Abstract
Strain SCT is an iodate-reducing bacterium isolated from marine sediment in Kanagawa Prefecture, Japan. In this study, we determined the draft genome sequence of strain SCT and compared it to complete genome sequences of other closely related bacteria, including Pseudomonas stutzeri. A phylogeny inferred from concatenation of core genes revealed that strain SCT was closely related to marine isolates of P. stutzeri. Genes present in the SCT genome but absent from the other analyzed P. stutzeri genomes comprised clusters corresponding to putative prophage regions and possible operons. They included pil genes, which encode type IV pili for natural transformation; the mer operon, which encodes resistance systems for mercury; and the pst operon, which encodes a Pi-specific transport system for phosphate uptake. We found that strain SCT had more prophage-like genes than the other P. stutzeri strains and that the majority (70%) of them were SCT strain-specific. These genes, encoded on distinct prophage regions, may have been acquired after branching from a common ancestor following independent phage transfer events. Thus, the genome sequence of Pseudomonas sp. strain SCT can provide detailed insights into its metabolic potential and the evolution of genetic elements associated with its unique phenotype.
Keywords: marine sediment, iodate-reducing bacterium, strain SCT, Pseudomonas stutzeri, genome analysis, comparative genomics, phylogeny, gene conservation
The genus Pseudomonas consists of gram-negative bacteria that inhabit a wide variety of environments, including the sea, soil, rhizospheres of plants, and the human microbiome (Aagot et al. 2001; Tekorienė 2008). Depending on the habitat, they employ various metabolic strategies, such as denitrification, nitrogen fixation, and metal reduction (Lovley 1993; Lloyd 2003). Pseudomonas spp. have thus been applied in bioremediation, for the removal or detoxification of environmental pollutants (Wasi et al. 2013). A 16S rRNA phylogenetic analysis split Pseudomonas into several subgroups (Anzai et al. 2000); one of them includes Pseudomonas stutzeri, Pseudomonas balearica, and Pseudomonas luteola. P. stutzeri is widespread and occupies diverse ecological niches, as listed exhaustively by Lalucat et al. (2006).
Iodine is an essential element for humans, as it is the main component of thyroid hormone and its deficiency causes goiter (Zava and Zava 2011). Historically, industrial iodine was extracted from ashes after burning seaweed, suggesting its abundance in marine environments (Zava and Zava 2011). Nowadays, iodine is taken up by ingesting marine products. At the same time, iodine in the oceans is involved in microbial metabolism (Winchester and Duce 1967; Amachi 2008). Strain SCT is a bacterium isolated from the slurry/sediment in Sagami Bay, Kanagawa Prefecture, Japan, by enrichment culture of iodate-reducing bacteria (Amachi et al. 2007). A phylogenetic analysis based on 16S rRNA gene sequencing indicated that strain SCT was most closely related to Pseudomonas stutzeri (Amachi et al. 2007).
Results by Amachi et al. (2007) suggest “SCT is a dissimilatory iodate-reducing bacterium and that its iodate reductase is induced by iodate under anaerobic growth conditions”. However, whereas the iodate-reducing activity of strain SCT has been extensively studied, its other potential functions remain largely unknown. The purpose of this study was to gain insights into the evolution and functional potential of strain SCT. To this end, we performed whole-genome shotgun sequencing of strain SCT and comparative genomic analyses with closely related bacteria including P. stutzeri.
Materials and Methods
Software
Bioinformatics analyses were conducted using Python version 3.5.5 and its molecular biology package Biopython version 1.72 (Cock et al. 2009). Statistical computing was implemented using R version 3.4.3, available at https://www.r-project.org/ (R Development Core Team 2008).
Sequence data
To compare strain SCT to the genome of Pseudomonas sp., we used 10 P. stutzeri and one P. balearica strains for which complete genome sequences were available through public databases. They included P. stutzeri Cr(VI)- and nitrate-reducing strain RCH2 (Chakraborty et al. 2017), naphthalene-degrading strain 19SMN4 (Rosselló-Mora et al. 1994), naphthalene-degrading strain CCUG 29243 (equivalent to AN10) (Brunet-Galmés et al. 2012), petroleum-emulsifying strain SLG510A3-8 (Hu et al. 2015), nitrogen-fixing and rhizosphere-associated strain DSM 4166 (Yu et al. 2011), type strain CGMCC 1.1803 (ATCC 17588) (Chen et al. 2011), nitrogen-fixing root-associated strain A1501 (Yan et al. 2008), highly transformable strain 28a24 (Smith et al. 2014), exopolysaccharide-producing strain 273 (Wu et al. 2017), natural transformation model strain DSM 10701 (JM300) (Busquets et al. 2012), and P. balearica DSM 6083T (Bennasar-Figueras et al. 2016). Based on a previous phylogenetic study (Özen and Ussery 2012), Pseudomonas mendocina ymp was used as an outgroup to root the phylogenetic tree. Genome sequence data in GenBank format (Benson et al. 2017) were retrieved/downloaded on 2018-02-24 from the National Center for Biotechnology Information (NCBI) reference sequence (RefSeq) database (Pruitt et al. 2007) using a set of scripts obtained from https://github.com/kblin/ncbi-genome-download. The final data set consisted of 13 Pseudomonas strains as shown in Table 1.
Table 1. Genomic features of Pseudomonas strains analyzed.
| Accessiona | Organism | Isolation source | Size (Mb) | bGC (%) | cCDS | dPhage | eConserved |
|---|---|---|---|---|---|---|---|
| This study | Pseudomonas sp. SCT | Marine sediment | 4.79 | 62.5 | 4520 | 184 | 4520 |
| GCF_000327065.1 | Pseudomonas stutzeri RCH2 | Cr(VI)-contaminated aquifer | 4.58 | 62.5 | 4353 | 43 | 3919 |
| GCF_000661915.1 | Pseudomonas stutzeri 19SMN4 | Marine sediment | 4.73 | 62.3 | 4533 | 109 | 3678 |
| GCF_000267545.1 | Pseudomonas stutzeri CCUG 29243 (=AN10) | Marine sediment | 4.71 | 62.7 | 4417 | 95 | 3578 |
| GCF_000195105.1 | Pseudomonas stutzeri SLG510A3-8 | Oil-contaminated soil | 4.65 | 64.0 | 4342 | 33 | 3547 |
| GCF_001038645.1 | Pseudomonas stutzeri DSM 4166 | Sorghum nutans rhizosphere | 4.69 | 64.0 | 4427 | 0 | 3560 |
| GCF_000219605.1 | Pseudomonas stutzeri CGMCC 1.1803 | Clinical specimen | 4.55 | 63.9 | 4242 | 23 | 3473 |
| GCF_000013785.1 | Pseudomonas stutzeri A1501 | Rice paddy soil | 4.57 | 63.9 | 4270 | 31 | 3461 |
| GCF_000590475.1 | Pseudomonas stutzeri 28a24 | Soil | 4.73 | 60.6 | 4317 | 27 | 2367 |
| GCF_001648195.1 | Pseudomonas stutzeri 273 | Marine sediment | 5.03 | 60.3 | 4687 | 77 | 2580 |
| GCF_000279165.1 | Pseudomonas stutzeri DSM 10701 | Soil | 4.17 | 63.2 | 3931 | 0 | 2463 |
| GCF_000818015.1 | Pseudomonas balearica DSM 6083 | Wastewater treatment plant | 4.38 | 64.0 | 4126 | 52 | 2522 |
| GCF_000016565.1 | Pseudomonas mendocina ymp | PCP-contaminated soil | 5.07 | 64.7 | 4714 | 68 | 1612 |
NCBI RefSeq accession number.
Percentage of G and C in the nucleotide sequence, defined as 100 × (G+C)/(A+T+G+C).
