 Pathogenic infection is one of the major causes of death in newborns.
Pathogenic infection is one of the major causes of death in newborns.
Abstract
Pathogenic infection is one of the major causes of death in newborns. Antibiotic based therapies are still the major mode of treatment for infection. Increased usage of antibiotics leads to selective evolution of microorganisms and causes diseases in adulthood. Attempts to develop alternatives to antibiotics did not yield much success. A recent viable trend is to identify novel probiotics that could alleviate problems associated with over usage of antibiotics. We screened three different Lactobacillus species to establish their efficacy in neonates in protecting against Salmonella challenge. The methodologies employed are metagenomics, metabonomics, transcriptional profiling, molecular assays and behavioral studies. Among the three probiotics used, only Lactobacillus rhamnosus GG (LGG) treatment of the neonates resulted in rescuing of 80% of the Salmonella-infected mice. We have shown that LGG (MTCC #1408) can prevent Salmonella mediated infection in neonates. In the current report, results from histopathology, gene expression, neutrophil infiltration, metabolite and metataxonomic profiling, and protein level data suggested that LGG treatment of the neonates enhanced anti-inflammatory cytokine expression and increased the gut barrier function. The current report establishes the potential use of LGG in clinical intervention of infectious diseases.
Introduction
In 2010, 7.6 million children under 5 years of age died globally, out of which 64%, i.e. about 4.9 million, died of infections such as pneumonia, sepsis, meningitis, typhoid and diarrhea. In India alone, 32% of about 1.7 million children died because of infectious disease1 and 93 million Salmonella infections happen globally every year.2Salmonella enterica serovar typhimurium (ST), a Gram-negative bacterium, is one of the leading causes of gastroenteritis.3 Gastroenteritis, known as infectious diarrhea, involves inflammation of the gut and severe diarrhea. Recent studies revealed that inflammation in the gut could give a nutritional advantage to Salmonella growth by increasing the availability of ethanolamine to provide a competitive advantage to Salmonella over other commensals.4 Reactive oxygen species from an inflamed gut reacts with luminal thiosulphate to produce tetrathionate which acts as a respiratory electron acceptor for Salmonella. This respiratory advantage helps Salmonella to proliferate in the gut.5 Inflammatory response helps in colonization of Salmonella by disrupting the stable gut microbiota.6,7 Studies with IL10 knockout mice exhibited more aggressive pathogenesis to S. typhimurium infection, supporting the fact that hyper-inflammation assists Salmonella in colonization.7 This result establishes anti-inflammatory immune modulation as a potential therapy for the treatment of Salmonella mediated pathogenesis.
Probiotics are commensal microorganisms which exert several physiologically beneficial effects on the host including anti-inflammatory responses. Lactobacillus rhamnosus strain GG (LGG) was initially isolated from newborn babies by Gorbach and Goldin.8 Recently LGG was used as a preventive intervention for the DSS induced inflammatory bowel disease (IBD) model in mice.8 Studies performed by Yan et al. revealed that LGG administration caused anti-inflammatory modulation to suppress the IBD.8 The study by Yan et al. further revealed that LGG treatment increased gut epithelial barrier functioning by decreasing gut permeability.8 LGG administration has been shown to increase IL-10 secretion in humans.9 Results from the studies indicated that LGG may be one the best candidates for the treatment of Salmonella-mediated pathogenesis.
In the current study we have used a neonate mouse model for ST challenge studies to establish the efficacy of LGG. ST usually causes gastroenteritis in humans and cattle but not in adult mice. Mice are resistant to Salmonella mediated gastroenteritis. In mice, only mild inflammation is observed following Salmonella infection.10,11 Streptomycin treated adult mice, however, are an established murine model to study Salmonella mediated gastroenteritis.12 But a streptomycin treated mouse model is not suitable for gut microbiota study as streptomycin treatment disrupts the gut microbiota. It has been recently shown that intestinal microbiota play critical roles in regulating infectivity and transmissivity of Salmonella infection.13
Administration of Salmonella to the neonates has been shown recently to cause full-scale pathogenesis and gastroenteritis.14 It is, therefore, of higher interest to study Salmonella pathogenesis and to develop a plausible treatment protocol in neonate mice. For this purpose, LGG treatment before a challenge with Salmonella in neonates is a potential way of preventing Salmonella infection by means of activating anti-inflammatory pathways. The current study clearly showed that LGG treatment, which began a day prior to Salmonella challenge on day 5, and continued for the rest of the study period could alleviate Salmonella infection by (a) reducing mortality, (b) increasing the molecular expression of genes associated with the host gut barrier and anti-inflammatory responses, (c) increasing the neuro-muscular strength of the mice and (d) altering gut microbiota. Because of practical difficulties, detailed cellular level validation to establish the exact mechanism of LGG action could not be reported in the current study. We have, however, correlated the tissue level immune response with that of the systemic response in the host by analysing metabolite profiling.
Experimental
Bacteria culture
Lactobacillus acidophilus (MTCC #10307), Lactobacillus rhamnosus GG (MTCC # 1408), and Salmonella enterica serovar typhimurium (MTCC #3232) were obtained from the Microbial Type Culture Collection and (MTCC). Lactobacillus delbrueckii subsp. Bulgaricus (strain DWT1) was received as a gift from Daflorn Private Limited, a Bulgarian probiotic company. All Lactobacillus strains were cultured in De Man, Rogosa and Sharpe agar. Salmonella was cultured in nutrient broth. Bismuth sulfite agar was used as a differential medium for Salmonella.
Animal experiments
BALB/c mice were used for the experiments. All animals were acquired from the NISER Animal Facility and all the procedures used for animal studies were approved by the Institute Animal Ethics Committee (IAEC), NISER that is approved by the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA) of the Government of India and co-housed in a specific pathogen free (SPF) facility. Male and female 6–8 week-old mice were kept for breeding in a cage in a 1 : 2 ratio. Neonates (irrespective of any gender bias as gender bias may not be an important factor at this age unlike adult mice) were challenged with the required amount of Salmonella. The dosage of the Salmonella challenge was determined from a dose titration between 103 CFU per mouse and 108 CFU per mouse and the challenge dose was selected as 106 CFU per mouse based on survival data as detailed in the Results section. Salmonella was gavaged once orally into mice on day 5 post birth. Probiotics were administered daily from postnatal day 4 (a day prior to Salmonella challenge) using a 24 gauze feeding tube.
Infection severity index scoring
The scoring method was adapted from Kim et al.15 The scoring was double-blinded to both scorers and to the introducer.
