Abstract
The evolution of mitochondrial genomes and their population-genetic environment among unicellular eukaryotes are understudied. Ciliate mitochondrial genomes exhibit a unique combination of characteristics, including a linear organization and the presence of multiple genes with no known function or detectable homologs in other eukaryotes. Here we study the variation of ciliate mitochondrial genomes both within and across 13 highly diverged Paramecium species, including multiple species from the P. aurelia species complex, with four outgroup species: P. caudatum, P. multimicronucleatum, and two strains that may represent novel related species. We observe extraordinary conservation of gene order and protein-coding content in Paramecium mitochondria across species. In contrast, significant differences are observed in tRNA content and copy number, which is highly conserved in species belonging to the P. aurelia complex but variable among and even within the other Paramecium species. There is an increase in GC content from ∼20% to ∼40% on the branch leading to the P. aurelia complex. Patterns of polymorphism in population-genomic data and mutation-accumulation experiments suggest that the increase in GC content is primarily due to changes in the mutation spectra in the P. aurelia species. Finally, we find no evidence of recombination in Paramecium mitochondria and find that the mitochondrial genome appears to experience either similar or stronger efficacy of purifying selection than the nucleus.
Keywords: Paramecium, mitochondria, telomeres, recombination, mutation spectrum, efficacy of purifying selection
Introduction
Mitochondrial genomes have played integral roles in furthering our understanding of relationships among species as well as revealing population structure and demographic history. As a consequence, we have obtained insights into the unique population-genetic properties of mitochondrial genomes. In most species, mitochondria are inherited uniparentally (but see Barr et al. 2005) and mitochondrial genomes are known to frequently undergo recombination in plants (Stadler and Delph 2002; Mackenzie 2007), fungi (Fritsch et al. 2014), as well as in animals (Piganeau et al. 2004; Tsaousis et al. 2005; Ladoukakis et al. 2011). Because of the unique mode of transmission, mitochondria have been suggested to have lower effective population sizes than their nuclear counterparts and therefore to accumulate more deleterious mutations (Lynch and Blanchard 1998; Neiman and Taylor 2009). In addition, mitochondrial genomes experience much higher spontaneous rates of mutation than their corresponding nuclear genomes in animals, but exhibit the opposite trend in plants (Lynch et al. 2006).
Unlike the relatively uniform and conserved properties of bilaterian metazoan mitochondrial genomes, mitochondrial genomes in other eukaryotes exhibit remarkable variation in genome structure and GC content, particularly among unicellular lineages. Mitochondrial genome structures range from hundreds of short linear segments (0.3–8.3 kb) in the ichtysporean Amoebidium parasiticum (Burger et al. 2003), an opisthokont, to many small (<10 kb) circular genomes in the diplonemid Diplonema papillatum (Vlcek et al. 2011), an excavate, to a single larger linear or circular chromosome, and a variety of other states (reviewed by Smith and Keeling 2015). In addition to variation in organization, mitochondria from unicellular lineages display widely diverse GC compositions, ranging from as low as 10% in some yeast (Smith 2012) to as high as 60% in Lobochlamys culleus (Borza et al. 2009), although most species are AT rich, with an average GC content of 35% (Smith 2012).
Much has been learned about the structure, evolution, and population-genetic environment of mitochondria in the main model systems, especially in plants and metazoans (Lynch 2007; Smith 2016). However, we lack such understanding of mitochondria of the majority of unicellular eukaryotes, where the bulk of eukaryotic phylogenetic diversity lies. We address this gap by surveying both within and between-species variation in mitochondrial genomes among multiple ciliate species belonging to the genus Paramecium.
In the large and morphologically and ecologically diverse ciliate lineage, mitochondrial genomes sampled so far are organized into large linear chromosomes, several tens of kilobases in length, with telomeres at the ends (Goddard and Cummings 1975; Morin and Cech 1988; Swart et al. 2012). However, few mitochondrial genomes have until now been fully sequenced among the ciliates, with two in the Paramecium genus (P. tetraurelia and P. caudatum; Barth and Berendonk 2011), and only a few others: Tetrahymena pyriformis (Burger et al. 2000), Euplotes minuta and Euplotes crassus (de Graaf et al. 2009), Oxytricha trifallax (Swart et al. 2012), Stentor coeruleus (Slabodnick et al. 2017), Ichthyophthirius multifiliis (Coyne et al. 2011), and the anaerobic ciliate Nyctotherus ovalis (de Graaf et al. 2011).
Although the Paramecium genus contains a number of distinct morphospecies, it is especially known for including a species complex consisting of multiple morphologically identical but sexually isolated species—the P. aurelia complex (Sonneborn 1975). Species in the P. aurelia complex are ancient, with the estimated time of divergence for the complex as a whole being on the order of 300 Myr (McGrath, Gout, Johri, et al. 2014), implying that the genus Paramecium itself is even more ancient. Interestingly, among Paramecium species, there is an increase of GC content in mtDNA in the branch leading to the P. aurelia complex (Burger et al. 2000; Barth and Berendonk 2011), allowing us to study the evolution of nucleotide composition across mitochondrial genomes that are structurally very similar.
The Paramecium species offer a particularly interesting system in which to study the evolution of mitochondrial genomes because of the unique population-genetic environment experienced by their cellular organelles. Paramecium cells are mitochondria-rich: Each individual cell in P. aurelia species is estimated to contain about 5,000 mitochondria, with about 8–10 genomes per mitochondrion (Beale and Tait 1981), which is much larger than in mammalian cells with 1,000–2,000 mitochondria (Kukat et al. 2011) and yeast cells with 20–30 mitochondria per cell (Visser et al. 1995). In addition, Paramecium lineages, like other ciliate species, possess two nuclei: the polyploid somatic nucleus (called the macronucleus), which divides amitotically where the bulk of transcriptional activity occurs, and the diploid germline nucleus (known as the micronucleus), which is transcriptionally silent, and which undergoes sexual reproduction. All Paramecium species can proliferate asexually for a limited number of generations, after which they senesce unless they undergo sexual reproduction, that is, conjugation. During asexual proliferation, Paramecium undergoes binary fission; in this process, mitochondria appear to double in length, replicate their genomes (Perasso and Beisson 1978), and are randomly distributed between the two daughter cells (Adoutte and Beisson 1972; Adoutte and Doussiere 1978). Mitochondria are therefore thought not to experience any bottlenecks during binary fission division.
During conjugation, Paramecium cells exhibit cytoplasmic inheritance (Koizumi and Kobayashi 1989), that is, despite the exchange of micronuclei between the two conjugants there is almost no exchange of cytoplasm and other organelles (reviewed by Meyer and Garnier 2002). Thus, mitochondria are uniparentally inherited. A distinct aspect of Paramecium mitochondrial biology is that Paramecium mitochondria appear to exist as independent structural units and do not undergo fusion, unlike the constant flux of organelle fusion and fission in other metazoan mitochondrial populations (Kiefel et al. 2006). Both uniparental inheritance of mitochondria and the absence of fusion in the cytoplasm suggest a lack of recombination among mitochondria genomes.
In this study, we further the understanding of the biology and the population-genetic environment of ciliate mitochondria by presenting the complete mitochondrial genomes of nine species belonging to the P. aurelia complex, four outgroup (relative to P. aurelia) species, and additional 5–10 isolates for each of four Paramecium species. Using phylogenetic and population-genetic analyses, we investigate variation in protein-coding genes, tRNA content, and the forces governing the evolution of nucleotide composition of mitochondrial genomes across the phylogeny. Finally, we estimate the recombination rate across the genome and address the controversy of whether mitochondrial genomes experience reduced efficacy of purifying selection in comparison to their nuclear counterparts.
Results
Sequencing and Assembly of Mitochondrial Genomes and Detection of Single Nucleotide Polymorphisms
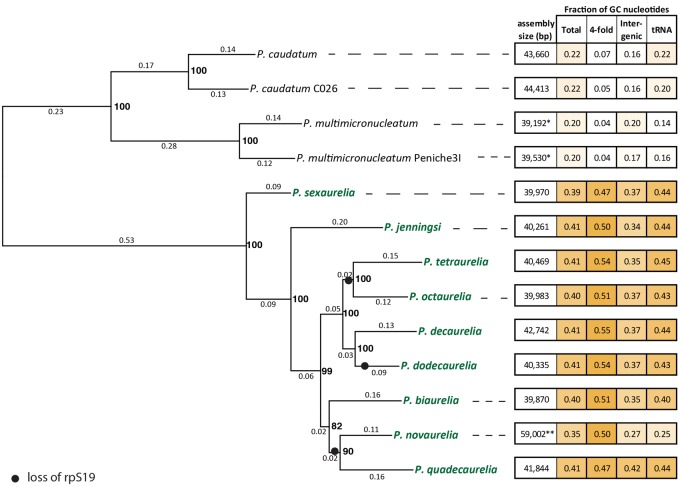
We assembled complete mitochondrial genomes of seven species belonging to the P. aurelia complex: P. biaurelia, P. tetraurelia, P. sexaurelia, P. octaurelia, P. novaurelia, P. decaurelia, P. dodecaurelia, P. quadecaurelia, and P. jenningsi. In addition, we analyzed previously reported complete mitochondrial genomes of P. tetraurelia and P. sexaurelia belonging to the P. aurelia complex, as well as four outgroup species: P. caudatum, P. caudatum-C026, P. multimicronucleatum, and P. multimicronucleatum-Peniche3I (fig. 1; isolates sequenced by Johri et al. [2017]). We note that P. caudatum-C026 and P. multimicronucleatum-Peniche3I were initially sampled as individual isolates belonging to the P. caudatum and P. multimicronucleatum species (based on morphological criteria), respectively. However, the analysis of the mitochondrial sequences revealed that they are highly diverged from the reference strains (see below) and are therefore almost certainly separate species and were treated as such in subsequent analyses. For all seven new mitochondrial genomes sequenced and assembled in this study, Illumina reads were assembled using SPAdes (Bankevich et al. 2012), and mitochondrial contigs were identified by BLAST searches against the publicly available P. caudatum and P. tetraurelia mitochondrial sequences (see the Materials and Methods section for more details).
Fig. 1.
—Mitochondrial phylogeny and change in nucleotide composition across Paramecium species. The species in green belong to the Paramecium aurelia complex. Numbers on branches indicate total number of substitutions per site and bold numbers on nodes show bootstrap values. Black solid circles show the inferred event of loss of the ribosomal protein rpS19. The numbers on the right show the total assembly size (in bp) and mean GC content of the whole mitochondrial genome (Total), at 4-fold degenerate sites, at intergenic regions, and of tRNAs, respectively. Starred (*) numbers indicate incomplete assembly of mitochondrial genomes.
In addition, we examined sequenced mitochondrial genomes of ten isolates of both P. tetraurelia and P. sexaurelia, and five isolates of each P. caudatum and P. multimicronucleatum (supplementary fig. 1, Supplementary Material online) sampled worldwide (Johri et al. 2017). Illumina paired-end reads from these isolates were mapped to the assembled reference genomes, and single nucleotide polymorphisms (SNPs) were called as in Johri et al. (2017). In this study, all mitochondrial genomes of individual isolates were also assembled de novo (supplementary figs. 2–4, Supplementary Material online) in order to examine large-scale genome organization mapping.