Number of protein-coding DNA sequences (CDSs).
Number of CDSs in phage-like regions predicted using PHASTER.
Number of SCT genes conserved in each genome inferred using LS-BSR.
Genome sequencing, assembly, and annotation of strain SCT
Strain SCT was grown aerobically in Lysogeny broth (LB) medium. Genomic DNA was extracted using a DNeasy blood and tissue kit (Qiagen, Hilden, Germany). According to the manufacturer’s protocols, an Illumina paired-end library (with an average insert size of 550 bp) was prepared, and whole-genome sequencing was performed using an Illumina MiSeq sequencing platform (Illumina, San Diego, CA) at the National Institute for Environmental Studies. The sequencer produced 1,084,678 paired-end reads (2 × 300 bp).
The paired-end reads were processed as follows: PhiX contaminations were removed using bbduk (https://sourceforge.net/projects/bbmap/). bbduk was run with the following parameters: “ref= phiX.fasta k=31 hdist=1” to remove all reads that had a 31-mer match to phiX (NCBI RefSeq accession: NC_001422), allowing one mismatch. After that, adapters were removed and reads were trimmed using Trimmomatic version 0.36 (Bolger et al. 2014) by typing “java -jar trimmomatic-0.36.jar PE -phred33 input_forward.fq input_reverse.fq output_forward_paired.fq output_forward_unpaired.fq output_reverse_paired.fq output_reverse_unpaired.fq ILLUMINACLIP:$adaptors:2:30:10 SLIDINGWINDOW:4:30 LEADING:3 TRAILING:3 MINLEN:100” (http://www.usadellab.org/cms/?page=trimmomatic). De novo assembly was conducted using SPAdes version 3.9.0 (Bankevich et al. 2012) with the following parameters “–careful–cov-cutoff auto”. After low-coverage contigs were disregarded, the resulting assembly consisted of 35 contigs containing 4,791,932 bp, with a G+C content of 62.5% and sequencing coverage of 56.5x.
Protein-coding DNA sequences (CDSs) were predicted and functional annotations (gene and product names) were assigned using Prokka version 1.11, which coordinates a suite of existing bioinformatics tools and databases for annotation of prokaryotic genome sequences (Seemann 2014), incorporating Prodigal (Hyatt et al. 2010), BLAST+ (Altschul et al. 1997), and HMMER (http://hmmer.org/). Prokka was run with the following parameters: “–kingdom Bacteria–compliant” (https://github.com/tseemann/prokka). We then performed similarity searches of all the predicted protein sequences against the UniRef90 sequence database (UniRef90 Release 2016_08 consisting of 44,448,796 entries) using the BLASTP program with an E-value cutoff of 1e-5, and assigned functional annotations from the most similar (best hit) protein sequences. BlastKOALA (Kanehisa et al. 2016) was used to assign KEGG Orthology identifiers to the protein sequences obtained by BLAST searches, for which taxonomy group information on “Bacteria” and the KEGG Genes database of “family_eukaryotes + genus_prokaryotes” were selected at https://www.kegg.jp/blastkoala/.
Search for mobile genetic elements
Mobile genetic elements such as phages were searched in the Pseudomonas genomes. The PHASTER search tool (Arndt et al. 2017) was used to identify putative prophage regions in the 13 Pseudomonas genomes analyzed. A new search of phage sequences in the SCT genome was performed at http://phaster.ca/, and pre-calculated results for the other 12 Pseudomonas genomes were obtained from http://phaster.ca/submissions.
Phylogenetic analysis
To infer phylogenetic relationships among the 13 Pseudomonas strains, we used single-copy core genes, which are shared by all genomes and contain only a single copy from each genome (and thus contain orthologs, but not paralogs). The core genes were built using the Roary pan-genomic analysis pipeline (Page et al. 2015) with a default parameter. All nucleotide alignments of the core genes were done in MAFFT (Katoh et al. 2002) and then concatenated by Roary. We used RAxML version 8.2.11 (Stamatakis 2014) for maximum likelihood-based inference of a phylogenetic tree on the concatenated sequence alignment under the GTR+CAT model. RAxML was run as follows: “raxmlHPC-PTHREADS -f a -x 12345 -p 12345 -# 100 -m GTRCAT -s ./core_gene_alignment.phy -n outfile”. The resulting tree was drawn using FigTree version 1.4.3, available at http://tree.bio.ed.ac.uk/software/figtree/.
Gene conservation analysis
To assess the conservation of SCT protein-coding genes in the other Pseudomonas genomes, we used the large-scale blast score ratio (LS-BSR) (Sahl et al. 2014). Briefly, the LS-BSR pipeline performed a TBLASTN search using the protein sequence of strain SCT as a query and the whole nucleotide sequence of each of the Pseudomonas strains as a database, and calculated the BSR. The obtained BSR value ranged from 0 (no sequence similarity) to 1 (maximal sequence similarity) and was used as a measure of the degree of conservation of SCT genes in the other Pseudomonas genomes. The ’$prefix_bsr_matrix.txt’ file contains the BSR value for each gene in each genome, and the ’$prefix_dup_matrix.txt’ file was used to determine gene presence and absence in each genome (https://github.com/jasonsahl/LS-BSR).
Data availability
The draft genome sequence of Pseudomonas sp. strain SCT has been deposited at GenBank/EMBL/DDBJ under BioProject number PRJDB5044, BioSample number SAMD00059319, and accession number BDJA00000000 (accession range: BDJA01000001-BDJA01000035). The version described in this paper is the first version, BDJA01000000. The raw reads have been deposited in the DDBJ Sequence Read Archive (DRA) under Submission DRA007938, Experiment DRX156286, and Run DRR165667.
Supplementary Table S1 has been deposited via the GSA Figshare portal. The Python scripts used in this study are available through the Github repository https://github.com/haradama/PSCT. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7829321.
Results and Discussion
Phylogeny
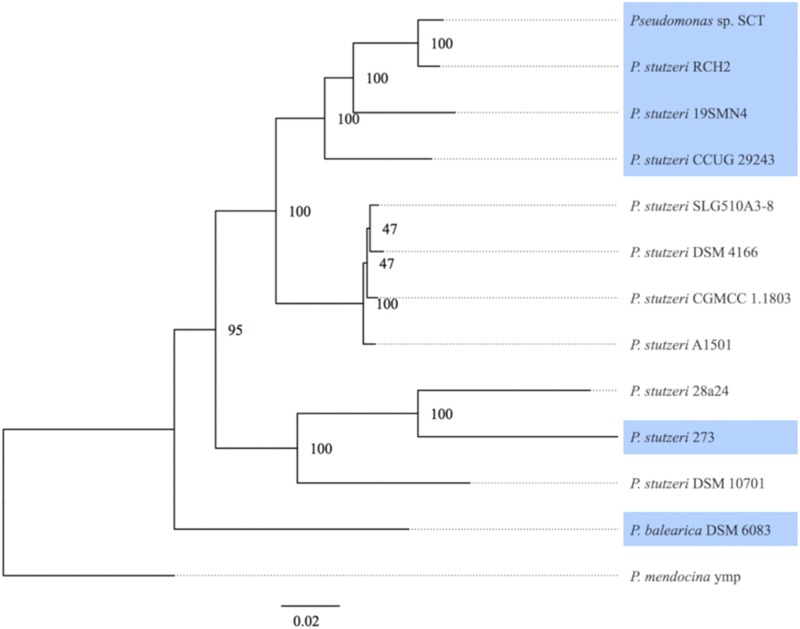
An accurate phylogenetic tree of a group of organisms provides a valid inference of its evolutionary history, gene gain, and loss events (Song et al. 2017). The subgroups of the genus Pseudomonas, including the P. stutzeri group, have been defined based on phylogenetic analyses of 16S rRNA gene sequences (Anzai et al. 2000). Recent studies have demonstrated that 16S rRNA gene sequences do not contain enough phylogenetic signals to distinguish closely related bacteria such as strains within the same species (Fox et al. 1992; Özen and Ussery 2012). To attain higher phylogenetic resolution, we inferred the phylogenetic relationship of SCT and other Pseudomonas strains based on 53 conserved core genes. Figure 1 shows the maximum likelihood phylogenetic tree based on the concatenated nucleotide sequence alignment of core genes. Core genome phylogeny with 100% bootstrap support indicated that strain SCT and P. stutzeri strain RCH2 (Chakraborty et al. 2017) formed a monophyletic group or clade that included also, in decreasing order of relevance, P. stutzeri strain 19SMN4 (Rosselló-Mora et al. 1994) and P. stutzeri strain CCUG 29243 (Brunet-Galmés et al. 2012). Results suggest that SCT belongs to P. stutzeri and is the sister strain to P. stutzeri strain RCH2.
