Abstract
Recent studies report serine ADP-ribosylation on nucleosomes during the DNA damage response. We unveil histone H3 serine 10 as the primary acceptor residue for chromatin ADP-ribosylation and find that specific histone acetylation marks block this activity. Our results provide a molecular explanation for the well-documented phenomenon of rapid deacetylation at DNA damage sites and support the combinatorial application of PARP and HDAC inhibitors for the treatment of PARP-dependent cancers.
Protein ADP-ribosylation is a critical feature of multiple DNA damage response (DDR) pathways1. Upon sensing DNA strand breaks, poly(ADP-ribose) polymerase (PARP) enzymes ADP-ribosylate a host of local substrates, including PARPs and histones, initiating chromatin remodeling and DNA repair factor recruitment1–4. Recently, a PARP½ stimulating protein, histone PARylation factor 1 (HPF1), was identified that activates PARP for serine ADP-ribosylation (Ser-ADPr) and that is required for DNA damage-induced histone ADP-ribosylation5,6. Given the clinical and biological significance of PARP11,7–9, it is necessary to fully define the molecular features that regulate the activity of this enzyme. Of particular interest is ADP-ribosylation of histones, which represent major trans-substrates for Ser-ADPr by PARP1/HPF15. Chromatin exhibits extensive and dynamic post-translational modification (PTM) landscapes7,10; however, the interplay between these modifications and histone ADP-ribosylation is poorly understood. Therefore, we envisioned biochemical and mammalian cell-based approaches that would enable strict control over the PTM profiles on both PARP enzymes and their nucleosome substrates.
To delineate PARP½ histone substrate preferences, we employed recombinant enzymes and mononucleosomes in NAD+ cofactor-based ADP-ribosylation assays (Supplementary Fig. 1). We observed nucleosome ADP-ribosylation by PARP1 and PARP2 almost exclusively on histone H3 in an HPF1-dependent manner (Fig. 1a, Lane 4 and Supplementary Fig. 2)5. Further analysis revealed H3S10 to be the primary chromatin target of both PARP1/HPF1 and PARP2/HPF1 (Fig. 1b and Supplementary Fig. 3), while H3S28, which resides in a similar Arg-Lys-Ser motif, serves as a secondary acceptor residue. This is consistent with a very recent report that demonstrated H3S10 is a more efficient ADP-ribose acceptor than H3S28 in mammalian cells11. We suspect background signal in the H3S10A/H3S28A sample (Fig. 1b, Lane 4) represents previously identified12, low abundance histone Ser-ADPr sites. Importantly, we verified the presence of a serine-linked ADP-ribose moiety within the H3K9/S10 (‘K-S’) substrate motif by treating ADP-ribosylation assay products with the O-linked-ADP-ribose hydrolase, ARH313 (Supplementary Fig. 4). Additionally, in the course of these initial biochemical experiments, we noticed that isolated HPF1 is a target for ADP-ribosylation by PARP½ (Fig. 1a,b, Lane 4 and Supplementary Fig. 5). While the site(s) of ADP-ribosylation on HPF1 remain to be determined (as does its functional effects), we note that the protein contains several ‘K-S’ motifs, which seem good candidates for the PARP-dependent modification.
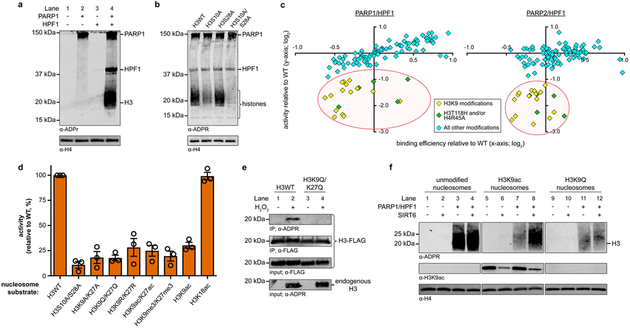
Figure 1. DNA damage-induced H3S10 ADP-ribosylation is blocked by H3K9 acetylation.
