Abstract
Background
Human mannose binding lectin (MBL) and dendritic cell-associated C-type lectin-1 (Dectin-1) are the two prototypical PRRs of innate immunity, whose direct role in recurrent vulvovaginal infections (RVVI) defense has been defined. Previously, MBL insufficiency was proposed as a possible risk factor for the rapid progression of RVVI while, Dectin-1 was found to be playing an active role in the defense. However, the complete genetic bases for the observed low MBL levels are still lacking as our previous studies in harmony with others demonstrated the un-expected genotype–phenotype patterns. This suggested the presence of unidentified regulatory variants that may modulate sMBL levels and risk of RVVI. Therefore, the present study was designed for more inclusive locus-wide MBL2 analysis and for the possible non-linear interaction analysis of two PRRs that may impact RVVI susceptibility.
Methods
The present study has extended the previous findings by investigating (1) the role of chosen additional SNPs falling in the 5′ near region relating to sMBL levels and RVVI susceptibility, using polymerase chain reaction-restriction fragment length polymorphism, (2) interactions among SNPs within gene by comprehensive locus-wide haplotype analyses of two MBL2 blocks, (3) gene–gene interaction analyses between two PRRs, using multifactor dimensionality reduction.
Results
rs11003124_G, rs7084554_C, rs36014597_G, and rs11003123_A were observed as the minor alleles in the representative North Indian cohort. RVVI cases and its types showed an appreciably high frequency of C allele, its homozygosity and heterozygosity, explaining the observed dominant mode of inheritance of rs7084554 polymorphism in contributing 1.81 fold risk of RVVI. The rs36014597 polymorphism showed the overdominant mode of inheritance, which further depicts that the carrier of a heterozygous genotype of this polymorphism had more extreme phenotype than either of its homozygous carriers in developing 4.07 fold risk of RVVI. sMBL levels significantly varied for rs11003124, rs36014597 and rs11003123 polymorphisms in bacterial vaginosis, while for rs7084554 polymorphism in mixed infection. Independent analysis of 5′ and 3′ haplotype blocks suggested the risk-modifying effect of all the 5′ additional variants, Y/X secretor polymorphism and 3′-UTR SNP i.e. rs10824792. Combined 5′/3′ haplotype analyses depicted the importance of rs36014597; an additional 5′ variant, Y/X and rs10824792 polymorphisms from both the blocks in regulating sMBL levels and RVVI risk. Three gene–gene interaction models involving uni-variant, bi-variant and tri-variant appeared as significant predictors of RVVI risk with cross-validation consistency of 10/10, 9/10 and 5/10, respectively.
Conclusions
The study presented a low-cost reproducible screening design for additional 5′ variants i.e. rs11003124, rs7084554, rs36014597 and rs11003123 of MBL2 that can act as markers of susceptibility for RVVI or any other diseases. Two additional 5′ variants of MBL2 i.e. rs7084554 and rs36014597 were suggested as novel molecular markers that may contribute to RVVI risk by varying sMBL levels. Variants of two blocks were found to have more of a combined effect than the independent effect in modulating RVVI susceptibility and sMBL levels. The study presented weak synergistic interaction between MBL2 and CLEC7A in association with RVVI risk. The preliminary data will establish the foundation for the investigation of within gene and between genes interaction analyses towards RVVI susceptibility.
Electronic supplementary material
The online version of this article (10.1186/s13578-019-0300-4) contains supplementary material, which is available to authorized users.
Keywords: Additional 5′ variants, 3′ UTR SNP, Innate immunity, Haplotype, Gene–gene interaction, Multifactor dimensionality reduction (MDR), Reproductive infectious diseases
Background
Three universal pathological conditions of recurrent vulvovaginal infection (RVVI) are bacterial vaginosis (BV), vulvovaginal candidiasis (VVC) and trichomoniasis [1, 2]. Despite the fine knowledge regarding organismal and non-organismal pathogenesis factors, RVVI remains one of the most enigmatic mucosal problems worldwide due to the presence of 20–30% asymptomatic cases (i.e. healthy women with vaginal microbiota composition same as that of RVVI), some of which also had predisposing non-organismal causes [3]. Moreover, women without any known predisposing factors have also been documented to acquire RVVI [4]. Therefore, identification of causal factors that modulate propensity to RVVI in women is much needed. Mannose binding lectin (MBL), encoded by MBL2 mapped to 10q21.1 has commonly been referred to as an acute phase protein whose serum levels increases following infections [5]. It is an ideal pattern recognition receptor that binds to specific sugars on pathogen’s surface, consequently causing pathogen removal by complement activation, opsonisation and/or phagocytosis [5]. The serum or vaginal fluid levels of MBL have been assessed in RVVI cases by different studies, suggesting its key involvement in pathogenesis of RVVI [6–13].
The MBL levels have been shown to be determined by single nucleotide polymorphisms (SNPs) in coding and promoter region of MBL2. The former includes three SNPs, present in exon 1 at rs5030737 (codon 52), rs1800450 (codon 54) and rs1800451 (codon 57), collectively called as MBL2 structural variations [14]. The codon 54 and 57 SNPs leads to the substitution of glycine with dicarboxylic acids, while codon 52 leads to the substitution of arginine with cysteine in the collagenous region of a monomeric protein resulting in variant monomers [15, 16]. These variant monomers dramatically affect the circulating levels of higher order functional MBL oligomers [17]. In addition to these, three other promoter region variations i.e. rs11003125 (L/H), rs7096206 (Y/X) and rs7095891 (P/Q) have been functionally authenticated to alter MBL2 transcription, clearly depicting the importance these genetic variations regarding MBL expression and its circulating levels [16, 18].
The linkage disequilibrium (LD) has been described in the literature between structural and promoter polymorphisms leading to the formation of seven standard haplotypes. These standard haplotypes include HYPA, LYPA, LYQA, LYPB, LXPA, LYQC and HYPD. These haplotypes are commonly referred to as the secretor haplotypes because they regulate sMBL levels [19]. However, additional secretor haplotypes including HXPA, LYQB, HYQA, HYQB, HXQB, LXPB, LXQB, and LYPD have also been reported by various studies in different populations [11, 20–23]. The novel haplotypes have been suggested to be observed, due to genetic heterogeneity between different populations and selective advantage in response to environmental pressures like infections or geographic location [24, 25].
From the past, the structural polymorphisms of MBL2 remain the centre point of most of the investigations including RVVI. As for instance, the association of MBL2 structural polymorphisms with RVVI have been documented in different populations [6–8, 26–30]. Our previous study has also elucidated the involvement of standard MBL2 haplotypes in modulating sMBL levels and RVVI susceptibility [11]. However, our study in harmony with others found an un-expected correlation pattern of genotypes with phenotypes, suggesting the presence of unrecognised regulatory elements of MBL2 [11, 19, 31]. This implicates the need for further inclusive locus-wide MBL2 analysis to reveal unrecognised regulatory variants that may modulate sMBL levels and risk of infectious diseases. For this selection of variants can be made based on putative functional effects e.g. altering transcription, translation or miRNA binding.
Our previous study sorted out 12 putative functionally important SNPs of MBL2 using in silico analysis [32]. From this, evaluation of MBL2 3′-UTR SNPs has depicted a novel association of rs10824792 SNP with low sMBL levels and RVVI risk [13]. However, the role of selected SNPs falling in the 5′ near region is still pending to be elucidated. Moreover, high serum levels of Dectin-1, another important innate immune molecule, have also been shown to play an active role in defense against RVVI [12]. Dectin-1, encoded by CLEC7A mapped to 12p13.2, is a collaborative PRR that team up with other PRRs via Syk pathway to generate optimal immune responses [33]. Both MBL and Dectin-1 are the essential innate immune components. Therefore to know the relationship between two, when co-activated against same pathogenic stimuli, would be of interest. Our previous investigation suggested the effect of rs3901533 CLEC7A SNP in modulating sMBL levels and RVVI susceptibility, suggesting RVVI a multi-factorial phenotype [12]. Though, we did not find any correlation between two proteins, finding the relationship between two genes i.e. MBL2 and CLEC7A is still pending to be elucidated, as the importance of genes before proteins have already been stated.
From this background, the present study was planned to elucidate (1) the role of selected SNPs falling in the 5′ near region relating to sMBL levels and RVVI susceptibility, using a conventional approach (2) interactions among SNPs within gene by comprehensive locus-wide haplotype analyses of MBL2, based on the LD pattern obtained across the genomic structure, using genotyped data of MBL2 variants evaluated in this study and reported previously. (3) gene–gene interaction analyses between two PRRs i.e. MBL and Dectin-1, using multifactor dimensionality reduction method. The preliminary data will establish the foundation for the investigation of within gene and between genes interaction analyses towards RVVI susceptibility.
Materials and methods
Study participants
The present study recruited RVVI cases (n = 258, mean age 29.33 years, ± S.D. 8.32) attending Bebe Nanki Mother and Child Care Centre, Department of Obstetrics and Gynaecology, Government Medical College, Amritsar (Pb) and were referred by the gynecologist. These cases were clinically pre-diagnosed with RVVI with minimum 4 documented recurrent experiences in a year, with frequent complaints of smelly discharge, itching, vaginal sores as well as pelvic pain. The controls (n = 203, mean age 29.33 years, ± S.D. 8.17) were matched to cases by age and had, by self-report, no recurrent history of vaginal infection. Participants were excluded if they were known carriers of HIV or any other chronic conditions, under chemotherapy or taking immunosuppressive medications. All the participants provided informed consent in writing. The Institutional Ethics Committee of Guru Nanak Dev University, Amritsar (Punjab), India, approved (Approval no. 06/HG dated 02/01/2015) the study protocol.
Samples and RVVI categorisation
Two types of samples were collected in the present study i.e. vaginal discharge and peripheral blood samples. Vaginal discharge samples from 200 RVVI cases were carried to the laboratory and subjected to standard diagnostic methods specified in European (IUSTI/WHO) guidelines on vaginal discharge management [34] as reported previously [35]. This categorised 200 RVVI cases into three major categories of RVVI i.e. Bacterial Vaginosis (BV; n = 97), vulvovaginal candidiasis (VVC; n = 62) and Mixed Infections (MI; n = 41) i.e. cases with both BV and VVC. However, 58 RVVI cases could not be processed, hence categorised, as these participants were either menstruating or were not willing to give vaginal samples. The peripheral blood samples (5 ml), collected from all the participants, were further processed for serum and DNA isolation by standard methodology [11–13].
SNPs selection
In silico analyses demonstrated twelve SNPs of MBL2 with possible functional consequences to structure and expression of MBL [32]. These twelve putative functional SNPs included rs11003125 (L/H), rs11003124, rs7084554, rs36014597, rs7096206 (Y/X), rs11003123, rs7095891 (P/Q), rs1800450 (codon 54), rs10082466, rs2165813, rs2099903 and rs2099902. Of these, the association of all the SNPs except rs11003124, rs7084554, rs36014597, rs11003123 and rs10082466 with RVVI has been reported previously [11, 13]. Thus, rs11003124, rs7084554, rs36014597 and rs11003123 SNPs except rs10082466 were investigated in the present study to assess their role in RVVI and its types. Hence, all the SNPs of MBL2 that were prioritised by in silico analyses were validated in relation to RVVI except one 3′UTR SNP i.e. rs10082466. This SNP could not be evaluated as different PCR approaches used for genotyping could not be standardised. Moreover, SNPs flanking rs10082466 SNP were either not functional or of low frequency, so the selection of other regions for sequencing seems expensive, hence not opted.
Genotype analyses by polymerase chain reaction-restriction fragment length polymorphism (PCR–RFLP)
The present study standardised a simple economical method i.e. PCR–RFLP, for the genotyping of four MBL2 SNPs including rs11003124, rs7084554, rs36014597, and rs11003123.
Primer designing and PCR
NCBI’s Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/), a freely available online software was used to design primers for PCR amplification. MBL2 gene sequence (NCBI Reference Sequence: NC_000010.11) flanking the given SNP, was used as an input for the software. The best primer pair was custom-synthesized from Bioserve Biotechnologies (Hyderabad, India). A reaction mixture (20 µl) consisting of template DNA, dNTPs (0.025 mM), Taq DNA polymerase (0.3 U) and Taq buffer with 15 mM MgCl2 (1X) was used for each PCR. Each PCR was performed in a thermal cycler (Applied Biosystems, Life Technologies, USA) with stipulated conditions. The particulars of primers sequence, concentrations of specific primers as well as the amplification conditions intended for all the PCRs have been provided in Table 1. The amplified products were analysed on 1.5% (w/v) EtBr-stained agarose gel (Himedia, India), after electrophoretic separation at 100 V, with gel viewer (Alpha imager, USA).
Table 1.