Cytokine analysis
Colon tissues were collected on day 8 post-infection from treated and un-treated mice. The colon was washed with 1× PBS and visible fat tissues were removed. The samples were churned in lysis buffer (Tris-hydrochloric acid, sodium chloride, and Triton X-100 in distilled water) containing a 1× protease inhibitor cocktail (PIC) (Himedia, India). The mixture was centrifuged and the supernatant was collected and used for further analysis. The protein level abundance of genes for TNF, IL-10, and IL-4 was measured by ELISA. An ELISA assay kit was purchased from BD Biosciences, USA and the assay was performed using the manufacturer's protocol. For absorbance reading, Multiskan GO instrument (Thermo Fisher Scientific, USA) was used. For normalization, protein concentration was measured using the Bradford assay (BD Biosciences, USA). The protocol was adopted from an earlier report as described in Kim et al.15
RNA extraction and cDNA preparation
Colonic tissues were stored in RNALater (Sigma Aldrich, USA). The tissues were churned using RLT buffer and further RNA extraction was done using an RNeasy Mini Kit (Qiagen, Germany) as described in the manufacturer's protocol. cDNA was synthesized from the RNA using an AffinityScript Herc. II RT-PCR kit (Agilent Technologies, USA).
Primer designing
Primers for genes were designed using Primer BLAST.16 The species-specific primer was designed against the 16s rRNA gene. The 16s rRNA sequence of the species of interest was taken from NCBI Nucleotide Database.17 Then it was aligned with the 16s rRNA database in BLAST.18 The top 50 hits were taken and aligned in MUSCLE.19 The unique sequence of interest was searched for. The 3′-OH was fixed as base G or C in the sequence of interest and the base was extended towards the 5′ end until the melting temperature (Tm) of 55 °C is reached. The species specificity of the primers was checked using blast.
qRT PCR
The qRT PCR was done using SYBR green chemistry using a GoTaq qPCR Kit (Promega, USA). The final volume of the reaction mixture was 25 μl per well. The relative gene expression values were calculated using the ΔΔCt method. The standard curve to determine Salmonella CFU count in the fecal sample was evaluated by spiking known CFU of ST in the fecal samples from un-infected (control) mice; spiking of ST was increased from 103 CFU per mouse up to 108 CFU per mouse with multiples of 10 CFU for every interval. Salmonella chromosomal DNA was isolated from spiked fecal samples (n = 3) and copy number was determined by PCR using the Salmonella specific primer. qRT-PCR was used to determine the fold change of select genes by comparing the expression of genes for the treated and un-treated samples. The primer sequences used for various genes are enlisted in Table 2.
Table 2. List of forward (_F) and reverse (_R) primers for genes used in this report for qPCR studies.
| Gene | Primer pair |
| ST_F | 5′-AGGCGTGGCTTCCGGACG-3′ |
| ST_R | 5′-GAAGGCACCAATCCATCTCTGGATTC-3′ |
| T-bet_F | 5′-AGGATGTTTGTGGATGTGGT-3′ |
| T-bet_R | 5′-ACATATAAGCGGTTCCCTGG-3′ |
| GATA3_F | 5′-CTCCTTGCTACTCAGGTGAT-3′ |
| GATA3_R | 5′-GAGAGAGGAATCCGAGTGTG-3′ |
| Muc2_F | 5′-GTGATTGTGTTTCAGGCTCC-3′ |
| Muc2_R | 5′-GTACATGGCAAAAGTCCCAC-3′ |
| cldn-3_F | 5′-AAGATCCTCTATTCTGCGCC-3′ |
| cldn-3_R | 5′-TTCATCGACTGCTGGTAGTG-3′ |
| Il-1B_F | 5′-GCCACCTTTTGACAGTGATG-3′ |
| Il-1B_R | 5′-GCCTGAAGCTCTTGTTGATG-3′ |
| Ocln_F | 5′-GTTGAACTGTGGATTGGCAG-3′ |
| Ocln_R | 5′-AAGATAAGCGAACCTTGGCG-3′ |
| Cldn1_F | 5′-TGCCCCAGTGGAAGATTTACT-3′ |
| Cldn1_R | 5′-CTTTGCGAAACGCAGGACAT-3′ |
| Il-10_F | 5′-TGGGAGGGGTTCTTCCTTGG-3′ |
| Il-10_R | 5′-GGGAACCCTCTGAGCTGCTG-3′ |
| Tlr4_F | 5′-CGCTGCCACCAGTTACAGAT-3′ |
| Tlr4_R | 5′-AGGAACTACCTCTATGCAGGGAT-3′ |
| Nos2_F | 5′-GCAACTACTGCTGGTGGTGACAG-3′ |
| Nos2_R | 5′-GGAAGTGAAGCGTTTCGGGATCTG-3′ |
MPO activity assay
The myeloperoxidase (MPO) assay is done to evaluate neutrophil infiltration. MPO is a peroxidase enzyme that is expressed abundantly in neutrophil granulocytes. It is a lysosomal protein stored in the azurophilic granules of the neutrophil. Myeloperoxidase contains a heme pigment which causes its green color in secretions rich in neutrophils, such as pus and some forms of mucus. Myeloperoxidase catalyzes the production of hypochlorous acid (HClO) from hydrogen peroxide (H2O2) and the chloride anion (Cl-, or the equivalent from a non-chlorine halide). The MPO assay was done as described elsewhere.15 50 mM solution of potassium phosphate buffer was prepared by adding solution B (K2HPO4, 8.7 g in 1 L of dH2O) to solution A (KH2PO4, 6.8 g in 1 L of dH2O) until a pH of 6.0 was achieved. 25 mg colonic tissue was churned in 2 ml of HTAB buffer (5 g HTAB in 1 L of potassium phosphate buffer) and the supernatant was collected following centrifugation. o-Dianisidine dihydrochloride (o-dianisidine) solution was prepared by mixing 16.7 mg of o-dianisidine dihydrochloride (Himedia, India), 90 mL of dH2O, and 10 mL of potassium phosphate buffer. 50 μL of diluted H2O2 (4 μL of 30% H2O2 diluted in 96 μL of dH2O) was added to the o-dianisidine solution. 50 μL of the collected supernatant was taken in triplicate for each biological sample and replicated. 200 μL of o-dianisidine solution was added to each well and the absorbance at 450 nm was measured. MPO unit per mg of protein was calculated using following formula.[ΔA(t2 – t1)]/[Δmin × (1.13 × 10–2)]/0.625.
Histology
Tissue samples from the ileum, colon, liver and spleen were collected and stored in 4% paraformaldehyde solution for H&E staining. Tissues were processed for paraffin embedding, and multiple 5-micron sections were prepared. For staining, slides were de-paraffinized by dipping in xylene and hydrated with deionized water followed by hematoxylin (Sigma Aldrich, USA) and eosin staining (Himedia, India). For better visibility of the nucleus, bluing solution (1% MgCl2 and 0.07% sodium bicarbonate) was used. Slides were thoroughly washed in water, dehydrated through sequential alcohol grading, then cleared in xylene and mounted with permanent mounting media (Vector Labs, USA). Stained slides were observed under a Leica DM500 light microscope, and representative images were taken at 4×, 10× and 40× magnifications.
