Abstract
As a member of four and a half LIM domain proteins, FHL3 gene-encoded protein (FHL3) plays an important role in skeletal muscle development and contraction. In this study, we determined the potential role of FHL3 in the proliferation and differentiation of primary satellite cells in chicken. RT-qPCR results showed that FHL3 mRNA was highly expressed in skeletal muscle in 12 chicken tissues. Four cell proliferation assays (CCK8 assay; EDU staining assay; flow cytometric detection of cell cycle assay; and detection of cell proliferation marker genes Ki67 and N-Ras assay) revealed that FHL3 knockdown had no effect on the proliferation rate of chicken satellite cells. FHL3 knockdown promoted the differentiation of satellite cells into myotubes, as evidenced by increased fusion index, number of nuclei per myotube, Myog, Myh7, Myf5, and Mrf4 mRNA expressions, and myog and myosin heavy chain protein expressions of myogenic markers (P < 0.05). These results showed that the FHL3 was a negative regulator of the differentiation and fusion of chicken satellite cells into myotubes. However, FHL3 expression was increased during the differentiation of chicken satellite cells into myotubes. The study suggested that FHL3 might have different functions in chicken myotubes compared with that in chicken satellite cells.
Keywords: FHL3, Chicken, Skeletal muscle satellite cell, Proliferation, Differentiation
Introduction
Skeletal muscle is the most abundant muscle tissue in animals and accounts for 45–60% of the body weight in adult animals. Skeletal muscle is composed of multinucleated myofibroblasts, which are formed by the differentiation of myoblasts. The process from muscle progenitor cells to the formation of muscle fibers, called myogenesis, is regulated by a series of myogenic regulatory factors, including myogenic factor 5 (Myf5), myogenic differentiation antigen (MyoD), myogenic (MyoG) and myogenic regulatory factors 4 (Mrf4) (Kablar et al. 2003; Kassar-Duchossoy et al. 2004; Rudnicki et al. 1993). Skeletal muscle satellite cells (SMSCs) are mononuclear myogenic stem cells with proliferation and differentiation potential. SMSCs undergo proliferation, differentiation and fusion to form myotubes and fuse into skeletal muscle fibers, which promote muscle fibers to grow and proliferate (Mauro 1961). At present, a large number of muscle regulatory factors have been discovered, such as paired box gene 3/7, hepatocyte growth factor and transforming growth factor-beta (Mcpherron et al. 1997; Velleman and Mcfarland 1999). Skeletal muscle plays important role in initiating movements, supporting respiration, and maintaining homeostasis; losing skeletal muscle function results in muscle aging and diseases, including cancers and diabetes (Glass 2003; Vinciguerra et al. 2010). Besides, skeletal muscle has significant economic benefits in the production of meat animals. Therefore, the exploration on new regulation mechanisms of skeletal muscle development may contribute to the improvements in the animal productivity.
FHL3 belongs to a family of proteins containing four semi-LIM structures, which has five members, namely, FHL1, FHL2, FHL3, FHL4, and ACT (Fimia et al. 2000). Four and a half LIM proteins serve as transcriptional regulators in actin and cytoskeleton and have a vital function in regulating muscle development (Mcgrath et al. 2006). FHL3 can interact with myogenic determinants, actin, transcription factors, and cell cycle regulators through its LIM domain and regulate myocyte differentiation, cytoskeletal structure, skeletal muscle formation, and the expression of certain genes (Coghill et al. 2003; Morgan and Madgwick 1999; Takahashi et al. 2005). In the study on C2C12 (Cottle et al. 2007), it was found that FHL3 negatively controlled muscle development that impaired the transcriptional activity of MyoD, which leads to differentiation process of myocytes restrained. FHL3 is conserved in multiple species. Its function has only been studied in mice and humans (Cottle et al. 2007; Huang et al. 2010), but the regulatory mechanisms in chicken skeletal muscle have not been reported.
Based on the above studies, we hypothesized that FHL3 played a role in the regulation of the proliferation and differentiation in avian SMSCs. In this study, the chicken primary SMSCs were transfected by constructed FHL3-siRNA and detected through immunofluorescence staining, real-time PCR, and western blotting so as to explore the effects of FHL3 on skeletal muscle proliferation and differentiation in chicken.