Genome Structure, Organization, and Telomeric Repeats
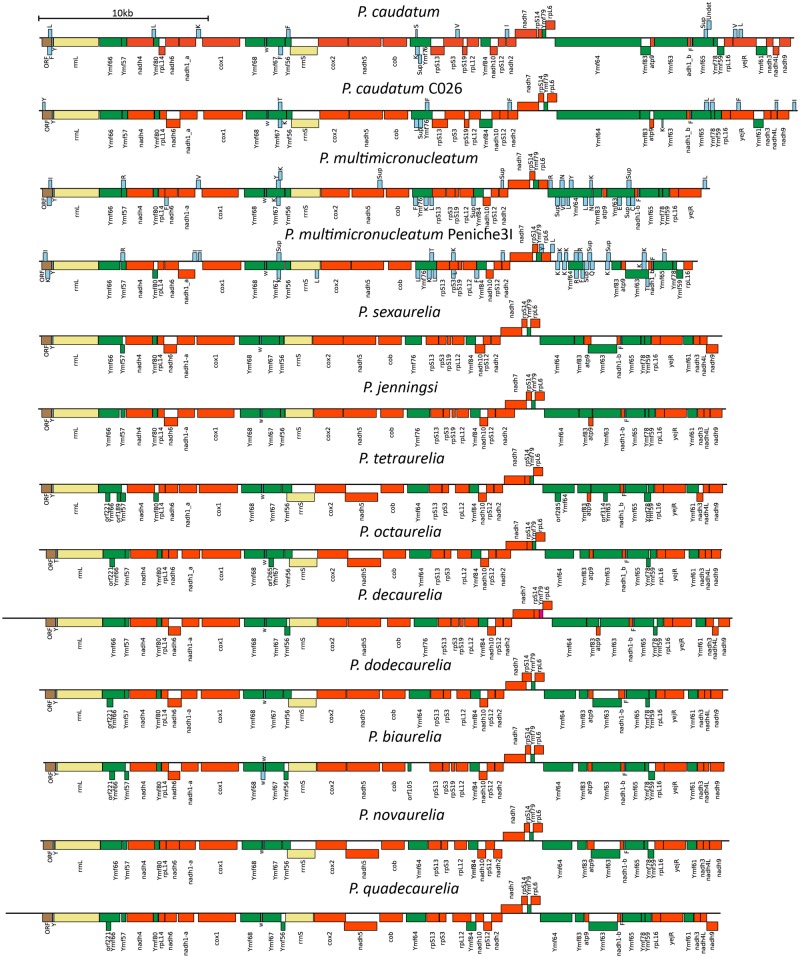
All Paramecium mitochondrial genomes are linear (Goddard and Cummings 1975; Morin and Cech 1988; Swart et al. 2012). Our assemblies show that they are ∼40 kb in the P. aurelia species and ∼44 kb in P. caudatum and P. caudatum-C026. Telomeric repeats and gene content observed at ends of the assembled genomes imply that the full lengths of the linear contigs have been assembled in most species with the exception of the P. multimicronucleatum and P. multimicronucleatum-Peniche3I mitocontigs, which appear to be missing small portions of the 3′-end of the chromosome (fig. 2). The length of the assembled contigs in P. multimicronucleatum and P. multimicronucleatum-Peniche3I suggests an overall size closer to that observed in P. aurelia than to the larger mitochondrial genomes in the P. caudatum lineage. We also note that the raw assemblies for two of the P. aurelia species contain extensions (supplementary figs. 3, 5, and 6, Supplementary Material online). In P. novaurelia, an additional ∼18 kb is present at the 5′-end of the mitocontig, whereas in P. quadecaurelia, a small, ∼1-kb extension is seen at the 3′-end. However, the read coverage over these regions is very different from the rest of the mitocontigs, suggesting either misassembly or heterogeneity within cell populations. We therefore ignored these extensions in subsequent analysis. The organization of the genomes is very similar, with gene order preserved almost perfectly across all species (fig. 2).
Fig. 2.
—Structure of mitochondrial genomes in Paramecium. All Paramecium mitochondrial genomes are linear with 24 protein-coding genes (shown in reddish orange), 2 rRNA genes (yellow), lineage-specific genes referred to as Ymf genes (in green) and varying numbers and content of tRNA genes (shown in light blue).
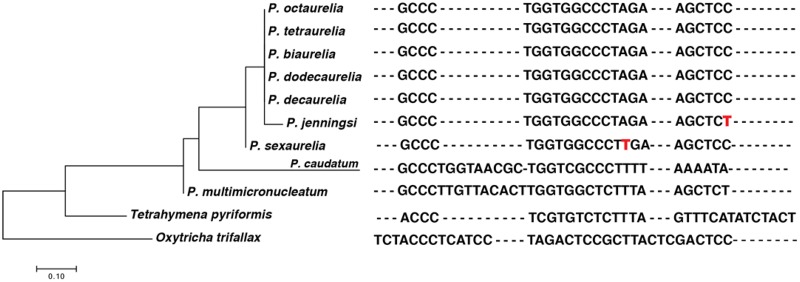
We also identified telomeric repeats based on the sequences at the end of assembled mitocontigs (see the Materials and Methods section for more details). In most P. aurelia species, we identified almost identical repeats, with a 23-bp consensus sequence GCCCTGGTGGCCCTAGAAGCTCC (fig. 3). However, in P. jenningsi and P. sexaurelia the telomere repeat motif is the same length, but differs from the consensus sequence in other species at one nucleotide position. Of note, these two species are the earliest diverging ones within the P. aurelia complex species included in our analysis. We observed even more divergent telomeric sequences in P. caudatum (GCCCTGGTAACGCTGGTCGCCCTTTTAAAATA) and P. multimicronucleatum (GCCCTTGTTACACTTGGTGGCTCTTTAAGCTCT). In these species, the core telomeric repeat sequence has been expanded by an additional 10 bp of sequence not present in P. aurelia. The Paramecium core telomeric repeat is broadly similar to that of Tetrahymena (ACCCTCGTGTCTCTTTA; fig. 3), the other Oligohymenophorean genus for which mitochondrial genomes are available, but distinct from what is observed in distantly branching ciliates such as the spirotrichean O. trifallax (fig. 3).
Fig. 3.
—Telomeric repeat sequences in the Paramecium genus. Nucleotides in red show the single base pair differences among the Paramecium aurelia species. The phylogenetic tree on the left is built using the telomeric repeat sequences.
Fig. 4.
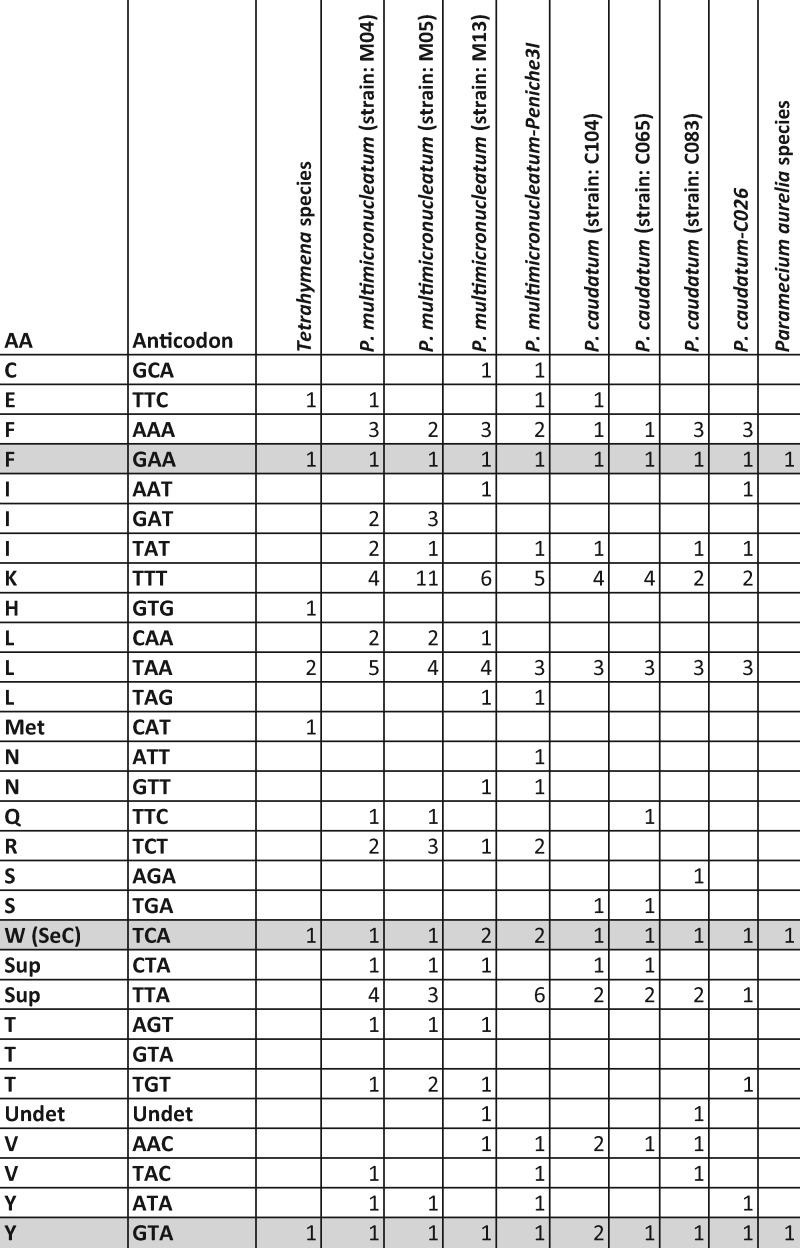
—tRNA content variation in Paramecium caudatum and Paramecium multimicronucleatum strains. tRNAs shaded in gray are present in all Paramecium and Tetrahymena species. tRNAs whose anticodons were ambiguous are displayed as “Undet.”
Examination of existing RNA-seq data sets for several of the species revealed no evidence for RNA editing in Paramecium mitochondria (data not shown), in concordance with previous reports (Orr et al. 1997).
Variation in Protein-Coding Gene Content between and within Species
All Paramecium mitochondrial genomes contain a core set of 15 genes (fig. 2) involved in electron transport and ATP synthesis (atp9, cob, cox1, cox2, nadh1, nadh2, nadh3, nadh4, nadh4L, nadh5, nadh6, nadh7, nadh9, and nadh10), the heme maturase YejR, and 8 ribosomal protein genes (rpL6, rpL12, rpL14, rpL16, rpS3, rpS12, rpS13, and rpS14). One ribosomal protein subunit rpS19 was found to be absent from P. tetraurelia, P. octaurelia, P. novaurelia, P. dodecaurelia, and P. quadecaurelia and therefore appears to have been independently lost from mitochondrial genomes at least three times along the P. aurelia phylogeny (fig. 1). Remarkably, we also find a presence–absence polymorphism of rpS19 within P. sexaurelia isolates, with the open-reading frame (ORF) not being identifiable in 2 out of 11 isolates studied (supplementary fig. 10, Supplementary Material online). Although this might suggest a mitochondrial gene in the process of transfer to the nucleus, the closest hit in the nuclear genome has an E-value of 0.27, implying either a complete loss or perhaps transfer to a mitochondrial plasmid, which has not been captured in existing assemblies.
Paramecium genomes also contain 16 ciliate-specific genes of unknown function, called Ymf genes (Burger et al. 2000), named according to the Commission on Plant Gene Nomenclature (Price and Reardon 2001). Each sequenced strain contains all of these genes, with the exception of P. multimicronucleatum strain M13 in which Ymf80 and Ymf83 appear to have fused with nadh4 and Ymf64, respectively. In addition, several ORFs originally identified in P. tetraurelia are observed in only one or a few additional P. aurelia mitochondrial genomes, typically overlapping longer ORFs: orf189, orf285, and orf314 restricted to the three P. tetraurelia isolates; orf221 present in P. tetraurelia, P. octaurelia, and P. biaurelia; orf265 in P. biaurelia; and orf78 found only in P. tetraurelia isolate A.
In addition, we found a previously unidentified ORF larger than 100 amino acids, which appears to be present in all Paramecium mitochondrial genomes and is located at the very 5′-end of the chromosome, immediately before the large ribosomal RNA. This ORF has homology to the clustered regularly interspaced short palindromic repeats (CRISPR)-associated endoribonuclease Cas6; its functional significance is unclear at present. Interestingly, an ORF homologous to CRISPR-associated helicase (Csf4) also has been identified in mitochondrial genomes of multiple bivalve species (Milani et al. 2013) and has been shown to produce a functional protein product (Milani et al. 2014). Thus, the ORF found in Paramecium mitochondrial genomes could potentially be functional and a case of horizontal gene transfer from some of the endosymbiotic bacteria (Preer 1969; Fokin and Gortz 2009) often associated with Paramecium. Sequencing of more ciliate mitochondrial genomes should provide deeper insights into its origin.