PCR-RFLP protocol for the genotyping of MBL2 additional 5′ variants, PCR primers and conditions
| MBL2 SNPs | Primer type | Primer sequence | Working primer conc. (p mol/µl) | PCR Conditions for all reactions | Product size (bp) |
|---|---|---|---|---|---|
| rs11003124a | Forward | 5′-GCTGGCTTATGCCTGTTAGC-3′ | 0.05 | 95 °C for 5 min, followed by 35 cycles (94 °C for 30 s, 53 °C for 30 s, and 72 °C for 45 s) and a final elongation at 72 °C for 5 min | 524 |
| Reverse | 5′-CTGCTGAGGTTTCTTAGGGGG-3′ | 0.05 | |||
| rs7084554a | Forward | 5′-AAGGAGGGGTTCATCTGTGC-3′ | 0.15 | 551 | |
| Reverse | 5′-TGGGAGGAGGATTCAAGGCA-3′ | 0.15 | |||
| rs36014597b | Forward | 5′-TCCTGCCAGAAAGTAGAGAGGT-3′ | 0.15 | 585 | |
| Reverse | 5′-TGGCCTCTAGCTGGGGATTT-3′ | 0.15 | |||
| rs11003123a | Forward | 5′-CTGGTTCCCCCTTTTCTCCC-3′ | 0.10 | 506 | |
| Reverse | 5′-TGCACGGTCCCATTTGTTCT-3′ | 0.10 |
aDenote SNPs reported at forward strand
bDenote SNPs reported at Reverse strand
Choice of restriction enzymes and RFLP
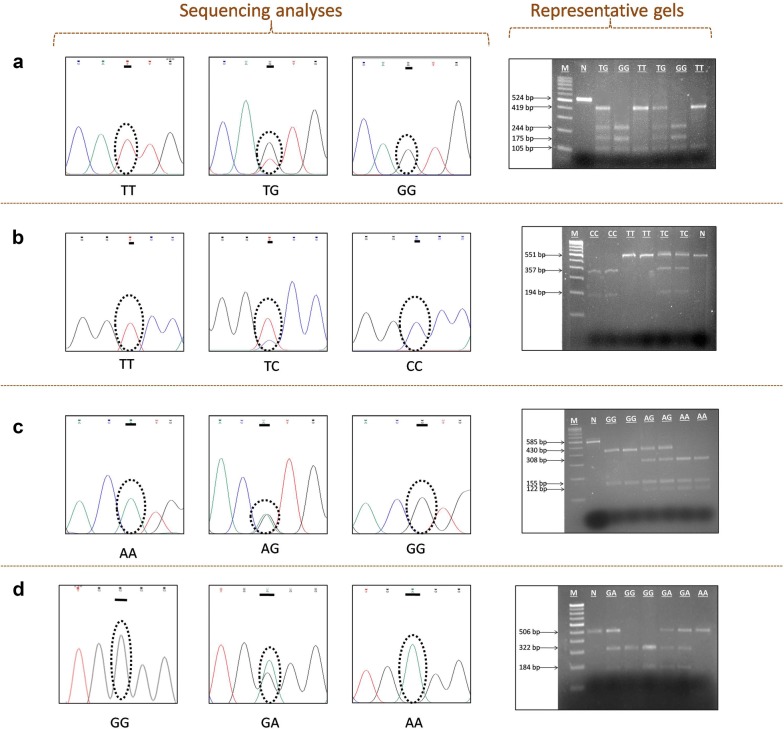
The choice of restriction enzymes to differentiate among variant, wild and heterozygous genotypes for SNPs including rs11003124, rs7084554, rs36014597, and rs11003123 was made with the help of online software NEBcutter v 2.0 (http://nc2.neb.com/NEBcutter2/). Restriction enzyme (IU) along with cut smart NEBuffer (1×) was used for the restriction analysis of amplified PCR product. RFLP conditions along with restriction endonucleases used for the genotyping of each SNP are provided in Table 2. The pattern of restriction digestion was visualised on gel documentation system after electrophoresis on 2.5% (w/v) agarose gel at 100 V. DNA sample of known genotype was used as positive control, while negative control contained all components of restriction digestion mixture except respective restriction enzyme. The PCR products were ascertained by analysing the restriction digestion pattern (Fig. 1). About 10% of the samples with representative genotypes were also confirmed by Sanger sequencing (Fig. 1).
Table 2.
PCR-RFLP protocol for the genotyping of MBL2 additional 5’ variants, restriction digestion conditions and expected restriction fragments size for MBL2 variants
| MBL2 SNP | Enzyme | Restriction site | RFLP conditions | Wild-type homozygotes (bp) | Variant homozygotes (bp) | Heterozygotes (bp) |
|---|---|---|---|---|---|---|
| rs11003124 | TspRI | 5′…NNCASTGNN▼…3′ 3′…▲NNGTSACNN…5′ |
22 h at 65 °C | 419 + 105 | 244 + 175 + 105 | 419 + 244 + 175 + 105 |
| rs7084554 | BanII | 5′…GRGCY▼C…3′ 3′…C▲YCGRG…5′ |
4 h at 37 °C | 551 | 357 + 194 | 551 + 357 + 194 |
| rs36014597 | NLaIII | 5′…CATG▼…3′ 3′…▲GTAC…5′ |
4 h at 37 °C | 308 + 155 + 122 | 430 + 155 | 430 + 308 + 155 + 122 |
| rs11003123 | BseYI | 5′…C▼CCAGC…3′ 3′…GGGTC▲G…5′ |
4 h at 37 °C | 322 + 184 | 506 | 506 + 322 + 184 |
Single letter code of restriction sites: R = A or G, Y = C or T, N = A or C or G or T, S = C or G
Fig. 1.
The additional 5′ variants of MBL2. Sequencing analyses using forward primer of PCR–RFLP indicating the presence of a rs11003124, b rs7084554, c rs36014597 and d rs11003123. Representative agarose gels showing restriction fragment digestion pattern of respective polymorphisms. M: 100 bp DNA ladder. N: negative control
Haplotype (within a gene) and Gene–gene interaction analyses
For haplotype analyses, SNPs evaluated in the present study along with previously reported SNPs of MBL2 were considered, to get hold of the entire MBL2 locus. This include four commonly known secretor polymorphisms [three promoter i.e. rs11003125 (L/H), rs7096206 (Y/X) and rs7095891 (P/Q) and one exonic SNP i.e. rs1800450 (codon 54)], Six 3′UTR polymorphisms [rs10824792, rs2120132, rs2120131, rs2165813, rs2099903 and rs2099902] along with the SNPs evaluated in the present study. The four MBL2 SNPs i.e. rs11003124, rs7084554, rs36014597, and rs11003123, evaluated in the present study are referred as additional 5′ near gene variants or simply as additional 5′ variants to differentiate them from standard secretor polymorphisms falling in 5′ near gene reported previously. Thus, fourteen SNPs including rs11003125 (L/H), rs11003124, rs7084554, rs36014597, rs7096206 (Y/X), rs11003123, rs7095891 (P/Q), rs1800450 (A/B), rs10824792, rs2120132, rs2120131, rs2165813, rs2099903 and rs2099902 across the MBL2 locus, from 5′ to 3′ direction were used for haplotype analyses in the present study (Table 3). For gene–gene interactions analyses, total 17 i.e. 14 MBL2 SNPs and three previously reported CLEC7A SNPs i.e. rs3901533, rs11053597 and rs11053593 were considered.
Table 3.
Features of SNPs used for haplotype analyses across the MBL2 locus
| dbSNP Identifiera | SNP position (5′ → 3′)b | Chromosome positionb | Secretorc | Region | Nucleotide change |
|---|---|---|---|---|---|
| rs11003125 | − 618 | 52,772,254 | L/H (aka − 550) | 5′ near gene | C/G |
| rs11003124 | − 495 | 52,772,131 | 5′ near gene | T/G | |
| rs7084554 | − 417 | 52,772,053 | 5′ near gene | T/C | |
| rs36014597 | − 404 | 52,772,040 | 5′ near gene | A/G | |
| rs7096206 | − 289 | 52,771,925 | Y/X (aka − 221) | 5′ near gene | G/C |
| rs11003123 | − 138 | 52,771,774 | 5′ near gene | G/A | |
| rs7095891 | − 65 | 52,771,701 | P/Q (aka + 4) | 5′ near gene | C/T |
| rs1800450 | + 161 | 52,771,475 | A/B (codon 54) | Exon1 | G/A |
| rs10824792 | + 4873 | 52,766,446 | 3′UTR | T/C | |
| rs2120132 | + 5037 | 52,766,280 | 3′UTR | T/C | |
| rs2120131 | + 5058 | 52,766,258 | 3′UTR | T/G | |
| rs2165813 | + 5090 | 52,766,224 | 3′UTR | G/A | |
| rs2099903 | + 5217 | 52,766,097 | 3′UTR | C/A | |
| rs2099902 | + 5225 | 52,766,089 | 3′UTR | T/C |
aNCBI dbSNP database (http://www.ncbi.nlm.nih.gov/SNP)
bNCBI contig accession number NT_030059.14 (NCBI build 142, locus ID 4153)
cDesignation extensively used in literature, is also followed in the present study, these four markers form the ‘secretor haplotypes’
Serum MBL concentration
The Serum MBL (sMBL) concentration was determined with enzyme-linked immunosorbent assay (Human MBL ELISA kit, Ray Biotech, USA) following the manufacturer’s instructions, as reported previously [11, 13]. Briefly, 100 μl of standard, blank and 4000 fold pre-diluted serum sample was added to the respective wells of microtitre plate, pre-coated with anti-human MBL antibody and incubated for 2.5 h at room temperature. After 3–4 washings with 1× wash buffer, 100 μl of biotinylated anti-human MBL antibody was added and the plate was incubated at room temperature for 1 h. Following incubation, unbound biotin conjugated anti-human antibody was removed by 3–4 washings with 1X wash buffer and 100 μl streptavidin-HRP was added to all the wells, followed by incubation at room temperature for 45 min. After 3–4 washings, 100 μl of 3,3,5,5′-tetramethylbenzidine (TMB) substrate was added and incubated for 30 min at room temperature in the dark. The blue color was developed in proportion to the amount of MBL present in the sample. The reaction was stopped with 50 μl of stop solution (0.2 M H2SO4) that changes the color from blue to yellow. The intensity of the color was measured at 450 nm by microplate reader (BIO-RAD, iMark™, USA). The assay was calibrated using MBL (25 ng/ml) standard provided in the kit, which was used to prepare different standard dilutions i.e. 8.33, 2.778, 0.926, 0.309, 0.103, 0.034 ng/ml, as instructed in the kit. The assay diluent A, provided in the kit was used as blank and to prepare standard dilutions. All these different standard concentrations were used to obtain MBL standard curve, hence standard equation, from which, the concentration of MBL in the serum samples was determined. The minimum detection sensitivity of the ELISA kit was 0.03 ng/ml.
Statistical analysis
To achieve a minimum of 80% power for the present study, required sample size was calculated with Genetic Association Study (GAS) power calculator (http://csg.sph.umich.edu/abecasis/gas_power_calculator/), considering assumptions that are 30% countrywide prevalence of abnormal vaginal discharge, 10% MAF, 1.5 odds ratio (OR) and 5% error rate (α = 0.05). Standard adds up was carried out to compute allelic as well as genotypic frequencies of different SNPs in cases and controls. These frequencies were further compared by odds ratio statistics with MedCalc software v 9.3.9.0 (MedCalc Software, Ostend, Belgium). The major allele and its subsequent homozygous genotype were selected as the reference (OR = 1). Deviation of each marker from Hardy–Weinberg equilibrium (HWE) was tested with SNPStats (https://www.snpstats.net/snpstats/start.htm). The best genetic models for each polymorphism was selected based on the smallest Akaike’s Information Criterion (AIC) as well as the Bayesian Information Criterion (BIC) values given by SNPStats. The interpretation of Linkage Disequilibrium (LD) between each polymorphism was done using Haploview v 4.2 (http://www.broad.mit.edu/mpg/haploview/). Haplotypes were inferred with PHASE software v 2.1.1. (http://stephenslab.uchicago.edu/phase/download.html) using genotypic data and for each analysis, the most common haplotype observed in the cohort of the present study was selected as a reference (OR = 1). One-way ANOVA (analyses of variance) with subsequent Tukey’s multiple comparison post hoc test were performed to compare serum biomarkers within group consisting of more than two categories. Comparison of serum biomarkers of cases with respective controls and between two categories of a group was done by Student’s t test. Gene–gene interactions analysis was performed with the help of Multifactor Dimensionality Reduction (MDR) software v 3.0.2 (http://www.multifactordimensionalityreduction.org/). Statistical package for social sciences (SPSS) v 16.0 (SPSS Inc., Chicago, IL) was used to perform all these statistical analyses unless mentioned. The p-value ≤ 0.05 were considered to be statistically significant for all data analyses.
