Abstract
In this genome report, we describe the sequencing and annotation of the genome of the wine grape Carménère (clone 02, VCR-702). Long considered extinct, this old French wine grape variety is now cultivated mostly in Chile where it was imported in the 1850s just before the European phylloxera epidemic. Genomic DNA was sequenced using Single Molecule Real Time technology and assembled with FALCON-Unzip, a diploid-aware assembly pipeline. To optimize the contiguity and completeness of the assembly, we tested about a thousand combinations of assembly parameters, sequencing coverage, error correction and repeat masking methods. The final scaffolds provide a complete and phased representation of the diploid genome of this wine grape. Comparison of the two haplotypes revealed numerous heterozygous variants, including loss-of-function ones, some of which in genes associated with polyphenol biosynthesis. Comparisons with other publicly available grape genomes and transcriptomes showed the impact of structural variation on gene content differences between Carménère and other wine grape cultivars. Among the putative cultivar-specific genes, we identified genes potentially involved in aroma production and stress responses. The genome assembly of Carménère expands the representation of the genomic variability in grapes and will enable studies that aim to understand its distinctive organoleptic and agronomical features and assess its still elusive extant genetic variability. A genome browser for Carménère, its annotation, and an associated blast tool are available at http://cantulab.github.io/data.
Keywords: genome assembly, heterozygosity, haplotype phasing, structural variation, Vitis vinifera
Carménère (also known as Grand Vidure) is a historically and economically important wine grape (Vitis vinifera L.) cultivar with distinctive organoleptic and agronomical features (Huamán-Castilla et al. 2017). Carménère is an old French cultivar, which is thought to be derived from a cross between Cabernet Franc and Gros Cabernet (Boursiquot et al. 2009). It was widely planted in the Bordeaux regions of Graves and Médoc before the aphid-like soil-born pest Phylloxera vastatrix devastated French vineyards in the 19th century. While almost extinct in France due to poor fruit set and late ripening, Carménère has well adapted to the Chilean climate and soil where it has become the flagship red wine grape with more than 10,000 hectares planted in most valleys throughout the country (Servicio Agrícola y Ganadero, https://www.sag.gob.cl/, 2016). Brought to Chile in the 1850s with other Bordeaux grapes just before phylloxera hit Europe, it was wrongly identified as Merlot until 1994 when the French ampelographer Jean Michel Boursiquot and, few years later, DNA fingerprinting (Hinrichsen et al. 2001) determined that the “Merlot Chileno” was instead Carménère (Pszczólkowski 2004; Richards 2006). A similar situation happened in Italy where it was confused with Cabernet Franc until 1991 (Caló et al. 1991). Carménère success is due to the peculiarity of its wines, which are deeply colored, with well-structured tannins, and distinctive aroma and flavor, that combine green, herbaceous features with fruity, spicy, berry-like notes (Casaubon et al. 2006; Fernández et al. 2007; Domínguez and Agosin 2010). Unlike most red wine cultivars, Carménère berries accumulates high concentration of methoxypyrazines, mainly 3-isobutyl-2-methoxypyrazine (IBMP) (Belancic and Agosin 2007), which confer the characteristic vegetal attributes in the resulting wines. The genetic bases of the phenological and compositional differences between Carménère and related varieties, such as Merlot, Cabernet Sauvignon, and Cabernet Franc, are not known.
Extensive structural variation between grape genotypes leads to significant unshared gene content between cultivars, which has been shown to contribute to varietal phenotypic characteristics (Venturini et al. 2013; Da Silva et al. 2013; Gambino et al. 2017; Zhou et al. 2018; Minio et al. 2019). Cultivar-specific genes have been discovered by whole-genome or transcriptome comparative analyses (Venturini et al. 2013; Da Silva et al. 2013; Gambino et al. 2017; Zhou et al. 2018; Minio et al. 2019). However, despite the relatively small genome size estimated at about 500 Mbp, the assembly of grape genomes is difficult because of the high level of heterozygosity (Minio et al. 2017). We recently reported that contiguous and accurate assemblies of grape genomes can be generated by assembling long Single-Molecule Real-Time (SMRT) sequencing reads using FALCON-Unzip, a diploid-aware assembler (Chin et al. 2016; Minio et al. 2017). In this work, we sequenced, assembled, and annotated the genome of Carménère clone 02 (VCR-702). As part of this project, we tested different combinations of assembly parameters, including variable sequencing coverage, to optimize the FALCON-Unzip pipeline and achieve the optimal contiguity and completeness of the assembly. Comparisons with other publicly available grape genomes identified structural variations and gene content differences in the Carménère genome.
Methods & Materials
Library preparation and sequencing
With permission from Vivai Cooperativi Rauscedo (Italy), we collected 1-2 cm-wide young leaves from Carménère clone 02 (equivalent to clone VCR 702) vines, maintained at Foundation Plant Services (FPS, University of California, Davis). High-molecular-weight genomic DNA (gDNA) was isolated using the method described in Chin et al. (2016). Genetic identity was confirmed with a standard set of microsatellite markers (Hinrichsen et al. 2001; This et al. 2004). DNA purity was evaluated with a Nanodrop 2000 spectrophotometer (Thermo Scientific, Hanover Park, IL, USA), DNA quantity with the DNA High Sensitivity kit on a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA), and DNA integrity by pulsed-field gel electrophoresis. gDNA was cleaned with 0.45x AMPure PB beads (Pacific Biosciences, Menlo Park, CA, USA) before library preparation. SMRTbell template was prepared with 15 µg of sheared DNA using SMRTbell Template Prep Kit (Pacific Biosciences, Menlo Park, CA, USA) following the manufacturer’s instructions. For size selection, 30 µl of SMRTbell template were loaded on a Sage Blue Pippin (Sage Science, Beverly, MA, USA) and size-selected with a cutoff range of 17-50 Kbp. The size-selected library was cleaned with 1x AMPure PB beads. After DNA damage repair, the library were cleaned again with 1x AMPure PB beads. A total of 62 SMRT cells were sequenced on a PacBio RS II using P6/C4 chemistry, generating 6,615,332 reads for a total of ∼56 Gbp. DNA-seq libraries were prepared using the Kapa LTP library prep kit (Kapa Biosystems, Wilmington, MA, USA) and evaluated for quantity and quality with the High Sensitivity chip on a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). A total of 100,190,577 DNA fragments were sequenced into 2x150bp reads on an Illumina HiSeq4000 (DNA Technology Core Facility, University of California, Davis).
