Abstract
In Saccharomyces cerevisiae, the meiosis-specific axis proteins Hop1 and Red1 are present nonuniformly across the genome. In a previous study, the meiosis-specific VMA1-derived endonuclease (VDE) was used to examine Spo11-independent recombination in a recombination reporter inserted in a Hop1/Red1-enriched region (HIS4) and in a Hop1/Red1-poor region (URA3). VDE-initiated crossovers at HIS4 were mostly dependent on Mlh3, a component of the MutLγ meiotic recombination intermediate resolvase, while VDE-initiated crossovers at URA3 were mostly Mlh3-independent. These differences were abolished in the absence of the chromosome axis remodeler Pch2, and crossovers at both loci became partly Mlh3-dependent. To test the generality of these observations, we examined inserts at six additional loci that differed in terms of Hop1/Red1 enrichment, chromosome size, and distance from centromeres and telomeres. All six loci behaved similarly to URA3: the vast majority of VDE-initiated crossovers were Mlh3-independent. This indicates that, counter to previous suggestions, levels of meiotic chromosome axis protein enrichment alone do not determine which recombination pathway gives rise to crossovers during VDE-initiated meiotic recombination. In pch2∆ mutants, the fraction of VDE-induced crossovers that were Mlh3-dependent increased to levels previously observed for Spo11-initiated crossovers in pch2∆, indicating that Pch2-dependent processes play an important role in controlling the balance between MutLγ-dependent and MutLγ-independent crossovers.
Keywords: meiosis, recombination, budding yeast, PCH2, MLH3
During meiosis, the crossover products of recombination form stable links between homologous chromosomes of different parental origin (homologs), to enable their proper segregation during the meiotic divisions (reviewed in Zickler and Kleckner 1999; Whitby 2005). Meiotic recombination is initiated by DNA double strand breaks (DSBs) that are formed by the meiosis-specific Spo11 protein (Bergerat et al. 1997; Keeney 2001). In budding yeast, Spo11 DSBs are unevenly distributed in the genome. Most DSB-rich regions correlate with domains that are enriched for the meiosis-specific chromosome axis proteins, Red1 and Hop1, which play an important role in DSB formation (Hollingsworth and Ponte 1997; Blat et al. 2002; Pan et al. 2011; Panizza et al. 2011; Smagulova et al. 2011; Baker et al. 2014). Pch2, a conserved hexameric AAA+ ATPase, remodels Hop1 to maintain its non-uniform distribution (San-Segundo and Roeder 1999; Chen et al. 2014). In budding yeast pch2 mutants, Hop1 persists longer and is more uniformly distributed on chromosomes; this is accompanied by a delay in meiotic progression and changes in the level and the distribution of COs and late-forming DSBs (Börner et al. 2008; Joshi et al. 2009; Zanders and Alani 2009; Lambing et al. 2015; Subramanian et al. 2016; Subramanian et al. 2019). Similar phenotypes are observed in mouse and Arabidopsis mutants lacking Pch2 homologs (Wojtasz et al. 2009; Roig et al. 2010; Lambing et al. 2015).
Meiotic DSBs are also important for homolog colocalization, pairing and synapsis (Keeney et al. 1997; Romanienko and Camerini-Otero 2000; Baudat et al. 2013). Current thinking is that most DSBs are repaired either by a synthesis-dependent strand annealing pathway that forms non-crossovers (NCOs), or by a pathway that forms double Holiday junction (dHJ) intermediates that are resolved as crossovers (COs) by the MutLγ (Mlh1-Mlh3 and Exo1) meiosis-specific resolvase (Schwacha and Kleckner 1994; Wang et al. 1999; Khazanehdari and Borts 2000; Kirkpatrick et al. 2000; Tsubouchi and Ogawa 2000; Allers and Lichten 2001b; Allers and Lichten 2001a; Hoffmann et al. 2003; Argueso et al. 2004; Bishop and Zickler 2004; Nishant et al. 2008; Zakharyevich et al. 2010; Al-Sweel et al. 2017). In budding yeast, COs and NCOs are formed at similar levels, suggesting that roughly equal fractions of DSBs are repaired by these two pathways (Martini et al. 2006; Mancera et al. 2008). Apart from these two major pathways, a minor pathway uses mitotic resolvases (structure-selective nucleases, SSNs: Mus81-Mms4, Yen1 and Slx1-4) to form both NCOs and COs (De Los Santos et al. 2003; Argueso et al. 2004; Lynn et al. 2007; Jessop and Lichten 2008; De Muyt et al. 2012; Zakharyevich et al. 2012; Agostinho et al. 2013; Oke et al. 2014). While the proteins and enzymatic activities contributing to each of these pathways has been the subject of considerable study (reviewed in Ehmsen and Heyer 2008; Hunter 2015; Manhart and Alani 2016), the question of what roles local chromosome environment might play in pathway choice remains much less explored. Medhi et al. (2016) addressed this question using a meiosis-specific endonuclease, VDE, that cleaves a recognition sequence (VRS) at high efficiency regardless of chromosomal context (Gimble and Thorner 1992; Gimble and Thorner 1993; Nogami et al. 2002; Fukuda et al. 2003; Medhi et al. 2016; this work). Like Spo11 DSBs, VDE DSBs are processed to form single-stranded overhangs that recruit the Rad51 and Dmc1 proteins that perform strand invasion and homology search (Bishop et al. 1992; Fukuda et al. 2003; Fukuda and Ohya 2006). Medhi et al. inserted a VRS-containing recombination reporter at two loci: HIS4, present in a region with high levels of both Spo11 DSBs and Hop1 binding; and URA3, in a region with low levels of Spo11 DSBs and Hop1 binding (Pan et al. 2011; Panizza et al. 2011). Most COs at HIS4 were Mlh3-dependent, while COs at URA3 were Mlh3-independent. In pch2∆ mutants, Hop1 occupancy at HIS4 was reduced, as were the fraction of COs that were Mlh3-dependent, while at URA3 the fraction of COs that were Mlh3-dependent increased. Based on these findings, Medhi et al. suggested that the local chromosome structure, in particular levels of Hop1 enrichment, may be an important determinant of CO pathway choice.
