Abstract
Background
It is important to explore renewable alternatives (e.g. biofuels) that can produce energy sources to help reduce reliance on fossil oils, and reduce greenhouse gases and waste solids resulted from fossil oils consumption. Camelina sativa is an oilseed crop which has received increasing attention due to its short life cycle, broader adaptation regions, high oil content, high level of omega-3 unsaturated fatty acids, and low-input requirements in agriculture practices. To expand its Camelina production areas into arid regions, there is a need to breed for new drought-tolerant cultivars. Leaf cuticular wax is known to facilitate plant development and growth under water-limited conditions. Dissecting the genetic loci underlying leaf cuticular waxes is important to breed for cultivars with improved drought tolerance.
Results
Here we combined phenotypic data and single nucleotide polymorphism (SNP) data from a spring C. sativa diversity panel using genotyping-by-sequencing (GBS) technology, to perform a large-scale genome-wide association study (GWAS) on leaf wax compositions. A total of 42 SNP markers were significantly associated with 15 leaf wax traits including major wax components such as total primary alcohols, total alkanes, and total wax esters as well as their constituents. The vast majority of significant SNPs were associated with long-chain carbon monomers (carbon chain length longer than C28), indicating the important effects of long-chain carbon monomers on leaf total wax biosynthesis. These SNP markers are located on genes directly or indirectly related to wax biosynthesis such as maintaining endoplasmic reticulum (ER) morphology and enabling normal wax secretion from ER to plasma membrane or Golgi network-mediated transport.
Conclusions
These loci could potentially serve as candidates for the genetic control involved in intracellular wax transport that might directly or indirectly facilitate leaf wax accumulation in C. sativa and can be used in future marker-assisted selection (MAS) to breed for the cultivars with high wax content to improve drought tolerance.
Keywords: Leaf cuticular wax, Intracellular wax transport, Genome-wide association studies (GWAS), Camelina sativa
Background
Camelina sativa L. Crantz (2n = 40, genome size ~ 782 Mb), belonging to Brassicaceae (Cruciferae) family, is an economically crop originated from southeastern Europe and southwestern Asia [1, 2]. After being cultivated in Europe and North America for centuries until 1950s, C. sativa was replaced by another higher-yielding oilseed crop, rapeseed. Recently, C. sativa revived public interest due to its exceptional level of omega-3 essential fatty acids, favorable agronomic characteristics, and potential to be a biofuel resource [3]. The oil content in C. sativa (36–47%) is twice as that of soybean (18–22%) [4] and its unsaturated fatty acids levels account for over 90% of total oil content, among which omega-3 α-linolenic essential fatty acid can reach up to 40% of total oil content [3]. These oil quality characteristics and advantageous agronomic traits attributes including early maturity [2], low-input requirements for water, nutrients and pesticides [2, 5], broader adaptability to diverse environments [1] and resistance against insects and pathogens [6], make C. sativa an ideal alternative resource for biofuel and animal feedstock for the development of sustainable agriculture. Despite these potentials, only limited breeding efforts have been carried out on C. sativa and very few registered varieties and advanced breeding lines are available so far due to low genetic diversity discovered within small set of released germplasms [7], which mainly bred for higher yield and/or higher oil composition without interest in abiotic stress resistance. Therefore, in order to breed for C. sativa, improvements need to be made in biotic and abiotic tolerance as well as in seed yield, oil content, and fatty acids composition.
Plant cuticle forms the first line in plant defense to protect plants from UV irradiation and water loss [8]. Dehydration avoidance is o of the mechanisms that plant species evolved to reduce plant productivity damage under drought stress, which includes depositing leaf cuticular wax to avoid non-stomatal water loss and improve leaf water retention capacity (LWRC) a common assessment for characterizing drought tolerance in plants [9, 10]. The cuticular wax in plant species consists of several major wax components such as alkanes (ALK), aldehydes (ALD), ketones, primary and secondary alcohols, and wax esters (WE), which are derivatives of very long-chain fatty acids (VLCFAs) [9, 11]. The cuticular wax biosynthesis pathway includes two steps: 1) the fatty acid elongase-mediated extension of the C16 and C18 fatty acids to VLCFA chains (C20-C34); 2) the conversion of VLCFAs to the major wax components by the decarbonylation (or the alkane) pathway and the acyl reduction (or primary alcohol) pathway in the endoplasmic reticulum (ER). The alkane pathway mediates the production of ALD, secondary alcohols, ALK, and ketones, while the primary alcohol pathway produces primary alcohols (ALC) and WE. Previous studies showed positive correlations between leaf cuticular wax loads and leaf water retention capacity (LWRC) in plant species such as Arabidopsis thaliana [12], maize [8], rice [13], banana [9], creeping bentgrass [14], and mulberry trees [15]. Many genes involved in cuticular wax biosynthesis have been characterized and cloned in Arabidopsis [16], maize [8], and rice [13] to reduce water loss under water deficit conditions. In C. sativa, the overexpression of an Arabidopsis gene (MYB96) conferred drought resistance via cuticular wax accumulation [17]. However, few studies have been conducted to provide a comprehensive characterization of potential genes related to the cuticular wax biosynthesis.
The quantity and compositions of epicuticular wax components vary significantly among different species, genotypes and organs [18]. In C. sativa, large content variations in cuticular wax components were found in different genotypes, with ALC, ALK, and WE accounting for 86% of the total wax content, followed by free fatty acids (FA), aldehydes (ALD), alkylguaiacols (AG), and methylalkylresorcinols (MAR), which together accounting for less than 5% of total wax content [19, 20]. Each major wax component has their own predominant constituents. For example, C24 (ALC24), C26 (ALC26) and C28 (ALC28) together accounted for 84% of the total ALC, C29 (ALK29), C32 (ALK32), and C35 (ALK35) were the predominant ALK constituents, accounting for 90% of the total ALK, while C40 (WE40), C42 (WE42), C44 (WE44), and C46 (WE46) homologs accounted for the majority in WE [20]. In addition, a moderate to high heritability was also found in major wax components and their constituents [20], demonstrating the possible effectiveness to improve genetic gain of these traits under certain selection pressure.
With the rapid development of next-generation sequencing (NGS) technologies, marker-assisted selection (MAS) can be used to accelerate genetic improvements in breeding programs [21]. QTL mapping identifies putative molecular markers underlying alleles/genes that are controlling quantitative traits, and can be used in MAS. Genome-wide association studies (GWAS) is one of the powerful tools to overcome limitations in traditional QTL mapping and could dissect the genetic architecture of complex traits in crop species [22]. To date, GWAS was used to identify candidate loci associated with various traits in plant species such as A. thaliana [23], soybean [24], maize [25], rice [26] and Brassica napus [27].
In the current study, we aim to apply GWAS analyses using the algorithm of the Settlement of MLM Under Progressively Exclusive Relationship (SUPER) in a C. sativa diversity panel, which consists of accessions originally from different geographical regions, to detect positive alleles (genes) potentially related to major leaf wax components and their constituents. This study could lay a foundation in future molecular breeding efforts to improve drought tolerance in C. sativa via improving leaf wax accumulation and reducing water loss under water limitation conditions.
Results
Phenotyping in wax biosynthesis related traits
Tomasi et al. [20] completed phenotyping of 50 traits related to cuticular wax components and their constituents in a spring C. sativa diversity panel. The total leaf wax content (wax_total) was mainly composed of 8 major wax components: free fatty acids (FA), alkanes (ALK), aldehydes (ALD), alkylguaiacols (AG), methylalkylresorcinols (MAR), wax esters (WE), primary alcohols (ALC), β-sitosterol. These major wax components were composed of 41 predominant constituents (i.e. monomers with different numbers of carbon such as ALC22, ALC24, ALC26, etc.) [20]. As a result, significant positive correlations were found between leaf total wax content (wax-total) and the amount of major wax components with p-value smaller than 0.0001 [20]. High heritability was also found in ALC and its predominant constituents (i.e. ALC24, ALC26 and ALC28) as well as for ALK and its predominant constituents (i.e. ALK29, ALK31, and ALK33) [20].