Results
Genetic analyses of additional 5′ variants in RVVI relative to controls
All the additional 5′ variants conformed to HWE (p > 0.05) except rs11003123 polymorphism. rs11003124_G, rs7084554_C, rs36014597_G, and rs11003123_A were found to be minor alleles in North Indian cohort (Table 4). Of all the evaluated additional 5′ variants, allelic and genotypic frequencies significantly varied for rs7084554 and rs36014597 polymorphisms only (Tables 4 and 5). The C allele of rs7084554 was found to be significantly (p = 0.009; OR = 1.54; 95% CI 1.11–2.13) more prevalent in RVVI cases than controls. The frequency of TC heterozygous genotype (p = 0.002) and CC homozygous genotype (p = 0.396) of rs7084554 was high in RVVI cases than controls. Excluding the recessive model of inheritance, all the other models were found to be significant for rs7084554 SNP. However, the dominant genetic model was found to be best, with least AIC = 629.5 and BIC = 641.9 values, depicting that C allele carrier (either in homozygous or heterozygous state) had a greater risk of developing RVVI than non-C carriers (p = 0.002; OR = 1.81; 95% CI 1.22–2.68). Moreover, significantly (p < 0.0001; OR = 2.23; 95% CI 1.62–3.07) higher prevalence of G allele of rs36014597 SNP was observed in RVVI cases comparative to controls. Also, a considerably high prevalence of AG genotype (p < 0.0001) was observed in RVVI cases than controls. The overdominant model of inheritance, with the lowest AIC = 590.5 and BIC = 602.9 values, was found to be best, of all the models that showed significant association with rs36014597 polymorphism. The overdominant model depicts that the carrier of heterozygous genotype will have more extreme phenotype than that of either of its homozygous carriers in developing RVVI risk (p < 0.0001; OR = 4.07; 95% CI 2.69–6.17).
Table 4.
Distribution of allele frequencies of MBL2 additional 5′ variants in RVVI cases and controls
| Alleles | All subjects | RVVI cases | Controls | RVVI cases vs controls | |
|---|---|---|---|---|---|
| (N = 461) | (N = 258) | (N = 203) | OR (95% CI) | p-value | |
| Freq (%) | Freq (%) | Freq (%) | |||
| rs11003124 | |||||
| T | 714 (77.44) | 400 (77.51) | 314 (77.33) | 1.00 | |
| G | 208 (22.55) | 116 (22.48) | 92 (22.66) | 0.98 (0.72–1.35) | 0.94 |
| rs7084554 | |||||
| T | 724 (78.52) | 389 (75.38) | 335 (82.51) | 1.00 | |
| C | 198 (21.47) | 127 (24.61) | 71 (17.48) | 1.54 (1.11–2.13) | 0.009* |
| rs36014597 | |||||
| A | 694 (75.27) | 356 (68.99) | 338 (83.25) | 1.00 | |
| G | 228 (24.72) | 160 (31.00) | 68 (16.74) | 2.23 (1.62–3.07) | 1.2 × 10−6** |
| rs11003123 | |||||
| G | 615 (66.70) | 343 (66.47) | 272 (66.99) | 1.00 | |
| A | 307 (33.30) | 173 (33.52) | 134 (33.01) | 1.02 (0.77–1.34) | 0.86 |
OR odds ratio, CI confidence intervals
* Indicates significant p values (p < 0.01)
** Indicates highly significant values (p < 0.001)
Table 5.
Distribution of genotypic frequencies along with inheritance models of MBL2 5′ additional variants in RVVI cases and controls
| Genetic models | Genotype | VVI cases | Controls | OR (95% CI) | p-value | AIC | BIC |
|---|---|---|---|---|---|---|---|
| (N = 258) | (N = 203) | ||||||
| Freq (%) | Freq (%) | ||||||
| rs11003124 | |||||||
| Codominant | T/T | 155 (60.11) | 125 (61.60) | 1.00 | |||
| T/G | 90 (34.92) | 64 (31.50) | 1.14 (0.76–1.69) | 0.53 | 639.4 | 655.9 | |
| G/G | 13 (5.00) | 14 (6.90) | 0.75 (0.34–1.65) | 0.58 | |||
| Dominant | T/T | 155 (60.10) | 125 (61.60) | 1.00 | |||
| T/G-G/G | 103 (39.91) | 78 (38.41) | 1.07 (0.73–1.56) | 0.74 | 638.4 | 650.8 | |
| Recessive | T/T-T/G | 245 (95.23) | 189 (93.12) | 1.00 | |||
| G/G | 13 (5.00) | 14 (6.90) | 0.72 (0.33–1.56) | 0.4 | 637.8 | 650.2 | |
| Overdominant | T/T-G/G | 168 (65.10) | 139 (68.50) | 1.00 | |||
| T/G | 90 (34.90) | 64 (31.50) | 1.17 (0.79–1.73) | 0.45 | 637.9 | 650.3 | |
| Log-additive | – | – | – | 0.99 (0.73–1.34) | 0.95 | 638.5 | 650.9 |
| rs7084554 | |||||||
| Codominant | T/T | 147 (56.97) | 143 (70.42) | 1.00 | |||
| T/C | 95 (36.82) | 49 (24.11) | 1.90 (1.25–2.89) | 0.002* | 631 | 647.6 | |
| C/C | 16 (6.21) | 11 (5.42) | 1.41 (0.63–3.15) | 0.396 | |||
| Dominant | T/T | 147 (57.21) | 143 (70.41) | 1.00 | |||
| T/C-C/C | 111 (43.12) | 60 (29.60) | 1.81 (1.22–2.68) | 0.002* | 629.5 | 641.9 | |
| Recessive | T/T-T/C | 242 (93.80) | 192 (94.60) | 1.00 | |||
| C/C | 16 (6.21) | 11 (5.41) | 1.15 (0.52–2.55) | 0.72 | 638.4 | 650.8 | |
| Overdominant | T/T-C/C | 163 (63.21) | 154 (75.90) | 1.00 | |||
| T/C | 95 (36.81) | 49 (24.12) | 1.85 (1.22–2.79) | 0.003* | 629.7 | 642.1 | |
| Log-additive | – | – | – | 1.50 (1.09–2.07) | 0.01* | 632 | 644.4 |
| rs36014597 | |||||||
| Codominant | A/A | 110 (42.60) | 147 (72.41) | 1.00 | |||
| A/G | 136 (52.70) | 44 (21.70) | 4.20 (2.75–6.41) | 1.08 × 10−10** | 592 | 608.5 | |
| G/G | 12 (4.70) | 12 (5.90) | 1.33 (0.57–3.08) | 0.497 | |||
| Dominant | A/A | 110 (42.60) | 147 (72.41) | 1.00 | |||
| A/G-G/G | 148 (57.42) | 56 (27.60) | 3.61 (2.42–5.38) | 8.8 × 10−10** | 596 | 608.4 | |
| Recessive | A/A-A/G | 246 (95.31) | 191 (94.12) | 1.00 | |||
| G/G | 12(4.70) | 12 (5.90) | 0.77 (0.34–1.77) | 0.54 | 638.1 | 650.5 | |
| Overdominant | A/A-G/G | 122 (47.31) | 159 (78.32) | 1.00 | |||
| A/G | 136 (52.70) | 44 (21.70) | 4.07 (2.69–6.17) | 1.1 × 10−10** | 590.5 | 602.9 | |
| Log-additive | – | – | – | 2.42(1.72–3.41) | 1.2 × 10−6** | 610.8 | 623.2 |
| rs11003123 | |||||||
| Codominant | G/G | 100 (38.81) | 84 (41.42) | 1.00 | |||
| G/A | 143 (55.41) | 104 (51.22) | 1.16(0.79–1.70) | 0.46 | 639.5 | 656 | |
| A/A | 15 (5.82) | 15 (7.41) | 0.84 (0.39–1.82) | 0.61 | |||
| Dominant | G/G | 100 (38.81) | 84 (41.42) | 1.00 | |||
| G/A-A/A | 158 (61.22) | 119 (58.61) | 1.12 (0.77–1.62) | 0.57 | 638.2 | 650.6 | |
| Recessive | G/G-G/A | 243 (94.22) | 188 (92.61) | 1.00 | |||
| A/A | 15 (5.81) | 15 (7.40) | 0.77 (0.37–1.62) | 0.5 | 638 | 650.4 | |
| Overdominant | G/G-A/A | 115 (44.61) | 99 (48.80) | 1.00 | |||
| G/A | 143 (55.41) | 104 (51.21) | 1.18 (0.82–1.71) | 0.37 | 637.7 | 650.1 | |
| Log-additive | – | – | – | 1.03 (0.76–1.40) | 0.85 | 638.5 | 650.9 |
Italics values indicate low AIC/BIC value
AIC Akaike’s Information Criterion, BIC Bayesian Information Criterion
* Indicates (p ≤ 0.01)
** Indicates (p ≤ 0.001)
Genetic analyses of additional 5′ variants in RVVI types relative to controls
For rs7084554, significantly higher prevalence of C allele was observed in BV (p = 0.03) and MI (p = 0.02) than controls (Table 6). Heterozygosity of C allele was observed to be appreciably higher in VVC (p = 0.007) and MI (p = 0.01) than controls. For rs36014597 polymorphism, G allele was significantly more prevalent in BV (p = 0.0001), VVC (p = 0.004) and MI cases (p = 0.0002) as compared to controls. Heterozygosity for G allele was observed to considerably more prevalent in BV (p = 0.0001), VVC (p = 0.0001) and MI cases (p = 0.0007) comparative to controls. However, no significant difference in genotypic as well as allelic distribution was observed for rs11003124 and rs11003123 variants. Also, no homozygote of the minor allele of all the additional 5′ variants was observed in VVC.
Table 6.