To test the generality of the above suggestion, we inserted the same VRS recombination reporter at six new loci with varying Hop1 occupancy in their vicinity and found that VDE-initiated meiotic COs at all six new loci were predominantly Mlh3-independent. Moreover, as previously seen for inserts at URA3 (Medhi et al. 2016), pch2∆ mutation increased the fraction of COs that were Mlh3-dependent. These results indicate that, contrary to our previous suggestion, local Hop1 occupancy levels alone do not determine the mechanism of JM resolution during the formation of VDE-induced meiotic crossovers. They also suggest that, at most loci, VDE DSBs are repaired differently than are Spo11 DSBs.
Materials and Methods
Yeast strains
All strains (Table S1) used in this study are of SK1 background (Kane and Roth 1974) and were constructed by transformation or genetic crosses. The recombination reporter cassette with the VRS (cleavable) or VRS-103 (uncleavable) site in the ARG4 gene (Medhi et al. 2016) were inserted by ends-out transformation (for VRS-containing inserts and for VRS-103 inserts at FIR1 and HSP30, Figure S1A) or by ends-in transformation (for VRS-103 constructs at CCT6, RIM15, IMD3 and TRK2, Figure S1B) at six different locations, using primers listed in Table S2. Ends-in transformation was used for inserts at divergently transcribed loci to minimize effects on expression caused by disruption of 5′ untranslated regions. Transformation was performed with overlapping DNA fragments as illustrated in Figure S1. The VRS-arg4 and VRS-103-arg4 constructs are 5.5kb and 8.6kb long, respectively, with ∼3kb sequence homology around the VRS site. This size difference, along with HindIII site differences, enables the detection of the parental and recombinant chromosomes on Southern blots (see Figure 2, below).
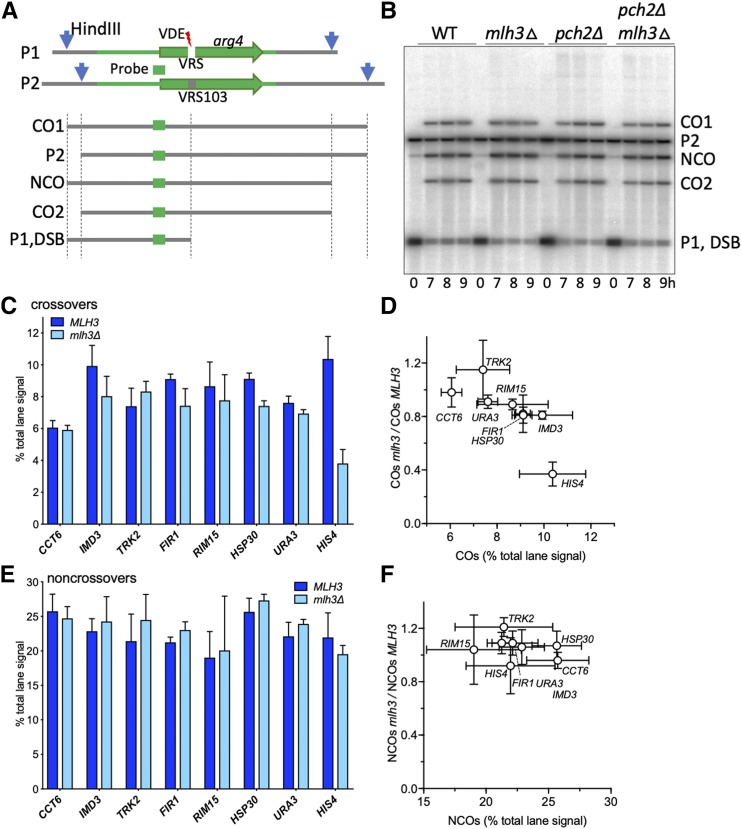
Figure 2.
VDE-initiated crossovers at most loci are MutLγ-independent. (A) Strategy for detection of VDE-initiated COs and NCOs. A cartoon of the VRS and VRS-103 inserts is shown, illustrating: white box—VRS sequences; blue arrows—HindIII restriction sites; green lines—sequences shared between the two inserts, with ARG4 coding sequences shown as a green arrow; green box—sequences used for Southern blot probes. Digestion with HindIII and PI-SceI (VDE) distinguishes parental (P1 and P2), CO and NCO products. VDE-cut inserts are not distinguished from parent P1 in these digests, but can be distinguished in digests with HindIII alone (Medhi et al. 2016). (B) Representative Southern blot containing DNA from strains with inserts at RIM15. (C) VDE-initiated COs in MLH3 and mlh3∆ cells. CO frequencies, average signal of CO1 and CO2 for 8 and 9 h samples from three independent experiments for inserts at HIS4 and from two independent experiments for inserts at all other loci. Data for inserts at URA3 and for two experiments with inserts at HIS4 are from Medhi et al. (2016). (D) fraction of COs that are MutLγ-independent (ratio of CO frequencies in mlh3∆ vs. MLH3), plotted as a function of CO frequencies in MLH3 strains. CO frequencies in MLH3 and mlh3∆ differ significantly only for inserts at HSP30 and HIS4 (adjusted p values of 0.003 and 0.0001, respectively) (E,F) VDE-initiated NCOs, details as in (B) and (C); frequencies in MLH3 and mlh3∆ do not differ significantly at any locus (adjusted p values ≥ 0.05). Error bars in all panels denote standard deviation. See Figure S2C for summary plots with CO and NCO values for all genotypes.
Growth and sporulation
Strains were grown in pre-sporulation SPS medium and transferred to sporulation medium as described (Goyon and Lichten 1993), with the inclusion of 10µM CuSO4 in sporulation medium to induce VDE expression (Medhi et al. 2016). DNA samples were collected and processed as described (Allers and Lichten 2000; Jessop et al. 2005; Jessop et al. 2006).
DNA extraction and Southern hybridization
DNA was extracted from samples using the CTAB extraction method (Allers and Lichten 2000; Oh et al. 2009). Genomic DNA was digested with HindIII or HindIII and PI-SceI, run on agarose gels, blotted, probed and analyzed as described (Medhi et al. 2016).
Cytology
Cells were collected, stained with DAPI, and scored by epifluoresence microscopy to follow nuclear divisions as described (Kaur et al. 2018).