Histopathology scoring
The following method was used for scoring histological samples. All the slides were given ID and were distributed to the lab members (they were blind to the sample types); the distributor of the slides was also blind to the sample types. The median score from the different representatives was taken. The scoring method was adapted as described before.20
Ambulation test
The ambulation test is designed to check the balance while walking. Pups were placed on a white rough surface where they were visible from the side as well as the top. Gently pups were prodded so that they will be induced to walk. The scoring method was adapted from Feather-Schussler et al.21
Grip strength test
The pup of interest was placed on a wire mesh. For approximately 5 s the pup was allowed to adjust to the environment. The mesh was slowly rotated until it attained 180 degrees. The approximate angle at which the pups got detached from the mesh was recorded. If mice remained attached until 180 degrees, then the hanging impulse was calculated by multiplying the time for which a mouse stayed attached (in seconds) with the weight of the mouse (in grams).21
Grasping reflex
The mouse was held by its neck like when it is carried by the dam. Each paw of the mouse was stroked with a bluntly rounded plastic card. The number of paws with which it grasped the card was measured.21
It is known that Salmonella infection leads to disbalance or loss of strength for gripping and grasping. The tests for ambulation, grip strength and grasping were designed to establish whether LGG could prevent such behaviour following Salmonella challenge.
Sample preparation for metabolomics
Functional characterization of serum metabolites is performed to see whether the immune alterations observed in the gut following Salmonella challenge with or without LGG can affect the metabolic profile at the systemic level of the host. For the serum extraction, the blood of the neonates was collected using the cardiac puncture method and serum was extracted by centrifuging blood at 1300g for 10 min. Abundant proteins in the serum were removed by passing it through pre-rinsed (7 times washed) Amicon Ultra-2 ml 3000 MWCO filters (Merck, Millipore, USA) at 4 °C and 12 000g centrifugation. A total of 700 μL of sample containing D2O, pH maintenance buffer and DSS as the reference sample was taken in 5 mm Shigemi tubes.
NMR data acquisition
All experiments were performed at 298 K on a Bruker 9.4T (400 MHz) AVANCE-III Nanobay liquid-state NMR spectrometer equipped with a 5 mm broadband (BBO) probe. A presaturation technique was used with a moderate relaxation delay of 5 seconds to ensure complete water saturation. Offset optimization was performed using real-time ‘gs’ mode for each sample. Topspin 2.1 was used to record and process the acquired spectra.
Metabolomic analysis of NMR data
The NMR data were analyzed in ChenomX (Canada) which derived the concentrations of metabolites in the serum using a Bayesian approach. The raw spectrum was phase and baseline corrected. Then through the profiler different concentrations of metabolites were obtained. The concentration file was then analyzed in Metaboanalyst.22,23 The sample was log transformed for normalization. Fold change analysis was performed for each of the samples in comparison with the untreated control. 2 log fold up- or down-regulated metabolites were considered significant. Then PCA was performed to see the differential clustering of samples and a 2D PCA plot was generated. A heat map was generated using Ward clustering algorithm where Euclidean distance measurement was used. Then pathway analysis was performed to obtain the associated pathways. Fisher's exact test was used to do over-representative analysis and out-degree centrality algorithm was used to do pathway topology analysis. All the analyses performed related to metabolomics were done in Metaboanalyst.
Cecal DNA extraction
A cecal sample was collected from day 8 post-infected neonate mice and stored at –80 °C. DNA from the cecal sample was isolated using a QIAamp DNA Stool Mini Kit (Qiagen, Germany). The same method was followed as that described in the manufacturer's protocol.
16s rRNA sequencing (V3–V4 metagenomics)
V3–V4 regions of 16S rRNA gene from cecal DNA samples were amplified using the primer pair V3F: 5′-CCTACGGGNBGCASCAG-3′ and V4R: 5′-GACTACNVGGGTATCTAATCC-3′. Then amplicons were sequenced using paired end (250bpX2) sequencing in an Illumina Miseq platform and sequencing depths of 393 503 ± 151 816 reads (mean ± SD) were generated. Subsequently, a FASTQ quality check was performed where the base quality, base composition and GC content were checked. For all the samples more than 90% of the sequences were having Phred quality scores of more than 30. For all the samples GC content was nearly 30–60%. Conserved regions from the paired end reads were removed. Using FLASH program, a consensus V3–V4 region sequence was constructed after trimming the unwanted sequences from the original paired-end data. Pre-processed reads from all the samples were pooled and clustered into Operational Taxonomic Units (OTUs) using the de novo clustering method based on their sequence similarity using UCLUST program. QIIME was used for OTU generation and taxonomic mapping.24 A representative sequence was identified for each OTU and aligned against a Greengenes core set of sequences using PyNAST program.25,26 Furthermore, we aligned these representative sequences against reference chimeric data sets. Then taxonomic classification was performed using the RDP classifier against SILVA OTUs database.
Microbiota compositional profiling and analysis
The biome file containing all phylogenetic information was used for further analysis in MicrobiomeAnalyst.27 The features were filtered for minimum counts of 2 and 20% prevalence. The lowest variance i.e. 10% was also filtered. The data were scaled using total sum scaling algorithm. A stacked bar plot with percentage abundance was generated. A heat map for phylum level data was generated using Ward clustering algorithm and Euclidean distance measurement. Chao1 diversity measure was used to calculate alpha diversity of each sample. The beta diversity measurement was performed using an ordination based method, principal coordinates analysis, where the distance method was the Bray-Curtis index and the statistical method used was permutational MANOVA (PERMANOVA).
Microbiota functional profiling and analysis
For microbiota functional profiling, closed reference OTU picking was performed and taxonomy based mapping was done using Greengenes taxa 13.8 database. After obtaining the biome file it was further processed in PICRUST for metagenomic prediction.28 The predicted metagenome was used to obtain the KEGG orthologue functional category. The resulting biome file was analyzed in MicrobiomeAnalyst for further clustering and diversity analysis.
Statistical analysis
All the graphs were plotted using GraphPad Prism 7.0a. The statistical package in Prism was used for statistical analysis of the data to perform the t-test (to compare any 2 data sets) or ANOVA (to compare more than two datasets) as described in the text.