Population structure and linkage disequilibrium (LD)
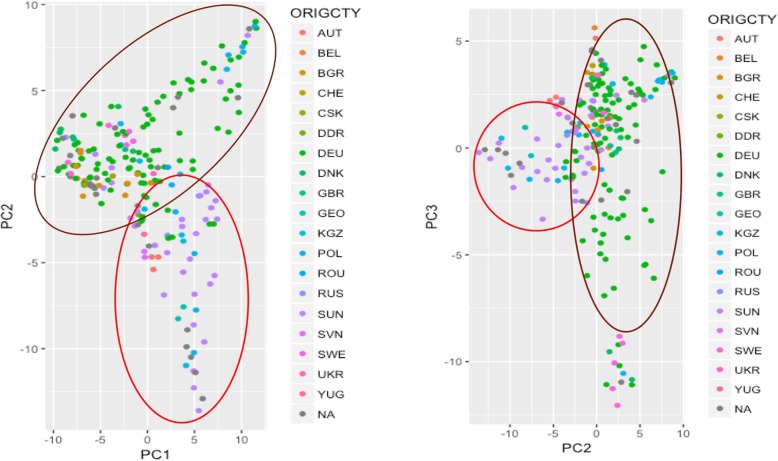
The 213 C. sativa accessions were sequenced and genotyped using GBS. After sequencing, data processing and SNP filtering, a total of 6192 high-quality SNPs were physically mapped across 20 chromosomes with an average marker density of 101.77 kb per chromosome. Detailed information regarding raw reads, filtered reads, filtered SNPs were provided by Luo et al. [28]. Population structure analysis [28] showed a sharp peak at K = 2 when the number of clusters (K) was plotted against ΔK, meaning that the optimal number of clusters was 2 and the population could be clustered into two subpopulations. In accordance with the population structure analysis, principle component analysis (PCA) results also showed two clearly divergent groups (Fig. 1). Basic statistics results for the two subpopulations regarding major wax components were provided in Table 1 that showed the variation in leaf wax traits between the two geographically separated subpopulations. Linkage disequilibrium (LD) decay varied across chromosomes ranging from 1736 to 3885 kb at r2 < 0.2 (Table 2). The average pairwise r2 and LD decay at r2 < 0.2 for the entire genome was approximately 0.07 and 2591 kb, respectively. The distribution of r2 with respect to the physical distance for each chromosome is presented in Table 2. The slowest LD decay was observed for chromosome 10 (3885 kb), followed by chromosome 18 (3599 kb) and chromosome 12 (3364 kb).
Fig. 1.
Principle component analysis (PCA) demonstrates two subgroups based on different geographical origins of the spring panel of Camelina sativa
Table 1.
Summary table of basic statistics among two subpopulations and significance of differences in eight major wax components and leaf total wax content
| Pop1 | Pop2 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Min | Max | Median | No. | Mean | SD | Min | Max | Median | No. | Mean | SD | |
| FA_total | 0.96 | 4.65 | 2.26 | 55 | 2.39 | 0.84 | 0.91 | 3.20 | 2.06 | 70 | 2.09 | 0.63 |
| ALK_total | 23.10 | 119.00 | 44.40 | 55 | 46.90 | 15.10 | 25.50 | 70.50 | 42.60 | 70 | 43.70 | 9.23 |
| ALC_total | 52.50 | 119.00 | 75.10 | 55 | 75.10 | 10.80 | 51.50 | 93.20 | 67.30 | 70 | 69.50 | 8.85 |
| ALD_total | 0.13 | 1.44 | 0.59 | 55 | 0.64 | 0.30 | 0.13 | 1.17 | 0.54 | 70 | 0.56 | 0.25 |
| AG_total | 1.96 | 7.77 | 3.22 | 55 | 3.56 | 1.21 | 1.55 | 7.13 | 3.81 | 70 | 3.93 | 1.20 |
| MAR_total | 1.20 | 4.30 | 2.27 | 55 | 2.32 | 0.62 | 0.98 | 4.67 | 2.78 | 70 | 2.77 | 0.75 |
| WE_total | 33.40 | 108.00 | 53.30 | 55 | 56.30 | 15.50 | 32.90 | 80.40 | 53.20 | 70 | 54.00 | 10.40 |
| Sitosterol | 0.06 | 0.88 | 0.19 | 55 | 0.24 | 0.17 | 0.06 | 0.64 | 0.20 | 70 | 0.24 | 0.15 |
| Wax_total | 152.00 | 288.00 | 200.00 | 55 | 205.00 | 29.10 | 158.00 | 250.00 | 194.00 | 70 | 195.00 | 22.60 |
Table 2.
Details of LD Decay distance observed at R2 < 0.2 on different chromosomes and the entire genome in Camelina sativa
| Chromosome | Size (Mb) | Mean r2 | LD decay distance (Kb) at r2 < 0.2 | Count |
|---|---|---|---|---|
| 1 | 22.86 | 0.06 | 2462.534 | 3506 |
| 2 | 27.50 | 0.06 | 3094.724 | 2885 |
| 3 | 28.19 | 0.08 | 2152.146 | 3974 |
| 4 | 29.72 | 0.07 | 2773.029 | 4081 |
| 5 | 34.76 | 0.05 | 2914.815 | 5176 |
| 6 | 26.00 | 0.06 | 2766.698 | 2922 |
| 7 | 33.05 | 0.06 | 2401.861 | 6601 |
| 8 | 27.63 | 0.06 | 2068.315 | 6793 |
| 9 | 37.52 | 0.04 | 2401.333 | 4918 |
| 10 | 25.04 | 0.07 | 3885.220 | 1672 |
| 11 | 49.58 | 0.07 | 3038.294 | 8268 |
| 12 | 32.05 | 0.13 | 3364.167 | 5533 |
| 13 | 23.49 | 0.08 | 1736.596 | 4044 |
| 14 | 31.47 | 0.05 | 2383.700 | 6093 |
| 15 | 29.96 | 0.06 | 2179.102 | 4503 |
| 16 | 28.91 | 0.08 | 2193.280 | 6621 |
| 17 | 34.94 | 0.09 | 2693.883 | 4034 |
| 18 | 20.62 | 0.07 | 3599.897 | 2294 |
| 19 | 25.84 | 0.10 | 1793.181 | 5132 |
| 20 | 29.83 | 0.05 | 1927.620 | 5473 |
| Whole genome | 598.96 | 0.07 | 2510.551 | 102,614 |
| Average | 29.95 | 0.07 | 2591.520 | 4726 |
Genome-wide association analysis (GWAS) in Camelina sativa
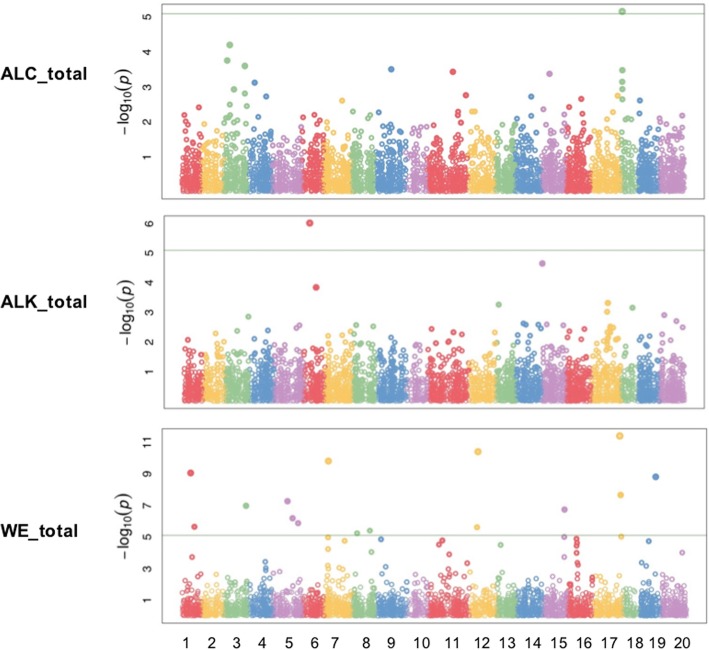
A core set of 125 C. sativa accessions were selected for GWAS analyses. In order to reduce false positive rates and improve computational power, GAPIT MLM_SUPER was used to perform analysis. Bonferroni correction was used to retain significant SNPs with the p-value smaller than 8.0e-6. A total of 42 SNP markers were significantly associated with 15 out of 50 phenotypic traits. Among eight major wax components, three of them (WE_total, ALK_total, and ALC_total) generated significant SNP hits. Sixteen SNPs, one SNP and one SNP were significantly associated with WE_total, ALK_total and ALC_total, respectively (Fig. 2, Table 3). However, no SNP was significantly associated with FA and its constituents, β-sitosterol as well as total wax content.
Fig. 2.
Manhattan plots of GWAS results showing significant SNPs associated with total wax esters (WE_total), total alkanes (ALK_total) and total primary alcohols (ALC_total) in spring Camelina sativa diversity panel. X-axis shows the distribution of SNPs across 20 chromosomes while y-axis shows Bonferroni corrections threshold. The SNPs in triangle, rectangular and circle shared significance among different trait
Table 3.