Statistical analysis
GraphPad Prism was used for comparisons of mean values, using two-tailed t-tests with the Holm-Šídák correction for multiple comparisons.
Data availability
All strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of this article are represented fully within the article, tables, figures, and supplementary figures, tables and files. Data underlying graphs in all figures and chromosome coordinates in Figure 1 are in File S1, available at Figshare (https://doi.org/10.25387/g3.7800728). Other supplementary files, available at the same URL, include:
Figure S1: Construction of inserts
Figure S2: Additional data and analyses. Includes cells completing meiosis I, timing of VDE DSBs, combined CO and NCO data, and NCO/CO ratios
Table S1: Strain genotypes
Table S2: Primers for all reporter inserts.
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7800728.
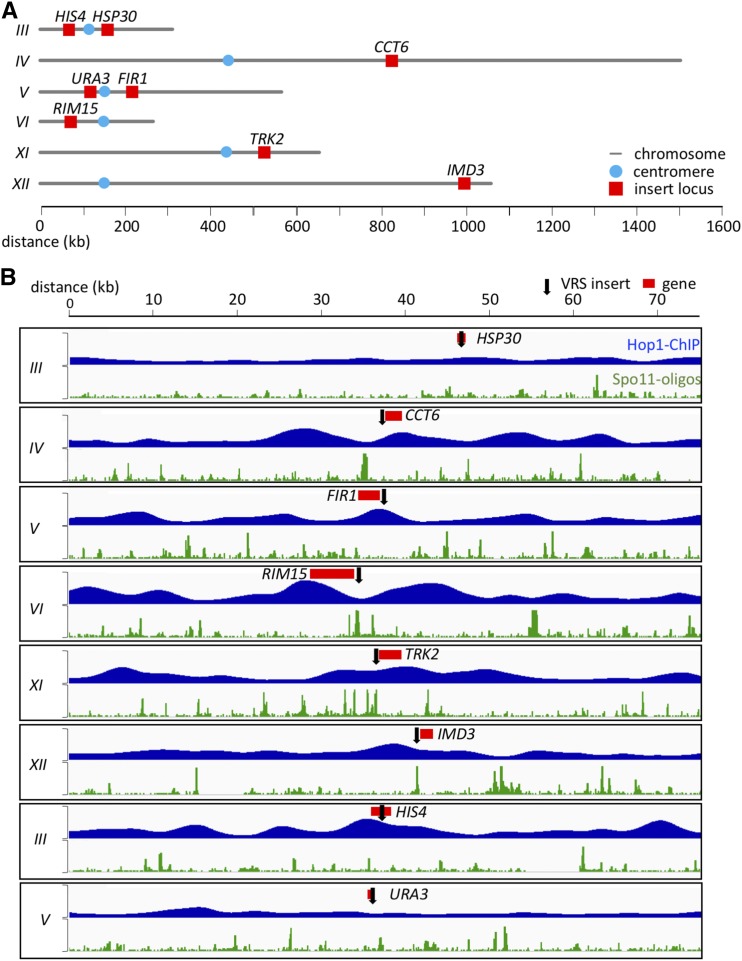
Figure 1.
Insert loci examined. Inserts at HIS4 and URA3 were previously studied by Medhi et al. (2016). (A) Locations of insert loci are illustrated (red). Blue circles denote centromere locations. (B) Maps of regions surrounding insert loci. Red—coding region of gene used to identify each insert; black arrow—site of VRS insert. Blue plots show relative Hop1 occupancy levels in mid meiosis, using smoothed ChIP-chip data from (Panizza et al. 2011); vertical scale = 0-7, decile-normalized ChIP/WCE. Green plots show relative DSB levels, using Spo11-oligo reads from Pan et al. (2011); vertical scale = 0-15 hits per million/base-pair. Chromosome coordinates and average Hop1 occupancy and Spo11-oligo reads in 2, 10, and 20 kb regions around each insert are given in File S1.
Results and Discussion
VDE-initiated COs are Mlh3-independent at most insert sites
To further test the hypothesis that Hop1-enrichment determines the MutLγ-dependence of meiotic CO formation, six new sites were selected for VRS reporter insertion, one (HSP30) with regional Hop1 levels (average Hop1 occupancy over 10-20kb around the insert location) similar to those at URA3, four (CCT6, RIM15, TRK2 and IMD3) with Hop1 levels similar to those at HIS4, and one (FIR1) with intermediate Hop1 levels (Figure 1B, File S1). Since it has been previously shown that Spo11-DSBs are reduced near centromeres and telomeres (Pan et al. 2011) and CO formation is regulated differently on longer and shorter chromosomes (Joshi et al. 2009; Zanders and Alani 2009), the new sites were selected such that they were on chromosomes of different sizes and were at varying distances from centromeres and telomeres (Figure 1A, File S1). At each site, recombination products can be differentiated on Southern blots (Figure 2A, B), as was previously used to quantify DSBs, COs and NCOs (Medhi et al. 2016).
Meiotic progression of all WT and mlh3∆ strains was similar, with most cells completing the first meiotic division by 7-8h post-induction (Figure S2A). In addition, VDE-initiated DSBs appeared and disappeared with levels and timing similar to those previously seen at HIS4 and URA3 (Figure S2B; Medhi et al. 2016).
