT | 10 | 5 | ||
| T | 29 | 22 | 0.003* | |
| р2 | 0.58 | 0.56 | ||
| I/D (ACE) | II | 25 | 25 | 0.97 |
| ID | 51 | 52 | ||
| DD | 24 | 23 | ||
| D | 49 | 49 | 0.85 | |
| р2 | 0.65 | 0.44 | ||
| G894T (NOS3) | GG | 39 | 49 | 0.03* |
| GT | 46 | 41 | ||
| TT | 15 | 10 | ||
| T | 38 | 31 | 0.008* | |
| р2 | 0.78 | 0.38 | ||
| G215C (TP53) | CC | 10 | 9 | 0.62 |
| CG | 39 | 43 | ||
| GG | 51 | 48 | ||
| C | 29 | 31 | 0.53 | |
| р2 | 0.35 | 0.94 | ||
| G634C (VEGF) | CC | 19 | 8 | 0.001* |
| CG | 44 | 36 | ||
| GG | 37 | 56 | ||
| C | 41 | 26 | 0.001* | |
| р2 | 0.13 | 0.13 | ||
| C936T (VEGF) | CC | 43 | 64 | 0.001* |
| CT | 44 | 30 | ||
| TT | 13 | 6 | ||
| T | 35 | 21 | 0.001* | |
| р2 | 0.71 | 0.13 | ||
| 4G/5G (SERPINE-1) | 4G/4G | 30 | 32 | 0.61 |
| 4G/5G | 55 | 51 | ||
| 5G/5G | 15 | 17 | ||
| 5G | 42 | 43 | 0.99 | |
| р2 | 0.08 | 0.57 | ||
n number of people in the group
1Chi-square test with Yates’ correction or Fisher’s exact test and the level of significance (p) were obtained by comparing the genotype and allele frequency in the controls and patients with RPL
2The level of significance achieved while estimating the compliance with Hardy–Weinberg equilibrium
*Statistically significant difference between the study groups
Statistically significant differences were revealed for four markers of the seven investigated polymorphic variants: C677T MTHFR, G894T NOS3, G634C VEGF, and C936T VEGF (Table 3).
The TT genotype and T allele of the C677T polymorphic variant of the MTHFR gene were associated with a high risk of RPL. However, the C allele and CC genotype indicate a protective effect against this pathology (Table 4).
Table 4.
Results of the odds ratio of the polymorphic variants of the MTHFR, NOS3, and VEGF genes
| SNPs (genes) | Genotypes/alleles | OR (95% CI) | p value |
|---|---|---|---|
| C677T (MTHFR) | T allele vs. C allele | 1.49 (1.15–1.94) | 0.003* |
| TT vs. CC+CT genotype | 2.01 (1.08–3.75) | 0.03* | |
| CT vs. CC+TT genotype | 1.27 (0.91–1.78) | 0.15 | |
| C allele vs. T allele | 0.67 (0.52–0.87) | 0.003* | |
| CC vs. TT+CT genotype | 0.66 (0.47–0.91) | 0.03* | |
| G894T (NOS3) | T allele vs. G allele | 1.39 (1.09–1.77) | 0.009* |
| TT vs. GG+GT genotype | 1.50 (0.92–2.46) | 0.14 | |
| GT vs. GG+TT genotypes | 1.27 (0.91–1.76) | 0.19 | |
| G allele vs. T allele | 0.72 (0.57–0.92) | 0.009* | |
| GG vs. TT+GT genotype | 0.66 (0.47–0.92) | 0.02* | |
| G634C (VEGF) | C allele vs. G allele | 1.97 (1.53–2.52) | 0.001* |
| CC vs. GG+GC genotype | 2.59 (1.57–4.27) | 0.0002* | |
| GC vs. GG+CC genotype | 1.41 (1.01–1.98) | 0.04* | |
| G allele vs. C allele | 0.51 (0.40–0.65) | 0.001* | |
| GG vs. CC+GC genotype | 0.46 (0.33–0.65) | 0.001* | |
| C936T (VEGF) | T allele vs. C allele | 2.06 (1.58–2.68) | 0.001* |
| TT vs. CC+CT genotype | 2.44 (1.35–4.41) | 0.004* | |
| CT vs. CC+TT genotype | 1.85 (1.31–2.60) | 0.0006* | |
| C allele vs. T allele | 0.49 (0.37–0.63) | 0.001* | |
| CC vs. TT+CT genotype | 0.42 (0.30–0.58) | 0.001* |
*Statistically significant differences between the alleles and genotypes
The G894T allelic variant of the NOS3 gene showed a protective effect against the development of RPL. Thus, the 894T allele may be predisposing to the occurrence of this pathology (Table 4).
A statistically significant high frequency of the 634C allele and CC and CG genotypes of the G634C polymorphic variant of the VEGF gene, as well as the 936T allele and CT and TT genotypes of the C936T polymorphic markers of the VEGF gene, were significantly more frequent in the RPL group (Tables 3 and 4). Therefore, the CT and TT genotypes of the C936T allelic variant of the VEGF gene, as well as the CC and GC genotypes of the G634C polymorphism of the VEGF gene, are related to the risk of RPL in Tomsk women.
Thus, the G894T polymorphic variant of the NOS3 gene, C677T of the MTHFR gene, and C936T and G634C of the VEGF gene can be considered as genetic factors associated with an increased predisposition to RPL and are involved in the development of hypercoagulation and endothelial dysfunction in Russians during pregnancy.
Gene–gene interaction in studied RPL candidate genes and RPL risks with different genotype combinations
The gene–gene interaction of the RPL candidate genes was analyzed by MDR. Because of MDR analysis, two models were identified to predict the risk of RPL (Table 5).
