 Capsaicin, a natural active ingredient of green and red peppers, has been demonstrated to exhibit anti-cancer properties in several malignant cell lines.
Capsaicin, a natural active ingredient of green and red peppers, has been demonstrated to exhibit anti-cancer properties in several malignant cell lines.
Abstract
Capsaicin, a natural active ingredient of green and red peppers, has been demonstrated to exhibit anti-cancer properties in several malignant cell lines. Excision repair cross-complementary 1 (ERCC1) has a leading role in the nucleotide excision repair (NER) process because of its involvement in the excision of DNA adducts. Erlotinib (TarcevaR) is a selective epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor that has demonstrated clinical activity in non-small cell lung cancer (NSCLC) cells. However, whether capsaicin and erlotinib could induce synergistic cytotoxicity in NSCLC cells through modulating ERCC1 expression is unknown. In this study, capsaicin decreased the ERCC1 expression in an AKT inactivation dependent manner in two human lung adenocarcinoma cells, namely, A549 and H1975. Enhancement of AKT activity by transfection with constitutive active AKT vectors increased the ERCC1 protein level as well as the cell survival by capsaicin. Moreover, capsaicin synergistically enhanced the cytotoxicity and cell growth inhibition of erlotinib in NSCLC cells, which were associated with the down-regulation of ERCC1 expression and inactivation of AKT in A549 and H1975 cells. Together, these results may provide a rationale to combine capsaicin with erlotinib for lung cancer treatment.
Introduction
Lung cancer is the leading cause of cancer deaths, and specifically, non-small cell lung cancer (NSCLC) accounts for most of the lung cancer-related deaths.1–4 Previous studies have indicated that the epidermal growth factor receptor (EGFR) is often overexpressed5 in NSCLC, and EGFR signaling activation can enhance cell proliferation, anti-apoptosis, angiogenesis, and metastasis, and then lead to poor disease prognosis.6,7 Erlotinib, an EGFR tyrosine kinase inhibitor (TKI), functions by reversibly inhibiting the EGFR through competitively binding at the ATP site in the tyrosine kinase domain, which results in downregulating the downstream proliferative signaling pathways.8,9 Erlotinib has been approved to prolong the survival of patients with advanced NSCLC after chemotherapy.10 The good tumor responses to erlotinib occur more frequently in patients who have never smoked and were women, are higher in adenocarcinoma than other cancer types.11
Capsaicin (trans-8-methyl-N-vanillyl-6-nonenamide) is the major pungent principle component found in chili peppers of the plant genus Capsicum.12 Capsaicin has an in vitro anti-proliferative effect on breast cancer,13 prostate cancer,14 colon adenocarcinoma,15 gastric cancer,16 hepatocellular carcinoma,17 small cell lung cancer,18 leukemic cancer cells,19 head and neck cancer,20 and many others. In addition, capsaicin inhibits AKT, providing a possible pathway whereby capsaicin sensitizes to sorafenib (a multi-kinase inhibitor) in hepatocellular carcinoma cells.21 Capsaicin enhances apoptosis and restricts benzo(a)pyrene induced lung tumorigenesis in Swiss albino mice.22 In this study, we investigated the cytotoxic effects of a combined treatment using erlotinib and capsaicin on NSCLC cells.
Previous studies indicated that capsaicin caused sister chromatid exchanges and micronuclei production in human lymphocytes23 and stimulated DNA strand breaks in human neuroblastoma and breast cancer cells.24,25 The capacity of DNA repair is linked to a genetic predisposition for lung cancer and to treatment outcomes in platinum-based chemotherapy.26 Excision repair cross-complementary 1 (ERCC1) is one of the key enzymes in the nucleotide excision repair (NER) pathway27 because of its involvement in the excision of DNA adducts.28 In human cells, the damaged DNA strand is cleaved by ERCC1-XPF (xeroderma pigmentosum-F) on the 5′ side during the NER process.29 The RNA levels of ERCC1 are highly correlated with NER activity in blood lymphocytes.30 A previous study showed that high ERCC1 levels are associated with clinical resistance to platinum-based chemotherapy in human NSCLC.31 Until now, the involvement of ERCC1 in the capsaicin induced cytotoxic effect by NSCLC cell lines is unclear.
Capsaicin has been used as a medication and it has other pharmacological properties including anti-cancer and anti-inflammatory effects.32–34 In this paper, the human NSCLC cell lines were selected to investigate the anti-cancer effects of capsaicin and further clarify its mechanisms. In this study, we wanted to explore the molecular mechanism of capsaicin in regulating the ERCC1 expression to enhance the cytotoxic effect of erlotinib on human lung cancer cells. Using A549 and H1975 human lung adenocarcinoma cancer cell lines, we found that the decrease in the ERCC1 expression by capsaicin was correlated with the enhancement in the sensitivity to erlotinib. These results may provide a rationale to combine capsaicin with erlotinib for lung cancer treatment.
Materials and methods
Cell lines and chemicals
Human lung carcinoma cells A549 and H1975 were obtained from the American Type Culture Collection (Manassas, VA) and the cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2 in a RPMI-1640 complete medium supplemented with sodium bicarbonate (2.2%, w/v), l-glutamine (0.03%, w/v), penicillin (100 units per mL), streptomycin (100 μg mL–1), and fetal calf serum (10%). The cell lines were routinely tested to confirm that they were free of Mycoplasma. Capsaicin (purity 90%) was purchased from Fluorochem (Pamplona, España). Erlotinib was purchased from Genentech (South San Francisco, CA, USA). Cycloheximide and actinomycin D were purchased from Sigma-Aldrich (St Louis, MO, USA). MG132, lactacystin, and LY294002 were purchased from Calbiochem-Novabiochem (San Diego, CA, USA). Actinomycin D, lactacystin, MG132, and LY294002 were dissolved in dimethyl sulfoxide (DMSO). Cycloheximide was dissolved in Milli-Q-purified water (Millipore, Billerica, MA, USA).
Western blot analysis
After different treatments, equal amounts of proteins from each set of experiments were subjected to western blot analysis as previously described.35 The specific phospho-AKT(Ser473) and phospho-AKT(Thr308) antibodies were purchased from Cell Signaling (Beverly, MA, USA). Rabbit polyclonal antibodies against ERCC1 (FL-297) (sc-10785), AKT(H-136) (sc-8312), and Actin (I-19) (sc-1616) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Plasmid and transfection
Exponentially growing human lung cancer cells (106) were plated for 18 h, and then constitutively active AKT (AKT-CA), which harbored a consensus myristylation domain that replaced the 4–129 amino acids of wild-type AKT, were transfected into cells using Lipofectamine (Invitrogen). The sense-strand sequences of small interfering RNA (siRNA) duplexes used for ERCC1 and scrambled (as a control) were 5′-GGAGCUGGCUAAGAUGUGU-3′ and 5′-GCGCGCUUUGUAGGATTCG-3′ (Dharmacon Research, Lafayette, CO). Cells were transfected with siRNA duplexes (200 nM) by using Lipofectamine 2000 (Invitrogen) for 24 h.