Results
LGG treatment rescues Salmonella-infected neonates and reduces pathogenesis
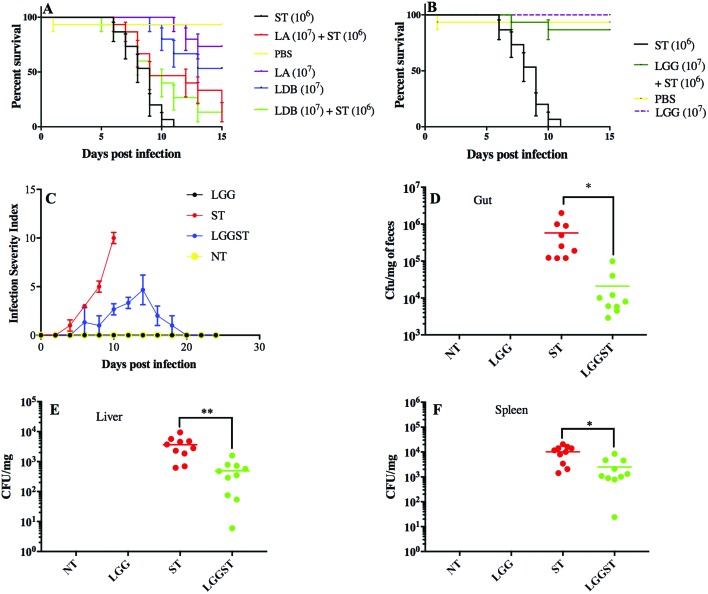
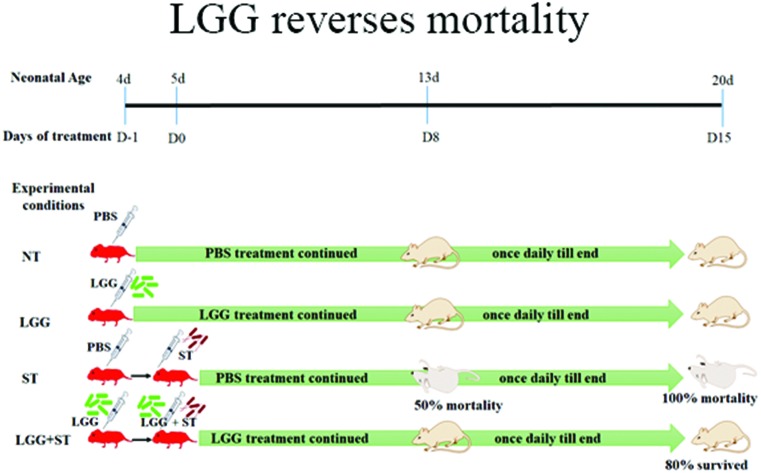
Doses of 1 × 107 CFU per mouse for each of Lactobacillus acidophilus (LA) and Lactobacillus delbrueckii subsp. Bulgaricus DWT1 (LDB) were chosen for treatment as described elsewhere.8,20 Treatment of BALB/c mice with LA and LDB showed nearly 50% lethality in neonates. A daily dose of LA of 1 × 107 CFU mL–1 and LDB of 1 × 107 CFU mL–1 could not rescue Salmonella infected neonates. These results suggested that LA and LDB treatment was not potent enough to alleviate Salmonella infection and, moreover, LA and LDB were lethal to half of the animals. Higher doses of these probiotics were toxic to neonates (Fig. 1A). Treatment with LGG, in contrast, could protect the neonates from Salmonella infection (Fig. 1B). Salmonella infection in neonates was standardized based on the survivability data of different Salmonella doses as described in the Materials and methods section. The percentage of mouse surviving against days post infection at different dosages of Salmonella was evaluated for Salmonella challenge dose titration between 103 CFU per mouse and 108 CFU per mouse. Challenge doses from 103–105 CFU per mouse took 15–20 days to set the infection, while doses above 107 CFU per mouse set the infection in 3 days in neonate mice. Results with a challenge dose of 106 CFU per mouse took around 8–10 days to set Salmonella infection. We selected the Salmonella challenge dose of 106 CFU per mouse to have sufficient time to study the effects of challenge on neonates as well as give enough room to study the effects of probiotic treatment. The current report revealed that around 80% of the neonates have been rescued from Salmonella infection following treatment with LGG (Fig. 1B). The data shown are the average of 15 neonates taken randomly from different littermates to minimize the litter biases. The dose of 1 × 107 CFU mL–1 of LGG was chosen based on previous studies.8 No deaths were observed when only LGG was administered orally. Three out of 15 Salmonella challenged neonate mice died following treatment with LGG while all Salmonella challenged pups died without LGG treatment by day 11 post challenge. Since the studies involved neonates from a very early age (day 3 onwards), methodologies to distinguish the pups based on gender are not very reliable and since the studies were completed before they reached the age of weaning (3 weeks), we did not attempt to separate the pups in the groups into males and females, because the effects would have been minimal based on gender difference for this infection-protection study.
Fig. 1. LGG treatment rescues neonates from Salmonella challenge. A Kaplan–Meier plot of survivability of neonates (A, B). A: Neonates challenged with Salmonella at 106 CFU per mouse showing 50% mortality on day 8 post infection. Lactobacillus acidophilus (LA) at 107 CFU per mouse dosage was toxic to neonates. Lactobacillus delbrueckii subspecies bulgaricus (LDB) at 107 CFU per mouse was also toxic to neonates. All mice (n = 15) that were treated with either LA or LDB on day 4 and continued until the end died by day 15 following Salmonella (107 CFU per mouse) infection on day 5. B: Lactobacillus rhamnosus GG (LGG) at a dosage of 107 CFU per mouse could protect 80% of mice beyond day 15. C: Infection severity index calculated for neonates in each interval post infection and treatment. D: ST count using the culture method in caecal matter of neonates on day 8 pi (P value <0.05). E: ST count using the culture method in the liver of neonates on day 8 post infection (pi) (P value <0.01). F: ST count using the culture method in the spleen on day 8 pi (P value <0.05). Unpaired t-test was performed to calculate the significant values for (D, E, and F). Each group consisted of n = 15 neonatal mice.
Scoring for infection severity (see the Materials and methods section) was done for LGG treated and un-treated neonates in the ileum, colon, liver and spleen. Salmonella-infected mice showed signs of diarrhea on 4–6 days post infection. But mice treated with LGG delayed the onset of diarrhea on days 13–16 post-infection. Salmonella-infected mice showed rectal bleeding, rectal prolapse and also blood in feces. The LGG treated Salmonella challenged mice did not show any of those symptoms (Fig. 1C).
It has been reported previously that Salmonella infiltrates the gut epithelium that leads to infection in multiple organs.14 Finally, multiple organ failures led to death of the neonates. It is, therefore, important to regulate the systemic inflammation for controlling the pathogenesis. Treatment with LGG showed a significant reduction in the Salmonella CFU count in the total gut tissue, liver and spleen (P < 0.01) (Fig. 1D–F). This observation suggested a systemic effect of LGG treatment. In the cecal content, Salmonella count was also significantly low in the LGG treated sample (P < 0.05) (Fig. 1D). In neonate mice, the average Salmonella count found in the gut following Salmonella challenge was similar to the challenge dose i.e. 106 CFU per mouse. Following treatment with LGG in Salmonella challenged mice, the count reduced to 104 CFU per mouse.