Table 5.
Characteristics of the gene–gene interaction models of the investigated polymorphic markers
| Model | Tr. Bal. Acc | Ts. Bal. Acc | Se | Sp | CV Cons | p value |
|---|---|---|---|---|---|---|
| C936T и G634C VEGF | 0.6797 | 0.6739 | 0.6034 | 0.7559 | 10/10 | 0.001 |
| 5G/4G SERPINE-1; I/D ACE; G894T NOS3; C677T MTHFR; C936T и G634C VEGF | 0.8792 | 0.5914 | 0.8793 | 0.8644 | 9/10 | 0.001 |
Tr. Bal. Acc training balanced accuracy, Ts. Bal. Acc tested balanced accuracy, Se model sensitivity, Sp specificity, CV Cons cross-validation
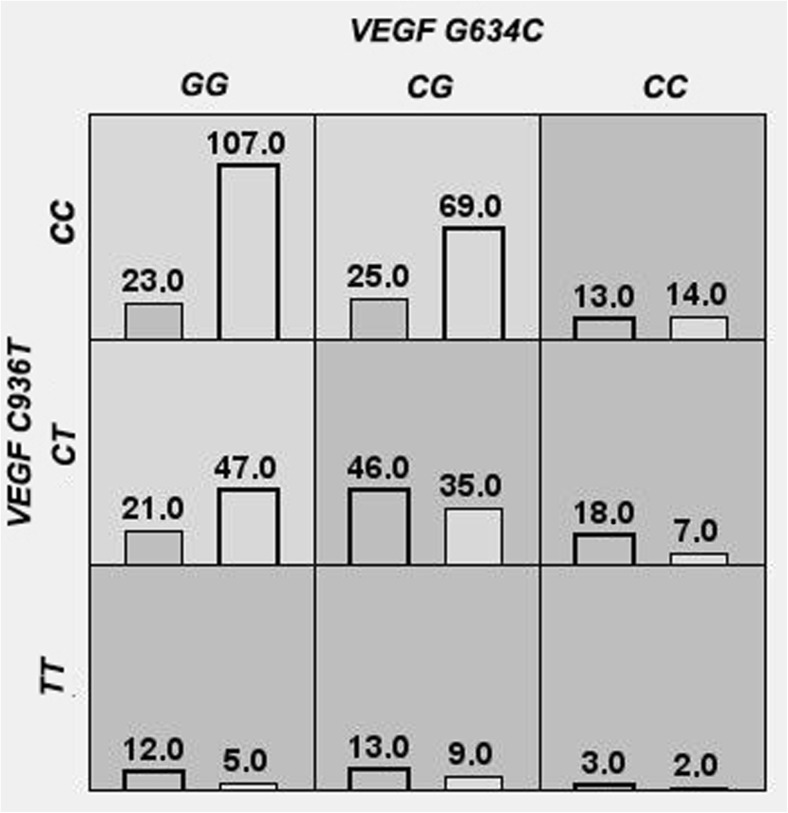
The combination of two allelic variants C936T and G634C of the VEGF gene (Fig. 1) demonstrates 10 out of 10 consistent cross-validations (p = 0.001). The balance accuracy of the entire model was 67.97%, the sensitivity was 60.34%, and the specificity was 75.59% (p < 0.0001).
Fig. 1.
Distribution of the genotype combinations of polymorphic variants C936T and G634C of the VEGF genes in the group of RPL patients and in the controls: dark gray cells, RPL high-risk genotypes; gray cells, low-risk genotypes; left columns, patients with RPL; the right columns, controls
Figure 1 shows that the СС/СС, CG/CT, CC/CT, GG/TT, CG/TT, and CC/TT (VEGF G634C/C936T, respectively) genotype combinations are predisposing factors for RPL development in Tomsk women. GG/CC, GG/CT, and CG/CC combinations are a low-risk factor for the development of RPL.
To assess the significance of this model, we analyzed LD between the investigated SNPs of the VEGF gene. It was determined that, in the study groups, C936T (rs3025039) and G634C (rs2010963) loci of the VEGF gene were not linked to each other (D′ = 0.05l, r2 = 0.002 for the control group; D′ = 0.012, r2 = 0.001 for the RPL group).
Notably, while comparing the study groups, statistically significant differences were shown for five of the nine genotype combinations (Table 6). А significant increase in the frequency of the 936CT-634CC genotype combinations was detected in the RPL group compared with that in the controls. This genotype combination increased the risk of RPL by more than fourfold (OR = 4.75), while the odds ratio of each of these genotypes (936CT and 634CC) was significantly lower (OR = 1.85 and 2.59, respectively) (Table 3). The risky genotype combinations of 936CT-634CG (OR = 2.67) and 936TT-634GG (OR = 4.30) were also found. The genotype combinations 936CC-634GG and 936CC-634CG reduced the risk of RPL and showed a protective effect (OR = 0.55 and OR = 0.27, respectively) (Table 6).
Table 6.
Frequency of the statistically significant genotype combinations of the VEGF gene in the RPL and control groups
| Genotype combination (C936T-G634C) | RPL group (n = 253) | Controls (n = 339) | р level | OR (95% confidence interval (CI)) |
|---|---|---|---|---|
| Percentage | ||||
| CC-GG | 13.2 | 36.3 | 0.001 | 0.55 (0.33–0.91) |
| CC-CG | 14.4 | 23.4 | 0.025 | 0.27 (0.16–0.44) |
| CT-CG | 26.4 | 11.9 | 0.0001 | 2.67 (1.64–4.35) |
| CT-CC | 10.3 | 2.4 | 0.005 | 4.75 (1.95–11.61) |
| TT-GG | 7.4 | 1.7 | 0.008 | 4.30 (1.49–12.41) |
n number of people in the group
Interestingly, in the study groups, there was no genotype combination that included all mutant alleles (936TT-634CC). It can be assumed that individuals who inherit all mutant alleles from these polymorphic variants are less likely to survive. However, this assumption requires further investigation in a more representative sample.