Quantitative real-time polymerase chain reaction (PCR)
PCRs were performed using an ABI Prism 7900HT according to the manufacturer's instructions. Amplification of specific PCR products was performed using the SYBR Green PCR Master Mix (Applied Biosystems). For each sample, the data were normalized to the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The primers in this study were: ERCC1 was amplified using primers with the sequence of 5′-CCCTGGGAATTTGGCGACGTAA-3′ (forward) and 5′-CTCCAGGTACCGCCCAGCTTCC-3′ (reverse); GAPDH forward primer, 5′-CATGAGAAGTATGACAACAGCCT-3′; GAPDH reverse primer, 5′-AGTCCTTCCACGATACCAAAGT-3′. Analysis was performed using the comparative Ct value method. For each sample, the data were normalized to the housekeeping gene gapdh.
MTS assay
In vitro 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenol)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay was performed. Cells were cultured at 5000 per well in 96-well tissue culture plates. To assess cell viability, drugs were added after plating. At the end of the culture period, 20 μL of MTS solution (CellTiter 96 Aqueous One Solution Cell Proliferation Assay; Promega, Madison, WI, USA) was added; the cells were incubated for further 2 h, and the absorbance was measured at 490 nm using an ELISA plate reader (Biorad Technologies, Hercules, CA).
Combination index analysis
Erlotinib and capsaicin were combined at a ratio of 1 : 5 or 1 : 20, and the effect of the combined treatment on cell viability was examined by the MTS assay. To calculate a combination index (CI), the computer software Calcusyn (Biosoft, Oxford, UK) was used, taking the entire shape of the cell viability curve into account for calculating whether a combination was synergistic (CI < 0.9), additive (CI = 0.9–1.1), or antagonistic (CI > 1.1).36 The CI values at a fraction affected (FA) of 0.90, 0.75, 0.50 were used to calculate between the three independent experiments.
Trypan blue dye exclusion assay
Cells were treated with capsaicin and/or erlotinib for 24, 48, and 72 h. After treatment, 500 cells were harvested, and the proportion of dead cells was determined by a hemocytometer by counting the number of cells stained with trypan blue. Trypan blue dye can be excluded from living cells, but is able to penetrate dead cells. The dead cells were calculated as follow: trypan blue (+) cells ratio (%) = (stained cell number/total cell number) × 100.
Statistical analyses
For each protocol, three or four independent experiments were performed. Results were expressed as the mean ± SEM. Statistical calculations were performed using SigmaPlot 2000 software (Systat Software, San Jose, CA). Differences in the measured variables between the experimental and control groups were assessed by unpaired t-test. P < 0.05 was considered statistically significant.
Results
Capsaicin decreased the viability of NSCLC cells
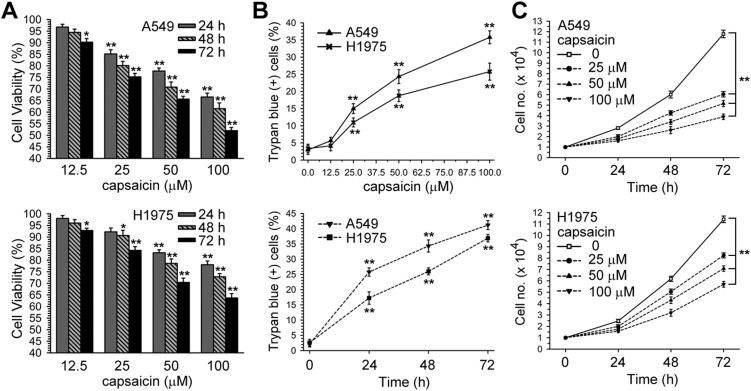
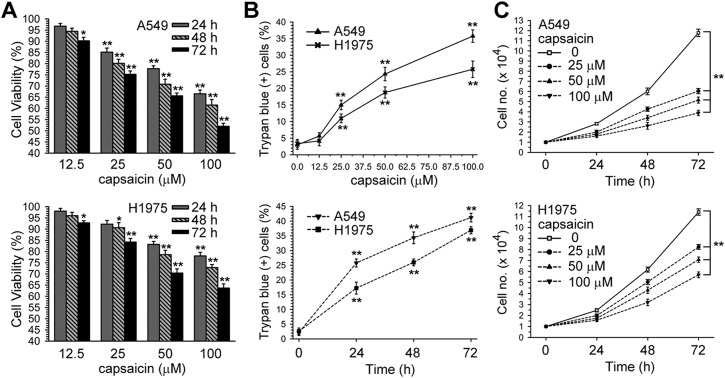
According to the Chakraborty et al. study, capsaicin (12.5–100 μM) inhibits NSCLC-induced endothelial cell migration;37 therefore, we wanted to know whether a range of concentrations of capsaicin could affect the viability of NSCLC cells. Cell viabilities were determined after A549 and H1975 cells were incubated with a vehicle (0.1% DMSO) or different concentrations of capsaicin for 24, 48, 72 h by the MTS assay, and were expressed as percent against control, which was taken as 100%. In Fig. 1A and B, it can be seen that capsaicin decreased the cell viability and induced cell death in a time and dose dependent manner. Moreover, capsaicin inhibited cell growth in A549 and H1975 cells (Fig. 1C).
Fig. 1. Dose and time-response curves of capsaicin for cell survival in A549 or H1975 cells. (A) A549 or H1975 cells were treated with various concentrations of capsaicin (12.5–100 μM) for 24, 48, and 72 h. Cell viability was determined by MTS assay. (B) After cells had been treated with various concentrations of capsaicin for 24 h (upper panel), or capsaicin (50 μM) for 24, 48, and 72 h (lower panel), both unattached and attached cells were collected and stained with trypan blue dye, and the number of dead cells were manually counted. The percentage of trypan blue-positive cells represented the population of dead cells, and the standard error (SE) was from three independent experiments. (C) After cells had been treated with various concentrations of capsaicin for 24, 48, and 72 h, both unattached and attached cells were collected and stained with trypan blue dye, and the numbers of living cells were manually counted. *p < 0.05, **p < 0.01 using Student's t-test for comparison between the cells treated with or without capsaicin.