COs in VRS inserts ranged from ∼6% of total lane signal at CCT6 to ∼10.3% at HIS4 (Figure 2C). As previously reported (Medhi et al. 2016), NCOs were recovered in substantial excess over COs at all insert loci (Figure 2E), with NCO/CO ratios ranging from 2.1 to 4.8 (mean = 3.1 ± 0.8; Figure S2D). The marked excess of NCOs over COs seen for VDE-initiated events differs from what is seen with Spo11-initiated events, where COs and NCOs are produced at similar levels (Martini et al. 2006; Mancera et al. 2008; Zakharyevich et al. 2012). In contrast to what was seen for VRS inserts at HIS4, where COs were reduced dramatically in mlh3∆ mutants (to ∼40% of wild-type levels), COs in the same sequences inserted at all other loci were only modestly affected, with COs in mlh3∆ ranging from ∼80% to ∼115% of wild type (mean = 91 ± 12%; Figure 2D); NCOs were similarly unaffected (Figure 2E, F). These results indicate that, in contrast to Spo11-initiated COs, which are reduced about twofold in mlh3∆ mutants (Wang et al. 1999; Khazanehdari and Borts 2000; Kirkpatrick et al. 2000; Tsubouchi and Ogawa 2000; Hoffmann et al. 2003; Argueso et al. 2004; Nishant et al. 2008; Al-Sweel et al. 2017; Chakraborty et al. 2017), most COs at the VDE break sites are formed independent of MutLγ, irrespective of the chromosome size, distance from centromere or telomere, or Hop1-enrichment in their vicinity. Thus, at most insert loci in otherwise wild-type cells, VDE-initiated recombination differs from Spo11-initiated recombination and more closely resembles mitotic recombination, in that NCOs are in excess over COs (Esposito 1978; Lichten and Haber 1989; Ira et al. 2003; Dayani et al. 2011) and, with the exception of those formed in inserts at HIS4, VDE-initiated COs are largely MutLγ-independent.
VDE-initiated COs are partially Mlh3-dependent in pch2Δ mutants
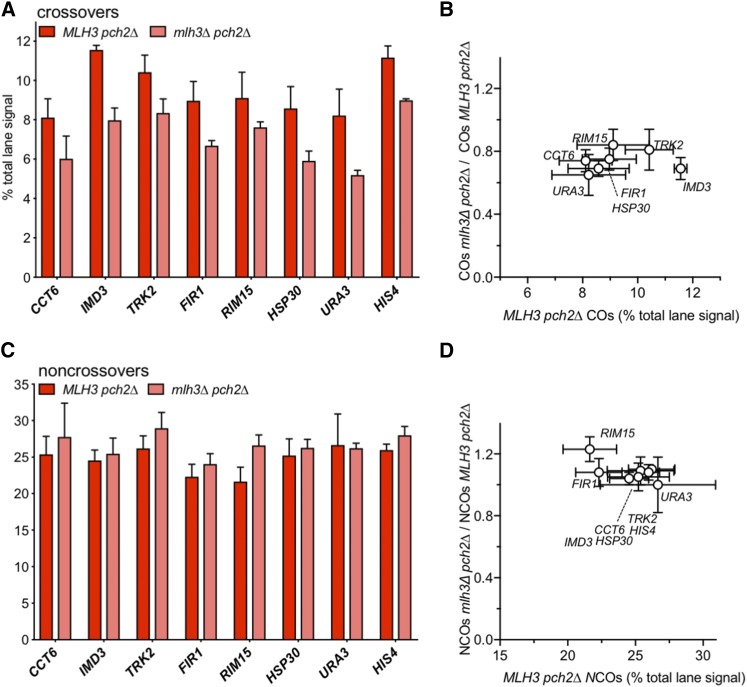
In pch2 mutants, meiotic axis proteins are redistributed, with less pronounced differences in Hop1 occupancy distributions measured either cytologically (Börner et al. 2008; Joshi et al. 2009) or by chromatin-immunoprecipitation (Medhi et al. 2016; Subramanian et al. 2019). Previously, we found that the absence of Pch2 did not substantially alter overall NCO or CO levels at HIS4 and URA3, but the Mlh3-dependence of CO formation was affected at both loci, with Mlh3-independent COs increasing at HIS4 and decreasing at URA3. Because the six new VRS insert loci studied here are similar to URA3, in that most VDE-initiated COs are Mlh3-independent, we wanted to see if COs at these loci also displayed increased Mlh3-dependence in pch2∆ mutants.
Consistent with previous findings (Börner et al. 2008), meiotic divisions were delayed in pch2∆ and pch2∆ mlh3∆ mutants relative to wild type (Figure S2A). Frequencies of NCOs at all eight VRS insert loci in the pch2∆ were similar to those seen in wild type (Figures 3C and S2C; pch2∆/PCH2 = 111 ± 10%), as were COs (Figures 3A and S2C; pch2∆/PCH2 = 113 ± 16%). Loss of Mlh3 did not substantially affect NCOs (Figure 3C; pch2∆ mlh3∆/pch2∆ MLH3 = 114 ± 14%). However, in pch2∆ mlh3∆ double mutants, COs were reduced 20–35% relative to pch2∆ MLH3 (Figure 3B; average pch2∆ mlh3∆/pch2∆ = 74 ± 7%), as was previously observed for inserts at URA3 and HIS4 (Medhi et al. 2016). A quantitatively similar MutLγ-dependence has also been observed for Spo11-initiated COs in pch2∆ mutants, both genome-wide (pch2∆ mlh3∆ / pch2∆ = 73%; Chakraborty et al. 2017) and for individual genetic intervals (pch2∆ mlh3∆ / pch2∆ = ∼75%, calculated from combined data of Nishant et al. 2008; Zanders and Alani 2009; Al-Sweel et al. 2017; Chakraborty et al. 2017). Thus, the absence of Pch2 increases the MutLγ-dependence of VDE-initiated COs at most loci, while decreasing the MutLγ-dependence of VDE-initiated COs at HIS4 and of Spo11-initiated COs.
Figure 3.
VDE-initiated crossovers in pch2∆ mutants are partially MutLγ-dependent. (A) VDE-initiated COs in MLH3 pch2∆ and mlh3∆ pch2∆ cells. (A) CO frequencies, average signal of CO1 and CO2 for 8 and 9 h samples from two independent experiments. For inserts at CCT6, IMD3, FIR1 and RIM15, 9 h values are from a single experiment. Data for inserts at HIS4 and URA3 are from Medhi et al. (2016). (B) fraction of COs that are MutLγ-independent (ratio of CO frequencies in mlh3∆ vs. MLH3), plotted as a function of CO frequencies in MLH3 strains. CO frequencies in MLH3 pch2∆ and mlh3∆ pch2∆ differ significantly for inserts at all loci (adjusted p values ≤ 0.03) except CCT6 and RIM15 (adjusted p values of 0.06 and 0.07, respectively). (C, D) VDE-initiated NCOs, as in panels (A) and (B). NCO frequencies in MLH3 pch2∆ and mlh3∆ pch2∆ do not differ significantly for any locus (adjusted p values ≥ 0.05). Error bars in all panels denote standard deviation. See Figure S2C for summary plots with CO and NCO values for all genotypes.