A model comprising six polymorphic variants of the SERPINE-1, ACE, NOS3, MTHFR, and VEGF genes (Table 7) showed a cross-validation consistency of 9 of 10 (p = 0.001). The total balance accuracy was 87.19%, with a sensitivity of 87.93% and a specificity of 86.44% (p < 0.0001).
Table 7.
Frequency of statistically significant combinations of the VEGF, MTHFR, SERPINE-1, NOS3, and ACE locus genotypes in the study groups
| Gene combinations C936T-G634C-C677T-4G/5G-G894T-I/D | RPL group (n = 253) | Controls (n = 339) | p value (Fisher’s ratio test) | |
|---|---|---|---|---|
| Percentage | ||||
| A | 936CC-634GG-677CT-4G/5G-894GG-ID | 0.6 | 3.4 | 0.015 |
| B | 936CT-634CG-677TT-4G/5G-894GT-II | 2.9 | 0.3 | 0.041 |
| C | 936CT-634CG-677TT-4G/5G-894GT-DD | 5.2 | 0.7 | 0.016 |
| D | 936CC-634GG-677CC-4G/5G-894GG-II | 0 | 2.7 | 0.029 |
n number of people in the group
Within this model, 258 of 729 possible combinations of genotypes were found in the study groups. Thirty-four combinations found in at least one of the study sample groups with a frequency of more than 1% were selected for further analysis. Statistically significant differences were shown for 4 of the 34 analyzed genotype combinations (Table 7). The odds ratio (OR) and confidence interval (95% CI) could be calculated only for three combinations of genotypes because, in both groups, the number of observations was different from 0 [34]. In the RPL group compared with the control group, we detected an increase in the frequency of B and C combinations. It was found that the risk of RPL was increased by almost ninefold in the C genotype combination and fivefold in the case of the B genotype combination. The A genotype combination had a protective effect because it reduced the risk of this disease (Table 7).
While analyzing this interaction model of the studied loci, a few significant genotype combinations were obtained. This can be explained by the small sample size that arises in multiple testing due to the increase in the number of subgroups as the number of interacting genes in the training sample increases. The inclusion of the 4G/5G allelic variant of the SERPINE-1 gene and I/D polymorphism of the ACE gene is notable because they did not demonstrate the association with RPL during the analysis at the level of individual loci.
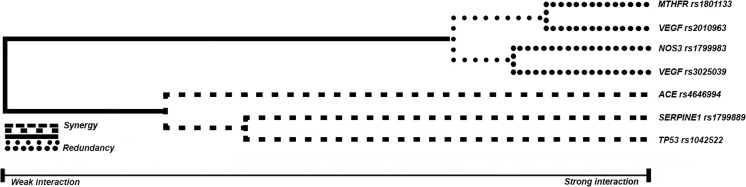
The MDR software capacity makes it possible to present the contribution of each SNP to the risk of RPL development in the form of a dendrogram and assess the nature of gene–gene interactions (Fig. 2).
Fig. 2.
Dendrogram of gene–gene interactions of MTHFR C677T, SERPINE-1 5G/4G, ACE I/D, NOS3 G894T, TP53 G215C, VEGF C936T loci, and G634C in cases of RPL. Note: The long lines in the dendrogram describe a weak relationship between the markers. The short lines connecting the two predictors reflect the strong interaction between the markers. The type of each line indicates the type of interaction: the dashed line represents synergistic interactions—i.e., complementarity between the SNPs of the studied genes; the solid line represents an independent effect, and the dotted line describes an additive effect—i.e., the effect of the polymer of each predictor
Based on this dendrogram, gene–gene interactions with an apparent synergic effect were found between the I/D markers of the ACE gene, 5G/4G of the SERPINE-1 gene, and G215C of the TP53 gene. At the same time, gene–gene interactions between the C677T locus of the MTHFR gene, G894T of the NOS3 gene, and C936T and G634C of the VEGF gene were additive in the risk of developing RPL in Russian women.
Discussion
As already mentioned in this paper, the T allele and TT genotype of the C677T polymorphic variant of the MTHFR gene were associated with an increased risk of RPL. It should be noted that, despite the large number of studies devoted to the connection between the C677T allelic variant of the MTHFR gene and RPL, there remains no unanimous consent on this issue. According to several meta-analyses, the C677T genetic variant contributes to the RPL susceptibility [14, 15]. However, other studies have not found a link between the C677T allelic variant of the MTHFR gene and idiopathic RPL [35, 36]. The study conducted in the Slovak and Romani populations confirmed the variable distribution of the C677T polymorphism in different ethnic groups, as well as its various effects on the clinical phenotype of patients with obstetric complications [37]. It is known that the non-synonymous replacement of C677T of the MTHFR gene leads to a significant decrease in the activity of MTHFR due to the replacement of alanine with valine in the place of the binding of this enzyme to flavin-adenine dinucleotide cofactor. This variant is of a great interest because of some pathological conditions leading to the accumulation of homocysteine in the body and damage to vascular endothelia, followed by intravascular coagulation and the development of several gestational complications, including RPL [16].
