Abstract
Sleep remains a major mystery of biology. In particular, little is known about the mechanisms that account for the drive to sleep. In an unbiased screen of more than 12,000 Drosophila lines, we identified a single gene, nemuri, that induces sleep. The NEMURI protein is an antimicrobial peptide that can be secreted ectopically to drive prolonged sleep (with resistance to arousal) and to promote survival after infection. Loss of nemuri increased arousability during daily sleep and attenuated the acute increase in sleep induced by sleep deprivation or bacterial infection. Conditions that increase sleep drive induced expression of nemuri in a small number of fly brain neurons and targeted it to the sleep-promoting, dorsal fan-shaped body. We propose that NEMURI is a bona fide sleep homeostasis factor that is particularly important under conditions of high sleep need; because these conditions include sickness, our findings provide a link between sleep and immune function.
We spend one-third of our lives sleeping, yet the purpose and mechanisms of sleep remain a major research challenge in neuroscience (1, 2). Sleep is broadly regulated by two major processes: the circadian system that times daily sleep and the homeostatic system that drives the need to sleep. Mechanisms that generate circadian clocks are now understood on a fundamental level (3, 4), and the neurochemistry associated with sleep and wake states has also been well studied (5, 6). However, we still know very little about the mechanisms that generate the homeostatic drive to sleep. Studies of sleep homeostasis frequently rely on depriving organisms of sleep and assaying the subsequent recovery sleep that results from the increased drive. Prolonged wakefulness promotes accumulation of sleep-promoting signals, some of which may be secreted. Thus, transfer of cerebrospinal fluid from sleep-deprived animals to rested animals triggers sleep (7), although the nature of the secreted factors is debatable. Adenosine is thought to be one such signal, but the role of adenosine and adenosine receptors is limited to specific aspects of sleep (8). Sleep drive also increases during sickness, but it is unclear whether increased sleep under these conditions is mediated by the same mechanisms that increase sleep upon sleep deprivation (9, 10).
Forward genetic screens (11) allow unbiased identification of molecular factors relevant for a particular biological phenomenon. Genetic screens for sleep mutants in Drosophila have successfully identified several molecules that modulate sleep and wakefulness (12–20). Because the screens were generally loss-of-function in nature and were targeted toward isolation of short-sleeping mutants, they identified factors whose loss reduces sleep. However, where tested, overexpression of these factors does not increase sleep above wildtype levels (14, 17, 21), which suggests that they are insufficient to induce sleep on their own. In addition, several fly sleep-regulating genes are required broadly in the brain (14, 17, 21), perhaps serving to modulate neural activity in response to upstream sleep-inducing signals. The paucity of sleep-inducing signals in any organism prompted us to undertake a screen for such molecules.
Identification of nemuri as a sleep-inducing molecule from a gain-of-function genetic screen
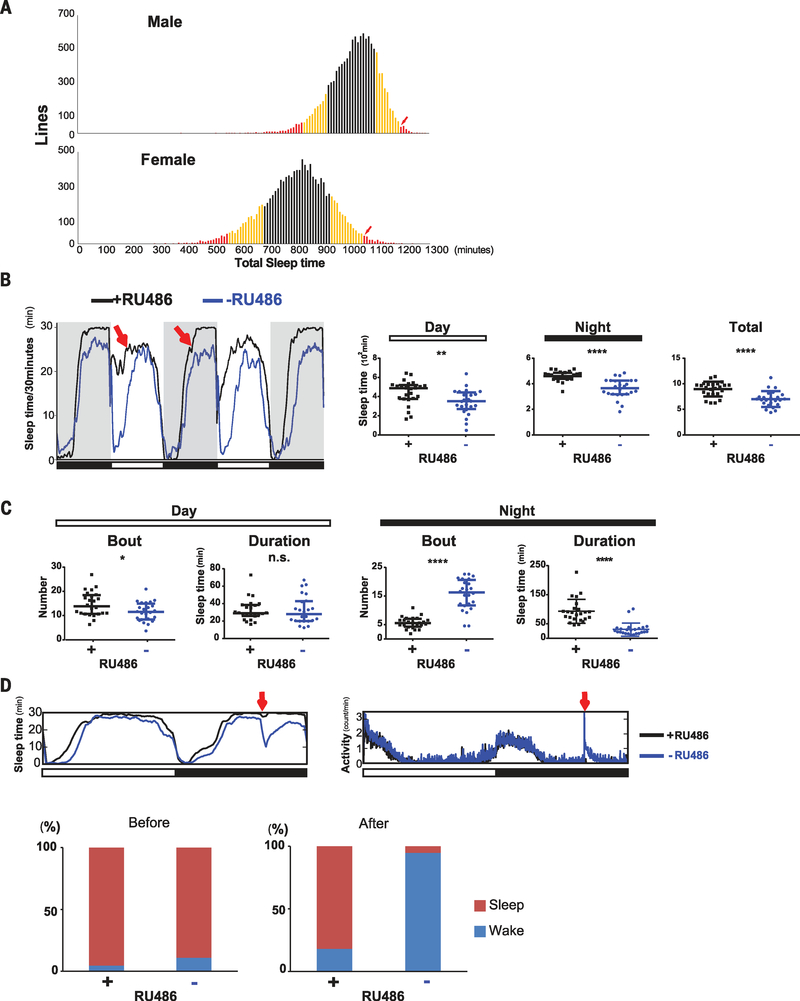
To identify genetic factors that promote sleep, we conducted an unbiased and genome-wide gain-of-function screen in Drosophila. We used an inducible pan-neuronal Gal4 driver, elav-GeneSwitch (elav-GS) (22), to drive expression of random genes tagged with Gal4-responsive upstream activating sequences (UASs). Enhancer- promoter (EP) collections (23), UAS-cDNA libraries in FlyORF (24), and the Bangalore Fly Resource Center (BFRC) (25) were the sources for flies carrying UAS insertions, each of which was crossed to elav-GS-carrying flies to allow pan-neuronal induction of UAS-tagged genes with the GS activator RU486 (trade name for mifepristone) (26). To circumvent potential developmental defects, we fed offspring RU486 only during the adult stage in the course of the sleep assay. In total, we screened 12,198 lines (table S1), which correspond to 8015 Drosophila genes. Although overexpression of most genes did not affect sleep, some of the lines displayed a decrease or an increase in sleep (Fig. 1A). We further tested lines in which both males and females had sleep times at least two standard deviations away from the average sleep time for all lines. Rescreens of candidate lines identified only one that showed a significant increase in sleep relative to controls (fig. S1A). Upon induction of elav-GS with RU486, the total amount of sleep in this line increased in both males and females and was reflected in increases in both daytime and nighttime sleep (fig. S1B).
