Abstract
A large portion of the Drosophila melanogaster genome is contained within heterochromatic regions of chromosomes, predominantly at centromeres and telomeres. The remaining euchromatic portions of the genome have been extensively characterized with respect to gene organization, function and regulation. However, it has been difficult to derive similar data for sequences within centromeric (centric) heterochromatin because these regions have not been as amenable to analysis by standard genetic and molecular tools. Here we present an updated genetic and molecular analysis of chromosome 3L centric heterochromatin (3L Het). We have generated and characterized a number of new, overlapping deficiencies (Dfs) which remove regions of 3L Het. These Dfs were critically important reagents in our subsequent genetic analysis for the isolation and characterization of lethal point mutations in the region. The assignment of these mutations to genetically-defined essential loci was followed by matching them to gene models derived from genome sequence data: this was done by using molecular mapping plus sequence analysis of mutant alleles, thereby aligning genetic and physical maps of the region. We also identified putative essential gene sequences in 3L Het by using RNA interference to target candidate gene sequences. We report that at least 25, or just under 2/3 of loci in 3L Het, are essential for viability and/or fertility. This work contributes to the functional annotation of centric heterochromatin in Drosophila, and the genetic and molecular tools generated should help to provide important insights into the organization and functions of gene sequences in 3L Het.
Keywords: centromeric heterochromatin, essential genes, functional annotation
Heterochromatin is the darkly-staining, densely-compacted chromatin of higher eukaryotes that occupies defined positions in homologous chromosomes (Heitz 1928, 1929). Constitutive heterochromatin is generally found near centromeres and in telomeres, remains densely packaged throughout the cell cycle, and consists largely of moderately and highly repetitive sequences (Lohe et al. 1993; Hoskins et al. 2007; Smith et al. 2007).
In Drosophila melanogaster, about 60 Mb of DNA (roughly 1/4 of the genome) in females is found within heterochromatic regions, while in males, the 40Mb Y chromosome is almost entirely heterochromatic (Adams et al. 2000; Hoskins et al. 2002). However, while there has been extensive genetic and molecular characterization of genes in Drosophila euchromatin, genes in the heterochromatic regions of the genome have been much more difficult to study. Some obstacles to mapping and assembling heterochromatic sequences include an absence of significant meiotic recombination, a paucity of prominent cytological landmarks, and the high repetitive sequence content within heterochromatin.
A defining property of heterochromatin is its ability to variably “silence”, in a mosaic fashion, euchromatic genes relocated immediately next to or within heterochromatin, a phenomenon called position effect variegation (PEV) (reviewed in Eissenberg and Reuter 2009). Genetic screens for second-site suppressors and enhancers of PEV have identified a large number of modifier genes, many of which encode known enzymatic products or structural components associated with establishment/maintenance of heterochromatin (Eissenberg et al. 1990; Reuter et al. 1990; Tschiersch et al. 1994; Schotta et al. 2004). Moreover, heterochromatic regions often contain signature patterns of histone modifications and bound proteins similar to those found in some silenced euchromatic gene regions, including the presence of HP1a, Su(var)3-9 and Su(var)3-7 proteins, as well as histones trimethylated at residues H3K9 and H4K20 (Kharchenko et al. 2011; Riddle et al. 2011).
Although heterochromatin has striking gene silencing properties, genetic analysis has demonstrated that these chromosomal regions contain active genes whose expression is essential for fly development and fertility. A large number of these genes reside in the centromeric heterochromatin of the autosomes–chromosome 2 (Hilliker and Holm 1975; Hilliker 1976; Myster et al. 2004; Coulthard et al. 2010) and chromosome 3 (Marchant and Holm 1988a,b; Schulze et al. 2001, 2005). In addition, a few essential genes are located on the X (Hilliker and Appels 1982; Mével-Ninio et al. 2007) and Y (Carvalho et al. 2015; Chang and Larracuente 2018) chromosomes, as well as a number on 4, the “dot” chromosome, which has several properties consistent with a heterochromatic environment (Riddle and Elgin 2018).
It is interesting that heterochromatic genes can in turn be repressed when placed in euchromatic locations, strongly suggesting that these genes require a heterochromatic environment for their expression (Wakimoto and Hearn 1990; Eberl et al. 1993; Howe et al. 1995). This hypothesis is further supported by several genetic and molecular studies which suggest that reduced dosage of the HP1a gene, which encodes a product required for heterochromatin integrity, leads to the reduced expression of some heterochromatic genes (Clegg et al. 1998; Sinclair et al. 2000; Lu et al. 2000; Schulze et al. 2005).
The first genetic screens for lethal mutations in chromosome 3 pericentric heterochromatin identified a minimum of 10 essential loci in 3L Het and 2 loci in 3R Het (Marchant and Holm 1988a and b). Subsequent genetic analysis generated more 3L Het deficiencies, additional alleles of already defined 3L Het genes, as well as mutations in new essential 3L and 3R Het loci, some of which were identified molecularly (for a review, see Fitzpatrick et al. 2005).
Data providing a corresponding physical map of chromosome 3 centric heterochromatin has come from genome sequencing as well as from cytological studies., Progress from the Drosophila Heterochromatin Genome project has provided an extensive physical map from DNA sequencing data, including numerous predicted gene models in centric heterochromatin (Smith et al. 2007; Hoskins et al. 2007, 2015). Comprehensive ChIP-array analysis of histone modifications and chromosomal proteins has also demonstrated different chromatin packaging profiles for heterochromatic vs. euchromatic domains (Riddle et al. 2011).
Koryakov et al. (2002) used various chromosomal rearrangements and heterochromatin banding analysis to produce a cytological map of the essential 3L and 3R Het loci defined by Marchant and Holm (1988b). This group later refined their map using the Suppressor of Under-Replication (SuUR) mutation, which allows visualization of chromatin landmarks in the otherwise rather amorphous heterochromatin (Koryakov et al. 2003). More recently, Andreyeva et al. (2007) used FISH mapping of BAC clones containing known essential 3L and 3R Het genes on polytene chromosomes from SuUR- Su(var)3-9 double mutants to generate a high-resolution map of heterochromatic gene models and highly repetitive satellite sequences.
A comprehensive analysis of essential genes in the centric heterochromatin of chromosome 2 has been provided by extensive genetic and molecular analyses by Hilliker (Coulthard et al. 2003, 2010) and others (Myster et al. 2004). In particular, Coulthard et al. (2010) further refined the genetic and molecular profile of the distal segment of proximal 2R Het, reducing the number of complementation groups defined by Myster et al. (2004) and identifying six additional essential loci in 2R Het via RNAi analysis . Their results complement well the extensive physical map generated by genome sequencing (Hoskins et al. 2007, 2015) and cytological analysis (Rossi et al. 2007; Dimitri et al. 2009).
Here we report on our work to obtain a comprehensive genetic map of chromosome 3L Het. We focused on the identification of essential genetic loci and their alignment with predicted gene models on the physical map, with the overall aim of obtaining a functional annotation of essential genes in this region. This and our previous work has also facilitated the characterization of a number of interesting essential genes in centric heterochromatin e.g., Schulze et al. (2005), Li et al. (2009), Hallson et al. (2008, 2012), Sinclair et al. (2009), as well as genes described in this report.
Briefly, we focused initially on compiling a collection of new genetic lesions, followed by genetic deficiency mapping and a large scale inter se complementation analysis to define genetic loci. Next, we used a PCR-based mapping approach to localize putative heterochromatic sequences from the available genome assembly to defined heterochromatic intervals, thereby allowing us to identify candidate gene(s) for each essential complementation group. We then identified essential loci present by correlating gene sequences to lethal complementation groups, coupled with RNA interference studies (see also Hallson et al. 2012 for examples). Most of the original lethal 3L Het complementation groups identified by Marchant and Holm have now been defined at the molecular level and we have also identified additional essential loci.
Our analysis has revealed a surprisingly high percentage of essential genes in 3L Het. The work also provides an expanded repertoire of genetic reagents, such as deletions (especially those with molecularly defined breakpoints), as well as novel lethal mutations (EMS-, P-element- and radiation-induced); these will be valuable tools for developing a comprehensive profile of gene functions in chromosome 3 heterochromatin.
Materials and Methods
Fly strains and crosses
Flies were raised on standard yeast-molasses-cornmeal medium and fly strains were maintained at 18°. Crosses were performed at 25° unless otherwise stated and all crosses were scored to completion. If no transheterozygous progeny emerged, or if the number of transheterozygous progeny was less than 5% of those expected from balancer classes (minimally, 100 balancer heterozygotes were examined per cross), the combination was considered lethal. In cases where the transheterozygotes were greater than 5% but substantially less than 50% of those expected, the combination was considered semi-lethal.
Unless otherwise stated, genetic strains used have been reported elsewhere. The G series of EMS-induced lines, as well as Df(3L)FX3, DF(3L)FX53, Df(3L)FX33 and Df(3L)MX18 were created in the Deitcher laboratory (Vilinsky et al. 2002; Schulze et al. 2005; D. Deitcher personal communication). The Z series of EMS mutations was obtained from the Zuker collection (Koundakjian et al. 2004). EMS lines 2-30 and 1-166-38, as well as the γ-radiation-induced deficiencies Df(3LR)6B-29, Df(3L)1-166, Df(3L)9-56, Df(3L)2-30 and Df(3L)8-80 were generated by Marchant and Holm (1988 a). The P-element alleles CH(3)4, CH(3)119, CH(3)4d, CH(3)53 and CH(3)7 were generated by Zhang and Spradling (1994) and were generously provided by P. Zhang. Df(3L)O-1, Df(3L)K2 and Df(3L)γ-28 were generated as described in Schulze et al. (2001).