LGG treatment reduces inflammation in Salmonella-infected neonates
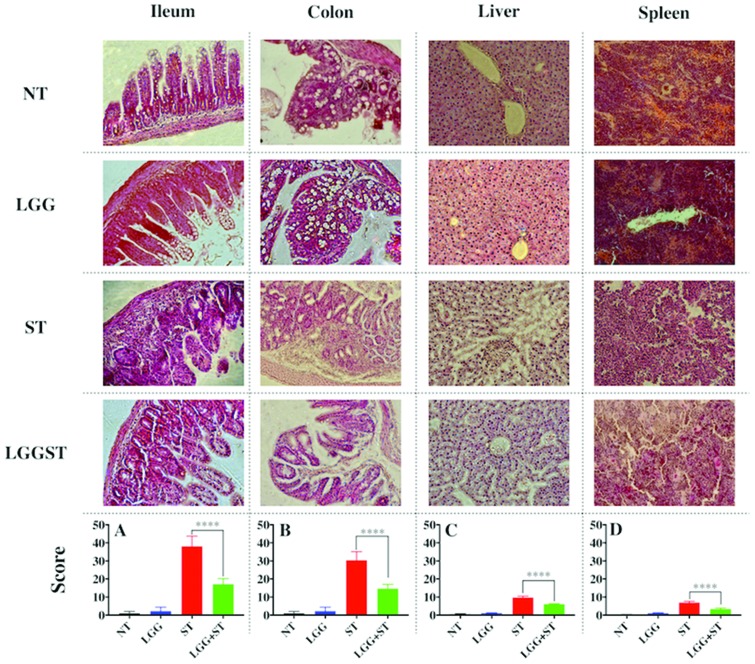
The ileum and colon of the Salmonella-infected mice showed severe immune cell infiltration (Fig. 2). Fig. 2 presents histology images and scores for the ileum, colon, liver and spleen of untreated (NT), LGG treated, ST challenged and LGG treated but ST challenged mice. Results indicated severe villi atrophy (Fig. 2). The Salmonella challenged pups had a smaller length of villi left compared to the uninfected pups. The LGG treated sections showed less, or in some cases, no immune cell infiltration and also the ileum and colon were less damaged. The goblet cells were fully destroyed in Salmonella-infected mice (Fig. 2). The Salmonella-infected mice also displayed severe liver damage compared to LGG treated mice (Fig. 2). In the spleen of Salmonella-infected mice, lymphoid sheath immune cell clusters were observed whereas LGG treatment revealed very little sheath development (Fig. 2). Damage to the liver and spleen, as determined from the histology scores, confirmed the presence of Salmonella and exhibited signs of systemic inflammation. In all cases, histopathology scores from the LGG treated group were significantly less than the histopathology scores obtained from the un-treated group (P < 0.0001) (Fig. 2A–D).
Fig. 2. LGG treatment decreases Salmonella mediated pathology of the gut, liver and spleen. The figure presents the eosin and hematoxylin staining of the corresponding samples. One of the representative images of each sample i.e. time matched (day 8) untreated (NT), day 8 post infected Salmonella (ST), day 8 post-treatment of LGG (LGG), and LGG to Salmonella infected (LGG + ST) pups. Histopathology scores of all collective images of the ileum (A), colon (B), liver (C), and spleen (D) (P value <0.001). Unpaired t-test was performed to calculate the significant value in (A)–(D). Histology scores are the average of n = 3 mice per condition.
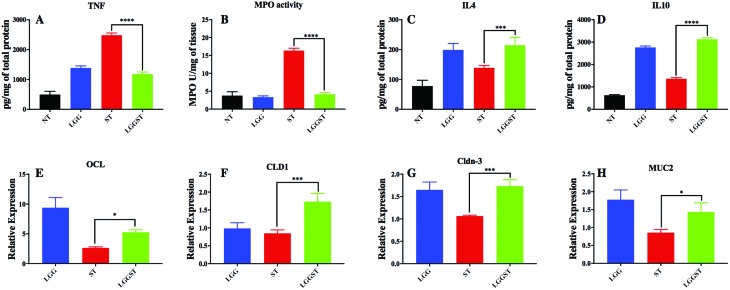
Current results revealed that TNFα, a proinflammatory marker, was significantly high following Salmonella challenge, while it came back to normal levels in mice that were treated with LGG followed by Salmonella challenge (Fig. 3A). In the current study, we also observed that both MPO activity (Fig. 3B) and IL-8 gene expression (Table 1) are higher in Salmonella-infected mice. It was reported that iNOS expression increases in the presence of Salmonella infection because of macrophage infiltration.29 In the current study, we established that the expression of iNOS gene was significantly low in (a) LGG treated control mice and (b) Salmonella challenged but LGG treated neonates (Table 1). Current results further revealed a higher expression level of IL-1B gene, an inflammatory cytokine, following challenge with Salmonella. The expression of IL-1B is significantly reduced following LGG treatment in Salmonella challenged mice (Table 1). The anti-inflammatory cytokine IL-4 was also seen to be highly expressed in LGG treated pups (Fig. 3C). We also showed that IL10 is significantly overexpressed at mRNA and protein levels in LGG treated mice than Salmonella-infected mice (Table 1, Fig. 3D). It is known that the SP-1 gene cluster of Salmonella activates IL-1B through inflammasome activation.30 High GATA3 expression again indicates a Th2 modulation by LGG treatment in the gut. Salmonella mediated host response required both TLR4 and its adapter, MyD88. Mice lacking functional TLR4 showed more susceptibility to Salmonella infection.31,32 Results from the current report suggested increased expression of TLR4 following challenge with ST in neonates. The reduction in TLR4 expression in ST challenged but LGG treated mice further correlated with the fact that the Salmonella count has been reduced (Table 1). In the current study, we found that the expressions of the gut epithelial barrier junction proteins such as Occludin-1, Claudin-1, and Claudin-3 were significantly increased in the LGG treated group compared to the un-treated control group (Fig. 3D, E and F). The Mucin-2 gene expression was also significantly increased in LGG treated mice (Fig. 3G). The results revealed an increase in barrier function or a decrease in gut permeability.
Fig. 3. LGG treatment decreases inflammation and increases gut barrier function and gene expression. A: Cytokine level for TNF in gut tissue (P value <0.001). B: Myeloperoxidase activity in day 8 pi gut tissue (P value <0.001). C: Cytokine level for IL4 in day 8 pi gut tissue (P value <0.001). D: Cytokine level for IL10 in day 8 pi gut tissue (P value <0.001). E: Occludin gene expression level with respect to NT (P value <0.05). F: Claudin1 gene expression level with respect to NT in day 8 pi gut tissue (P value <0.001). G: Claudin3 gene expression level with respect to NT in day 8 pi gut tissue (P value <0.001). H: Mucin2 gene expression level with respect to NT in day 8 pi gut tissue (P value <0.05). Data presented are the average of n = 3 mice per condition.