Spo11-initiated COs are reduced about twofold in mutants lacking MutLγ; this is thought to reflect unbiased JM resolution by SSNs to form both COs and NCOs, as opposed to MutLγ-mediated biased JM resolution as COs in wild type (Argueso et al. 2004; Zakharyevich et al. 2012). If the same holds true for pch2 mutants, the ∼25% reduction in COs seen in pch2∆ mlh3∆ would suggest that about half of the COs formed in pch2MLH3 cells are the products of MutLγ-mediated resolution, regardless of whether they were initiated by VDE or by Spo11. It therefore appears that Pch2, or processes regulated by it, prevents VDE-initiated events from forming MutLγ-dependent COs.
Summary and concluding remarks
In this study, we examined VDE-initiated meiotic recombination in a recombination reporter inserted at six loci in addition to the two loci (HIS4 and URA3) originally examined by Medhi et al. (2016). With the exception of HIS4, VDE-initiated COs at all insert loci were largely Mlh3-independent, regardless of whether inserts were at loci in Hop1-enriched or Hop1-depleted regions of the genome. Therefore, our previous hypothesis, that local Hop1 occupancy determines mechanisms of CO formation, is inaccurate, at least for VDE-initiated recombination, in that it was based on analysis of inserts at a locus (HIS4) that appears to be exceptional. We currently do not understand why the genetic dependence of VDE-induced CO formation at HIS4 differs from that seen at the other loci examined, but cannot rule out other effects or interactions on the basis of current data.
The observation that VDE-initiated COs at most insert loci are Mlh3-independent, in turn, raises the question of whether or not VDE-initiated recombination events that occur in cells undergoing meiosis can be properly described as being “meiotic”. VDE-initiated NCOs are recovered in excess of COs (2 to 5-fold, average 3.2 ± 0.1), which is reminiscent of, although less than, the 5 to 20-fold excess of NCOs over COs seen in budding yeast mitotic recombination (Esposito 1978; Lichten and Haber 1989; Ira et al. 2003; Bzymek et al. 2010; Dayani et al. 2011). VDE-initiated DSB processing also resembles DSB processing in the mitotic cell cycle, in that break ends are continuously resected over time (Lee et al. 1998; Neale et al. 2002; Johnson et al. 2007), unlike the constrained resection seen with Spo11 DSBs (Mimitou et al. 2017). Finally, unlike Spo11, VDE frequently cuts both sister chromatids in a single meiosis (Gimble and Thorner 1992; Gimble and Thorner 1993; Medhi et al. 2016), and gene conversion of both sister chromatids is associated with a reduced CO/NCO ratio among HO endonuclease-initiated meiotic recombinants (Malkova et al. 2000). Further studies will be necessary to determine which of these or other factors are responsible for the marked Mlh3-independence of VDE-initiated COs at seven of the eight insert locations examined, and why the majority of VDE-initiated COs at HIS4 are Mlh3-dependent.
In contrast, in pch2∆ strains, VDE-initiated COs show the same Mlh3-dependence as Spo11-initiated COs, regardless of wild-type Hop1 occupancy levels around insert loci. It therefore seems unlikely that Hop1 redistribution in pch2∆ mutants is the only factor responsible for the increased Mlh3-dependence of COs at most insert loci and the decreased Mlh3-dependence of COs at HIS4. Homolog synapsis, recombinant formation and meiotic divisions are all delayed in pch2∆ mutants; pch2∆ mutants also display a more even distribution of the Zip1 central element protein along chromosomes and reduced CO interference (Börner et al. 2008; Joshi et al. 2009; Zanders and Alani 2009). These or other pch2∆ mutant defects might delay either recruitment of factors promoting MutLγ action at Spo11-initiated events or implementation of CO interference, thus increasing the window of opportunity for these factors to act at VDE-initiated events. It also has been suggested that Pch2-dependent remodeling affects the stability of recombination intermediates (Deshong et al. 2014), and it is possible that this might differentially affect events not initiated by Spo11. Exogenous DNA damage is unable to fully rescue spo11 mutant phenotypes in several organisms (Thorne and Byers 1993; Celerin et al. 2000; Romanienko and Camerini-Otero 2000; Loidl and Mochizuki 2009; Carofiglio et al. 2018), and budding yeast responds differently during meiosis to DSBs induced by Spo11 and to DSBs formed by exogenous DNA damage (Cartagena-Lirola et al. 2008; reviewed in Longhese et al. 2008). Our current data suggest that Pch2 might implement mechanisms that distinguish Spo11-initiated recombination events from those initiated by other forms of DNA damage.
In summary, the data presented here indicate that VDE-initiated recombination events are treated differently than are those initiated by Spo11 during wild-type meiosis. VDE-initiated events produce an excess of NCOs over COs and, at seven of eight loci examined, form COs by MutLγ-independent mechanisms, and thus their outcome more closely resembles those of DSB repair events that occur during the mitotic cell cycle. We conclude that the full spectrum of meiotic recombination processes that occur at Spo11-initiated DSBs do not occur at VDE-initiated DSBs, and, by inference, DSBs formed during meiosis by other nucleases. Thus, our findings call for caution in the use of DSBs formed by this nuclease, or by other exogenous means, for inferring factors that control normal meiotic recombination.
Acknowledgments
We thank Jean Paul Ouyan, Seyoun Kim, and Matan Cohen for help in strain construction, and Jasvinder Ahuja, Needhi Bhalla, Matan Cohen, Julia Cooper, and Martin Xaver for comments and discussion. This work was supported by the Intramural Research Program of the NIH through the Center for Cancer Research at the National Cancer Institute.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7800728.