X-ray mutagenesis and genetic screens
Three- to five-day old KG03264/KG03264 males (carrying a P{SUPor-P} insert at cytological position 80C1 near the nrm locus) were treated with 4,000 rads of X-ray radiation, allowed to recover for 24 hr, and crossed en masse to TM3 Sb Ser/TM6 Hu Tb virgin females. Approximately one thousand single F1 KG03264{w+}*/TM3 Sb Ser or KG03264{w+}*/TM6 Hu Tb males (where * indicates a mutagenized third chromosome) were then crossed to virgin females carrying a 3L Het deficiency (Df(3L)FX3/TM3 Ser or Df(3L)MX18 /TM3 Ser), and the resulting progeny were examined for lethality or visible phenotypes. Stocks of putative lethal mutations were established from KG03264{w+}*/TM3 Ser siblings. To further map the newly-isolated lesions, all mutations were separately crossed to flies bearing a subset of smaller 3L Het deficiencies (Df(3L)1-166, Df(3L)γ28, Df(3L)K2, Df(3L)9-56, Df(3L)FX33), and to at least two existing alleles of each lethal complementation group (except where only a single allele existed).
We then conducted a second X-ray screen, designed to isolate lethal lesions in both 3L Het and 3R Het. In this study, 3- to 5-day old e1/e1 males were treated with 4,000 rads of X-ray radiation and crossed en masse to TM3 e1 Sb Ser/TM6 e1 Hu Tb virgin females. Single F1 e1*/TM3 e1 Sb Ser males (where * indicates a mutagenized third chromosome) were then crossed to Df(3LR)6B-29/TM3 e1 Ser females, and stocks of putative lethal mutations established from e1*/TM3 e1 Ser siblings. To further map the newly isolated lesions, we crossed all mutations to the subset of smaller heterochromatic deficiencies, as described above, with the addition of Df(3R)10-65 and Df(3R)XM3, as well as to alleles of each lethal complementation group.
Screen of the EMS Zuker collection
We obtained 3,400 strains (balanced with TM6 e1 Tb) from the Zuker collection that contained putative third chromosome mutations generated with EMS, because it had been reported that approximately one third of the Zuker strains contained third chromosome lethal mutations (Koundakjian et al. 2004). We tested each balanced line for lethality with different 3L or 3R Het Df/TM6 Tb strains: Df(3LR)6B-29, Df(3L)FX53, Df(3L)Delta1AK and Df(3R)XM3. Confirmed lethal mutations were then further mapped by using Df(3L)γ28, Df(3L)K2, Df(3L)9-56, Df(3L)FX33, Df(3R)XM3 and Df(3R)10-65, each balanced over TM6B Tb. In addition, we tested the lethals for allelism with representative alleles of appropriate lethal complementation groups.
Single embryo PCR for detection of homozygous mutant embryos, deficiency mapping, and sequencing of mutant alleles
We isolated embryos homozygous for chromosomal deficiencies and other induced mutations using the methodology described in Hallson et al. (2008; 2012). In brief, using DNA from homozygous deficiency strains, along with PCR mapping primers for candidate gene sequences (see Table S1), we tested for the presence or absence of PCR products in homozygous mutant embryos from each strain. The absence of a PCR product indicated that a specific gene sequence mapped to the region covered by the deficiency.
For sequence analysis, DNA was isolated from homozygous mutant embryos, all exon regions were PCR amplified, and purified PCR products were then sequenced to look for corresponding DNA lesions that might result in mutant phenotypes (see Hallson et al. 2008, 2012). DNA sequencing was performed by UBC NAPS or Genewiz or Macrogen sequencing services. Mutations were identified by comparing sequences to the reference sequences available on Flybase using the BLAST algorithm (Altschul et al. 1990) and multiple protein alignments were carried out using CLUSTALW (Larkin et al. 2007).
RNA interference experiments
Transgenic UAS-RNAi lines, expressing inverted repeats (IRs) under the control of Gal4, were obtained from the Vienna Drosophila RNAi Center (VDRC: http://stockcenter.vdrc.at/control/main), Drosophila Transgenic RNAi Project (TriP: http://www.flyrnai.org/TRiP-HOME.html) and the Fly Stocks of the National Institute of Genetics (NIG-fly; http://www.shigen.nig.ac.jp/fly/nigfly/index.jsp). We crossed UAS-IR-bearing males to virgin females, initially held at 29° for at least two days, bearing UAS-Dcr-2/UAS-Dcr-2; tub-Gal4/TM3 Sb. Actin and tubulin driver stocks were used to induce ubiquitous expression of the IR transgenes and a copy of UAS-Dcr2 (a component of the RNAi pathway) was included to sensitize the RNAi interference pathway and enhance knockdown. We maintained crosses at 29° and then examined them for lethality, sterility and visible phenotypes in flies expressing the IR sequence. Crosses producing UAS-IR driven progeny, but at proportions substantially less than 50% of those expected from the balancer classes, were deemed semi-lethal (due to the incomplete penetrance/ expression of RNA, some “escapers” may survive).
Data Availability
A comprehensive and representative set of stocks have been sent to Dr Kevin Cook at the Bloomington Drosophila stock center. Supplemental files available at FigShare. Table S1 lists primers used in mapping. Table S2 lists EMS mutations identified. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7828301.
Results
As a first step in our analysis, we generated and mapped a large number of additional lethal lesions (putative Dfs and/or point mutations) within 3L and 3R Het. To do this, we conducted two X-ray mutagenesis screens, and in parallel screened 3,400 EMS-mutagenized 3rd chromosomes from the Zuker collection (Koundakjian et al. 2004), as outlined in Materials and Methods.
From these large-scale genetic screens, we isolated 91 novel lesions (46 from the X-ray screens, 45 from the Zuker lines), and of these, 60 mapped to 3L Het and 14 to 3R Het (Table 1). The remaining 17 mutations may be located outside of 3rd chromosome heterochromatin, possibly due to the presence of second site lethal mutations outside of 3L Het, see below.
Table 1. Summary of results from EMS and X-ray screens.
| Mutagen | 3L lesions | 3R Lesions | Total number of lesions | Number of chromosomes | Df used for screening | Frequency of lesions |
|---|---|---|---|---|---|---|
| X-ray #1 | 14 | 0 | 14 | ∼1000 | FX3, MX18 | 1.40% |
| X-ray #2 | 25 | 7 | 32 | 3829 | 6B-29 | 0.84% |
| EMS | 21 | 7 | 45* | 3400 | 6B-29, Delta1-AK, FX53, MX3 | 1.32% |
Screens for lethal lesions in 3L and 3R Het induced by X-rays
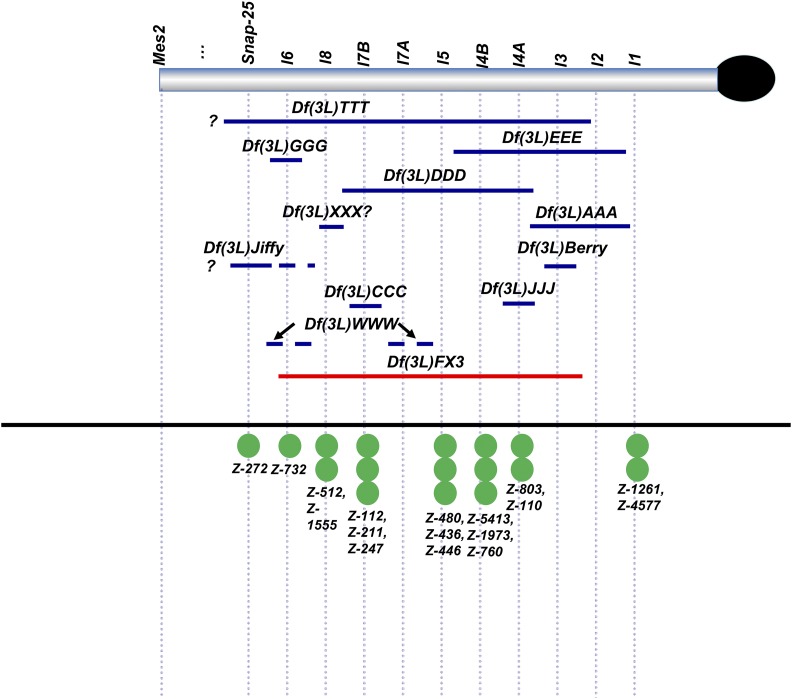
Our first X-ray screen with the large Df(3L)FX3 and Df(3L)MX18 deficiencies identified 14 lesions in 3L Het (X-ray #1 in Table 1). To position the new lesions more definitively, we tested them for complementation with deficiencies removing specific intervals of 3L Het and with alleles of previously identified essential loci. Of the aforementioned 14 lesions, 11 failed to complement Df(3L)FX3 (see Figure 1). The remaining three mutations (Df(3L)Lola, Df(3L)BAC and Df(3L)ZZZ) were Minute-like in phenotype in combination with Df(3L)FX3 or Df(3L)MX18, but it was not possible to map these lesions further. The combined frequency of lethal and Minute-like lesions generated in this screen was 1.4%. The putative X-ray-induced lesions varied in size; some were large and spanned several lethal complementation groups, corresponding to many megabases of DNA (e.g., Df(3L)TTT) (Figure 1), whereas others were either small deletions/other rearrangements or possibly point mutations affecting single genes (e.g., Df(3L)GGG) (Figure 1).
Figure 1.
Genetic map of EMS mutants and deficiencies isolated from our initial screens of Zuker lines and first X-ray screen. Blue lines - newly discovered deficiencies; red lines - deficiencies used to screen for new lesions; green circles - newly-mapped EMS lesions from the Zuker collection. Dashed lines on deficiencies indicate semi-lethality with alleles of intersecting complementation groups. Df(3L)WWW may be a discontinuous lesion affecting at least two genes (lethal 6 and lethal 7A).