Figure 1.
Maximum likelihood tree obtained from a concatenated nucleotide sequence alignment of core genes for the 13 Pseudomonas strains. The horizontal bar at the base of the figure represents 0.02 substitutions per nucleotide site. Bootstrap support values for each of the branches of the tree are indicated based on 100 bootstrap replicates. Blue represents strains isolated from water environments such as marine sediments.
The earliest branching lineage in this tree (Figure 1) was P. balearica, followed by the P. stutzeri clade. The latter contained three subgroups: the first one comprised strains SCT, RCH2, 19SMN4, and CCUG 29243; the second comprised strains SLG510A3-8, DSM 4166, CGMCC 1.1803, and A1501; and the third comprised strains 28a24, 273, and DSM 10701. A previous study revealed that distinct subgroups for the P. stutzeri clade could be accredited to ecotype status resulting from niche-specific adaptations; accordingly, the first subgroup contained marine isolates and the second subgroup contained soil/sludge isolates (Sharma et al. 2015). Given the primary niche of the first subgroup strains (SCT, RCH2, 19SMN4, and CCUG 29243), and a comparison to other strains in the P. stutzeri clade (Table 1), this phylogeny suggests that adaptation to marine/aquifer and soil/rhizosphere environments might have evolved after divergence of these subgroups from a common ancestor.
Genome features
Genome features can reflect not only phylogenetic positions but also lifestyles or ecological niches, as indicated by free-living soil bacteria with large G+C-rich genomes and obligatory intracellular symbionts with small G+C-poor genomes (Dutta and Paul 2012). Table 1 shows the genome features (size, G+C content, and CDS number) of the 13 Pseudomonas strains included in this analysis. Genome size ranged from 4.17 Mb to 5.07 Mb with a median of 4.69 Mb, G+C content ranged from 60.3 to 64.7% with a median of 63.2%, and CDS numbers ranged from 3,931 to 4,714 with a median of 4,353. The genome features of strain SCT were thus in line with those of other Pseudomonas strains. The G+C content for the first subgroup (ranging from 62.3 to 62.7%) in our phylogenetic tree (Figure 1), was lower than that of the second subgroup (ranging from 63.9 to 64.0%).
In contrast to previous studies showing a positive correlation between genome size and G+C content for sequenced bacterial genomes (McCutcheon et al. 2009; Dutta and Paul 2012), a correlation between genome size and G+C content for the 13 Pseudomonas strains used in this study was weakly negative (Pearson’s product-moment correlation coefficient, r = -0.34 and p-value = 0.25; Spearman’s rank correlation coefficient, rho = -0.33 and p-value = 0.27).
Mobile genetic elements
Horizontal transfer of DNA occurs generally via three different mechanisms: conjugation, transformation, and transduction (Kwong et al. 2000). It is now widely accepted that mobile genetic elements, such as plasmids and phages, contribute to the evolution of bacteria and the spreading of virulence and drug resistance in microbial communities (Frost et al. 2005).
Putative prophage regions have been detected in the genomes of P. stutzeri strains CCUG 29243 (Brunet-Galmés et al. 2012) and DSM 10701 (Busquets et al. 2012), as well as P. balearica DSM 6083 (Bennasar-Figueras et al. 2016). The PHASTER phage search tool identified six putative prophage regions in distinct contig sequences (contig accession numbers: BDJA01000001, BDJA01000002, BDJA01000003, BDJA01000004, BDJA01000005, and BDJA01000006) in the genome of strain SCT. The lengths of the putative prophage regions were 11,644 bp (BDJA01000001), 37,073 bp (BDJA01000002), 33,444 bp (BDJA01000003), 5,886 bp (BDJA01000004), 36,693 bp (BDJA01000005), and 18,683 (BDJA01000006). BLASTN best hits in the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) using the putative prophage regions as query sequences identified the following taxa: P. balearica DSM 6083 (accession: CP007511.1; 75% query coverage and 89% identity) for BDJA01000001, P. stutzeri RCH2 (accession: CP003071.1; 4% query coverage and 100% identity) for BDJA01000002, uncultured Caudovirales phage clone 3S_12 (accession: MF417932.1; 70% query coverage and 92% identity) for BDJA01000003, Pseudomonas aeruginosa PA7 (accession: CP000744.1; 54% query coverage and 79% identity) for BDJA01000004, P. stutzeri DW21 (accession: CP027543.1; 78% query coverage and 90% identity) for BDJA01000005, and P. stutzeri 19SMN4 (accession: CP007509.1; 40% query coverage and 94% identity) for BDJA01000006. A total of 184 CDSs were encoded in the six putative prophage regions: 13 CDSs for BDJA01000001 (locus_tag range: PSCT_00201 to PSCT_00214), 54 CDSs for BDJA01000002 (locus_tag range: PSCT_01140 to PSCT_01193), 50 CDSs for BDJA01000003 (locus_tag range: PSCT_01861 to PSCT_01910), 8 CDSs for BDJA01000004 (locus_tag range: PSCT_02604 to PSCT_02611), 35 CDSs for BDJA01000005 (locus_tag range: PSCT_02824 to PSCT_02859), and 24 CDSs for BDJA01000006 (locus_tag range: PSCT_03081 to PSCT_03104). Among the 13 Pseudomonas strains analyzed, the largest number of phage-like genes was found in strain SCT with 184, followed by P. stutzeri strain 19SMN4 with 109, and then P. stutzeri strain CCUG 29243 with 95. These three strains, belonging to the first subgroup of the P. stutzeri clade in our phylogeny (Figure 1), were isolated from marine sediments (Table 1). This result is consistent with previous findings reporting the frequent detection and routine isolation of Pseudomonas phages in marine environments (Jiang et al. 1998; Watkins et al. 2018).
Plasmids can confer their hosts resistance to antibiotics and heavy metals (Popowska and Krawczyk-Balska 2013). P. stutzeri strains isolated from polluted environments tend to contain plasmids (Ginard et al. 1997) and some have been reported to harbor plasmid-encoded silver (Haefeli et al. 1984) or mercury resistance (Barbieri et al. 1989). P. stutzeri strain RCH2 (Chakraborty et al. 2017) contained three plasmids pPSEST01, pPSEST02, and pPSEST03 (12,763 bp, 9,865 bp, and 2,804 bp, respectively, in length), and strain 19SMN4 (Rosselló-Mora et al. 1994) contained one plasmid, pLIB119 (107,733 bp in length). The four plasmids were not similar to each other based on all-by-all BLASTN searches with a cutoff E value of 1e-5. Given the presence of plasmids in these two strains and their absence in the other Pseudomonas strains analyzed, our phylogeny (Figure 1) suggests that these plasmids may have been acquired independently by each lineage, along the branch leading to the ancestor of either strain RCH2 or strain 19SMN4.