Fig. 1. nemuri (nur) overexpression promotes sleep.
(A) Summary of the gain-of-function screen. A UAS insertion in each line was driven by elav-GS, which was induced by adding RU486 to the food. Average sleep time (minutes per 24 hours; x axis) is shown for males (top) and females (bottom). Lines within one standard deviation (SD) of the average sleep time are indicated in black, one to two SDs in yellow, and two or more SDs in red. An arrow indicates the line termed nemuri. (B) Left: Sleep profile of elav-GS/UAS-nemuri flies +RU486 (black) or −RU486 (blue) obtained by video tracking. Red arrows indicate longer sleep in the presence of RU486 (black line) in nur-overexpressing flies. Right: Median ± interquartile minutes of sleep during daytime, night, and total 24-hour period in +RU486 flies versus vehicle controls; n = 24 females for each group. **P < 0.01, ****P < 0.0001 (Mann-Whitney U test). (C) Sleep architecture assayed through video tracking. Median ± interquartile number of sleep bouts and bout duration is shown for the daytime (left) and night (right) in +RU486 flies and vehicle controls; n = 24 females for each group. *P < 0.05, ****P < 0.0001 (Mann-Whitney U test); n.s., not significant. (D) Top: Sleep/wake patterns in the Drosophila activity monitoring system of flies subjected to a mechanical stimulus at ZT20 (red arrow). Sleep (left) and activity (right) are shown for nur overexpressors (black) and control (blue) flies (n = 32 females per group). Bottom: Bar graphs indicate the percentage of RU486-treated (+) or control (−) flies in sleep (red) versus wake (blue) states, before (left) and after (right) the stimulus.
The line with the increased sleep phenotype contains a UAS-regulated transgene (#DP2629 at BFRC) corresponding to a previously uncharacterized gene, CG31813 (www.ncbi.nlm.nih.gov, accession number NP_723832/AHN54420), which we named nemuri (nur; Japanese word for “sleep”). In situ analysis of nur mRNA in adult Drosophila brains revealed that the gene was overexpressed broadly upon induction of the UAS insertion by elav-GS (fig. S1C).
To determine whether nur overexpression affected circadian rhythms (27), we measured locomotor activity in elav-GS/UAS-nur flies in constant darkness (fig. S2A). Although the period was slightly short in nur-overexpressing flies [22.8 ± 0.3 hours (mean ± SD)] relative to that in control animals (23.2 ±0.3 hours), behavioral rhythms were as clearly defined in nur-overexpressing flies as in controls (fig. S2B). Thus, overexpression of nur has minimal effect on circadian rhythms but strong effects on sleep.
Increases of sleep length and depth as a result of nur overexpression
For higher-resolution analysis of sleep induced by nur, we evaluated sleep with a video tracking system (28). Sleep in flies occurs largely during the nighttime, but it also manifests as a mid-day siesta that is more pronounced in males. Video analysis confirmed that nur overexpression increased both daytime and nighttime sleep relative to control flies not treated with RU486 (Fig. 1B). Overexpression of nur increased the number of sleep bouts during the day with no change in bout duration (Fig. 1C). On the other hand, during the night, the number of sleep bouts was decreased, but the bout duration was much longer in nur-overexpressing flies (Fig. 1C), indicating that nur overexpression consolidates nighttime sleep.
To test whether nur affects sleep depth, which is measured as the extent to which sleeping animals are resistant to external stimuli, we subjected flies to a mechanical arousing stimulus (29) in the middle of the night at zeitgeber time (ZT) 20 (ZT0, lights on; ZT12, lights off). Although >94% of control flies awoke upon stimulation, only ~18% of nur-overexpressing flies were aroused (Fig. 1D). The strong mechanical stimulus used in this assay typically elicits the consistent response seen in controls, so the very low response in experimental animals indicated that nur increases sleep depth. After the mechanical stimulus, the few aroused nur-overexpressing flies were sluggish and had reduced locomotor speed relative to control animals (fig. S3A). However, locomotor ability in nur-overexpressing flies was not impaired, as these flies moved faster than controls after both lights on and lights off (fig. S3, B and C). The reason for the increased speed is unclear, but it could reflect better consolidation of wake activity as a result of increased sleep. Even 1 day of nur overexpression induced sleep (Fig. 2) but did not impair food intake or affect lifespan (fig. S4).