Summary genome-wide association study (GWAS) of loci significantly associated with different leaf major wax components and their constituents in a spring Camelina sativa diversity panel (Please see the independent excel table)
| SNP | Chr. | Position | Alleles | P.value | Bonferroni Corrected | MAF | FDR Adjusted P-values | Nearest Gene | Gene Annotation | Protein Function | Associated Traits |
|---|---|---|---|---|---|---|---|---|---|---|---|
| S1_10,108,457 | 1 | 10,108,457 | A/G | 5.64E-10 | 3.49E-06 | 0.5281690141 | 0.00000 | LOC104786434 | clavaminate synthase-like protein At3g21360 | metal ion binding; oxidoreductase activity | WE48 |
| 3.17E-10 | 1.96E-06 | 0.00000 | WE46 | ||||||||
| 1.12E-06 | 6.93E-03 | 0.00346 | WE44 | ||||||||
| 2.74E-11 | 1.69E-07 | 0.00000 | WE42 | ||||||||
| 8.99E-10 | 5.57E-06 | 0.00000 | WE_total | ||||||||
| S1_14,592,616 | 1 | 14,592,616 | G/A | 2.24E-06 | 1.39E-02 | 0.1150234742 | 0.00106 | LOC104707605 | uncharacterized | NA | WE_total |
| 6.92E-07 | 4.28E-03 | 0.00043 | AG19 | ||||||||
| S3_25,592,310 | 3 | 25,592,310 | T/C | 1.07E-07 | 6.61E-04 | 0.0915492958 | 0.00008 | LOC104778100 | uncharacterized | NA | WE_total |
| 3.89E-09 | 2.41E-05 | 0.00001 | AG19 | ||||||||
| S5_17,084,259 | 5 | 17,084,259 | A/C | 3.46E-06 | 2.14E-02 | 0.9319248826 | 0.00513 | LOC104789323 | uncharacterized | NA | WE44 |
| 3.49E-09 | 2.16E-05 | 0.00001 | WE42 | ||||||||
| 5.48E-08 | 3.40E-04 | 0.00005 | WE_total | ||||||||
| 3.75E-07 | 2.32E-03 | 0.00029 | AG19 | ||||||||
| S5_23,163,110 | 5 | 23,163,110 | A/G | 1.08E-06 | 6.70E-03 | 0.9084507042 | 0.00112 | LOC104789501 | 40S ribosomal protein S11–1-like | RNA binding | WE42 |
| 6.48E-07 | 4.01E-03 | 0.00036 | WE_total | ||||||||
| S5_23,163,133 | 5 | 23,163,133 | A/C | 1.08E-06 | 6.70E-03 | 0.9084507042 | 0.00112 | LOC104789501 | 40S ribosomal protein S11–1-like | RNA binding | WE42 |
| 6.48E-07 | 4.01E-03 | 0.00036 | WE_total | ||||||||
| S5_29,786,454 | 5 | 29,786,454 | T/C | 3.25E-09 | 2.01E-05 | 0.0704225352 | 0.00000 | LOC104789700 | uncharacterized | NA | WE48 |
| 2.31E-08 | 1.43E-04 | 0.00003 | WE46 | ||||||||
| 1.32E-06 | 8.19E-03 | 0.00068 | WE_total | ||||||||
| S6_7,999,118 | 6 | 7,999,118 | A/G | 3.82E-08 | 2.37E-04 | 0.0046948357 | 0.00024 | LOC104780231 | brefeldin A-inhibited guanine nucleotide-exchange protein 5 | vesicle trafficking, PAMP-triggered immunity (PTI), effector-triggered immunity (ETI), and salicylic acid (SA)-regulated immunity | ALK33 |
| 9.75E-07 | 6.04E-03 | 0.00604 | ALK_total | ||||||||
| S7_24,153,024 | 7 | 24,153,024 | C/T | 1.20E-09 | 7.43E-06 | 0.929577465 | 0.00000 | LOC104704901 | probable purine permease 14 | NA | AG19 |
| S7_4,441,246 | 7 | 4,441,246 | T/C | 4.65E-06 | 2.88E-02 | 0.1384976526 | 0.00221 | LOC109125528 | uncharacterized | NA | WE48 |
| S7_4,787,171 | 7 | 4,787,171 | A/G | 7.15E-11 | 4.43E-07 | 0.9201877934 | 0.00000 | LOC104700327 | B-box zinc finger protein 25-like | negative regulator of seedling photomorphogenesis | WE48 |
| 2.58E-09 | 1.60E-05 | 0.00000 | WE46 | ||||||||
| 1.60E-10 | 9.91E-07 | 0.00000 | WE_total | ||||||||
| S7_9,579,071 | 7 | 9,579,071 | G/T | 4.26E-07 | 2.64E-03 | 0.117370892 | 0.00029 | LOC104700816 | uncharacterized | NA | AG19 |
| S8_12,332,278 | 8 | 12,332,278 | C/T | 5.84E-07 | 3.62E-03 | 0.765258216 | 0.00090 | LOC109126081 | RNA-directed DNA polymerase homolog | RNA-directed 5′-3′ RNA polymerase activity; RNA-directed DNA polymerase activity | Mar25 |
| S8_21,155,371 | 8 | 21,155,371 | C/T | 4.36E-09 | 2.70E-05 | 0.8004694836 | 0.00000 | LOC104709776 | glutathione S-transferase T3-like | conjunction of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles and detoxification against certain herbicides | WE48 |
| 5.08E-08 | 3.15E-04 | 0.00004 | WE46 | ||||||||
| 9.12E-07 | 5.64E-03 | 0.00112 | WE42 | ||||||||
| 3.98E-06 | 2.47E-02 | 0.00164 | WE_total | ||||||||
| S8_23,054,597 | 8 | 23,054,597 | T/C | 2.07E-06 | 1.28E-02 | 0.061032864 | 0.00107 | LOC104709847 | cytochrome P450 81D1-like (pseudo) | metal binding | AG19 |
| S8_5,912,798 | 8 | 5,912,798 | A/C | 6.67E-06 | 4.13E-02 | 0.9084507042 | 0.00283 | LOC104706385 | uro-adherence factor A-like | cell wall anchored protein--erythrocyte binding activity | WE48 |
| 5.83E-06 | 3.61E-02 | 0.00226 | WE_total | ||||||||
| S9_7,943,388 | 9 | 7,943,388 | A/G | 3.68E-06 | 2.28E-02 | 0.9014084507 | 0.00190 | LOC104710709 | protein arginine methyltransferase NDUFAF7 homolog, mitochondrial | assembly or stability of mitochondrial NADH:ubiquinone oxidoreductase complex | WE48 |
| S11_13,254,172 | 11 | 13,254,172 | T/G | 6.98E-06 | 4.32E-02 | 0.0633802817 | 0.00393 | LOC109124846 | vacuolar protein sorting-associated protein 25-like | delivery of transmembrane proteins into the lumen of the lysosome for degradation | WE46 |
| S11_47,072,592 | 11 | 47,072,592 | A/G | 1.17E-06 | 7.27E-03 | 0.9014084507 | 0.00073 | LOC104726883 | peroxisomal membrane protein PEX14 | peroxisome movement | WE48 |
| 3.33E-06 | 2.06E-02 | 0.01032 | ALC32 | ||||||||
| 3.68E-08 | 2.28E-04 | 0.00011 | ALC34 | ||||||||
| 2.61E-07 | 1.62E-03 | 0.00081 | ALD30 | ||||||||
| S11_47,072,593 | 11 | 47,072,593 | A/T | 1.17E-06 | 7.27E-03 | 0.00073 | WE48 | ||||
| S11_49,518,817 | 11 | 49,518,817 | C/T | 4.14E-06 | 2.57E-02 | 0.6267605634 | 0.00513 | LOC104727408 | GATA transcription factor 5-like | regulation of some light-responsive genes | WE44 |
| S12_10,333,667 | 12 | 10,333,667 | C/G | 3.76E-10 | 2.33E-06 | 0.9460093897 | 0.00000 | LOC104731181 | glutamate--cysteine ligase, chloroplastic-like | resistance to fungal and bacterial pathogens-regulation of salicylic acid (SA) and phytoalexin (camalexin) production. | WE48 |