Gene annotations
The genome of Pseudomonas sp. strain SCT contained 4,520 CDSs, of which 1,089 are currently annotated as unknown functions (i.e., product names of “hypothetical proteins”), 2,709 are annotated by UniProtKB (Boutet et al. 2016), 509 by PFAM (Finn et al. 2014), 310 by CDD (Marchler-Bauer et al. 2011), and 20 by HAMAP (Lima et al. 2009). Of the 4,520 proteins, 4,401 (97.5%) had matches with 4,329 unique records in the UniRef90 database, and 2,543 (56.3%) were assigned to the 1,991 unique KEGG Orthology identifiers. Among the 4,520 CDSs, length in amino acids (Laa) ranged from 30 to 2,842 with a median of 272, and G+C content at the third codon position (GC3) ranged from 33 to 100%, with a median of 79%. G+C content varies more widely at the third codon position than at the first or second positions, which are constrained by protein-coding requirements (Sharp et al. 2005). The corresponding data for the SCT genes are shown in Supplementary Table S1.
The following paragraphs detail the functional annotations assigned to SCT genes by Prokka (gene and product names) as well as the UniRef90 and KEGG databases. The SCT genome contains genes encoding cytochrome c and related proteins such as cytochrome c oxidase subunits. A cluster of genes for type II secretion system proteins M, L, K, J, I, H, G, F, and E (locus_tag range: PSCT_03513 to PSCT_03521) was also identified.
Previously, P. stutzeri strains were reported as capable of degrading aromatic hydrocarbons, such as phenol, naphthalene, toluene, and xylenes (Rosselló-Mora et al. 1994; Nie et al. 2016; Brown et al. 2017; Singh and Tiwary 2017). Here, the SCT genome appeared to contain genes involved in hydrocarbon degradation, including benA and benB for large and small subunits of benzoate 1,2-dioxygenase (KEGG: K05549 and K05550); catA, which encodes “catechol 1,2-dioxygenase [EC:1.13.11.1]” (KEGG: K03381); and a CDS annotated as “glyoxalase-like domain protein” or “catechol 2,3-dioxygenase” (locus_tag: PSCT_01283). Indeed, some of these genes formed the cluster benABCDK-catBCA-benE (contig accession number: BDJA01000005; locus_tag range: PSCT_03059 to PSCT_03067). These results suggest that strain SCT can potentially metabolize aromatic hydrocarbons.
Denitrifying bacteria reduce nitrate (NO3-) to nitrite (NO2-), nitric oxide (NO), nitrous oxide (N2O), and finally dinitrogen (N2) (Carlson and Ingraham 1983). The organization of the denitrification genes has been extensively investigated in P. stutzeri, in which nos genes (for N2O reduction), nir genes (for NO2- reduction), and nor genes (for NO reduction) are adjacent and far from nar genes (for NO3- reduction) on the chromosome (Baker et al. 1998; Arai et al. 2003; Yan et al. 2005). In the SCT genome, the nos, nir, and nor genes were also adjacent (contig accession number: BDJA01000001; locus_tag range: PSCT_00772 to PSCT_00802) and far from the nar genes (contig accession number: BDJA01000010; locus_tag range: PSCT_04176 to PSCT_04184). The gene cluster narLXK-uspA-narTGHJI-cbf2-moaA-moaB-moeA (contig accession number: BDJA01000010; locus_tag range: PSCT_04176 to PSCT_04188) is a possible operon consisting of nar genes and genes encoding the molybdenum cofactor (Padilla et al. 2017). The products of the moaA, moaB, and moeA genes (locus_tag: PSCT_04186, PSCT_04187, and PSCT_04188) are “cyclic pyranopterin monophosphate synthase”, “molybdenum cofactor biosynthesis protein B”, and “molybdopterin molybdenumtransferase”, respectively. The corresponding definitions in the KEGG database (K03639, K03638, and K03750) are “GTP 3’,8-cyclase [EC:4.1.99.22]”, “molybdopterin adenylyltransferase [EC:2.7.7.75]”, and “molybdopterin molybdotransferase [EC:2.10.1.1]”. The genome also contains a cluster of three genes, moaE-moaD-moaC (contig accession number: BDJA01000006; locus_tag: PSCT_03192, PSCT_03193, and PSCT_03194), whereby moaE encodes “molybdopterin synthase catalytic subunit [EC:2.8.1.12]” (KEGG: K03635), moaD encodes “molybdopterin synthase sulfur carrier subunit” annotated also as “molybdopterin converting factor, small subunit” (UniProt: L0GQS7), and moaC encodes “cyclic pyranopterin monophosphate synthase [EC:4.6.1.17]” (KEGG: K03637). Molybdopterin is a cofactor essential for nitrate reductase activity (Almeida et al. 2017). Pseudomonas spp. such as P. stutzeri, P. aeruginosa, and P. denitrificans possess the ability to reduce nitrate, although reduction rates vary among species (Carlson and Ingraham 1983). Strain SCT has been reported to use nitrate or nitrite as an electron acceptor (Amachi et al. 2007).
Bacteria, such as Pseudomonas putida KT2440 and P. stutzeri TS44, possess genes whose products allow resistance to and metabolism of arsenic compounds (Andres and Bertin 2016). We report that the SCT genome contains genes putatively involved in such processes; these include several genes encoding ArsR family transcriptional regulators; spxA (locus_tag: PSCT_01329) and arsC (locus_tag: PSCT_01343 and PSCT_04450); genes encoding “arsenate reductase [EC:1.20.4.1]” (KEGG: K00537 and K03741), arsA (locus_tag: PSCT_02478), which encodes “arsenite/tail-anchored protein-transporting ATPase [EC:3.6.3.16 3.6.3.-]” (KEGG: K01551); and ubiG (locus_tag: PSCT_04351), which encodes “arsenite methyltransferase [EC:2.1.1.137]” (KEGG: K07755). There is a cluster of six genes azr-ywlE-aseR-arsC1-czcO-CDS (contig accession number: BDJA01000003; locus_tag range: PSCT_01732 to PSCT_01737); of these, azr encodes “arsenical resistance protein ArsH” (KEGG: K11811), ywlE encodes “arsenate reductase [EC:1.20.4.1]” (KEGG: K03741), aseR and arsC1 encode “ArsR family transcriptional regulator, arsenate/arsenite/antimonite-responsive transcriptional repressor” (KEGG: K03892), czcO encodes “putative oxidoreductase CzcO”, and the CDS encodes “arsenite transporter” (KEGG: K03325). These results suggest that strain SCT can potentially resist and metabolize arsenic compounds.
Gene conservation
Conservation of SCT protein genes in the genome of all Pseudomonas strains (Table 1) was determined using the gene screen method with the TBLASTN tool in the LS-BSR pipeline. Of the 4,520 SCT genes, 1,908 (42%) were conserved in all 11 P. stutzeri strains, and 1,318 (29%) were conserved in all 13 Pseudomonas strains examined. Among P. stutzeri strains, more SCT genes were conserved in the three strains belonging to the first subgroup (RCH2, 19SMN4, and CCUG 29243; 3,578 to 3,919 genes) than in the strains belonging to the second subgroup (SLG510A3-8, DSM 4166, CGMCC 1.1803, and A1501; 3,461 to 3,547 genes), or the third subgroup (28a24, 273, and DSM 10701; 2,367 to 2,580 genes). Thus, the conservation of SCT genes in the Pseudomonas strains analyzed reflects their phylogenetic relationships (Figure 1).