ERCC1 mRNA and protein levels were decreased after capsaicin exposure
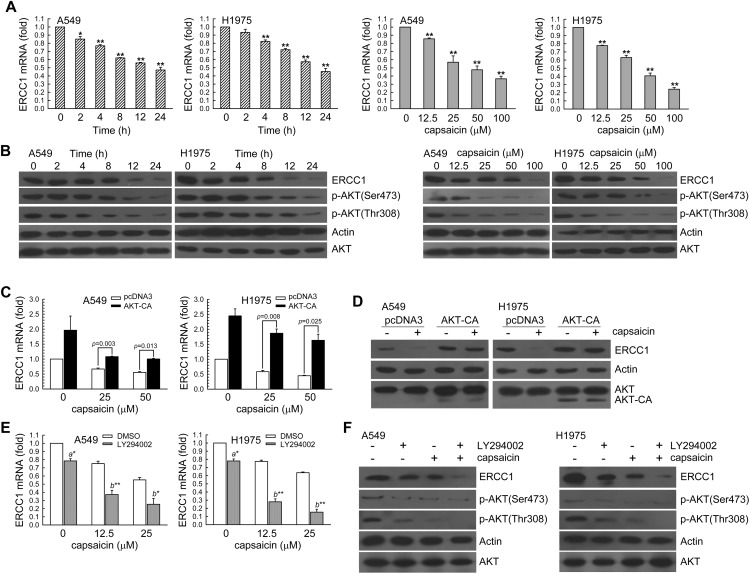
To determine whether ERCC1 expression was associated with the effect of capsaicin, we first assessed A549 or H1975 cells treated with capsaicin (50 μM) for 2–24 h or various concentrations of capsaicin (12.5, 25, 50, 100 μM) for 24 h. Then, the real-time PCR and western blot analysis was used for the determination of the ERCC1 mRNA and protein levels, respectively. In Fig. 2A and B, it can be seen that capsaicin reduced ERCC1 mRNA and protein expression in a time and dose-dependent manner; this was accompanied with a decrease in the phospho-AKT(Ser473) and phospho-AKT(Thr308) protein levels. To investigate the role of AKT activation in regulating ERCC1 expression, we examined whether the enhancement of AKT kinase activity could inhibit the down-regulation of ERCC1 expression in capsaicin-exposed A549 and H1975 cells. In Fig. 2C and D, we can see that A549 or H1975 cells were transiently transfected with a plasmid carrying AKT-CA, a constitutively active form of AKT. Compared to transfection with the control vector-pcDNA3, transfection with AKT-CA recused the capsaicin reduced ERCC1 mRNA and protein levels. However, once these cells were pretreated with a PI3K inhibitor (LY294002), the ERCC1 mRNA and protein levels in capsaicin-exposed A549 or H1975 cells would further decrease (Fig. 2E and F). Therefore, capsaicin down-regulated the ERCC1 expression in an AKT kinase inactivation manner.
Fig. 2. Capsaicin decreased the ERCC1 expression in a dose and time-dependent manner. (A) Left panel, A549 or H1975 cells (106) were cultured in complete medium for 18 h and then exposed to capsaicin (50 μM) for 2, 4, 8 12, or 24 h. Right panel, various concentrations of capsaicin (12.5–100 μM) were added to cells for 24 h in complete medium. The total RNA was isolated and subjected to real-time PCR for ERCC1 mRNA expression. The results (mean ± SEM) were from three independent experiments. *p < 0.05, **p < 0.01 using Student's t-test for comparison between the cells treated with or without capsaicin. (B) After treatment as above, the cell extracts were examined by western blot for determination of ERCC1, phospho-AKT(Ser473), phospho-AKT(Thr308), actin, and AKT protein levels. Capsaicin decreased ERCC1 expression via AKT inactivation in A549 and H1975 cells. (C and D) A549 or H1975 cells (5 × 105) were transfected with the AKT-CA expression vector for 24 h prior to treatment with capsaicin in complete medium for 24 h. The results (mean ± SEM) were from 3 independent experiments. **p < 0.01, using Student's t-test for comparison between cells treated with capsaicin in pcDNA3 or AKT-CA-transfected cells. (E and F) LY294002 (10 μM) was added to A549 or H1975 cells for 1 h before capsaicin treatment for 24 h. The results (mean ± SEM) were from four independent experiments. **p < 0.01 using Student's t-test for comparison between the cells treated with capsaicin–DMSO or a capsaicin–LY294002 combination. After treatment, the cell extracts were examined via real-time PCR (C, E) and western blot (D, F) for the determination of ERCC1 mRNA and protein levels, respectively.
Down-regulation of ERCC1 expression involved in regulating capsaicin-induced cytotoxicity and growth inhibition in NSCLC cells
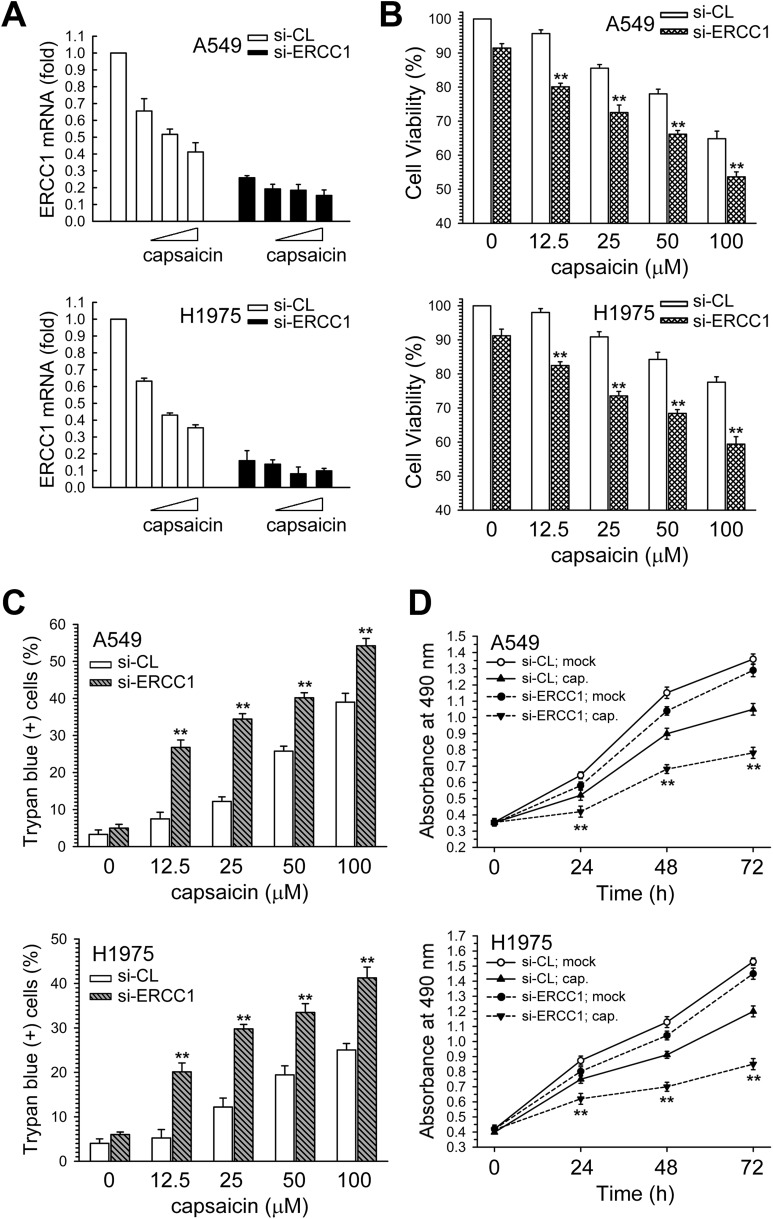
Next, the role of the decreased ERCC1 expression and AKT kinase inactivation in the cytotoxic effect of capsaicin was examined. We next examined the effect of siRNA-mediated ERCC1 knockdown on capsaicin-induced cytotoxicity and cell growth inhibition in NSCLC cells. At 24 h post-transfection, real-time PCR analysis showed a further decrease in the ERCC1 mRNA in capsaicin-treated A549 and H1975 cells (Fig. 3A). Furthermore, the suppression of ERCC1 expression by si-ERCC1 RNA resulted in an increased sensitivity to capsaicin as compared to si-control transfected cells (Fig. 3B and C), and more inhibition of cell growth was induced by the combination of ERCC1 siRNA and capsaicin than by capsaicin alone in A549 or H1975 cells (Fig. 3D). Therefore, the down-regulation of ERCC1 expression could enhance the capsaicin-induced cytotoxicity and growth inhibition in NSCLC cells.