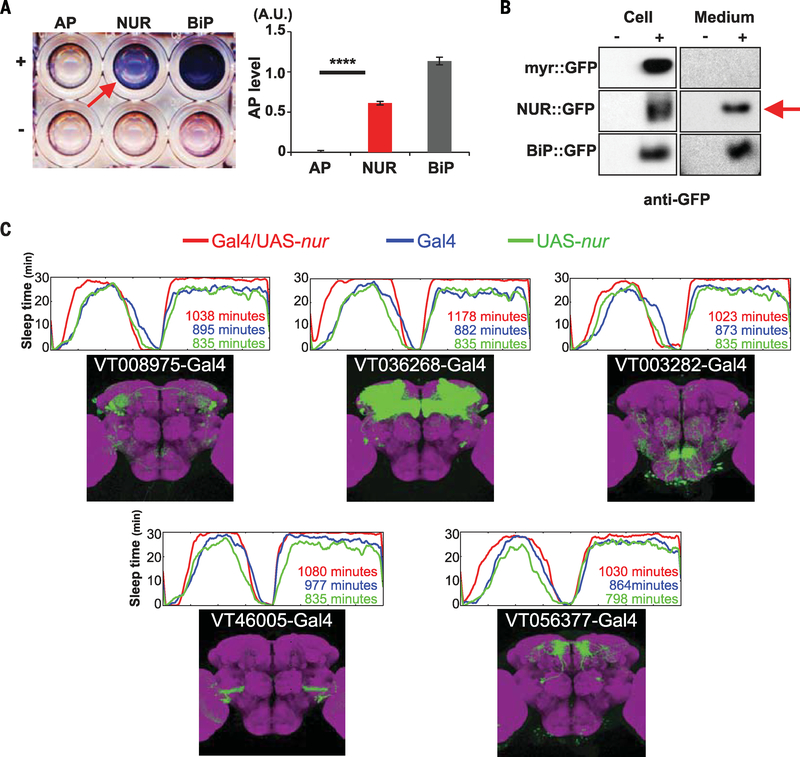
Fig. 2. NUR is a secreted protein and its ectopic expression induces sleep.
(A) SEAP assay. Left: Culture medium from S2R+ cells expressing alkaline phosphatase (AP), NUR::AP, or a fusion of AP with the immunoglobulin binding protein (BiP), BiP::AP, used as a secreted control, with (+) or without (−) induction by Cu2+ in a 96-well plate. The arrow indicates NUR expression in the media. Right: Quantification of the SEAP assay by measuring absorbance at 595 nm for each sample (A.U., absorbance units). ****P < 0.0001 (Student t test). Error bars denote SEM. (B) Protein immunoblot of S2R+ cells (left) or conditioned medium (right) using antibodies to GFP. Cells expressed myr::GFP (negative control), NUR::GFP, or BiP::GFP (positive control); with (+) or without (−) induction by Cu2+.The arrow indicates the detection of NUR::GFP in the medium. (C) Sleep profiles and expression patterns of Gal4 lines ectopically expressing nur in various subsets of neurons in the brain. Upper panels show the sleep pattern of each genotype: Gal4/UAS-nur (red), Gal4 control (blue), and UAS-nur control (green). The average number of minutes of total sleep in eight female flies is indicated. Lower panels show the expression pattern of each Gal4 line, as determined by expression of UAS-mCD8::GFP (green) and costaining with nc82 (neuropile marker; magenta) (images from Brainbase: https://braingazer.org/).
Characterization of NUR as a secreted sleep-inducing molecule
The sequence of NUR predicts an N-terminal signal peptide, followed by a short open reading frame containing many arginines and glycines and no transmembrane region, indicating that it is a secreted protein (fig. S5A). To test this, we transfected NUR tagged with alkaline phosphatase at the C terminus (NUR::AP) into Drosophila cultured cells (S2R+) and collected extracellular medium for evaluation in a secreted alkaline phosphatase (SEAP) assay (30). NUR::AP was present in the extracellular medium (Fig. 2A). In addition, we transfected a construct encoding a fusion protein of NUR with green fluorescent protein (NUR::GFP) into S2R+ cells and confirmed the presence of NUR::GFP in the medium (Fig. 2B and fig. S5B).
Given that NUR can be secreted, we tested whether it increases sleep when expressed ectopically by crossing UAS-nur to each of 159 Gal4 lines [(31) and VDRC, http://stockcenter.vdrc.at/] that sparsely label various subsets of neurons (table S2). We found that 51Gal4 lines crossed with UAS-nur showed an increase of sleep (>100 min) relative to Gal4 or UAS-nur control lines (see Fig. 2C and table S2 for representative examples). These findings support the idea that NUR acts non-cell-autonomously to promote sleep.