a, ADP-ribosylation of recombinant mononucleosomes by recombinant PARP1 in the absence and presence of HPF1 (cofactor = unlabeled NAD+). b, Recombinant PARP1/HPF1 ADP-ribosylation activity on recombinant mononucleosomes substrates wherein known H3 Ser-ADPr sites were mutated as indicated (cofactor = unlabeled NAD+). For biotin-NAD+ cofactor-based assays and product quantification see Supplementary Fig. 3. c, Binding efficiency and ADP-ribosylation activity (relative to wild type nucleosomes) for each member of the mononucleosome library in PARP1/HPF1 (left) and PARP2/HPF1 (right) assays. The red circle indicates all significant (two-tailed P-value ≤ 0.01; n = 3 from three independent assays) hits that exhibit a > 2-fold effect on activity. d, Quantification of recombinant PARP1/HPF1 ADP-ribosylation activity on recombinant, mutated/modified mononucleosome substrates as indicated for each lane. Replicate western blots used for quantification can be found in Supplementary Fig. 9 and 26. Center bar represents the mean and error bars represent standard error of the mean (n = 3 from three independent assays). e, ADP-ribosylation analysis of H3-Flag constructs from H2O2-treated U2OS cells. f, Recombinant PARP1/HPF1 ADP-ribosylation assays on the indicated recombinant, modified/mutated mononucleosomes substrates in the presence of the recombinant SIRT6 deacetylase (cofactor = unlabeled NAD+). Both ADP-ribosylation and H3K9 deacetylation activities are shown. Uncropped blots and replicates can be found in Supplementary Fig. 26. Unlabeled NAD+ cofactor-based assays and live cell assays were repeated once to ensure reproducibility.
In the nuclear environment, PARPs are likely to encounter a wide range of pre-existing or DDR-induced chromatin landscapes upon activation at DNA damage sites14. With this in mind, we utilized a DNA-barcoded mononucleosome library that comprises unique histone PTMs, mutations, and variants15 to identify nucleosome modifications that modulate PARP1/HPF1 and PARP2/HPF1 binding and activity (Supplementary Fig. 6). While no significant (>2-fold) binding preferences were observed, both paralogs exhibited 2- to 6-fold decreases in activity relative to wild type substrates on 20 modified nucleosomes (Fig. 1c). These negative effectors fell into two distinct classes: (i) mutations to residues at the dyad axis and (ii) modifications at H3K9, the residue directly upstream (the ‘−1’ site) relative to the primary serine target (Supplementary Fig. 7). Remarkably, all nucleosomes in our library screen with a modification at H3K9 (15 members) met our repressive effector criteria for both PARP1 and PARP2 (Supplementary Dataset 1). Immunoblot-based validation assays confirmed that the dyad axis mutations have an inhibitory effect on PARP enzyme activity, which is in agreement with previous reports that PARP1 interacts with this region of the nucleosome16 (Supplementary Fig. 8). We also generated a series of designer nucleosomes to further investigate the dependence of H3 Ser-ADPr on the ‘−1’ lysine residue (Fig. 1d and Supplementary Figs. 8 and 9). Collectively, our biochemical data indicate that −1 lysine mutations and common PTMs, particularly acetylation (which has both steric and electrostatic effects), disrupt Ser-ADPr by PARP½. Additionally, we verified that this negative regulation is specific to Lys-9 modifications by showing that H3K18ac has no effect on PARP1/HPF1 activity while H3K9ac is sufficient to abolish H3S10ADPr.
Next, we developed a mammalian cell-based assay to explore the effect of histone acetylation on Ser-ADPr in the DDR. Robust histone ADP-ribosylation was observed in U2OS cells following hydrogen peroxide (H2O2) treatment as previously described6 (Supplementary Fig. 10). Further immunoblot analysis of the chromatin fraction indicated that the predominant ADP-ribosylated species corresponded to histone H3. FLAG-tagged H3 mutants were then introduced into U2OS cells, which could be isolated via histone immunoprecipitation (IP) after H2O2 treatment and subsequently probed with a pan ADP-ribosylation detection reagent (Supplementary Fig. 11). This workflow revealed that H2O2-induced chromatin ADP-ribosylation is dependent upon the presence of Ser-10 (Supplementary Fig. 12). We also confirmed that ‘−1’ lysine acetylation blocks Ser-ADPr by PARP½, as the acetyllysine mimic H3K9Q/K27Q was refractory to ADP-ribosylation in our cellular DNA damage assays (Fig. 1e, Lane 4).