In this study, the connection between RPL and the G894T allelic variant of the NOS3 gene was also recorded. It is known that the G894T polymorphism of the NOS3 gene is a substitution of guanine for thymine at position 894 of exon 7 of the NOS3 gene, leading to the replacement of glutamine by asparagine in the oxygenase domain of the enzyme and change in its catalytic activity [28]. It has been shown that the G894T variant of the NOS3 gene causes a decrease in NO synthase activity, bioavailability of nitric oxide (NO), and NO decrease in plasma [38]. The pathogenetic role of endogenous NO in RPL patients is associated with the development of endothelial dysfunction, both in the maternal body and fetoplacental complex [17]. Thus, significant differences in NO synthase activity and the NO level in the plasma and endometrium of women with RPL are shown when comparing these parameters with the control group [39, 40]. Considering the significant role of NO synthase and nitric oxide in angiogenesis, vascular tone control, and antithrombotic effects in the endometrium during decidualization and in the placenta in the early stages of pregnancy, changes in the gene coding for endothelial NO synthase can serve as potential factors of predisposition to RPL [17].
Several meta-analyses revealed a close relationship between the G894T variant of the NOS3 gene and idiopathic RPL [10, 17, 18]. In the Chinese, there was a significant reduction in the 894G allele frequency in patients with RPL [41], corresponding to data obtained in Indian populations [38], Koreans [42], and the present study. Additionally, Shin et al. found that the 894TT genotype of the NOS3 gene was significantly more common in the RPL group than in the control group (OR = 2.39; CI 1.25–4.58; p = 0.008) [42]. We also recorded a higher frequency of this genotype in the RPL group than in the control group (15% and 10%, respectively), but these differences were not statistically significant. It is important to note that the results of some studies devoted to the analysis of the association of G894T polymorphism of NOS3 gene with RPL do not reveal the connection between this genetic marker and this pathology [20, 43].
As shown above in this study, association with RPL has also been established for G634C and C936T markers of the VEGF gene. The data obtained indicate that the T allele and CT and TT genotypes of the C936T polymorphism of the VEGF gene, as well as the CC and GC genotypes and allele of the G634C polymorphism of the VEGF gene, are RPL risk factors in the Russian population. Similar results were illustrated in the paper written by Xu et al. [21]. The meta-analysis involving 1832 patients with RPL and 2271 women with good obstetric history demonstrated the connection between RPL and the C allele (OR = 1.16, CI 1.03–1.31, p = 0.01) and CC genotype (OR = 1.36, CI 1.06–1.74, p = 0.02) of the G634C polymorphic variant. Additionally, it was found that the C allele (OR = 0.72, CI 0.56–0.93, p = 0.01) and CC genotype (OR = 0.69, CI 0.53–0.89, p < 0.005) of the C936T variant have protective properties for the development of RPL [35]. At the same time, in some scientific studies, the links between the C936T and G634C variants of the VEGF gene and LPR were not found [44–46].
It is known that the vascular endothelial growth factor (VEGF) plays a key role in regulating angiogenesis because it increases vascular permeability and is the main chemotactic and mitogenic stimulus for endotheliocytes. The above-listed properties determined the significance of VEGF in both the embryonic development processes and maternal vascular adaptation to pregnancy [47, 48]. An association was found between the G634C and C936T polymorphic variants of the VEGF gene located respectively in the 5′ and 3′-untranslated regions of this locus and the variability of its transcript level [49, 50]. It was shown that the violation of VEGF gene expression in the placental tissue can lead to perfusion abnormalities in the endometrium, resulting in several unfavorable pregnancy outcomes, including RPL, fetal death, fetal growth retardation, and pre-eclampsia [51].
When we compared the RPL and control groups, no statistically significant differences were found in the distribution of the allele and genotype frequencies of the G215C variants of the TP53 gene, 4G/5G polymorphism of the SERPINE-1 gene, and I/D polymorphism of the ACE gene. Accordingly, gene–gene interaction analysis was chosen in this study to identify the possible joint effects of these “nonsignificant” SNPs in association with RPL. Using MDR, we demonstrated the complementary genetic interaction and contributed risk of different genotype combinations in the ACE-SERPINE-1 system on RPL (Tables 5 and 7, Fig. 2).
It was established that the product of the SERPINE-1/PAI-1 gene is a key regulator of proteolysis and remodeling of the maternal tissues during the invasion of trophoblast [22]. Homozygotes for the 4G allele have more PAI-1 in plasma than heterozygotes and homozygotes for the 5G allele. Hyperexpression of PAI-1 may act as a major inhibitor of fibrinolysis and lead to the weakening of fibrinolytic function, which may be one of the causes of early pregnancy loss [52, 53]. Based on the data of the meta-analysis performed by Chen and colleagues, the 4G/4G genotype predisposes to the development of RPL in Asians (OR = 2.12; CI 1.20–3.76) and Africans (OR = 4.48; CI 2.38–8.43) but not in Europe and the USA. The study included 3864 women with RPL and 2208 women with a normal pregnancy [23]. These data were confirmed by the studies of Subrt and colleagues, who established that 4G/4G homozygotes predominate among women with RPL who have 2 miscarriages (37.5%) and 3–8 miscarriages (37.7%) compared with the controls (13.5%) (p = 0.002) [54]. However, in a recent case–control study, Kim et al. could not prove the relationship between the -675 4G/5G polymorphic variant of the SERPINE-1 gene and RPL [55].