Communicating editor: K. McKim
Literature Cited
- Agostinho A., Meier B., Sonneville R., Jagut M., Woglar A., et al. , 2013. Combinatorial regulation of meiotic Holliday junction resolution in C. elegans by HIM-6 (BLM) helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 nucleases. PLoS Genet. 9: e1003591 (erratum: PLoS Genet. 9: 10.1371/annotation/d8c73205-151d-4e22-89c6-3aa574037d10). 10.1371/journal.pgen.1003591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sweel N., Raghavan V., Dutta A., Ajith V. P., Di Vietro L., et al. , 2017. mlh3 mutations in baker’s yeast alter meiotic recombination outcomes by increasing noncrossover events genome-wide. PLoS Genet. 13: e1006974 (erratum: PLOS Genet. 13: e1007067). 10.1371/journal.pgen.1006974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T., Lichten M., 2000. A method for preparing genomic DNA that restrains branch migration of Holliday junctions. Nucleic Acids Res. 28: e6 10.1093/nar/28.2.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T., Lichten M., 2001a Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57. 10.1016/S0092-8674(01)00416-0 [DOI] [PubMed] [Google Scholar]
- Allers T., Lichten M., 2001b Intermediates of yeast meiotic recombination contain heteroduplex DNA. Mol. Cell 8: 225–231. 10.1016/S1097-2765(01)00280-5 [DOI] [PubMed] [Google Scholar]
- Argueso J. L., Wanat J., Gemici Z., Alani E., 2004. Competing crossover pathways act during meiosis in Saccharomyces cerevisiae. Genetics 168: 1805–1816. 10.1534/genetics.104.032912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. L., Walker M., Kajita S., Petkov P. M., Paigen K., 2014. PRDM9 binding organizes hotspot nucleosomes and limits Holliday junction migration. Genome Res. 24: 724–732. 10.1101/gr.170167.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F., Imai Y., de Massy B., 2013. Meiotic recombination in mammals: localization and regulation. Nat. Rev. Genet. 14: 794–806. 10.1038/nrg3573 [DOI] [PubMed] [Google Scholar]
- Bergerat A., de Massy B., Gadelle D., Varoutas P. C., Nicolas A., et al. , 1997. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386: 414–417. 10.1038/386414a0 [DOI] [PubMed] [Google Scholar]
- Bishop D. K., Park D., Xu L., Kleckner N., 1992. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69: 439–456. 10.1016/0092-8674(92)90446-J [DOI] [PubMed] [Google Scholar]
- Bishop D. K., Zickler D., 2004. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117: 9–15. 10.1016/S0092-8674(04)00297-1 [DOI] [PubMed] [Google Scholar]
- Blat Y., Protacio R. U., Hunter N., Kleckner N., 2002. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111: 791–802. 10.1016/S0092-8674(02)01167-4 [DOI] [PubMed] [Google Scholar]
- Börner G. V., Barot A., Kleckner N., 2008. Yeast Pch2 promotes domainal axis organization, timely recombination progression, and arrest of defective recombinosomes during meiosis. Proc. Natl. Acad. Sci. USA 105: 3327–3332. 10.1073/pnas.0711864105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzymek M., Thayer N. H., Oh S. D., Kleckner N., Hunter N., 2010. Double Holliday junctions are intermediates of DNA break repair. Nature 464: 937–941. 10.1038/nature08868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carofiglio F., Sleddens-Linkels E., Wassenaar E., Inagaki A., van Cappellen W. A., et al. , 2018. Repair of exogenous DNA double-strand breaks promotes chromosome synapsis in SPO11-mutant mouse meiocytes, and is altered in the absence of HORMAD1. DNA Repair (Amst.) 63: 25–38. 10.1016/j.dnarep.2018.01.007 [DOI] [PubMed] [Google Scholar]
- Cartagena-Lirola H., Guerini I., Manfrini N., Lucchini G., Longhese M. P., 2008. Role of the Saccharomyces cerevisiae Rad53 checkpoint kinase in signaling double-strand breaks during the meiotic cell cycle. Mol. Cell. Biol. 28: 4480–4493. 10.1128/MCB.00375-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celerin M., Merino S. T., Stone J. E., Menzie A. M., Zolan M. E., 2000. Multiple roles of Spo11 in meiotic chromosome behavior. EMBO J. 19: 2739–2750. 10.1093/emboj/19.11.2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty P., Pankajam A. V., Lin G., Dutta A., Krishnaprasad G. N., et al. , 2017. Modulating crossover frequency and interference for obligate crossovers in Saccharomyces cerevisiae meiosis. G3 (Bethesda) 7: 1511–1524. 10.1534/g3.117.040071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Jomaa A., Ortega J., Alani E. E., 2014. Pch2 is a hexameric ring ATPase that remodels the chromosome axis protein Hop1. Proc. Natl. Acad. Sci. USA 111: E44–E53. 10.1073/pnas.1310755111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayani Y., Simchen G., Lichten M., 2011. Meiotic recombination intermediates are resolved with minimal crossover formation during return-to-growth, an analogue of the mitotic cell cycle. PLoS Genet. 7: e1002083 10.1371/journal.pgen.1002083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T., Hunter N., Lee C., Larkin B., Loidl J., et al. , 2003. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164: 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A., Jessop L., Kolar E., Sourirajan A., Chen J., et al. , 2012. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell 46: 43–53. 10.1016/j.molcel.2012.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshong A. J., Ye A. L., Lamelza P., Bhalla N., 2014. A quality control mechanism coordinates meiotic prophase events to promote crossover assurance. PLoS Genet. 10: e1004291 10.1371/journal.pgen.1004291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehmsen K. T., Heyer W. D., 2008. Biochemistry of meiotic recombination: formation, processing, and resolution of recombination intermediates. Genome Dyn. Stab. 3: 91–164. 10.1007/7050_2008_039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito M. S., 1978. Evidence that spontaneous mitotic recombination occurs at the two-strand stage. Proc. Natl. Acad. Sci. USA 75: 4436–4440. 10.1073/pnas.75.9.4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T., Nogami S., Ohya Y., 2003. VDE-initiated intein homing in Saccharomyces cerevisiae proceeds in a meiotic recombination-like manner. Genes Cells 8: 587–602. 10.1046/j.1365-2443.2003.00659.x [DOI] [PubMed] [Google Scholar]
- Fukuda T., Ohya Y., 2006. Recruitment of RecA homologs Dmc1p and Rad51p to the double-strand break repair site initiated by meiosis-specific endonuclease VDE (PI-SceI). Mol. Genet. Genomics 275: 204–214. 10.1007/s00438-005-0078-4 [DOI] [PubMed] [Google Scholar]