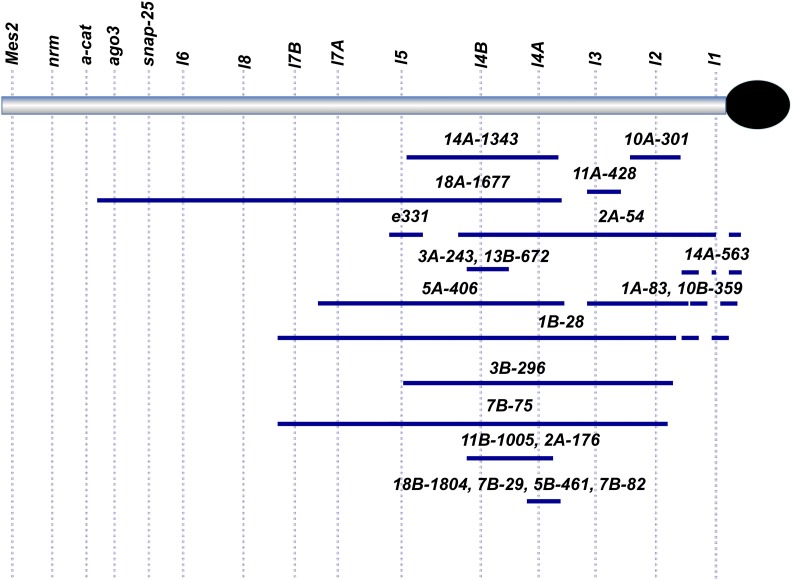
In a subsequent larger-scale X-ray screen (X-ray #2 in Table 1), we screened the mutagenized chromosomes for failure to complement Df(3LR)6B-29, a large deficiency removing heterochromatic segments from both 3L and 3R (Marchant and Holm op. cit.). In total, we isolated 32 lesions in this second screen: 25 on 3L and 7 on 3R. Of the 25 putative 3L Het deficiencies isolated in our second X-ray screen, 21 were used for further analysis (see Figure 2). We then performed inter se complementation tests of each of these lesions in combination with various other 3L Het deficiencies and in combination with representative alleles of existing complementation groups, in order to position the new lesions along the chromosome arm (see Figure 2 for a summary).
Figure 2.
Map of deficiencies generated in a more intensive, second X-ray screen. Positioning was also obtained by crossing new deficiency strains to alleles affecting essential loci and other deficiencies. Note that of the 25 deficiencies isolated, 2 were not used further (not shown) and 1 was viable with alleles of all representative complementation groups we tested in 3L Het.
Screening for lesions induced by EMS in 3L and 3R Het
As described in Materials and Methods, an initial screen of recessive lethal lines from the Zuker EMS collection (Koundakjian et al. 2004) involved testing each strain for lethality in combination with Df(3LR)6B-29, a deficiency that removes much of chromosome 3 heterochromatin (Marchant and Holm 1988a). All putative 3L and 3R Het lethal mutations identified in this analysis were tested with four other deficiencies that remove segments of 3rd chromosome centric heterochromatin (Df(3L)Delta1-AK, Df(3L)FX53, Df(3L)FX3, and Df(3R)XM3). Of the 45 3L Het EMS lesions identified (Table 1), 44 were lethal or semi-lethal in combination with one of the four deficiencies. The single lesion (Z-4493) that was viable in trans with all four deficiencies was characterized by a curly wings phenotype when tested in combination with Df(3R)XM3. Three other lesions also caused visible phenotypes (black deposits at leg joints, abnormal positioning of legs and curled wings) and were also semi-lethal when hemizygous over particular deficiencies.
Of the 45 EMS mutations identified (designated with an asterisk in Table 1), 21 could be assigned unequivocally to 3L Het, based on complementation tests with Df(3L)FX53, Df(3L)Delta 1-AK or lethal 1 alleles (see Table S2). Seventeen of these 21 lesions were allelic with previously identified complementation groups, 2 failed to complement Df(3L)Delta 1-AK (distal to SNAP-25), 1 (Z-855) was semi-lethal in combination with Df(3L)FX53, and 1 failed to complement Df(3L)K2 and Df(3L)1-166. An additional seven lesions were found to be allelic with complementation groups in 3R Het.
None of the remaining 17 lesions could be assigned to known lethal complementation groups in either 3L or 3R Het. It is likely that at least some of the additional mutations are allelic to second-site mutations carried by the deletion chromosomes used for complementation analysis, as accumulation of lethal mutations on permanently heterozygous chromosomes has been well-documented (Mukai 1964).
Our analyses revealed a few inconsistencies with respect to the previously published 3L Het map (Fitzpatrick et al. 2005), as well as one erroneous allele assignment. Our revised genetic map now places Snap-25 and lethal 6 distal to lethal 8, which in turn was placed distal to lethal 7A and 7B (Figure 2). This gene order is now consistent with the physical map provided by genome sequencing (Hoskins et al. 2015), and this order was also confirmed by our mapping and sequencing of lethal complementation groups corresponding to these published gene models. Finally, allele G1e was previously incorrectly assigned to lethal 7B rather than lethal 4B.
Refining the 3L Het physical map
In order to align the genetic and physical maps of 3L Het, we mapped candidate essential genes to specific genetic intervals as defined by our deficiency maps. Using DNA from homozygous deficiency strains and primers for a set of candidate genes, we tested for the presence or absence of PCR products in flies homozygous for a given deficiency strain. The absence of a PCR product indicated that a specific gene sequence was removed by the deficiency. We then refined our analysis by using reference sequences in the physical map as anchor points for our genetic map, limiting the number of candidate genes required for testing with each deficiency.
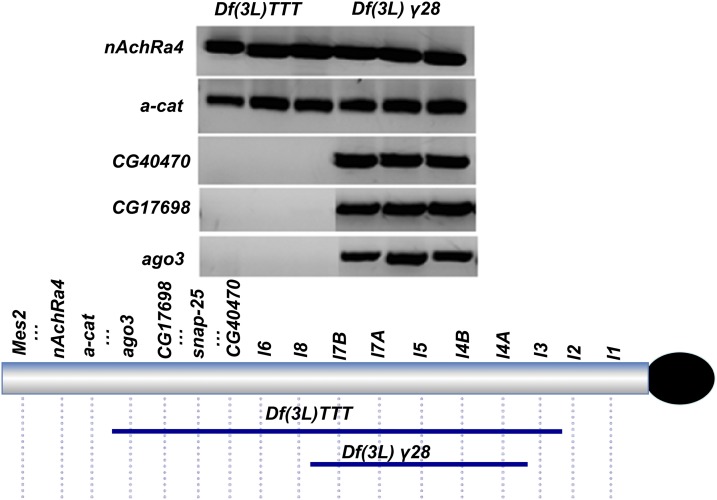
In order to better align the genetic complementation map to the published heterochromatic sequence in Release 6 (Hoskins et al. 2015), we also mapped endpoints of several large and/or distal deficiencies by PCR relative to 3L Het candidate gene sequences. Since to date, few mutations have been mapped to the distal segment of 3L Het (e.g., between Snap-25 and Mes 2), we were especially interested in determining the distal-most extent of Df(3L)TTT. PCR analysis of homozygous Df(3L)TTT embryos showed that this deficiency removes the CG40470, CG17698 and ago3 loci, but not the α-catenin or the n-AchRA4 genes (Figure 3). From our complementation analysis and single embryo PCR mapping data, we conclude that Df(3L)TTT deletes a region predicted to contain at least 3 Mb of DNA. A summary of our refined map is presented in Figure 4.
Figure 3.
Determining the distal extent of Df(3L)TTT by gene-specific PCR. CG40470 is a gene located between SNAP-25 and l6. Prior genetic analysis confirmed that Df(3L)TTT failed to complement alleles of essential genes located in the 3L Het segment extending from lethal 3 to SNAP-25. Df(3L)gamma-28, which does not delete the regions tested, was used as a positive control.
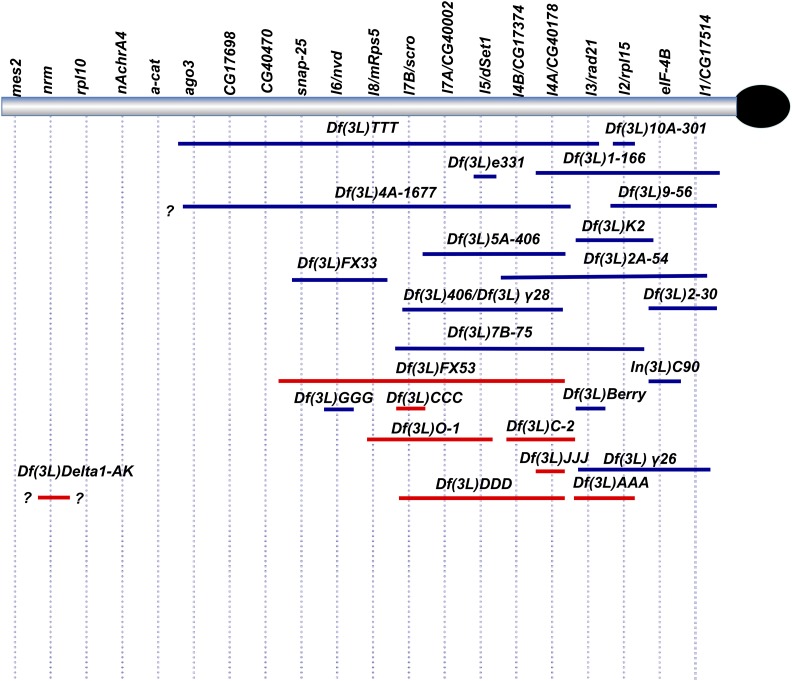
Figure 4.
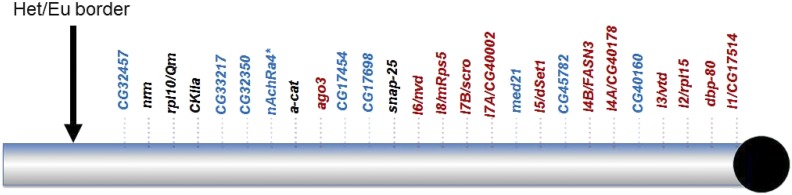
Refined map of 3L Het incorporating genetic and molecular data from this study. Blue marks deficiencies in which extent was determined genetically and molecularly. Red marks deficiencies in which limits were determined genetically. “?” denotes boundary limits that have not been molecularly defined.