A total of 451 genes were present in the SCT genome but absent from the other 10 P. stutzeri genomes analyzed; they are referred here as the “SCT strain-specific gene set”. They included 254 hypothetical proteins as well as clusters of genes corresponding to putative prophages or possible operons (Supplementary Table S1). The SCT strain-specific genes may have been acquired following separation from common ancestors (gained on the branch leading to the SCT ancestor) and may be associated with SCT strain-specific phenotypic properties (e.g., iodate reduction and living in marine environments).
Sequence statistics (e.g., Laa and GC3) were used to compare SCT strain-specific genes and other genes in the SCT genome. The median Laa value for SCT strain-specific genes (183 aa) was smaller than that for the other genes (280 aa), the difference being highly significant according to a Wilcoxon rank sum test (p-value < 2.2e-16). Thus, in general, the SCT strain-specific genes tended to be shorter than the remaining genes in the SCT genome. The median value of GC3 (G+C content at the third codon position) was lower for the SCT strain-specific genes (70%) than for the other genes (79%). Again, the difference was significant according to a Wilcoxon rank sum test (p-value < 2.2e-16). Thus, in general, GC3 tended to be lower for the strain-specific genes than for the remaining genes in the SCT genome. For example, GC3 values for a cluster of five genes (contig accession number: BDJA01000002; locus_tag range: PSCT_01674 to PSCT_01678) were lower (ranging from 33 to 41%) than those for the flanking genes (>50%). BLASTP best hits in the UniRef90 database for the flanking genes were identified as belonging to Pseudomonas taxa, whereas those for the five genes were unknown (PSCT_01676 and PSCT_01678) or did not belong to the Pseudomonas genus; i.e., Halorhodospira halochloris (Class: Gammaproteobacteria; Order: Chromatiales) for PSCT_01674, Inquilinus limosus (Class: Alphaproteobacteria) for PSCT_01675, and Alteromonas confluentis (Class: Gammaproteobacteria; Order: Alteromonadales) for PSCT_01677. Studies have revealed that genes acquired by recent horizontal/lateral transfer often bear unusual nucleotide compositions (Lawrence and Ochman 1997) and are usually rich in A+T (Daubin et al. 2003). Thus, nucleotide composition such as total and positional G+C content (GC3) of genes have been used to detect horizontally transferred genes in various complete bacterial genomes (Becq et al. 2010).
Some P. stutzeri strains are competent for natural genetic transformation (Lorenz and Sikorski 2000), a process whereby DNA is taken up from external environments and is heritably integrated into the genome. This ability enables the bacterium to adapt to various conditions and, not surprisingly, P. stutzeri has been found in a wide range of environments (Lalucat et al. 2006). Natural transformation of P. stutzeri requires a competence phase and the formation of functional type IV pili (Meier et al. 2002). Genes for natural transformation (comA, exbB, and pil genes for type IV pili) were found in P. stutzeri strains CCUG 29243 (Brunet-Galmés et al. 2012) and DSM 10701, a model organism for natural transformation (Chakraborty et al. 2017). The SCT genome contains two distinct pil gene clusters for type IV pilus assembly protein. One cluster consists of pilQ-pilP-pilO-pilN-pilM genes (contig accession number: BDJA01000004; locus_tag range: PSCT_02297 to PSCT_02301; KEGG: K02666, K02665, K02664, K02663, and K02662). Another cluster is SCT strain-specific, and consists of pilY1-pilX-pilW-pilV-fimT-fimT genes for “type IV pilus assembly protein” and “type IV fimbrial biogenesis protein FimT” (contig accession number: BDJA01000006; locus_tag range: PSCT_03390 to PSCT_03395; KEGG: K02674, K02673, K02672, K02671, K08084, and K08084). GC3 values for the six SCT strain-specific genes pilY1-pilX-pilW-pilV-fimT-fimT were lower (ranging from 53 to 61%) than those for the flanking genes (>73%). Taxa of the BLASTP best hits in the UniRef90 database for the six genes were unknown (PSCT_03390 and PSCT_03392) or did not belong to P. stutzeri; i.e., Microbulbifer agarilyticus (Class: Gammaproteobacteria; Order: Alteromonadales) for PSCT_03391, Pseudomonas taeanensis for PSCT_03393, Marinimicrobium agarilyticum (Class: Gammaproteobacteria; Order: Alteromonadales) for PSCT_03394, and Thiobacillus (Class: Betaproteobacteria) for PSCT_03395. Evidence suggests that P. aeruginosa minor pilins PilV, PilW, and PilX require PilY1 for inclusion in surface pili and vice versa (Nguyen et al. 2015). A comprehensive list describing conservation of type IV pili accessory and assembly proteins (FimT, FimU, PilV, PilW, PilX, PilY1, PilY2, and PilE) among P. aeruginosa strains was produced by Asikyan et al. (2008). Present results suggest that strain SCT may be competent for natural genetic transformation and has been subjected to horizontal gene transfer events via natural transformation.
Extensively studied bacterial resistance systems for mercury are clustered in an operon (mer operon), which varies in structure, and consists of genes encoding functional proteins for regulation (merR), transport (merT, merP, and/or merC, merF), and reduction (merA) (Nascimento and Chartone-Souza 2003) of mercury compounds. The marine bacterium P. stutzeri 273, which belongs to the third subgroup in our phylogeny (Figure 1), has been shown to be resistant to Hg2+ and to be capable of removing it; specifically, genes encoding MerT, MerP, MerA, and MerD appear essential for bacterial mercuric resistance (Zheng et al. 2018). SCT strain-specific genes for mercuric resistance/transport/reductase were found in two distinct gene clusters (contig accession numbers: BDJA01000012 and BDJA01000013): merR-merT-merP-merA-podJ-merR (contig accession number: BDJA01000012; locus_tag range: PSCT_04381 to PSCT_04386) and merR-merT-merP-merF-merA-ahpD-comR-dehH2 (contig accession number: BDJA01000013; locus_tag range: PSCT_04402 to PSCT_04409). The first one contains the genes merR for “MerR family transcriptional regulator, mercuric resistance operon regulatory protein” (KEGG: K08365), merT for “mercuric ion transport protein” (KEGG: K08363), merP for “periplasmic mercuric ion binding protein” (KEGG: K08364), merA for “mercuric reductase [EC:1.16.1.1]” (KEGG: K00520), podJ for “localization factor PodJL”, and merR for “mercuric resistance operon regulatory protein”. The second one consists of five mer genes (CDS encoding “membrane transport protein MerF” is located between merP and merA) flanked by comR, which encodes “TetR/AcrR family transcriptional regulator, transcriptional repressor for nem operon” (KEGG: K16137) and dehH2 encoding “2-haloacid dehalogenase [EC:3.8.1.2]” (KEGG: K01560). These results provide genetic evidence of potential mercury resistance and its metabolism by the marine bacterium Pseudomonas sp. strain SCT. Moreover, this newly discovered capacity of strain SCT makes the bacterium attractive for the bioremediation of mercury-contaminated environments.
Previous studies have reported the molecular mechanism of 2-haloacid dehalogenase from Pseudomonas spp. including P. putida (Kawasaki et al. 1994; Liu et al. 1995; Wang et al. 2018) and described known haloacid-dehalogenating bacteria and their dehalogenases (Adamu et al. 2016). Of the seven genes annotated as “haloacid dehalogenase” in the SCT genome, three (locus_tag: PSCT_04345, PSCT_04409, and PSCT_04536) were SCT strain-specific. The SCT strain-specific gene (locus_tag: PSCT_04345) encoding “2-haloacid dehalogenase [EC:3.8.1.2]” (KEGG: K01560) is flanked by a CDS (locus_tag: PSCT_04346) encoding “molybdenum cofactor biosynthesis protein A” and merA (locus_tag: PSCT_04347), which encodes “mercuric reductase”. Both of these genes are not SCT strain-specific, although the flanking genes yjlD-CDS-oprD-ubiG-pstS1-pstC-pstA-pstB (locus_tag range: PSCT_04348 to PSCT_04355) are SCT strain-specific.