Antimicrobial activity and prosurvival effects of NUR
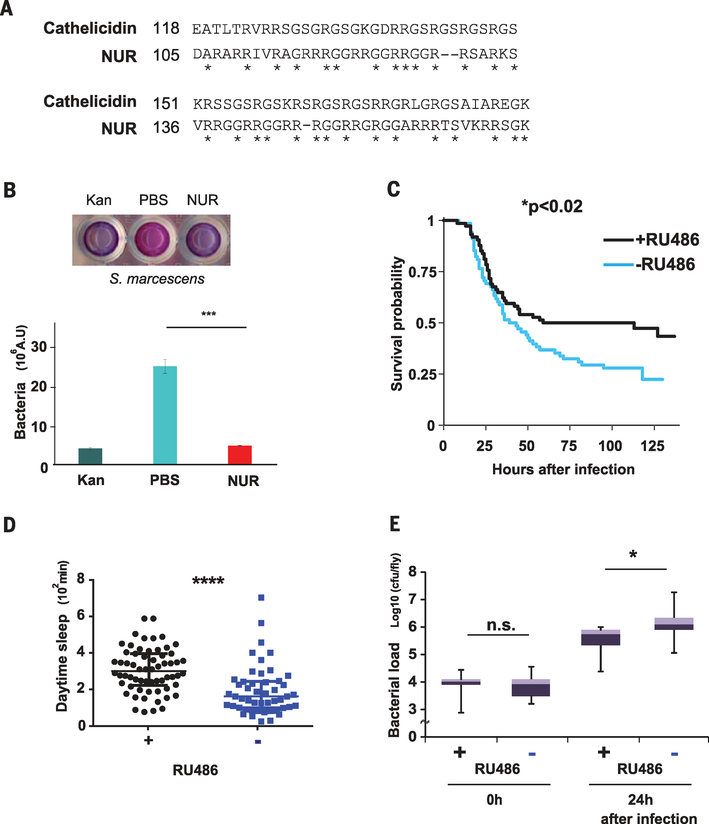
NUR contains a domain of cationic amino acids characteristic of several antimicrobial peptides (AMPs) (32). Indeed, NUR has sequence similarity to an AMP, cathelicidin, isolated from fish (33) (Fig. 3A). To test for antimicrobial activity of NUR, we used the Alamar Blue cell viability assay (34) and examined survival of bacteria treated with NUR. NUR was very effective in this assay, killing microbes (Serratia marcescens or Escherichia coli) at concentrations considered physiological for AMPs (Fig. 3B and fig. S6, A and B). In vivo experiments confirmed the antimicrobial activity of NUR (fig. S6C).
Fig. 3. nur encodes an antimicrobial peptide that promotes survival upon expression in neurons.
(A) Alignment between NUR and a fish cathelicidin. The C-terminal domain of NUR shows sequence similarity with a Greenland cod (Gadus ogac) cathelicidin. Note the overlap of glycine and arginine residues, indicated by asterisks. Amino acid abbreviations: A, Ala; D, Asp; E, Glu; G, Gly; I, Ile; K, Lys; L, Leu; R, Arg; S, Ser; T, Thr; V, Val. (B) Top: Alamar Blue cell viability assay; S. marcescens in PBS alone (center), with kanamycin (left), or with NUR protein (right). Blue color indicates lack of bacteria. Bottom: The assay was quantified by measuring absorbance at 595 nm (n = 3). ***P < 0.001 (Student t test). Error bars denote SEM. (C) Kaplan-Meier plot depicting survival of infection with S. marcescens in nur-overexpressing flies (+RU486) and controls (−RU486); n = 68 and 74 females, respectively. *P < 0.02 (log rank test). (D) Daytime sleep in nur-overexpressing flies (black) and controls (blue) after infection at ZT18 the previous night; n = 49 control and 62 +RU486-fed female flies, respectively. Median ± interquartile is shown. ****P < 0.0001 (Mann-Whitney U test). Note that daytime sleep values are lower than in other figures (fig. S1B). This effect was also observed in uninfected controls and may be caused by the CO2 anesthesia necessary for infecting flies. (E) Bacterial load in control flies (−) and nur overexpressors (+) at indicated times after infection; n = 9 for each. Median ± interquartile is shown; cfu, colony-forming units. *P < 0.05 (Mann-Whitney U test).
AMPs promote survival of organisms upon infection, largely by killing microbes, but in some cases also by modulating other aspects of the immune response (35). Because sleep also facilitates recovery from illness (36) and even enhances survival of flies after bacterial infection (37), we tested whether sleep induction by NUR contributes to host defense. We induced NUR expression in neurons and subjected the flies to bacterial infection with two different bacterial species, S. marcescens and Streptococcus pneumoniae (37, 38). Flies overexpressing NUR survived significantly longer than controls (Fig. 3C and fig. S7A), whereas RU486, used to induce NUR overexpression, had no effect on survival when fed to control flies (fig. S7B). Flies with increased NUR also showed higher amounts of daytime sleep than controls after infection and had a lower bacterial load, consistent with the previous finding that sleep enhances clearance of bacteria (37) (Fig. 3, D and E, and fig. S7C). Thus, induction of sleep may represent another mechanism by which AMPs combat infection.
AMPs are typically also expressed in the periphery and, as discussed below, nur is expressed in fly bodies. Because the fat body is a major site of AMP expression, we tested whether increased expression of NUR in this tissue would promote survival. Expression of transgenic NUR in the fat body did not increase daily sleep or survival after infection (fig. S8, A and B). Consistently, it also did not reduce bacterial load (fig. S8C), possibly because endogenous amounts of NUR in the fat body are saturating for this purpose. These findings indicate that effects of increased NUR on sleep and survival are specific to the brain.