The Alu-element of the ACE gene, consisting of 287 base pairs, is associated with the expression of angiotensin-converting enzyme (ACE) in various tissues: the minimum level of this enzyme occurs in carriers of the I/I genotype, whereas its maximum content is noted in individuals with the D/D genotype. It is suggested that the influence of this polymorphic variant on the variability of the ACE level and function of the renin–angiotensin–aldosterone system is associated with a change in the regulation of ACE gene expression [18]. Additionally, an association was demonstrated between the D allele of the ACE gene and increased resistance of the uterine arteries, a marker of fetal growth restriction [56]. There are also data on the association between the I/D variant of the ACE gene and RPL. Thus, women with the DD or ID genotype in the investigated allelic variant were reported to be more prone to develop RPL in early pregnancy [22]. According to the meta-analysis including 1264 patients with RPL and 845 women in the control group, the D allele was found to increase the risk of RPL in Asian populations by 1.7 fold (OR = 1.69, CI 1.06–2.36) and in representatives of European ethnicity by twofold (OR = 2.06, CI 1.46–2.91) [20]. Another meta-analysis of the relationship between the I/D polymorphism of the ACE gene and RPL in 1766 women with RPL and 1591 women with a normal pregnancy established that the genotypes of DD and ID are predisposing factors of RPL development in Europeans and East Asians (OR = 1.81, 95% CI 1.23–2.66, p = 0.003 and OR = 1.50, 95% CI 1.25–1.80, p < 0.001, respectively) [24].
It is well known that ACE plays an important role in the control of the fibrinolytic process [57]. The D/D genotype the ACE gene enhances the production of angiotensin II from angiotensin I and is associated with high levels of circulating PAI-1 due to reduced levels of fibrinolysis [57, 58]. Both the PAI-1 4G and ACE D variants may compromise placental formation and trophoblast invasion because of increased PAI-1 expression and concomitant reduced fibrinolytic activity [59, 60]. In this context, the results of a study by Buchholz et al. (2003) are of great interest. They demonstrated that the risk of RPL development before the 25th week of pregnancy significantly increases in women of European ethnicity who are homozygous for the D allele of the ACE gene and the SERPINE-1 4G allele [61].
There are some limitations in our study. We could not measure the serum levels of the vascular endothelial growth factor, methylenetetrahydrofolate reductase, and endothelial nitric oxide synthase in the patient cases and control women; therefore, the functionality of this association could not be addressed as well as genotype–phenotype correlations. The sample size was relatively small, thereby attenuating the power of the statistical significance. Thus, the MDR method was introduced to identify gene–gene interactions that are associated with idiopathic RPL. The MDR method reduces high-dimensional genetic data into a single dimension, permitting gene–gene or gene–environmental interactions to be detected in relatively small sample sizes. Larger genetic studies in different populations are warranted to confirm these findings.
In conclusion, we observed that the genotypes 677TT of MTHFR gene, 936TT, 936CT, and 634CC, 634GC of the VEGF gene, and allele 894T of the NOS3 gene are associated with the predisposition to RPL in the Russian population. One of the strengths of our study is that it was sufficiently powered and that RPL cases and control women were matched according to ethnicity, reducing the problems of ethnic differences inherent in genetic association studies. Another strength is the significant role of the additive and epistatic effects of the gene–gene interactions of the SNPs of the SERPINE-1, ACE, NOS3, MTHFR, and VEGF genes on RPL. The above results indicate a more informative assessment of the risk of developing miscarriage by the analysis of a combination of genotypes of several allelic variants compared with data obtained at the level of individual polymorphic markers. These studies support the hypothesis that vasculogenesis dysfunction and thrombophilia during embryogenesis, which are promoted by gene alteration, may affect pregnancy and the predisposition to RPL. Our study added further evidence that RPL is a polygenic disease, implying that these polymorphisms are potential markers for RPL Further studies of gene–gene interactions in the structure of the susceptibility to RPL can be the basis to gradually fill the gaps in the missing heritability of this pathology.
Funding information
The work received financial support from the Russian Foundation for Basic Research (grant no. 18-44-700007).
Compliance with ethical standards
This study was approved by the Research Ethics Committee of the Institute for Medical Genetics of Tomsk city. Informed consent was obtained from all individual participants included in the study.
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Page JM, Silver RM. Genetic causes of recurrent pregnancy loss. Clin Obstet Gynecol. 2016;59:498–508. doi: 10.1097/GRF.0000000000000217. [DOI] [PubMed] [Google Scholar]
- 2.Rull K, Nagirnaja L, Laan M. Genetics of recurrent miscarriage: challenges, current knowledge, future directions. Front Genet. 2012;3(34):1–13. doi: 10.3389/fgene.2012.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sergi C, Al Jishi T, Walker M. Factor V Leiden mutation in women with early recurrent pregnancy loss: a meta-analysis and systematic review of the causal association. Arch Gynecol Obstet. 2015;291(3):671–679. doi: 10.1007/s00404-014-3443-x. [DOI] [PubMed] [Google Scholar]
- 4.Kolte AM, Nielsen HS, Steffensen R, et al. Inheritance of the 8.1 ancestral haplotype in recurrent pregnancy loss. Evol Med Public Health. 2015;2015(1):325–331. doi: 10.1093/emph/eov031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi X, Xie X, Jia Y, Li S. Maternal genetic polymorphisms and unexplained recurrent miscarriage: a systematic review and meta-analysis. Clin Genet. 2017;91(2):265–284. doi: 10.1111/cge.12910. [DOI] [PubMed] [Google Scholar]
- 6.Yu W, Gwinn M, Clyne M, Yesupriya A, Khoury MJ. A navigator for human genome epidemiology. Nat Genet. 2008;40(2):124–125. doi: 10.1038/ng0208-124. [DOI] [PubMed] [Google Scholar]
- 7.Arias-Sosa LA, Acosta ID, Lucena-Quevedo E, Moreno-Ortiz H, Esteban-Pérez C, Forero-Castro M. Genetic and epigenetic variations associated with idiopathic recurrent pregnancy loss. J Assist Reprod Genet. 2018;35(3):355–366. doi: 10.1007/s10815-017-1108-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pereza N, Ostojić S, Kapović M, Peterlin B. Systematic review and meta-analysis of genetic association studies in idiopathic recurrent spontaneous abortion. Fertil Steril. 2017;107(1):150–159. doi: 10.1016/j.fertnstert.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Trifonova EA, Ganzha OA, Gabidulina TV, et al. Genetic factors in the development of recurrent miscarriage: an overview of the data of meta-analyses. Obstet Ginecol. 2017. 10.18565/aig.2017.4.14-20.