MBL2 additional 5′ variant’s distribution and comparison of genotypes and alleles in RVVI types with controls
| No. (%) of controls | No. (%) of VVI types | Genotype comparison | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BV vs HC | VVC vs HC | MI vs HC | ||||||||
| HC (N = 203) | BV (n = 97) | VVC (n = 62) | MI (n = 41) | OR (95% CI) | p-value | OR (95% CI) | p-value | OR (95% CI) | p-value | |
| rs11003124 | ||||||||||
| Genotypes | ||||||||||
| TT | 125 (61.51) | 64 (65.91) | 36 (58.12) | 23 (56.12) | 1 | 1 | 1 | |||
| TG | 64 (31.52) | 27 (27.82) | 26 (41.91) | 15 (36.51) | 0.82 (0.47–1.41) | 0.48 | 1.41(0.78–2.53) | 0.25 | 1.27 (0.62–2.60) | 0.50 |
| GG | 14 (6.81) | 6 (6.11) | 0 (0.00) | 3 (7.31) | 0.83 (0.30–2.28) | 0.72 | – | NA | 1.16 (0.30–4.37) | 0.82 |
| Alleles | ||||||||||
| T | 314 (77.31) | 155 (79.81) | 98 (79.12) | 61 (74.31) | 1 | 1 | 1 | |||
| G | 92 (22.62) | 39 (20.12) | 26 (20.91) | 21 (25.62) | 0.85 (0.56–1.30) | 0.47 | 0.90 (0.55–1.47) | 0.69 | 1.17 (0.67–2.03) | 0.56 |
| rs7084554 | ||||||||||
| Genotypes | ||||||||||
| TT | 143 (70.40) | 59 (60.82) | 35 (56.41) | 21 (51.21) | 1 | 1 | 1 | |||
| TC | 49 (24.10) | 28 (28.81) | 27 (43.52) | 17 (41.42) | 1.38 (0.79–2.41) | 0.24 | 2.25 (1.23 4.09) | 0.007** | 2.36 (1.15–4.83) | 0.01** |
| CC | 11 (5.41) | 10 (10.30) | 0 (0.00) | 3 (7.30) | 2.20 (0.88–5.46) | 0.08 | – | NA | 1.85 (0.47–7.20) | 0.37 |
| Alleles | ||||||||||
| T | 335 (82.50) | 146 (75.21) | 97 (78.21) | 59 (71.90) | 1 | 1 | 1 | |||
| C | 71 (17.40) | 48 (24.72) | 27 (21.72) | 23 (28.01) | 1.55 (1.02–2.34) | 0.03* | 1.31 (0.79–2.15) | 0.28 | 1.83 (1.06–3.17) | 0.02* |
| rs36014597 | ||||||||||
| Genotypes | ||||||||||
| AA | 147 (72.40) | 41 (42.21) | 29 (46.77) | 18 (36.51) | 1 | 1 | 1 | |||
| AG | 44 (21.61) | 50 (51.51) | 33 (53.21) | 19 (46.32) | 4.07 (2.39–6.94) | 3.7 × 10−7*** | 3.80(2.08–6.94) | 1.7 × 10−5*** | 3.52 (1.70–7.29) | 0.0007*** |
| GG | 12 (5.91) | 6 (6.12) | 0 (0.00) | 4 (9.70) | 1.79 (0.63–5.06) | 0.27 | – | NA | 2.72 (0.79–9.34) | 0.111 |
| Alleles | ||||||||||
| A | 338 (83.20) | 132 (68.12) | 81 (65.31) | 49 (59.70) | 1 | 1 | 1 | |||
| G | 68 (16.71) | 62 (31.91) | 33 (26.62) | 27 (32.91) | 2.33 (1.56–3.47) | 3.6 × 10−5*** | 2.02(1.25–3.27) | 0.004** | 2.73 (1.60–4.68) | 0.0002*** |
| rs11003123 | ||||||||||
| Genotypes | ||||||||||
| GG | 84 (41.31) | 36 (37.11) | 26 (41.91) | 16 (39.01) | 1 | 1 | 1 | |||
| GA | 104 (51.22) | 53 (54.62) | 36 (582) | 21 (51.21) | 1.18 (0.71–1.98) | 0.50 | 1.11 (0.62–1.99) | 0.70 | 1.06 (0.52–2.15) | 0.87 |
| AA | 15 (7.38) | 8 (8.21) | 0 (0.00) | 4 (9.71) | 1.24 (0.48–3.19) | 0.64 | – | NA | 1.4 (0.41–4.76) | 0.59 |
| Alleles | ||||||||||
| G | 272 (66.91) | 125 (64.4) | 88 (70.91) | 53 (64.62) | 1 | 1 | 1 | |||
| A | 134 (33.12) | 69 (35.5) | 36 (29.12) | 29 (35.31) | 1.12 (0.78–1.60) | 0.53 | 0.83 (0.53–1.28) | 0.40 | 1.11(0.67–1.82) | 0.67 |
OR odds ratio, CI confidence intervals
* Indicates significant p values (p ≤ 0.05)
** Indicates high significant values (p ≤ 0.01)
*** Indicates highly significant values (p ≤ 0.001)
Genotype–phenotype association of additional 5′ variants
The stratification of previously measured sMBL levels was made on the basis of the observed genotypes of additional 5′ variants in cases and controls (Table 7). In BV, overall significant difference between genotypic sMBL levels was observed for rs11003124 (p = 0.026), rs36014597 (p = 0.01) and rs11003123 (p = 0.006) polymorphism. Further analysis of rs11003124 polymorphism indicated that TT (p = 0.032) and TG (p = 0.021) genotypes contributed considerably low measured sMBL levels than GG genotype. For rs36014597 polymorphism, AA (p = 0.007) and AG (p = 0.014) genotypes were significantly contributing low sMBL levels comparative to GG genotype. For rs11003123 polymorphism, considerably low sMBL levels were observed for GA (p = 0.01) genotype than AA genotype. In MI, an overall significant difference between genotypic sMBL levels was observed for rs7084554 (p = 0.01) polymorphism only. Further analysis of this polymorphism indicated that sMBL levels contributed by TT genotype were significantly (p = 0.013) different from levels contributed by TC genotype. Data analysis for genotypic sMBL levels in controls, RVVI and VVC revealed no significant difference in sMBL levels of studied 5′ variants. In addition, for the same genotypes, cases were found to have considerably low sMBL levels than controls.
Table 7.
Distribution of sMBL levels in RVVI, RVVI types and controls, stratified on the basis of MBL2 additional 5′ variant’s genotypes
| Genotypes | Controls | RVVI | p-value (Controls vs. RVVI)† | BV | p-value (Controls vs. BV)† | VVC | p-value (Controls vs. VVC)† | MI | p-value (Controls vs. MI)† | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Mean ± SD | n | Mean ± SD | n | Mean ± SD | n | Mean ± SD | n | Mean ± SD | |||||
| rs11003124 | ||||||||||||||
| TT | 125 | 1080.5 ± 465.50 | 155 | 744.07 ± 495.75 | 1.8 × 10−8*** | 64 | 678.61 ± 390.79a | 1.5 × 10−8*** | 36 | 442.98 ± 184.28 | 2.7 × 10−24*** | 23 | 634.42 ± 293.56 | 2.8 × 10−7*** |
| TG | 64 | 1049.9 ± 430.30 | 90 | 795.44 ± 531.28 | 0.002** | 27 | 626.25 ± 326.24b | 1.5 × 10−5*** | 26 | 458.20 ± 230.82 | 1.1 × 10−12*** | 15 | 887.35 ± 648.44 | 0.368 |
| GG | 14 | 1313.5 ± 520.39 | 13 | 962.81 ± 496.91 | 0.084 | 6 | 1111.8 ± 678.54ab | 0.472 | 0 | 0.00 ± 0.00 | NA | 3 | 850.39 ± 25.19 | 0.152 |
| p-value | †† | 0.144 | †† | 0.290 | †† | 0.026* | † | 0.774 | †† | 0.234 | ||||
| rs7084554 | ||||||||||||||
| TT | 143 | 1112.4 ± 525.32 | 147 | 719.73 ± 478.38 | 1.4 × 10−10*** | 59 | 624.45 ± 325.85 | 2.0 × 10−13*** | 35 | 447.24 ± 207.63 | 7.3 × 10−23*** | 21 | 500.00 ± 104.30f | 9.7 × 10−25*** |
| TC | 49 | 1316.7 ± 679.60 | 95 | 865.74 ± 602.85 | 7.7 × 10−5*** | 28 | 748.64 ± 483.93 | 2.1 × 10−4*** | 27 | 452.23 ± 201.68 | 1.4 × 10−11*** | 17 | 929.32 ± 665.41f | 0.046* |
| CC | 11 | 1069.6 ± 422.36 | 16 | 833.60 ± 321.42 | 0.111 | 10 | 797.20 ± 368.89 | 0.134 | 0 | 0.00 ± 0.00 | NA | 3 | 850.39 ± 25.19 | 0.40 |
| p-value | †† | 0.080 | †† | 0.097 | †† | 0.218 | † | 0.925 | †† | 0.01** | ||||
| rs36014597 | ||||||||||||||
| AA | 147 | 1165.5 ± 577.79 | 110 | 720.49 ± 478.04 | 2.8 × 10−10*** | 41 | 634.69 ± 302.67c | 1.1 × 10−12*** | 29 | 455.00 ± 207.48 | 1.7 × 10−21*** | 18 | 669.20 ± 311.55 | 2.4 × 10−6*** |
| AG | 44 | 1049.0 ± 440.84 | 136 | 796.61 ± 527.34 | 0.005** | 50 | 680.18 ± 417.03d | 7.0 × 10−5*** | 33 | 444.32 ± 202.85 | 2.8 × 10−11*** | 19 | 887.19 ± 640.10 | 0.325 |
| GG | 12 | 1297.6 ± 530.97 | 12 | 899.07 ± 428.27 | 0.325 | 6 | 1164.3 ± 670.40cd | 0.05* | 0 | 0.00 ± 0.000 | NA | 4 | 842.52 ± 430.05 | 0.145 |
| p-value | †† | 0.293 | †† | 0.328 | †† | 0.01** | † | 0.839 | † | 0.416 | ||||
| rs11003123 | ||||||||||||||
| GG | 84 | 1086.4 ± 466.75 | 100 | 754.57 ± 508.33 | 8.7 × 10−6*** | 36 | 780.92 ± 474.43 | 0.001*** | 26 | 448.89 ± 199.68 | 1.9 × 10−16*** | 16 | 646.30 ± 343.61 | 0.001*** |
| GA | 104 | 1066.1 ± 430.02 | 143 | 769.09 ± 512.02 | 2.6 × 10−6*** | 53 | 598.29 ± 322.96e | 3.7 × 10−12*** | 36 | 449.89 ± 208.86 | 1.1 × 10−20*** | 21 | 872.59 ± 607.82 | 0.177 |
| AA | 15 | 1143.8 ± 549.47 | 15 | 856.24 ± 384.30 | 0.108 | 8 | 1062.2 ± 604.69e | 0.746 | 0 | 0.00 ± 0.000 | NA | 4 | 896.06 ± 346.68 | 0.409 |
| p-value | †† | 0.816 | †† | 0.767 | †† | 0.006** | † | 0.985 | †† | 0.365 | ||||
NA not applicable
* p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001
†Independent t-test for patient vs. control as well as within groups
††One Way ANOVA with overall p-value followed by Post Hoc Tukey test that further indicates values with similar letter are significantly different
ap = 0.032, b p = 0.021, c p = 0.007, d p = 0.014, e p = 0.01, f p = 0.013
Genotypes with n < 3 are not included in statistical analysis
Linkage disequilibrium analyses
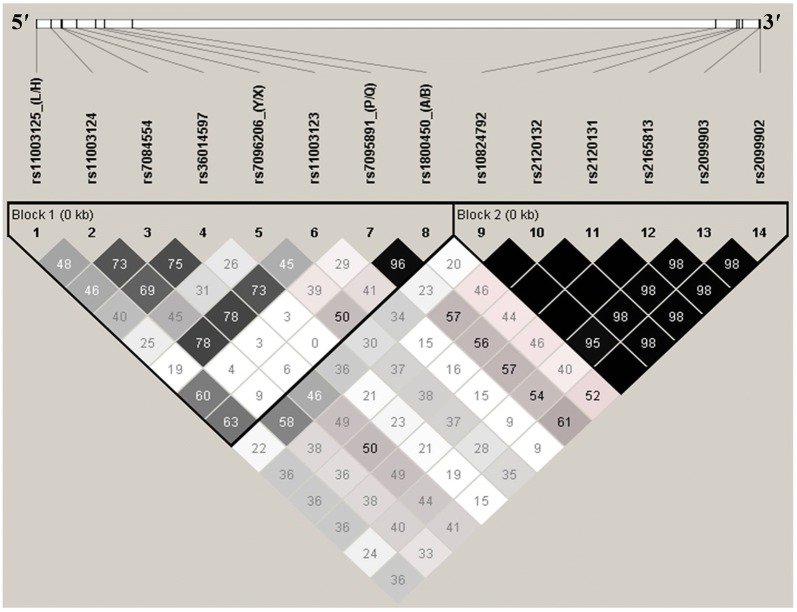
The LD pattern of 14 variants across MBL2 was determined using their genotyped data. As LD is likely to decrease with increase in physical distance, the LD plot with two blocks was observed in the cohort of the present study for MBL2 (Fig. 2). The secretor polymorphisms along with additional 5′ near gene variants form the 5′ block. The six 3′UTR variants form the 3′ block of MBL2. The markers of one block were not in LD with the markers of other. The LD analysis of 5′ block of MBL2 indicated nearly complete LD between P/Q and codon 54 variants with D′ of 0.96 and thus co-inherited together. SNP pairs including rs11003124/rs11003123, rs7084554/rs11003123, rs7084554/rs36014597, rs11003124/rs7084554 and rs36014597/rs11003123 were in strong LD with D′ of 0.78, 0.78, 0.75, 0.73 and 0.73, respectively. In addition, SNP pairs including rs11003124/rs36014597, LH/AB and LH/PQ showed fairly high LD with D′ of 0.69, 0.63 and 0.6. The LD analysis indicated that all the SNPs of 3′ block are in strong LD with each other and are co-inherited together [13].
Fig. 2.
LD pattern of MBL2 variants in all participants. Block 1 represents the 5′ block variants. Block 2 represents the 3′ block variants. D′ is scaled between white diamond: D′ = 0 i.e. complete Linkage equilibrium, shades of grey: 0 < D′ < 1, Black diamond: D′ = 1 i.e. complete LD. Numbers in diamonds are D′-value expressed as a percentile. Black diamond without numbers represents a D′ of 100
Haplotypes analyses
The LD analyses confirmed two haplotype blocks i.e. 5′ haplotype block and 3′ haplotype block. Both separate and combined analysis of each block was performed. The distribution of 3′ block haplotypes in cases (RVVI and its types) and controls has been reported previously [13]. This analysis showed the presence of three common haplotypes i.e. TTTGCT (3′H-1), CCGAAC (3′H-2) and CTTGCT (3′H-3) with frequency ≥ 0.05 (5%) either in cases or controls. Five rare haplotypes i.e. TTTGAT (3′H-4), CCGACT (3′H-5), CTTGAT (3′H-6), CCTAAT (3′H-7) and CTTGCC (3′H-8) were also observed, but excluded from the previous analysis due to their very low frequency (< 0.05).