Fig. 3. Down-regulation of ERCC1 expression enhanced the capsaicin-induced cytotoxicity and growth inhibition. (A) A549 or H1975 cells were transfected with siRNA duplexes (200 nM) specific to ERCC1 or scrambled (control) in complete medium for 24 h prior to the treatment with capsaicin in complete medium for 24 h. The total RNA was isolated and subjected to real-time PCR for ERCC1 mRNA expression. (B and C) After the above-mentioned treatment, cytotoxicity was determined by MTS assay and trypan blue exclusion assay. (D) After the cells were transfected with si-ERCC1 or si-scrambled RNA, the cells were treated with capsaicin (25 μM) for 24, 48, and 72 h, after which living cells were determined by MTS assay. The results (mean ± SEM) were from three independent experiments. **p < 0.01 using Student's t-test for comparison between the cells treated with capsaicin in si-ERCC1 RNA or si-scrambled RNA-transfected cells.
AKT kinase signaling pathway mediated capsaicin-induced cytotoxicity
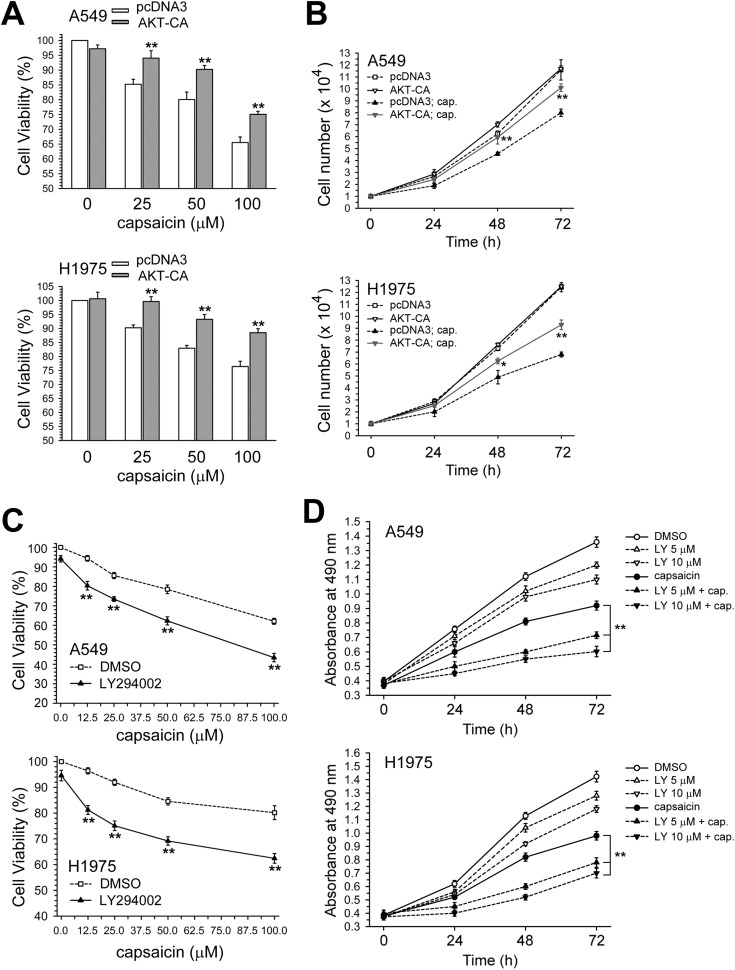
In Fig. 4A, the enforced expression of the AKT-CA vector could rescue A549 and H1975 cell viability after being decreased by capsaicin, and also reverse the growth inhibition effect (Fig. 4B). In contrast, blocking the AKT kinase activity by LY294002 caused further significant decreases in cell viability in capsaicin-exposed A549 or H1975 cells, compared with capsaicin treatment alone (Fig. 4C). Co-treatment with LY294002 could more effectively inhibit cell growth than with either drug alone (Fig. 4D). Taken together, the inactivation of the AKT kinase and down-regulation of the ERCC1 expression were involved in capsaicin-induced cytotoxicity and growth inhibition in NSCLC cells.
Fig. 4. Enhancement of AKT activity by AKT-CA vector transfection decreased the cytotoxicity induced by capsaicin. (A) After the cells were transfected with pcDNA3 or AKT-CA expression vector, the cells were treated with capsaicin for 24 h. Cytotoxicity was determined by MTS assay. The results (mean ± SEM) were from three independent experiments. (B) After the cells were transfected with a pcDNA3 or AKT-CA expression vector, the cells were treated with capsaicin (50 μM) for 24, 48, and 72 h, after which living cells were determined by trypan blue exclusion assay. *p < 0.05, **p < 0.01 using Student's t-test for comparison between the cells treated with capsaicin in AKT-CA or pcDNA3 vector-transfected cells. (C) Cells were pretreated with LY294002 (5 μM) for 1 h and then co-treated with various concentration of capsaicin for 24 h. Cytotoxicity was determined by MTS assay. **p < 0.01 using Student's t-test for comparison between the cells pretreated with or without LY294002 in capsaicin exposed cells. (D) Cells were treated with capsaicin (25 μM) and/or LY294002 (5, 10 μM) for 1–3 days, after which living cells were determined by MTS assay. **p < 0.01 using Student's t-test for comparison between cells treated with capsaicin alone or with a capsaicin and LY294002 combination.