Materials and methods
Isolation and culture of chicken skeletal muscle satellite cells
Posthatch chickens (7-day-old male commercial generation Avian broiler chicks) were purchased from Wenjiang Charoen Pokphand Livestock & Poultry Co., Ltd. The pectoralis muscle was removed and used for preparation of primary myogenic cultures. About 5 g of muscle was finely minced and treated with 0.1% collagenase I (Sigma, St. Louis, MO, USA) followed by 0.25% trypsin (Hyclone, Logan, UT, USA) to release cells. Then, the cell suspension was subjected to percoll density centrifugation to separate satellite cells from contaminating myofibril debris and nonmyogenic cells. Cells were plated in 25 cm3 cell culture bottles with complete medium [DMEM/F12 (Invitrogen, Carlsbad, CA, USA) + 15% FBS (Gibco, Grand Island, NY, USA) + 1% penicillin–streptomycin (Solarbio, Beijing, China) + 3% chicken embryo extraction]. The cells cultured at 37 °C and 5% CO2 with saturating humidity, which were allowed to proliferate in growth medium for 2–4 days, and the medium was refresh every 24 h. To induce differentiation, SMSCs were grown to 90% confluence in growth medium, and then replaced with differentiation medium composed of DMEM, 2% horse serum (Hyclone) and 1% penicillin–streptomycin, and the medium was refresh every 24 h.
Knockdown of FHL3 mRNA in chicken skeletal satellite cells
The small interfering RNA (siRNA)-mediated knockdown was conducted in 6-well plates. When the cells reached approximately 70% confluence, cells in each well were transfected with FHL3 siRNAs. The FHL3 siRNA sequences were 5′-GCAAUGACUGCUACUGCAATT-3′ (sense) and 5′-UUGCAGUAGCAGUCAUUGCTT-3′ (anti-sense). The FHL3 siRNA was a chemically synthesized siRNA in the desalted, preannealed duplex form (Sangon biotech, Shanghai, China). Cell transfection was performed using the reagent protocol Lipofectamine 3000 (Invitrogen). The Lipofectamine 3000 and siRNA were diluted with optim-MEM culture medium. The diluted siRNA and Lipofectamine 3000 were mixed evenly and placed at room temperature for 10 min. The composite was added to the cell culture plate and was mixed in the culture plate. Knockdown efficiency was estimated by quantitative RT-qPCR and western blot of FHL3.
Quantitative RT-PCR
Total RNA from cells was extracted using TRI Reagent TRIzol reagent (Invitrogen), according to the manufacturer’s instructions. The RNA 6000 Nano chip assay was used with a Bio analyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) to assess the RNA integrity, quality, and quantity. All total RNA samples were stored at − 80 °C. Reverse transcription was performed using 2 µg of total RNA by PrimeScript RT Master Mix Perfect Real Time (Takara, Dalian, China) according to the manufacturer’s instructions. Real-time PCR primers were designed by Primer Premier 5 (Table 1) and the mRNA abundance of each gene was determined using the CFX96-Touch™ real-time PCR detection system (Bio-Rad, Hercules, CA, USA). Real-time PCR reactions were performed in triplicate in a volume of 10 µL containing 1 µL of cDNA, 0.5 µL of reverse and forward primers (10 µM) for each gene, 3 µL of double-distilled H2O, and 5 µL of SYBR® Premix Ex TaqTM II (Bio-Rad). The gene expression was normalized to β-actin, according to the 2 − ΔΔCT method (Livak and Schmittgen 2001).
Table 1.
Primers used for RT-qPCR in this study
| Gene | Forward primer (5′ → 3′) | Reverse primer (5′ → 3′) | Product/bp |
|---|---|---|---|
| MyoG | GGAGGCTGAAGAAGGTGAACG | GCTCGATGTACTGGATGGCG | 117 |
| Myh7 | GAAGGAGACCTCAACGAGATGG | ATTCAGGTGTCCCAAGTCATCC | 138 |
| MRF4 | CTTCCTCTGGAGTTGCTCTACC | TAAGTCCACAGGGTTCAGTAGG | 66 |
| Myf5 | CCTCATGTGGGCTTGCAAA | CCTTCCGCCGGTCCAT | 59 |
| FHL3 | AATGGCCCATACTGCATCCC | TGGTAGTGGCGATCCTCGTA | 116 |
| Ki67 | TACTTCGAGCAGTTCGGCAC | CTCGAACTCGATGAAGGCGT | 93 |
| N-Ras | TCAGCCAAAACGAGACAGGG | TGCAGGACAACCCCATACAG | 127 |
| β-actin | GTCCACCGCAAATGCTTCTAA | TGCGCATTTATGGGTTTTGTT | 78 |
Cell counting kit-8 (CCK-8) assay
Chicken satellite cells were transfected with the negative control siRNA or FHL3 siRNA in 96-well plates according to the method described above and the complete culture medium without cells was added to the standard blank hole (6 holes were taken for each group). After 0, 24, 48, and 72 h of culturing, 10 μl cck-8 (Bestbio, Shanghai, China) reagents were added to each well and placed in the culture box for 2 h. A value of each well under the wavelength of 450 nm was determined, and the changes of cell proliferation in each group were observed.