Distribution of MBL2 5′ block haplotypes
Total fifty-nine 5′ block haplotypes from 5′H-1 to 5′H-59, with frequencies ≥ 0.001 (0.1%) either in cases or controls, were observed in the present study. An overall significant difference (global p = 0.01) were observed in distributions of all the observed 5′ block haplotypes among case–control groups (Additional file 1). Only six haplotypes i.e. HTTAXGQB (5′H-1), HTTAYGQB (5′H-2), LTTAXGQB (5′H-3), LTTAYGPA (5′H-4), LGCGYAPA (5′H-5) and HTTAYAPA (5′H-6) with frequency ≥ 0.05 (5%, either in cases or controls) were observed and are referred as common haplotypes, while the rest haplotypes were considered as rare and thus excluded from the present analysis due to their very low frequency. The case–control comparison of common haplotypes depicted considerably low prevalence of haplotype 5′H-2 in RVVI and MI cases than controls (Table 8). Also, the frequency of haplotype 5′H-4 was observed to be significantly low in RVVI and BV cases than controls. Moreover, the other common haplotypes did not show any statistical differences in their observed frequencies between cases and controls.
Table 8.
Distribution and comparison of 5′ block haplotypes of MBL2 in cases and controls
| Haplotypes (5′ → 3′) | No. (%) of controls | No. (%) of cases | Haplotype comparison | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total RVVI cases | Clinical categories of RVVI | RVVI vs controls | BV vs controls | VVC vs controls | MI vs controls | ||||||||
| (n = 406) | (n = 516) | BV (n = 194) | VVC (n = 124) | MI (n = 82) | OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value | |
| Common | |||||||||||||
| HTTAXGQB (5′H-1) | 52 (12.80) | 98 (18.99) | 33 (17.01) | 20 (16.12) | 16 (19.51) | 1 | 1 | 1 | 1 | ||||
| HTTAYGQB (5′H-2) | 57 (14.03) | 50 (9.68) | 19 (9.79) | 16 (12.90) | 6 (7.31) | 0.46 (0.28–0.77) | 0.003* | 0.52 (0.26–1.03) | 0.062 | 0.72 (0.34–1.55) | 0.415 | 0.34 (0.12–0.94) | 0.03* |
| LTTAXGQB (5′H-3) | 39 (9.60) | 65 (12.59) | 27 (13.91) | 17 (13.70) | 7 (8.53) | 0.88 (0.52–1.48) | 0.643 | 1.09 (0.56–2.10) | 0.795 | 1.13 (0.52–2.44) | 0.749 | 0.58 (0.21–1.55) | 0.281 |
| LTTAYGPA (5′H-4) | 51 (12.56) | 51 (9.88) | 12 (6.18) | 14 (11.29) | 10 (12.19) | 0.53 (0.31–0.88) | 0.01* | 0.37 (0.17–0.79) | 0.01* | 0.71 (0.32–1.56) | 0.399 | 0.63 (0.26–1.53) | 0.315 |
| LGCGYAPA (5′H-5) | 24 (5.91) | 64 (12.40) | 19 (9.79) | 14 (11.29) | 14 (17.07) | 1.41 (0.79–2.52) | 0.238 | 0.55 (0.28–1.09) | 0.089 | 1.51 (0.65–3.50) | 0.329 | 1.89 (0.79–4.50) | 0.147 |
| HTTAYAPA (5′H-6) | 17 (4.18) | 26 (5.03) | 11 (5.67) | 6 (4.83) | 3 (3.65) | 0.81 (0.40–1.63) | 0.557 | 1.01 (0.42–2.44) | 0.965 | 0.91 (0.31–2.65) | 0.874 | 0.57 (0.14–2.21) | 0.419 |
Global p-value for case/control haplotype association was 0.01
* p ≤ 0.01
Distribution of combined 5′/3′ block haplotypes
The combined haplotype analyses included genotypic data of only 218 participants i.e. 109 cases and 109 controls from the total cohort of the present study, as the same number of participants, were used for the evaluation of 3′ block variants as reported previously [13]. A global significant difference (p = 0.01) in combined 5′/3′ haplotype frequency was observed. Total 86 combined haplotypes, with frequencies ≥ 0.001 either in cases or controls, were observed (Additional file 2). Of these, haplotypes i.e. LTTAYGQB/CCGAAC (5′H-10/3′H-2), LGCGYAPA/TTTGCT (5′H-5/3′H-1), LTTAXGQB/CTTGCT (5′H-3/3′H-3), HTTAXGQB/TTTGCT (5′H-1/3′H-1), HTTAYGPA/TTTGCT (5′H-9/3′H-1), HTTAXGQB/CCGAAC (5′H-1/3′H-2), HTTAXGPA/CTTGCT (5′H-35/3′H-3) and HTTAXGQB/TTTGCT (5′H-60/3′H-1) with frequency ≥ 0.05 (5%, either in cases or controls) were observed and are referred as common haplotypes, while the rest were considered as rare haplotypes and excluded from the present analysis. Considerably higher frequency of 5′H-60/3′H-1 haplotype was observed in BV cases than controls (Table 9). Moreover, the other combined common MBL2 haplotypes did not show any statistical differences in their observed frequencies between cases and controls.
Table 9.
Distribution and comparison of combined 5′/3′ haplotypes of MBL2 in cases and controls
| Haplotypes (5′ → 3′) | No. (%) of controls | No. (%) of cases | Haplotype comparison | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total RVVI cases | Clinical categories of RVVI | RVVI vs controls | BV vs controls | VVC vs controls | MI vs controls | ||||||||
| (n = 218) | (n = 218) | BV (n = 112) | VVC (n = 56) | MI (n = 50) | OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value | |
| Common | |||||||||||||
| 5′H-10/3′H-2 | 24 (11.00) | 23 (10.55) | 9 (8.03) | 8 (14.2) | 6 (12.00) | 1 | 1 | 1 | 1 | ||||
| 5′H-5/3′H-1 | 12 (5.50) | 16 (7.33) | 9 (8.03) | 3 (5.35) | 4 (8.00) | 1.39 (0.54–3.56) | 0.492 | 2.00 (0.63–6.34) | 0.2395 | 0.75 (0.16–3.35) | 0.706 | 1.33 (0.31–5.64) | 0.695 |
| 5′H-3/3′H-3 | 15 (6.88) | 10 (4.58) | 4 (3.57) | 3 (5.35) | 3 (6.00) | 0.69 (0.26–1.86) | 0.469 | 0.71 (0.18–2.72) | 0.618 | 0.60 (0.13–2.62) | 0.497 | 0.80 (0.17–3.68) | 0.774 |
| 5′H-1/3′H-1 | 14 (6.42) | 10 (4.58) | 10 (8.92) | 0 (0.00) | 0 (0.00) | 0.74 (0.27–2.01) | 0.561 | 1.90 (0.62–5.81) | 0.257 | – | NA | – | NA |
| 5′H-9/3′H-1 | 15 (6.88) | 8 (3.66) | 4 (3.57) | 2 (3.57) | 2 (4.00) | 0.55 (0.19–1.56) | 0.265 | 0.71 (0.18–2.72) | 0.618 | 0.40 (0.07–2.14) | 0.284 | 0.53 (0.09–2.99) | 0.475 |
| 5′H-1/3′H-2 | 9 (4.12) | 11 (5.04) | 4 (3.57) | 5 (8.92 | 2 (4.00) | 1.27 (0.44–3.64) | 0.649 | 1.18 (0.29–4.83) | 0.812 | 1.66 (0.43–06.45) | 0.459 | 0.88 (0.15–5.24) | 0.896 |
| 5′H-35/3′H-3 | 4 (1.83) | 11 (5.04) | 5 (4.46) | 3 (5.35) | 3 (6.00) | 2.86 (0.79–10.31) | 0.106 | 3.33 (0.72–15.26) | 0.121 | 2.25 (0.41–12.28) | 0.349 | 3.00 (0.52–17.15) | 0.216 |
| 5′H-60/3′H-1 | 3 (1.37) | 11 (5.04) | 7 (6.25) | 3 (5.35) | 1 (2.00) | 3.82 (0.94–15.49) | 0.060 | 6.22 (1.31–29.44) | 0.021* | 3.00 (0.50–17.95) | 0.228 | 1.33 (0.11–15.20) | 0.816 |
Global p-value for case/control haplotype association was 0.01
NA not applicable
* p ≤ 0.05; HTTAYGPA (5′H-9), LTTAYGQB (5′H-10), HTTAXGPA (5′H-35), HTTAXGQB (5′H-60)
Haplotype-phenotype associations
The previously measured sMBL levels were stratified in cases and controls on the basis of independent and combined haplotypes of two blocks. The distribution and comparisons of sMBL levels of 3′ block haplotypes in cases and control groups have been reported previously, which suggested that 3′ block haplotypes may alter sMBL levels and susceptibility to RVVI [13].
Distribution of sMBL levels in 5′ block haplotypes
sMBL levels were segregated according to 5′ block common haplotypes in cases and controls (Table 10). In controls, an overall significant difference (p = 0.004) was observed among sMBL of different haplotypes. Further analysis has shown that 5′H-1 (HTTAXGQB; p = 0.002) and 5′H-4 (LTTAYGPA; p = 0.048) haplotypes were conferring significantly low levels comparative to 5′H-2 (HTTAYGQB) haplotype, whereas other haplotypes in controls did not show any significant difference among sMBL levels. In RVVI cases, sMBL levels of different haplotypes showed an overall significant difference (p = 0.008). Further analysis has shown that 5′H-3 (LTTAXGQB) haplotype were accounted for significantly (p = 0.029) low levels than 5′H-2 haplotype, whereas other haplotypes in RVVI cases did not show any significant difference among sMBL levels. In RVVI types i.e. BV, VVC and MI, no overall as well as in particular significant difference was observed among sMBL levels of different haplotypes. Furthermore, significantly (p < 0.05) low sMBL levels were accounted by various haplotypes of RVVI and its types comparative to corresponding haplotypes in controls.
Table 10.
Distribution of sMBL levels in cases and controls, stratified on the basis of 5′ block haplotypes of MBL2
| Haplotypes | Controls | RVVI | p-value (controls vs RVVI)† | BV | p-value (controls vs BV)† | VVC | p-value (controls vs VVC)† | MI | p-value (controls vs MI)† | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Mean ± SD | n | Mean ± SD | n | Mean ± SD | n | Mean ± SD | n | Mean ± SD | |||||
| 5′H-1 | 52 | 929.13 ± 328.26a | 98 | 796.14 ± 497.42 | 0.05* | 33 | 756.96 ± 518.41 | 0.095 | 20 | 495.04 ± 146.90 | 6.5 × 10−11*** | 16 | 774.02 ± 512.29 | 0.268 |
| 5′H-2 | 57 | 1229.7 ± 457.22ab | 50 | 964.66 ± 694.09c | 0.024* | 19 | 711.44 ± 421.49 | 4.1 × 10−5*** | 16 | 568.28 ± 288.36 | 2.1 × 10−8*** | 6 | 912.34 ± 720.02 | 0.132 |
| 5′H-3 | 39 | 1053.3 ± 521.53 | 65 | 666.02 ± 450.61c | 1.2 × 10−4*** | 27 | 571.21 ± 222.96 | 3.8 × 10−6*** | 17 | 419.29 ± 282.35 | 3.2 × 10−7*** | 7 | 701.91 ± 401.10 | 0.098 |
| 5′H-4 | 51 | 1001.6 ± 333.61b | 51 | 703.54 ± 505.09 | 0.001*** | 12 | 763.35 ± 554.86 | 0.177 | 14 | 372.70 ± 182.16 | 1.7 × 10−11*** | 10 | 691.51 ± 367.44 | 0.01** |
| 5′H-5 | 24 | 1075.7 ± 353.79 | 64 | 896.52 ± 548.16 | 0.077 | 19 | 883.99 ± 511.06 | 0.210 | 14 | 563.78 ± 384.49 | 1.8 × 10−4*** | 14 | 990.78 ± 618.07 | 0.643 |
| 5′H-6 | 17 | 1188.7 ± 461.46 | 26 | 631.93 ± 248.25 | 1.5 × 10−4*** | 11 | 715.82 ± 302.77 | 0.003** | 6 | 487.56 ± 220.87 | 1.1 × 10−4*** | 3 | 663.52 ± 23.84 | 2.4 × 10−4*** |
| p-value†† | 0.004* | 0.008* | 0.333 | 0.272 | 0.697 | |||||||||
* p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001
†Independent t-test between patients and controls as well as within groups
††One Way ANOVA with overall p-value followed by Post Hoc Tukey test that further indicates values with similar letter are significantly different
ap = 0.002, b p = 0.048, c p = 0.029
Distribution of sMBL levels in combined 5′/3′ block haplotypes
sMBL levels were segregated according to combined 5′/3′ block common haplotypes in cases and controls (Table 11). In controls, only an overall significant difference (p = 0.032) was observed among sMBL of different haplotypes. In RVVI cases, an overall significant difference (p = 0.002) was observed among sMBL of different haplotypes. Further analysis has shown that 5′H-10/3′H-2 (LTTAYGQB/CCGAAC) haplotype accounted for significantly low sMBL levels than 5′H-5/3′H-1 (LGCGYAPA/TTTGCT; p = 0.033) and 5′H-1/3′H-2 (HTTAXGQB/CCGAAC; p = 0.010) haplotypes. Also, 5′H-35/3′H-3 (HTTAXGPA/CTTGCT) haplotype was contributing significantly (p = 0.021) low sMBL levels than 5′H-1/3′H-2 haplotype whereas, no other significant differences were observed. In BV and VVC cases, different haplotypes did not show any in particular significant difference among sMBL levels, though overall significant difference (p = 0.050) was found in BV. In MI cases, an overall significant difference (p = 0.006) was observed among sMBL levels of different haplotypes. Further analysis has shown that 5′H-10/3′H-2 (LTTAYGQB/CCGAAC; p = 0.008) and 5′H-35/3′H-3 (HTTAXGPA/CTTGCT; p = 0.016) haplotypes accounted for significantly low sMBL levels than 5′H-5/3′H-1 (LGCGYAPA/TTTGCT) haplotype. Furthermore, significantly (p < 0.05) low sMBL levels were accounted by various haplotypes of RVVI and its types comparative to respective haplotypes in controls.