Capsaicin and erlotinib exerts a synergistic cytotoxic effect on NSCLC cells
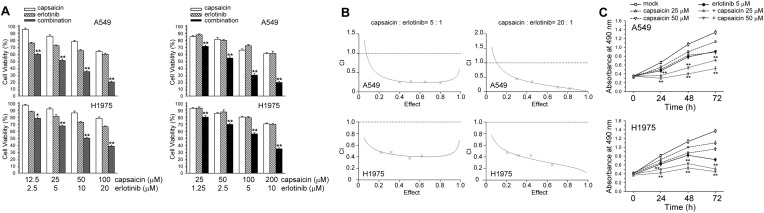
We attempted to determine whether capsaicin could enhance the cytotoxic effect of erlotinib through down-regulating the ERCC1 expression in NSCLC cells. The effect of combined treatment on cell viability was examined by the MTS assay. The combined treatment with capsaicin and erlotinib for 24 h resulted in a greater loss of cell viability in A549 and H1975 cells than the treatment with either erlotinib or capsaicin (Fig. 5A). CI allows the quantification of synergism or antagonism for two drugs where CI of 1 indicates an additive effect, whereas a CI < 1 or CI > 1 indicates synergism or antagonism, respectively. The CI values for capsaicin and erlotinib were <1, indicating that the combined treatment had a synergistic effect (Fig. 5B). In addition, A549 and H1975 cells were exposed to capsaicin and/or erlotinib, and cell proliferation was determined 1–3 days after exposure. The capsaicin and erlotinib co-treatment had a greater cell growth inhibition effect than either treatment alone (Fig. 5C).
Fig. 5. Capsaicin co-treatment with erlotinib synergistically enhanced cytotoxicity. (A) Erlotinib and capsaicin were combined at a ratio of 1 : 5 or 1 : 20 and the MTS assay was used to analyze cell viability. (B) The mean CI values at a fraction affected (FA) of 0.50, 0.75, 0.90 for erlotinib and capsaicin combined treatment were averaged for three independent experiments. (C) Cells were treated with capsaicin (25, 50 μM) and/or erlotinib (5 μM) for 1–3 days, after which living cells were determined by MTS assay. **p < 0.01 using Student's t-test for comparison between cells treated with a drug alone or with a capsaicin/erlotinib combination. **p < 0.01 using Student's t-test for comparison between cells treated with a drug alone or with the capsaicin/erlotinib combination.
Capsaicin down-regulated the ERCC1 protein and the mRNA level in erlotinib-treated human lung cancer cells
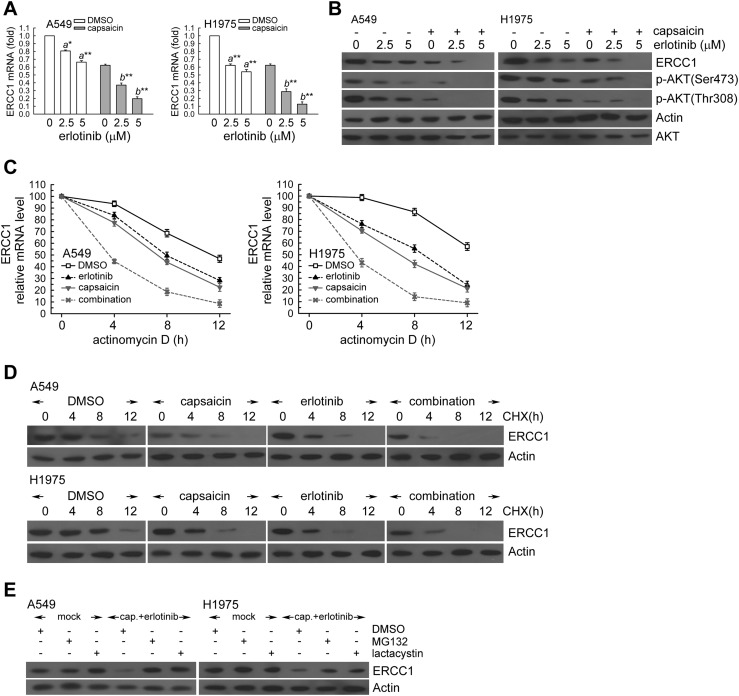
In order to assess the mechanism of the synergistic effect, we hypothesized that capsaicin would affect the ERCC1 expression in erlotinib-treated NSCLC cells. To test this hypothesis, A549 and H1975 cells were exposed to capsaicin and/or erlotinib for 24 h. In Fig. 6A and B, capsaicin decreased the ERCC1 mRNA and protein levels in erlotinib-treated A549 and H1975 cells. Moreover, capsaicin decreased the phospho-AKT kinase protein levels in erlotinib-treated NSCLC cells (Fig. 6B).
Fig. 6. Capsaicin decreased the ERCC1 protein and mRNA levels in erlotinib-exposed NSCLC cells. (A) A549 or H1975 cells (106) were cultured in complete medium for 18 h and then were exposed to capsaicin (25 μM) and erlotinib (2.5, 5 μM) for 24 h. After treatment, total RNA was isolated and subjected to real-time PCR for ERCC1 mRNA expression. The means ± standard deviation (SD) from four independent experiments. a** denotes p < 0.01, a* denotes p < 0.05 using Student's t-test for comparison between the cells treated with or without erlotinib. b** denotes p < 0.01, respectively, using Student's t-test for comparison between the cells treated with capsaicin alone or capsaicin and erlotinib combined. (B) After treatment as the above, cell extracts were examined by western blot for the determination of ERCC1, phospho-AKT(Ser473), actin, and AKT protein levels. (C) A549 or H1975 cells were treated with capsaicin (25 μM) and/or erlotinib (5 μM) for 12 h in the presence or absence of actinomycin D (2 μg mL–1) for 4, 8, or 12 h; total RNA was isolated and subjected to real-time PCR for ERCC1 mRNA expression. (D) The cells were exposed to capsaicin (25 μM) and/or erlotinib (5 μM) for 12 h followed by co-treatment with cycloheximide (CHX; 0.1 mg mL–1) for 4, 8, or 12 h. Whole-cell extracts were collected for western blot analysis. Capsaicin treatment triggers 26S proteasome-mediated proteolysis of ERCC1. (E) Capsaicin (25 μM) and erlotinib (5 μM) was added to A549 or H1975 cells for 18 h, then the cells were co-treated with 26S proteasome inhibitor MG132 (10 μM) or lactacystin (10 μM) for 6 h. Whole cell extracts were collected for western blot analysis.