Genome assembly
Assembly of SMRT reads was performed with a customized FALCON-Unzip pipeline (v.2017.06.28-18.01; Chin et al. 2016), whose codes can be found at https://github.com/andreaminio/FalconUnzip-DClab. Prior to error correction, repeats were marked using the TANmask and REPmask modules from the DAmasker (Myers 2014). Repeats were marked also on error-corrected reads before assembly with FALCON. This additional repeat masking step increased assembly contiguity by 20% and decreased the computational time required for assembly by about 6%. To test the impact of sequencing coverage on FALCON assembly, raw SMRT reads were down-sampled randomly using seqtk (v.1.2-r101-dirty; https://github.com/lh3/seqtk) at theoretical 100x, 75x, 50x, 25x, 10x and 5x coverages. All datasets were assembled with FALCON without masking of corrected reads for all coverage combinations and with masking for full dataset down to 50x of coverage. As sequencing coverage influences error correction, we repeated the assembly on datasets created by down-sampling the error-corrected reads at 25x, 20x, 15x, 10x, and 5x coverages. Hybrid error correction was also tested to improve sequence accuracy of low-coverage dataset with short reads. Hybrid error correction was performed using LoRDEC (v.0.7 with GATB v.1.2.2; Salmela and Rivals 2014) with 1, 5 or 9 iterations over 50x, 25x, 10x and 5x datasets. FALCON-Unzip was performed on all 82 datasets with multiple assembly parameters (i.e., read length retention threshold, self-alignment diagonal bands of width, read correlation rate, k-mer size and number of hits) for a total of 1,027 independent assemblies that were evaluated for contiguity and completeness (Table S1). Gene space completeness in the assembly was assessed by alignment of the complete PN40024 V1 genes (http://genomes.cribi.unipd.it/DATA/) on primary contigs using GMAP (v.2015-09-29; Wu and Watanabe 2005) with parameters “-B 4 -x 30 -f2” and discarding mappings with translocations. Haplotype phasing was carried out with Unzip and default parameters (Chin et al. 2013). Primary contigs and haplotigs were polished with Arrow (from ConsensusCore2 v.3.0.0). Primary contigs were scaffolded using SSPACE Longreads (v.1.1; Boetzer and Pirovano 2014), followed by gap closing with PBJelly (PBsuite v.15.8.4; English et al. 2012, 2014). Gene space completeness of the final assembly was assessed with BUSCO (v.3; Simão et al. 2015).
Genome annotation
Repetitive sequences were identified with RepeatMasker (v.open-4.0.6; Smit et al. 2013) using a custom V. vinifera repeat library described in Minio et al. (2019). Repeats were masked prior to gene prediction. Protein-coding genes were predicted with EVM (v.1.1.1; Haas et al. 2008) using as input: (i) ab initio predictions from SNAP (v.2006-07-28; Korf 2004), Augustus (v.3.0.3; Stanke et al. 2006), GeneMark-ES (v.4.32; Lomsadze et al. 2005), GlimmerHMM (v.3.0.4; Majoros et al. 2004), and GeneID (v.1.4.4; Parra et al. 2000) trained on Cabernet Sauvignon; (ii) ab initio predictions of Augustus trained on BUSCO dataset; (iii) as experimental evidence, proteins from Swissprot viridiplantae (downloaded on 2016.03.15), mapped with Exonerate (v.2.2.0; Slater and Birney 2005); (iv) as transcriptional evidence, Vitis ESTs and flcDNAs (downloaded on 2016.03.15), Vitis vinifera PN40024 V1 CDS (http://genomes.cribi.unipd.it/DATA/), Tannat (TSA GAKH01.1) and Corvina (TSA PRJNA169607) transcriptomes, and Cabernet Sauvignon corrected Iso-Seq reads (SRP132320); (v) PASA (v.2.1.0; Haas et al. 2003, 2008; Campbell et al. 2006) predicted gene models based on the transcriptional evidences from (iv). Functional annotations were assigned based on homology with proteins in the RefSeq plant protein database (downloaded on 2017.01.17) and on functional domains identified with InterProScan (v.5; Jones et al. 2014; Table S2). Gene content variability between cultivars was assessed by alignment of the Carménère genes onto the PN40024 genome 12X.v2 (Canaguier et al. 2017) and Cabernet Sauvignon clone 08 genome (Chin et al. 2016) using GMAP (v.2015-09-29; Wu and Watanabe 2005) with identity and coverage >80%. Homology with Corvina, Tannat, and Nebbiolo transcripts was determined by blastn search of hits with reciprocal identity and coverage greater than 80%. Structural comparisons between assemblies were performed with MUMMER (v.4.0; Marçais et al. 2018) and variant impacts were annotated with SnpEff (v.4.3m; Cingolani et al. 2012).