EDU assay
After knockdown the FHL3, the proliferation of satellite cells was tested using a Cell-Light EdU Apollo 567 in vitro Flow Cytometry Kit (Ribobio, Guangzhou, China). The cells were exposed to 50 μM Edu for 2 h at 37 °C following the manufacturer’s instructions. The EdU-stained cells were visualized by fluorescence microscopy (Nikon, Tokyo, Japan). The analysis of cell proliferation was performed using images of randomly selected fields obtained from the fluorescence microscope. We performed four repeats for each group, and three images were used to calculate the cell proliferation rate in each repeats.
Characterization of myotubes
To clearly distinguish the nuclei from the myotubes, the differentiated chicken satellite cells were washed with PBS and fixed with 4% paraformaldehyde for 10 min. The cells were then stained with Gemisa (Invitrogen) dyeing for 10 min. The total number of nuclei and the number of nuclei within the myotubes were counted using NIH ImageJ software. Muscle cells with three or more nuclei were regarded as myotubes. The fusion index is calculated as the number of nuclei in the myotubes divided by the total number of nuclei in the cell. The average number of nuclei in the myotubes is calculated as the number of nuclei in the myotubes divided by the total number of myotubes.
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min and permeated with 0.25% Triton X-100 for 10 min at room temperature. Cells were then blocked with 1% bovine serum albumin (BSA) in PBST (PBS + 0.05% Tween-20) and incubated with anti-myosin heavy chain (MHC) antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 1:200 dilution at 4 °C overnight. The anti-MHC antibody was detected by incubating the cells with anti-mouse IgG FITC antibody (Abcam, San Francisco, CA, USA) at 1:200 dilution at room temperature for 1 h. Cell nuclei were stained by incubating the cells in 1 mg/ml DAPI (Sigma) for 1 min at room temperature. Fluorescence was detected with a eclipse E600 florescence microscope (Nikon).
Western blot analysis
Cells were washed with PBS and lysed in RIPA lysed buffer. Next, 200 μg of total protein were separated by 12% SDS–polyacrylamide gel electrophoresis (SDS–PAGE), and transferred to a PVDF membrane (Millipore Corporation, Billerica, MA, USA). The PVDF membrane was incubated with 5% defatted milk powder at room temperature for 1 h, followed by incubation with the following specific primary antibodies at 4 °C overnight. The following primary antibodies were used: anti-FHL3 (Santa Cruz Biotechnology); anti-MHC (Santa Cruz Biotechnology); anti-MyoG (Santa Cruz Biotechnology) and anti-β-actin (Santa Cruz Biotechnology). The PVDF membrane was then rinsed with TBST and stained with the appropriate HRP-labeled secondary antibody for 1 h at room temperature. After washing with TBST, the strip was coated with enhanced chemiluminescence (Amersham Pharmacia Biotech, Piscataway, NJ, USA) with a Kodak imager (Eastman Kodak, Rochester, NY, USA). The relative expression of protein was analyzed with Quantity One software.
Flow cytometric analysis
To detect cell cycle, satellite cells were cultured in 12-well plates. When the cell density reached 50%, FHL3 siRNA and negative siRNA were transfected into satellite cells, respectively. After 48 h of transfection, cells were harvested and fixed overnight in 70% ethanol at 4 °C. Then, we stained the fixed cells by 50 μg/mL propidium iodide solution (Sigma) with 10 μg/mL RNase A (Takara) and 0.2% (v/v) Triton X-100 (Sigma) contained and incubated at 37 °C in the dark for 30 min. Subsequently, flow cytometry analysis was conducted on a BD Accuri C6 flow cytometer (BD Biosciences, USA) and FlowJo7.6 software was used for data processing.
Statistical analyses
All statistical analyses were performed using SPSS 17.0 (SPSS Inc., Chicago, IL, USA). Data are presented as least squares means ± standard error of the mean (SEM), and values were considered statistically different at P < 0.05.