Disruption of sleep in nur mutants
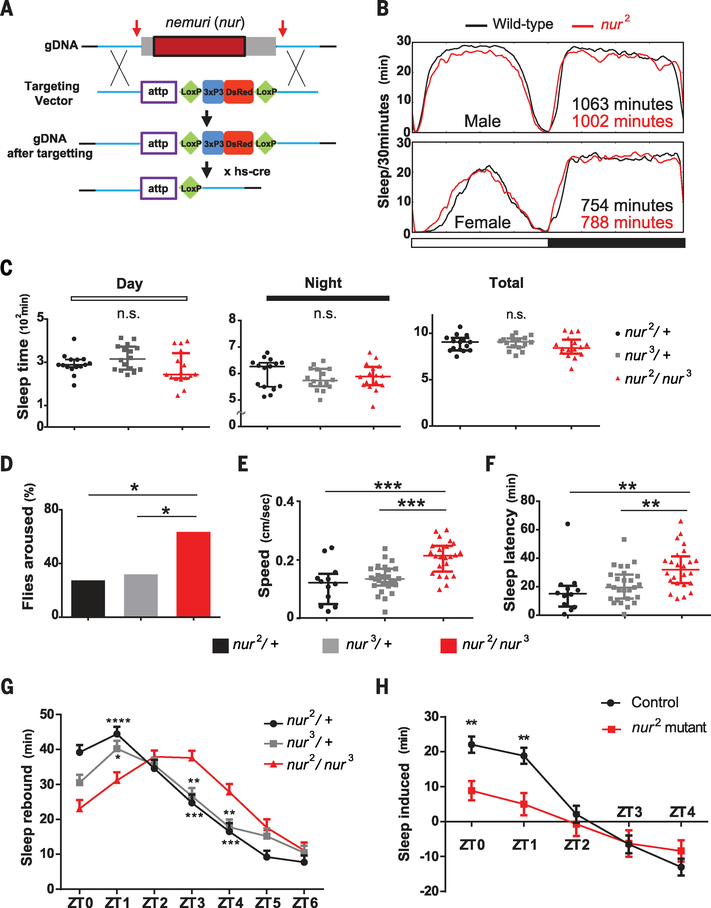
NUR can induce sleep, but can sleep be induced in the absence of NUR? To address this question, we knocked out the nur gene with CRISPR/Cas9 technology, replacing the single nur exon with a site for future site-specific recombination (attP), and an eyeless promoter-DsRed flanked by LoxP sites (Fig. 4A). We isolated two independent lines, nur2 and nur3, and confirmed successful gene targeting by polymerase chain reaction (PCR) and Southern blotting (fig. S9, A to C). We also verified complete absence of the nur gene in each line (fig. S9D).
Fig. 4. Requirement of nur for sleep depth and for acute sleep induction after sleep deprivation or infection.
(A) Gene targeting to knock out nur using CRISPR/Cas9. Arrows indicate regions targeted by guide RNA. Homology arms (~1 kbp; blue) are flanked by attp-Loxp-3xP3 (eyeless promoter)–DsRed-Loxp sequences. After the targeting, hs-cre was crossed in to flip out the Loxp-3xP3-DsRed-LoxP sequence. (B) Sleep pattern of nur2 mutants or wild-type controls (iso31) in males and females. Horizontal white and black bars correspond to light and dark cycles. (C) Day, night, and total sleep in the genotypes indicated; n = 15, 16, and 16 females for nur2/+, nur3/+, and nur2/nur3, respectively. Median ± interquartile is shown in all cases. P > 0.05 (Mann-Whitney U test). (D) Arousal threshold assay using a light pulse of 10 s in the middle of the night. Shown are proportions of flies awakened by the stimulus; n = 44, 54, and 73 females for nur2/+, nur3/+, and nur2/nur3, respectively. *P < 0.05 (Fisher exact test). (E and F) Speed after awakening (E) and latency to sleep (F) of flies awakened by light stimuli in (D). Speed in (E) was measured by video tracking after awakening until they fell asleep again. Median ± interquartile is shown; n = 12, 27, and 24 females for nur2/+, nur3/+, and nur2/nur3, respectively. ***P < 0.001, **P < 0.01 (Kruskal-Wallis test with Dunn post hoc comparison). (G) Recovery sleep after 6 hours of sleep deprivation. Rebound was calculated as the mean ± SEM net change in sleep relative to the baseline day before deprivation; n = 64 females for each genotype. (H) Infection-induced sleep, calculated as the mean ± SEM net change in sleep in the morning after infection relative to the corresponding baseline; n = 45 and 37 females for W1118 control and nur2 homozygous mutants, respectively. In (G) and (H), sleep values are plotted against zeitgeber time. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (two-way analysis of variance followed by Bonferroni multiple comparison test).
There was no consistent effect on sleep in nur2 or nur3 mutants, each of which was backcrossed four times into the iso31 wild-type background (39) used commonly for sleep analysis, and no effect on sleep in the trans-heterozygotes (Fig. 4, B and C, and fig. S10, A to C). However, consolidation of daytime sleep was decreased, particularly in males, such that the number of bouts was increased and bout duration was decreased (fig. S11). In subsequent experiments, which revealed recessive effects of mutant nur on specific aspects of sleep, we further controlled for background by comparing the sleep behavior of flies heterozygous for nur2 or nur3 with transheterozygotes or homozygotes (Fig. 4, C to H, and figs. S10C, S12, and S13).
To determine whether nur affected sleep quality, we used an arousal threshold assay. As noted above, the mechanical stimulus used to test arousal in nur overexpressors is strong and awakens virtually all wild-type flies. We expected that nur mutants might be hyperarousable, so we sought a milder stimulus to distinguish arousal phenotypes. We subjected flies to brief light pulses in the middle of the night and found that nur mutants were indeed aroused more easily than control flies (Fig. 4D). Video tracking to assess speed of movement after the light stimulus showed that awakened nur mutants moved faster than aroused control animals (Fig. 4E and fig. S12A). The nur mutants also took longer to fall asleep again once they had been aroused (Fig. 4F). Thus, nur is required for sleep depth and for reinitiation of sleep after arousal. We found that nur mutants were also more arousable by a strong odor (3-octanol; fig. S12B).
We evaluated sleep homeostasis in nur mutants by assaying their response to 6 hours of sleep deprivation in the latter half of the night (40, 41). The kinetics of sleep recovery the following morning were altered, such that sleep increases were delayed in nur mutants, although overall sleep amounts were not affected over the 12-hour day (Fig. 4G and fig. S13A). We surmise that nur mutant flies are impaired in initiating sleep rebound and thereby recover sleep more slowly after sleep deprivation.