| 1.97E-10 | 1.22E-06 | 0.00000 | WE46 | ||||||||
| 1.68E-06 | 1.04E-02 | 0.00347 | WE44 | ||||||||
| 4.13E-11 | 2.56E-07 | 0.00000 | WE_total | ||||||||
| 1.26E-06 | 7.80E-03 | 0.00071 | AG19 | ||||||||
| S12_20,455,684 | 12 | 20,455,684 | G/A | 2.72E-07 | 1.68E-03 | 0.1408450704 | 0.00056 | LOC104732333 | uncharacterized | NA | Mar25 |
| S12_9,157,483 | 12 | 9,157,483 | G/T | 2.39E-06 | 1.48E-02 | 0.8920187793 | 0.00106 | LOC104731044 | kinesin-like protein KIN-7I | somatic cell cytokinesis, cell-plate formation and its expansion | WE_total |
| S13_1,532,622 | 13 | 1,532,622 | G/A | 6.46E-06 | 4.00E-02 | 0.7699530516 | 0.00400 | LOC104734437 | probable lysine-specific demethylase ELF6 | transcriptional gene regulation; repressor of the photoperiodic flowering pathway and of FT. | Mar25 |
| S13_21,456,713 | 13 | 21,456,713 | A/G | 1.70E-06 | 1.05E-02 | 0.7769953052 | 0.00211 | LOC104738457 | uncharacterized | NA | Mar25 |
| S13_5,037,723 | 13 | 5,037,723 | C/A | 6.43E-07 | 3.98E-03 | 0.0751173709 | 0.00112 | LOC104735215 | U-box domain-containing protein 52-like | protein ubiquitination | WE42 |
| S15_11,699,414 | 15 | 11,699,414 | C/T | 2.51E-09 | 1.55E-05 | 0.896713615 | 0.00002 | LOC104746477 | uncharacterized | NA | Mar25 |
| S15_25,879,428 | 15 | 25,879,428 | C/T | 2.44E-06 | 1.51E-02 | 0.9107981221 | 0.00137 | LOC104748799 | uncharacterized | NA | WE48 |
| 5.39E-06 | 3.34E-02 | 0.00334 | WE46 | ||||||||
| 1.85E-07 | 1.14E-03 | 0.00013 | WE_total | ||||||||
| S16_10,454,057 | 16 | 10,454,057 | T/C | 2.37E-06 | 1.47E-02 | 0.1103286385 | 0.00210 | LOC104750305 | ERAD-associated E3 ubiquitin-protein ligase HRD1B-like | protein ubiquitination | WE42 |
| S16_11,083,391 | 16 | 11,083,391 | C/T | 8.64E-10 | 5.35E-06 | 0.931924883 | 0.00000 | LOC104750339 | leucine-rich repeat receptor-like protein CLAVATA2 | membrane component | AG19 |
| S16_2,915,369 | 16 | 2,915,369 | T/C | 6.87E-07 | 4.25E-03 | 0.1009389671 | 0.00425 | LOC104749495 | DEAD-box ATP-dependent RNA helicase 21-like | ribosome biogenesis, mRNA degradation and translation initiation. | ALC26 |
| S16_9,912,650 | 16 | 9,912,650 | T/C | 1.31E-07 | 8.11E-04 | 0.112676056 | 0.00012 | LOC104753537 | uncharacterized | NA | AG19 |
| S16_9,912,651 | 16 | 9,912,651 | G/A | 7.71E-08 | 4.77E-04 | 0.08685446 | 0.00010 | LOC104753537 | uncharacterized | NA | AG19 |
| S16_9,912,652 | 16 | 9,912,652 | G/A | 1.31E-07 | 8.11E-04 | 0.112676056 | 0.00012 | LOC104753537 | uncharacterized | NA | AG19 |
| S17_32,490,691 | 17 | 32,490,691 | T/A | 2.56E-11 | 1.59E-07 | 0.0704225352 | 0.00000 | LOC104758647 | protein SPA1-RELATED 4-like | Repressor of photomorphogenesis in the blue light | WE48 |
| 7.71E-11 | 4.77E-07 | 0.00000 | WE46 | ||||||||
| 1.02E-06 | 6.32E-03 | 0.00346 | WE44 | ||||||||
| 4.17E-12 | 2.58E-08 | 0.00000 | WE_total | ||||||||
| 3.65E-07 | 2.26E-03 | 0.00226 | ALK35 | ||||||||
| 1.46E-09 | 9.04E-06 | 0.00000 | AG19 | ||||||||
| S17_33,558,126 | 17 | 33,558,126 | A/T | 7.83E-10 | 4.85E-06 | 0.8849765258 | 0.00000 | LOC104758812 | probable acyl-activating enzyme 18, peroxisomal | peroxisomal activation of 2,4-dichlorophenoxybutyric acid (2,4-dB) | WE48 |
| 2.97E-08 | 1.84E-04 | 0.00003 | WE46 | ||||||||
| 2.23E-08 | 1.38E-04 | 0.00002 | WE_total | ||||||||
| S17_3,778,452 | 17 | 3,778,452 | A/G | 2.77E-06 | 1.71E-02 | 0.882629108 | 0.00214 | LOC104755111 | U11NAU12 small nuclear ribonucleoprotein 65 kDa protein | branch-point site recognition for normal plant development | WE42 |
| 5.08E-06 | 3.15E-02 | 0.00242 | AG19 | ||||||||
| S18_2,191,451 | 18 | 2,191,451 | C/A | 6.97E-06 | 4.32E-02 | 0.3755868545 | 0.04316 | LOC104760305 | WUSCHEL-related homeobox 8-like | basal embryo development after fertilization | ALC_total |
| S19_10,881,936 | 19 | 10,881,936 | G/C | 6.85E-06 | 4.24E-02 | 0.103286385 | 0.00283 | LOC104765950 | F-box protein At5g07610-like | NA | WE48 |
| 2.56E-06 | 1.59E-02 | 0.00176 | WE46 | ||||||||
| 3.63E-06 | 2.24E-02 | 0.00249 | WE42 | ||||||||
| S19_19,179,520 | 19 | 19,179,520 | G/T | 4.70E-09 | 2.91E-05 | 0.8708920188 | 0.00000 | LOC109131004 | histone deacetylase 6-like | protein deacetylase activity; degradation of misfoleded proteins | WE48 |
| 1.61E-07 | 9.99E-04 | 0.00012 | WE46 | ||||||||
| 1.57E-09 | 9.73E-06 | 0.00000 | WE_total | ||||||||
| S19_4,005,938 | 19 | 4,005,938 | C/G | 2.19E-06 | 1.36E-02 | 0.8544600939 | 0.00735 | LOC104764661 | probable DNA helicase MCM8 (pseudo) | DNA repair during meiosis | AG23 |
| 2.21E-06 | 1.37E-02 | 0.00228 | MAR25 | ||||||||
| S20_6,023,906 | 20 | 6,023,906 | G/A | 1.59E-08 | 9.85E-05 | 0.1408450704 | 0.00005 | LOC104769581 | magnesium-chelatase subunit ChlH, chloroplastic | chlorophyll synthesis,plastid-to-nucleus retrograde signaling and abscisic acid (ABA) perception | MAR25 |
GWAS for primary alcohols (ALC), aldehydes (ALD) and their predominant constituents
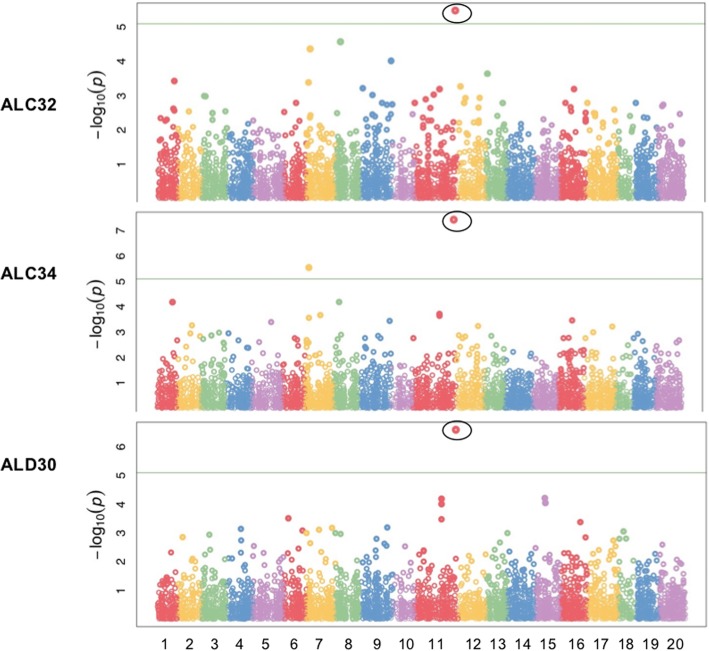
A high heritability (0.77) was found for ALC_total and a moderate to high heritability ranging from 0.38 to 0.86 were found for the predominant ALC constituents as described by Tomasi et al. [20]. ALC_total, ALC26, ALC32 and ALC34 shared the same significant SNP marker on chromosome 11, which was located on a gene functioning peroxisomal import machinery in peroxisome movement, which could contribute to cuticular wax accumulation by maintaining ER network [29] (Fig. 3, Table 3). This SNP was also significantly associated with ALD30 (heritability of 0.53), which was the only constituent under ALD that carrying a significant SNP (Fig. 3, Table 3).
Fig. 3.