Correlation of gene models with lethal complementation groups
Our molecular analysis of deficiency strains combined with deletion mapping of essential genes allowed us to narrow the number of candidate genes that could potentially correspond to each genetically identified locus on our map. In an attempt to determine the molecular identity of each vital locus, we amplified and sequenced the entire coding region of each candidate gene from DNA homozygous for mutant alleles representative of a given complementation group. The presence of a significant sequence change (e.g., predicted deleterious effects on gene product) relative to the control background sequence, led us to conclude that a given complementation group corresponded to the candidate gene in which the change was found. In certain cases, we were also able to rescue flies containing appropriate mutations transheterozygous to deficiencies removing the candidate gene by expressing a Gal4 driven cDNA transgene of the corresponding gene of interest. Essential genes identified using this method are listed in Table 2; with putative or confirmed functions of these genes included in Table 6. Taken together with the RNAi results described below, the data from these analyses have allowed us to assign a molecular identity for each lethal complementation group present on our genetic map (shown in Figure 4).
Table 2. Known lethal complementation groups in 3L Het, and their molecular assignment based on the DNA lesions identified in mutants. (*These designations based on those used in Marchant and Holm 1988a,b). More comprehensive data for lethal 6 (nvd) will be reported in Syrzycka et al. manuscript in preparation.
| *Lethal complementation group: | Mutant Allele(s): | Mutagen: | Gene Affected: | Resulting change: | Source |
|---|---|---|---|---|---|
| lethal 1 | In(3L)C90 | γ-ray | CG17514 | Genes missing | Marchant and Holm (1988b) |
| eIF-4B/CG10837 | |||||
| Df(3L)2-30 | γ-ray | eIF-4B/CG10837 | Gene missing | Marchant and Holm (1988b) | |
| Z-4577 | EMS | CG17514 | W1092 > STOP | Zuker collection | |
| (Koundakjian et al. 2004) | |||||
| Z-1261 | EMS | unidentified | unidentified | Zuker collection | |
| (Koundakjian et al. 2004) | |||||
| lethal 2 | See Schulze et al. (2005) | Various | RpL15/CG17420 | See Schulze et al. (2005) | Schulze et al. (2005) |
| lethal 3 | See Hallson et al. (2008) | Various | rad21/vtd (cohesin) CG17436 | See Hallson et al. (2008) | Hallson et al. (2008) |
| lethal 4A | CH(3)53 | P-element | CG40178 | P insertion in intron | Zhang and Spradling (1994) |
| CH(3)4d | P-element | CG40178 | P insertion in intron | Zhang and Spradling (1994) | |
| Z-110 | EMS | CG40178 | L326 > STOP | Zuker Collection | |
| (Koundakjian et al. 2004) | |||||
| G8 | EMS | CG40178 | W377 > STOP | Deitcher laboratory | |
| (Vilinsky et al. 2002) | |||||
| G47 | EMS | CG40178 | K171 > STOP | Deitcher laboratory | |
| (Vilinsky et al. 2002) | |||||
| G6 | EMS | CG40178 | K180 > STOP | Deitcher laboratory | |
| (Vilinsky et al. 2002) | |||||
| lethal 4B | G10 | EMS | CG17374 | A1463 > T | Deitcher laboratory |
| (Vilinsky et al. 2002) | |||||
| G11 | EMS | CG17374 | R1542 > STOP | Deitcher laboratory | |
| (Vilinsky et al. 2002) | |||||
| G30 | EMS | CG17374 | G1780 > E | Deitcher laboratory | |
| (Vilinsky et al. 2002) | |||||
| Z-5413 | EMS | CG17374f | G1779 > R | Zuker Collection | |
| (Koundakjian et al. 2004) | |||||
| Z-760 | EMS | CG17374 | I240 > S | Zuker Collection | |
| (Koundakjian et al. 2004) | |||||
| Z-1973 | EMS | CG17374 | A1060 > V | Zuker Collection | |
| (Koundakjian et al. 2004) | |||||
| 3#69 | EMS | CG17374 | R67 > S | D. Sinclair | |
| (unpublished) | |||||
| lethal 5 | See Hallson et al. (2012) | Various | Set1/CG40351 | See Hallson et al. (2012) | Hallson et al. (2012) |
| lethal 7A | fsa2l(3) | EMS | CG40002 | Gene missing | Kennison and Tamkun |
| (unpublished)+ | |||||
| lethal 7B | G43 | EMS | Scro/CG17594 | L250 > STOP | Deitcher laboratory |
| (Vilinsky et al. 2002) | |||||
| Z-211 | EMS | Scro/CG17594 | P insertion of “A” nucleotide leading to frameshift at a.a. 293 | Zuker Collection | |
| (Koundakjian et al. 2004) | |||||
| Z-247 | EMS | Scro/CG17594 | P insertion of “A” nucleotide leading to frameshift at a.a. 293 | Zuker Collection | |
| (Koundakjian et al. 2004) | |||||
| lethal 8 | K125 | P-element | mRpS5/CG40049 | P insertion in intron 2 | Current study |
| Z-1555 | EMS | mRpS5/CG40049 | ATG to ATA | Zuker Collection | |
| (Koundakjian et al. 2004) | |||||
| Z-512 | EMS | mRpS5/CG40049 | G201 > E | Zuker Collection | |
| (Koundakjian et al. 2004) | |||||
| G18 | EMS | mRpS5/CG40049 | No lesion detected | Deitcher Laboratory | |
| (Vilinsky et al. 2002) |
Table 6. Essential genes in 3L Het identified to date.
| Essential genes in 3L Het. (centromere outwards): | Determined essential based on: | Sources: |
|---|---|---|
| CG17514/lethal1 | Lethality (muts/RNAi)/Male and fem. sterility (RNAi) | this study |
| dbp80 | Male and fem. sterility (RNAi) | this study, Yan et al., 2014 |
| rpl15/lethal2 | Lethality (muts) | this study, Schulze et al. 2005 |
| vtd/lethal3 | Lethality (muts/RNAi) | Hallson et al. 2008, this study (muts), Vass et al. 2003 (RNAi) |
| CG40160 | Lethality (RNAi) | this study |
| CG40178/lethal4A | Lethality (muts/RNAi) | this study |
| FASN3/lethal4B | Lethality (muts/RNAi) | this study |
| CG45782 | Male sterility (RNAi) | this study |
| dSet1/lethal5 | Lethality (RNAi/muts) | Hallson et al. 2012, this study |
| med21 | Lethality (RNAi) | this study |
| CG40002/lethal7A | Lethality (muts) | this study |
| scro/lethal7B | Lethality (RNAi/muts) | this study |
| mRps5/lethal8 | Lethality (muts/RNAi) | this study, Yan et al., 2014 |
| nvd/lethal6 | Lethality (muts/RNAi) | Yoshiyama et al. 2006 (RNAi), Syrzycka et al. in prep |
| snap-25 | Lethality (muts/RNAi) | Vilinsky et al. 2002 (muts), this study (RNAi) |
| CG17698 | Lethality (RNAi) | this study |
| CG17454 | Lethality (RNAi) | this study |
| ago3 | Female sterility (muts) | this study, Li et al. 2009 (muts) |
| alpha-Cat | Lethality (muts, RNAi) | Sarpal et al. 2012 (muts), this study (RNAi) |
| nAChRalpha4* | Lethality (RNAi) | this study |
| CG32350 | Lethality (RNAi) | this study |
| CG33217 | Lethality (RNAi) | this study |
| CKIIalpha | Lethality (muts) | Lin et al. 2002 (muts) |
| rpL10/Qm | Lethality (muts, RNAi) | Cook et al. 2012 (muts), this study (RNAi) |
| nrm | Lethality (muts) | Kania et al. 1993, Kania and Bellen 1995 (muts) |
| CG32457 | Lethality/Sterility (RNAi) | this study |
%The sequences of these genes are very similar and are likely to result from annotation issues or more improbably, very recent duplications.
Potential essential gene identified based on failure to recover males in a single RNAi line. Note that this result is not entirely conclusive, as this RNAi line also has 1 predicted off-target.
While most of the gene assignments summarized in Tables 2 and 6 were straightforward, the molecular and genetic characterizations of lethal 1 and lethal 7A were slightly more complicated; these are therefore described in more detail below.
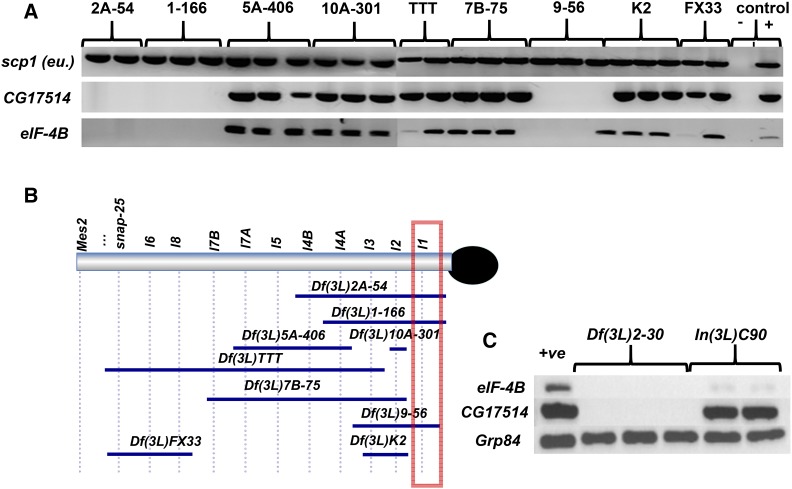
Molecular characterization of lethal 1
lethal 1 is the proximal-most genetically identified essential locus in 3L Het (see Figures 4 and 5). The locus was originally identified in mutagenesis screens conducted by Marchant and Holm (1988b) and it has been defined as a putative trithorax group (trxG) gene (Schulze et al. 2001). Numerous lethal 1 alleles were isolated by Marchant and Holm (1988) and Schulze et al. (2001), suggesting that it may encode a large mutagenesis target; however, only two of the original mutations remain: Df(3L)2-30 and In(3L)C90. Df(3L)2-30 is associated with a duplication of proximal heterochromatic material between h48 and h50 (Koryakov et al. 2002), but it also likely carries a small deletion not detected by cytological analysis. In(3L)C90 is an inversion with a heterochromatic breakpoint near or within lethal 1, and a euchromatic breakpoint in 62D2-7 (Lindsley and Hardy 1992). We have also identified two additional putative lethal 1 alleles, Z-1261 and Z-4577, both of which fail to complement deficiencies that lack the gene. In combination, these two alleles produce a small number of sterile transheterozygotes with a Minute-like phenotype, and extra wing vein material (Table 3). The conventional Minute phenotype is haplo-abnormal and thus dominant, and is characterized by developmental delay, shorter and thinner bristles, roughened/reduced eyes, misrotated genitalia, and, often, semi-sterility; this phenotype is most often caused by defects in global protein synthesis (see Marygold et al. 2007).