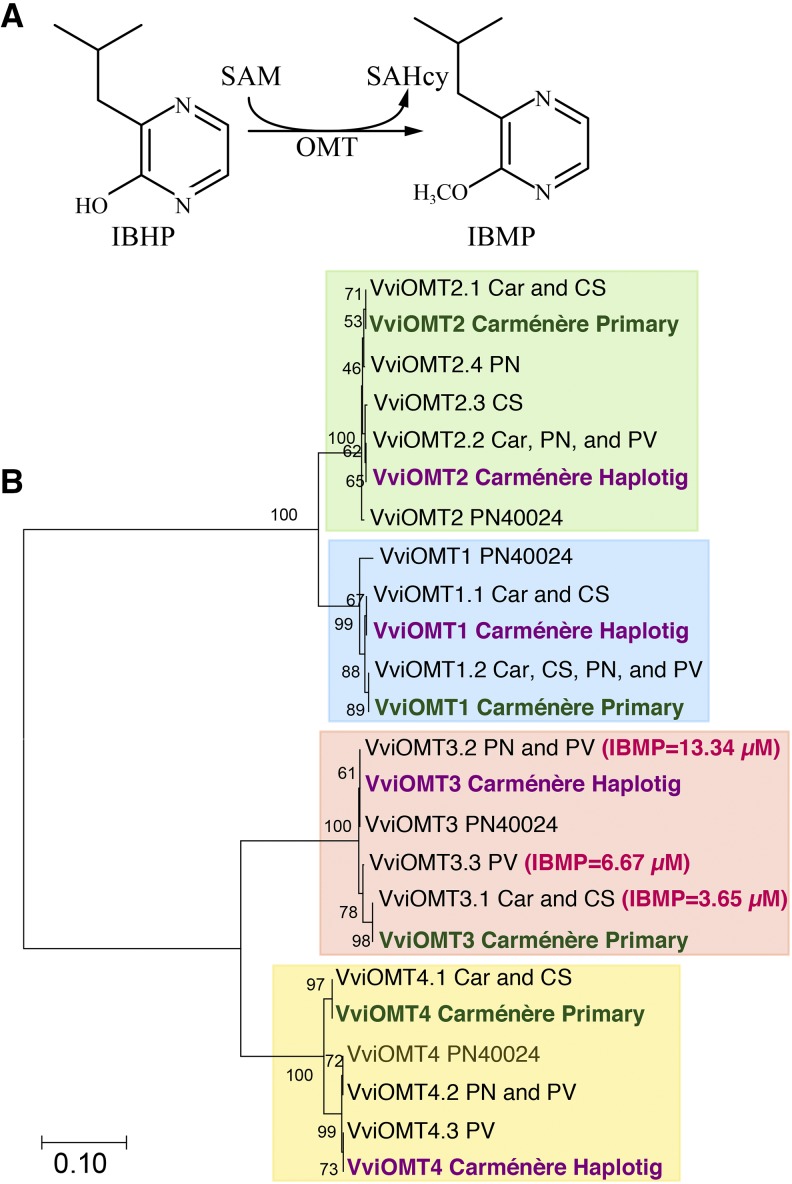
The phylogenetic tree illustrating the relation between the different VviOMT alleles was obtained using the Neighbor-Joining method (Saitou and Nei 1987). The percentage of replicate trees in which the associated sequences clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. The analysis involved 330 amino acid positions. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA7 (Kumar et al. 2016).
Data availability
Raw sequences are available at NCBI (Bioproject PRJNA517468). Other relevant data, such as genome sequence, gene and protein sequences, gene and repeat coordinates and annotation, along with a genome browser and a blast tool, are available at http://cantulab.github.io/data.html. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7666886.
Results and Discussion
Assembly of the Carménère genome
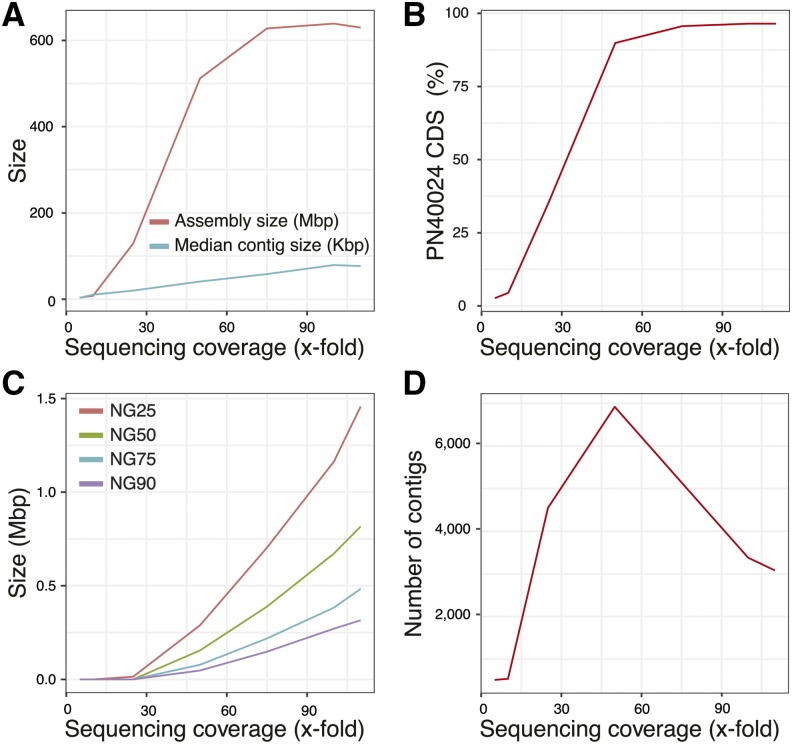
The genome of Vitis vinifera cv. Carménère clone 02 was sequenced at 115x coverage using Single Molecule Real Time (SMRT; Pacific Biosciences) technology. The long reads (N50 = 13.1 Kbp) were assembled into primary contigs and haplotigs using the diploid-aware assembler FALCON-Unzip (Chin et al. 2016). As detailed in the Methods section, to optimize assembly we systematically tested the effect of sequencing coverage, type of error correction, and assembly parameters on genome contiguity and completeness (Figure 1; Table S1). We also tested the addition of an extra step of repeat masking of the error-corrected reads prior to the assembly step. The most contiguous assembly was obtained with full SMRT reads dataset, non-hybrid error correction, masking of repeats in corrected reads, and a minimum corrected reads retention threshold of 7.5 Kbp (Table 1). While optimal assembly and gene space completeness were achieved already at coverage 75x and 50x, respectively (Figure 1A&B), assembly contiguity increased exponentially with increasing coverage (Figure 1C&D). These results suggest that even more contiguous assemblies could have been produced at sequencing coverage greater than 115x likely because of the larger number of long reads used in the assembly. These results also show that under 25x of coverage, assembly contiguity and gene space completeness are compromised even when reads are error-corrected with over 100x coverage of long reads or a hybrid error correction using short reads is applied. The most contiguous and complete primary assembly was scaffolded into 1,411 scaffolds covering 622.8 Mbp (N50 = 1.04 Mbp) with a maximum length of 5.9 Mbp and with as few as 857 Kbp in gaps (0.14%). As expected, haplotigs were more fragmented and covered only 420.3 Mbp (7,969 contigs, N50 = 89.6 Kbp) (Table 1). The assembly contained 93.3% of the complete universal single-copy orthologs (BUSCO) genes. As observed previously in Cabernet Sauvignon (Chin et al. 2016) and Chardonnay (Zhou et al. 2018), the size of the assembly constructed with FALCON-Unzip is larger than the expected genome size (∼500 Mbp) likely due to the retention of both copies of some heterozygous regions in the primary contigs. Nonetheless, the total assembly size (primary + haplotigs of 1.04 Gbp) was twice the expected haploid genome size, which suggests that sequences of all homologous chromosomes are represented in the final assembly. This was confirmed by the presence in the total assembly of an average of 2.07 ± 0.86 copies for each of the PN40024 genes.