Manhattan plots of GWAS results showing significant SNPs associated with primary alcohols constituents (ALC32 &ALC34) and aldehydes constituents (ALD30) in spring Camelina sativa diversity panel. X-axis shows the distribution of SNPs across 20 chromosomes while y-axis shows Bonferroni corrections threshold. The SNPs in triangle, rectangular and circle shared significance among different traits
GWAS for alkanes (ALK) and its predominant constituents
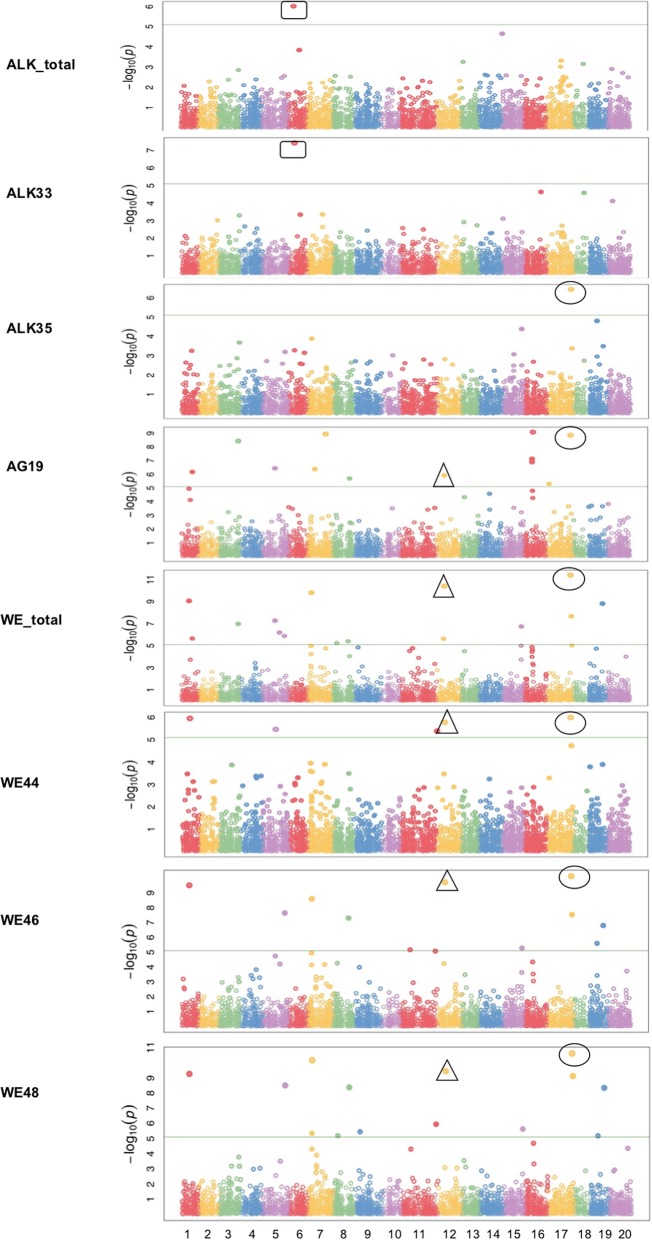
A high heritability (0.77) was observed for total ALK, and a wide range of heritability was estimated for its predominant constituents [20]. ALK_total and ALK33 shared the same significant SNP on chromosome 6 (Fig. 4, Table 3). This SNP was located on ARF-GEF gene, which encodes brefeldin A-inhibited guanine nucleotide-exchange factors (GEF) and activates the auxin response factors (ARF) by exchanging bound GDP for free GTP. This gene plays an important role in vesicle formation and trafficking, which are required for wax secretion from epidermal cells [30–32]. A six-SNP region spanning around the position at 15.68 Mb on chromosome 6 was also found to be associated with ALK_total even if it did not reach the significant threshold level (Fig. 4). Another SNP underlying the gene encoding constitutive photomorphogenic 1 (COP1/SPA) E3 ubiquitin-protein ligase, was significantly associated with ALK35. This SNP was reported to be mainly related to abiotic stress tolerance [33], and its function in photomorphogenesis repressing was also reported [34] (Fig. 4, Table 3).
Fig. 4.
Manhattan plots of GWAS results showing significant SNPs associated with wax esters constituents (WE44, WE46 &WE48), alkanes constituents (ALK33 &ALK35) and alkylguaiacols constituent (AG19) in spring Camelina sativa diversity panel. X-axis shows the distribution of SNPs across 20 chromosomes while y-axis shows Bonferroni corrections threshold. The SNPs in triangle, rectangular and circle shared significance among different traits
GWAS for wax esters (WE) and its predominant constituents
Tomasi et al. [20] estimated a high heritability (0.77) for WE_total and a moderate to high heritability ranging from 0.65 to 0.86 were estimate for six constituents [20]. Sixteen SNPs were identified to be significantly associated with WE_total and some of these SNPs were also associated with WE42, WE44, WE46 and WE48 (Fig. 4, Table 3). These SNPs included three (S17_32,490,691, S11_49,518,817, and S7_4,787,171) that are related to photomorphogenesis repressor, one SNP (S12_10,333,667) that is related to glutamate-cysteine ligase [35], and one SNP (S17_33,558,126) that is related to peroxisomal activation of 2,4-dichlorophenoxybutyric acid (2,4-D), which was reported to increase and maintain water content under citrus fruit epicuticular wax layer during the post-harvest storage period [36]. Some of these SNPs also showed significant associations with other wax sub-constituents, e.g. SNP S17_32,490,691 was found to be significantly associated with both ALK35 and AG19, and SNP S12_10,333,667 was found to be significantly associated with AG19 (Table 3).
GWAS for alkylguaiacols (AG) and its predominant constituents
A high heritability ranging from 0.77 to 0.87 was found for all the three predominant constituents under AG class [20]. Thirteen SNP and one SNP were significantly associated with AG19 and AG23, respectively. However, most of these SNP neither were located on coding regions (CDS) nor near to characterized genes. Another SNP was found to be significantly associated with AG19, WE_total, and several WE constituents (Fig. 4, Table 3). This SNP is located on glutamate-cysteine ligase, which mainly confers resistance to fungal and bacterial pathogens via the regulation of salicylic acid (SA) and phytoalexin (camalexin) production [37] but also plays an important role in maintaining ER morphology and secretory membrane traffic via glutathione biosynthesis [35]. Other than these, there was a four-SNP region spanning from 9.9 Mb to 11 Mb on chromosome 16 found to be significantly related to AG19 (Fig. 4, Table 3), but no gene functions were annotated in this region.
GWAS for Methylalkylresorcinols (MAR) and its predominant constituents
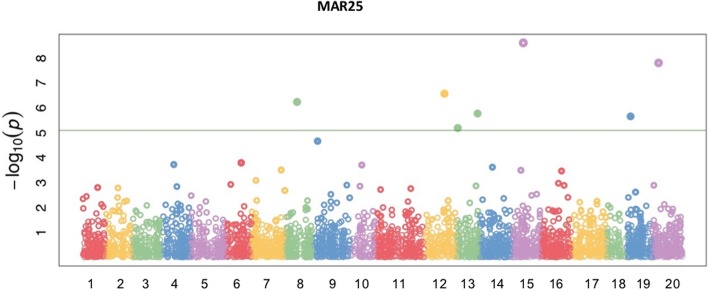
Seven significant SNP markers were controlling the phenotypic variations in MAR25 with moderate heritability (0.56). One of the seven SNPs was located on the gene controlling chlorophyll synthesis (Fig. 5, Table 3). Several SNPs, clustered in region spanning 54 bp on chromosome 13 (Fig. 5, Table 3), were positioned on gene encoding lysine-specific demethylase ELF6, which represses the photoperiodic flowering pathway and flowering time [38, 39].
Fig. 5.
Manhattan plots of GWAS results showing the significant SNPs associated with methylalkylresorcinols constituent (MAR25) in spring Camelina sativa diversity panel. X-axis shows the distribution of SNPs across 20 chromosomes while y-axis shows Bonferroni corrections threshold. The SNPs in triangle, rectangular and circle shared significance among different traits
Discussion
Few molecular breeding and genetics studies have been conducted on C. sativa so far. The published genetic/genomic Camelina studies were mostly preliminary with small molecular markers coverage that are not enough to be employed in marker-assisted selection. These studies included two genetic maps constructed using recombinant inbred lines (RIL) populations and amplified fragment length polymorphism (AFLP) and simple sequence repeats (SSR) markers [40], and updated with SNP markers [1]. Using these genetic maps, QTLs associated with seed yield, fatty acid compositions and oil content were identified [40]. Recently, a simple marker-trait association study was conducted using 20 winter-type C. sativa accessions [41]. On the top of these studies, our study is the first GWAS study in C. sativa that based on high-density SNP markers coverage over the 20 chromosomes, and could pave the foundation for marker-assisted breeding in this promising feedstock oilseed crop.
Population differentiation and linkage disequilibrium (LD)
The number of studies on population genetics of C. sativa is limited. Different populations have shown different patterns of population structure and diversity [1, 3, 5, 42], but these studies were based on small population size, small number of molecular markers and/or laborious genotyping technologies. Our results revealed two clearly separated subgroups, which corresponds to the available geographical information in the spring C. sativa diversity panel [28]. In general, the extent of LD is affected by the mating system, the breeding history (e.g. the occurrence of bottlenecks) and the genetic diversity in different germplasms and species studied [43]. LD decay is more rapid in outcrossing species and/or germplasms with higher genetic diversity. R2 value of 1 in LD means perfect linkage or predictability at one locus with another. The mean pairwise r2 value in our study is 0.07, which is greater than Brassica napus with a previously estimated mean r2 value of 0.037 [44], confirming the higher overall level of LD than B. napus. In C. sativa, we observed that LD decay below a critical level (r2 = 0.2) ranges from 1736 kb to 3885 kb on different chromosomes, with an average value of 2591 kb for the whole genome. These values are greater than B. napus where LD decay was between 300 and 1000 kb depending on the germplasm collections [44], or 250 kb in A. thaliana [45]. That is not surprising since C. sativa is an inbreeding species and most accessions collected were originated from limited regions in Europe and Asia. It was reported that LD decay could vary across different germplasms, for example, LD decay was observed less than 1 kb for maize landraces [46], 2 kb for diverse inbred maize lines [47], and can reach up to100 kb for commercial elite maize inbred lines [48]. Therefore, more C. sativa germplasm form different resources are needed to comprehensively compare and estimate LD in Camelina species.
GWAS for major wax components and their constituents
Table 3 shows that the clear majority of traits that associated significantly with SNPs were found to elongate carbon chain longer than C28. This was in agreement with a previous finding that significant correlations were found between LWRC and cuticular wax components with carbon chain length longer than C28 [9]. These mainly include major components and constituents in ALC (r = 0.730) and WE (r = 0.597) [9]. This may explain why our GWAS study of wax biosynthesis variations resulted in the most majority of significant SNPs associated with ALC and WE components and constituents.