Results
Expression of FHL3 in different tissues of chickens
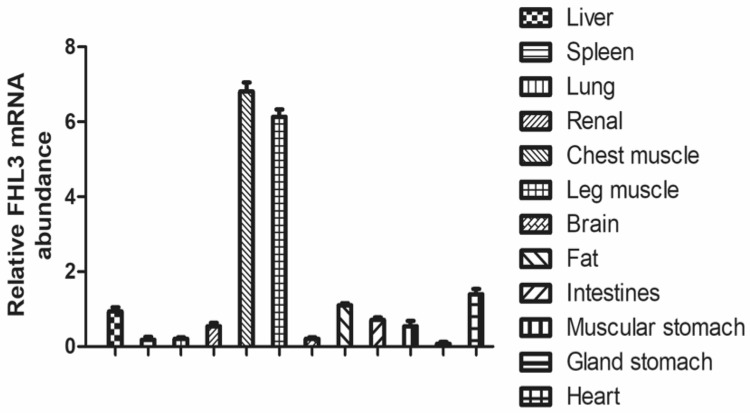
The expression of FHL3 mRNA in skeletal muscle in 12 different chicken tissues or organs was mainly detected by RT-qPCR (Fig. 1). Relatively expression levels of FHL3 mRNA in different tissues and organs were decreased according to the following order: heart, fat, liver, intestine and kidney and FHL3 mRNA level was extremely low in lung, spleen and stomach.
Fig. 1.
FHL3 mRNA expression in twelve different tissues and organs of chicken. Data are expressed as means ± SEM (N = 3)
Knockdown of FHL3 expression in chicken SMSCs
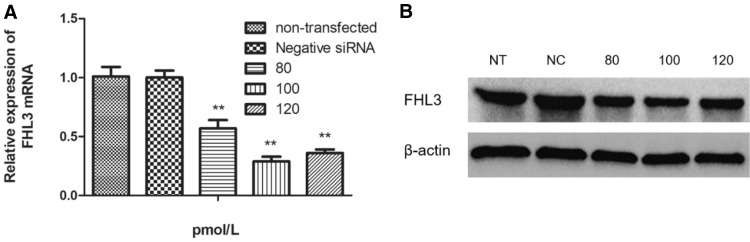
Chicken SMSCs were transfected with 3 different concentrations of siRNA targeting FHL3 mRNA to reduce FHL3 gene expression. The final concentrations in 6-well plate were respectively 80, 100, and 120 pmol/L. Cells were respectively transfected with 80, 100, and 120 pmol/L FHL3 siRNA for 48 h and the interference efficiencies were quantitatively detected to be 54.68%, 71.24% and 43.28% by real-time PCR (Fig. 2a). Western blot also indicated that the transfection with 100 pmol/L siRNA realized the best interference efficiency (Fig. 2b). In subsequent experiments, the final siRNA concentration in the 6-well plate was selected to be 100 pmol/L.
Fig. 2.
Interference efficiencies of 3 different concentrations of FHL3-siRNA. a Relative expression levels of FHL3 mRNA at 48 h after transfection. b The protein levels of FHL3 examined by a western blot analysis. β-actin was used as loading control. Data are expressed as means ± sem (N = 3). **Means extremely significant difference (P < 0.01)
Interference with FHL3 had no effect on chicken SMSCs proliferation
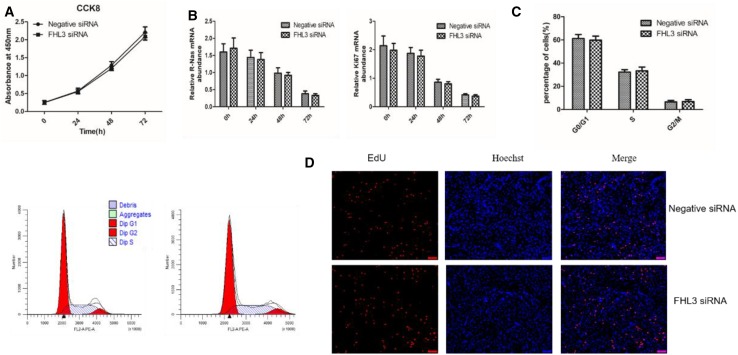
To determine the potential role of FHL3 in chicken satellite cell proliferation, the same number of SMSCs were transfected with negative siRNA and FHL3 siRNA for 0, 24, 48 and 72 h when the cell density reached 70%. The effect of interference FHL3 on SMSC proliferation was detected by CCK8. The number of cells displayed almost linear growth in 72 h (Fig. 3a). The proliferation rate of satellite cells transfected with FHL3-siRNA was not different from those cells which transfected with negative siRNA. To further detect the effect of FHL3 on satellite cell proliferation, the expressions of satellite cell proliferation-related genes ki67 and N-Ras were detected by RT-qPCR (Fig. 3b). The results showed that mRNA of ki67 and N-Ras showed no significant change. In addition, we performed cell cycle analysis and EDU assays after 48-h knockdown of FHL3 in satellite cells. Cell cycle analysis showed that the number of S and G2/M phase cells was no significantly different compared with the negative siRNA group (Fig. 3c). Similarly, compared to the control group, FHL3 siRNA group showed no difference in the number of EDU-stained cells (Fig. 3d). The results showed that FHL3 had no effect on the proliferation of chicken SMSCs.