Paramecium Species Are Highly Diverged and Contain Cryptic Species Complexes
We found most species to be fairly evolutionarily distant from each other, as measured by the average number of nonsynonymous (dN) and synonymous (ds) site substitutions per site (supplementary fig. 11, Supplementary Material online) between all pairs of species, using yn00, PAML (Yang 2007; version 4.9a). Parameciumdecaurelia and P. dodecaurelia (average ds: 0.85; average dN: 0.05; average total divergence: 0.23) are the closest species pair, followed by P. tetraurelia and P. octaurelia (average ds: 0.93; average dN: 0.09; average total divergence: 0.27). All other species pairs exhibit on average ds > 1.0, that is, synonymous sites in protein-coding genes have on average undergone more than 1 substitution each since the time of divergence. We found that mean ds between P. caudatum and P. caudatum-C026 is ∼2.0 (average dN: 0.07; average total divergence: 0.28), implying that these two isolates possibly represent separate species. A similar observation confirms that P. multimicronucleatum-Peniche3I and P. multimicronucleatum are probably separate species (average ds = 1.97; average dN: 0.10; average total divergence: 0.26). Our observations suggest the possibility of cryptic species complexes in both P. caudatum and P. multimicronucleatum, as previously suspected (Hori et al. 2006; Tarcz et al. 2012). We also note that several additional P. multimicronucleatum isolates most closely related to P. multimicronucleatum-Peniche3I might also be separate species as they are fairly divergent from the reference strain as well as P. multimicronucleatum-Peniche3I (supplementary fig. 1, Supplementary Material online). The phenomenon of species complexes containing numerous morphologically identical species, already known for P. aurelia (Sonneborn 1937) and T. pyriformis (Gruchy 1955), is therefore probably much more widespread among ciliates than previously appreciated.
Selective Pressures on Ciliate-Specific Mitochondrial Genes
The 16 ciliate-specific mitochondrial genes found in all Paramecium species have orthologs in Tetrahymena species as well as in Oxytricha. Thus, these genes have been preserved for a long evolutionary time, and yet have diverged sufficiently that no known homologs exist in well-studied species in other eukaryotic kingdoms. Across the Paramecium species, they appear to be on average faster evolving (supplementary fig. 12 and table 1, Supplementary Material online) with average dN/ds ≅ 0.11, in comparison to genes that encode enzyme components of the respiratory chains (average dN/dS ≅ 0.04) and ribosomal proteins (average dN/ds ≅ 0.06). The dN/dS values here were obtained for each gene under the assumption that dN/dS remains constant across the phylogeny using CODEML, PAML (Yang 2007; version 4.9a). Ciliate-specific Ymf genes therefore exhibit a relatively higher rate of evolution than other genes, with as little as 30% sequence identity between P. aurelia, P. caudatum, and P. multimicronucleatum, similar to observations in Tetrahymena (Moradian et al. 2007), as well as with higher values of πn/πs in P. tetraurelia, P. sexaurelia, P. caudatum, and P. multimicronucleatum (supplementary fig. 13, Supplementary Material online).
The higher rate of evolution of Ymf genes can either be explained by relaxed purifying selection at the sequence level, or recurrent positive selection over long periods of time, or a combination of both. To distinguish between these possibilities, we performed a McDonald–Kreitman test (MK test) in the species P. tetraurelia, P. sexaurelia, P. caudatum, and P. multimicronucleatum. As most of these species are highly diverged from each other, we used ancestral reconstruction over the set of all 13 taxa to first infer ancestral nucleotides for each internal node. The numbers of synonymous (DS) and nonsynonymous (DN) changes were then inferred along each terminal branch leading to each of the four species mentioned above. We tested for positive selection by determining whether the proportion of nonsynonymous fixed differences relative to synonymous ones was significantly larger than the proportion of nonsynonymous relative to synonymous polymorphic differences. None of the genes in P. tetraurelia was found to have undergone positive selection by this criterion (Holm–Bonferroni corrected P ≥ 0.05 in each case). In P. sexaurelia, cox1, Ymf76, and Ymf63 are the only genes that showed significant positive selection, whereas in P. caudatum, only Ymf64 exhibits positive selection (supplementary table 2, Supplementary Material online). With very few SNPs available in P. multimicronucleatum, the MK test was not significant for any gene. Nevertheless, we find no statistical enrichment for number of genes evolving under positive selection among the Ymf relative to other genes in P. sexaurelia (P = 0.56; Fisher’s exact test) and P. caudatum (P = 0.40; Fisher’s exact test), suggesting that the majority of them are likely evolving fast due to relaxed sequence constraints.
It is possible that most of the Ymf genes are simply highly diverged genes that perform the same functions as genes found in other mitochondrial genomes, but are not identifiable by sequence. In order to gain some insight into their possible functions, we used HHPred (Soding et al. 2005) to predict their homologs and found that parts of Ymf genes display homology to standard mitochondrial genes, encoding ribosomal subunits and proteins involved in electron transport. About 37% of sequence from Ymf59 was found to be homologous to rpS10, ∼22% sequence of Ymf61 to a 30S ribosomal protein S24E, ∼28% of Ymf64 sequence to a 30S rpS3, ∼79% of Ymf65 sequence to the NADH subunit, and ∼11% of Ymf67 to cytochrome c oxidase, subunit P. Ymf59 and Ymf64 in Oxytricha were also previously found to be homologous to rpsS10 and rpS3, respectively (Swart et al. 2012).
tRNA Content Variation across and within Species
Although the number and identity of protein-coding genes are generally the same across the genus, tRNAs display significant variation. All species in the P. aurelia complex have only three tRNA loci, at highly conserved locations (fig. 2), most of the time consisting of a Y, F, and W tRNAs (the latter recognizing the UGA stop codon that has been reassigned to tryptophan in ciliate mitochondria). In stark contrast, large and highly variable sets of tRNA are predicted in mitochondrial genomes in the P. caudatum and P. multimicronucleatum lineages (fig. 3), with as many as 38 tRNA predictions in P. multimicronucleatum M05, and with individual P. caudatum and P. multimicronucleatum isolates having different tRNAs at multiple loci in their mitochondrial genomes. These tRNAs often overlap protein-coding genes (in contrast to what is observed in P. aurelia), in particular Ymf genes. Of note, the three tRNAs found in the P. aurelia species are also present in all outgroup species. The absolute numbers of the tRNAs found should be taken with caution, as some of the predicted tRNAs overlap each other as well as Ymf64 in the P. multimicronucleatum strains. However, even if these are excluded, the number of tRNAs in both P. caudatum and P. multimicronucleatum strains is still large and variable and consists of tRNAs not found in the P. aurelia species. This is a unique finding among mitochondrial genomes and should be investigated more thoroughly in future studies to better understand the origin of the tRNAs.
Mutation Spectrum and GC-Content Variation within the Paramecium Genus
Perhaps, the most remarkable discontinuity in Paramecium mitochondrial genomes is the difference between the high GC content observed in species of the P. aurelia complex—∼40%—and the low GC content in the outgroup species—∼20% (supplementary figs. 7–9, Supplementary Material online). Very low GC content is also observed in mitochondrial genomes of other ciliates, for instance, in the five Tetrahymena species (∼18–21%), in Icthyophthirius multifiliis (∼16%) (Coyne et al. 2011), and in O. trifallax (∼24%) suggesting a lineage-specific increase in GC content along the branch leading to P. aurelia species. In contrast, nuclear genomes have relatively similar levels of GC content across all Paramecium species (McGrath, Gout, Doak, et al. 2014). The low GC content of P. caudatum and P. multimicronucleatum mtDNA is marked by a highly biased codon usage (Barth and Berendonk 2011), with most synonymous positions exhibiting a strong bias for A or T nucleotides (supplementary fig. 14, Supplementary Material online). Indeed, the difference in GC content is most pronounced at 4-fold degenerate sites, with the GC content at such sites being as low as 3.5% in P. multimicronucleatum and as high as 54.5% in P. decaurelia (fig. 1). The 0-fold redundant sites in the outgroup species have much higher GC content (∼26–37%), and the GC content (31–36%) of the rRNAs is similar across the entire genus suggesting strong selection on rRNA nucleotide content.
At least three forces may be responsible for determining the GC content at 4-fold degenerate sites: 1) mutational processes, 2) codon usage bias due to selection, and/or 3) genome-wide selection for higher/lower GC content. We therefore examined GC composition in other regions of the genomes, such as tRNAs and intergenic regions, which do not experience selection for codon usage. Both tRNAs and intergenic regions have elevated GC content in the P. aurelia species (∼40–45% and ∼34–37%) relative to P. caudatum and P. multimicronucleatum (∼14–22% and ∼16–20%, respectively; fig. 1), suggesting that either differences in mutational bias or selection for genome-wide GC content is primarily responsible for changes in nucleotide composition across the genome.
To distinguish between mutation and selection as the possible explanations for these observations, we evaluated the mutational pressures acting on Paramecium species, using two approaches. First, we used the mitochondrial mutation spectrum obtained from mutation-accumulation (MA) studies of P. tetraurelia (Sung et al. 2012) to calculate the AT mutation bias, m = v/u, where v is the mutation rate from G/C to A/T and u is the rate of mutation from A/T to G/C (table 1). We found m ≅ 1.0, showing little mutation bias and corresponding to expected equilibrium GC content of 51.3% (table 1). This is very close to the GC content of 4-fold redundant sites (54%) in P. tetraurelia. We also analyzed existing sequencing data from MA experiments in P. biaurelia and P. sexaurelia (Long, Doak, et al. 2018; supplementary table 3, Supplementary Material online). These data imply slight mutation bias toward GC in P. biaurelia and P. sexaurelia, corresponding to expected equilibrium GC percentages (Sueoka 1993) of 55.8% and 67.0%, respectively. Given the relatively small number of mutations (a few tens) identified in each set of MA experiments, there is considerable uncertainty in these estimates. Nonetheless, these data do not present evidence that 4-fold degenerate sites in P. aurelia mitochondria are evolving away from mutation equilibrium via selection.
Table 1.
Mutation Bias toward AT (m) in the Mitochondrial Genomes of Paramecium and Other Species, Where v and u Are Mutation Rates from G/C to A/T, and A/T to G/C, respectively.
| Species | Type of Study | Total Number of Base-Substitution Mutations or SNPs | AT Mutation Bias (m = v/u) | Expected Equilibrium GC Content [1/(1 + m)] | Whole-Genome GC Content | 4-Fold Site GC Content | S (avg) | S (4-fold) |
|---|---|---|---|---|---|---|---|---|
| Parameciumtetraurelia1 | MA | Not known | 0.88 | 0.53 | 0.42 | 0.54 | 0.197 | −0.013 |
| Parameciumtetraurelia | Population | 37 | 0.95 | 0.51 | 0.42 | 0.54 | 0.163 | −0.047 |
| Parameciumbiaurelia2 | MA | 55 | 0.79 | 0.56 | 0.40 | 0.51 | 0.278 | 0.085 |
| Parameciumsexaurelia2 | MA | 87 | 0.49 | 0.67 | 0.39 | 0.47 | 0.504 | 0.368 |
| Parameciumsexaurelia | Population | 112 | 1.56 | 0.39 | 0.39 | 0.47 | 0.000 | −0.137 |
| Parameciumcaudatum | Population | 96 | 5.02 | 0.17 | 0.22 | 0.07 | −0.151 | 0.397 |
| Other species from previous studies | ||||||||
| Saccharomyces cerevisiae3 | MA | 13 | 0.00 | 1.00 | 0.17 | 0.08 | >>0 | >>0 |
| Caenorhabditis elegans4,5 | MA | 25 | 3.79 | 0.21 | 0.24 | 0.14 | −0.073 | 0.224 |
| Daphnia pulex6 | MA | 6 | 2.48 | 0.29 | 0.38 | 0.30 | −0.182 | −0.026 |
| Caenorhabditis briggsae7 | MA | 15 | 18.10 | 0.05 | 0.25 | 0.14 | −0.780 | −0.451 |
| Drosophila melanogaster8 | MA | 28 | 381.25 | 0.00 | 0.18 | 0.06 | −1.923 | −1.386 |
note.—Values of m <1 indicate that mutation spectrum is biased toward G/C. S = 4Nes (or 2Nes) for diploids (or haploids) represents the strength of selection favoring A/T composition, where negative values of S represent selection favoring G/C. S is calculated using the equation, PAT = 1/(1 + m−1e−S), where PAT is the fraction of AT nucleotides. 1: Sung et al. (2012); 2: Long et al. (2018); 3: Lynch et al. (2008); 4: Denver et al. (2000); 5: Konrad et al. (2017); 6: Xu et al. (2012); 7: Howe et al. (2010); 8: Haag-Liautard et al. (2008).