Figure 5.
Mapping of elF-4B and CG17514. A) Molecular positioning eIF-4B and CG17514 in the lethal 1 region. B) CG17514 and eIF-4B are deleted by Df(3L)2-30, and eIF-4B, but not CG17514, is deleted in the In(3L)C90 strain.
Table 3. lethal 1 inter se complementation matrix. MAll transheterozygous progeny were Minute-like. % expected = number of observed progeny / number of expected progeny based on balancer classes (i.e., O/E)*100%. *In addition to deleting lethal 1, Df(3L)9-56, Df(3L)1-166, Df(3L)e*#54 and Df(3LR)6B-29 remove the lethal 2 (rpL15) locus and are Minute because of this (data not shown).
| Z -1261 | % expected | Z-4577 (W1092STOP) | % expected | |
|---|---|---|---|---|
| Df(3LR)6B-29* | 20M / 162 total | 49% | 2 M / 195 total | 4% |
| Df(3L)1-166* | 11M / 148 total | 30% | 8 M / 182 total | 18% |
| Df(3L)9-56* | 1M / 111 total | 4% | 0 / 186 total, i.e., LETHAL | 0% |
| Df(3L)e*#54* | 5M / 214 total | 9% | 5 M / 249 total | 8% |
| In(3L)C90 | 1M / 129 total | 3% | 19 M / 280 total | 27% |
| Df(3L)2-30 | 24M sterile / 203 | 47% | 11M sterile / 147 total | 30% |
| Z- 4577 | 6M sterile / 229 total | 11% | Homozygous lethal | N/A |
| Z- -1261 | Homozygous lethal | N/A | 6M sterile / 229 total | 11% |
In single embryo PCR mapping (Figure 5A) two potential candidate genes were absent from Df(3L)2-30: eIF-4B (eukaryotic Initiation Factor 4B or CG10837), a member of the eukaryotic initiation factor complex required for translational activation, and CG17514, a large translation activator. However, In(3L)C90 deletes eIF-4B but not CG17514 (Figure 5C). This was somewhat surprising to us, because lethal 1 is quite mutable, and because CG17514 encodes a much longer protein (2,630 aa) than eIF-4B (459 aa); it therefore seemed more likely to us that lethal 1 would correspond to CG17514. One possible explanation for the presence of CG17514 exon sequences in In(3L)C90 could be that only regulatory regions are mutated, and thus CG17514 expression could still be affected.
Indeed, subsequent sequence analysis of mutant alleles provided evidence to identify CG17514 as corresponding to lethal 1: the EMS-induced lethal 1 allele Z-4577 contains a mutation predicted to change of residue W1092 in the CG17514 product to a stop codon, strongly suggesting that CG17514 corresponds to the lethal 1 locus.
In order to obtain additional support for our contention that lethal 1 corresponds to CG17514, we carried out RNAi knockdown of CG17514; in this analysis, we crossed flies containing UAS responsive CG17514 IRs to flies carrying the tub-GAL4 transgene at 29° in order to activate targeting RNAi. Expression of 2 out of the 3 CG17514 RNAi lines, VDRC 47269 and BL34355, resulted in reduced viability and complete lethality respectively. Escaper progeny expressing RNAi from the BDRC 47269 line display a Minute-like phenotype (thin bristles) and are sterile (Table 4). Taken together with the aforementioned data, these results indicate that lethal 1 coincides with CG17514.
Table 4. RNAi experiments expressing CG17514 RNAi-activating IRs with tub-driven GAL4 (at 29 C) suggest that CG17514 is essential. However, viable progeny emerge when using the eIF-4B RNAi line (VDRC 31364). *Escapers emerging from the VDRC 47269 CG17514 RNAi cross display a Minute phenotype and are sterile.
| Female parent | Balancer progeny | RNAi driven progeny | % of expected RNAi flies |
|---|---|---|---|
| CG17514 RNAi / CG17514 RNAi (VDRC 47269) | 195 | 17* | 9% (semi-lethal) |
| CG17514 RNAi / CG17514 RNAi (BL34355) | 303 | 0 | 0% (lethal) |
| CG17514 RNAi / CG17514 RNAi (VDRC 47268) | 151 | 105 | 63% (viable) |
| eIF-4B RNAi /eIF-4B RNAi (VDRC 31364) | 196 | 109 | 71% (viable) |
| eIF-4B RNAi /CyO (BL57305) | 324 | 123 | >100% (viable) |
However, these results do not exclude an essential role for the eIF-4B gene as well, given its presumed key role in protein. It is quite possible that our screens did not isolate any mutations in this smaller target gene, although results from RNAi experiments do not suggest that eIF-4B is essential either (see below).
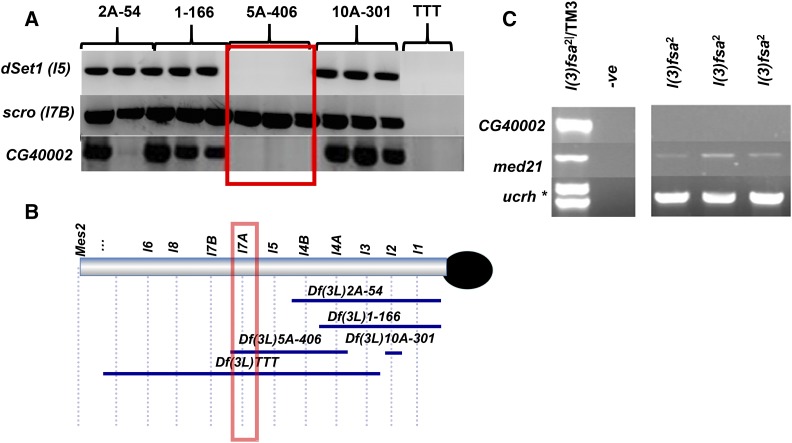
Molecular characterization of lethal 7A
We currently possess only a single EMS induced allele of lethal 7A, l(3)fsa2 (Fitzpatrick et al. 2005). This allele displays a lethal phase during late embryogenesis, with no obvious cuticle defects (Fitzpatrick et al. 2005). Molecular and genetic mapping show that lethal 7A is distal to lethal 5 and proximal to lethal 7B (scarecrow) (see e.g., Figure 6A). Two gene models, CG40002 and CG40472 (an apparent pseudogene of CG40002), appear the most likely candidates for lethal 7A. The CG40002 gene encodes a 94 a.a. protein that is a member of the NADH dehydrogenase complex (a.k.a. Complex I). The NADH dehydrogenase complex is a conserved complex present throughout Eukarya and is the first major complex in the electron transport chain (Ohnishi et al. 2018). Given the important role of Complex I in the oxidative production of ATP, it would not be surprising if a gene encoding any of its subunits were essential.
Figure 6.
Mapping of lethal 7A to CG40002. A) Single embryo PCR mapping positions CG40002 in the lethal 7A region. B) sePCR of DNA from l(3)fsa2mutants using CG40002, med21 and ucrh primers. The asterisk marks a band that is amplified only from l(3)fsa2/TM3 animals and likely corresponds to a polymorphic ucrh sequence found on this particular TM3 balancer chromosome.
There are also two other possible candidate gene models, med21 and ucrh, in the vicinity. However, upon PCR mapping, we found that only the CG40002 gene has been deleted in DNA from homozygous l(3)fsa2 mutant embryos, while med21 and ucrh remain present (Figure 6C). This strongly suggests that lethal 7A corresponds to CG40002.
RNAi knockdown of heterochromatic genes
Some 3L Het genes that were predicted to have important functions based on the existence of conserved human homologs did not yield mutant alleles in our screens. In order to test for additional essential genes in 3L Het, we performed RNAi knockdown analysis using relevant RNAi lines available from the VDRC, TRiP or NIG collections, which allowed us to target most 3L Het gene models. Each RNAi line was crossed to UAS-Dcr2/UAS-Dcr2; tub-Gal4/TM3 Sb at 29°; UAS-Dcr2 was used to optimize the RNAi pathway and therefore increase the likelihood of detecting gene knockdown effects. The results identify 9-10 additional essential genes in 3L Het (10 if nAchRalpha4 is included, see Discussion below) not previously identified in mutagenesis studies (Tables 5 and 6). As positive controls, we used some RNAi transgenes targeting known lethal complementation groups, and these were found to be effective.