- 10.Tang W, Zhou X, Chan Y, Wu X, Luo Y. p53 codon 72 polymorphism and recurrent pregnancy loss: a meta-analysis. J Assist Reprod Genet. 2011;28(10):965–969. doi: 10.1007/s10815-011-9618-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen H, Yang X, Wang Z. Association between p53 Arg72Pro polymorphism and recurrent pregnancy loss: an updated systematic review and meta-analysis. Reprod BioMed Online. 2015;31(2):149–153. doi: 10.1016/j.rbmo.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Li X, Liu Y, Zhang R, Tan J, Chen L, Liu Y. Meta-analysis of the association between plasminogen activator inhibitor-1 4G/5G polymorphism and recurrent pregnancy loss. Med Sci Monit. 2015;21:1051–1056. doi: 10.12659/MSM.892898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang B, Dai B, Zhang X, Wang Z. Vascular endothelial growth factor and recurrent spontaneous abortion: a meta-analysis. Gene. 2012;507(1):1–8. doi: 10.1016/j.gene.2012.06.049. [DOI] [PubMed] [Google Scholar]
- 14.Cao Y, Xu J, Zhang Z, Huang X, Zhang A, Wang J, Zheng Q, Fu L, du J. Association study between methylenetetrahydrofolate reductase polymorphisms and unexplained recurrent pregnancy loss: a meta-analysis. Gene. 2013;514:105–111. doi: 10.1016/j.gene.2012.10.091. [DOI] [PubMed] [Google Scholar]
- 15.Chen H, Yang X, Lu M. Methylenetetrahydrofolate reductase gene polymorphisms and recurrent pregnancy loss in China: a systematic review and meta-analysis. Arch Gynecol Obstet. 2016;293(2):283–290. doi: 10.1007/s00404-015-3894-8. [DOI] [PubMed] [Google Scholar]
- 16.Ren A, Wang J. Methylenetetrahydrofolate reductase C677T polymorphism and the risk of unexplained recurrent pregnancy loss: a meta-analysis. Fertil Steril. 2006;86(6):1716–1722. doi: 10.1016/j.fertnstert.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 17.Pereza N, Peterlin B, Volk M, Kapović M, Ostojić S. A critical update on endothelial nitric oxide synthase gene variations in women with idiopathic recurrent spontaneous abortion: genetic association study, systematic review and meta-analyses. Mol Hum Reprod. 2015;21(5):466–478. doi: 10.1093/molehr/gav008. [DOI] [PubMed] [Google Scholar]
- 18.Su MT, Lin SH, Chen YC. Genetic association studies of angiogenesis- and vasoconstriction-related genes in women with recurrent pregnancy loss: a systematic review and meta-analysis. Hum Reprod Update. 2011;17(6):803–812. doi: 10.1093/humupd/dmr027. [DOI] [PubMed] [Google Scholar]
- 19.Cao Y, Zhang Z, Xu J, Wang J, Yuan W, Shen Y, du J. Genetic association studies of endothelial nitric oxide synthase gene polymorphisms in women with unexplained recurrent pregnancy loss: a systematic and meta-analysis. Mol Biol Rep. 2014;41(6):3981–3989. doi: 10.1007/s11033-014-3266-7. [DOI] [PubMed] [Google Scholar]
- 20.Yang C, Fangfang W, Jie L, Yanlong Y, Jie W, Xuefei L, Xuerong Z, Yanling H. Angiotensin-converting enzyme insertion/deletion (I/D) polymorphisms and recurrent pregnancy loss: a meta-analysis. J Assist Reprod Genet. 2012;29(1):1167–1173. doi: 10.1007/s10815-012-9870-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu X, Du C, Li H, et al. Association of VEGF genetic polymorphisms with recurrent spontaneous abortion risk: a systematic review and meta-analysis. PLoS One. 2015;10. 10.1371/journal.pone.0123696. [DOI] [PMC free article] [PubMed]
- 22.Su MT, Lin SH, Chen YC, Kuo PL. Genetic association studies of ACE and PAI-1 genes in women with recurrent pregnancy loss. Thromb Haemost. 2013;109(1):8–15. doi: 10.1160/TH12-08-0584. [DOI] [PubMed] [Google Scholar]
- 23.Chen H, Nie S, Lu M. Association between plasminogen activator Inhibitor-1 gene polymorphisms and recurrent pregnancy loss: a systematic review and meta-analysis. Am J Reprod Immunol. 2015;73(4):292–300. doi: 10.1111/aji.12321. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z, Wang P, Wang X. Significant association between angiotensin-converting enzyme gene insertion/deletion polymorphism and risk of recurrent miscarriage: a systematic review and meta-analysis. Metabolism. 2013;62(9):1227–1238. doi: 10.1016/j.metabol.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 25.Aschard H, Lutz S, Maus B, Duell EJ, Fingerlin TE, Chatterjee N, Kraft P, van Steen K. Challenges and opportunities in genome-wide environmental interaction (GWEI) studies. Hum Genet. 2012;131(10):1591–1613. doi: 10.1007/s00439-012-1192-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marian AJ. Elements of ‘missing heritability’. Curr Opin Cardiol. 2012;27(3):197–201. doi: 10.1097/HCO.0b013e328352707d. [DOI] [PubMed] [Google Scholar]