- Gimble F. S., Thorner J., 1992. Homing of a DNA endonuclease gene by meiotic gene conversion in Saccharomyces cerevisiae. Nature 357: 301–306. 10.1038/357301a0 [DOI] [PubMed] [Google Scholar]
- Gimble F. S., Thorner J., 1993. Purification and characterization of VDE, a site-specific endonuclease from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 268: 21844–21853. [PubMed] [Google Scholar]
- Goyon C., Lichten M., 1993. Timing of molecular events in meiosis in Saccharomyces cerevisiae: stable heteroduplex DNA is formed late in meiotic prophase. Mol. Cell. Biol. 13: 373–382. 10.1128/MCB.13.1.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E. R., Shcherbakova P. V., Kunkel T. A., Borts R. H., 2003. MLH1 mutations differentially affect meiotic functions in Saccharomyces cerevisiae. Genetics 163: 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N. M., Ponte L., 1997. Genetic interactions between HOP1, RED1 and MEK1 suggest that MEK1 regulates assembly of axial element components during meiosis in the yeast Saccharomyces cerevisiae. Genetics 147: 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N., 2015. Meiotic recombination: the essence of heredity. Cold Spring Harb. Perspect. Biol. 7: a016618 10.1101/cshperspect.a016618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G., Malkova A., Liberi G., Foiani M., Haber J. E., 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115: 401–411. 10.1016/S0092-8674(03)00886-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L., Allers T., Lichten M., 2005. Infrequent co-conversion of markers flanking a meiotic recombination initiation site in Saccharomyces cerevisiae. Genetics 169: 1353–1367. 10.1534/genetics.104.036509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L., Lichten M., 2008. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol. Cell 31: 313–323. 10.1016/j.molcel.2008.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L., Rockmill B., Roeder G. S., Lichten M., 2006. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of Sgs1. PLoS Genet. 2: e155 10.1371/journal.pgen.0020155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R., Borde V., Neale M. J., Bishop-Bailey A., North M., et al. , 2007. Excess single-stranded DNA inhibits meiotic double-strand break repair. PLoS Genet. 3: e223 10.1371/journal.pgen.0030223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N., Barot A., Jamison C., Börner G. V., 2009. Pch2 links chromosome axis remodeling at future crossover sites and crossover distribution during yeast meiosis. PLoS Genet. 5: e1000557 10.1371/journal.pgen.1000557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane S. M., Roth R., 1974. Carbohydrate metabolism during ascospore development in yeast. J. Bacteriol. 118: 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H., Ahuja J. S., Lichten M., 2018. Methods for controlled protein depletion to study protein function during meiosis. Methods Enzymol. 601: 331–357. 10.1016/bs.mie.2017.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S., 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52: 1–53. 10.1016/S0070-2153(01)52008-6 [DOI] [PubMed] [Google Scholar]
- Keeney S., Giroux C. N., Kleckner N., 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. 10.1016/S0092-8674(00)81876-0 [DOI] [PubMed] [Google Scholar]
- Khazanehdari K. A., Borts R. H., 2000. EXO1 and MSH4 differentially affect crossing-over and segregation. Chromosoma 109: 94–102. 10.1007/s004120050416 [DOI] [PubMed] [Google Scholar]
- Kirkpatrick D. T., Ferguson J. R., Petes T. D., Symington L. S., 2000. Decreased meiotic intergenic recombination and increased meiosis I nondisjunction in exo1 mutants of Saccharomyces cerevisiae. Genetics 156: 1549–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambing C., Osman K., Nuntasoontorn K., West A., Higgins J. D., et al. , 2015. Arabidopsis PCH2 mediates meiotic chromosome remodeling and maturation of crossovers. PLoS Genet. 11: e1005372 10.1371/journal.pgen.1005372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. E., Moore J. K., Holmes A., Umezu K., Kolodner R. D., et al. , 1998. Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94: 399–409. 10.1016/S0092-8674(00)81482-8 [DOI] [PubMed] [Google Scholar]
- Lichten M., Haber J. E., 1989. Position effects in ectopic and allelic mitotic recombination in Saccharomyces cerevisiae. Genetics 123: 261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J., Mochizuki K., 2009. Tetrahymena meiotic nuclear reorganization is induced by a checkpoint kinase-dependent response to DNA damage. Mol. Biol. Cell 20: 2428–2437. 10.1091/mbc.e08-10-1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese M. P., Guerini I., Baldo V., Clerici M., 2008. Surveillance mechanisms monitoring chromosome breaks during mitosis and meiosis. DNA Repair (Amst.) 7: 545–557. 10.1016/j.dnarep.2007.09.006 [DOI] [PubMed] [Google Scholar]
- Lynn A., Soucek R., Borner G. V., 2007. ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 15: 591–605. 10.1007/s10577-007-1150-1 [DOI] [PubMed] [Google Scholar]
- Malkova A., Klein F., Leung W. Y., Haber J. E., 2000. HO endonuclease-induced recombination in yeast meiosis resembles Spo11-induced events. Proc. Natl. Acad. Sci. USA 97: 14500–14505. 10.1073/pnas.97.26.14500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E., Bourgon R., Brozzi A., Huber W., Steinmetz L. M., 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454: 479–485. 10.1038/nature07135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manhart C. M., Alani E., 2016. Roles for mismatch repair family proteins in promoting meiotic crossing over. DNA Repair (Amst.) 38: 84–93. 10.1016/j.dnarep.2015.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini E., Diaz R. L., Hunter N., Keeney S., 2006. Crossover homeostasis in yeast meiosis. Cell 126: 285–295. 10.1016/j.cell.2006.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medhi D., Goldman A. S., Lichten M., 2016. Local chromosome context is a major determinant of crossover pathway biochemistry during budding yeast meiosis. eLife 5 10.7554/eLife.19669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E. P., Yamada S., Keeney S., 2017. A global view of meiotic double-strand break end resection. Science 355: 40–45. 10.1126/science.aak9704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale M. J., Ramachandran M., Trelles-Sticken E., Scherthan H., Goldman A. S., 2002. Wild-type levels of Spo11-induced DSBs are required for normal single-strand resection during meiosis. Mol. Cell 9: 835–846. 10.1016/S1097-2765(02)00498-7 [DOI] [PubMed] [Google Scholar]