Table 11.
Distribution of sMBL levels in cases and controls, stratified on the basis of combined 5′/3′ block haplotypes of MBL2
| Haplotypes | Controls | RVVI | p-value (controls vs RVVI)† | BV | p-value (controls vs BV)† | VVC | p-value (controls vs VVC)† | MI | p-value (controls vs MI)† | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Mean ± SD | n | Mean ± SD | n | Mean ± SD | n | Mean ± SD | n | Mean ± SD | |||||
| 5′H-10/3′H-2 | 24 | 836.70 ± 293.17 | 23 | 434.33 ± 190.36ab | 1.7 × 10−6*** | 9 | 466.93 ± 253.23 | 0.002** | 8 | 348.26 ± 113.77 | 1.9 × 10−7*** | 6 | 502.68 ± 148.28d | 0.001*** |
| 5′H-5/3′H-1 | 12 | 957.48 ± 453.28 | 16 | 857.95 ± 549.84a | 0.614 | 9 | 976.38 ± 598.13 | 0.935 | 3 | 296.06 ± 345.25 | 0.036* | 4 | 1042.5 ± 282.10d e | 0.733 |
| 5′H-3/3′H-3 | 15 | 1267.1 ± 418.30 | 10 | 714.96 ± 513.21 | 0.007** | 4 | 792.13 ± 657.76 | 0.090 | 3 | 541.73 ± 635.47 | 0.02* | 3 | 785.30 ± 260.96 | 0.076 |
| 5′H-1/3′H-1 | 14 | 1261.6 ± 612.37 | 10 | 795.10 ± 425.08 | 0.050* | 10 | 795.10 ± 425.08 | 0.050* | 0 | 0.00 ± 0.00 | NA | 0 | 0.00 ± 0.00 | NA |
| 5′H-9/3′H-1 | 15 | 1228.8 ± 545.30 | 8 | 729.92 ± 407.04 | 0.034* | 4 | 788.98 ± 463.87 | 0.160 | 2 | 796.85 ± 645.86 | NA | 2 | 544.88 ± 75.72 | NA |
| 5′H-1/3′H-2 | 9 | 999.78 ± 162.15 | 11 | 969.51 ± 530.29bc | 0.861 | 4 | 1113.4 ± 138.52 | 0.251 | 5 | 934.80 ± 788.63 | 0.864 | 2 | 768.50 ± 311.79 | NA |
| 5′H-35/3′H-3 | 4 | 1070.9 ± 362.48 | 11 | 389.29 ± 194.90c | 3.6 × 10−4*** | 5 | 363.78 ± 281.19 | 0.013* | 3 | 346.46 ± 51.56 | 0.026* | 3 | 466.14 ± 127.39e | 0.042* |
| 5′H-60/3′H-1 | 3 | 915.49 ± 320.84 | 11 | 735.61 ± 320.19 | 0.405 | 7 | 578.27 ± 174.35 | 0.057 | 3 | 576.38 ± 110.23 | 0.158 | 1 | 1159.05 ± 0.00 | NA |
| p-value†† | 0.032* | 0.002** | 0.050* | 0.269 | 0.006** | |||||||||
Haplotypes with n < 3 are not included in statistical analysis
* p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001
†Independent t-test between patients and controls as well as within groups
††One Way ANOVA with overall p-value followed by Post Hoc Tukey test that further indicates values with similar letter are significantly different
ap = 0.033, b p = 0.010, c p = 0.021, d p = 0.008, e p = 0.016
Gene–gene interaction analyses
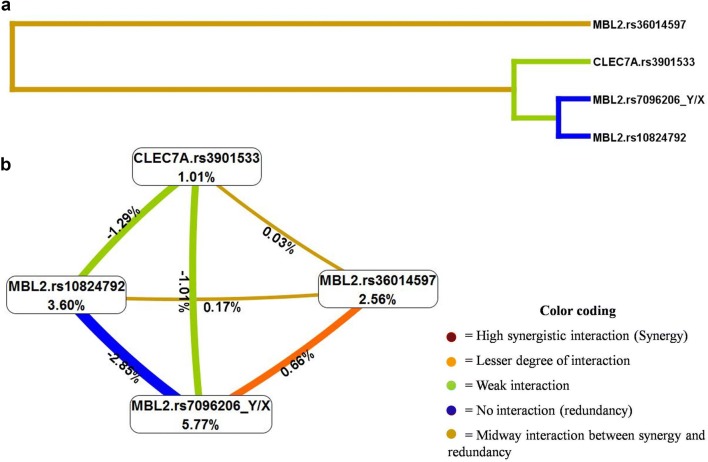
Multifactor dimensionality reduction (MDR) method was used to study interaction between all the seventeen SNPs of two genes i.e. MBL2 and CELC7A. Three gene–gene interaction models involving uni-variant, bi-variant and tri-variant appeared as significant (p < 0.001) predictors of RVVI risk (Table 12). Among 17 SNPs, one-way interaction model including MBL2 Y/X polymorphism was found to have maximum cross-validation consistency (CVC) of 10/10 with testing balance accuracy of 63.70%. Two-way interaction model (MBL2.rs36014597 and MBL2.Y/X) and three-way interaction model (MBL2.Y/X, MBL2.rs10824792 and CLEC7A.rs3901533) also showed association with RVVI risk but had comparatively low CV consistency i.e. 9/10 and 5/10, respectively relative to the uni-variant model. MDR analysis provides dendrogram to interpret the nature of possible interaction between SNPs (Fig. 3a). It was found that MBL2.Y/X, MBL2.rs10824792 and CLEC7A.rs3901533 belonging to one group had shown a weak synergistic interaction in predicting RVVI risk. This group along with MBL2.rs36014597 showed an intermediate level of association between synergy and redundancy in predicting RVVI susceptibility. MDR analysis also provides a graphical model of entropy interaction (Fig. 3b). It was found that entropy-based analysis was positive (0.03%) for a pairwise effect of CLEC7A.rs3901533 and MBL2.rs36014597 indicating synergy while negative for pair CLEC7A.rs3901533 and MBL2.Y/X (− 1.01%) as well as pair CLEC7A.rs3901533 and MBL2.rs10824792 (− 1.29%) indicating redundancy towards RVVI susceptibility.
Table 12.
Multifactor dimensionality reduction (MDR) analysis for the identification of best gene–gene interaction models for RVVI cases and controls
| Models | SNPs interaction | Training balance accuracy | Testing balance accuracy | Cross validation consistency | OR (95% CI) | χ2 value | p-value |
|---|---|---|---|---|---|---|---|
| 1 | MBL2.rs7096206 (Y/X) | 0.637 | 0.637 | 10/10 | 3.25 (1.84–5.74) | 17.15 | < 0.0001* |
| 2 | MBL2.rs36014597, MBL2.rs7096206 (Y/X) | 0.657 | 0.623 | 9/10 | 5.33 (2.71–10.50) | 26.05 | < 0.0001* |
| 3 | MBL2.rs7096206 (Y/X), MBL2.rs10824792, CLEC7A.rs3901533 | 0.702 | 0.637 | 5/10 | 5.73 (3.15–10.45) | 35.10 | < 0.0001* |
* Indicates significant at p ≤ 0.001
Fig. 3.
a The dendrogram representing the nature of possible interaction between SNPs by MDR analysis. b Interaction entropy model by MDR
Discussion
The present study is the first report presenting the frequency distribution of the four additional 5′ near gene variants of MBL2 in North Indian cohort, which depicted rs11003124_G, rs7084554_C, rs36014597_G, rs11003123_A to be minor alleles. The distribution of these variants was in agreement with all the different populations of the 1000 Genomes Project (Phase 3) except African population owing to its population substructure, high genetic diversity and less LD between genetic loci (Additional file 3) [36]. The two SNPs i.e. rs7084554 and rs36014597 were found to be significantly predisposing individuals to RVVI and its types, while the other two variants i.e. rs11003124 and rs11003123 was not found to be associated with the disease condition. Significantly higher prevalence of C allele, its homozygosity and heterozygosity were observed in RVVI cases and its types as compared to controls, explaining the observed dominant mode of inheritance of rs7084554 polymorphism that is increasing the risk of RVVI. In addition, the best fit model of rs36014597 polymorphism showed the overdominant mode of inheritance, which further depicts that the carrier of the heterozygous genotype of this polymorphism had more extreme phenotype than either of its homozygous carriers in developing RVVI risk as complemented by the observed results of genotype distribution. The literature search did not reveal any articles that have evaluated the role of select 5′ additional variants of MBL2 in association with RVVI or its types. However, these polymorphisms have been evaluated in relation to leprosy and malaria with no significant associations [23, 37]. An association of rs11003124 and rs11003123 SNPs have been shown with the increased risk of leprosy and hepatocellular carcinoma in patients with hepatitis B-related cirrhosis, respectively [37, 38].
Linking genotypes of these polymorphisms with phenotypes showed that for the same genotypes, cases accounted significantly low levels than controls. Furthermore, genotypic sMBL levels significantly varied for rs11003124, rs36014597 and rs11003123 polymorphisms in BV, while for rs7084554 polymorphism in MI. However, sMBL levels for these particular loci did not vary as expected from their genetic association analysis, possibly due to the small size of the respective groups, as the study was underpowered for RVVI categories. Therefore, increasing the sample size may ascertain the role of these SNPs in susceptibility to BV and MI. To date, no studies have examined the genotype–phenotype correlation of these polymorphisms. However, a single and recent study depicted no significant difference in genotypic sMBL levels of rs11003124 polymorphism in bakery workers with work-related respiratory symptoms [39]. Thus, of the 14 screened polymorphisms of MBL2, the single variant analysis showed five polymorphisms including rs7096206 (Y/X), rs7084554, rs36014597, rs10824792, and rs2099903 have been found to be associated with RVVI risk [11, 13]. For the association studies, the haplotype-based analysis has been suggested to be more influential approach, which can help avoid the risk of misinterpretation of individual SNPs analysis [40]. Therefore, haplotypes were constructed on the basis of linkage disequilibrium analysis as 5′ and 3′ block haplotypes. These blocks were evaluated independently as well as in combination, as it’s the different combinations of amino acids in a polypeptide chain that collectively determine the protein structure and function.
Independent analyses of 5′ block haplotypes have shown a significantly low prevalence of common haplotypes i.e. HTTAYGQB (5′H-2) in RVVI and MI cases as well as haplotype LTTAYGPA (5′H-4) in RVVI and BV cases relative to controls. This depicts that the major alleles (marked by underline) of 5′ additional variants and Y/X secretor polymorphism are collectively conferring protection against RVVI, BV and MI cases. Evaluation of MBL2 3′ block haplotypes analyses showed the presence of three common haplotypes i.e. TTTGCT (3′H-1), CCGAAC (3′H-2) and CTTGCT (3′H-3) in RVVI cases and controls. Independent analyses 3′ block haplotypes showed the risk effect of 3′H-3, the haplotype including the minor allele of rs10824792 SNP only in RVVI [13].
Combined 5′/3′ haplotypes analyses have shown significantly high prevalence of 5′H-60/3′H-1 (HTTAXGQB/TTTGCT) haplotype in BV cases than controls, showing X allele as an important marker for conferring risk of disease development. Thus, independent analysis of two blocks suggested the risk-modifying effect of 5′ additional variants, Y/X secretor polymorphism and 3′-UTR SNP i.e. rs10824792. However, complete haplotype analysis depicted only Y/X polymorphism as an important marker for determining disease risk. It also suggested the possibility of unrevealed regulating variants that may be masking the effect of others. Therefore, for further clarification, haplotype-phenotype correlation analysis was carried out.