Given its immune function and sleep-inducing properties, we sought to determine whether nur contributes to the sleep increases that normally occur during sickness—for instance, after bacterial infection in Drosophila (42). We infected flies with bacteria at ZT18 (6 hours after lights off) and found that the morning after infection, control flies showed a significantly larger increase in sleep than nur mutants (Fig. 4H and fig. S13, B and C).
Up-regulation of nur upon sleep deprivation
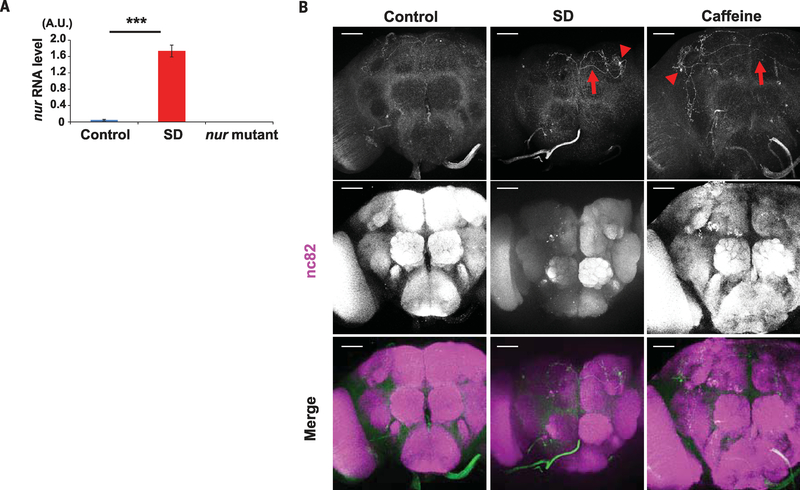
To characterize the cells that produce nur in vivo, we generated nurGal4 lines. We replaced nur coding sequences with Gal4 by scarless genome engineering with CRISPR (http://flycrispr.molbio.wisc.edu/scarless), thereby placing Gal4 expression under the control of the endogenous nur promoter (fig. S14A). nurGal4 did not drive brain expression of a Gal4-responsive GFP reporter under unperturbed conditions, and so we considered the possibility that nur expression was regulated by external factors. CG31813, the transcript corresponding to the nur locus, accumulates upon administration of various types of chemicals or stress agents such as cadmium, zinc, caffeine, and paraquat [(43) and http://flybase.org/reports/FBgn0051813.html]. To determine whether sleep loss induced the expression of nur, we subjected flies to sleep deprivation. Indeed, nur RNA expression was increased in adult heads after sleep deprivation, whereas little nur expression was detected in normal conditions (Fig. 5A). The expression of nur RNA also increased after infection in the fly body, albeit with delayed timing (fig. S15). Increased sleep at earlier time points after infection could involve up-regulation of other aspects of NUR function (e.g., release).
Fig. 5. Increased nur mRNA expression after sleep deprivation.
(A) Expression of nur mRNA in adult fly heads; n = 3. qPCR for nur expression used actin as a normalization control. ***P < 0.001 (Student t test). Error bars denote SEM. (B) Brains of nurGal4/UAS-CD4::tdGFP flies stained with anti-GFP (green) (top), nc82 (neuropil marker; magenta) (middle), or merged (bottom) in control (left), sleep-deprived (SD) (center), or caffeine-fed (right) animals. Arrows indicate GFP-positive projections; arrowheads indicate GFP-positive cell bodies. Scale bars, 50 μm.
Consistent with accumulation of nur mRNA after sleep deprivation (Fig. 5A), we found that nurGal4 flies expressed a sensitive form of GFP, CD4::tdGFP, in the brain after the flies were sleep-deprived either mechanically or through caffeine feeding. We detected CD4::tdGFP in only a single neuron in each brain hemisphere, indicating that nur expression in the brain is limited (Fig. 5B). nurGal4 expression was also observed in the same cells after infection, but only in ~30% of flies, and again with delayed timing (fig. S15). GFP-positive tracts of nur+ neurons were detected in the area of the dorsal fan-shaped body (dFSB) (fig. S14B), which is a major sleep-promoting structure in Drosophila (44–48). To determine whether nur neurons are required for sleep after stress, we used the infection assay. Silencing of nur neurons by expression of an inward-rectifying potassium channel (Kir) attenuated sleep increases after infection (fig. S14C).
Localization of NUR at the dorsal fan-shaped body in response to sleep deprivation
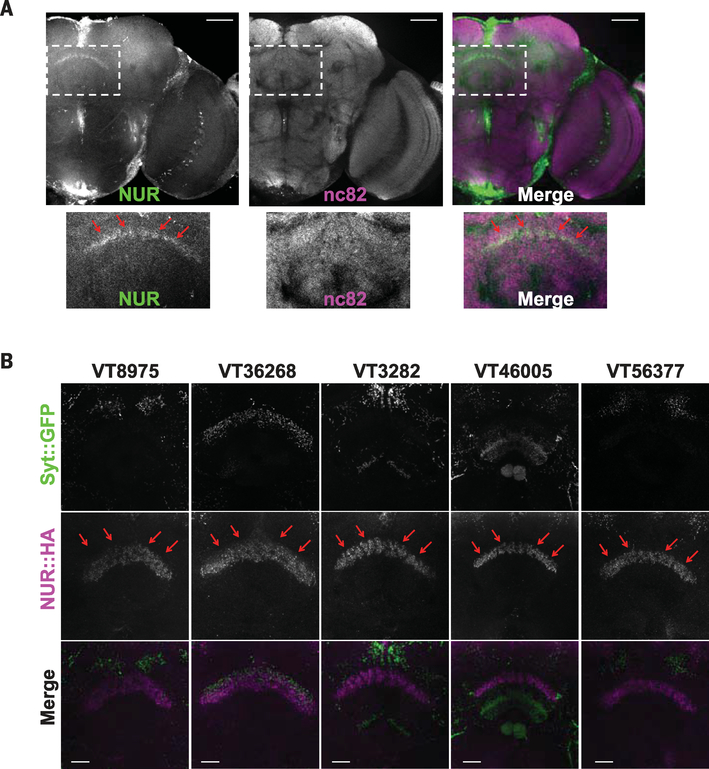
To address the regulation of NUR protein in the brain, we raised antibodies against NUR (fig. S16A). NUR antibodies recognized NUR transfected in S2 cells (fig. S16B) by immunoblotting and recognized NUR overexpressed in fly brains by immunohistochemistry (fig. S16C). The immunohistochemistry supported the quantitative PCR (qPCR) analysis and Gal4 expression (Fig. 5, A and B) in that the NUR antibody did not detect a specific signal in nur mutants or in unperturbed wild-type flies (fig. S16D), although some background staining was evident in both. After sleep deprivation, endogenous NUR was detected in the dFSB (Fig. 6A). Overexpression by a pan-neuronal driver, elav-GS, also showed greater accumulation of NUR in the dFSB as well as in the sleepregulating mushroom bodies (22, 49) (fig. S16C), indicating the dFSB as a possible site of its action/localization.