Capsaicin and erlotinib co-treatment decreased the ERCC1 mRNA and protein stability
Next, we examined possible mechanisms for post-transcriptional regulation of ERCC1 transcripts under capsaicin and/or erlotinib treatment. In the presence of actinomycin D, the capsaicin treatment showed lower levels of ERCC1 mRNA relative to the untreated cells (Fig. 6C). Moreover, ERCC1 protein levels were progressively reduced with time in the presence of cycloheximide (an inhibitor of de novo protein synthesis) (Fig. 6D). However, capsaicin treatment significantly promoted ERCC1 degradation after cycloheximide treatment compared to untreated cells (Fig. 6D). Therefore, the ERCC1 protein was less stable after capsaicin treatment in A549 and H1975 cells. Interestingly, after co-treatment, erlotinib enhanced the ERCC1 mRNA and protein instability in capsaicin-treated A549 and H1975 cells (Fig. 6C and D). Next, we hypothesized that the capsaicin and erlotinib combination induced the down-regulation of ERCC1 protein levels through the ubiquitin-26S proteasome-mediated proteolysis of ERCC1. In Fig. 6E, the 26S proteasome inhibitors MG132 or lactacystin were added for the final 6 h before harvesting in capsaicin and erlotinib-treated A549 and H1975 cells. The result showed that both MG132 and lactacystin restored the decreased ERCC1 protein levels induced by capsaicin and erlotinib (Fig. 6E). Taken together, the capsaicin and erlotinib combination decreased the ERCC1 levels by enhancing the ERCC1 mRNA instability and triggering the ERCC1 protein degradation in A549 and H1975 cells.
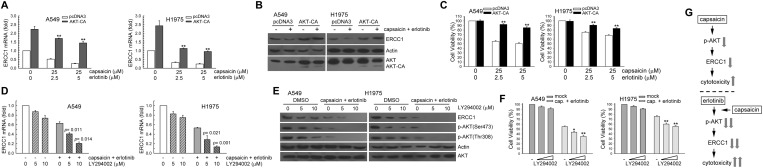
Transfection with AKT-CA vectors increased the ERCC1 protein level as well as the cell survival by erlotinib and capsaicin
We investigated whether capsaicin and erlotinib combination-mediated ERCC1 down-regulation was correlated with AKT kinase inactivation in NSCLC cells. In Fig. 7A–C, the enforced AKT-CA vector expression could increase the ERCC1 mRNA and protein expression in capsaicin and erlotinib cotreated A549 and H1975 cells; also, it recused the viability after being decreased by capsaicin and erlotinib. In contrast, the inactivation of AKT kinase activity by pretreatment with LY294002 further decreased the ERCC1 expression and cell viability in capsaicin and erlotinib cotreated A549 and H1975 cells (Fig. 7D–F). Taken together, capsaicin enhanced the erlotinib-induced cytotoxicity by AKT kinase inactivation mediated ERCC1 down-regulation in A549 and H1975 cells (Fig. 7G).
Fig. 7. Enhancement of AKT activity restored the suppressed ERCC1 protein expression and cell survival in capsaicin and erlotinib-exposed A549 and H1975 cells. (A and B) AKT-CA (3 μg) or pcDNA3 (3 μg) expression plasmids were transfected into cells using lipofectamine. After expression for 24 h, the cells were treated with capsaicin (25 μM) and erlotinib (5 μM) for an additional 24 h, and total RNA was isolated and subjected to real-time PCR for ERCC1 mRNA expression, and western blot for determination of ERCC1 protein levels. (C) After treatment as in (A). Cytotoxicity was determined by assessment with the MTS assay. The means ± standard deviation (SD) from four independent experiments. ** denotes p < 0.01, using Student's t-test to compare cells treated with capsaicin and erlotinib in AKT-CA vs. pcDNA3-transfected cells. (D) A549 or H1975 cells were pretreated with LY294002 for 1 h and then co-treated with capsaicin (12.5 μM) and erlotinib (2.5 μM) for 24 h. Total RNA was isolated and subjected to real-time PCR for ERCC1 mRNA expression. (E) After treatment as the above, the whole-cell extracts were collected for western blot analysis. (F) After treatment as in (D). Cytotoxicity was determined by the MTS assay. **p < 0.01 using Student's t-test for comparison between the cells treated with capsaicin/erlotinib–DMSO or a capsaicin/erlotinib–LY294002 combination. (G) Upper panel, inactivation of the AKT kinase and down-regulation of the ERCC1 expression involved in capsaicin-induced cytotoxicity. Lower panel, capsaicin enhanced NSCLC cells sensitivity to erlotinib as a result of AKT inactivation mediated ERCC1 decrease in NSCLC cells.
Discussion
Capsaicin, a homovanillic acid derivate (8-methyl-N-vanillyl-6-nonenamide), is the most abundant pungent ingredient found in pepper plants (genus Capsicum, family Solanaceae);33 it exhibits anti-proliferative actions inhibiting the progression of many types of tumors through various mechanisms.38 For example, capsaicin can induce a cell-cycle arrest or apoptosis or may inhibit proliferation in a variety of cancer cells through the generation of reactive oxygen species (ROS) and persistent disruption of the mitochondrial membrane potential.39 In hepatocellular carcinoma, capsaicin inhibits proliferation through the induction of autophagy and apoptosis.17 Capsaicin induced apoptosis in liver hepatocellular cells involves the increase in intracellular calcium concentrations, expression of heme oxygenase-1,40 production of ROS and activation of caspase-3.41 In SK-Hep-1 cells, capsaicin efficiently induced apoptosis through a Bcl-2 down-regulation and caspase-3-dependent mechanism.42 In this study, capsaicin induced cytotoxic effects in NSCLC cells via ERCC1 down-regulation and AKT inactivation.
Capsaicin has been investigated for its effects on mutagenicity and genotoxicity in vitro and in vivo, but the study results are contradictory.43 Previous studies have shown that high ERCC1 levels in several cancers are associated with the resistance to platinum-based treatment31,44 and that the transfection of ERCC1 into an ERCC1-deficient Chinese hamster ovary cell line could restore the repairability of DNA adducts, countering the platinum-resistance.45 In contrast, depletion of ERCC1 restored the sensitivity to cisplatin in the ovarian cancer cell line.46 Our findings have demonstrated that MKK1/2-ERK1/2 signals participate in maintaining ERCC1 protein expression in human lung cancer cells.47 In addition, a previous study has shown that the radiation-induced ERCC1 protein expression is also dependent on the ERK1/2 pathway in DU145 prostate carcinoma cells.48 The present study has revealed that the inactivation of AKT is correlated with the protein and mRNA downregulation of ERCC1 in capsaicin-exposed NSCLC cells. Enhancement of the AKT activity restored the cell viability decreased by capsaicin. The down-regulation of the ERCC1 expression was involved in regulating the capsaicin-induced cytotoxicity and growth inhibition in human lung cancer cells. Therefore, capsaicin induced cytotoxic effects in human lung cancer cells lines via ERCC1 down-regulation and AKT inactivation in human lung cancer cells.