Next, derived singleton alleles at 4-fold degenerate sites were used to quantify the number of G/C to A/T mutations relative to A/T to G/C mutations and thus infer an estimate of AT mutation bias from population data. Again, we find that m is close to 1.0 (i.e., no mutation bias toward A/T) in both P. tetraurelia and P. sexaurelia, with the predicted GC content under mutation equilibrium being remarkably close to that of their 4-fold degenerate sites (table 1). These results suggest that the composition of 4-fold degenerate sites is mostly determined by mutation. The same can be seen by calculating the strength of selection (S = 4Nes) favoring A/T at 4-fold degenerate sites. As S usually takes values between 0.1 and 4.0 (Lynch 2007; Long, Sung, et al. 2018) and we obtain much lower magnitudes (0.0–0.5) in the P. aurelia species (table 1), we infer negligible or weak selection at 4-fold degenerate sites, consistent with absence of codon bias at the third position.
In contrast to the P. aurelia species, we infer a significant AT mutation bias in P. caudatum (table 1), yielding a predicted GC content at equilibrium of ∼17%, with the observed GC content in intergenic and tRNA regions (16% and 20%) being remarkably close to this value. These calculations suggest that genome-wide GC content in P. caudatum mitochondria is mostly governed by mutation bias, and that there has been a major shift in mutation bias along the branch leading to the P. aurelia species.
A possible confounding factor affecting these analyses is the presence of chunks of mitochondrial sequence in the nuclear genome, also known as NUMTs (nuclear mitochondrial DNA sequences). In general, the nuclear dualism of ciliates would be expected to result in few to no NUMTs being present in significant quantities as the highly polyploid macronucleus contributes nothing to future generations whereas the germline micronucleus is transcriptionally silent and heterochromatinized (thus less susceptible to insertions of NUMTs). Nevertheless, the possibility that NUMTs are present in Paramecium genomes cannot be dismissed, especially given that no micronucleus assemblies are available and even the macronucleus ones are not entirely complete. To account for this possibility, we carried out a parallel reanalysis of our data using a more conservative set of SNPs for all species, derived by mapping all sequenced reads to the mitochondrial reference genome and the macronuclear reference genome simultaneously, and subsequently excluding all sites that exhibit heterozygous genotypes (if NUMTs are the source of these variants, their genotypes would be heterozygous). We found that most variant positions and their allele frequencies do not change (supplementary table 4, Supplementary Material online) and that πn/πs values for all genes are highly correlated (R2 of 0.88, 0.99, and 0.67 in P. tetraurelia, P. sexaurelia, P. caudatum, respectively; supplementary fig. 15, Supplementary Material online). Most importantly, only a very slight change in the inferred mutation spectrum from population data is observed between the two sets of analyses, with the AT mutation bias in the conservative SNP set being 1.102 in P. tetraurelia (using 41 SNPs), 2.058 in P. sexaurelia (using 99 SNPs), and 6.479 in P. caudatum (using 30 SNPs).
Recombination in Paramecium Mitochondrial Genomes
Next, we sought to evaluate evidence for the occurrence of recombination in Paramecium mitogenomes by performing multiple tests for its presence. First, we evaluated the relationship between linkage disequilibrium (LD), calculated by r2 (Hill and Robertson 1968) and distance between sites. We find that r2does not decrease with distance in P. tetraurelia, P. sexaurelia, and P. caudatum (P. multimicronucleatum isolates lacked sufficient number of polymorphic sites; supplementary fig. 16, Supplementary Material online), consistent with the expectation under no recombination. Next, we conducted the four-gamete test (FGT) (Hudson and Kaplan 1985), which detects recombination by searching for pairs of polymorphic sites with all four segregating haplotypes and assumes that such pairs of sites must have arisen via recombination (under the infinite-sites model). We found all four gametes at 2 pairs of sites (1.45 × 10−5 of all pairs) in P. tetraurelia, 312,781 pairs (2.14 × 10−2 of all pairs) in P. sexaurelia, 42,647 pairs (1.21 × 10−2 of all pairs) in P. caudatum, and 0 pairs of sites in P. multimicronucleatum. The observed variation between species correlates well with levels of total sequence diversity and indicates the bias in power to detect recombination toward species with more sequence variation. Although results from the FGT suggest the presence of some recombination, the probability of finding all four gametes does not increase with the distance between the pairs of sites (slope= −7.64 × 10−9, P = 0.765 for P. sexaurelia; slope = −1.39 × 10−8, P = 0.792 for P. caudatum), contrary to the expectation under recombination, and is inconsistent with other distance-based analyses. Violation of the infinite-sites model (i.e., recurrent mutation at the same site) can result in the presence of pairs of sites with all four alleles. The FGT works best for species and genomes whose recombination rate is much larger than the mutation rate, which may not be the case in Paramecium mitochondria.
In order to test whether the presence of four gametes was more likely to be caused by recombination or mutation, we used LDhat (McVean et al. 2002) to test for recombination and estimate 2Ner, using values of 2Neμ estimated by nucleotide diversity as calculated by Johri et al. (2017) (supplementary table 5, Supplementary Material online). Here, r is the recombination rate, μ is the mutation rate, and Ne is the effective population size of the species under consideration. 2Ner estimated under the finite-sites mutation model (Wakeley 1997; Hudson 2001) was found to be 0.0 for all four species, and is consistently 0.0 in all nonoverlapping windows (1,000 bp) spanning the genome. We also used permutation tests (McVean et al. 2002) to detect recombination. The idea behind these tests is that in the absence of recombination the relative position of SNPs would not change inferred values of statistics used to measure recombination. Thus, in the absence of recombination, statistics like sum-of-distance between sites with all four gametes (G4) (Meunier and Eyre-Walker 2001), and correlation between r2 or D′ and physical distance, would not be significantly different from an expectation obtained by randomly shuffling SNPs and computing these statistics. We find no statistical significance for the presence of recombination using permutation tests (supplementary table 5, Supplementary Material online) and thus conclude that there is no recombination in Paramecium mitochondria. It should be noted that the final sets of SNPs used to infer the presence of recombination consist of 526 SNPs in P. tetraurelia, 5,401 SNPs in P. sexaurelia, 3,353 SNPs in P. caudatum, and 545 SNPs in P. multimicronucleatum. These sets of SNPs are not only large in absolute terms, but are also uniformly distributed across the genome (supplementary fig. 16, Supplementary Material online), providing a lot of power to detect any recombination.
Efficacy of Selection in Mitochondria versus the Nucleus
Finally, we asked whether mitochondrial genes experience weaker efficacy of purifying selection compared with nuclear genes, as would be expected due to smaller effective population size and the lack of recombination in the mitochondria. We did so by comparing multiple statistics in each of the two genomes. Because our statistics included divergence at synonymous sites which are saturated for most species pairs, we conducted these analyses primarily on P. tetraurelia where values of divergence were calculated with respect to the closest outgroup species (ds), P. biaurelia, for which we had available sequences in both the nucleus and mitochondria. Average ds values for the set of nuclear and mitochondrial genes were found to be 0.885 and 1.174, respectively (table 2), a small but significant difference. In order to control for differences in ds driving the patterns, we also conducted all analyses restricted to genes with ds < 1.0. For the set of genes with ds < 1.0, the average value of ds is not significantly different between the two genomes, 0.636 among the nuclear and 0.658 among the mitochondrial genes. A potential caveat of comparing the efficacy of purifying selection between all genes present in the mitochondria versus nucleus is that we might instead be measuring differences in strength of purifying selection. In order to correct for that, we also compared nuclear genes with similar functions to those in the mitochondria. We compared the 14 mitochondrial genes with ∼13–27 (of total 87) nuclear genes involved in the oxidative phosphorylation (OXPHOS) pathway, as well as the 8 ribosomal genes in the mitochondria with ∼77–430 (of total 585) nuclear genes that encode for structural components of the ribosomes.
Table 2.
NI of Mitochondrial and Nuclear Genes in Paramecium tetraurelia, Calculated Using Multiple Statistics
| All (excluding Ymf) | dS < 1.0 | OXPHOS | Ribosomal | |
|---|---|---|---|---|
| dS | ||||
| Nuclear | 0.885 | 0.636 | 0.774 | 0.404 |
| Mitochondrial | 1.174 | 0.658 | 1.324 | 1.581 |
| P | 2.74 × 10−6 | 0.807 | 0.035 | 4.22 × 10−5 |
| dN/dS | ||||
| Nuclear | 0.079 | 0.086 | 0.029 | 0.088 |
| Mitochondrial | 0.082 | 0.142 | 0.052 | 0.044 |
| P | 0.307 | 0.210 | 0.208 | 0.208 |
| πn/πs | ||||
| Nuclear | 0.235 | 0.235 | 0.200 | 0.092 |
| Mitochondrial | 0.163 | 0.180 | 0.127 | 0.062 |
| P | 0.183 | 0.618 | 0.223 | 0.387 |
| NIπ [(πn/πs)/(dN/dS)] | ||||
| Nuclear | 4.018 | 3.436 | 6.712 | 1.653 |
| Mitochondrial | 2.294 (2.495) | 1.218 | 2.384 | 2.248 |
| P | 0.124 (0.537) | 0.024 | 0.422 | 0.262 |
| NI [(Pn/Ps)/(Dn/Ds)] | ||||
| Nuclear | 2.145 | 2.271 | 6.087 | 2.873 |
| Mitochondrial | 1.528 (1.855) | 1.294 | 1.696 | 2.195 |
| P | 0.272 (0.817) | 0.208 | 0.018 | 0.072 |
| NIDoS [Dn/(Dn+Ds)] – [Pn/(Pn+Ps)] | ||||
| Nuclear | −0.041 | −0.053 | −0.227 | −0.136 |
| Mitochondrial | 0.026 (0.024) | 0.046 | 0.001 | 0.073 |
| P | 0.272 (0.738) | 0.208 | 0.010 | 0.072 |
note.—All P values (corrected for multiple testing by Holm–Bonferroni method) represent comparisons between nuclear and mitochondrial statistics. Statistically significant scores (P < 0.05) are underlined. Values in round brackets refer to analyses done with the set of genes excluding Ymf genes.
Average dN/ds is similar or slightly lower for genes in the nucleus (mt = 0.082, nuc = 0.079, P = 0.307; table 2), with the exception of the well-conserved ribosomal genes, which have lower dN/ds in the mitochondria (mt = 0.044, nuc = 0.088, P = 0.208). None of these differences in dN/ds is significant, suggesting that the average amount of purifying selection experienced by genes in the nucleus is not significantly different from that experienced by those in the mitochondria.