Independent analyses of 5′ block haplotypes with sMBL levels showed significant difference in sMBL levels of different haplotypes, in controls and RVVI cases. Further analysis depicted, 5′H-1 (HTTAXGQB) haplotype accounted for significantly low levels than 5′H-2 (HTTAYGQB) haplotype in controls. Also, 5′H-3 (LTTAXGQB) haplotype accounted for significantly low levels than 5′H-2 haplotype in RVVI. These results again suggested the contribution of Y/X variant in modulating sMBL levels in line with the above findings of the present study. Moreover, stratification of sMBL levels based on 3′ block haplotypes suggested 3′H-3, the haplotype with the minor allele of rs10824792 SNP only, accounted for significantly low sMBL levels, and RVVI risk [13]. Considering this, it is possible that both 5′ and 3′ haplotypes contribute to sMBL levels, but at this point, it is still tentative whether these blocks have an independent impact on the sMBL levels or they have combined effect.
Therefore, sMBL levels were further correlated with combined 5′/3′ haplotypes. Low sMBL levels were observed in various haplotypes of RVVI and its types as compared to respective haplotypes in controls. Moreover, an overall significant difference was observed among sMBL levels of different haplotypes in controls, RVVI, BV, and MI cases. Analyses in these groups showed that 5′H-10/3′H-2 haplotype having LYQB a low secretor haplotype in 5′ block in combination with the minor allele haplotype of 3′ block was contributing low level than 5′H-5/3′H-1 i.e. haplotype having LYPA a high secretor haplotype in 5′ block and the major allele haplotype of 3′ block in RVVI and MI cases. Thus, the inclusion of additional variants in haplotype analysis solved the previously reported discrepancy where a high secretor haplotype i.e. LYPA accounted for significantly low sMBL levels than low secretor haplotype i.e. LYQB in all cases types and controls [11]. Also, 5′H-35/3′H-3 haplotype having low secretor haplotype ‘HXPA’ and 3′ block haplotype ‘CTTGCT’ with only risk allele ‘C’ of SNP rs10824792 was significantly contributing low sMBL levels than 5′H-5/3′H-1 haplotype with high secretor haplotype ‘LYPA’ and major allele haplotype of 3′ block in MI cases. Thus, correlation of combined haplotypes with sMBL levels signifies the importance of Y/X and rs10824792 polymorphisms from both the blocks in regulating sMBL levels and RVVI risk.
Thus, permutation analysis of haplotype with phenotype suggested that variants of two haplotype blocks have more of a combined effect than the independent effect in regulating sMBL levels and hence RVVI risk. These results are in consonance with the studies suggesting that there are additional 5′ variants as well as SNPs in 3′ haplotype block along with secretor polymorphisms modifies MBL function and its circulating levels and further increases risk of diseases [31, 41–43]. However, further functional studies are needed to confirm these observations and the proposed functional mechanism of these polymorphisms observed by in silico analyses towards disease development. Other than these findings, some other significant associations also observed in the present study that was unexplainable due to unexpected pattern of the variants, suggesting the possibility of other additional variants falling outside the region of observed haplotype blocks.
Our previous investigation suggested the effect of rs3901533 CLEC7A SNP in modulating sMBL levels, suggesting RVVI, a polygenic phenotype. Therefore, the interactions between genes i.e. MBL2 and CLEC7A were assessed using multifactor dimensionality reduction (MDR) analysis. Three gene–gene interaction models involving uni-variant, bi-variant and tri-variant appeared as significant predictors of RVVI risk. Of which, the one-way interaction model including MBL2 Y/X polymorphism with maximum cross-validation consistency (CVC) of 10/10 was found to be the best model for susceptibility to RVVI for pooled RVVI patients and controls. This strongly suggested MBL2 Y/X polymorphism as the fundamental candidate genetic variant that determining RVVI risk. In addition, two-way interaction model (MBL2.rs36014597 and MBL2.Y/X) and three-way interaction model (MBL2.Y/X, MBL2.rs10824792 and CLEC7A.rs3901533) also showed association with RVVI risk, though with comparatively low CV consistency to uni-variant model. This further suggests the contribution of both MBL2 (5′ and 3′ block) and CLEC7A genes in the pathogenesis of the RVVI. As per our finest knowledge, this is the first interaction study that found significant associations of genetic interaction in susceptibility to RVVI. However, in the rule as regards with CVC, the three loci model was not the best model for vulnerability to RVVI. The dendrograms and interaction entropy models also suggested a weak synergistic interaction between the variants of three loci model in predicting RVVI risk. In consonance with these findings, our previous results did not show any significant correlation pattern between sMBL and sDectin-1 levels in cases and controls [12]. This further implies, an independent effect of MBL and Dectin-1 to counter the same antigenic stimuli i.e. RVVI. However, analyses of additional putative functional variants especially in CLEC7A may reveal a synergy between the two molecules. A single study has shown that opsonophagocytosis of pathogens mediated by MBL leads to an increased intracellular expression of Dectin-1 [44]. This study suggested the inhibitory effect on the phagocytosis of C. albicans by neutrophils owing to the Dectin-1 blockage was completely compensated by the exogenous MBL, attributed to its direct role in opsonophagocytosis of pathogens, which is further supported by several receptors e.g. calreticulin and complement receptor 1 present on phagocytes surface that bind to MBL and mediate uptake and phagocytosis of MBL-pathogen complex, independent of complement activation. This further indicates that in the case of infections by C. albicans, the interplay between the MBL and Dectin-1 would have a compensatory characteristic, which in turn change the view of synergism mechanism of these molecules in specific situations.
Further, estimation of serum MBL levels was preferred in the present study than methods like mRNA expression. This is because the gene mRNA expression profile does not exactly reflect its phenotypic level in the serum. This difference may possibly be due to protein translational failure or failure of higher order proteins due to variant monomers formation. The same was depicted by a recent study that found low sMBL levels in RVVC cases with high MBL mRNA expression [10]. In addition, proteins with variant allele are suggested to be functionally inactive and are more likely to degrade relative to the proteins with no variations [45]. The functional implication of these variations on phagocytosis or complement activation by MBL warrants further investigations.
Conclusions
The study presented a low-cost reproducible screening design for additional 5′ variants i.e. rs11003124, rs7084554, rs36014597 and rs11003123 of MBL2 that can act as markers of susceptibility for vulvovaginal infections or any other diseases. Evaluation of these variants revealed two SNPs i.e. rs7084554 and rs36014597 that are significantly predisposing individuals to RVVI and its types by altering sMBL levels. The LD analyses of the SNP map of MBL2 indicated two haplotype blocks inside the gene. Permutation analysis of haplotype with phenotype suggested that variants of two blocks have more of a combined effect than the independent effect in regulating sMBL levels and hence RVVI risk, in which associated variants from both the blocks played the crucial role. In consonance with literature, sMBL levels did not exactly associate with standard secretor haplotypes, signifying the role of additional regulating variants of MBL2, which possibly be altering sMBL levels. Thus, inclusion of additional regulating variants of MBL2 in the present study has helped to solve these inconsistencies. The study presented weak synergistic interaction between MBL2 and CLEC7A in association with RVVI risk. However, analyses of additional putative functional variants especially in CLEC7A may possibly have simplified the nature of interactions among the polymorphisms and genes studied and might have provided a better understanding of the pathway implicated in the pathogenesis of RVVI. All these preliminary findings of the present study, demand further in-depth functional investigations to clarify the connection observed within and between genes in susceptibility to RVVI. Such kind of studies with larger data-sets will provide invaluable data to authenticate the therapeutic possibilities of MBL for RVVI and its types. Once validated, the day is not so far when different MBL formulations will be available over the counter for the RVVI treatment. Because, MBL replacement therapy has already passed the phase I clinical trials to aid patients with MBL paucity [46–49]. While, recombinant MBL production, Phase II and phase III trials are presently in progress. However, in contrast to MBL, more extensive research is needed to dissect the complete and specific role of Dectin-1 in RVVI, prior to judging its therapeutic potential.
Additional files
Additional file 1. Distribution and comparison of 5′ block haplotypes of MBL2 in cases and controls.
Additional file 2. Distribution and comparison of combined 5′/3′ haplotypes of MBL2 in cases and controls.
Additional file 3. MBL2 SNPs frequencies in present study population (North Indian) and modern human populations in human genomes variation project i.e. The 1000 Genomes Project (Phase 3).
Acknowledgements
The authors express their sincere gratitude to the University Grants Commission, New Delhi, Govt. of India for providing Senior Research fellowship to NK.
Authors’ contributions
NK reviewed the literature, was involved in the design, performing experiments, analysis, interpretation, and drafted the manuscript. SS participated in sample collection and RVVI diagnosis. MK, JS, SS contributes in the experimental design, data analysis, manuscript editing, and supervision. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
The present study was commenced after getting approval from the Institutional Ethics Committee (Approval no. 06/HG dated 02/01/2015) of Guru Nanak Dev University, Amritsar (Punjab), India, in accordance with Indian Council of Medical Research guidelines (ICMR, 2006) modified from Declaration of Helsinki (2004). Voluntary informed consent in written was attained from all the participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Namarta Kalia, Email: kalianamarta62@gmail.com.
Jatinder Singh, Phone: 0183-2258802-09, Email: jatinderarora2009@gmail.com.
Sujata Sharma, Email: sujata_sharma09@yahoo.com.
Manpreet Kaur, Phone: 0183-2258802-09, Email: dr.manpreetdhuna@gmail.com.