Fig. 3.
Effects of FHL3 knockdown on the proliferation of chicken satellite cells. a Cell proliferation rate was estimated by measuring the number of viable cells at 0, 24, 48 and 72 h after transfection. Absorbance at 450 nm on the y-axis represented the number of viable cells. b The mRNA expression of Ki67 and N-Ras at 0, 24, 48 and 72 h detected by RT-qPCR. c Cell cycle was analyzed through flow cytometry. d Edu assays for cells transfected with negative or FHL3 siRNA. EdU (red) fluorescence indicates proliferation. Nuclei are indicated by Hoechst (blue) fluorescence. All photomicrographs are at 100 × magnification. Data are expressed as means ± sem (N = 3)
FHL3 knockdown promoted the differentiation of chicken SMSCs
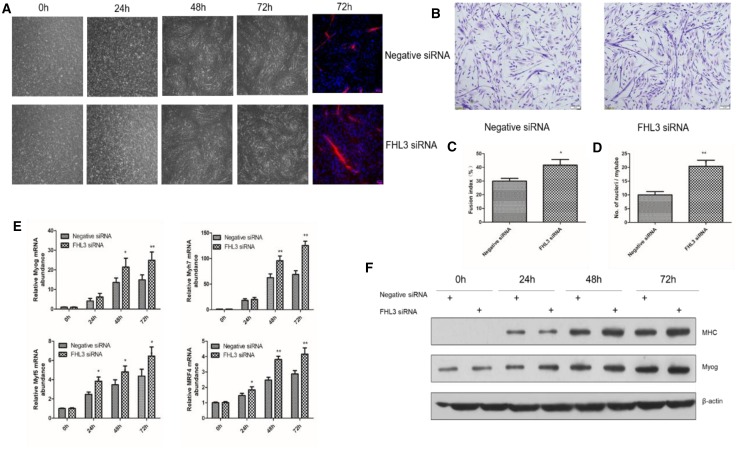
To investigate whether FHL3 affected the differentiation chicken SMSCs, SMSCs were transfected respectively with negative siRNA and FHL3-siRNA when the cell density reached approximately 70%. Morphological observations demonstrated that the satellite cells transferred with FHL3 siRNA formed larger and more myotubes than the negative control group (Fig. 4a). To determine the potential role of FHL3 in the differentiation of satellite cells, satellite cells transfected with FHL3 siRNAs or negative siRNA were induced to differentiate and their differentiation status was assessed by measuring the fusion index and the average number of nuclei per myotube. After 72-h of differentiation, more satellite cells transfected with FHL3 siRNAs formed myotubes than those transfected with negative siRNA (41% vs 29%, Fig. 4b, c). Myotubes formed from FHL3 siRNAs-transfected satellite cells had more nuclei on average than those from negative siRNA-transfected satellite cells (18 vs 11 nuclei/myotube; Fig. 4d).
Fig. 4.
Effects of FHL3 knockdown on the differentiation of chicken satellite cells. Chicken satellite cells were transfected with FHL3-siRNA or negative control siRNA and cultured in differentiation medium for the following experiments. a The morphology of cells transfected at 0, 24, 48 and 72 h was observed under the light microscope, and cells at the far right were stained with anti-myosin heavy chain antibody. b After cells differentiation for 72 h, Gemisa staining was used to observe the morphology and the number of nuclei, myotubes in cells. c Fusion index at 72 h of differentiation. The fusion index is calculated as the percentage of total nuclei in cells with three or more nuclei. d The average number of nuclei per muscle tube at 72 h of differentiation. e The relative mRNA expression of Myh7, Myog, MRF4 and Myf5 measured at 0, 24, 48 and 72 h of differentiation. f Western blots detected the protein relative expression levels at 0, 24, 48 and 72 h in Myhc and Myog. β-actin was used as loading control. Data are expressed as means ± sem (N = 3). *P < 0.05; **P < 0.01 versus “Negative siRNA”
To further determine the differentiation effects of FHL3 on chicken satellite cells, we also measured the mRNA levels of 4 myogenic markers including Myog, Myh7, MRF4 and Myf5 and the protein levels of 2 myogenic markers including Myog and myosin heavy chain (MHC) at 0, 24, 48, and 72 h of differentiation. The cells transfected with FHL3 siRNA had higher mRNA expressions of Myog, Myh7, MRF4 and Myf5 than the cells transfected with negative siRNA at 24, 48, and 72 h of differentiation (Fig. 4e). Consistently, the former also had higher expressions of Myog and MHC proteins than the latter at 24, 48, and 72 h of differentiation (Fig. 4f). The myogenic marker expression data further suggested that FHL3 knockdown promoted the differentiation of chicken satellite cells into myotubes.