Bacterial uptake of phosphate occurs via Pi-specific transport (Pst) systems, which are multi-subunit ABC transporters encoded by a four-gene operon, pstSCAB (Gebhard et al. 2009). A previous study reported the existence of two operons encoding two distinct Pst systems named pst1 and pst2 and that pst2 was present in all Pseudomonas spp. including P. aeruginosa, P. fluorescens, P. putida, P. syringae, and P. stutzeri strains DSM 4166, A1501, and DSM 10701, whereas pst1 was not (Lidbury et al. 2016). The SCT genome contains two pstSCAB operons, of which one is SCT strain-specific. The SCT strain-specific pstSCAB operon (contig accession number: BDJA01000012; locus_tag: PSCT_04352, PSCT_04353, PSCT_04354, and PSCT_04355) is flanked by ubiG, which encodes “ubiquinone biosynthesis O-methyltransferase” also annotated as “arsenite methyltransferase [EC:2.1.1.137]” (KEGG: K07755). Another pstSCAB operon (contig accession number: BDJA01000001; locus_tag: PSCT_00287, PSCT_00288, PSCT_00289, and PSCT_00290) is flanked by phoU (locus_tag: PSCT_00291), which encodes “phosphate-specific transport system accessory protein PhoU”. Lidbury et al. (2016) hypothesized that Pst2 was essential for efficient uptake of Pi in Pseudomonas spp. and that Pst1 and Pst2 possessed different kinetic parameters.
Of the 184 proteins encoded on the putative prophage regions in the SCT genome mentioned above, 129 were SCT strain-specific: 3/13 for BDJA01000001, 52/54 for BDJA01000002, 50/50 for BDJA01000003, 8/8 for BDJA01000004, 6/35 for BDJA01000005, and 10/24 for BDJA01000006. The majority (129/184, 70%) of the putative prophage genes are SCT strain-specific. It is likely that the genes in the cluster (i.e., prophage regions) were gained during the same phage transfer event, rather than by several independent events. Our results suggest that phage-mediated gene transfer events have occurred since separation from common ancestors (on the branch leading to the SCT ancestor).
Conclusion
We report a draft genome assembly for strain SCT, an iodate-reducing bacterium isolated from a marine environment. Phylogenetic analysis indicates that strain SCT belongs to the species Pseudomonas stutzeri and is closely related to marine isolates of P. stutzeri including strain RCH2. The SCT genome contains genes putatively involved in hydrocarbon degradation, nitrogen metabolism, and arsenic resistance and metabolism. Gene conservation analysis identified a set of genes present in the SCT genome but absent from the other P. stutzeri genomes analyzed. This SCT strain-specific gene set included (i) the pil gene cluster encoding minor pilins of the type IV pilus system for natural transformation, (ii) mer gene clusters encoding resistance systems for mercury, (iii) the pst gene cluster encoding Pst systems for uptake of phosphate, and (iv) gene clusters corresponding to putative prophage regions. These results suggest that strain SCT has evolved in marine environments and those polluted by hydrocarbons and heavy metals (e.g., arsenic and mercury) and has been subjected to horizontal gene transfer events via natural transformation and (phage-mediated) transduction. Accordingly, strain SCT has potential in bioremediation of hydrocarbon- and heavy-metal-polluted environments. Finally, bioinformatics analyses of the Pseudomonas sp. strain SCT genome sequence have identified a number of new gene targets, whose function will be revealed by future experimental testing.
Acknowledgments
We gratefully thank the members of the Institute for Advanced Biosciences, Keio University, Japan, for their insightful discussions. Computational resources were provided by the Data Integration and Analysis Facility, National Institute for Basic Biology. This work was supported, in part, by research funds from Keio University, the Yamagata prefectural government, and the City of Tsuruoka.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7829321.
Communicating editor: D. Baltrus
Literature Cited
- Aagot N., Nybroe O., Nielsen P., Johnsen K., 2001. An altered Pseudomonas diversity is recovered from soil by using nutrient-poor Pseudomonas-selective soil extract media. Appl. Environ. Microbiol. 67: 5233–5239. 10.1128/AEM.67.11.5233-5239.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamu A., Wahab R. A., Huyop F., 2016. l-2-Haloacid dehalogenase (DehL) from Rhizobium sp. RC1. Springerplus 5: 695 10.1186/s40064-016-2328-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida S., Sousa C., Abreu V., Diniz C., Dorneles E., et al. , 2017. Exploration of Nitrate reductase metabolic pathway in Corynebacterium pseudotuberculosis. Int. J. Genomics 2017: 9481756 10.1155/2017/9481756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., et al. , 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amachi S., 2008. Microbial contribution to global iodine cycling: volatilization, accumulation, reduction, oxidation, and sorption of iodine. Microbes Environ. 23: 269–276. 10.1264/jsme2.ME08548 [DOI] [PubMed] [Google Scholar]
- Amachi S., Kawaguchi N., Muramatsu Y., Tsuchiya S., Watanabe Y., et al. , 2007. Dissimilatory iodate reduction by marine Pseudomonas sp. strain SCT. Appl. Environ. Microbiol. 73: 5725–5730. 10.1128/AEM.00241-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres J., Bertin P. N., 2016. The microbial genomics of arsenic. FEMS Microbiol. Rev. 40: 299–322. 10.1093/femsre/fuv050 [DOI] [PubMed] [Google Scholar]
- Anzai Y., Kim H., Park J.-Y., Wakabayashi H., Oyaizu H., 2000. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. Evol. Microbiol. 50: 1563–1589. 10.1099/00207713-50-4-1563 [DOI] [PubMed] [Google Scholar]
- Arai H., Mizutani M., Igarashi Y., 2003. Transcriptional regulation of the nos genes for nitrous oxide reductase in Pseudomonas aeruginosa. Microbiology 149: 29–36. 10.1099/mic.0.25936-0 [DOI] [PubMed] [Google Scholar]
- Arndt D., Marcu A., Liang Y., Wishart D. S., 2017. PHAST, PHASTER and PHASTEST: Tools for finding prophage in bacterial genomes. Brief. Bioinform bbx121, 1–8. 10.1093/bib/bbx121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asikyan M. L., Kus J. V., Burrows L. L., 2008. Novel proteins that modulate type IV pilus retraction dynamics in Pseudomonas aeruginosa. J. Bacteriol. 190: 7022–7034. 10.1128/JB.00938-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. C., Ferguson S. J., Ludwig B., Page M. D., Richter O. M. H., et al. , 1998. Molecular genetics of the genus Paracoccus: metabolically versatile bacteria with bioenergetic flexibility. Microbiol. Mol. Biol. Rev. 62: 1046–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., et al. , 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19: 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri P., Galassi G., Galli E., 1989. Plasmid-encoded mercury resistance in a Pseudomonas stutzeri strain that degrades o-xylene. FEMS Microbiol. Ecol. 5: 375–383. 10.1111/j.1574-6968.1989.tb03393.x [DOI] [Google Scholar]