- 27.Cole BS, Hall MA, Urbanowicz RJ, et al. Analysis of gene-gene interactions. Curr Protoc Hum Genet. 2017. 10.1002/cphg.45. [DOI] [PubMed]
- 28.Nasr HB, Dimassi S, M’hadhbi R, Debbabi H, Kortas M, Tabka Z, Chahed K. Functional G894T (rs1799983) polymorphism and intron-4 VNTR variant of nitric oxide synthase (NOS3) gene are susceptibility biomarkers of obesity among Tunisians. Obes Res Clin Pract. 2016;10(4):465–475. doi: 10.1016/j.orcp.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 29.Blasiak J, Smolarz B. Plasminogen activator inhibitor-1 (PAI-1) gene 4G/5G promoter polymorphism is not associated with breast cancer. Acta Biochim Pol. 2000;47(1):191–199. [PubMed] [Google Scholar]
- 30.Kalita J, Somarajan BI, Kumar B, Mittal B, Misra UK. A study of ACE and ADD1 polymorphism in ischemic and hemorrhagic stroke. Clin Chim Acta. 2011;412(7–8):642–646. doi: 10.1016/j.cca.2010.12.022. [DOI] [PubMed] [Google Scholar]
- 31.El-Aziz TAA, Mohamed RH. Influence of MTHFR C677T gene polymorphism in the development of cardiovascular disease in Egyptian patients with rheumatoid arthritis. Gene. 2017;610:127–132. doi: 10.1016/j.gene.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 32.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 33.dbSNP; https://www.ncbi. nlm.nih.gov/snp
- 34.Chung Y, Lee SY, Elston RC, Park T. Odds ratio based multifactor-dimensionality reduction method for detecting gene–gene interactions. Bioinformatics. 2007;23(1):71–76. doi: 10.1093/bioinformatics/btl557. [DOI] [PubMed] [Google Scholar]
- 35.Yousefian E, Kardi MT, Allahveisi A. Methylenetetrahydrofolate reductase C677T and A1298C polymorphism in Iranian women with idiopathic recurrent pregnancy losses. Iranian Red Crescent Med J. 2014;16(7):1–4. doi: 10.5812/ircmj.16763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isaoglu U, Ulug P, Delibas IB, et al. The association between inherited thrombophilia and recurrent pregnancy loss in Turkish women. Clin Exp Obstet Gynecol. 2014;41(2):177–181. [PubMed] [Google Scholar]
- 37.Bozikova A, Gabrikova D, Pitonak J, Bernasovska J, Macekova S, Lohajova-Behulova R. Ethnic differences in the association of thrombophilic polymorphisms with obstetric complications in Slovak and Roma (Gypsy) populations. Genet Test Mol Biomarkers. 2015;19(2):98–102. doi: 10.1089/gtmb.2014.0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parveen F, Faridi RM, Alam S, Agrawal S. Genetic analysis of eNOS gene polymorphisms in association with recurrent miscarriage among North Indian women. Reprod BioMed Online. 2011;23(1):124–131. doi: 10.1016/j.rbmo.2011.03.022. [DOI] [PubMed] [Google Scholar]
- 39.Banerjee P, Ghosh S, Dutta M, Subramani E, Khalpada J, RoyChoudhury S, et al. Identification of key contributory factors responsible for vascular dysfunction in idiopathic recurrent spontaneous miscarriage. PLoS One. 2013;8. 10.1371/journal.pone.0080940. [DOI] [PMC free article] [PubMed]
- 40.Najafi T, Novin MG, Ghazi R, Khorram O. Altered endometrial expression of endothelial nitric oxide synthase in women with unexplained recurrent miscarriage and infertility. Reprod BioMed Online. 2012;25(4):408–414. doi: 10.1016/j.rbmo.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo L, Li DH, Wei SG, Zhang HB, Li SB, Zhao J. Polymorphisms in the endothelial nitric oxide synthase gene associated with recurrent miscarriage. Genet Mol Res. 2013;12(3):3879–3886. doi: 10.4238/2013.September.23.6. [DOI] [PubMed] [Google Scholar]
- 42.Shin SJ, Lee HH, Cha SH, Kim JH, Shim SH, Choi DH, Kim NK. Endothelial nitric oxide synthase gene polymorphisms (− 786T> C, 4a4b, 894G> T) and haplotypes in Korean patients with recurrent spontaneous abortion. Eur J Obstet Gynecol Reprod Biol. 2010;152(1):64–67. doi: 10.1016/j.ejogrb.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 43.Zammiti W, Mtiraoui N, Mahjoub T. Lack of consistent association between endothelial nitric oxide synthase gene polymorphisms, homocysteine levels and recurrent pregnancy loss in Tunisian women. Am J Reprod Immunol. 2008;59(2):139–145. doi: 10.1111/j.1600-0897.2007.00551.x. [DOI] [PubMed] [Google Scholar]