Expression of FHL3 mRNA in chicken SMSCs increased during differentiation
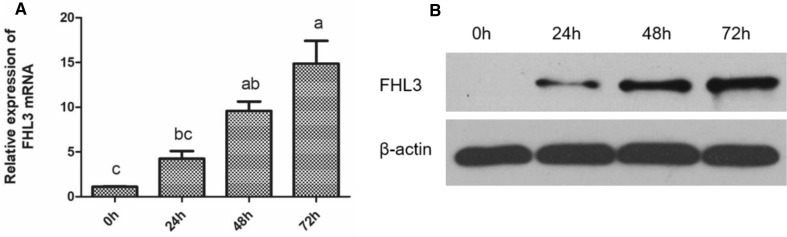
To determine the role of FHL3 in differentiation into myotubes in chicken SMSCs, we compared the expression level of FHL3 mRNA in differentiated chicken skeletal muscle satellite cells in 0, 24, 48, and 72 h. It is worth noting that during the differentiation of chicken satellite cells into myotubes, the FHL3 mRNA expression was significantly increased with time (Fig. 5a). Western blot analysis showed that FHL3 protein expression gradually increased during this differentiation (Fig. 5b).
Fig. 5.
Expressions of mRNA and protein levels of FHL3 in chicken satellite cells during differentiation. Chicken satellite cells were induced to differentiate 0 h, 24 h, 48 h, and 72 h in differentiation medium. a The mRNA expression of FHL3 after induced differentiation. Bars not sharing the same letter labels are different (P < 0.05; N = 3 independent cell cultures). b The protein level of FHL3 from 3 independent cell cultures after induced differentiation. β-actin was used as loading control
Discussion
In this study, we determined the expression pattern of FHL3 in chicken. FHL3 was highly expressed in skeletal muscle compared with other tissues and the result was consistent with the study on FHL3 in mice and human beings (Cottle et al. 2007; Lee et al. 1998). The high expression of FHL3 in chicken skeletal muscle indicated that FHL3 was important in the growth, maintenance and function of chicken skeletal muscle. We also studied the potential regulation mechanism of FHL3 on the proliferation and differentiation in chicken satellite cells. After FHL3 was silenced, the proliferation rate of SMSCs had no significant difference compared with the control group, indicating that FHL3 has no direct regulatory effect on the proliferation in SMSCs. In differentiated chicken satellite cells interfered with FHL3, muscle differentiation-related genes Myh7, MyoG, Myf5, and Mrf4 exhibited significantly higher expression levels of mRNA than the negative control group. Meanwhile, the fusion rate of the myotubes and the average number of nuclei in the myotubes were significantly higher than those in the control group. This experiment demonstrated that the down-regulation of FHL3 promoted the formation of muscle tube in satellite cells and that FHL3 played a negative role in the differentiation of chicken skeletal muscle satellite cells.