Figure 1.
Impact of sequencing coverage on (A) assembly size, (B) completeness of the gene space, (C) assembly contiguity, and (D) assembly fragmentation. Best assemblies at each coverage were plotted.
Table 1. Summary statistics of the Carménère genome assembly.
| Primary assembly | Haplotigs | |
|---|---|---|
| Assembly length | 622,795,289 bp | 420,345,460 bp |
| Number of sequences | 1,411 | 7,969 |
| Average length | 441,386 bp | 52,748 bp |
| Maximum length | 5,905,621 bp | 743,383 bp |
| N50 length (index) | 1,039,379 bp (168) | 89,565 bp (1029) |
| Total gap length | 0.14% | 0.00% |
| Repetitive content | 308.9 Mbp (49.6%) | 192 Mbp (45.7%) |
| Number of genes | 40,684 | 32,425 |
Annotation of the Carménère genome
Forty eight percent of the assembled sequences in the primary scaffolds and haplotigs were classified as repetitive, mostly due to LTR transposable elements of the Gypsy (23.6% of repetitive content) and Copia (8.4% of repetitive content) families. A total of 73,109 protein-coding genes were found in the assembly, 40,684 in the primary assembly, and 32,425 in the haplotigs. The predicted transcriptome represented 95% of the BUSCO genes. All genes had at least one homolog plant protein in the RefSeq database, 69,918 (95.6%) had an InterPro match, 53,556 (73.3%) were assigned a gene ontology (GO) term, and 8,449 were associated with an enzyme code (EC; Table S2). One of the key aromatic compounds in Carménère are methoxypyrazines (MP), which impart the characteristic herbaceous, green, vegetal sensory attributes to Carménère wines. With wide variability among clones, Carménère grapes can accumulate high IBMP concentrations (5.0 to 44.4 ng/L; Belancic and Agosin 2007). The last step of the MP biosynthesis pathway consists in the conversion of 3-isobutyl-2-hydroxypyrazine (IBHP) into IBMP by a S-adenosyl-l-Met (SAM)-dependent O-methyltransferase (OMT; Figure 2A). Four VviOMT genes have been found in the grape genome, among which VviOMT3 is considered the major determinant of IBMP production during berry ripening (Dunlevy et al. 2013; Guillaumie et al. 2013). We could identify in the Carménère genome all four members of the VviOMT gene family. For each VviOMT, both alleles were represented in the Carménère assembly, one in the primary contigs and one in the haplotigs, confirming the completeness of the diploid assembly (Figure 2B; Table S3). Interestingly, the two alleles of VviOMT3 were polymorphic, with one allele closer to Cabernet Sauvignon (VviOMT3.1), likely derived from Cabernet Franc, and one closer to the allele found in Pinot Noir and Petit Verdot (VviOMT3.2). The latter allele was shown to be a strong IBMP producer in vitro, which may explain the greater accumulation of IBMP in Carménère than in Cabernet Sauvignon (Belancic and Agosin 2007). Interestingly, VviOMT3.2 was not found in a Carménère clone cultivated in Bordeaux (Guillaumie et al. 2013), which may explain the variability in IBMP accumulation among Carménère clones (Belancic and Agosin 2007). Because OMT expression was shown to play a critical role in IBMP accumulation (Guillaumie et al. 2013), further genetic analysis of the two alleles across multiple Carménère clones should be combined with gene expression measurements during ripening in order to determine the role of the different alleles in the accumulation of IBMP in Carménère grapes.