Our results indicate that H3K9ac, an abundant mark in active regions of the genome10, must be removed for H3 Ser-ADPr to occur. Indeed, previous reports implicate multiple histone deacetylases in DNA repair pathways17,18 and show that H3K9 is rapidly deacetylated in response to DNA damage19. In particular, SIRT6, which directly interacts with and activates PARP1 via its mono-ADP-ribosylation activity, also exhibits H3K9 deacetylation activity to further stimulate repair through an unresolved mechanism17,20. We found that addition of active SIRT6 to our biochemical ADP-ribosylation assays restored H3 Ser-ADPr by PARP1/HPF1 on H3K9ac-containing nucleosomes (Fig. 1f, Lanes 7–8). This effect was not observed in the presence of a non-hydrolyzable acetylation mimic at this position (H3K9Q; Fig. 1f, Lanes 11–12), confirming that SIRT6 nucleosome deacetylation activity is required for activation of PARP1/HPF1 towards these substrates. Notably, we also found that SIRT6 is a substrate for ADP-ribosylation by PARP1/HPF1 (Supplementary Fig. 13), which confirms an interaction between these proteins in our assay. Thus, we propose that one function of deacetylases such as SIRT6 at DNA strand break sites is to prime histone substrates for efficient Ser-ADPr in PARP1-dependent repair pathways.
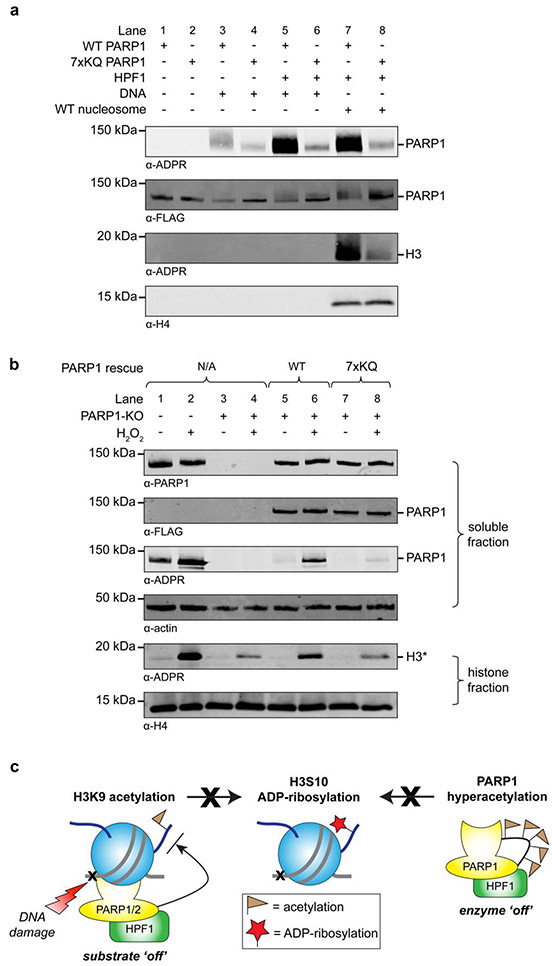
Analysis of MS-based proteomics data5,21 revealed that greater than 60% of annotated Ser-ADPr sites (171 out of 276) contain a ‘−1’ lysine residue (Supplementary Fig. 14). The prevalence of this K-S motif, together with our biochemical findings on H3 Ser-ADPr, suggests that ‘−1’ lysine acetylation may be a conserved regulatory feature of protein ADP-ribosylation. To investigate this idea, we analyzed an intrinsically disordered segment of the PARP1 automodification domain (AM peptide, residues 494–524), which is a substrate for Ser-ADPr, whilst also being a region of hyperacetylation by p3005,22. Indeed, we found that −1 lysine acetylation (K498/K506/K518ac) on the AM peptide blocks Ser-ADPr (<10% activity relative to wild type substrate), while a majority of activity is retained on an AM peptide with acetylation at other lysines (K512/K521/K524ac, Supplementary Fig. 15). Next, a recombinant PARP1 construct that mimics the hyperacetylated enzyme (‘7xKQ’; for protein folding characterization see Supplementary Fig. 16) was generated for biochemical assays, which revealed that auto-ADP-ribosylation was drastically reduced regardless of HPF1 stimulation (Fig. 2a, Lanes 3–6). Notably, in the context of full-length PARP1, auto-ADP-ribosylation of aspartate, glutamate, serine, and lysine residues has been reported5,21. We propose that the reduction in activity we observe reflects a mechanism wherein acetylation not only directly blocks ADP-ribosylation of lysines and adjacent serine residues, but also negatively regulates PARP1 enzymatic activity. This mechanism is in agreement with previous evidence that acetylation and ADP-ribosylation within the AM domain modulate PARP1 function22,23.