In recent years, it has been reported that FHL3 can inhibit gene transcription in non-muscle cells (Takahashi et al. 2005). FHL3 inhibited the transcription activity of MyoD by forming a transcription complex with MyoD, thus restraining the differentiation and fusion of C2C12 cells into myotubes (Cottle et al. 2007). Our experiment showed that FHL3 could inhibit the differentiation and fusion of SMSCs. The result was in accordance with the role of FHL3 in C2C12 myoblasts. Besides, muscle LIM protein promoted myogenesis by interacting with the muscle basic helix–loop–helix (bHLH) transcription factors MyoD, MRF4, and myogenin (Kong et al. 1997). Alison et al. illuminated that FHL2 can interact with Hand1 via bHLH domain. Overexpression of FHL2 can cause the binding of FHL2 protein with β-catenin, thus reducing the expression of LEF/TCF and promoting the differentiation of C2C12 cells into myotubes (Hill and Riley 2004). Different from MLP and FHL2, FHL3 controlled myoblast differentiation negatively. In this study, the interference with FHL3 up-regulated the expression of MyoD and other key genes of muscle cells differentiation, thus promoting the differentiation of SMSCs into myotubes. The molecular regulation mechanism of FHL3-mediated myotubes in the differentiation of satellite cells is completely different from that of muscle differentiation mediated by other FHL protein families or LIM domain proteins. It is supposed that the binding of FHL3 to bHLH protein blocks the heterodimerization of MyoD and E12, which hinders the transcription activity of MyoD and finally leads to the negative regulation of FHL3 in muscle development (Jen et al. 1992). In addition, MyoD and MyoG are essential myogenic regulatory factors in the process of muscle differentiation and functions in the differentiation and fusion of muscle cells (Megeney et al. 1996; Sassoon et al. 1989). The lack of MyoD gene can give rise to diseases such as muscular atrophy (Wu et al. 2002). Therefore, we speculated that FHL3 bound to MyoD protein to form a complex through the LIM domain and functioned in its potent negative co-transcriptional regulation. The complex inhibited the transcription activity of MyoD, thus ultimately leading to the negative regulation of chicken satellite cells into myotubes.
Myogenesis is a strictly controlled program. Precocious differentiation of satellite cells may bring about the dysfunction in the formation of muscle fibers (Bentzinger et al. 2012; Schuster-Gossler et al. 2007), therefore, the negative regulation of factors is non-negligible in the formation of normal muscle fibers. FHL3 may be one of the important factors for early satellite cell differentiation during skeletal muscle development. Meanwhile, FHL3 protein increased gradually during the differentiation of satellite cells into myotubes. The result seems to be contrary to the fact that FHL3 inhibits the differentiation of myotubes. The difference may be ascribed to the different roles of FHL3 in the satellite cells and the myotubes. One known function of the Lim domain FHL3 in myotubes or myofibers is that it can be colocalized with α7β1 integrin receptor at the periphery of Z-discs, suggesting its role in the mechanical stabilization of muscle cells (Samson et al. 2004). Therefore, the increased expression of FHL3 in myotubes is required for this function in skeletal muscle. In addition, the LIM domain in FHL3 protein is a classical domain of protein–protein interactions (Lee et al. 1998). The specific mechanism of muscle development regulation by FHL3 is still not fully understood and deserves further study. The mechanism of the differentiation regulation of satellite cells into myotubes by FHL3 deserves further study.
In summary, the study demonstrated that chicken FHL3 was predominantly expressed in skeletal muscle. The results also suggested that FHL3 in chicken satellite cells might control the differentiation and fusion of these cells into myotubes.
Acknowledgements
This research was supported by Natural Science Foundation of China (31402069). We thank Toppaper (http://www.toppaper.cn) for editing the English text of a draft of this manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no conflicts of interest.
References
- Bentzinger CF, Wang YX, Rudnicki MA. Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biol. 2012;4(2):441. doi: 10.1101/cshperspect.a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghill ID, Brown S, Cottle DL, McGrath MJ, Robinson PA, Nandurkar HH, Dyson JM, Mitchell CA. FHL3 is an actin-binding protein that regulates alpha-actinin-mediated actin bundling: FHL3 localizes to actin stress fibers and enhances cell spreading and stress fiber disassembly. J Biol Chem. 2003;278(26):24139–24152. doi: 10.1074/jbc.M213259200. [DOI] [PubMed] [Google Scholar]
- Cottle DL, Mcgrath MJ, Cowling BS, Coghill ID, Brown S, Mitchell CA. FHL3 binds MyoD and negatively regulates myotube formation. J Cell Sci. 2007;120(8):1423–1435. doi: 10.1242/jcs.004739. [DOI] [PubMed] [Google Scholar]