Figure 2.
Phylogenetic tree of O-methyltransferases (OMTs). (A) Graphical representation of the last and critical step of the methoxypyrazine biosynthesis pathway, where a S-adenosyl-l-Met (SAM)-dependent O-methyltransferase (OMT) converts 3-isobutyl-2-hydroxypyrazine (IBHP) into 3-isobutyl-2-methoxypyrazine (IBMP) producing S-Adenosyl-l-homo-Cys (SAHcy). (B) Phylogenetic tree illustrating the relation between the different alleles of the VviOMT clade. The VviOMT clade was divided into 4 subclades as reported in Guillaumie et al. (2013) (VviOMT1, VviOMT2, VviOMT3 and VviOMT4). The percentage of replicate trees in the bootstrap test clustering the associated sequences are shown next to the branches. Sequences of the different alleles identified in Carménère genome annotation are indicated in bold, green for the allele reported in the primary sequences and violet for the one in the haplotigs. These sequences were compared to PN40024 annotation and the VviOMT alleles reported for different genotypes in Guillaumie et al. (2013). The acronyms reported indicate the original genotype: Carménère (Car), Cabernet Sauvignon (CS), Pinot Noir (PN), Petit Verdot (PV); where more than one genotype was sharing the same allele, a coma separated list is reported. The amount of IBMP produced in vitro by the three recombinant VviOMT3 proteins is indicated in parenthesis (Supplementary data from Guillaumie et al. 2013).
Sequence and structural heterozygosity in the Carménère genome
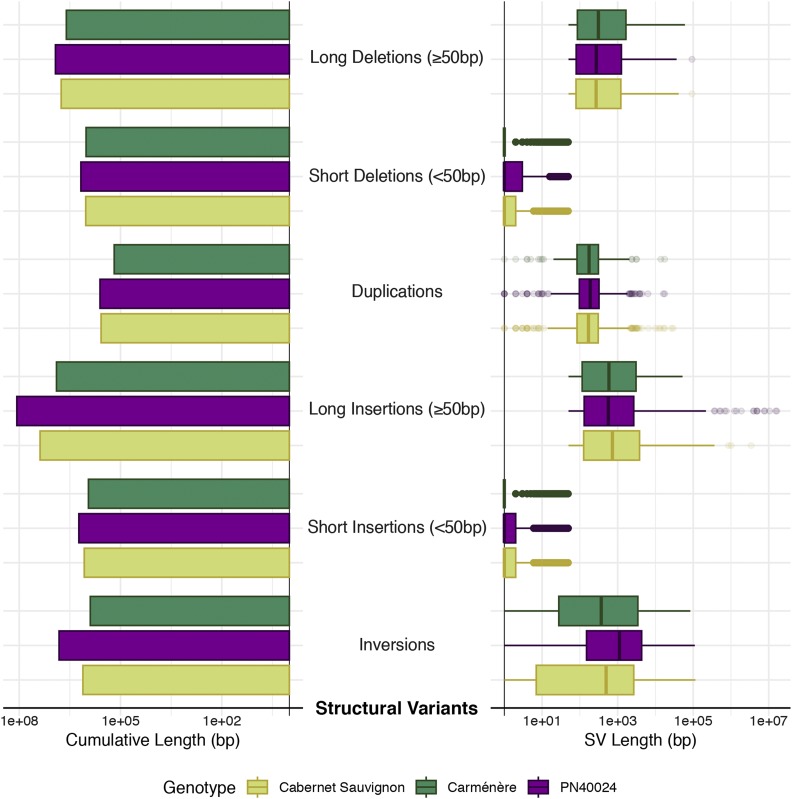
Comparisons between the primary assembly and haplotigs revealed 1,506,269 SNPs (2.41 SNPs/Kbp), 1,127,746 INDELs (<50bp), and 6,159 deletion/insertion events (≥50bp), representing a total variation of 15.3 Mbp between the two haplotypes (2.5% and 3.6% of the primary assembly and haplotigs, respectively; Table 2; Figure 3). A total of 50,619 SNPs were identified in coding sequences (28,032 non-synonymous and 22,587 synonymous), resulting in the introduction of premature stop codons in 421 genes. Among the affected genes were four genes involved in the phenylpropanoid/flavonoid biosynthetic pathway: a 4-coumarate-CoA ligase, a chalcone synthase, a flavonoid 3′,5′-hydroxylase, and a flavonol synthase (Castellarin et al. 2012). The large deletions/inversions (≥50bp) involved 63 complete genes. A larger number of potentially hemizygous genes (2,844 sequences; identity and coverage >80%) was found by alignment of the haplotigs’ genes onto the primary assembly.