The mechanism of the reduced cell viability by capsaicin in cancer cells involves the increase in intracellular Ca2+ levels,49,50 the generation of ROS,17 disruption of the mitochondrial membrane potential50 and activation of transcription factors, such as STATs (signal transducer and activator of transcription protein family).17 Capsaicin induced apoptosis in A549 in a p53-dependent manner, in which p53, activated by ROS-induced DNA damage, trans-activated Bax, which is the pro-apoptotic family member of Bcl-2.51 Brown et al. showed that capsaicin has anti-proliferative activity against human small cell lung cancer in cell cultures and nude mice models via the E2F pathway.18 Ying et al.52 demonstrated that capsaicin induced apoptosis in human osteosarcoma cells via the activation of the AMPK signaling pathway and c-Jun NH2-terminal kinases. In addition, the combination of capsaicin with 5-FU synergistically suppressed the tumor growth in the cholangiocarcinoma xenograft to a greater extent than 5-FU alone. This effect probably occurs via the PI3K/AKT/mTOR pathway activation.53 Hong et al. suggested that this was a promising strategy for the development of combination drugs for cholangiocarcinoma.53 In this study, a combined treatment with capsaicin enhanced the AKT inactivation and down-regulated the expression of ERCC1 in erlotinib-exposed A549 and H1975 cells, and subsequently resulted in synergistic cytotoxic effects in NSCLC cells.
Overall, these results show that capsaicin enhanced NSCLC cells are sensitive to erlotinib as a result of AKT inactivation mediated ERCC1 decrease (Fig. 7G). Although further study is required to evaluate the effect of capsaicin and erlotinib combination in vivo, it represents a promising and attractive strategy for the treatment of NSCLC. These results may provide additional insights into the potential synergistic effects of capsaicin in combination with erlotinib on NSCLC.
Conflicts of interest
The authors declare that they have no conflict of interest.
Acknowledgments
This study was funded by grants from the Ministry of Science and Technology, Taiwan, Grant MOST 107-2314-B-415-007 and 107-2314-B-002-236.
References
- Pfister D. G., Johnson D. H., Azzoli C. G., Sause W., Smith T. J., Baker Jr. S., Olak J., Stover D., Strawn J. R., Turrisi A. T., Somerfield M. R. J. Clin. Oncol. 2004;22:330–353. doi: 10.1200/JCO.2004.09.053. [DOI] [PubMed] [Google Scholar]
- Chen W., Zheng R., Baade P. D., Zhang S., Zeng H., Bray F., Jemal A., Yu X. Q., He J. CA-Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. CA-Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Molina J. R., Yang P., Cassivi S. D., Schild S. E., Adjei A. A. Mayo Clin. Proc. 2008;83:584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon D. S., Brandt R., Ciardiello F., Normanno N. Crit. Rev. Oncol. Hematol. 1995;19:183–232. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- Arteaga C. L. J. Clin. Oncol. 2003;21:289s–291s. doi: 10.1200/JCO.2003.10.523. [DOI] [PubMed] [Google Scholar]
- Raymond E., Faivre S., Armand J. P., Drugs, 2000, 60Suppl 1 , 15 –23 , ; discussion 41–12 . [DOI] [PubMed] [Google Scholar]
- Steins M. B., Reinmuth N., Bischoff H., Kindermann M., Thomas M. Onkologie. 2010;33:704–709. doi: 10.1159/000322214. [DOI] [PubMed] [Google Scholar]
- Nguyen K. S., Neal J. W. Biol.: Targets Ther. 2012;6:337–345. doi: 10.2147/BTT.S26558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felip E., Rosell R. Drugs Today. 2006;42:147–156. doi: 10.1358/dot.2006.42.3.957358. [DOI] [PubMed] [Google Scholar]
- Kris M. G., Natale R. B., Herbst R. S., Lynch Jr. T. J., Prager D., Belani C. P., Schiller J. H., Kelly K., Spiridonidis H., Sandler A., Albain K. S., Cella D., Wolf M. K., Averbuch S. D., Ochs J. J., Kay A. C. J. Am. Med. Assoc. 2003;290:2149–2158. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- Monsereenusorn Y., Kongsamut S., Pezalla P. D. Crit. Rev. Toxicol. 1982;10:321–339. doi: 10.3109/10408448209003371. [DOI] [PubMed] [Google Scholar]
- Chou C. C., Wu Y. C., Wang Y. F., Chou M. J., Kuo S. J., Chen D. R. Oncol. Rep. 2009;21:665–671. [PubMed] [Google Scholar]
- Sanchez A. M., Malagarie-Cazenave S., Olea N., Vara D., Chiloeches A., Diaz-Laviada I. Apoptosis. 2007;12:2013–2024. doi: 10.1007/s10495-007-0119-z. [DOI] [PubMed] [Google Scholar]
- Kim M. Y., Trudel L. J., Wogan G. N. Anticancer Res. 2009;29:3733–3740. [PubMed] [Google Scholar]
- Lo Y. C., Yang Y. C., Wu I. C., Kuo F. C., Liu C. M., Wang H. W., Kuo C. H., Wu J. Y., Wu D. C. World J. Gastroenterol. 2005;11:6254–6257. doi: 10.3748/wjg.v11.i40.6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Tan M., Xie Z., Feng B., Zhao Z., Yang K., Hu C., Liao N., Wang T., Chen D., Xie F., Tang C. Free Radical Res. 2016;50:744–755. doi: 10.3109/10715762.2016.1173689. [DOI] [PubMed] [Google Scholar]
- Brown K. C., Witte T. R., Hardman W. E., Luo H., Chen Y. C., Carpenter A. B., Lau J. K., Dasgupta P. PLoS One. 2010;5:e10243. doi: 10.1371/journal.pone.0010243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Nakazato T., Yamato K., Miyakawa Y., Yamada T., Hozumi N., Segawa K., Ikeda Y., Kizaki M. Cancer Res. 2004;64:1071–1078. doi: 10.1158/0008-5472.can-03-1670. [DOI] [PubMed] [Google Scholar]
- Le T. D., Jin D., Rho S. R., Kim M. S., Yu R., Yoo H. Yonsei Med. J. 2012;53:834–841. doi: 10.3349/ymj.2012.53.4.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bort A., Spinola E., Rodriguez-Henche N., Diaz-Laviada I. Oncotarget. 2017;8:87684–87698. doi: 10.18632/oncotarget.21196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandakumar P., Kamaraj S., Jagan S., Ramakrishnan G., Devaki T. Int. Immunopharmacol. 2013;17:254–259. doi: 10.1016/j.intimp.2013.05.015. [DOI] [PubMed] [Google Scholar]