- Fimia GM, De Cesare D, Sassone-Corsi P. A family of LIM-only transcriptional coactivators: tissue-specific expression and selective activation of CREB and CREM. Mol Cell Biol. 2000;20(22):8613–8622. doi: 10.1128/MCB.20.22.8613-8622.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass DJ. Molecular mechanisms modulating muscle mass. Trends Mol Med. 2003;9(8):344–350. doi: 10.1016/S1471-4914(03)00138-2. [DOI] [PubMed] [Google Scholar]
- Hill AA, Riley PR. Differential regulation of Hand1 homodimer and Hand1-E12 heterodimer activity by the cofactor FHL2. Mol Cell Biol. 2004;24(22):9835–9847. doi: 10.1128/MCB.24.22.9835-9847.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wang JF, Xia WR, Zou MJ, Cai X, Xu DG. Identification of the transactivation domain of the human FHL3. Mol Bio (Mosk) 2010;44(2):335–339. [PubMed] [Google Scholar]
- Jen Y, Weintraub H, Benezra R. Overexpression of Id protein inhibits the muscle differentiation program: in vivo association of Id with E2A proteins. Genes Dev. 1992;6(8):1466–1479. doi: 10.1101/gad.6.8.1466. [DOI] [PubMed] [Google Scholar]
- Kablar B, Krastel K, Tajbakhsh S, Rudnicki MA. Myf5 and MyoD activation define independent myogenic compartments during embryonic development. Dev Biol. 2003;258(2):307–318. doi: 10.1016/S0012-1606(03)00139-8. [DOI] [PubMed] [Google Scholar]
- Kassar-Duchossoy L, Gayraud-Morel B, Gomès D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5:MyoD double-mutant mice. Nature. 2004;431(7007):466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- Kong Y, Flick MJ, Kudla AJ, Konieczny SF. Muscle LIM protein promotes myogenesis by enhancing the activity of MyoD. Mol Cell Biol. 1997;17(8):4750–4760. doi: 10.1128/MCB.17.8.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Tsui SK, Chan KK, Kotaka M, Li HY, Chim SS, Waye MM, Fung KP, Lee CY. Chromosomal mapping of a skeletal muscle specific LIM-only protein FHL3 to the distal end of the short arm of human chromosome 1. Somat Cell Mol Genet. 1998;24(3):197–202. doi: 10.1023/B:SCAM.0000007122.03392.4b. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mauro A. Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol. 1961;9(2):493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcgrath MJ, Cottle DL, Nguyen MA, Dyson JM, Coghill ID, Robinson PA, Holdsworth M, Cowling BS, Hardeman EC, Mitchell CA. Four and a half LIM protein 1 binds myosin-binding protein C and regulates myosin filament formation and sarcomere assembly. J Biol Chem. 2006;281(11):7666–7683. doi: 10.1074/jbc.M512552200. [DOI] [PubMed] [Google Scholar]
- McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387(6628):83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- Megeney LA, Kablar B, Garrett K, Anderson JE, Rudnicki MA. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10(10):1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- Morgan MJ, Madgwick AJ. The LIM proteins FHL1 and FHL3 are expressed differently in skeletal muscle. Biochem Biophys Res Commun. 1999;255(2):245–250. doi: 10.1006/bbrc.1999.0179. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75(7):1351–1359. doi: 10.1016/0092-8674(93)90621-V. [DOI] [PubMed] [Google Scholar]
- Samson T, Smyth N, Janetzky S, Wendler O, Müller JM, Schüle R, von der Mark H, von der Mark K, Wixler V. The LIM-only proteins FHL2 and FHL3 interact with alpha- and beta-subunits of the Muscle alpha7beta1 integrin receptor. J Biol Chem. 2004;279(27):28641–28652. doi: 10.1074/jbc.M312894200. [DOI] [PubMed] [Google Scholar]
- Sassoon D, Lyons G, Wright WE, Lin V, Lassar A, Weintraub H, Buckingham M. Expression of two myogenic regulatory factors myogenin and MyoD1 during mouse embryogenesis. Nature. 1989;341(6240):303–307. doi: 10.1038/341303a0. [DOI] [PubMed] [Google Scholar]
- Schuster-Gossler K, Cordes R, Gossler A. Premature myogenic differentiation and depletion of progenitor cells cause severe muscle hypotrophy in Delta1 mutants. Proc Natl Acad Sci USA. 2007;104(2):537–542. doi: 10.1073/pnas.0608281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Matsumoto C, Ra C. FHL3 negatively regulates human high-affinity IgE receptor beta-chain gene expression by acting as a transcriptional co-repressor of MZF-1. Biochem J. 2005;386:191–200. doi: 10.1042/BJ20040775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velleman SG, Mcfarland DC. Influence of fibroblast growth factor, insulin-like growth factor I, and transforming growth factor beta on satellite cell type I collagen expression and localization during differentiation. Cytobios. 1999;100(394):101–110. [PubMed] [Google Scholar]
- Vinciguerra M, Musaro A, Rosenthal N. Regulation of muscle atrophy in aging and disease. Adv Exp Med Biol. 2010;694:211–233. doi: 10.1007/978-1-4419-7002-2_15. [DOI] [PubMed] [Google Scholar]
- Wu Z, Jin H, Gu Y. The effect of MyoD family proteins on muscular atrophy induced by brachial plexus injury in rats. Chin Med J. 2002;82(8):561–563. [PubMed] [Google Scholar]