Two ways were hypothesized to form intracellular wax transport: direct transfer from ER to the plasma membrane and Golgi-mediated exocytosis [49]. A few SNPs, located on genes functioning in peroxisomal movement were identified to be significantly associated with wax components and their constituents (Fig. 5, Table 3). This was in agreement with previous findings that peroxisomes help maintain ER morphology and enable its normal functioning in cuticular wax biosynthesis in Arabidopsis [29], a closely related species to C. sativa.
A SNP located on the gene encoding ARF-GEF was identified to be significantly associated with ALK-related constituents (Fig. 5, Table 3). This is not surprising because ARF-GEFs were reported to act at Golgi membranes, regulating COP1-coated vesicle formation, a function important for ER-Golgi transport [30, 31]. Previous researchers found that very-long-chain (VLC) alkanes, ketones and alcohols synthesized in ER must be trafficked to the plasma membrane to form cuticular waxes and protect plant cuticles [32]. The wax secretion process requires Golgi network-mediated vesicle trafficking, in which ARF-GEFs play an important role. Another SNP (S12_10,333,667) associated with WE_total and its predominant constituents (Fig. 4, Table 3) was related to glutamate-cysteine ligase (GCL), the first enzyme in the glutathione (GSH) biosynthesis pathway. GSH pathway was reported not only to reduce oxidative damage and maintain an intracellular redox environment in response to plant stress [36] but also maintain ER morphology and secretory membrane traffic [35], which were important for intracellular wax transport in cuticular wax biosynthesis [49].
As for a few significant SNPs positioned on genes potentially related to photomorphogenesis repressing or photoperiodic flowering (e.g. COP1/SPA E3 ubiquitin-protein ligase, ELF6, etc.), it remains unknown that whether the light signaling system regulates plant growth and development that has overlapping effects on wax biosynthesis or not. Clustered around significant SNPs that were associated with leaf wax traits, other SNPs, even if not significant, still worthy the investigation with in-depth studies. These SNPs could be located near the genes of functions, or on functioning regulators to mediate the activities of genes. The SNPs with no annotated functions were also likely to be located on novel genes, even if false positive detections may occur. Further validation studies are required to understand the function of these genes or QTL regions.
Conclusion
This study presents the first GWAS study on C. sativa, a promising oilseed crop for food, feed and fuel uses. As many as 50 phenotypic traits related to leaf wax accumulation in Camelina were used to identify putative SNPs associated with these traits. The significant SNPs were positioned in genes directly or indirectly related to cuticular wax accumulation, which might help improve drought tolerance under water deficit. These identified SNPs could provide hints for future molecular breeding studies as potential breakthroughs for the selection of drought tolerance C. sativa cultivars. However, more relevant functional genomics, genetics, and validation studies are needed to understand the functions of these alleles/genes.
Materials and methods
Plant materials and phenotyping
Phenotypic data of the spring C. sativa panel was produced by our research group in 2017 [20]. Briefly, the plant materials we used for current GWAS study came from a spring C. sativa diversity panel of C. sativa accessions, which consisted of 125 different genotypes and were grown under greenhouse conditions in USDA Arid-Land Agricultural Research Center (ALARC) in Maricopa, AZ in a randomized complete block design (RCBD) with three replications. In brief, the seeds of each accession were planted in pot filled with Sunshine Mix #1/LC1 (Sun Gro Horticulture, Canada), and seedling were regularly watered and fertilized with 20–20-20 fertilizer (Scotts Miracle-Grow, USA). After 35 days of planting, three leaf subsamples were collected from seventh to twelfth leaf of basal rosette. Waxes were extracted following Tomasi el al. [19] protocol with hexane (Sigma-Aldrich, USA), and three internal standards (nonadecanoic acid, tetracosane and tricosano) were added to the hexane-leaves mixture. After 45 s, leaves were removed from the hexane and leaf area were determined using a flatbed scanner and ImageJ software. The hexane extracts including waxes were evaporated to dryness under N2 gas. Samples were re-dissolved in equal amounts of N,O-bis-(trimethylsilyl)-trifluoroacetamide (BSTFA, Sigma-Aldrich, USA) and hexane ad transferred into GC vials. Vials were loaded onto the Agilent 7890A gas chromatograph equipped with a 5975C mass spectrometer Tomasi et al. [19, 20]. Waxes were characterized and quantified by characteristic quadrupole electron impact mass spectra and internal standard and leaf surface areas [19, 20].
Genotyping-by-sequencing (GBS) of Camelina sativa accessions
Briefly as described by Luo et al. [28], DNA extraction was conducted in C. sativa lyophilized leaf tissue (~ 0.13 g) using Qiagen Plant DNeasy 96 kit following the manufacturer’s protocol. DNA concentration and quality were determined using Quantifluor (Promega, Inc.) and a Synergy H1 plate reader. The PstI restriction enzyme was used to construct GBS libraries [50]. Library construction and Illumina sequencing were done by the University of Cornell Genomic Diversity Facility. Raw sequence data was analyzed using an HTCondor /Directed Acyclic Graph (DAG) workflow [51] integrated into the TASSEL v5.0 GBS v2 pipeline [52] following the pipeline steps. The HTCondor job files and DAG workflow are available at https://github.com/danforthcenter/camelina. Raw reads were filtered using a minimum base quality score of 20 (kmerLength = 64, minKmerL = 20, mnQS = 20, mxKmerNum = 100,000,000). The quality-filtered reads were aligned to the C. sativa genome using BWA MEM [53]. SNPs were called from the alignments with the standards as follows: maxTagsCutSite = 64, mnLCov = 0.1, mnMAF = 0.01. Only biallelic SNPs were filtered by vcftools with missing data smaller than 20% [54]. The VCF file was converted to HAPMAP format using TASSEL. The resulting SNPs were further filtered by disregarding the ones with MAF < 0.05 for GWAS study.
Population genetic analyses and linkage disequilibrium (LD)
Population structure was estimated using a Bayesian Markov Chain Monte Carlo (MCMC) model implemented in STRUCTURE v2.3.4 [55]. Five independent runs were performed for each assumed population numbers (k) ranging from 1 to 10. Burn-in time and MCMC replication number were both set to 100,000 for each run. Structure Harvester [56] was used to find the most likely K value, which was determined by the log probability of the data (LnP(D)) and ΔK based on the rate of change in (LnP(D) between successive K values [57]. Principle component analysis (PCA) performed in R with the package ggplot2 [58] was also used in population structure analysis. Linkage disequilibrium (LD) between SNPs on each chromosome and on an overall level were estimated using r2 from TASSEL5.0 [52]. For the clustered subpopulations, basic summary statistics were conducted to compare major wax categories: FA_total, ALC_total, ALK_total, ALD_total, AG_total, MAR_total, WE_total, Sitosterol, and Wax_total.
Genome-wide association studies
Association analyses were performed in R [59] to identify loci controlling wax related traits using the SUPER algorithm [60] implemented in the Genomic Association and Prediction Integrated Tool (GAPIT) package [61]. This method proceeds by extracting a subset of testing SNPs from the total dataset and using pseudo quantitative trait nucleotides (QTNs) that are not in LD with these testing SNPs to define relatedness [60] among the population. This method retains the computational advantage of factored spectrally transformed linear mixed models (FaST-LMM) without impairing the statistical power even when compared to other methods using the entire SNP dataset [60]. To improve the accuracy, we implemented SUPER into mixed linear model (MLM) to perform MLM-SUPER analysis. Manhattan and quantile-quantile (Q-Q) plots were generated using the R package qqman [62]. The Bonferroni correction, negative log (0.05/n), was used as a threshold for significance of associations between SNPs and traits of interests, where n was the total number of SNPs used in the association analysis [57, 63]. Genes within ~ 50 kb upstream and downstream to the associated SNPs were selected for annotation.
In silico mapping of SNPs and candidate gene identification
Physical mapping of significantly associated SNPs and functional annotation of the predicted genes harboring these SNPs were performed using the C. sativa genome browser (https://www.ncbi.nlm.nih.gov/genome/gdv/browser/?context=genome&acc=GCF_000633955.1) submitted by Agriculture & AgriFood Canada. For associated SNPs that mapped to intron regions or gene promoter regions, we included polymorphisms in a 2.0-kb region surrounding the SNP with highest statistical association to the trait. Functional annotation of the genes was performed in the BLAST2GO [64] and UniProt database [65].
Acknowledgements
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U. S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Funding
The authors gratefully thank the financial support from the United States Department of Agriculture and National Institute of Food and Agriculture award (grant no. 2016–67009-25639).
Availability of data and materials
The ddRAD Sequencing data from Camalina sativa leaf was deposited in NCBI Sequence Read Archive (SRA) under accession numbers ranging from SRX4286823-SRX4286866.