- Marques S., Oliveira N. G., Chaveca T., Rueff J. Mutat. Res. 2002;517:39–46. doi: 10.1016/s1383-5718(02)00040-2. [DOI] [PubMed] [Google Scholar]
- Richeux F., Cascante M., Ennamany R., Saboureau D., Creppy E. E. Arch. Toxicol. 1999;73:403–409. doi: 10.1007/s002040050680. [DOI] [PubMed] [Google Scholar]
- Yoon J. H., Ahn S. G., Lee B. H., Jung S. H., Oh S. H. Biochem. Pharmacol. 2012;83:747–757. doi: 10.1016/j.bcp.2011.12.029. [DOI] [PubMed] [Google Scholar]
- Friedberg E. C. Nat. Rev. Cancer. 2001;1:22–33. doi: 10.1038/35094000. [DOI] [PubMed] [Google Scholar]
- Reardon J. T., Vaisman A., Chaney S. G., Sancar A. Cancer Res. 1999;59:3968–3971. [PubMed] [Google Scholar]
- de Laat W. L., Appeldoorn E., Jaspers N. G., Hoeijmakers J. H. J. Biol. Chem. 1998;273:7835–7842. doi: 10.1074/jbc.273.14.7835. [DOI] [PubMed] [Google Scholar]
- Evans E., Moggs J. G., Hwang J. R., Egly J. M., Wood R. D. EMBO J. 1997;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel U., Dybdahl M., Frentz G., Nexo B. A. Mutat. Res. 2000;461:197–210. doi: 10.1016/s0921-8777(00)00051-3. [DOI] [PubMed] [Google Scholar]
- Lord R. V., Brabender J., Gandara D., Alberola V., Camps C., Domine M., Cardenal F., Sanchez J. M., Gumerlock P. H., Taron M., Sanchez J. J., Danenberg K. D., Danenberg P. V., Rosell R. Clin. Cancer Res. 2002;8:2286–2291. [PubMed] [Google Scholar]
- Deal C. L., Schnitzer T. J., Lipstein E., Seibold J. R., Stevens R. M., Levy M. D., Albert D., Renold F. Clin. Ther. 1991;13:383–395. [PubMed] [Google Scholar]
- Bley K., Boorman G., Mohammad B., McKenzie D., Babbar S. Toxicol. Pathol. 2012;40:847–873. doi: 10.1177/0192623312444471. [DOI] [PubMed] [Google Scholar]
- Kim C. S., Kawada T., Kim B. S., Han I. S., Choe S. Y., Kurata T., Yu R. Cell. Signalling. 2003;15:299–306. doi: 10.1016/s0898-6568(02)00086-4. [DOI] [PubMed] [Google Scholar]
- Ko J. C., Ciou S. C., Jhan J. Y., Cheng C. M., Su Y. J., Chuang S. M., Lin S. T., Chang C. C., Lin Y. W. Mol. Cancer Res. 2009;7:1378–1389. doi: 10.1158/1541-7786.MCR-09-0051. [DOI] [PubMed] [Google Scholar]
- Peters G. J., van der Wilt C. L., van Moorsel C. J., Kroep J. R., Bergman A. M., Ackland S. P. Pharmacol. Ther. 2000;87:227–253. doi: 10.1016/s0163-7258(00)00086-3. [DOI] [PubMed] [Google Scholar]
- Chakraborty S., Adhikary A., Mazumdar M., Mukherjee S., Bhattacharjee P., Guha D., Choudhuri T., Chattopadhyay S., Sa G., Sen A., Das T. PLoS One. 2014;9:e99743. doi: 10.1371/journal.pone.0099743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Laviada I., Rodriguez-Henche N. Prog. Drug Res. 2014;68:181–208. doi: 10.1007/978-3-0348-0828-6_8. [DOI] [PubMed] [Google Scholar]
- Bu H. Q., Cai K., Shen F., Bao X. D., Xu Y., Yu F., Pan H. Q., Chen C. H., Du Z. J., Cui J. H. Neoplasma. 2015;62:582–591. doi: 10.4149/neo_2015_070. [DOI] [PubMed] [Google Scholar]
- Joung E. J., Li M. H., Lee H. G., Somparn N., Jung Y. S., Na H. K., Kim S. H., Cha Y. N., Surh Y. J. Antioxid. Redox Signaling. 2007;9:2087–2098. doi: 10.1089/ars.2007.1827. [DOI] [PubMed] [Google Scholar]
- Huang S. P., Chen J. C., Wu C. C., Chen C. T., Tang N. Y., Ho Y. T., Lo C., Lin J. P., Chung J. G., Lin J. G. Anticancer Res. 2009;29:165–174. [PubMed] [Google Scholar]
- Jung M. Y., Kang H. J., Moon A. Cancer Lett. 2001;165:139–145. doi: 10.1016/s0304-3835(01)00426-8. [DOI] [PubMed] [Google Scholar]
- Chanda S., Erexson G., Riach C., Innes D., Stevenson F., Murli H., Bley K. Mutat. Res. 2004;557:85–97. doi: 10.1016/j.mrgentox.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Altaha R., Liang X., Yu J. J., Reed E. Int.Int. J. Mol. Med.J. Mol. Med. 2004;14:959–970. [PubMed] [Google Scholar]
- Lee K. B., Parker R. J., Bohr V., Cornelison T., Reed E. Carcinogenesis. 1993;14:2177–2180. doi: 10.1093/carcin/14.10.2177. [DOI] [PubMed] [Google Scholar]
- Selvakumaran M., Pisarcik D. A., Bao R., Yeung A. T., Hamilton T. C. Cancer Res. 2003;63:1311–1316. [PubMed] [Google Scholar]
- Ko J. C., Su Y. J., Lin S. T., Jhan J. Y., Ciou S. C., Cheng C. M., Chiu Y. F., Kuo Y. H., Tsai M. S., Lin Y. W. Lung Cancer. 2010;69:155–164. doi: 10.1016/j.lungcan.2009.10.013. [DOI] [PubMed] [Google Scholar]
- Yacoub A., Park J. S., Qiao L., Dent P., Hagan M. P. Int. J. Radiat. Biol. 2001;77:1067–1078. doi: 10.1080/09553000110069317. [DOI] [PubMed] [Google Scholar]
- Wu C. C., Lin J. P., Yang J. S., Chou S. T., Chen S. C., Lin Y. T., Lin H. L., Chung J. G. Mutat. Res. 2006;601:71–82. doi: 10.1016/j.mrfmmm.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Ip S. W., Lan S. H., Lu H. F., Huang A. C., Yang J. S., Lin J. P., Huang H. Y., Lien J. C., Ho C. C., Chiu C. F., Wood W., Chung J. G. Hum. Exp. Toxicol. 2012;31:539–549. doi: 10.1177/0960327111417269. [DOI] [PubMed] [Google Scholar]
- Chakraborty S., Mazumdar M., Mukherjee S., Bhattacharjee P., Adhikary A., Manna A., Khan P., Sen A., Das T. FEBS Lett. 2014;588:549–559. doi: 10.1016/j.febslet.2013.11.040. [DOI] [PubMed] [Google Scholar]
- Ying H., Wang Z., Zhang Y., Yang T. Y., Ding Z. H., Liu S. Y., Shao J., Liu Y., Fan X. B. Mol. Cell. Biochem. 2013;384:229–237. doi: 10.1007/s11010-013-1802-8. [DOI] [PubMed] [Google Scholar]
- Hong Z. F., Zhao W. X., Yin Z. Y., Xie C. R., Xu Y. P., Chi X. Q., Zhang S., Wang X. M. PLoS One. 2015;10:e0121538. doi: 10.1371/journal.pone.0121538. [DOI] [PMC free article] [PubMed] [Google Scholar]