Table 1. Fold change values of significantly and differentially expressed genes as reported for treatment with LGG, challenge with ST and treatment with LGG following challenge with ST. Fold change values are computed with respect to time matched untreated (control) neonatal mice.
| Fold change values (±SD) |
p-Values |
|||||
| LGG | ST | LGGST | LGGST vs. ST | LGGST vs. LGG | ST-LGG | |
| NOS2 | 4.7 ± 2.72 | 226.46 ± 70.7 | 13.5 ± 1.67 | 0.0004 | ns | 0.0003 |
| IL1B | 18.24 ± 3.09 | 100.46 ± 8.06 | 3.72 ± 2.73 | <0.0001 | 0.0184 | <0.0001 |
| T-bet | 0.74 ± 0.33 | 1.73 ± 0.37 | 0.79 ± 0.27 | 0.0144 | ns | 0.0102 |
| TLR4 | 0.04 ± 0.01 | 16.48 ± 6.27 | 0.03 ± 0.03 | 0.0009 | ns | 0.0009 |
| IL10 | 5.61 ± 0.16 | 3.55 ± 0.17 | 7.22 ± 0.64 | <0.0001 | 0.0019 | 0.0004 |
| GATA3 | 1.71 ± 0.22 | 0.55 ± 0.06 | 1.04 ± 0.18 | 0.0159 | 0.0023 | <0.0001 |
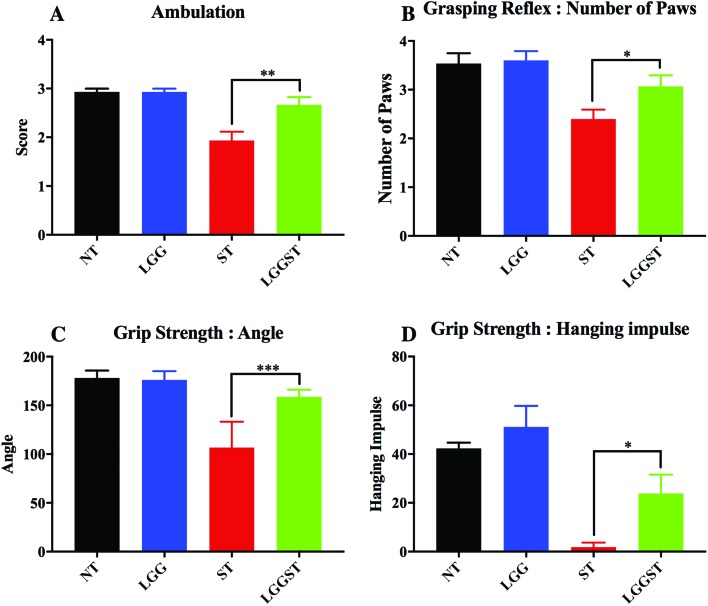
It has been described earlier that ambulation is related to stress and anxiety.33 The current report revealed that the Salmonella-infected mice were having low ambulation scores. This may be because of the defective transition in the limb movement. The effects are found to be reversed following treatment with LGG (Fig. 4A). In the untreated control and LGG treated pups, the grasping reflex test showed that, in most of the cases, the Salmonella-infected mice were defective in both of the hind paw reflexes. Conversely, LGG treatment rescued the mice from the hind paw grasping defect (Fig. 4B). Data in Fig. 4C and D revealed that Salmonella-infected mice could not show strong grip strength. LGG treatment, however, could rescue the neonates from defective grip strength. Salmonella-infected pups were detaching from the mesh at nearly a 100° angle. Untreated control and LGG treated pups were falling at a 180° angle. LGG treated Salmonella infected pups showed a reduced angle of inclination at which pups were falling. Hanging impulse was almost zero in the case of Salmonella-infected pups. LGG treatment significantly increased the hanging impulse in these pups.
Fig. 4. LGG treatment rescues the neonates from Salmonella mediated strength related defects. A: Ambulation test for day 5 pi neonates (P value <0.01). B: Grasping reflex test for day 8 pi neonates (P value <0.05). C: Grip strength test, measurement of detachment angle (P value <0.001). D: Grip strength test, measurement of hanging impulse (P value <0.05). Each experiment was done with n = 5 neonatal mice.
LGG treatment and Salmonella infection alters the microbiota of the gut
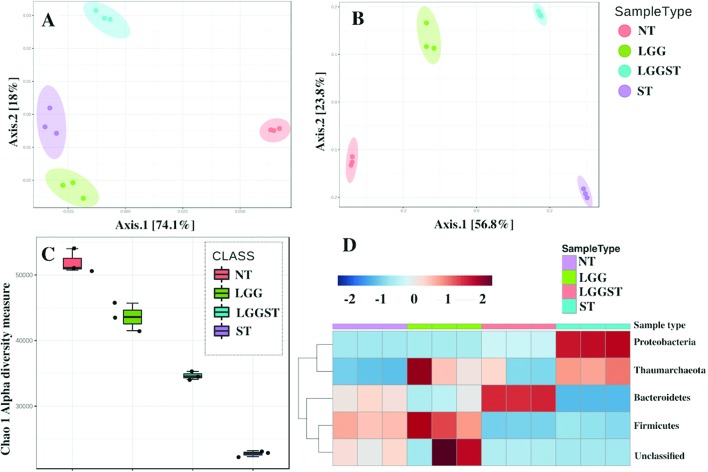
Treatment with LGG following challenge with Salmonella changed both the structure and function of gut microbiota as shown in Fig. 5A and B. Beta diversity graphs (Fig. 5A and B) present different clustering of microbiota in both functional and structural levels. Salmonella-infected gut revealed low alpha diversity of gut microbiota (Fig. 5C). LGG treatment, however, showed an increase in the alpha diversity (Fig. 5C). KEGG orthologue based functional classification also differentially clustered different groups suggesting that microbiota are different both structurally and functionally.
Fig. 5. LGG treatment alters the host microbiota. A: Functional beta diversity plot from PICRUST predicted metagenome for all day 8 pi caecal samples. B: Structural beta diversity plot for day 8 pi caecal samples from 16s rRNA marker based analysis. C: Chao1 alpha diversity measurement for day 8 pi caecal samples. D: Heat map presenting the change in phylum level in the day 8 pi caecal sample. Metagenomic analysis data are presented as the average of n = 5 neonatal mice per condition.
Fig. 5D presents phylum level profiling of microbiota for various treatment or challenge conditions. Proteobacteria mainly consist of Gram-negative bacteria. This phylum includes a wide range of pathogens including Salmonella. The results explain the pathogenicity of Salmonella in infected mice. Thaumarchaeota mainly involves extreme thermophiles, and thus does not have a known function in explaining disease severity. The phylum, Firmicutes, mainly consists of Gram-positive bacteria. Firmicutes have been established to inhibit inflammation, which possibly explains the anti-inflammatory effects of LGG.34 A meta-analysis report revealed that a lower Bacteroidetes level is associated with higher incidence of inflammatory bowel disease.35 This gives an idea of the inflammation regulatory role of Bacteroidetes. In LGG treated Salmonella-infected mice, Bacteroidetes have increased by nearly 2 log. This may explain how LGG could regulate the inflammation in Salmonella-infected neonates.