Table 5. Summary of RNAi experiments targeting genes in 3L Het. Column 1: The 3L Het genes targeted; Column 2: The identifier numbers of RNAi lines, appended with abbreviated prefixes indicating where they were sourced from (V - VDRC, BL - Bloomington Stock Center or NIG - National Institute of Genetics (Japan)); Column 3: Effects of driving RNAi on survival to adulthood and fertility (V - viable, L -lethal, SL - semi-lethal - i.e., substantially less than 50% of expected progeny, F - fertile, S - sterile, IN - inconclusive, NT - not tested, M - male and FE – female % - for semi-lethal lines, percentage of RNAi progeny emerging relative to that expected from balancer classes); Column 4: Phenotypes observed in multiple RNAi-driven flies, where applicable; and Column 5: Predicted function based on homologous protein structure or functional studies.
| 3L Het gene targeted (from proximal to distal) | RNAi lines used: | Effects of driving RNAi lines: | Phenotypes observed: | Encoded function targeted: |
|---|---|---|---|---|
| CG17514/lethal1 | V47268, V47269, BL34355 | V/F, SL (M: 9%, FE: 8%)/S, L | Thin thoracic bristles, tergite defects, body elongation, held out/blistered wings. | Translational activator. |
| eIF-4B | V31364, BL57305 | V/F, V/F | Thin or missing thoracic bristles (V31364). | Translational initiation factor (may have an accessory role). |
| dbp80 | BL3468, V109742 | SL (M: 17%, FE: 18%)/S, L | Kinked bristles, tergite defects, wings held out or appear blistered. | RNA helicase. May be required for nuclear export of RNA. |
| rpl15/lethal2 | not tested | N/A | Component of the ribosome. | |
| vtd/lethal3 | not tested | N/A | Sister chromosome cohesion, gene expression. | |
| CG40228 | V110000 | SL (M: 11%, FE: 12%)/F | Ectopic thorax bristles, melanotic spots, flipped out wings. | Transcriptional elongation factor. Component of ELL? |
| CG42598% | V109076 | V/F | Flies appear wider, segments elongated, flipped out wings. | Unknown. |
| CG41284% | V109076 | V/F | Flies appear wider, segments elongated, flipped out wings. | Unknown. |
| CG40160 | V109559, BL44268 | L, V/F | Peptidase. | |
| CG40178/lethal4A | V110089, BL44578 | L, L | Thioredoxin, chaperone. | |
| FASN3 (CG17374)/lethal4B | CG17374-2M, 3M | L, L | Fatty acid synthase. | |
| CG45782 | V109249, BL38367, V109094, BL40876 | V/S (M only), V/F, L, L | Sucrose transporter. | |
| Ucrh/UQCR-11 | not tested | N/A | Oxidative phosphorylation. | |
| med21 (CG17397) | BL34731, V31940 V109982, V13667 | L, V, L, L | Transcriptional initiation, component of mediator. | |
| dSet1/lethal5 | V40682, V40683, V10833, V45267 | L, L, L, L | H3K4 di- and tri-methylase. | |
| CG40002%/ND-AGGG | V109239, BL43285 | V/F, V/F | Oxidative phosphorylation. | |
| CG40472% | nothing available | N/A | Oxidative phosphorylation. | |
| scro/lethal7B | V33902 | L | Cell-specific transcription factor. | |
| mRps5/lethal8 | BL36202 | L | Mitochondrial ribosomal protein. | |
| nvd/lethal6 | not tested | N/A | Ecdysone biosynthesis, molting/metamorphosis. | |
| snap-25 | BL27306, BL34377 | V/F, L | SNARE protein, exocytosis. | |
| CG40045 | V109167 | V/F | E2 ubiquitin conjugating enzyme. | |
| CG40470 | not tested | N/A | Peptidase. | |
| CG43968 | nothing available | N/A | Unknown, may be secreted. | |
| CG17698 | V35634, V105884 | V/F, L | Curved/kinked thoracic bristles. | Calmodulin dependent kinase, calcium signaling. |
| CG40298 | nothing available | N/A | Unknown. | |
| CG17454 | V20458, BL43199 | L, L | Spliceosome component. | |
| ago3 | not tested | N/A | piRNA biogenesis, germline silencing of transposons. | |
| alpha-Cat | V19182, V20123 | L, L | Cellular adhesion, adherens junction formation. | |
| CG32230/ND-MLRQ | V9804, V10316, V101482 | V/F, V/F, V/F | Melanin deposits on some flies. | Oxidative phosphorylation. |
| nAChRa4 | V11392, BL31985, V12441 | SL/F (M: 0%, FE: 9%), V/F, SL (M: 19%; FE: 29%)/F | n-acetylcholine receptor, neurotransmission. | |
| CG34031 | V109096 | V/F | Unknown. | |
| vps11/CG32350 | V24731, V107420 | L, V/F | Vacuolar protein sorting, lysosomes/late endosome function. | |
| CG33217 | V47818, V103309, BL43315, BL43167 | L, L, L, L | Unknown. | |
| CKIIalpha | not tested | N/A | Diurnal rhythm regulation, involved in cell signaling. | |
| rpL10/Qm | V19083, V19084 | L, L | Component of the ribosome. | |
| nrm | V44176, BL52945 | V/F, V/F | Adhesion at neuromuscular junctions. | |
| CG32457 | V15966, V15968 | L, SL (M: 0%, FE: 10%)/F | Wings often not folded out in male escapers. | Unknown. |
| CG32461 | V26149, V26150, V108971 | V/F. V/F, V/F | Unknown. |
%The sequences of these genes are very similar and are likely to result from annotation issues or more improbably, very recent duplications.
Summary of essential genes in 3L heterochromatin
The essential 3L Het genes identified either through isolation/analysis of mutant alleles or RNAi analysis are shown in Figure 7 and listed in Table 6. Overall, our data suggest that at least 25/39 genes (roughly 2/3) of genes in 3L Het are essential (26/39 if we include nAchRalpha4 as a putative lethal), an unexpectedly large proportion relative to the roughly 1/3 of euchromatic genes that are predicted to be essential based on analyses of specific euchromatic regions (see Discussion). In addition, it is possible that our estimate of essential 3L genes is conservative and the number of essential genes in this region may be higher. We aren’t able to readily estimate whether more genes may have been missed in our mutagenesis screens (see Hilliker et al. 1981 for a discussion of this point), and there is currently an absence of genetic reagents (RNAi constructs, other) for studying some 3L Het gene models.
Figure 7.
Map of essential genes present in 3L Het (not to scale). Blue- genes identified by us as essential through RNAi screening; red- genes identified by us as essential through mutants; black – genes identified by others as essential. *marks the uncertain status of nAchRa4, as discussed above.
Unassigned lethal mutations generated in the current study
A number of lethal mutations characterized in our study have not yet been assigned to known complementation groups but may be located in 3L Het. For example, the Df(3L)XXX allele from the initial X-ray screen fails to complement Df(3L)γ-28, but it does complement Df(3L)1-166 and Df(3L)FX33 (see Figure 4, Df(3L)XXX not shown on map), suggesting that an additional essential gene may reside between the lethal 7B /scarecrow and lethal 8 loci. There are also several EMS lesions that fail to complement Df(3LR)6B-29, but which have not been assigned to any known complementation group. While some of these may be alleles of second-site mutations on the Df(3LR)6B-29 chromosome, outside of 3L Het, others may represent alleles of unidentified essential 3L Het loci e.g., Z-855 and Z-5460 clearly map to 3L Het, but they complement alleles of all known essential 3L Het complementation groups. In addition, Z-1326 and Z-1605 were identified on the basis of failure to complement Df(3L)Delta1-AK and remain unassigned; however, while these alleles may correspond to distally located 3L Het genes, it is also possible that they are alleles of euchromatic genes at the heterochromatic/euchromatic border which have not yet been well characterized. Further work is necessary to determine the basis of all of the unassigned mutations; however, the next release of an updated Drosophila genome sequence should make it possible to identify and characterize more of these uncharacterized loci.
Discussion
The work reported here links a detailed genetic map of 3L Het with the physical map available on Flybase. In the process, we have been able to assign molecular identities to most of our lethal complementation groups in 3L Het, which in the past have only been studied at a genetic level. Since full characterization of any genome is incomplete without an analysis of heterochromatin, our work provides an initial platform for the functional annotation of essential genes residing within these fascinating, yet poorly understood, regions of the genome. Our collection of mutant strains will be useful for studying the function of heterochromatic genes that have resisted characterization due to their location in a genetically less tractable region of the genome.
Using RNAi knockdown analyses, we have identified an additional group of essential 3L Het genes not identified in mutagenesis screens. Lethality associated with RNAi gene targeting provides good evidence that a gene is essential: the possibility of RNAi off-target effects seems unlikely because for all but one of the genes tested for RNAi knockdown effects, there were no detectable homologies to the 19 b.p. gene regions used in the RNAi transgene design (data not shown); moreover, off-targeting effects in flies have been reported as being very low (Dietzl et al. 2007). The single exception involved two VDRC RNAi lines targeting the nAChRalpha4 gene; each of these exhibits a single predicted 19 b.p. off-target dsRNA; however, the majority of dsRNAs available for this gene do result in gene specific targeting, and therefore this single off-target RNA may be inconsequential. A homozygous viable allele of nAChRalpha4 has recently been reported to exhibit circadian defects (Shi et al. 2014). However, since this allele is associated with a single amino acid substitution and was obtained in a screen for viable modifier mutations, it is likely not a null allele. If the effects on viability we observe by expressing nAChRalpha4 RNAi lines result from direct gene-targeting, the nAchRalpha4 protein is likely essential and may have additional biological roles.
There remain some 3L Het gene models for which we have neither RNAi lines nor mutants, so at this point it is not possible to define whether any of these genes are essential. Furthermore, the observed viability when expressing available RNAi lines for certain genes does not prove that these genes are non-essential; an RNAi line may be poorly-expressed or substantial maternal effects may allow survival and thus obscure the effects of RNAi targeting. Indeed, gene expression studies showed that several RNAi lines tested only modestly reduced expression of the target genes (data not shown). Finally, while it seems likely that eIF-4B should also be essential, based on its presumed cell function, it is noteworthy that our results, as well as other RNAi analyses to date (http://www.genomernai.org/v17/fullPagePhenotypes/gene/3355041) are not consistent with an essential role for this protein (although again, similar caveats apply).