References
- 1.Mulu W, Yimer M, Zenebe Y, Abera B. Common causes of vaginal infections and antibiotic susceptibility of aerobic bacterial isolates in women of reproductive age attending at Felegehiwot referral Hospital, Ethiopia: a cross sectional study. BMC Women’s Health. 2015;15(1):42. doi: 10.1186/s12905-015-0197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh J, Kalia N, Kaur M. Recurrent vulvovaginal infections: etiology, diagnosis, treatment and management. In: Singh PP, editor. Infectious diseases and your health. Singapore: Springer; 2018. pp. 257–289. [Google Scholar]
- 3.Ma B, Forney LJ, Ravel J. Vaginal microbiome: rethinking health and disease. Annu Rev Microbiol. 2012;66:371–389. doi: 10.1146/annurev-micro-092611-150157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford LL, Ravel J, Bruno V. Understanding vulvovaginal candidiasis through a community genomics approach. Curr Fungal Infect Rep. 2013;7:126–131. doi: 10.1007/s12281-013-0135-0. [DOI] [Google Scholar]
- 5.Dean MM, Minchinton RM, Heatley S, Eisen DP. Mannose binding lectin acute phase activity in patients with severe infection. J Clin Immunol. 2005;25(4):346–352. doi: 10.1007/s10875-005-4702-1. [DOI] [PubMed] [Google Scholar]
- 6.Babula O, Lazdane G, Kroica J, Ledger WJ, Witkin SS. Relation between recurrent vulvovaginal candidiasis, vaginal concentrations of mannose-binding lectin, and a mannose-binding lectin gene polymorphism in Latvian women. Clin Infect Dis. 2003;37:733–737. doi: 10.1086/377234. [DOI] [PubMed] [Google Scholar]
- 7.Liu F, Liao Q, Liu Z. Mannose-binding lectin and vulvovaginal candidiasis. Int J Gynecol Obstet. 2006;92:43–47. doi: 10.1016/j.ijgo.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 8.Milanese M, Segat L, De Seta F, Pirulli D, Fabris A, Morgutti M, Crovella S. MBL2 genetic screening in patients with recurrent vaginal infections. Am J Reprod Immunol. 2008;59:146–151. doi: 10.1111/j.1600-0897.2007.00549.x. [DOI] [PubMed] [Google Scholar]
- 9.Henić E, Thiel S, Mårdh PA. Mannan-binding lectin in women with a history of recurrent vulvovaginal candidiasis. Eur J Obstet Gynecol Reprod Biol. 2010;148:163–165. doi: 10.1016/j.ejogrb.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 10.Ghazanfari M, Falahati M, Fattahi A, Bazrafshan F, Nami S, Hosseinzadeh M, Heydarikohan F, Ghelman M, Khoshmirsafa M, Farzanegan A. Is MBL serum concentration a reliable predictor for recurrent vulvovaginal candidiasis? Mycoses. 2019;62:106–111. doi: 10.1111/myc.12723. [DOI] [PubMed] [Google Scholar]
- 11.Kalia N, Singh J, Sharma S, Arora H, Kaur M. Genetic and phenotypic screening of mannose-binding lectin in relation to risk of recurrent vulvovaginal infections in women of North India: a prospective cohort study. Front Microbiol. 2017;8:75. doi: 10.3389/fmicb.2017.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalia N, Kaur M, Sharma S, Singh J. A comprehensive in silico analysis of regulatory SNPs of human CLEC7A gene and its validation as genotypic and phenotypic disease marker in recurrent vulvovaginal infections. Front Cell Infect Microbiol. 2018;8:65. doi: 10.3389/fcimb.2018.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalia N, Singh J, Sharma S, Kaur M. SNPs in 3′-UTR region of MBL2 increases susceptibility to recurrent vulvovaginal infections by altering sMBL levels. Immunobiology. 2019;224:42–49. doi: 10.1016/j.imbio.2018.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Turner MW. The role of mannose-binding lectin in health and disease. Mol Immunol. 2003;40:423–429. doi: 10.1016/S0161-5890(03)00155-X. [DOI] [PubMed] [Google Scholar]
- 15.Sumiya M, Super M, Tabona P, Levinsky RJ, Arai T, Turner MW, Summerfield JA. Molecular basis of opsonic defect in immunodeficient children. Lancet. 1991;337:1569–1570. doi: 10.1016/0140-6736(91)93263-9. [DOI] [PubMed] [Google Scholar]
- 16.Madsen HO, Garred P, Thiel S, Kurtzhals JA, Lamm LU, Ryder LP, Svejgaard A. Interplay between promoter and structural gene variants control basal serum level of mannan-binding protein. J Immunol. 1995;155:3013–3020. [PubMed] [Google Scholar]
- 17.Larsen F, Madsen HO, Sim RB, Koch C, Garred P. Disease associated mutations in human mannose-binding lectin compromise oligomerization and activity of the final protein. J Biol Chem. 2004;279:21302–21311. doi: 10.1074/jbc.M400520200. [DOI] [PubMed] [Google Scholar]
- 18.Juliger S, Luckner D, Mordmuller B, May J, Weierich A, Lell B, Luty A, Kremsner PG, Kun JF. Promoter variants of the human mannose binding lectin gene show different binding. Biochem Biophys Res Commun. 2000;275:617–622. doi: 10.1006/bbrc.2000.3343. [DOI] [PubMed] [Google Scholar]
- 19.Madsen HO, Satz ML, Hogh B, Svejgaard A, Garred P. Different molecular events result in low protein levels of mannan-binding lectin in populations from southeast Africa and South America. J. Immunol. 1998;161:3169–3175. [PubMed] [Google Scholar]
- 20.Boldt ABW, Petzl-Erler ML. A new strategy for mannose-binding lectin gene haplotyping. Hum Mutat. 2002;19(3):296–306. doi: 10.1002/humu.10051. [DOI] [PubMed] [Google Scholar]
- 21.Verdu P, Barreiro LB, Patin E, Gessain A, Cassar O, Kidd JR, et al. Evolutionary insights into the high worldwide prevalence of MBL2 deficiency alleles. Hum Mol Gen. 2006;15:2650–2658. doi: 10.1093/hmg/ddl193. [DOI] [PubMed] [Google Scholar]
- 22.Oudshoorn AM, van den Dungen FA, Bach KP, Koomen I, Fetter WP, Catsburg A, et al. Mannose-binding lectin in term newborns and their mothers: genotypic and phenotypic relationship. Hum Immunol. 2008;69:344–348. doi: 10.1016/j.humimm.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 23.Jha AN, Sundaravadivel P, Singh VK, Pati SS, Patra PK, Kremsner PG, et al. MBL2 variations and malaria susceptibility in Indian populations. Infect Immun. 2014;82:52–61. doi: 10.1128/IAI.01041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi K, Gordon J, Liu H, Sastry KN, Epstein JE, Motwani M, Laursen I, Thiel S, Jensenius JC, Carroll M, Ezekowitz RAB. Lack of mannose-binding lectin-A enhances survival in a mouse model of acute septic peritonitis. Microbes Infect. 2002;4:773–784. doi: 10.1016/S1286-4579(02)01597-6. [DOI] [PubMed] [Google Scholar]
- 25.Soborg C, Madsen HO, Andersen ÅB, Lillebaek T, Kok-Jensen A, Garred P. Mannose-binding lectin polymorphisms in clinical tuberculosis. J Infect Dis. 2003;188:777–782. doi: 10.1086/377183. [DOI] [PubMed] [Google Scholar]
- 26.Giraldo PC, Babula O, Gonçalves AK, Linhares IM, Amaral RL, Ledger WJ, Witkin SS. Mannose-binding lectin gene polymorphism, vulvovaginal candidiasis, and bacterial vaginosis. Obstet Gynecol. 2007;109:1123–1128. doi: 10.1097/01.AOG.0000260386.17555.a5. [DOI] [PubMed] [Google Scholar]
- 27.De Seta F, Maso G, Piccoli M, Bianchini E, Crovella S, De Santo D, Caterina P, Guaschino S. The role of mannose-binding lectin gene polymorphisms in women with recurrent bacterial vaginosis. Am J Obstet Gynecol. 2007;197:613-e1. doi: 10.1016/j.ajog.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 28.Donders GG, Babula O, Bellen G, Linhares IM, Witkin SS. Mannose-binding lectin gene polymorphism and resistance to therapy in women with recurrent vulvovaginal candidiasis. BJOG Int J Obstet Gynaecol. 2008;115:1225–1231. doi: 10.1111/j.1471-0528.2008.01830.x. [DOI] [PubMed] [Google Scholar]
- 29.Wojitani MD, de Aguiar LM, Baracat EC, Linhares IM. Association between mannose-binding lectin and interleukin-1 receptor antagonist gene polymorphisms and recurrent vulvovaginal candidiasis. Arch Gynecol Obstet. 2012;285:149–153. doi: 10.1007/s00404-011-1920-z. [DOI] [PubMed] [Google Scholar]
- 30.Velazquez-Hernandez N, Aguilar-Duran M, Perez-Alamos AR, Estrada-Martinez S, Salas-Pacheco JM, Sanchez-Anguiano LF, Sandoval-Carrillo AA, Lazalde-Medina B, Alvarado-Esquivel C. Lack of association between mannose-binding lectin 2 codons 54 and 57 gene polymorphisms and cervicovaginal infections in mexican women. Int J Biomed Sci. 2017;13:79. [PMC free article] [PubMed] [Google Scholar]
- 31.Bernig T, Breunis W, Brouwer N, Hutchinson A, Welch R, Roos D, Kuijpers T, Chanock S. An analysis of genetic variation across the MBL2 locus in Dutch Caucasians indicates that 3′ haplotypes could modify circulating levels of mannose-binding lectin. Hum Genet. 2005;118:404–415. doi: 10.1007/s00439-005-0053-5. [DOI] [PubMed] [Google Scholar]
- 32.Kalia N, Sharma A, Kaur M, Kamboj SS, Singh J. A comprehensive in silico analysis of non-synonymous and regulatory SNPs of human MBL2 gene. Springerplus. 2016;5:811. doi: 10.1186/s40064-016-2543-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown GD. Dectin-1: a signalling non-TLR pattern-recognition receptor. Nat Rev Immunol. 2006;6:33–43. doi: 10.1038/nri1745. [DOI] [PubMed] [Google Scholar]
- 34.Sherrard J, Donders G, White D, Jensen JS. European (IUSTI/WHO) guideline on the management of vaginal discharge, 2011. Int J STD AIDS. 2011;22:421–429. doi: 10.1258/ijsa.2011.011012. [DOI] [PubMed] [Google Scholar]
- 35.Kalia N, Singh J, Sharma S, Kamboj SS, Arora H, Kaur M. Prevalence of vulvovaginal infections and species specific distribution of vulvovaginal candidiasis in married women of North India. Int J Curr Microbiol Appl Sci. 2015;4(8):253–266. [Google Scholar]
- 36.Campbell MC, Tishkoff SA. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genomics Hum Genet. 2008;9:403–433. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang DF, Huang XQ, Wang D, Li YY, Yao YG. Genetic variants of complement genes ficolin-2, mannose-binding lectin and complement factor H are associated with leprosy in Han Chinese from Southwest China. Hum Genet. 2013;132(6):629–640. doi: 10.1007/s00439-013-1273-8. [DOI] [PubMed] [Google Scholar]
- 38.Wang PS, Kuai J, Li H, Wang CG, Shi BJ, Zhong L. Mannose-binding lectin 2 rs11003123 polymorphism is associated with the development of hepatocellular carcinoma in patients with hepatitis B-related cirrhosis in the Chinese population. Hepatobiliary Pancreat Dis Int. 2016;15(3):282–288. doi: 10.1016/S1499-3872(16)60050-4. [DOI] [PubMed] [Google Scholar]
- 39.Kim M, Yoon MK, Kim SH, Park HS. Association of MBL with work-related respiratory symptoms in bakery workers. Allergy Asthma Immunol Res. 2017;9(1):85–91. doi: 10.4168/aair.2017.9.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris RW, Kaplan NL. On the advantage of haplotype analysis in the presence of multiple disease susceptibility alleles. Genet Epidemiol. 2002;23(3):221–233. doi: 10.1002/gepi.10200. [DOI] [PubMed] [Google Scholar]
- 41.Bernig T, Boersma BJ, Howe TM, Welch R, Yadavalli S, Staats B, Mechanic LE, Chanock SJ, Ambs S. The mannose-binding lectin (MBL2) haplotype and breast cancer: an association study in African-American and Caucasian women. Carcinogenesis. 2007;28:828–836. doi: 10.1093/carcin/bgl198. [DOI] [PubMed] [Google Scholar]
- 42.Collard CD, Shernan SK, Fox AA, Bernig T, Chanock SJ, Vaughn WK, Takahashi K, Ezekowitz AB, Jarolim P, Body SC. The MBL2 ‘LYQA secretor’ haplotype is an independent predictor of postoperative myocardial infarction in whites undergoing coronary artery bypass graft surgery. Circulation. 2007;116:1–106. doi: 10.1161/CIRCULATIONAHA.106.679530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zanetti KA, Haznadar M, Welsh JA, Robles AI, Ryan BM, McClary AC, Bowman ED, Goodman JE, Bernig T, Chanock SJ, Harris CC. 3′-UTR and functional secretor haplotypes in mannose-binding lectin 2 are associated with increased colon cancer risk in African Americans. Cancer Res. 2012;72:1467–1477. doi: 10.1158/0008-5472.CAN-11-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li D, Dong B, Tong Z, Wang Q, Liu W, Wang Y, et al. MBL-mediated opsonophagocytosis of Candida albicans by human neutrophils is coupled with intracellular Dectin-1-triggered ROS production. PLoS ONE. 2012;7:e50589. doi: 10.1371/journal.pone.0050589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Super M, Gillies SD, Foley S, Sastry K, Schweinle JE, Silverman VJ, Ezekowitz RA. Distinct and overlapping functions of allelic forms of human mannose binding protein. Nat Genet. 1992;2:50–55. doi: 10.1038/ng0992-50. [DOI] [PubMed] [Google Scholar]
- 46.Valdimarsson H, Stefansson M, Vikingsdottir T, Arason GJ, Koch C, Thiel S, Jensenius JC. Reconstitution of opsonizing activity by infusion of mannan-binding lectin (MBL) to MBL-deficient humans. Scand J Immunol. 1998;48:116–123. doi: 10.1046/j.1365-3083.1998.00396.x. [DOI] [PubMed] [Google Scholar]
- 47.Valdimarsson H. Infusion of plasma-derived mannan-binding lectin (MBL) into MBL-deficient humans. Biochem Soc Trans. 2003;31:768–769. doi: 10.1042/bst0310768. [DOI] [PubMed] [Google Scholar]
- 48.Valdimarsson H, Vikingsdottir T, Bang P, Saevarsdottir S, Gudjonsson JE, Oskarsson O, Christiansen M, Blou L, Laursen I, Koch C. Human plasma-derived mannose-binding lectin: a phase I safety and pharmacokinetic study. Scand J Immunol. 2004;59:97–102. doi: 10.1111/j.0300-9475.2004.01357.x. [DOI] [PubMed] [Google Scholar]
- 49.Bang P, Laursen I, Thornberg K, Schierbeck J, Nielsen B, Valdimarsson H, Koch C, Christiansen M. The pharmacokinetic profile of plasma-derived mannan-binding lectin in healthy adult volunteers and patients with Staphylococcus aureus septicaemia. Scand J Infect Dis. 2008;40:44–48. doi: 10.1080/00365540701522959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Distribution and comparison of 5′ block haplotypes of MBL2 in cases and controls.
Additional file 2. Distribution and comparison of combined 5′/3′ haplotypes of MBL2 in cases and controls.
Additional file 3. MBL2 SNPs frequencies in present study population (North Indian) and modern human populations in human genomes variation project i.e. The 1000 Genomes Project (Phase 3).
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.