LGG treatment alters the host metabolome
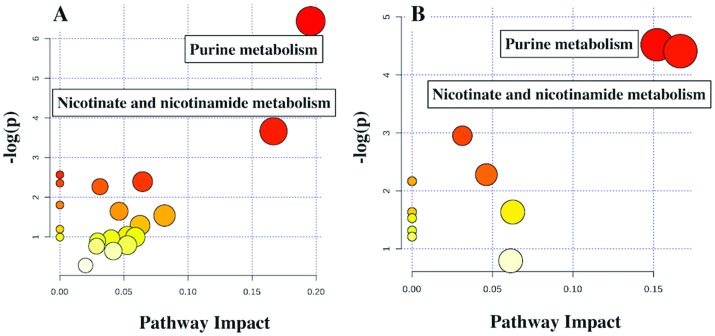
PCA, classification and heat map analysis (Fig. 5) revealed that Salmonella infected and LGG treated mice clustered differently and altered metabolite profiling. Further analyses suggested that the downregulated metabolites following challenge compared to the untreated control could reveal some meaningful biological pathways while no pathways could be identified from the list of upregulated metabolites in either LGG treated or LGG treated but ST challenged mice. In both LGG treated and Salmonella infected LGG treated mice, purine metabolism and nicotinate and nicotinamide metabolism were downregulated (Fig. 6A and B).
Fig. 6. LGG treatment downregulates purine, nicotinate and nicotinamide metabolism pathways. A: Pathway analysis for downregulated metabolites in LGG treated day 8 pi in blood serum of neonates. B: Pathway analysis for downregulated metabolites in Salmonella challenged LGG treated day 8 pi in blood serum of neonates. Because of low volume, serum from 6 neonatal mice per treatment condition was pooled and the results presented are the average of 3 such pools (each of n = 6) per condition.
Discussion
The current study is novel because perhaps this is the first report revealing the effect of LGG in protecting neonate mice against Salmonella challenge. The only other study in a related area that is available revealed that colonization of LGG in neonatal mice could reduce susceptibility to colitis in adulthood.8 The current report established for the first time the efficacy and mechanism by which LGG can alleviate Salmonella infection in neonate mice. Neutrophil infiltration marks the inflammation of the intestine. Neutrophil phagosome complexes kill pathogens by expressing Myeloperoxidase (MPO), which helps in generating oxygen free radicals. Salmonella infection causes massive neutrophil infiltration leading to diarrhea.36 MPO activity was also significantly decreased as compared to Salmonella challenge (Fig. 3B). Studies with IL10 knockout mice showed higher pathogenesis and larger mortality than normal mice, suggesting that IL-10 played an important role in controlling Salmonella infection.7 Reports from gene expression and cytokine level studies showed an increase in IL-10 expression following treatment with LGG (Table 1, Fig. 3D) to promote anti-inflammation.
Salmonella infection destroyed the gut epithelium structure and severely damaged the gut barrier function. ST challenge caused an increase in gut permeability, which led to increased inflammation in the gut.14 It was reported earlier that treatment with LGG could (a) increase the expression of tight junction proteins, and (b) decrease gut permeability to compromise barrier function.37 In this report, we established that transcriptional expression levels of the tight junction proteins are present in higher amount in LGG treated pups that were either challenged or not challenged with ST (Fig. 3E–G).
Salmonella mediated infection leads to a stage of multiple organ failure and sepsis which causes fatigue and muscle weakness. This accounts for strength related defects. The behavioural tests such as the grip strength test and the ambulation test showed clearly that Salmonella-infected pups are weaker in strength as established in the current report. LGG treatment recovered mice from infection and subsequently from strength related defects (Fig. 4A, C, and D). Defects in lower limb grasping, (Fig. 4B) in Salmonella-infected pups are probably due to lower abdominal pain caused by severe gut inflammation following ST challenge. LGG treatment could significantly alleviate the defects.
Loss of gut microbial diversity was associated with inflammation and infectious diseases.38 Probably the inflammatory burst causes a decrease in microbial diversity in the gut in Salmonella-infected mice. LGG treatment was shown to restore the microbiota diversity in Salmonella-infected pups, which may be due to the regulated inflammation in the gut (Fig. 5C). The decrease in microbial diversity in mice treated with LGG alone can be explained by Th2 modulation through LGG, which increased mucus, host defence peptide and IgA secretion.
Nicotinate metabolism and nicotinamide metabolism were related to inflammation and, in fact, drugs like Saikosaponins had been reported to inhibit inflammation by regulating this pathway.39 This study hinted at a possible mechanism by which LGG treatment can regulate the inflammation (Fig. 6A and B). It was also reported that upregulation of purine metabolism led to the production of a large amount of uric acid that can disrupt the gut epithelial barrier and might increase the gut permeability.40 The current study also revealed an increase in the gut epithelial barrier function upon decreasing gut permeability in LGG treated mice (Fig. 6A and B). Downregulation of purine metabolism can, therefore, be a possible mechanism by which LGG treatment decreases gut permeability. Further studies are needed to establish the detailed mechanism; for example, IL-10 knockout mice can be used to check if the anti-inflammation is through IL-10 secretion. The current study revealed that LGG treatment rescued the pups from Salmonella mediated pathogenesis. Salmonella infected neonate gut tissue showed higher expression of inflammatory markers in contrast with the Salmonella infected pups following treatment with LGG, which showed a reduction in inflammatory markers and an increase in the expression of anti-inflammatory genes. Results from gut histology and MPO revealed that the Salmonella-infected gut was severely damaged. Similarly, the junction protein expression was also low in Salmonella-infected mice but increased expression was seen following treatment with LGG. Microbiota diversity in the Salmonella infected gut was low and can be restored following treatment with LGG.
Conclusions
The current results from our host metabolomic study showed the potential pathways by which LGG treatment may be playing a crucial role in reducing inflammation and increasing gut epithelial barrier function. The current study showed potential functional pathways by which LGG treatment could rescue the neonate mice following Salmonella challenge. The current report is novel as it has established the role of an important probiotic in protecting neonates against Salmonella infection. Salmonella infection in neonates is a global problem that requires a solution. The current study in neonates has the potential to lead to further clinical studies to explore the possibility.
Conflicts of interest
There are no conflicts to declare.