Fig. 6. NUR is induced by sleep loss and is localized to the dFSB area of the brain.
(A) Expression of endogenous NUR after sleep deprivation. Staining is for anti-NUR (green) and nc82 (magenta). The insets in the upper images are magnified below. Red arrows indicate expression of NUR in the dFSB. Scale bars, 50 μm. (B) Ectopic expression of NUR by different Gal4 drivers. Each Gal4 line was crossed with flies carrying UAS-syt::GFP and UAS-NUR::HA. Staining is with anti-GFP (top), anti-HA (middle), or merge (bottom). Note NUR expression in the dFSB area. Scale bars, 20 μm.
To determine whether localization of NUR to the dFSB is required for its sleep-inducing effects, we selected nine Gal4 lines from the ectopic expression analysis that expressed GFP relatively sparsely in the fly brain (Fig. 2C and table S2) and assayed NUR expression. Because the NUR antibody staining was not strong, we crossed each Gal4 line with UAS-nur::HA (hemagglutinin) to provide an epitope tag for effective detection. Prominent NUR expression in the dFSB was observed with five Gal4 lines that had increased sleep (Fig. 6B). Crossing each of these to synaptotagmin::GFP, which labels axonal projections, revealed that almost all projected near the dFSB (Fig. 6B). Conversely, four Gal4 lines that did not show increased sleep upon nur expression (table S2) showed little NUR expression or axonal projections in the region of the dFSB (fig. S17).
The dFSB is a neuropil structure in which the sleep-promoting projections arise from cell bodies labeled by the 23E10-Gal4 driver (44) and located in the superior medial protocerebrum. We considered the possibility that nur+ neurons signal through 23E10 neurons to promote sleep. However, NUR staining in the dFSB did not colocalize with 23E10 projections (fig. S18A), which is consistent with findings that the dFSB primarily houses the axons, rather than postsynaptic terminals, of 23E10 neurons (46, 50). Silencing of 23E10 neurons with the inward-rectifying potassium channel Kir also did not prevent increased sleep by pan-neuronally expressed NUR (fig. S18B). These findings indicate that 23E10+ neurons and nur+ neurons may innervate a common downstream target within the dFSB, or they may promote sleep through independent mechanisms.
Discussion
We identified a sleep-inducing molecule, nemuri (nur), that is required for deep sleep as well as for the induction of sleep after arousal, sleep deprivation, or infection (Fig. 4, D to H). NUR fits the criteria for a somnogen, a secreted molecule that transmits homeostatic sleep need. It increases when sleep need is high, and inducing its expression increases sleep.
From a screen of >12,000 lines, we found only one gene that increased sleep in Drosophila. Sleep-inducing molecules may be rare; this presumption is supported by genetic analysis of other sleep factors in flies (14, 17, 21) and also by data available for neurochemical systems that affect sleep and wake. In both flies and mammals, many classical neurotransmitters promote arousal-acetylcholine, dopamine, histamine, norepinephrine/octopamine-but only g-aminobutyric acid (GABA) is consistently associated with increases in sleep (5, 6). However, we acknowledge that our screen may have missed some sleep-inducing genes, either because they are small and less targetable by transposable elements or because they have small effects.
Other than adenosine, previously identified candidates for somnogens are cytokines such as interleukin-1 and tumor necrosis factor-α, which accumulate after prolonged wakefulness and appear to promote sleep (10, 51). In mammals, cytokines can induce production of AMPs (52, 53), but AMPs can also affect expression of cytokines (35). Expression of some AMPs has been associated with changes in sleep state in Drosophila (37, 54, 55), and we now identify an AMP that provides a mechanistic link between immune function and sleep. NUR is relevant for sickness or stress-induced sleep, which is seen in many organisms (56–58). In response to infection, NUR appears to kill microbes, most likely in the periphery, and increases sleep through its action in the brain. Several AMPs have multiple functions that help to combat infection (35), but the additional functions usually affect the immune system. The sleep-promoting role of NUR may be as important for host defense, given that increased sleep during sickness promotes survival (36–8, 59).