In terms of other potential caveats, lethal phenotypes resulting from RNAi knockdowns may not indicate that a gene is essential, because in some cases, genetic mutations knocking out the same gene do not show a lethal phenotype. This relatively rare phenomenon has been attributed to other factors such as genetic compensation (Rossi et al. 2015, and refs therein).
Other strategies could be employed to determine whether any of the remaining 3L Het genes are essential. For example, since transposon insertions near genes of interest are becoming increasingly available, these could serve as starting points for generating imprecise excisions that affect adjacent genes. In addition, genetic screens for mutations lethal in trans to the distal-most 3L Het deficiencies (Df(3L)TTT or Df(3L)1677) may yield more lethal mutations between ago3 and RpL10. Analysis of the latter 3L Het segment has proved difficult because deletions that remove the RpL10 locus exhibit dominant semi-lethality and female sterility (K. Fitzpatrick unpublished observations). Lastly, with the innovations of CRISPR technology, plus the availability of newer targeted genetic reagents (e.g., Mi{MIC} and Mi{Trojan-GAL4.0} alleles, see Flybase), it may be possible to extend the current work, although there could be issues with accessibility of DNA sequences when packaged in the chromatin environment of centric heterochromatin.
The findings from this and other studies indicate that although gene density is relatively low in centric heterochromatin, many genes located in this chromatin environment are functionally important. The essential genes identified in the current study appear to span a diverse set of molecular and developmental functions. We and others have noted that there is no correlation between developmental or other specific expression patterns and the location of genes within 3L and other centric Het regions. Although the size of an average heterochromatic gene is considerably larger than that of a typical euchromatic gene, neither is there any apparent correlation between heterochromatic gene size and whether a given gene is essential. While our mutagenesis experiments often produced mutations in larger 3L Het genes (based both on physical locus and gene product length), mutant alleles of some smaller essential genes (rpl15 and CG40002) were also isolated, and several other small essential genes (such as med21 and CG17454) were identified through RNAi experiments. Thus, although our results indicate an unexpectedly higher proportion of essential genes in 3L Het, there are no obvious properties that distinguish them from euchromatic genes.
As mentioned, our data suggest that approximately 2/3 of genes residing in 3L Het are required for developmental viability and/or adult fertility. In contrast, it is estimated that only about 1/3 of euchromatic genes are essential (Miklos and Rubin 1996; Ashburner et al. 1999; Dietzl et al. 2007; Chen et al. 2010). It remains to be determined whether the large proportion of essential genes in 3L Het is significant or results from the relatively small sample size: in 3R Het, only four of the 11 genes appear to be essential (our unpublished observations), while the overall proportion of essential genes in 2R and 2L Het remains to be established.
The work described here, and the genetic tools generated, provide a foundation for advancing our understanding of the organization, functions and regulation to genes in these unique chromosomal regions. Fly stocks from this work, including representative Dfs across 3L and 3R Het, lethal alleles of individual genes, as well as other reagents (cDNA transgene and RNAi lines) will be available from the Bloomington Drosophila Stock Center, Indiana.
Acknowledgments
We would like to thank Sarah Padilla, Kelsey Jefferson, Kate Auyeung, Sara Brown, Florence Yuen, Gary Tong, Sabrina Rayworth, Amanda Berscht and a number of talented undergraduate assistants for expert technical assistance. We also thank the Drosophila community for their support, including providing resources necessary for this work. We would particularly like to acknowledge Roger Hoskins, Gary Karpen, the Drosophila Heterochromatic Genome Project and Flybase for access to genome sequence data and physical maps of heterochromatin; Kevin Cook and the Bloomington Drosophila Stock Center; the Vienna Drosophila RNAi Center, National Institute of Japan, and the Transgenic RNAi Projects for RNAi lines; the Zuker and Deitcher laboratories for providing mutant stocks essential for this work; and the Drosophila Genomic Resource Center for cloned cDNAs. This research was funded by a Natural Sciences and Engineering Research Council (NSERC) grants to B.M. Honda and to A.J. Hilliker; a Canadian Institute of Health Research (CIHR) grant to A.J. Hilliker, B.M. Honda, and D.A.R. Sinclair; and NSERC doctoral fellowships to M.A. Syrzycka and G. Hallson.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7828301
Communicating editor: J. Birchler
Literature Cited
- Adams M. D., Celniker S. E., Holt R. A., Evans C. A., Gocayne J. D., et al. , 2000. The genomic sequence of Drosophila melanogaster. Science 287: 2185–2195. 10.1126/science.287.5461.2185 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Andreyeva E. N., Kolesnikova T. D., Demakova O. V., Mendez-Lago M., Pokholkova G. V., et al. , 2007. High-resolution analysis of Drosophila heterochromatin organization using SuUR Su(var)3–9 double mutants. Proc. Natl. Acad. Sci. USA 104: 12819–12824. 10.1073/pnas.0704690104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Misra S., Roote J., Lewis S. E., Blazej R., et al. , 1999. An exploration of the sequence of a 2.9-mb region of the genome of Drosophila melanogaster: The Adh region. Genetics 153: 179–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A. B., Vicosoa B., Russo C. A. M., Swenor B., Clark A. G., 2015. Birth of a new gene on the Y chromosome of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 112: 12450–12455. 10.1073/pnas.1516543112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. H., Larracuente A. M., 2018. Heterochromatin-enriched assemblies reveal the sequence and organization of the Drosophila melanogaster Y chromosome. Genetics. 10.1534/genetics.118.301765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhang Y. E., Long M., 2010. New genes in Drosophila quickly become essential. Science 330: 1682–1685. 10.1126/science.1196380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg N. J., Honda B. M., Whitehead I. P., Grigliatti T. A., Wakimoto B., et al. , 1998. Suppressors of position-effect variegation in Drosophila melanogaster affect expression of the heterochromatic gene light in the absence of a chromosome rearrangement. Genome 41: 495–503. 10.1139/g98-041 [DOI] [PubMed] [Google Scholar]
- Cook R. K., Christensen S. J., Deal J. A., Coburn R. A., Deal M. E., et al. , 2012. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol. 13: R21 10.1186/gb-2012-13-3-r21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulthard A. B., Alm C., Cealiac I., Sinclair D. A., Honda B. M., et al. , 2010. Essential loci in centromeric heterochromatin of Drosophila melanogaster. I: The right arm of chromosome 2. Genetics 185: 479–495. 10.1534/genetics.110.117259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulthard A. B., Eberl D. F., Sharp C. B., Hilliker A. J., 2003. Genetic analysis of the second chromosome centromeric heterochromatin of Drosophila melanogaster. Genome 46: 343–352. 10.1139/g03-010 [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., et al. , 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Dimitri P., Caizzi R., Giordano E., Accardo M. C., Lattanzi G., et al. , 2009. Constitutive heterochromatin: a surprising variety of expressed sequences. Chromosoma 118: 419–435. 10.1007/s00412-009-0211-y [DOI] [PubMed] [Google Scholar]
- Eberl D. F., Duyf B. J., Hilliker A. J., 1993. The role of heterochromatin in the expression of a heterochromatic gene, the rolled locus of Drosophila melanogaster. Genetics 134: 277–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J. C., James T. C., Foster-Hartnett D. M., Hartnett T., Ngan V., et al. , 1990. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 87: 9923–9927. 10.1073/pnas.87.24.9923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J. C., Reuter G., 2009. Cellular mechanism for targeting heterochromatin formation in Drosophila, pp. 1–47 in International Review of Cell and Molecular Biology, Vol. 273 chap 1., edited by Jeon K. W. Academic Press, Cambridge, MA. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick K. A., Sinclair D. A., Schulze S. R., Syrzycka M., Honda B. M., 2005. A genetic and molecular profile of third chromosome centric heterochromatin in Drosophila melanogaster. Genome 48: 571–584. 10.1139/g05-025 [DOI] [PubMed] [Google Scholar]
- Hallson G., Syrzycka M., Beck S. A., Kennison J. A., Dorsett D., et al. , 2008. The Drosophila cohesin subunit Rad21 is a trithorax group (trxG) protein. Proc. Natl. Acad. Sci. USA 105: 12405–12410. 10.1073/pnas.0801698105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallson G., Hollebakken R. E., Li T., Syrzycka M., Kim I., et al. , 2012. dSet1 is the main H3K4 Di- and tri-methyltransferase throughout Drosophila development. Genetics 190: 91–100. 10.1534/genetics.111.135863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz E., 1928. Das heterochromatin der moose, Bornträger, Berlin. [Google Scholar]
- Heitz E., 1929. Heterochromatin, chromocentren, chromomeren. Komm, Fischer, Jena, Germany. [Google Scholar]
- Hilliker A. J., Appels R., 1982. Pleiotropic effects associated with the deletion of heterochromatin surrounding rDNA on the X chromosome of Drosophila. Chromosoma 86: 469–490. 10.1007/BF00330122 [DOI] [PubMed] [Google Scholar]
- Hilliker A. J., Chovnick A., Clark S. H., 1981. The relative mutabilities of vital genes in Drosophila melanogaster. Drosophila. Inform. Serv. 56: 64–65. [Google Scholar]
- Hilliker A. J., Holm D. G., 1975. Genetic analysis of the proximal region of chromosome 2 of Drosophila melanogaster. I. Detachment products of compound autosomes. Genetics 81: 705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker A. J., 1976. Genetic analysis of the centromeric heterochromatin of chromosome 2 of Drosophila melanogaster: deficiency mapping of EMS-induced lethal complementation groups. Genetics 83: 765–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins, R. A., C. D. Smith, J. W. Carlson, A. B. Carvalho, A. Halpern et al., 2002 Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biology 3: RESEARCH0085.1–0085.6. [DOI] [PMC free article] [PubMed]