The efficacy of purifying selection can be measured by estimating the fraction of segregating nonsynonymous polymorphisms that undergo fixation. Such a measure can be calculated using the neutrality index (NI = [Pn/Ps]/[Dn/Ds], where Pn and Ps are the number of nonsynonymous and synonymous polymorphisms, respectively; Dn and Ds are the number of nonsynonymous and synonymous fixed differences, respectively). The NI is usually found to be larger than 1.0 for genes experiencing purifying selection because mildly deleterious variants are allowed to segregate among individuals, but rarely fix in populations. Higher values of the NI suggest that smaller proportion of segregating nonsynonymous variants are allowed to fix in the population, indicating stronger efficacy of purifying selection. The number of branch-specific substitutions at synonymous (Ds) and nonsynonymous (Dn) sites was calculated by ancestral reconstruction, using P. tetraurelia, P. biaurelia, P. sexaurelia, P. caudatum, and P. multimicronucleatum, for both the nucleus and the mitochondria, and restricted to sites for which changes could be confidently inferred along specific branches. The advantage of using ancestral reconstruction to infer Dn and Ds is that particular sites that are very fast evolving can be excluded from the analysis, thus minimizing problems arising due to saturation of divergence at synonymous sites. Comparing the distributions of values of NI in the mitochondrion and nucleus shows that overall there is no significant difference between the two sets when all genes are included (whether we include or exclude Ymf genes) in P. tetraurelia (table 2). However, for genes involved in OXPHOS, nuclear genes appear to be experiencing significantly decreased efficacies of purifying selection than mitochondrial genes (mt NI = 1.696, nuc NI = 6.087, P = 0.02).
We also calculated the NI as (πn/πs)/dN/ds) (Betancourt et al. 2012; denoted by NIπ below) for each gene with dN/ds obtained from pairwise comparison with respect to P. biaurelia. In this case all sites contribute to the analysis, but the maximum likelihood estimate of pairwise dN/ds can minimize biases due to saturation of ds. Again, we observe that NI of nuclear genes is either similar or higher than those of mitochondrial genes. Genes involved in OXPHOS in the nucleus have higher but not significantly different NIπ compared with those in the mitochondria (mt = 2.384, nuc = 6.712, P = 0.422).
In order to minimize statistical biases that can arise due to sampling, we also estimated a variation of the NI: NIDoS = Dn/(Dn + Ds) − Pn/(Pn+Ps) (Stoletzki and Eyre-Walker 2011), where more negative values represent stronger efficacy of purifying selection. As previously observed, we find no significant difference between the NI of genes in the nucleus versus mitochondria except for OXPHOS genes, where nuclear genes have significantly lower values of NIDoS than mitochondrial genes (mt NIDoS = 0.001, nuc NIDoS = −0.227, P = 0.01)
This suggests that there are more deleterious variants segregating in the nucleus than in the mitochondrion in P. tetraurelia. Overall, mitochondria appear to experience either similar or stronger efficacy of purifying selection than the nucleus.
Lastly, we used πn/πs as a proxy for the efficacy of recent purifying selection. This comparison can be extended to all four species (supplementary table 6, Supplementary Material online). We find that average πn/πs of mitochondrial (mt) and nuclear genes (nuc) is not significantly different in P. tetraurelia (mean mt = 0.163; mean nuc = 0.235; P = 0.18) and P. caudatum (mean mt = 0.135; mean nuc = 0.170; P = 0.22), respectively. However, mean πn/πs in the mitochondrial genes is significantly lower than that of nuclear genes in P. sexaurelia (mean mt = 0.051; mean nuc = 0.268; P < 2.2 × 10−16) and P. multimicronucleatum (mean mt = 0.099; mean nuc = 0.178; P = 1.87 × 10−3), respectively, suggesting that mitochondrial genes might be under stronger purifying selection than nuclear genes.
As a side note, values of NI for P. sexaurelia, P. caudatum, and P. multimicronucleatum were consistently found to be much less than 1.0, likely due to underestimation of changes at synonymous sites. We aimed to reduce the bias caused by saturation of DS by recalculating NI only in the mitochondrion using all 13 taxa (in order to break up longer branches). We continue to recover extremely low values of NIs (supplementary table 7, Supplementary Material online) and do not obtain values close to those obtained using NIπ, suggesting that the absolute values of NI can be misleading and have to be interpreted with caution.
Discussion
In this study, we greatly expand the set of sequenced ciliate mitochondrial genomes by presenting a wider sampling of the mitogenome diversity within the Paramecium genus. Using this wealth of sequence data, we characterize the diversity and conservation of genome organization and gene content, and we study in depth the population genetic characteristics such as mutational and selection pressures acting on mtDNA within the genus.
Ciliate-Specific Mitochondrial Genes
Paramecium mitochondrial genomes possess 16 lineage-specific ORFs (referred to as Ymf genes) that have no known homologs in nonciliate species and lack assigned functions, but are nonetheless conserved across Paramecium and Tetrahymena species. Other ciliates like Oxytricha have also been found to have unidentified ORFs (Swart et al. 2012), but not all Ymf genes have homologs identified in ciliates outside of Oligohymenophora (which contains both Tetrahymena and Paramecium). We find that Ymf genes are evolving faster, mostly due to relaxed purifying selection, as Ymf genes are not significantly more likely to undergo positive selection than other genes. Shedding light on the identity of the Ymf genes could possibly indicate entirely new sets of genes present in the ancestor of Oligohymenophorean mitochondrial genome.
We found that parts of five of the 16 Ymf genes show homology to standard mitochondrial proteins, especially ribosomal proteins. As ciliates are evolutionarily distant from most model organisms in the eukaryotic tree, it is possible that some Ymf genes are ribosomal subunits or genes belonging to the NADH complex and are simply not identifiable because of being highly diverged. Similar findings have been reported for other protozoan mitochondrial genomes in the past (de Graaf et al. 2009; Pombert et al. 2013; Burger et al. 2016; Skippington et al. 2017) as well as some nonprotist species like bivalve molluscs and cnidarians (Shao et al. 2006; McFadden et al. 2010; Kayal et al. 2012; Milani et al. 2013) that have diverged substantially from the most well-studied mammalian mitochondrial genomes. In some species, RNA editing (including insertions and deletions) can be substantial (Gray 2003), which can mask the proteins encoded from identification from genomic sequence data. We, however, found no evidence of RNA editing, consistent with a previous report in P. tetraurelia mitochondria (Orr et al. 1997), thus it is unlikely to account for the observed divergence. An interesting possibility is that most of the Ymf genes are either part of or interact with the ATP synthase complex. In Tetrahymena thermophila, the ATP synthase has been reported to form an unusual structure possessing completely novel subunits whose orthologs are not identifiable in other organisms, some of which are Ymf genes (Balabaskaran Nina et al. 2010). It is therefore possible that ciliates possess structurally unique proteins that perform relatively conventional functions in the mitochondria, but are difficult to identify based on other known sequences. Although the origin of these genes is unclear, it is intriguing that a set of such fast evolving genes is preserved across highly diverged species.
Change in Mutation Spectrum and Nucleotide Composition
We used a combination of previous MA studies and our population-genomics data to determine that the change in nucleotide composition of P. aurelia mitochondrial genomes toward higher GC is most likely the result of changes in mutational biases. Unfortunately, due to our modest sample sizes, some of the SNPs observed as singletons in our data may in fact be fairly common in the population at large. Our estimates of mutation spectra might thus be biased by selection. MA lines in P. caudatum would help further disentangle these forces. Because ciliates have very low mutation rates, their MA study requires a large number of generations to produce only a handful of mutations, making it very difficult to obtain precise estimates of the mutation spectrum. The most feasible strategy to more precisely estimate these spectra in Paramecium would thus be to acquire larger population samples in order to observe lower-frequency variants. We also note that although an alternative explanation for higher GC content in the P. aurelia species could be biased gene conversion, we found no evidence of mitochondrial recombination (including noncrossovers) in the P. aurelia species.
An interesting question raised by our results is how fast the mutation spectrum in mitochondria evolves across species (Lynch et al. 2008; Montooth and Rand 2008). Reanalyzing data from previous work on MA lines in mitochondria of other model organisms presents a number of interesting observations (table 1). First, within opisthokonts, there is a huge variation in mutation bias (m) in the mitochondria, ranging from nearly 0 (strongly biased toward GC) to values much larger than 1 (biased toward AT; table 1). However, most species except Saccharomycescerevisiae have a mutation bias toward A/T, consistent with most mitochondrial genomes being AT-rich. Second, across opisthokonts, we find that selection can have a significant impact on mitochondrial GC content. The effect of selection (S) can be observed by comparing observed genome-wide nucleotide composition (or specifically at 4-fold degenerate sites) with the expected composition under mutation equilibrium (table 1). Again, in most species selection favors higher G/C, but in S. cerevisiae there appears to be strong selection favoring A/T genome-wide. Thus, there might not be a universal direction for mutation bias or selection for nucleotide composition in mitochondria. Finally, closely related species in other lineages have been shown to have significant differences in mutation bias in the mitochondria as observed for example between Caenorhabditis elegans and Caenorhabditis briggsae (mutation bias recalculated by combining observations from Howe et al. [2010] and Konrad et al. [2017]). All of these observations suggest that it may not be such an unusual event for the mutational biases to shift away from AT along the long branch leading to the P. aurelia species. However, the exact nature of the biochemical mechanisms responsible for remains an intriguing open question for future research.
Changes in mutation spectra could occur due to environmental variables resulting in differential mutagenic pressures, or due to the differences in DNA repair processes. However, we note that nuclear GC composition is very similar across all Paramecium species; the change in GC composition in the P. aurelia complex has only occurred in the mitochondrial genome. This quite strongly suggests that environmental conditions are unlikely to account for the observed differences, otherwise we should expect to find concordant differences between nuclear genomes. In addition, P. caudatum and the P. aurelia species very often co-occur in the same lakes, sometimes even from the same sampling site. Changes in mitochondrial DNA repair systems along the P. aurelia branch are therefore more likely to be the explanation. One possibility is that the nuclear whole-genome duplication events that happened prior to the P. aurelia radiation have resulted in an expansion in the number and variety of DNA repair enzymes, which has resulted in changes in the DNA repair systems operating in mitochondria specifically. Indeed, a number of genes functioning in DNA repair processes are retained in more copies in the P. aurelia than in P. caudatum nuclear genomes (supplementary table 8, Supplementary Material online). However, whether any of these proteins have been neo- or subfunctionalized to play a role in mitochondrial DNA repairs cannot be determined at present due to the very limited knowledge of DNA repair mechanisms in ciliates in general, and in their mitochondria more specifically; further experimental studies will be needed to answer these questions.
Similar Efficacy of Purifying Selection Experienced by the Mitochondria and Nucleus
Mitochondrial genomes are often nonrecombining and are usually passed on via uniparental inheritance. Mitochondria are therefore expected to have lower effective population sizes than that of the nucleus within the same organism (Lynch and Blanchard 1998; Neiman and Taylor 2009). One consequence of reduced effective population sizes is an increased probability of segregation and fixation of slightly deleterious mutations. There have been multiple contradictory reports on whether mitochondrial genes experience stronger or weaker purifying selection than the nuclear genes. Studies examining a small number of protein-coding loci across a large number of species have concluded that mitochondria experience less effective purifying selection than the nucleus (Weinreich and Rand 2000; Betancourt et al. 2012; Popadin et al. 2013). However a recent study conducted on multiple individuals and whole genomes in Drosophila melanogaster and humans found no significant difference between the efficacy of purifying selection in the mitochondrial versus nuclear protein-coding genes (Cooper et al. 2015).
We confirm the absence of recombination in mitochondria of three species of Paramecium. Previous studies in P. tetraurelia (Adoutte et al. 1979; Barth et al. 2008) and P. primaurelia (Beale et al. 1972) had reached similar conclusions using a small number of markers. It should be noted that we do not detect recombination with extant sequence variation, which is consistent with the observed uniparental inheritance in Paramecium species. However, the lack of fusion of mitochondria presents a barrier to recombination within the cell. Thus, even if recombination was occurring at an extremely low rate, it might be difficult to detect it. Despite the lack of recombination in the mitochondria, the efficacy of selection in the mitochondria is similar to if not stronger than that of the nucleus.