Table 2. Structural variants identified in the Carménère primary scaffolds when compared to Carménère haplotigs, PN40024 (chromosomes), and Cabernet Sauvignon (primary assembly).
| Carménère Primary Scaffolds | ||||||
|---|---|---|---|---|---|---|
| Carménère Haplotigs | PN40024 | Cabernet Sauvignon | ||||
| Count | Total length | Count | Total length | Count | Total length | |
| SNPs | 1,506,269 | 1,506,269 | 3,917,352 | 3,917,352 | 2,449,007 | 2,449,007 |
| Short Insertions (<50bp) | 503,729 | 891,412 | 617,760 | 1,712,482 | 499,827 | 1,185,538 |
| Short Deletions (<50bp) | 624,017 | 1,046,233 | 452,437 | 1,489,269 | 408,465 | 1,073,441 |
| Long Insertions (≥50bp) | 3,412 | 7,875,979 | 11,436 | 117,627,739 | 6,394 | 24,115,347 |
| Long Deletions (≥50bp) | 2,747 | 4,021,742 | 6,986 | 8,466,055 | 4,599 | 5,708,870 |
| Duplication Contraction | 186 | 72,918 | 736 | 256,328 | 396 | 134,216 |
| Duplication Expansion | 229 | 80,725 | 541 | 148,123 | 405 | 241,869 |
| Inversions | 170 | 784,551 | 1,551 | 6,594,750 | 460 | 1,300,866 |
Figure 3.
Size and length distribution of structural variants identified in the Carménère primary scaffolds when compared to Carménère haplotigs, PN40024 (chromosomes), and Cabernet Sauvignon (primary assembly).
Genome structure and gene content comparison with other publicly available grapevine genomes and transcriptomes
The Carménère assembly was compared with the genome sequences of PN40024 (Jaillon et al. 2007; Canaguier et al. 2017) and Cabernet Sauvignon (Chin et al. 2016) to assess the extent and nature of the genetic diversity between these publicly available genomes. Direct comparison of genomic sequences identified many more variants between Carménère and PN40024 (3,917,352 SNPs; 1,070,197 <50bp INDELs) than between Carménère and Cabernet Sauvignon (2,449,007 SNPs; 908,292 <50bp INDELs) (Table 2; Figure 3). This result likely reflects the fact that Carménère and Cabernet Sauvignon share one of their parents (Bowers and Meredith 1997; Boursiquot et al. 2009). As expected, variants were detected at greater frequency in the intergenic space and introns than in exons in both comparisons (Table 2). SNPs and small INDELs were predicted to have deleterious impact on 8,988 and 7,835 Carménère genes when compared to PN40024 and Cabernet Sauvignon, respectively. We also identified large structural variants (SVs) between the three cultivars. A larger number of SVs was identified between Carménère and PN40024 (21,250 SVs) involving 133.1 Mbp (21.4%) of the Carménère primary assembly. Relative to the Cabernet Sauvignon genome, we could identify 12,254 SVs involving 31.5 Mbp of the Carménère assembly. Some of the large SVs intersected the gene space, which resulted in the absence of 494 and 253 Carménère genes in the PN40024 and Cabernet Sauvignon genomes, respectively. These SVs, and potentially additional undetected ones, may have contributed to the differences in gene content between Carménère and the other cultivars. About 2% of the Carménère genes (1,561) were not found in PN40024 and 0.61% (449) were not found in the Cabernet Sauvignon genome. A total of 198 genes were not found in any other available V. vinifera transcriptomes (Venturini et al. 2013; Da Silva et al. 2013; Gambino et al. 2017). These putative cultivar-specific genes comprised three sesquiterpene synthases, including two (-)-germacrene D synthases, which may be involved in terpenoid biosynthesis and grape aroma (Lücker et al. 2004). Carménère-specific genes also included: a sugar transporter ERD6-like 6 gene (early-responsive to dehydration) and an inositol-3-phosphate synthase-encoding gene, both potentially associated with water-deficit stress response (Büttner 2007; Yamada et al. 2010; Conde et al. 2015); seven Nucleotide Binding Site/Leucine-Rich Repeat (NBS-LRR) genes, five Serine/Threonine kinase genes, three LRR receptor-like kinase genes, that belong to three classes of resistance genes (Di Gaspero and Cipriani 2003; Kruijt et al. 2005). This level of unshared gene content is similar to what has been reported in previous works that compared other cultivars with PN40024 (Da Silva et al. 2013; Minio et al. 2019). Further work is necessary to determine whether some of these “private” Carménère genes contribute to its distinctive organoleptic and agronomical features. Further genetic analyses of Carménère will benefit from the availability of this high-quality genome assembly.
Acknowledgments
This work was funded by the Chilean Economic Development Agency (CORFO; Project 13CEI2-21852), Viña San Pedro, and Viña Concha y Toro and partially supported by the National Science Foundation Plant Genome Research grant #1741627. DC was also supported by the Louis P. Martini Endowment in Viticulture.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7666886.