Authors’ contributions
HAH designed the conception and experiment; PT conducted the phenotyping analyses, NF performed GBS analysis; ZL collected data and did GWAS analysis; ZL wrote the manuscript; NF, PT, HAH provided suggestions and comments for the manuscript; ZL, PT, NF and HAH revised the manuscript. All authors read and approved the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zinan Luo, Email: Lily.Luo@ARS.USDA.GOV.
Pernell Tomasi, Email: Pernell.Tomasi@ARS.USDA.GOV.
Noah Fahlgren, Email: NFahlgren@danforthcenter.org.
Hussein Abdel-Haleem, Email: Hussein.Abdel-Haleem@ARS.USDA.GOV.
References
- 1.Singh R, Bollina V, Higgins EE, Clarke WE, Eynck C, Sidebottom C, Gugel R, Snowdon R, Parkin IAP. Single-nucleotide polymorphism identification and genotyping in Camelina sativa. Mol Breeding. 2015;35:35. 10.1007/s11032-015-0224-62. [DOI] [PMC free article] [PubMed]
- 2.Kagale S, Koh CS, Nixon J, Bollina V, Clarke WE, Tuteja R, Spillane C, Robinson SJ, Links MG, Clarke C et al. The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat Commun 2014;5:3706. 10.1038/ncomms4706. [DOI] [PMC free article] [PubMed]
- 3.Ghamkhar K, Croser J, Aryamanesh N, Campbell M, Kon'kova N, Francis C. Camelina (Camelina sativa (L.) Crantz) as an alternative oilseed: molecular and ecogeographic analyses. Genome. 2010;53(7):558–567. doi: 10.1139/G10-034. [DOI] [PubMed] [Google Scholar]
- 4.Moser BR. Biodiesel from alternative oilseed feedstocks: camelina and field pennycress. Biofuels. 2012;3(2):193–209. doi: 10.4155/bfs.12.6. [DOI] [Google Scholar]
- 5.Manca A, Pecchia P, Mapelli S, Masella P, Galasso I. Evaluation of genetic diversity in a Camelina sativa (L.) Crantz collection using microsatellite markers and biochemical traits. Genet Resour Crop Evol. 2013;60(4):1223–1236. doi: 10.1007/s10722-012-9913-8. [DOI] [Google Scholar]
- 6.Seguin-Swartz G, Eynck C, Gugel RK, Strelkov SE, Olivier CY, Li JL, Klein-Gebbinck H, Borhan H, Caldwell CD, Falk KC. Diseases of Camelina sativa (false flax) Can J Plant Pathol. 2009;31(4):375–386. doi: 10.1080/07060660909507612. [DOI] [Google Scholar]
- 7.Berti M, Gesch R, Eynck C, Anderson J, Cermak S. Camelina uses, genetics, genomics, production, and management. Ind Crop Prod. 2016;94:690–710. doi: 10.1016/j.indcrop.2016.09.034. [DOI] [Google Scholar]
- 8.Li Li, Li Delin, Liu Sanzhen, Ma Xiaoli, Dietrich Charles R., Hu Heng-Cheng, Zhang Gaisheng, Liu Zhiyong, Zheng Jun, Wang Guoying, Schnable Patrick S. The Maize glossy13 Gene, Cloned via BSR-Seq and Seq-Walking Encodes a Putative ABC Transporter Required for the Normal Accumulation of Epicuticular Waxes. PLoS ONE. 2013;8(12):e82333. doi: 10.1371/journal.pone.0082333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sampangi-Ramaiah MH, Ravishankar KV, Seetharamaiah SK, Roy TK, Hunashikatti LR, Rekha A, Shilpa P. Barrier against water loss: relationship between epicuticular wax composition, gene expression and leaf water retention capacity in banana. Funct Plant Biol. 2016;43(6):492–501. doi: 10.1071/FP15296. [DOI] [PubMed] [Google Scholar]
- 10.Seo PJ, Park CM. Cuticular wax biosynthesis as a way of inducing drought resistance. Plant Signal Behav. 2011;6(7):1043–1045. doi: 10.4161/psb.6.7.15606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Lingyan, Ni Erdong, Yang Jiawei, Zhou Hai, Liang Hong, Li Jing, Jiang Dagang, Wang Zhonghua, Liu Zhenlan, Zhuang Chuxiong. Rice OsGL1-6 Is Involved in Leaf Cuticular Wax Accumulation and Drought Resistance. PLoS ONE. 2013;8(5):e65139. doi: 10.1371/journal.pone.0065139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu L, Guo JS, Zhu J, Zhou C. Enhanced expression of EsWAX1 improves drought tolerance with increased accumulation of cuticular wax and ascorbic acid in transgenic Arabidopsis. Plant Physiol Bioch. 2014;75:24–35. doi: 10.1016/j.plaphy.2013.11.028. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Xiaoyun, Li Linzhi, Xiang Jianhua, Gao Guofu, Xu Faxi, Liu Ailing, Zhang Xianwen, Peng Yan, Chen Xinbo, Wan Xiangyuan. OsGL1-3 is Involved in Cuticular Wax Biosynthesis and Tolerance to Water Deficit in Rice. PLoS ONE. 2015;10(1):e116676. doi: 10.1371/journal.pone.0116676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou M, Li DY, Li ZG, Hu Q, Yang CH, Zhu LH, Luo H. Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping Bentgrass. Plant Physiol. 2013;161(3):1375–1391. doi: 10.1104/pp.112.208702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ni Y, Sun ZY, Huang XZ, Huang CS, Guo YJ. Variations of cuticular wax in mulberry trees and their effects on gas exchange and post-harvest water loss. Acta Physiol Plant. 2015;37:112. 10.1007/s11738-015-1856-1.
- 16.Chen XB, Goodwin SM, Boroff VL, Liu XL, Jenks MA. Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and WAX production. Plant Cell. 2003;15(5):1170–1185. doi: 10.1105/tpc.010926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SB, Kim H, Kim RJ, Suh MC. Overexpression of Arabidopsis MYB96 confers drought resistance in Camelina sativa via cuticular wax accumulation. Plant Cell Rep. 2014;33(9):1535–1546. doi: 10.1007/s00299-014-1636-1. [DOI] [PubMed] [Google Scholar]
- 18.Zhang JY, Broeckling CD, Sumner LW, Wang ZY. Heterologous expression of two Medicago truncatula putative ERF transcription factor genes, WXP1 and WXP2, in Arabidopsis led to increased leaf wax accumulation and improved drought tolerance, but differential response in freezing tolerance. Plant Mol Biol. 2007;64(3):265–278. doi: 10.1007/s11103-007-9150-2. [DOI] [PubMed] [Google Scholar]
- 19.Tomasi P, Wang H, Lohrey GT, Park S, Dyer JM, Jenks MA, Abdel-Haleem H. Characterization of leaf cuticular waxes and cutin monomers of Camelina sativa and closely-related Camelina species. Ind Crop Prod. 2017;98:130–138. doi: 10.1016/j.indcrop.2017.01.030. [DOI] [Google Scholar]
- 20.Tomasi P, Dyer JM, Jenks MA, Abdel-Haleem H. Characterization of leaf cuticular wax classes and constituents in a spring Camelina sativa diversity panel. Ind Crop Prod. 2018;112:247–251. doi: 10.1016/j.indcrop.2017.11.054. [DOI] [Google Scholar]
- 21.Bai B, Wang L, Lee M, Zhang YJ, Rahmadsyah, Alfiko Y, Ye BQ, Wan ZY, Lim CH, Suwanto A et al. Genome-wide identification of markers for selecting higher oil content in oil palm. Bmc Plant Biol. 2017;17:93. 10.1186/s12870-017-1045-z. [DOI] [PMC free article] [PubMed]
- 22.Edwards D, Batley J, Snowdon RJ. Accessing complex crop genomes with next-generation sequencing. Theor Appl Genet. 2013;126(1):1–11. doi: 10.1007/s00122-012-1964-x. [DOI] [PubMed] [Google Scholar]
- 23.Verslues PE, Lasky JR, Juenger TE, Liu TW, Kumar MN. Genome-wide association mapping combined with reverse genetics identifies new effectors of low water potential-induced proline accumulation in Arabidopsis. Plant Physiol. 2014;164(1):144–159. doi: 10.1104/pp.113.224014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonah H, O'Donoughue L, Cober E, Rajcan I, Belzile F. Identification of loci governing eight agronomic traits using a GBS-GWAS approach and validation by QTL mapping in soya bean. Plant Biotechnol J. 2015;13(2):211–221. doi: 10.1111/pbi.12249. [DOI] [PubMed] [Google Scholar]
- 25.Li H, Peng ZY, Yang XH, Wang WD, Fu JJ, Wang JH, Han YJ, Chai YC, Guo TT, Yang N, et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat Genet. 2013;45(1):43–U72. doi: 10.1038/ng.2484. [DOI] [PubMed] [Google Scholar]
- 26.Zheng Xiao-Ming, Gong Tingting, Ou Hong-Ling, Xue Dayuan, Qiao Weihua, Wang Junrui, Liu Sha, Yang Qingwen, Olsen Kenneth M. Genome-wide association study of rice grain width variation. Genome. 2018;61(4):233–240. doi: 10.1139/gen-2017-0106. [DOI] [PubMed] [Google Scholar]
- 27.Gacek K, Bayer PE, Bartkowiak-Broda I, Szala L, Bocianowski J, Edwards D, Batley J. Genome-Wide Association Study of Genetic Control of Seed Fatty Acid Biosynthesis in Brassica napus. Front Plant Sci 2017;7:2062. 10.3389/fpls.2016.02062. [DOI] [PMC free article] [PubMed]