Figure 2. Acetylation of PARP1 regulates its auto- and trans-ADP-ribosylation activities.
a, Recombinant PARP1 ADP-ribosylation assays comprising wild type or 7xKQ PARP1 constructs, and different stimulatory factors (HPF1, free DNA, mononucleosomes) as indicated above each lane (cofactor = unlabeled NAD+). b, Immunoblot analysis of wild type and 7xKQ PARP1 construct auto- and trans-ADP-ribosylation activities in a PARP1 knockout (KO) U2OS cell line. Note: the H2O2-dependent ADP-ribosylation observed in the PARP1-knockout cell line represents PARP2 activity8. c, Model wherein acetylation of H3K9 (substrate) or the PARP1 AM domain (enzyme) blocks ADP-ribosylation of the H3S10 residue. Uncropped blots can be found in Supplementary Fig. 26. Unlabeled NAD+ cofactor-based assays and live cell assays were repeated once to ensure reproducibility.
We next investigated the effect of PARP1 hyperacetylation on its ability to catalyze Ser-ADPr of trans-substrates. We found that the 7xKQ PARP1 construct displayed a profound reduction in nucleosomal H3 ADP-ribosylation activity relative to the wild type enzyme in biochemical assays (Fig. 2a, Lanes 7–8). The mutant enzyme also showed diminished activity towards histone H1 (Supplementary Fig. 17)5, further demonstrating the inhibitory effect of PARP1 hyperacetylation. To explore this effect in mammalian cells, the CRIPSR/Cas9 system was utilized to generate two unique PARP1 knockout U2OS cell lines, which were found to be deficient in DDR-induced histone ADP-ribosylation (Fig. 2b, Lanes 1–4 and Supplementary Fig. 18). As expected, transfection of a wild type PARP1 construct into this cell line was able to rescue DNA damage-induced histone ADP-ribosylation (Fig. 2b, Lanes 5–8 and Supplementary Fig. 18). By contrast, expression of the 7xKQ mutant construct had no effect on H3 ADP-ribosylation levels in these cellular assays. Hence, AM domain acetylation is a PARP1 regulatory feature that controls Ser-ADPr of nucleosomes and likely other substrates (Fig. 2c).
Here we use sophisticated chemical biology tools to unveil multiple independent mechanisms through which protein acetylation antagonizes DNA damage-induced chromatin ADP-ribosylation. Given the high incidence of ‘−1’ lysine residues in known Ser-ADPr sites, we speculate that the acetylation-ADP-ribosylation PTM switch characterized herein may be a prevalent mode of regulation throughout the proteome. From a biomedical perspective, we note that both histone deacetylase and PARP inhibitors are FDA-approved chemotherapeutics, and previous reports have found that they act synergistically to target various cancers24,25. Our work lays out a potential explanation for this observation whereby combinatorial treatment would abrogate DNA damage-induced ADP-ribosylation through multiple pathways, which include disruption of the acetylation-ADP-ribosylation PTM switch. Building on this idea may lead to the development of more potent and effective therapeutic strategies, particularly in the area of known PARP-dependent diseases such as BRCA½-deficient malignancies.
Online Methods
Materials
Dimethylformamide (DMF), dichloromethane (DCM), and triisopropylsilane (TIS) were purchased from Sigma-Aldrich (Milwaukee, WI) and used without further purification. Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) was purchased from ThermoFisher Scientific (Waltham, MA). Fmoc amino acids were purchased from Novabiochem (Darmstadt, Germany) or Bachem (Torrance, CA). ChemMatrix Rink Amide resin and ChemMatrix Trityl-OH PEG resin was purchased from Biotage (Charlotte, NC). Trifluoroacetic acid (TFA) was purchased from Halocarbon (North Augusta, SC). Olaparib was purchased from Selleckchem (Houston, TX). Biotin-NAD+ was purchased from Trevigen (Gaithersburg, MD). All other chemicals were purchased from SigmaAldrich (Milwaukee, WI) and used without further purification. Analytical reversed-phase HPLC (RP-HPLC) was performed on an Agilent 1100 or 1260 series instrument employing a Vydac C18 column (5 μm, 4 × 150 mm) at a flow rate of 1 mL/min. Semi-preparative scale purifications were performed on an Agilent 1260 series instrument employing a Vydac C18 semipreparative column (12 μm, 10 mm × 250 mm) at a flow rate of 4 mL/min. Preparative RP-HPLC was performed on a Waters prep LC system comprised of a Waters 2545 Binary Gradient Module and a Waters 2489 UV detector. Purifications were carried out on a C18 Vydac 218TP1022 column (10 µM; 22 × 250 mm) at a flow rate of 18 mL/min. For all RP-HPLC purifications and analyses, 0.1% TFA in water (HPLC solvent A) and 90% acetonitrile, 0.1% TFA in water (HPLC solvent B) were used as the mobile phases. Electrospray ionization mass spectrometry (ESI-MS) analysis was conducted on a MicrOTOF-Q II ESI-Qq-TOF mass spectrometer (Bruker Daltonics). All FPLC steps described here were performed on an AKTA FPLC system (GE Healthcare). SDS-PAGE gels were imaged on an ImageQuant system (GE Healthcare) and western blots were performed using fluorescent secondary antibodies (see ‘Supplementary Table 1‘) and imaged on an Odyssey system (Li-Cor, Lincoln, NE).