Communicating editor: P. Morrell
Literature Cited
- Belancic A., Agosin E., 2007. Methoxypyrazines in grapes and wines of Vitis vinifera cv. Carmenere. Am. J. Enol. Vitic. 58: 462–469. [Google Scholar]
- Boetzer M., Pirovano W., 2014. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics 15: 211 10.1186/1471-2105-15-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursiquot J. M., Lacombe T., Laucou V., Julliard S., Perrin F. X., et al. , 2009. Parentage of Merlot and related winegrape cultivars of southwestern France: discovery of the missing link. Aust. J. Grape Wine Res. 15: 144–155. 10.1111/j.1755-0238.2008.00041.x [DOI] [Google Scholar]
- Bowers J. E., Meredith C. P., 1997. The parentage of a classic wine grape, Cabernet Sauvignon. Nat. Genet. 16: 84–87. 10.1038/ng0597-84 [DOI] [PubMed] [Google Scholar]
- Büttner M., 2007. The monosaccharide transporter(-like) gene family in Arabidopsis. FEBS Lett. 581: 2318–2324. 10.1016/j.febslet.2007.03.016 [DOI] [PubMed] [Google Scholar]
- Caló A., Stefano R. D., Costacurta A., Caló G., 1991. Caratterizzazione di Cabernet franc e Carménère (Vitis spp.) e chlarimenti sulla loro coltura in Italia. Riv. Vitic. Enol. 3: 3–25. [Google Scholar]
- Campbell M. A., Haas B. J., Hamilton J. P., Mount S. M., Buell C. R., 2006. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics 7: 327 10.1186/1471-2164-7-327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaguier A., Grimplet J., Di Gaspero G., Scalabrin S., Duchêne E., et al. , 2017. A new version of the grapevine reference genome assembly (12X.v2) and of its annotation (VCost.v3). Genom. Data 14: 56–62. 10.1016/j.gdata.2017.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaubon, G., A. Belancic, and E. Agosín, 2006 Develando los Aromas del Carménère. Rev. Vendimia 36–40.
- Castellarin S. D., Bavaresco L., Falginella L., Gonçalves M. I. V. Z., Di Gaspero G., 2012. Phenolics in Grape Berry and Key Antioxidants, pp. 89–110 in The Biochemistry of the Grape Berry, edited by Gerós H., Manuela Chaves M., Delrot S. BENTHAM SCIENCE PUBLISHERS, Sharjah, United Arab Emirates: 10.2174/978160805360511201010089 [DOI] [Google Scholar]
- Chin C.-S., Alexander D. H., Marks P., Klammer A. A., Drake J., et al. , 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10: 563–569. 10.1038/nmeth.2474 [DOI] [PubMed] [Google Scholar]
- Chin C.-S., Peluso P., Sedlazeck F. J., Nattestad M., Concepcion G. T., et al. , 2016. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13: 1050–1054. 10.1038/nmeth.4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T., et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde A., Regalado A., Rodrigues D., Costa J. M., Blumwald E., et al. , 2015. Polyols in grape berry: transport and metabolic adjustments as a physiological strategy for water-deficit stress tolerance in grapevine. J. Exp. Bot. 66: 889–906. 10.1093/jxb/eru446 [DOI] [PubMed] [Google Scholar]
- Da Silva C., Zamperin G., Ferrarini A., Minio A., Dal Molin A., et al. , 2013. The high polyphenol content of grapevine cultivar tannat berries is conferred primarily by genes that are not shared with the reference genome. Plant Cell 25: 4777–4788 (erratum Plant Cell https//.org/10.1105/tpc.17.00230). 10.1105/tpc.113.118810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez A. M., Agosin E., 2010. Gas chromatography coupled with mass spectrometry detection for the volatile profiling of Vitis Vinifera cv. Carménère wines. J. Chil. Chem. Soc. 55: 385–391. 10.4067/S0717-97072010000300025 [DOI] [Google Scholar]
- Dunlevy J. D., Dennis E. G., Soole K. L., Perkins M. V., Davies C., et al. , 2013. A methyltransferase essential for the methoxypyrazine-derived flavour of wine. Plant J. 75: 606–617. 10.1111/tpj.12224 [DOI] [PubMed] [Google Scholar]
- English A. C., Richards S., Han Y., Wang M., Vee V., et al. , 2012. Mind the Gap: Upgrading Genomes with Pacific Biosciences RS Long-Read Sequencing Technology. PLoS One 7: e47768 10.1371/journal.pone.0047768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English A. C., Salerno W. J., Reid J. G., 2014. PBHoney: identifying genomic variants via long-read discordance and interrupted mapping. BMC Bioinformatics 15: 180 10.1186/1471-2105-15-180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández K., Kennedy J. A., Agosin E., 2007. Characterization of Vitis vinifera L. Cv. Carménère Grape and Wine Proanthocyanidins. J. Agric. Food Chem. 55: 3675–3680. 10.1021/jf063232b [DOI] [PubMed] [Google Scholar]
- Gambino G., Dal Molin A., Boccacci P., Minio A., Chitarra W., et al. , 2017. Whole-genome sequencing and SNV genotyping of “Nebbiolo” (Vitis vinifera L.) clones. Sci. Rep. 7: 17294 10.1038/s41598-017-17405-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Gaspero G., Cipriani G., 2003. Nucleotide binding site/leucine-rich repeats, Pto-like and receptor-like kinases related to disease resistance in grapevine. Mol. Genet. Genomics 269: 612–623. 10.1007/s00438-003-0884-5 [DOI] [PubMed] [Google Scholar]
- Guillaumie S., Ilg A., Réty S., Brette M., Trossat-Magnin C., et al. , 2013. Genetic Analysis of the Biosynthesis of 2-Methoxy-3-Isobutylpyrazine, a Major Grape-Derived Aroma Compound Impacting Wine Quality. Plant Physiol. 162: 604–615. 10.1104/pp.113.218313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J., Delcher A. L., Mount S. M., Wortman J. R., Smith R. K., et al. , 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31: 5654–5666. 10.1093/nar/gkg770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J., Salzberg S. L., Zhu W., Pertea M., Allen J. E., et al. , 2008. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 9: R7 10.1186/gb-2008-9-1-r7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichsen P., Narváez C., Bowers J. E., Boursiquot J. M., Valenzuela J., et al. , 2001. Distinguishing Carmenère from Similar Cultivars by DNA Typing. Am. J. Enol. Vitic. 52: 396–399. [Google Scholar]