Our results seem to be in discordance with theoretical predictions according to which one would expect the efficacy of selection in the mitochondria to be lower than in the nucleus. One possibility is that because there are multiple copies of mitochondrial genomes within a cell, recessive deleterious mutations might not produce a phenotypic effect unless they reach sufficiently high frequencies. A subsequent bottleneck could result in either transmission of highly fit mitochondrial mutations, or possibly the most deleterious. This increase in variance of fitness of individual cells would result in efficient selection between cells, eventually leading to stronger purifying selection (Stewart et al. 2008; Wai et al. 2008; Ghiselli et al. 2013; Stewart and Larsson 2014). We might thus expect mitochondrial genes to experience more efficient purifying selection than the nuclear genes. However, the severity of mitochondrial bottleneck in Paramecium is not yet clear (see below). Another possibility is that we observe stronger efficacy of selection in the mitochondria because the magnitude of negative selection itself is stronger in the mitochondrial genes (Popadin et al. 2013), which differ from the average nuclear gene in several key ways (Adrion et al. 2016). Most mitochondrial genes are expressed at higher levels compared with nuclear genes (Havird and Sloan 2016). Mitochondrial encoded proteins (cox1 and cox2) that are part of the OXPHOS pathway are core enzyme catalytic subunits (Tsukihara et al. 1996; Zhang and Broughton 2013; Havird and Sloan 2016). Finally, most genes retained in the mitochondria encode for highly hydrophobic proteins and have a high GC content (Johnston and Williams 2016). Thus, finding comparable sets of genes between these two genomes is admittedly difficult (but see Lynch 1996; Lynch 1997).
Lastly, our results need to be evaluated in the light of Paramecium-specific life cycle. In Paramecium, there is almost no exchange of cytoplasm during conjugation (Koizumi and Kobayashi 1989; Meyer and Garnier 2002). Thus, it appears that both parents pass on their mitochondria to their respective offspring, without exchange or degradation of mitochondrial genomes. In addition, mitochondria double before binary fission (Perasso and Beisson 1978) and appear to be distributed randomly across the daughter cells. Evidence of the latter comes from experiments where P.aurelia cells were injected with two different mitochondrial genotypes, and were proliferated in nonselective media. Paramecium cells were observed to maintain the two populations stably if the two mitochondrial genotypes were equally fit when present as pure populations in cells (Adoutte and Beisson 1972; Adoutte et al. 1979). On the other hand, if one of the genotypes was less fit than the other, it would be lost, with the time of loss being proportional to the fitness difference (Adoutte and Doussiere 1978). These studies provide indirect evidence for equal and random distribution of mitochondria in the two daughter cells. Thus, mitochondria do not appear to undergo bottlenecks during any stage of the life cycle in Paramecium, and experience no associated reduction in effective population size. On the other hand, Paramecium species frequently undergo asexual reproduction, thereby reducing the nuclear effective population size. Therefore, effective population sizes of mitochondrial and nuclear genomes may be more similar in Paramecium than in many other organisms. Due to the combination of sexual and asexual reproduction, under equilibrium conditions, expected heterozygosity in the nucleus would take values between 4Neμn – 2Neμn. Similarly, in the mitochondria, expected hetrozygosity would be ∼2Neμm. Assuming that the ratio of divergence at silent sites can be used as a proxy for the ratio of mutation rate between the two compartments, it is possible to approximately estimate the ratio of effective population sizes of the two compartments as Ne(m)/Ne(n) = y × (πm/πn)/(dm/dn), where y would be some number between 1 and 2. For P. tetraurelia, for which we have relatively closer outgroup species and thus more reliable divergence estimates, our estimated range of Ne(m)/Ne(n) is 0.94–1.88. Although underestimation of neutral divergence in the mitochondria relative to the nucleus could skew our inference slightly, the above calculation suggests that effective population sizes of mitochondria may be similar or, larger than that of the nucleus in Paramecium.
We therefore conclude that our finding of similar or stronger efficacy of selection in the mitochondria relative to the nucleus in Paramecium may lie within theoretical expectations given Paramecium’s unique life cycle and mode of mitochondrial transmission. A better understanding of the Paramecium life cycle in the wild might help build more appropriate null expectations in the future. Our results suggest the possibility that unicellular eukaryotes in general may have larger mitochondrial than nuclear effective population sizes and more efficacious purifying selection in the mitochondria might be more common than believed.
Materials and Methods
Genome Sequencing and Assembly
Single isolates of P. jenningsi (strain: M), P. octaurelia, P. decaurelia (strain: 223), P. dodecaurelia (strain: 274), P. novaurelia (strain: TE), and P. quadecaurelia (strain: 328) were used to extract macronuclear DNA. DNA extraction, sequencing library preparation, and genome sequencing were previously described (Johri et al. 2017). Sequencing reads were assembled using SPAdes (Bankevich et al. 2012; version 3.5.0) after removing potential adapter sequence with Trimmomatic (Bolger et al. 2014; version 0.33). Mitochondrial contigs were identified from the resulting assemblies by BLAST (Altschul et al. 1997) searches against the published P. caudatum and P. tetraurelia mitochondrial genomes.
SNP Detection
Whole-genome sequencing raw reads from five to ten isolates of P. tetraurelia, P. sexaurelia, P. caudatum, and P. multimicronucleatum were downloaded from SRA (SRA accession: SRR8698631–SRR8698604; Bioproject: PRJNA525710; Biosample: SAMN11059622–SAMN11086832), and SNPs were called as described by Johri et al. (2017), using reference genomes of strain 99 for P. tetraurelia, strain 130 for P. sexaurelia, C104 for P. caudatum, and M04 for P. multimicronucleatum. Briefly, reads were trimmed using Trimmomatic (version 0.36) (Bolger et al. 2014) and mapped to reference genomes using bwamem (0.7.12) (Li and Durbin 2010) under default parameters. Duplicate reads were marked using picard (2.8.0) (https://broadinstitute.github.io/picard/). Sites were only considered for further analysis if the mapping quality was above 30, base quality was above 20, per-base alignment quality was above 15, and the sum of the depth of coverage for all individuals was about five times the number of individuals and less than twice the average population coverage. Variants were called using bcftools (Li et al. 2009) and filtered using vcftools (Danecek et al. 2011). Only those sites were considered whose quality value (–minQ) was above 20. Genotypes whose genotype quality score (–minGQ) was <30 or those that were supported by <4 reads (–minDP) were excluded or considered missing.
Mitochondrial Genome Annotation
Genome annotation was carried out as follows. Protein-coding genes were identified by generating all ORFs longer than 60 amino acids in all six frames, using the Mold, Protozoan, and Coelenterate Mitochondrial Code (i.e., UGA codes for W instead of being a stop codon) and all alternative start codons specific to Paramecium (AUU, AUA, AUG, AUC, GUG, and GUA), and retaining the longest ORFs associated with each stop codon. BlastP was then used to identify homologs of annotated mitochondrial proteins in P. tetraurelia and P. caudatum. Additional ORFs were identified by imposing the requirement that their length exceeds 100 amino acids, and subsequently annotated using BlastP against the nonredundant protein sequences (nr) database and HMMER3.0 (Eddy 2011) scans against the PFAM 27.0 database (Finn et al. 2014). tRNA genes were annotated with tRNAscan-SE (Schattner et al. 2005; version 1.21), using the “Mito/Chloroplast” source. rRNA genes were identified using Infernal (Nawrocki et al. 2009; version 1.1.1).
Identification of Telomeres
Telomeric repeats were identified as follows. The first and the last 200 bp of each de novo assembled mitochondrial genome were used as input to the MEME de novo motif finding program (Bailey et al. 2009; version 4.6.1), which was run with the following parameters: -maxw 25 -dna -nmotifs 5 -mod anr. The repetitive units defined that way were then manually aligned to each other and refined to arrive at final telomeric repeats comparable across all species.
RNA-Seq Analysis
For each species, sequencing reads were aligned against a combined Bowtie (Langmead et al. 2009) index containing both the nuclear and mitochondrial genomes using TopHat2 (Kim et al. 2013; version 2.0.8) with the following settings: –bowtie1 –no-discordant –no-mixed –microexon-search –read-realign-edit-dist 0 –read-edit-dist 4 –read-mismatches 4 –min-intron-length 10 –max-intron-length 1000000 –min-segment-intron 10 –min-coverage-intron 10. Custom python scripts were then used to identify sequence variants relative to the mitogenomes assemblies.
Building Phylogenetic Trees
Nucleotide sequences were extracted, aligned using MUSCLE, and concatenated. Missing data were encoded as “N.” RAxML was used to build the tree with GTRGAMMA as the substitution model. Bootstrap values for 1,000 replicates were obtained via the fast method recommended by RAxML with the following command line:
raxmlHPC -f a -s sequences.fasta –n sequences_boot -m GTRGAMMA -T 50 -p 31 -x 7777 -N 1000
Estimation of dN/dS, Dn, Ds, Pn, Ps, and πn/πs
dN/dS was estimated across the phylogeny using CODEML, PAML (Yang 2007; version 4.9a) and for the P. aurelia species it was also estimated pairwise with respect to the closest outgroup species (denoted as dN/dS) using yn00, PAML. πn/πs was obtained for all protein-coding genes in the four species—P. tetraurelia, P. sexaurelia, P. caudatum, and P. multimicronucleatum, where total number of changes in synonymous sites was >1. This filter was executed in order to reduce errors in πn/πs due to very low values of synonymous polymorphisms in a gene.
D n and Ds, the number of nonsynonymous and synonymous changes, were inferred by performing ancestral reconstruction at each site, and then counting branching-specific substitutions. Ancestral reconstruction (PAML, baseml, GTR model) was conducted over the phylogeny of all available taxa. For comparing statistics between mitochondrial and nuclear genes, the ancestral reconstruction was performed over the same set of taxa for both, that is, over P. tetraurelia, P. sexaurelia, P. caudatum, and P. multimicronucleatum. For all analyses that involved ancestral reconstruction, only sites whose posterior probability of the inferred ancestral state ≥ 0.85 were used in the analyses—this filter was imposed for counting synonymous and nonsynonymous polymorphisms (Ps and Pn, respectively) as well as divergent sites (Dn and Ds). There may be some concern that ancestral reconstruction could end up biasing the ratio of Dn/Ds as many more changes at synonymous sites might result in lower confidence in inferring ancestral states at synonymous but not nonsynonymous sites. Such a bias would increase Dn relative to Ds and thus decrease values of NI. Therefore, all analyses involving Dn and Ds were also performed including all sites, with no filter, and results remained unchanged.
Calculation of Multiple Estimators of NI and Statistical Tests
Several estimators of NI have been proposed in order to counter different biases. The estimators we used were calculated as follows:
Simple neutrality index, NI = (Pn/Ps)/(Dn/Ds) (Rand and Kann 1996).
NIπ = (πn/πs)/(dN/dS), where dN/dS was calculated pairwise, with respect to closest outgroup species (Betancourt et al. 2012).
NITG = Σi [Dsi.×Pni /(Psi + Dsi)] /Σi [Psi.×Dni /(Psi + Dsi)], where i is the ith gene (Tarone 1981; Greenland 1982).
NIDoS = [Dn/(Dn + Ds)] − [Pn/(Pn + Ps)] (Stoletzki and Eyre-Walker 2011).
For MK test (McDonald and Kreitman 1991), Fisher’s exact test in R (R-Core-Team 2014) was used to test significance. In all cases, P value was corrected by Bonferonni–Holm method (Holm 1979) for multiple tests, using R.
Mutation Spectrum from MA Lines
MA line experiments for P. tetraurelia had previously been published (Sung et al. 2012), and results from their analysis were used directly in this study.