- Becq J., Churlaud C., Deschavanne P., 2010. A benchmark of parametric methods for horizontal transfers detection. PLoS One 5: e9989 10.1371/journal.pone.0009989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasar-Figueras A., Salvà-Serra F., Jaén-Luchoro D., Seguí C., Aliaga F., et al. , 2016. Complete genome sequence of Pseudomonas balearica DSM 6083t. Genome Announc. 4: e00217–16. 10.1128/genomeA.00217-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D. A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Ostell J., et al. , 2017. GenBank. Nucleic Acids Res. 45: D37–D42. 10.1093/nar/gkw1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet E., Lieberherr D., Tognolli M., Schneider M., Bansal P., et al. , 2016, pp. 23–54 in “UniProtKB/Swiss-Prot, the Manually Annotated Section of the UniProt Knowledgebase: How to Use the Entry View.,” in Plant Bioinformatics. Methods in Molecular Biology, Vol. 1374, edited by Edwards D., Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- Brown L. M., Gunasekera T. S., Ruiz O. N., 2017. Draft genome sequence of Pseudomonas stutzeri Strain 19, an isolate capable of efficient degradation of aromatic hydrocarbons. Genome Announc. 5: e01373–17. 10.1128/genomeA.01373-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet-Galmés I., Busquets A., Peña A., Gomila M., Nogales B., et al. , 2012. Complete genome sequence of the naphthalene-degrading bacterium Pseudomonas stutzeri AN10 (CCUG 29243. J. Bacteriol. 194: 6642–6643. 10.1128/JB.01753-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busquets A., Peña A., Gomila M., Bosch R., Nogales B., et al. , 2012. Genome sequence of Pseudomonas stutzeri strain JM300 (DSM 10701), a soil isolate and model organism for natural transformation. J. Bacteriol. 194: 5477–5478. 10.1128/JB.01257-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson C. A., Ingraham J. L., 1983. Comparison of denitrification by Pseudomonas stutzeri, Pseudomonas aeruginosa, and Paracoccus denitrificans. Appl. Environ. Microbiol. 45: 1247–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R., Woo H., Dehal P., Walker R., Zemla M., et al. , 2017. Complete genome sequence of Pseudomonas stutzeri strain RCH2 isolated from a hexavalent chromium [Cr (VI)] contaminated site. Stand. Genomic Sci. 12: 23 10.1186/s40793-017-0233-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Yan Y., Zhang W., Lu W., Wang J., et al. , 2011. Complete genome sequence of the type strain Pseudomonas stutzeri CGMCC 1.1803. J. Bacteriol. 193: 6095 10.1128/JB.06061-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock P. J., Antao T., Chang J. T., Chapman B. A., Cox C. J., et al. , 2009. Biopython: freely available Phyton tools for computational molecular biology and bioinformatics. Bioinformatics 25: 1422–1423. 10.1093/bioinformatics/btp163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubin V., Lerat E., Perrière G., 2003. The source of laterally transferred genes in bacterial genomes. Genome Biol. 4: R57 10.1186/gb-2003-4-9-r57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta C., Paul S., 2012. Microbial lifestyle and genome signatures. Curr. Genomics 13: 153–162. 10.2174/138920212799860698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., et al. , 2014. Pfam: the protein families database. Nucleic Acids Res. 42: D222–D230. 10.1093/nar/gkt1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox G. E., Wisotzkey J. D., Jurtshuk J. R. P., 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42: 166–170. 10.1099/00207713-42-1-166 [DOI] [PubMed] [Google Scholar]
- Frost L. S., Leplae R., Summers A. O., Toussaint A., 2005. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3: 722–732. 10.1038/nrmicro1235 [DOI] [PubMed] [Google Scholar]
- Gebhard S., Ekanayaka N., Cook G. M., 2009. The low-affinity phosphate transporter PitA is dispensable for in vitro growth of Mycobacterium smegmatis. BMC Microbiol. 9: 254 10.1186/1471-2180-9-254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginard M., Lalucat J., Tümmler B., Römling U., 1997. Genome organization of Pseudomonas stutzeri and resulting taxonomic and evolutionary considerations. Int. J. Syst. Bacteriol. 47: 132–143. 10.1099/00207713-47-1-132 [DOI] [PubMed] [Google Scholar]
- Haefeli C., Franklin C., Hardy K., 1984. Plasmid-determined silver resistance in Pseudomonas stutzeri isolated from a silver mine. J. Bacteriol. 158: 389–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Nie Y., Geng S., Wu X.-L., 2015. Complete genome sequence of the petroleum-emulsifying bacterium Pseudomonas stutzeri SLG510a3–8. J. Bacteriol. 211: 1–2. [DOI] [PubMed] [Google Scholar]
- Hyatt D., Chen G.-L., LoCascio P. F., Land M. L., Larimer F. W., et al. , 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11: 119 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S. C., Kellogg C. A., Paul J. H., 1998. Characterization of marine temperate phage-host systems isolated from Mamala Bay, Oahu, Hawaii. Appl. Environ. Microbiol. 64: 535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Morishima K., 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428: 726–731. 10.1016/j.jmb.2015.11.006 [DOI] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K. I., Miyata T., 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30: 3059–3066. 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki H., Toyama T., Maeda T., Nishino H., Tonomura K., 1994. Cloning and sequence analysis of a plasmid-encoded 2-haloacid dehalogenase gene from Pseudomonas putida No. 109. Biosci. Biotechnol. Biochem. 58: 160–163. 10.1271/bbb.58.160 [DOI] [PubMed] [Google Scholar]
- Kwong S. M., Yeo C. C., Suwanto A., Poh C. L., 2000. Characterization of the endogenous plasmid from Pseudomonas alcaligenes NCIB 9867: DNA sequence and mechanism of transfer. J. Bacteriol. 182: 81–90. 10.1128/JB.182.1.81-90.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalucat J., Bennasar A., Bosch R., García-Valdés E., Palleroni N. J., 2006. Biology of Pseudomonas stutzeri. Microbiol. Mol. Biol. Rev. 70: 510–547. 10.1128/MMBR.00047-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J. G., Ochman H., 1997. Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 44: 383–397. 10.1007/PL00006158 [DOI] [PubMed] [Google Scholar]
- Lidbury I. D., Murphy A. R., Scanlan D. J., Bending G. D., Jones A. M., et al. , 2016. Comparative genomic, proteomic and exoproteomic analyses of three Pseudomonas strains reveals novel insights into the phosphorus scavenging capabilities of soil bacteria. Environ. Microbiol. 18: 3535–3549. 10.1111/1462-2920.13390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima T., Auchincloss A. H., Coudert E., Keller G., Michoud K., et al. , 2009. HAMAP: a database of completely sequenced microbial proteome sets and manually curated microbial protein families in UniProtKB/Swiss-Prot. Nucleic Acids Res. 37: D471–D478. 10.1093/nar/gkn661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.-Q., Kurihara T., Miyagi M., Esaki N., Soda K., 1995. Reaction mechanism of L-2- haloacid dehalogenase of Pseudomonas sp. YL. Identification of Asp10 as the active site nucleophile by 18O incorporation experiments. J. Biol. Chem. 270: 18309–18312. 10.1074/jbc.270.31.18309 [DOI] [PubMed] [Google Scholar]
- Lloyd J. R., 2003. Microbial reduction of metals and radionuclides. FEMS Microbiol. Rev. 27: 411–425. 10.1016/S0168-6445(03)00044-5 [DOI] [PubMed] [Google Scholar]
- Lorenz M. G., Sikorski J., 2000. The potential for intraspecific horizontal gene exchange by natural genetic transformation: sexual isolation among genomovars of Pseudomonas stutzeri. Microbiology 146: 3081–3090. 10.1099/00221287-146-12-3081 [DOI] [PubMed] [Google Scholar]
- Lovley D. R., 1993. Dissimilatory metal reduction. Annu. Rev. Microbiol. 47: 263–290. 10.1146/annurev.mi.47.100193.001403 [DOI] [PubMed] [Google Scholar]
- McCutcheon J. P., McDonald B. R., Moran N. A., 2009. Origin of an alternative genetic code in the extremely small and GC–rich genome of a bacterial symbiont. PLoS Genet. 5: e1000565 10.1371/journal.pgen.1000565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., et al. , 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39: D225–D229. 10.1093/nar/gkq1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier P., Berndt C., Weger N., Wackernagel W., 2002. Natural transformation of Pseudomonas stutzeri by single-stranded DNA requires type IV pili, competence state and comA. FEMS Microbiol. Lett. 207: 75–80. 10.1111/j.1574-6968.2002.tb11031.x [DOI] [PubMed] [Google Scholar]