- Huamán-Castilla N. L., Mariotti-Celis M. S., Perez-Correa J. R., 2017. Polyphenols of Carménère Grapes. Mini Rev. Org. Chem. 14: 176–186. 10.2174/1570193X14666170206151439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O., Aury J.-M., Noel B., Policriti A., Clepet C., et al. , 2007. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467. 10.1038/nature06148 [DOI] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H.-Y., Fraser M., Li W., et al. , 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30: 1236–1240. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I., 2004. Gene finding in novel genomes. BMC Bioinformatics 5: 59 10.1186/1471-2105-5-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruijt M., Kock M. J. D. D. E., de Wit P. J. G. M., 2005. Receptor-like proteins involved in plant disease resistance. Mol. Plant Pathol. 6: 85–97. 10.1111/j.1364-3703.2004.00264.x [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K., 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomsadze A., Ter-Hovhannisyan V., Chernoff Y. O., Borodovsky M., 2005. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 33: 6494–6506. 10.1093/nar/gki937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lücker J., Bowen P., Bohlmann J., 2004. Vitis vinifera terpenoid cyclases: Functional identification of two sesquiterpene synthase cDNAs encoding (+)-valencene synthase and (-)-germacrene D synthase and expression of mono- and sesquiterpene synthases in grapevine flowers and berries. Phytochemistry 65: 2649–2659. 10.1016/j.phytochem.2004.08.017 [DOI] [PubMed] [Google Scholar]
- Majoros W. H., Pertea M., Salzberg S. L., 2004. TigrScan and GlimmerHMM: Two open source ab initio eukaryotic gene-finders. Bioinformatics 20: 2878–2879. 10.1093/bioinformatics/bth315 [DOI] [PubMed] [Google Scholar]
- Marçais G., Delcher A. L., Phillippy A. M., Coston R., Salzberg S. L., et al. , 2018. MUMmer4: A fast and versatile genome alignment system. PLOS Comput. Biol. 14: e1005944 10.1371/journal.pcbi.1005944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minio A., Lin J., Gaut B. S., Cantu D., 2017. How Single Molecule Real-Time sequencing and haplotype phasing have enabled reference-grade diploid genome assembly of wine grapes. Front. Plant Sci. 8: 826 10.3389/fpls.2017.00826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minio A., Massonnet M., Figueroa-Balderas R., Vondras A. M., Blanco-Ulate B., et al. , 2019. Iso-Seq Allows Genome-Independent Transcriptome Profiling of Grape Berry Development. G3-Genes Genomes Genet. 9: 755–767. 10.1534/g3.118.201008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers G., 2014. Efficient Local Alignment Discovery amongst Noisy Long Reads, Algorithms in Bioinformatics, WABI 2014, edited by Brown D., Morgenstern B., (Lecture Notes in Computer Science), Vol. 8701, pp. 52–67. Springer, Berlin, Heidelberg: 10.1007/978-3-662-44753-6_5 [DOI] [Google Scholar]
- Parra G., Blanco E., Guigó R., 2000. GeneID in Drosophila. Genome Res. 10: 511–515. 10.1101/gr.10.4.511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pszczólkowski P., 2004. La invención del cv. Carménère (Vitis vinifera L) en Chile, desde la mirada de uno de sus actores. Universum (Talca) 19: 150–165. 10.4067/S0718-23762004000200010 [DOI] [Google Scholar]
- Richards P., 2006. The Wines of Chile, Octopus, London. [Google Scholar]
- Saitou N., Nei M., 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Salmela L., Rivals E., 2014. LoRDEC: Accurate and efficient long read error correction. Bioinformatics 30: 3506–3514. 10.1093/bioinformatics/btu538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Slater G. S. C., Birney E., 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6: 31 10.1186/1471-2105-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, A. F. A., R. Hubley, and P. Green, 2013 RepeatMasker Open-4.0. http://www.repeatmasker.org 2013–2015.
- Stanke M., Keller O., Gunduz I., Hayes A., Waack S., et al. , 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34: W435–W439. 10.1093/nar/gkl200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- This P., Jung A., Boccacci P., Borrego J., Botta R., et al. , 2004. Development of a standard set of microsatellite reference alleles for identification of grape cultivars. Theor. Appl. Genet. 109: 1448–1458. 10.1007/s00122-004-1760-3 [DOI] [PubMed] [Google Scholar]
- Venturini L., Ferrarini A., Zenoni S., Tornielli G. B., Fasoli M., et al. , 2013. De novo transcriptome characterization of Vitis vinifera cv. Corvina unveils varietal diversity. BMC Genomics 14: 41 10.1186/1471-2164-14-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. D., Watanabe C. K., 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875. 10.1093/bioinformatics/bti310 [DOI] [PubMed] [Google Scholar]
- Yamada K., Osakabe Y., Mizoi J., Nakashima K., Fujita Y., et al. , 2010. Functional analysis of an Arabidopsis thaliana abiotic stress-inducible facilitated diffusion transporter for monosaccharides. J. Biol. Chem. 285: 1138–1146. 10.1074/jbc.M109.054288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y. S., Minio A., Massonnet M., Solares E. A., Lyu Y., et al. , 2018. Structural variants, clonal propagation, and genome evolution in grapevine (Vitis vinifera). bioRxiv 508119 10.1101/508119 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw sequences are available at NCBI (Bioproject PRJNA517468). Other relevant data, such as genome sequence, gene and protein sequences, gene and repeat coordinates and annotation, along with a genome browser and a blast tool, are available at http://cantulab.github.io/data.html. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7666886.