- Nishant K. T., Plys A. J., Alani E., 2008. A mutation in the putative MLH3 endonuclease domain confers a defect in both mismatch repair and meiosis in Saccharomyces cerevisiae. Genetics 179: 747–755. 10.1534/genetics.108.086645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogami S., Fukuda T., Nagai Y., Yabe S., Sugiura M., et al. , 2002. Homing at an extragenic locus mediated by VDE (PI-SceI) in Saccharomyces cerevisiae. Yeast 19: 773–782. 10.1002/yea.872 [DOI] [PubMed] [Google Scholar]
- Oh S. D., Jessop L., Lao J. P., Allers T., Lichten M., et al. , 2009. Stabilization and electrophoretic analysis of meiotic recombination intermediates in Saccharomyces cerevisiae. Methods Mol. Biol. 557: 209–234. 10.1007/978-1-59745-527-5_14 [DOI] [PubMed] [Google Scholar]
- Oke A., Anderson C. M., Yam P., Fung J. C., 2014. Controlling meiotic recombinational repair - specifying the roles of ZMMs, Sgs1 and Mus81/Mms4 in crossover formation. PLoS Genet. 10: e1004690 (erratum: PLoS Genet. 10: e1004870). 10.1371/journal.pgen.1004690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Sasaki M., Kniewel R., Murakami H., Blitzblau H. G., et al. , 2011. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144: 719–731. 10.1016/j.cell.2011.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizza S., Mendoza M. A., Berlinger M., Huang L., Nicolas A., et al. , 2011. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146: 372–383. 10.1016/j.cell.2011.07.003 [DOI] [PubMed] [Google Scholar]
- Roig I., Dowdle J. A., Toth A., de Rooij D. G., Jasin M., et al. , 2010. Mouse TRIP13/PCH2 is required for recombination and normal higher-order chromosome structure during meiosis. PLoS Genet. 6: e1001062. 10.1371/journal.pgen.1001062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanienko P. J., Camerini-Otero R. D., 2000. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6: 975–987. 10.1016/S1097-2765(00)00097-6 [DOI] [PubMed] [Google Scholar]
- San-Segundo P. A., Roeder G. S., 1999. Pch2 links chromatin silencing to meiotic checkpoint control. Cell 97: 313–324. 10.1016/S0092-8674(00)80741-2 [DOI] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N., 1994. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell 76: 51–63. 10.1016/0092-8674(94)90172-4 [DOI] [PubMed] [Google Scholar]
- Smagulova F., Gregoretti I. V., Brick K., Khil P., Camerini-Otero R. D., et al. , 2011. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472: 375–378. 10.1038/nature09869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian V. V., MacQueen A. J., Vader G., Shinohara M., Sanchez A., et al. , 2016. Chromosome synapsis alleviates Mek1-dependent suppression of meiotic DNA repair. PLoS Biol. 14: e1002369 10.1371/journal.pbio.1002369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian V. V., Zhu X., Markowitz T. E., Vale-Silva L. A., San-Segundo P., et al. , 2019. Persistent DNA-break potential near telomeres increases initiation of meiotic recombination on short chromosomes. Nat Commun 10: 970 10.1038/s41467-019-08875-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne L. W., Byers B., 1993. Stage-specific effects of X-irradiation on yeast meiosis. Genetics 134: 29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H., Ogawa H., 2000. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol. Biol. Cell 11: 2221–2233. 10.1091/mbc.11.7.2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. F., Kleckner N., Hunter N., 1999. Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc. Natl. Acad. Sci. USA 96: 13914–13919. 10.1073/pnas.96.24.13914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby M. C., 2005. Making crossovers during meiosis. Biochem. Soc. Trans. 33: 1451–1455. 10.1042/BST0331451 [DOI] [PubMed] [Google Scholar]
- Wojtasz L., Daniel K., Roig I., Bolcun-Filas E., Xu H., et al. , 2009. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 5: e1000702 10.1371/journal.pgen.1000702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K., Ma Y., Tang S., Hwang P. Y., Boiteux S., et al. , 2010. Temporally and biochemically distinct activities of Exo1 during meiosis: double-strand break resection and resolution of double Holliday junctions. Mol. Cell 40: 1001–1015. 10.1016/j.molcel.2010.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K., Tang S., Ma Y., Hunter N., 2012. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149: 334–347. 10.1016/j.cell.2012.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanders S., Alani E., 2009. The pch2∆ mutation in baker’s yeast alters meiotic crossover levels and confers a defect in crossover interference. PLoS Genet. 5: e1000571 10.1371/journal.pgen.1000571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D., Kleckner N., 1999. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33: 603–754. 10.1146/annurev.genet.33.1.603 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of this article are represented fully within the article, tables, figures, and supplementary figures, tables and files. Data underlying graphs in all figures and chromosome coordinates in Figure 1 are in File S1, available at Figshare (https://doi.org/10.25387/g3.7800728). Other supplementary files, available at the same URL, include:
Figure S1: Construction of inserts
Figure S2: Additional data and analyses. Includes cells completing meiosis I, timing of VDE DSBs, combined CO and NCO data, and NCO/CO ratios
Table S1: Strain genotypes
Table S2: Primers for all reporter inserts.
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7800728.
Figure 1.
Insert loci examined. Inserts at HIS4 and URA3 were previously studied by Medhi et al. (2016). (A) Locations of insert loci are illustrated (red). Blue circles denote centromere locations. (B) Maps of regions surrounding insert loci. Red—coding region of gene used to identify each insert; black arrow—site of VRS insert. Blue plots show relative Hop1 occupancy levels in mid meiosis, using smoothed ChIP-chip data from (Panizza et al. 2011); vertical scale = 0-7, decile-normalized ChIP/WCE. Green plots show relative DSB levels, using Spo11-oligo reads from Pan et al. (2011); vertical scale = 0-15 hits per million/base-pair. Chromosome coordinates and average Hop1 occupancy and Spo11-oligo reads in 2, 10, and 20 kb regions around each insert are given in File S1.