MA experiments carried out in order to obtain nuclear mutation rates in P. biaurelia and P. sexaurelia (Long, Doak, et al. 2018) were reanalyzed as follows. Sequencing reads were assembled for each MA line individually and mitochondrial contigs identified as described above. A composite consensus mitochondrial genome sequence was then created from the individual assemblies by creating multiple sequence alignments of all mitochondrial contigs using MAFFT (Katoh and Standley 2013; version 7.221) and retaining the most frequent base for each alignment column (with the exception of telomeres, which were manually curated). Adapter-trimmed reads were then aligned in a 2 × 100 bp format against a combined Bowtie index, containing a combination of the nuclear and consensus mitochondrial genomes, allowing for up to three mismatches and retaining only unique reads. Putative mutations were identified by requiring that any variant is supported by at least three nonredundant read pairs on each strand, is supported by not more than four times more reads on one strand than on the other, and is also observed in ≥5% of reads covering a given position. Telomeric sequences were excluded due to an excessively high number of sequence variants observed in those regions.
Mutation Spectrum from Population Genomics Data
For each SNP, the ancestral allele was inferred by performing ancestral reconstruction on the 13-taxa phylogeny to predict the nucleotides on internal nodes (see above). The ancestral allele was used to infer the derived allele segregating in P. tetraurelia, P. sexaurelia, P. caudatum, and P. multimicronucleatum. This analysis was restricted to sites where the ancestral nucleotide was inferred with confidence score ≥0.90, where the derived allele was present in only a single individual, that is, was a singleton, and was at 4-fold degenerate site. Of these, we counted all mutations that were from G/C to A/T or from A/T to G/C, with respect to the total number of utilizable sites that were counted according to the same criteria as above. Care was also taken to remove all sites that were part of overlapping ORFs.
Calculation of Bias in Mutation Spectrum
Mutation rate of A/T → G/C (u) and G/C → A/T (v) mutations was calculated as follows:
u = (number of A/T → G/C mutations)/(total number of utilizable A/T sites).
v = (number of G/C → A/T mutations)/(total number of utilizable G/C sites).
Mutation bias toward A/T (m) was calculated as, m = v/u, and the expected equilibrium G/C content was calculated as 1/(1+m) following Lynch (2007).
S = 4Nes (or 2Nes), is the population-scaled strength of selection toward A/T nucleotides and can be calculated using the equation, PAT = 1/(1 + m−1e−S), where PAT is the observed fraction of A/T sites and S is the average selective advantage of A/T over G/C nucleotides (Bulmer 1991). Thus S = −ln[(m.(1−PAT))/PAT] = ln(PAT/PGC/v/u).
Recombination Analyses
All analyses to detect recombination were restricted to SNPs that were biallelic, homozygous, and had a known ancestral state. The statistic (r2) to measure LD was calculated as r2 = (fAa – fA × fa)2/[fA × fa × (1 − fA)×(1 − fa)]. The program “pairwise” in LDhat 2.2 (McVean et al. 2002) was used to infer recombination rates using the permuted composite likelihood test as well other permutation tests. These tests were performed under both the gene conversion (average tract length: 500) and crossover models with 2 values of θ (=4Neμ) for each species: The closest allowed θ value lower than that estimated from nucleotide diversity values, and the closest higher value.
Identifying Nuclear Genes Belonging to OXPHOS Pathway and Ribosomal Complex
KEGG (Kanehisa et al. 2017) was used to obtain the full list of genes that are part of complexes involved in OXPHOS (complexes I–V) for P. tetraurelia. A total number of 87 genes in P. tetraurelia were obtained, and their corresponding orthologs were identified in other species. For genes encoding proteins that are part of the ribosomal complex, we used the PANTHER (Mi et al. 2017) annotation obtained in a previous study (McGrath, Gout, Johri, et al. 2014) for all species and selected all genes that were structural constituents of ribosome. This allowed us to start with a set of 585 genes in P. tetraurelia, 564 in P. sexaurelia, and 213 genes in P. caudatum. However, analyses requiring orthologs from all three species were conducted with a smaller subset of genes.
Data Availability
Assembled mitogenomes and protein-coding gene annotations can be accessed through Zenodo (https://doi.org/10.5281/zenodo.2539699). Mitochondrial variant files can be accessed through Github (https://github.com/paruljohri/Paramecium_mitochondrial_SNPs).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Jean-Francois Gout for technical help, as well as for a critical reading of the manuscript. We thank Hongan Long for providing us with sequencing data for P. biaurelia and P. sexaurelia nuclear MA experiments. We also thank Jeffrey Palmer for critical reading of the manuscript and discussions about the project. We thank two anonymous reviewers and Fabrizio Ghiselli for greatly improving the manuscript. This work was financially supported by the National Science Foundation (MCB-1050161 and DEB-1257806).
Literature Cited
- Adoutte A, Beisson J.. 1972. Evolution of mixed populations of genetically different mitochondria in Paramecium aurelia. Nature 235(5338):393–396. [DOI] [PubMed] [Google Scholar]
- Adoutte A, Doussiere J.. 1978. Physiological consequences of mitochondrial antibiotic-resistant mutations in Paramecium: growth-rates, cytochromic defects and cyanide-insensitive respiration of mutant and erythromycin-treated wild-type strains. Mol Gen Genet. 161(2):121–134. [PubMed] [Google Scholar]
- Adoutte A, Knowles JK, Sainsard-Chanet A.. 1979. Absence of detectable mitochondrial recombination in Paramecium. Genetics 93(4):797–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrion JR, White PS, Montooth KL.. 2016. The roles of compensatory evolution and constraint in aminoacyl tRNA synthetase evolution. Mol Biol Evol. 33(1):152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, et al. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37(Web Server issue):W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balabaskaran Nina P, et al. 2010. Highly divergent mitochondrial ATP synthase complexes in Tetrahymena thermophila. PLoS Biol. 8(7):e1000418.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr CM, Neiman M, Taylor DR.. 2005. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 168(1):39–50. [DOI] [PubMed] [Google Scholar]
- Barth D, Berendonk TU.. 2011. The mitochondrial genome sequence of the ciliate Paramecium caudatum reveals a shift in nucleotide composition and codon usage within the genus Paramecium. BMC Genomics 12:272.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth D, Przyboś E, Fokin SI, Schlegel M, Berendonk TU.. 2008. Cytochrome b sequence data suggest rapid speciation within the Paramecium aurelia species complex. Mol Phylogenet Evol. 49(2):669–673. [DOI] [PubMed] [Google Scholar]
- Beale GH, Knowles JK, Tait A.. 1972. Mitochondrial genetics in Paramecium. Nature 235(5338):396–397. [DOI] [PubMed] [Google Scholar]
- Beale GH, Tait A.. 1981. International review of cytology In G.H. Bourne JFD, Jeon KW, editors. Mitochondrial genetics of Paramecium aurelia. Academic Press; p. 19–40. [Google Scholar]
- Betancourt AJ, Blanco-Martin B, Charlesworth B.. 2012. The relation between the neutrality index for mitochondrial genes and the distribution of mutational effects on fitness. Evolution 66(8):2427–2438. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borza T, Redmond EK, Laflamme M, Lee RW.. 2009. Mitochondrial DNA in the Oogamochlamys clade (Chlorophyceae): high GC content and unique genome architecture for Green algae(1). J Phycol. 45(6):1323–1334. [DOI] [PubMed] [Google Scholar]
- Bulmer M. 1991. The selection-mutation-drift theory of synonymous codon usage. Genetics 129(3):897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, et al. 2000. Complete sequence of the mitochondrial genome of Tetrahymena pyriformis and comparison with Paramecium aurelia mitochondrial DNA. J Mol Biol. 297(2):365–380. [DOI] [PubMed] [Google Scholar]
- Burger G, Forget L, Zhu Y, Gray MW, Lang BF.. 2003. Unique mitochondrial genome architecture in unicellular relatives of animals. Proc Natl Acad Sci U S A. 100(3):892–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Moreira S, Valach M.. 2016. Genes in hiding. Trends Genet. 32(9):553–565. [DOI] [PubMed] [Google Scholar]
- Cooper BS, Burrus CR, Ji C, Hahn MW, Montooth KL.. 2015. Similar efficacies of selection shape mitochondrial and nuclear genes in both Drosophila melanogaster and Homo sapiens. G3 (Bethesda) 5(10):2165–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne RS, et al. 2011. Comparative genomics of the pathogenic ciliate Ichthyophthirius multifiliis, its free-living relatives and a host species provide insights into adoption of a parasitic lifestyle and prospects for disease control. Genome Biol. 12(10):R100.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, et al. 2011. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf RM, et al. 2009. The mitochondrial genomes of the ciliates Euplotes minuta and Euplotes crassus. BMC Genomics 10:514.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf RM, et al. 2011. The organellar genome and metabolic potential of the hydrogen-producing mitochondrion of Nyctotherus ovalis. Mol Biol Evol. 28(8):2379–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Morris K, Lynch M, Vassilieva LL, Thomas WK. 2000. High Direct Estimate of the Mutation Rate in the Mitochondrial Genome of Caenorhabditis elegans. Science 289(5488):2342–2344. [DOI] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol. 7(10):e1002195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. 2014. Pfam: the protein families database. Nucleic Acids Res. 42(Database issue):D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokin SI, Gortz HD.. 2009. Diversity of Holospora bacteria in Paramecium and their characterization. Microbiol Monogr. 12:161–199. [Google Scholar]
- Fritsch ES, Chabbert CD, Klaus B, Steinmetz LM.. 2014. A genome-wide map of mitochondrial DNA recombination in yeast. Genetics 198(2):755–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiselli F, et al. 2013. Structure, transcription, and variability of metazoan mitochondrial genome: perspectives from an unusual mitochondrial inheritance system. Genome Biol Evol. 5(8):1535–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard JM, Cummings DJ.. 1975. Structure and replication of mitochondrial DNA from Paramecium aurelia. J Mol Biol. 97(4):593–609. [DOI] [PubMed] [Google Scholar]
- Gray MW. 2003. Diversity and evolution of mitochondrial RNA editing systems. IUBMB Life 55(4–5):227–233. [DOI] [PubMed] [Google Scholar]
- Greenland S. 1982. Interpretation of summary measures when interaction is present. Am J Epidemiol. 116:587–587. [Google Scholar]
- Gruchy DF. 1955. The breeding system and distribution of Tetrahymena pyriformis. J Protozool. 2(4):178–185. [Google Scholar]
- Haag-Liautard C, et al. 2008. Direct estimation of the mitochondrial DNA mutation rate in Drosophila melanogaster. PLoS Biol. 6(8):e204.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havird JC, Sloan DB.. 2016. The roles of mutation, selection, and expression in determining relative rates of evolution in mitochondrial versus nuclear genomes. Mol Biol Evol. 33(12):3042–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A.. 1968. Linkage disequilibrium in finite populations. Theor Appl Genet. 38(6):226–231. [DOI] [PubMed] [Google Scholar]
- Holm S. 1979. A simple sequentially rejective multiple test procedure. Scand J Stat. 6:65–70. [Google Scholar]
- Hori M, Tomikawa I, Przyboś E, Fujishima M.. 2006. Comparison of the evolutionary distances among syngens and sibling species of Paramecium. Mol Phylogenet Evol. 38(3):697–704. [DOI] [PubMed] [Google Scholar]
- Howe DK, Baer CF, Denver DR.. 2010. High rate of large deletions in Caenorhabditis briggsae mitochondrial genome mutation processes. Genome Biol Evol. 2:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. 2001. Two-locus sampling distributions and their application. Genetics 159(4):1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL.. 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111(1):147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston IG, Williams BP.. 2016. Evolutionary inference across eukaryotes identifies specific pressures favoring mitochondrial gene retention. Cell Syst. 2(2):101–111. [DOI] [PubMed] [Google Scholar]