Acknowledgments
We are thankful to Mr Madan Mohan Mallick, a technician at the Institute of Life Sciences, Bhubaneswar, Odisha, India for assistance in preparing the initial histological slides. This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
References
- Liu L., Johnson H. L., Cousens S., Perin J., Scott S., Lawn J. E., Rudan I., Campbell H., Cibulskis R., Li M., Mathers C., Black R. E. Lancet. 2012;379:2151–2161. doi: 10.1016/S0140-6736(12)60560-1. [DOI] [PubMed] [Google Scholar]
- Gunn J. S., Marshall J. M., Baker S., Dongol S., Charles R. C., Ryan E. T. Trends Microbiol. 2014;22:648–655. doi: 10.1016/j.tim.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.-M., Wang Y., Su L.-H., Chiu C.-H. Pediatr. Neonatol. 2013;54:147–152. doi: 10.1016/j.pedneo.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Thiennimitr P., Winter S. E., Winter M. G., Xavier M. N., Tolstikov V., Huseby D. L., Sterzenbach T., Tsolis R. M., Roth J. R., Baumler A. J. Proc. Natl. Acad. Sci. U. S. A. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter S. E., Thiennimitr P., Winter M. G., Butler B. P., Huseby D. L., Crawford R. W., Russell J. M., Bevins C. L., Adams L. G., Tsolis R. M., Roth J. R., Bäumler A. J. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C., Robertson M. L., Wickham M. E., Sekirov I., Champion O. L., Gaynor E. C., Finlay B. B. Cell Host Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Stecher B., Robbiani R., Walker A. W., Westendorf A. M., Barthel M., Kremer M., Chaffron S., Macpherson A. J., Buer J., Parkhill J., Dougan G., von Mering C., Hardt W.-D. PLoS Biol. 2007;5:e244. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F., Liu L., Cao H., Moore D. J., Washington M. K., Wang B., Peek R. M., Acra S. A., Polk D. B. Mucosal Immunol. 2016;10:117–127. doi: 10.1038/mi.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessi T., Sutas Y., Hurme M., Isolauri E. Clin. Exp. Allergy. 2000;30:1804–1808. doi: 10.1046/j.1365-2222.2000.00948.x. [DOI] [PubMed] [Google Scholar]
- Wallis T. S., Galyov E. E. Mol. Microbiol. 2000;36:997–1005. doi: 10.1046/j.1365-2958.2000.01892.x. [DOI] [PubMed] [Google Scholar]
- Santos R. L., Zhang S., Tsolis R. M., Kingsley R. A., Adams L. G., Baumler A. J. Microbes Infect. 2001;3:1335–1344. doi: 10.1016/s1286-4579(01)01495-2. [DOI] [PubMed] [Google Scholar]
- Barthel M., Hapfelmeier S., Quintanilla-Martinez L., Kremer M., Rohde M., Hogardt M., Pfeffer K., Russmann H., Hardt W. D. Infect. Immun. 2003;71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley T. D., Bouley D. M., Hoy Y. E., Gerke C., Relman D. A., Monack D. M. Infect. Immun. 2008;76:403–416. doi: 10.1128/IAI.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Dupont A., Torow N., Gohde F., Leschner S., Lienenklaus S., Weiss S., Brinkmann M. M., Kuhnel M., Hensel M., Fulde M., Hornef M. W. PLoS Pathog. 2014;10:e1004385. doi: 10.1371/journal.ppat.1004385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. J., Shajib M. S., Manocha M. M., Khan W. I. J. Visualized Exp. 2012;(60):pii: 3678. doi: 10.3791/3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T. L. BMC Bioinf. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K. D., Tatusova T., Maglott D. R. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T. L. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan B., Guha D., Naik A. K., Banerjee A., Tambat S., Chawla S., Senapati S., Aich P. Probiotics Antimicrob. Proteins. 2018 doi: 10.1007/s12602-018-9436-5. [DOI] [PubMed] [Google Scholar]
- Feather-Schussler D. N., Ferguson T. S. J. Visualized Exp. 2016;(117) doi: 10.3791/53569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Wishart D. S. Nat. Protoc. 2011;6:743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- Xia J., Sinelnikov I. V., Han B., Wishart D. S. Nucleic Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Pena A. G., Goodrich J. K., Gordon J. I., Huttley G. A., Kelley S. T., Knights D., Koenig J. E., Ley R. E., Lozupone C. A., McDonald D., Muegge B. D., Pirrung M., Reeder J., Sevinsky J. R., Turnbaugh P. J., Walters W. A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., Huber T., Dalevi D., Hu P., Andersen G. L. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis Jr. T. Z., Hugenholtz P., Keller K., Brodie E. L., Larsen N., Piceno Y. M., Phan R., Andersen G. L. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhariwal A., Chong J., Habib S., King I. L., Agellon L. B., Xia J. Nucleic Acids Res. 2017;45:W180–W188. doi: 10.1093/nar/gkx295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille M. G., Zaneveld J., Caporaso J. G., McDonald D., Knights D., Reyes J. A., Clemente J. C., Burkepile D. E., Vega Thurber R. L., Knight R., Beiko R. G., Huttenhower C. Nat. Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henard C. A., Vazquez-Torres A. Front. Microbiol. 2011;2:84. doi: 10.3389/fmicb.2011.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh D., Monack D. M., Smith M. R., Ghori N., Falkow S., Zychlinsky A. Proc. Natl. Acad. Sci. U. S. A. 1999;96:2396–2401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapfelmeier S., Stecher B., Barthel M., Kremer M., Muller A. J., Heikenwalder M., Stallmach T., Hensel M., Pfeffer K., Akira S., Hardt W. D. J. Immunol. 2005;174:1675–1685. doi: 10.4049/jimmunol.174.3.1675. [DOI] [PubMed] [Google Scholar]
- Vazquez-Torres A., Vallance B. A., Bergman M. A., Finlay B. B., Cookson B. T., Jones-Carson J., Fang F. C. J. Immunol. 2004;172:6202–6208. doi: 10.4049/jimmunol.172.10.6202. [DOI] [PubMed] [Google Scholar]
- Kastenberger I., Lutsch C., Herzog H., Schwarzer C. PLoS One. 2012;7:e34251. doi: 10.1371/journal.pone.0034251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D. N., St Amand A. L., Feldman R. A., Boedeker E. C., Harpaz N., Pace N. R. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhi F. BioMed Res. Int. 2016;2016:5828959. doi: 10.1155/2016/5828959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolis R. M., Adams L. G., Ficht T. A., Baumler A. J. Infect. Immun. 1999;67:4879–4885. doi: 10.1128/iai.67.9.4879-4885.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R. M., Myers L. S., Kurundkar A. R., Maheshwari A., Nusrat A., Lin P. W. Am. J. Pathol. 2012;180:626–635. doi: 10.1016/j.ajpath.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J. Y., Antonopoulos D. A., Kalra A., Tonelli A., Khalife W. T., Schmidt T. M., Young V. B. J. Infect. Dis. 2008;197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- Ma Y., Bao Y., Wang S., Li T., Chang X., Yang G., Meng X. Inflammation. 2016;39:1453–1461. doi: 10.1007/s10753-016-0377-4. [DOI] [PubMed] [Google Scholar]
- Chiaro T. R., Soto R., Zac Stephens W., Kubinak J. L., Petersen C., Gogokhia L., Bell R., Delgado J. C., Cox J., Voth W., Brown J., Stillman D. J., O'Connell R. M., Tebo A. E., Round J. L. Sci. Transl. Med. 2017;9(380):pii: eaaf9044. doi: 10.1126/scitranslmed.aaf9044. [DOI] [PMC free article] [PubMed] [Google Scholar]