- Hoskins R. A., Carlson J. W., Kennedy C., Acevedo D., Evans-Holm M., et al. , 2007. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science 316: 1625–1628. 10.1126/science.1139816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins R. A., Carlson J. W., Wan K. H., Park S., Mendez I., et al. , 2015. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 25: 445–458. 10.1101/gr.185579.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe M., Dimitri P., Berloco M., Wakimoto B. T., 1995. Cis-effects of heterochromatin on heterochromatic and euchromatic gene activity in Drosophila melanogaster. Genetics 140: 1033–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania A., Bellen H., 1995. Mutations in neuromusculin, a gene encoding a cell Roux Arch. Dev. Biol. 204: 259–270. 10.1007/BF00208493 [DOI] [PubMed] [Google Scholar]
- Kania A., Han P. L., Kim Y. T., Bellen H., 1993. Neuromusculin, a Drosophila gene expressed in peripheral neuronal precursors and muscles, encodes a cell adhesion molecule. Neuron 11: 673–687. 10.1016/0896-6273(93)90078-6 [DOI] [PubMed] [Google Scholar]
- Kharchenko P. V., Alekseyenko A. A., Schwartz Y. B., Minoda A., Riddle N. C., et al. , 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485. 10.1038/nature09725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koryakov D. E., Zhimulev I. F., Dimitri P., 2002. Cytogenetic analysis of the third chromosome heterochromatin of Drosophila melanogaster. Genetics 160: 509–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koryakov D. E., Domanitskaya E. V., Belyakin S. N., Zhimulev I. F., 2003. Abnormal tissue-dependent polytenization of a block of chromosome 3 pericentric heterochromatin in Drosophila melanogaster. J. Cell Sci. 116: 1035–1044. 10.1242/jcs.00283 [DOI] [PubMed] [Google Scholar]
- Koundakjian E. J., Cowan D. M., Hardy R. W., Becker A. H., 2004. The Zuker Collection: a resource for the analysis of autosomal gene function in Drosophila melanogaster. Genetics 167: 203–206. 10.1534/genetics.167.1.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., Mcgettigan P. A., et al. , 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Li C., Vagin V. V., Lee S., Xu J., Ma S., et al. , 2009. Collapse of germline piRNAs in the absence of argonaute3 reveals somatic piRNAs in flies. Cell 137: 509–521. 10.1016/j.cell.2009.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. M., Kilman V. L., Keegan K., Paddock B., Emery-Le M., et al. , 2002. A role for casein kinase 2alpha in the Drosophila circadian clock. Nature 420: 816–820. 10.1038/nature01235 [DOI] [PubMed] [Google Scholar]
- Lindsley D. L., Hardy R. W., 1992. Cytology of In(3L)TM8 and In(3L)TM9. DIS 71: 154. [Google Scholar]
- Lu B. Y., Emtage P. C., Duyf G. J., Hilliker A. J., Eissenberg J. C., 2000. Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila. Genetics 155: 699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe A. R., Hilliker A. J., Roberts P. A., 1993. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics 134: 1149–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant G. E., Holm D. G., 1988a Genetic analysis of the heterochromatin of chromosome 3 in Drosophila melanogaster. I. Products of compound autosome detachment. Genetics 120: 503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant G. E., Holm D. G., 1988b Genetic analysis of the heterochromatin of chromosome 3 in Drosophila melanogaster. II. Vital loci identified through EMS mutagenesis. Genetics 120: 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marygold S. J., Roote J., Reuter G., Lambertsson A., Ashburner M., et al. , 2007. The ribosomal protein genes and minute loci of Drosophila melanogaster. Genome Biol. 8: R216 10.1186/gb-2007-8-10-r216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mével-Ninio M., Pelisson A., Kinder J., Campos A. R., Bucheton A., 2007. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics 175: 1615–1624. 10.1534/genetics.106.068106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklos G. L. G., Rubin G. M., 1996. The role of the genome project in determining gene function: insights from model organisms. Cell 86: 521–529. 10.1016/S0092-8674(00)80126-9 [DOI] [PubMed] [Google Scholar]
- Mukai T., 1964. The genetic structure of natural populations of Drosophila melanogaster. I. Spontaneous mutation rate of polygenes controlling viability. Genetics 50: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myster S. H., Wang F., Cavallo R., Christian W., Bhotika S., et al. , 2004. Genetic and bioinformatic analysis of 41C and the 2R heterochromatin of Drosophila melanogaster: a window on the heterochromatin-euchromatin junction. Genetics 166: 807–822. 10.1534/genetics.166.2.807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T., Ohnishi S. T., Salerno J. C. 2018. Five decades of research on mitochondrial NADH-quinone oxidoreductase (complex I). J. Biol Chem. Oct 25: 1249–1264. [DOI] [PubMed] [Google Scholar]
- Reuter G., Giarre M., Farah J., Gausz J., Spierer A., et al. , 1990. Dependence of position-effect variegation in Drosophila on dose of a gene encoding an unusual zinc-finger protein. Nature 344: 219–223. 10.1038/344219a0 [DOI] [PubMed] [Google Scholar]
- Riddle N. C., Elgin S. C. R., 2018. The Drosophila Dot Chromosome: Where Genes Flourish Amidst Repeats. Genetics 210: 757–772. 10.1534/genetics.118.301146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle N. C., Minoda A., Kharchenko P. V., Alekseyenko A. A., Schwartz Y. B., et al. , 2011. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 21: 147–163. 10.1101/gr.110098.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A., Kontarakis Z., Gerri C., Nolte H., Hölper S., et al. , 2015. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524: 230–233. 10.1038/nature14580 [DOI] [PubMed] [Google Scholar]
- Rossi F., Moschetti R., Caizzi R., Corradini N., Dimitri P., 2007. Cytogenetic and molecular characterization of heterochromatin gene models in Drosophila melanogaster. Genetics 175: 595–607. 10.1534/genetics.106.065441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarpal R., Pellikka M., Patel R. R., Hui F. Y., Godt D., et al. , 2012. Mutational analysis supports a core role for Drosophila α-catenin in adherens junction function. J. Cell Sci. 125: 233–245. 10.1242/jcs.096644 [DOI] [PubMed] [Google Scholar]
- Schotta G., Lachner M., Sarma K., Ebert A., Sengupta R., et al. , 2004. A silencing pathway to induce H3–K9 and H4–K20 trimethylation at constitutive heterochromatin. Genes Dev. 18: 1251–1262. 10.1101/gad.300704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze S., Sinclair D. A., Silva E., Fitzpatrick K. A., Singh M., et al. , 2001. Essential genes in proximal 3L heterochromatin of Drosophila melanogaster. Mol. Gen. Genet. 264: 782–789. 10.1007/s004380000367 [DOI] [PubMed] [Google Scholar]
- Schulze S. R., Sinclair D. A., Fitzpatrick K. A., Honda B. M., 2005. A genetic and molecular characterization of two proximal heterochromatic genes on chromosome 3 of Drosophila melanogaster. Genetics 169: 2165–2177. 10.1534/genetics.103.023341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Yue Z., Kuryatov A., Lindstrom J. M., Sehgal A., 2014. Identification of Redeye, a new sleep-regulating protein whose expression is modulated by sleep amount. eLife 3: e01473 10.7554/eLife.01473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair D. A. R., Schulze S., Silva E., Fitzpatrick K. A., Honda B. M., 2000. Essential Genes in Autosomal Heterochromatin of Drosophila melanogaster. Genetica 109: 9–18. 10.1023/A:1026500620158 [DOI] [PubMed] [Google Scholar]
- Sinclair D. A. R., Syrzycka M., Macauley M. S., Rastgardani T., Komljenovic I., et al. , 2009. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc). Proc. Natl. Acad. Sci. USA 106: 13427–13432. 10.1073/pnas.0904638106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. D., Shu S. Q., Mungall C. J., Karpen G. H., 2007. The release 5.1 annotation of Drosophila melanogaster heterochromatin. Science 316: 1586–1591. 10.1126/science.1139815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschiersch B., Hofmann A., Krauss V., Dorn R., Korge G., et al. , 1994. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3–9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 13: 3822–3831. 10.1002/j.1460-2075.1994.tb06693.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vass S., Cotterill S., Valdeolmillos A. M., Barbero J. L., Lin E., et al. , 2003. Depletion of Drad21/Scc1 in Drosophila cells leads to instability of the cohesin complex and disruption of mitotic progression. Curr. Biol. 13: 208–218. 10.1016/S0960-9822(03)00047-2 [DOI] [PubMed] [Google Scholar]
- Vilinsky I., Stewart B. A., Drummond J., Robinson I., Deitcher D. L., 2002. A Drosophila SNAP-25 null mutant reveals context-dependent redundancy with SNAP-24 in neurotransmission. Genetics 162: 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakimoto B. T., Hearn M. G., 1990. The effects of chromosome rearrangements on the expression of heterochromatic genes in chromosome 2L of Drosophila melanogaster. Genetics 125: 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., Neumüller R. A., Buckner M., Ayers K., Li H., Hu Y., Yang-Zhou D., et al. , 2014. A regulatory network of Drosophila germline stem cell self-renewal. Developmental Cell 28: 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama T., Namiki T., Mita K., Kataoka H., Niwa R. 2006. Neverland is an evolutionally conserved Rieske-domain protein that is essential for ecdysone synthesis and insect growth. Development 133: 2565–74. 10.1242/dev.02428 [DOI] [PubMed] [Google Scholar]
- Zhang P., Spradling A. C., 1994. Insertional mutagenesis of Drosophila heterochromatin with single P elements. Proc. Natl. Acad. Sci. USA 91: 3539–3543. 10.1073/pnas.91.9.3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
A comprehensive and representative set of stocks have been sent to Dr Kevin Cook at the Bloomington Drosophila stock center. Supplemental files available at FigShare. Table S1 lists primers used in mapping. Table S2 lists EMS mutations identified. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7828301.