- Nascimento A. M., Chartone-Souza E., 2003. Operon mer: bacterial resistance to mercury and potential for bioremediation of contaminated environments. Genet. Mol. Res. 2: 92–101. [PubMed] [Google Scholar]
- Nguyen Y., Sugiman-Marangos S., Harvey H., Bell S. D., Charlton C. L., et al. , 2015. Pseudomonas aeruginosa minor pilins prime type IVa pilus assembly and promote surface display of the PilY1 adhesin. J. Biol. Chem. 290: 601–611. 10.1074/jbc.M114.616904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie H., Nie M., Yang Y., Zhao J., Zhang X., et al. , 2016. Characterization of phenol metabolization by P. stutzeri N2. Polycycl. Aromat. Compd. 36: 587–600. 10.1080/10406638.2015.1033434 [DOI] [Google Scholar]
- Özen A. I., Ussery D. W., 2012. Defining the Pseudomonas genus: where do we draw the line with Azotobacter? Microb. Ecol. 63: 239–248. 10.1007/s00248-011-9914-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla C. C., Bertagnolli A. D., Bristow L. A., Sarode N., Glass J. B., et al. , 2017. Metagenomic binning recovers a transcriptionally active Gammaproteobacterium linking methanotrophy to partial denitrification in an anoxic oxygen minimum zone. Front. Mar. Sci. 4: 23 10.3389/fmars.2017.00023 [DOI] [Google Scholar]
- Page A. J., Cummins C. A., Hunt M., Wong V. K., Reuter S., et al. , 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31: 3691–3693. 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popowska M., Krawczyk-Balska A., 2013. Broad-host-range incp-1 plasmids and their resistance potential. Front. Microbiol. 4: 44 10.3389/fmicb.2013.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K. D., Tatusova T., Maglott D. R., 2007. Ncbi reference sequences (refseq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35: D61–D65. 10.1093/nar/gkl842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team , 2008. R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Rosselló-Mora R. A., Lalucat J., García-Valdés E., 1994. Comparative biochemical and genetic analysis of naphthalene degradation among Pseudomonas stutzeri strains. Appl. Environ. Microbiol. 60: 966–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl J. W., Caporaso J. G., Rasko D. A., Keim P., 2014. The large-scale blast score ratio (LS-BSR) pipeline: a method to rapidly compare genetic content between bacterial genomes. PeerJ 2: e332 10.7717/peerj.332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T., 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30: 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Sharma A., Sangwan N., Negi V., Kohli P., Khurana J. P., et al. , 2015. Pan-genome dynamics of Pseudomonas gene complements enriched across hexachlorocyclohexane dumpsite. BMC Genomics 16: 313 10.1186/s12864-015-1488-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Bailes E., Grocock R. J., Peden J. F., Sockett R. E., 2005. Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res. 33: 1141–1153. 10.1093/nar/gki242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Tiwary B. N., 2017. Optimization of conditions for polycyclic aromatic hydrocarbons (PAHs) degradation by Pseudomonas stutzeri P2 isolated from Chirimiri coal mines. Biocatal. Agric. Biotechnol. 10: 20–29. 10.1016/j.bcab.2017.02.001 [DOI] [Google Scholar]
- Smith B. A., Dougherty K. M., Baltrus D. A., 2014. Complete genome sequence of the highly transformable Pseudomonas stutzeri strain 28a24. Genome Announc. 2: e00543–14. 10.1128/genomeA.00543-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Zheng S., Nguyen N., Wang Y., Zhou Y., et al. , 2017. Integrated pipeline for inferring the evolutionary history of a gene family embedded in the species tree: a case study on the STIMATE gene family. BMC Bioinformatics 18: 439 10.1186/s12859-017-1850-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., 2014. Raxml version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekorienė R., 2008. Distribution of the genus Pseudomonas bacteria in oil-polluted soil, water, polymeric materials, plant remnants and food products. Ekologija (Liet. Moksl. Akad.) 54: 143–148. 10.2478/V10055-008-0022-0 [DOI] [Google Scholar]
- Wang Y., Feng Y., Cao X., Liu Y., Xue S., 2018. Insights into the molecular mechanism of dehalogenation catalyzed by D-2-haloacid dehalogenase from crystal structures. Sci. Rep. 8: 1454 10.1038/s41598-017-19050-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasi S., Tabrez S., Ahmad M., 2013. Use of Pseudomonas spp. for the bioremediation of environmental pollutants: a review. Environ. Monit. Assess. 185: 8147–8155. 10.1007/s10661-013-3163-x [DOI] [PubMed] [Google Scholar]
- Watkins S. C., Sible E., Putonti C., 2018. Pseudomonas PB1-like phages: Whole genomes from metagenomes offer insight into an abundant group of bacteriophages. Viruses 10: E331. 10.3390/v10060331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winchester J. W., Duce R. A., 1967. The global distribution of iodine, bromine, and chlorine in marine aerosols. Naturwissenschaften 54: 110–113. 10.1007/BF00640572 [DOI] [PubMed] [Google Scholar]
- Wu S., Zheng R., Sha Z., Sun C., 2017. Genome sequence of Pseudomonas stutzeri 273 and identification of the exopolysaccharide EPS273 biosynthesis locus. Mar. Drugs 15: E218. 10.3390/md15070218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Yang J., Chen L., Yang F., Dong J., et al. , 2005. Structural and functional analysis of denitrification genes in Pseudomonas stutzeri A1501. Sci. China C Life Sci. 48: 585 10.1360/062005-45 [DOI] [PubMed] [Google Scholar]
- Yan Y., Yang J., Dou Y., Chen M., Ping S., et al. , 2008. Nitrogen fixation island and rhizosphere competence traits in the genome of root-associated Pseudomonas stutzeri A1501. Proc. Natl. Acad. Sci. USA 105: 7564–7569. 10.1073/pnas.0801093105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Yuan M., Lu W., Yang J., Dai S., et al. , 2011. Complete genome sequence of the nitrogen-fixing and rhizosphere-associated bacterium Pseudomonas stutzeri strain DSM4166. J. Bacteriol. 193: 3422–3423. 10.1128/JB.05039-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zava T.T., Zava D.T., 2011. Assessment of Japanese iodine intake based on seaweed consumption in Japan: a literature-based analysis. Thyroid Res. 4.1: 14. 10.1186/1756-6614-4-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng R., Wu S., Ma N., Sun C., 2018. Genetic and physiological adaptations of marine bacterium Pseudomonas stutzeri 273 to mercury stress. Front. Microbiol. 9: 682 10.3389/fmicb.2018.00682 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The draft genome sequence of Pseudomonas sp. strain SCT has been deposited at GenBank/EMBL/DDBJ under BioProject number PRJDB5044, BioSample number SAMD00059319, and accession number BDJA00000000 (accession range: BDJA01000001-BDJA01000035). The version described in this paper is the first version, BDJA01000000. The raw reads have been deposited in the DDBJ Sequence Read Archive (DRA) under Submission DRA007938, Experiment DRX156286, and Run DRR165667.
Supplementary Table S1 has been deposited via the GSA Figshare portal. The Python scripts used in this study are available through the Github repository https://github.com/haradama/PSCT. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7829321.