- 44.Lee HH, Hong SH, Shin SJ, Ko JJ, Oh D, Kim NK. Association study of vascular endothelial growth factor polymorphisms with the risk of recurrent spontaneous abortion. Fertil Steril. 2010;93(4):1244–1247. doi: 10.1016/j.fertnstert.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 45.Traina E, Daher S, Moron AF, Sun SY, Franchim CS, Mattar R. Polymorphisms in VEGF, progesterone receptor and IL-1 receptor genes in women with recurrent spontaneous abortion. J Reprod Immunol. 2011;88(1):53–57. doi: 10.1016/j.jri.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 46.Papazoglou D, Galazios G, Papatheodorou K, Liberis V, Papanas N, Maltezos E, Maroulis GB. Vascular endothelial growth factor gene polymorphisms and idiopathic recurrent pregnancy loss. Fertil Steril. 2005;83(4):959–963. doi: 10.1016/j.fertnstert.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 47.Lash GE, Cartwright JE, Whitley GS. The effects of angiogenic growth factors on extravillous trophoblast invasion and motility. Placenta. 1999;20:661–667. doi: 10.1053/plac.1999.0427. [DOI] [PubMed] [Google Scholar]
- 48.Yalcintepe SA, Silan F, Hacivelioglu SO, et al. Fetal VEGF genotype is more important for abortion risk than mother genotype. Int J Mol Cell Med. 2014;3(2):88–94. [PMC free article] [PubMed] [Google Scholar]
- 49.Amirchaghmaghi E, Rezaei A, Moini A, et al. Gene expression analysis of VEGF and its receptors and assessment of its serum level in unexplained recurrent spontaneous abortion. Cell J. 2015;4(16):538–545. doi: 10.22074/cellj.2015.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galazios G, Papazoglou D, Tsikouras P, Kolios G. Vascular endothelial growth factor gene polymorphisms and pregnancy. J Matern Fetal Neonatal Med. 2009;22(5):371–378. doi: 10.1080/14767050802645035. [DOI] [PubMed] [Google Scholar]
- 51.Andraweera PH, Dekker GA, Roberts CT. The vascular endothelial growth factor family in adverse pregnancy outcomes. Hum Reprod Update. 2012;18(4):436–457. doi: 10.1093/humupd/dms011. [DOI] [PubMed] [Google Scholar]
- 52.Sun L, Lv H, Wei W, Zhang D, Guan Y. Angiotensin-converting enzyme D/I and plasminogen activator inhibitor-1 4G/5G gene polymorphisms are associated with increased risk of spontaneous abortions in polycystic ovarian syndrome. J Endocrinol Investig. 2010;33(2):77–82. doi: 10.1007/BF03346557. [DOI] [PubMed] [Google Scholar]
- 53.Labied S, Blacher S, Carmeliet P, Noël A, Frankenne F, Foidart JM, Munaut C. Transient reduction of placental angiogenesis in PAI-1-deficient mice. Physiol Genomics. 2011;43(4):188–198. doi: 10.1152/physiolgenomics.00147.2010. [DOI] [PubMed] [Google Scholar]
- 54.Subrt I, Ulcova-Gallova Z, Cerna M, Hejnalova M, Slovanova J, Bibkova K, Micanova Z. Recurrent pregnancy loss, plasminogen activator inhibitor-1 (-675) 4G/5G polymorphism and antiphospholipid antibodies in Czech women. Am J Reprod Immunol. 2013;70(1):54–58. doi: 10.1111/aji.12099. [DOI] [PubMed] [Google Scholar]
- 55.Kim JJ, Choi YM, Lee SK, Yang KM, Paik EC, Jeong HJ, Jun JK, Han AR, Hong MA. The PAI-1 4G/5G and ACE I/D polymorphisms and risk of recurrent pregnancy loss: a case–control study. Am J Reprod Immunol. 2014;72(6):571–576. doi: 10.1111/aji.12302. [DOI] [PubMed] [Google Scholar]
- 56.Serrano NC, Díaz LA, Páez MC, Mesa CM, Cifuentes R, Monterrosa A, et al. Angiotensin-converting enzyme I/D polymorphism and pre-eclampsia risk: evidence of small-study bias. PLoS Med. 2006;3. 10.1371/journal.pmed.0030520. [DOI] [PMC free article] [PubMed]
- 57.Kim DK, Kim JW, Kim S, Gwon HC, Ryu JC, Huh JE, Choo JA, Choi Y, Rhee CH, Lee WR. Polymorphism of angiotensin converting enzyme gene is associated with circulating levels of plasminogen activator inhibitor-1. Arterioscler Thromb Vasc Biol. 1997;17:3242–3247. doi: 10.1161/01.atv.17.11.3242. [DOI] [PubMed] [Google Scholar]
- 58.Al Sallout RJ, Sharif FA. Polymorphisms in NOS3, ACE and PAI-1 genes and risk of spontaneous recurrent miscarriage in the Gaza Strip. Med Princ Pract. 2010;19(2):99–104. doi: 10.1159/000273067. [DOI] [PubMed] [Google Scholar]
- 59.Floridon C, Nielsen O, Hølund B, Sweep F, Sunde L, Thomsen SG, Teisner B. Does plasminogen activator inhibitor-1 (PAI-1) control trophoblast invasion? A study of fetal and maternal tissue in intrauterine, tubal and molar pregnancies. Placenta. 2000;21(8):754–762. doi: 10.1053/plac.2000.0573. [DOI] [PubMed] [Google Scholar]
- 60.Dossenbach-Glaninger A, van Trotsenburg M, Schneider B, Oberkanins C, Hopmeier P. ACE I/D polymorphism and recurrent first trimester pregnancy loss: interaction with SERPINE1 4G/5G and F13 Val34Leu polymorphisms. Br J Haematol. 2008;141(2):269–271. doi: 10.1111/j.1365-2141.2008.07058.x. [DOI] [PubMed] [Google Scholar]
- 61.Buchholz T, Lohse P, Rogenhofer N, et al. Polymorphisms in the ACE and PAI-1 genes are associated with recurrent spontaneous miscarriages. Hum Reprod. 2003;18(11):2473–2477. doi: 10.1093/humrep/deg474. [DOI] [PubMed] [Google Scholar]