- Johri P, et al. 2017. Population genomics of Paramecium species. Mol Biol Evol. 34(5):1194–1216. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K.. 2017. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45(D1):D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayal E, et al. 2012. Evolution of linear mitochondrial genomes in medusozoan cnidarians. Genome Biol Evol. 4(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefel BR, Gilson PR, Beech PL.. 2006. Cell biology of mitochondrial dynamics. Int Rev Cytol. 254:151–213. [DOI] [PubMed] [Google Scholar]
- Kim D, et al. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14(4):R36.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi S, Kobayashi S.. 1989. Microinjection of plasmid DNA encoding the A surface antigen of Paramecium tetraurelia restores the ability to regenerate a wild-type macronucleus. Mol Cell Biol. 9(10):4398–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad A, et al. 2017. Mitochondrial mutation rate, spectrum and heteroplasmy in Caenorhabditis elegans spontaneous mutation accumulation lines of differing population size. Mol Biol Evol. 34(6):1319–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukat C, et al. 2011. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc Natl Acad Sci U S A. 108(33):13534–13539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladoukakis ED, Theologidis I, Rodakis GC, Zouros E.. 2011. Homologous recombination between highly diverged mitochondrial sequences: examples from maternally and paternally transmitted genomes. Mol Biol Evol. 28(6):1847–1859. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10(3):R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26(5):589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H, Doak TG, Lynch M. 2018. Limited mutation-rate variation within the Paramecium aurelia species complex. G3 (Bethesda) 8(7):2523–2526. [DOI] [PMC free article] [PubMed]
- Long H, Sung W, et al. 2018. Evolutionary determinants of genome-wide nucleotide composition. Nat Ecol Evol. 2:237–240. [DOI] [PMC free article] [PubMed]
- Lynch M. 1996. Mutation accumulation in transfer RNAs: molecular evidence for Muller’s ratchet in mitochondrial genomes. Mol Biol Evol. 13(1):209–220. [DOI] [PubMed] [Google Scholar]
- Lynch M. 1997. Mutation accumulation in nuclear, organelle, and prokaryotic transfer RNA genes. Mol Biol Evol. 14(9):914–925. [DOI] [PubMed] [Google Scholar]
- Lynch M. 2007. The origins of genome architecture. Sunderland (MA: ): Sinauer Associates. [Google Scholar]
- Lynch M, Blanchard JL.. 1998. Deleterious mutation accumulation in organelle genomes. Genetica 102–103(1–6):29–39. [PubMed] [Google Scholar]
- Lynch M, et al. 2008. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci U S A. 105(27):9272–9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Koskella B, Schaack S.. 2006. Mutation pressure and the evolution of organelle genomic architecture. Science 311(5768):1727–1730. [DOI] [PubMed] [Google Scholar]
- Mackenzie SA. 2007. The unique biology of mitochondrial genome instability in plants In: Logan D, editor. Plant mitochondria. Oxford: Blackwell Publishing; p. 36–46. [Google Scholar]
- McDonald JH, Kreitman M.. 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351(6328):652–654. [DOI] [PubMed] [Google Scholar]
- McFadden CS, Sanchez JA, France SC.. 2010. Molecular phylogenetic insights into the evolution of Octocorallia: a review. Integr Comp Biol. 50(3):389–410. [DOI] [PubMed] [Google Scholar]
- McGrath CL, Gout JF, Doak TG, Yanagi A, Lynch M.. 2014. Insights into three whole-genome duplications gleaned from the Paramecium caudatum genome sequence. Genetics 197(4):1417–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath CL, Gout JF, Johri P, Doak TG, Lynch M.. 2014. Differential retention and divergent resolution of duplicate genes following whole-genome duplication. Genome Res. 24(10):1665–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G, Awadalla P, Fearnhead P.. 2002. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics 160(3):1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier J, Eyre-Walker A.. 2001. The correlation between linkage disequilibrium and distance: implications for recombination in hominid mitochondria. Mol Biol Evol. 18(11):2132–2135. [DOI] [PubMed] [Google Scholar]
- Meyer E, Garnier O.. 2002. Non-Mendelian inheritance and homology-dependent effects in ciliates. Adv Genet. 46:305–337. [DOI] [PubMed] [Google Scholar]
- Mi H, et al. 2017. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45(D1):D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Guerra D, Breton S, Passamonti M.. 2013. A comparative analysis of mitochondrial ORFans: new clues on their origin and role in species with doubly uniparental inheritance of mitochondria. Genome Biol Evol. 5(7):1408–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Maurizii MG, Nuzhdin SV, Passamonti M.. 2014. Paternally transmitted mitochondria express a new gene of potential viral origin. Genome Biol Evol. 6(2):391–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montooth KL, Rand DM.. 2008. The spectrum of mitochondrial mutation differs across species. PLoS Biol. 6(8):e213.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradian MM, Beglaryan D, Skozylas JM, Kerikorian V.. 2007. Complete mitochondrial genome sequence of three Tetrahymena species reveals mutation hot spots and accelerated nonsynonymous substitutions in Ymf genes. PLoS One. 2(7):e650.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin GB, Cech TR.. 1988. Mitochondrial telomeres: surprising diversity of repeated telomeric DNA sequences among six species of Tetrahymena. Cell 52(3):367–374. [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Kolbe DL, Eddy SR.. 2009. Infernal 1.0: inference of RNA alignments. Bioinformatics 25(10):1335–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman M, Taylor DR.. 2009. The causes of mutation accumulation in mitochondrial genomes. Proc Biol Sci. 276(1660):1201–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr AT, Rabets JC, Horton TL, Landweber LF.. 1997. RNA editing missing in mitochondria. RNA 3(4):335–336. [PMC free article] [PubMed] [Google Scholar]
- Perasso R, Beisson J.. 1978. Temporal pattern of mitochondrial multiplication during cell-cycle of Paramecium. Biol Cell. 32:275–290. [Google Scholar]
- Piganeau G, Gardner M, Eyre-Walker A.. 2004. A broad survey of recombination in animal mitochondria. Mol Biol Evol. 21(12):2319–2325. [DOI] [PubMed] [Google Scholar]
- Pombert J-F, et al. 2013. The complete mitochondrial genome from an unidentified Phalansterium species. Protist Genomics 1:25–32. [Google Scholar]
- Popadin KY, Nikolaev SI, Junier T, Baranova M, Antonarakis SE.. 2013. Purifying selection in mammalian mitochondrial protein-coding genes is highly effective and congruent with evolution of nuclear genes. Mol Biol Evol. 30(2):347–355. [DOI] [PubMed] [Google Scholar]
- Preer LB. 1969. Alpha, an infectious macronuclear symbiont of Paramecium aurelia. J Protozool. 16(3):570–578. [DOI] [PubMed] [Google Scholar]
- Price CA, Reardon EM.. 2001. Mendel, a database of nomenclature for sequenced plant genes. Nucleic Acids Res. 29(1):118–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R-Core-Team. 2014. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing. [Google Scholar]
- Rand DM, Kann LM.. 1996. Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Mol Biol Evol. 13(6):735–748. [DOI] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM.. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33(Web Server issue):W686–W689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Graf S, Chaga OY, Lavrov DV.. 2006. Mitochondrial genome of the moon jelly Aurelia aurita (Cnidaria, Scyphozoa): a linear DNA molecule encoding a putative DNA-dependent DNA polymerase. Gene 381:92–101. [DOI] [PubMed] [Google Scholar]
- Skippington E, Barkman TJ, Rice DW, Palmer JD.. 2017. Comparative mitogenomics indicates respiratory competence in parasitic Viscum despite loss of complex I and extreme sequence divergence, and reveals horizontal gene transfer and remarkable variation in genome size. BMC Plant Biol. 17(1):49.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabodnick MM, et al. 2017. The macronuclear genome of Stentor coeruleus reveals tiny introns in a giant cell. Curr Biol. 27(4):569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR. 2012. Updating our view of organelle genome nucleotide landscape. Front Genet. 3:175.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR. 2016. The past, present and future of mitochondrial genomics: have we sequenced enough mtDNAs? Brief Funct Genomics. 15(1):47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Keeling PJ.. 2015. Mitochondrial and plastid genome architecture: reoccurring themes, but significant differences at the extremes. Proc Natl Acad Sci U S A. 112(33):10177–10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J, Biegert A, Lupas AN.. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33(Web Server issue):W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneborn TM. 1937. Sex, sex inheritance and sex determination in Paramecium aurelia. Proc Natl Acad Sci U S A. 23(7):378–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneborn TM. 1975. Paramecium aurelia complex of 14 sibling species. Trans Am Microsc Soc. 94(2):155–178. [Google Scholar]
- Stadler T, Delph LF.. 2002. Ancient mitochondrial haplotypes and evidence for intragenic recombination in a gynodioecious plant. Proc Natl Acad Sci U S A. 99(18):11730–11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JB, et al. 2008. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 6(1):e10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JB, Larsson NG.. 2014. Keeping mtDNA in shape between generations. PLoS Genet. 10(10):e1004670.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoletzki N, Eyre-Walker A.. 2011. Estimation of the neutrality index. Mol Biol Evol. 28(1):63–70. [DOI] [PubMed] [Google Scholar]
- Sueoka N. 1993. Directional mutation pressure, mutator mutations, and dynamics of molecular evolution. J Mol Evol. 37(2):137–153. [DOI] [PubMed] [Google Scholar]
- Sung W, et al. 2012. Extraordinary genome stability in the ciliate Paramecium tetraurelia. Proc Natl Acad Sci U S A. 109(47):19339–19344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart EC, et al. 2012. The Oxytricha trifallax mitochondrial genome. Genome Biol Evol. 4(2):136–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarcz S, Potekhin A, Rautian M, Przyboś E.. 2012. Variation in ribosomal and mitochondrial DNA sequences demonstrates the existence of intraspecific groups in Paramecium multimicronucleatum (Ciliophora, Oligohymenophorea). Mol Phylogenet Evol. 63(2):500–509. [DOI] [PubMed] [Google Scholar]
- Tarone RE. 1981. On summary estimators of relative risk. J Chronic Dis. 34(9–10):463–468. [DOI] [PubMed] [Google Scholar]
- Tsaousis AD, Martin DP, Ladoukakis ED, Posada D, Zouros E.. 2005. Widespread recombination in published animal mtDNA sequences. Mol Biol Evol. 22(4):925–933. [DOI] [PubMed] [Google Scholar]
- Tsukihara T, et al. 1996. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science 272(5265):1136–1144. [DOI] [PubMed] [Google Scholar]
- Visser W, et al. 1995. Effects of growth conditions on mitochondrial morphology in Saccharomyces cerevisiae. Antonie Van Leeuwenhoek 67(3):243–253. [DOI] [PubMed] [Google Scholar]
- Vlcek C, Marande W, Teijeiro S, Lukes J, Burger G.. 2011. Systematically fragmented genes in a multipartite mitochondrial genome. Nucleic Acids Res. 39(3):979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai T, Teoli D, Shoubridge EA.. 2008. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat Genet. 40(12):1484–1488. [DOI] [PubMed] [Google Scholar]
- Wakeley J. 1997. Using the variance of pairwise differences to estimate the recombination rate. Genet Res. 69(1):45–48. [DOI] [PubMed] [Google Scholar]
- Weinreich DM, Rand DM.. 2000. Contrasting patterns of nonneutral evolution in proteins encoded in nuclear and mitochondrial genomes. Genetics 156(1):385–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, et al. 2012. High mutation rates in the mitochondrial genomes of Daphnia pulex. Mol Biol Evol. 29(2):763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang F, Broughton RE.. 2013. Mitochondrial-nuclear interactions: compensatory evolution or variable functional constraint among vertebrate oxidative phosphorylation genes? Genome Biol Evol. 5(10):1781–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Assembled mitogenomes and protein-coding gene annotations can be accessed through Zenodo (https://doi.org/10.5281/zenodo.2539699). Mitochondrial variant files can be accessed through Github (https://github.com/paruljohri/Paramecium_mitochondrial_SNPs).