- 28.Luo Z, Brock J, Dyer JM, Kutchan T, Schachtman D, Augustin M, Ge Y, Fahlgren N, Abdel-Haleem H. Genetic diversity and population structure of a Camelina sativa spring panel. Front Plant Sci. 2019;10:184. doi: 10.3389/fpls.2019.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamigaki A, Kondo M, Mano S, Hayashi M, Nishimura M. Suppression of peroxisome biogenesis factor 10 reduces Cuticular wax accumulation by disrupting the ER network in Arabidopsis thaliana. Plant Cell Physiol. 2009;50(12):2034–2046. doi: 10.1093/pcp/pcp152. [DOI] [PubMed] [Google Scholar]
- 30.Richter S, Geldner N, Schrader J, Wolters H, Stierhof YD, Rios G, Koncz C, Robinson DG, Jurgens G. Functional diversification of closely related ARF-GEFs in protein secretion and recycling. Nature. 2007;448(7152):488–U410. doi: 10.1038/nature05967. [DOI] [PubMed] [Google Scholar]
- 31.Teh OK, Moore I. An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature. 2007;448(7152):493–496. doi: 10.1038/nature06023. [DOI] [PubMed] [Google Scholar]
- 32.McFarlane HE, Watanabe Y, Yang WL, Huang Y, Ohlrogge J, Samuels AL. Golgi- and trans-Golgi network-mediated vesicle trafficking is required for wax secretion from epidermal cells. Plant Physiol. 2014;164(3):1250–1260. doi: 10.1104/pp.113.234583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim JY, Jang IC, Seo HS. COP1 Controls Abiotic Stress Responses by Modulating AtSIZ1 Function through Its E3 Ubiquitin Ligase Activity. Front Plant Sci. 2016;7:1182. 10.3389/fpls.2016.01182. [DOI] [PMC free article] [PubMed]
- 34.Luo Q, Lian HL, He SB, Li L, Jia KP, Yang HQ. COP1 and phyB physically interact with PIL1 to regulate its stability and Photomorphogenic development in Arabidopsis. Plant Cell. 2014;26(6):2441–2456. doi: 10.1105/tpc.113.121657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Au KKC, Perez-Gomez J, Neto H, Muller C, Meyer AJ, Fricker MD, Moore I. A perturbation in glutathione biosynthesis disrupts endoplasmic reticulum morphology and secretory membrane traffic in Arabidopsis thaliana. Plant J. 2012;71(6):881–894. doi: 10.1111/j.1365-313X.2012.05022.x. [DOI] [PubMed] [Google Scholar]
- 36.Ma QL, Ding YD, Chang JW, Sun XH, Zhang L, Wei QJ, Cheng YJ, Chen LL, Xu J, Deng XX. Comprehensive insights on how 2,4-dichlorophenoxyacetic acid retards senescence in post-harvest citrus fruits using transcriptomic and proteomic approaches. J Exp Bot. 2014;65(1):61–74. doi: 10.1093/jxb/ert344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hicks LM, Cahoon RE, Bonner ER, Rivard RS, Sheffield J, Jez JM. Thiol-based regulation of redox-active glutamate-cysteine ligase from Arabidopsis thaliana. Plant Cell. 2007;19(8):2653–2661. doi: 10.1105/tpc.107.052597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu JW, Rubio V, Lee NY, Bai SL, Lee SY, Kim SS, Liu LJ, Zhang YY, Irigoyen ML, Sullivan JA, et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell. 2008;32(5):617–630. doi: 10.1016/j.molcel.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jang S, Marchal V, Panigrahi KCS, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 2008;27(8):1277–1288. doi: 10.1038/emboj.2008.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gehringer A, Friedt W, Luhs W, Snowdon RJ. Genetic mapping of agronomic traits in false flax (Camelina sativa subsp sativa) Genome. 2006;49(12):1555–1563. doi: 10.1139/g06-117. [DOI] [PubMed] [Google Scholar]
- 41.Kim C, Lee JH, Chung YS, Choi SC, Guo H, Lee TH, Lee S. Characterization of twenty Camelina spp. accessions using single nucleotide polymorphism genotyping. Hortic Environ Biote. 2017;58(2):187–194. doi: 10.1007/s13580-017-0264-4. [DOI] [Google Scholar]
- 42.Vollmann J, Grausgruber H, Stift G, Dryzhyruk V, Lelley T. Genetic diversity in camelina germplasm as revealed by seed quality characteristics and RAPD polymorphism. Plant Breed. 2005;124(5):446–453. doi: 10.1111/j.1439-0523.2005.01134.x. [DOI] [Google Scholar]
- 43.Flint-Garcia SA, Thornsberry JM, Buckler ES. Structure of linkage disequilibrium in plants. Annu Rev Plant Biol. 2003;54:357–374. doi: 10.1146/annurev.arplant.54.031902.134907. [DOI] [PubMed] [Google Scholar]
- 44.Delourme R, Falentin C, Fomeju BF, Boillot M, Lassalle G, Andre I, Duarte J, Gauthier V, Lucante N, Marty A, et al. High-density SNP-based genetic map development and linkage disequilibrium assessment in Brassica napus L. BMC Genomics. 2013;14:120. doi: 10.1186/1471-2164-14-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nordborg M, Borevitz JO, Bergelson J, Berry CC, Chory J, Hagenblad J, Kreitman M, Maloof JN, Noyes T, Oefner PJ, et al. The extent of linkage disequilibrium in Arabidopsis thaliana. Nat Genet. 2002;30(2):190–193. doi: 10.1038/ng813. [DOI] [PubMed] [Google Scholar]
- 46.Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp mays L.) P Natl Acad Sci USA. 2001;98(16):9161–9166. doi: 10.1073/pnas.151244298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, Buckler ES. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci U S A. 2001;98(20):11479–11484. doi: 10.1073/pnas.201394398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ching A, Caldwell KS, Jung M, Dolan M, Smith OS, Tingey S, Morgante M, Rafalski AJ. SNP frequency, haplotype structure and linkage disequilibrium in elite maize inbred lines. BMC Genet. 2002;3:19. doi: 10.1186/1471-2156-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kunst L, Samuels AL. Biosynthesis and secretion of plant cuticular wax. Prog Lipid Res. 2003;42(1):51–80. doi: 10.1016/S0163-7827(02)00045-0. [DOI] [PubMed] [Google Scholar]
- 50.Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One. 2011;6(5):e19379. doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Couvares P, Kosar T, Roy A, Weber J, Wenger K. Workflow Management in Condor. In: Taylor I.J., Deelman E., Gannon D.B., Shields M. (eds) Workflows for e-Science. London: Springer; 2007. 10.1007/978-1-84628-757-2_22.
- 52.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23(19):2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 53.Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Earl DA, Vonholdt BM. Structure harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv Genet Resour. 2012;4(2):359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- 57.Xu LP, Hu KN, Zhang ZQ, Guan CY, Chen S, Hua W, Li JN, Wen J, Yi B, Shen JX, et al. Genome-wide association study reveals the genetic architecture of flowering time in rapeseed (Brassica napus L.) DNA Res. 2016;23(1):43–52. doi: 10.1093/dnares/dsv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ginestet C. ggplot2: elegant graphics for data analysis. J R Stat Soc a Stat. 2011;174:245. doi: 10.1111/j.1467-985X.2010.00676_9.x. [DOI] [Google Scholar]
- 59.Team RC . R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 60.Wang Qishan, Tian Feng, Pan Yuchun, Buckler Edward S., Zhang Zhiwu. A SUPER Powerful Method for Genome Wide Association Study. PLoS ONE. 2014;9(9):e107684. doi: 10.1371/journal.pone.0107684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang You, Liu Xiaolei, Wang Jiabo, Li Meng, Wang Qishan, Tian Feng, Su Zhongbin, Pan Yuchun, Liu Di, Lipka Alexander E., Buckler Edward S., Zhang Zhiwu. GAPIT Version 2: An Enhanced Integrated Tool for Genomic Association and Prediction. The Plant Genome. 2016;9(2):0. doi: 10.3835/plantgenome2015.11.0120. [DOI] [PubMed] [Google Scholar]
- 62.Turner SD: Qqman: an R package for visualizing GWAS results using Q-Q and Manhattan plots. 2014.
- 63.Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, de Andrade M, Feenstra B, Feingold E, Hayes MG, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43(6):519–U544. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36(10):3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bairoch A, Bougueleret L, Altairac S, Amendolia V, Auchincloss A, Puy GA, Axelsen K, Baratin D, Blatter MC, Boeckmann B, et al. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkn141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The ddRAD Sequencing data from Camalina sativa leaf was deposited in NCBI Sequence Read Archive (SRA) under accession numbers ranging from SRX4286823-SRX4286866.