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Sato, T. Suzuki, Y. Kato, M. Taki, T. Tomoda, A. Keene, and the Sehgal laboratory for helpful discussions and technical input on this work; T. Matsuda, C. Quake, A. Vigderman, H. Wang, Z. Yue, D. Chen, and C. Stein for technical assistance; A. King, D. Garbe, and D. Cavanaugh for selection of Gal4 lines; A. Parks for the list of EP lines; P. Haynes and C. Dubowy for feedback on the manuscript; M. Heisenberg, C.-H. Lee, and L. Prieto-Godino for providing flies; the Bloomington Drosophila Stock Center, VDRC, Centre for Cellular and Molecular Platforms, and FlyORF for providing fly stocks; DSHB for providing antibody; and NIG and DGRC for plasmid distribution.
Funding: A.S. is an Investigator of the Howard Hughes Medical Institute. J.A.W. is supported by NIH grant 1R01GM123783.
Footnotes
Competing interests: Authors declare no competing interests.
Data and materials availability: All data are available in the manuscript or the supplementary materials; raw data are available upon request.
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/363/6426/509/suppl/DC1
Materials and Methods
Figs. S1 to S18
Tables S1 to S3
References (60–63)
REFERENCES AND NOTES
- 1.Frank MG, Rev. Neurosci 17, 375–392 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Mignot E, PLOS Biol. 6, e106 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allada R, Chung BY, Annu. Rev. Physiol 72, 605–624 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohawk JA, Green CB, Takahashi JS, Annu. Rev. Neurosci 35, 445–462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crocker A, Sehgal A, Genes Dev 24, 1220–1235 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sehgal A, Mignot E, Cell 146, 194–207 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pappenheimer JR, Miller TB, Goodrich CA, Proc. Natl. Acad. Sci. U.S.A 58, 513–517 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bjorness TE, Greene RW, Curr. Neuropharmacol 7, 238–245 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryant PA, Trinder J, Curtis N, Nat. Rev Immunol 4, 457–467 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Imeri L, Opp MR, Nat. Rev Neurosci 10, 199–210 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Axelrod S, Saez L, Young MW, Methods Enzymol. 551, 3–27 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Kume K, Kume S, Park SK, Hirsh J, Jackson FR, J. Neurosci 25, 7377–7384 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cirelli C et al. , Nature 434, 1087–1092 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Stavropoulos N, Young MW, Neuron 72, 964–976 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Afonso DJ et al. , Curr. Biol 25, 1717–1726 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S et al. , Neuron 82, 151–166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi M, Yue Z, Kuryatov A, Lindstrom JM, Sehgal A, eLife 3, e01473 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rogulja D, Young MW, Science 335, 1617–1621 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfeiffenberger C, Allada R, PLOS Genet 8, e1003003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koh K et al. , Science 321, 372–376 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu MN et al. , Nat. Neurosci 13, 69–75 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joiner WJ, Crocker A, White BH, Sehgal A, Nature 441, 757–760 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Rørth P, Proc. Natl. Acad. Sci. U.S.A 93, 12418–12422 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bischof J, Sheils EM, Björklund M, Basler K, Nat. Protoc 9, 1607–1620 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guruharsha KG et al. , Cell 147, 690–703 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGuire SE, Mao Z, Davis RL, Sci. STKE 2004, pl6 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Tataroglu O, Emery P, Methods 68, 140–150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garbe DS et al. , Biol. Open 4, 1558–1568 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dubowy C et al. , Sleep 39, 1083–1095 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Özkan E et al. , Cell 154, 228–239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenett A et al. , Cell Rep. 2, 991–1001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zasloff M, Nature 415, 389–395 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Halldórsdóttir K, Árnason E, PeerJ 3, e976 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soscia SJ et al. , PLOS ONE 5, e9505 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai Y, Gallo RL, Trends Immunol. 30, 131–141 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allada R, Cirelli C, Sehgal A, Cold Spring Harb. Perspect. Biol 9, a027730 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuo TH, Williams JA, Sleep 37, 1077–1086 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuo TH, Williams JA, Sleep 37, 859–869 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryder E et al. , Genetics 167, 797–813 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hendricks JC et al. , Neuron 25, 129–138 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G, Science 287, 1834–1837 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Kuo TH, Pike DH, Beizaeipour Z, Williams JA, BMC Neurosci. 11, 17 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown JB et al. , Nature 512, 393–399 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Donlea JM, Pimentel D, Miesenböck G, Neuron 81, 860–872 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Donlea JM, Thimgan MS, Suzuki Y, Gottschalk L, Shaw PJ, Science 332, 1571–1576 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pimentel D et al. , Nature 536, 333–337 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ueno T et al. , Nat. Neurosci 15, 1516–1523 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Liu Q, Liu S, Kodama L, Driscoll MR, Wu MN, Curr. Biol 22, 2114–2123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pitman JL, McGill JJ, Keegan KP, Allada R, Nature 441, 753–756 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Cavanaugh DJ, Vigderman AS, Dean T, Garbe DS, Sehgal A, Sleep 39, 345–356 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Opp MR, Krueger JM, Brain Behav Immun. 47,1–3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Erdag G, Morgan JR, Ann. Surg 235, 113–124 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bando M et al. , Immunol. Cell Biol 85, 532–537 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Williams JA, Sathyanarayanan S, Hendricks JC, Sehgal A, Sleep 30, 389–400 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dissel S et al. , Brain Behav Immun 47, 75–85 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nelson MD et al. , Curr. Biol. 24, 2406–2410 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lenz O, Xiong J, Nelson MD, Raizen DM, Williams JA, Brain Behav Immun. 47, 141–148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hill AJ, Mansfield R, Lopez JM, Raizen DM, Van Buskirk C, Curr. Biol 24, 2399–2405 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toth LA, Tolley EA, Krueger JM, Proc. Soc. Exp. Biol. Med 203, 179–192 (1993). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.