Molecular cloning
All PCR amplification steps described here were performed using the PfuUltra II Fusion HS DNA Polymerase (Agilent Technologies) according to manufacturer’s protocols. All primers were ordered from Integrated DNA Technologies (IDT, Coralville, IA). All plasmids used in this study were sequence verified by GENEWIZ (South Plainfield, NJ). Amino acid and nucleosome DNA sequences can be found below (‘DNA/protein sequence information’).
601 DNA preparation
601 DNA was amplified with primers containing the desired 15 base pair overhangs from a parent plasmid harboring the 147 base pair 601 sequence. The PCR product was purified via a Qiagen (Valencia, CA) miniprep spin column protocol, ethanol precipitated, and resuspended to the desired DNA concentration (A260 ~20 or ~1 μg/μL) for nucleosome assembly.
PARP½
The human PARP1 gene was purchased from GE Healthcare and subcloned into Sf9 cell (pACEBac1) and mammalian cell (pCMV) expression plasmids via restriction enzyme digest cloning. Amino acid tags for purification, immunoprecipitation, and western blot identification were incorporated into the gene amplification primers. PARP1 automodification domain mutants were introduced via inverse PCR by incorporating desired mutations into primer overhangs. The linear product was then purified on an agarose gel, phosphorylated (PNK, New England Biolabs), and ligated (T4 DNA ligase, New England Biolabs) following standard procedures. The human PARP2 gene was synthesized by IDT and cloned into expression plasmids as described for PARP1.
HPF1
The human HPF1 gene was synthesized by IDT and cloned into an E. coli expression plasmid (SUMO-pET30) via restriction enzyme digest cloning. Amino acid tags for purification, immunoprecipitation, and western blot identification were incorporated into the gene amplification primers.
ARH3
The human ARH3 gene was synthesized by IDT and cloned into an E. coli expression plasmid (SUMO-pET30) via restriction enzyme digest cloning. Amino acid tags for purification, immunoprecipitation, and western blot identification were incorporated into the gene amplification primers.
Histone H3
All histone H3 mutations described here were introduced via inverse PCR by incorporating desired mutations into primer overhangs using a parent plasmid harboring the human H3.1 gene (E. coli expression plasmid: pET30). The linear product was then purified on an agarose gel, phosphorylated, and ligated following standard procedures. The mutants were then subcloned into a mammalian cell expression plasmid (pCMV) via the Gibson Assembly method (New England Biolabs) with primers that contain the 3xFlag tag for immunoprecipitation.
Recombinant protein preparation
Final purified recombinant proteins (PARP1-WT, PARP1–7xKQ, PARP2, HPF1, ARH3, and SIRT6) are shown on an SDS-PAGE gel in Supplementary Fig. 19.
PARP½
All PARP proteins described in this study were produced in Sf9 cells using a baculovirus expression system. The plasmids described above were used to generate bacmids following the manufacturer’s protocol (MultiBac, Geneva Biotech). To generate virus for protein production, bacmid transfection into Sf9 cells was carried out in 6-well plates. All transfection, viral amplification, and infection steps were performed in a sterile hood. Typically, 8 μg of bacmid were transfected into 1 × 106 attached Sf9 cells according to the manufacturer’s instructions (Baculovirus Expression Vector System, ThermoFisher Scientific). After transfection, cells were overlaid with 2 mL fresh medium (Sf-900III SFM, Thermo Fisher Scientific) and incubated at 27 °C for 96 h in the dark. The supernatant was collected for virus amplification and filtered through 0.22 µm to generate the P1 virus (2% FBS (v/v) was added). Between uses, all viral stocks were stored at 4 °C in the dark. Subsequent steps were carried out in medium with penicillin/streptomycin. To generate the P2 virus, 1 mL P1 virus was added to 10 mL of Sf9 cells in a sterile flask at 1.5 × 106 cells per mL. Cells were grown at 27 °C in suspension culture in the dark until they reached 50% viability as monitored by trypan blue staining. The culture supernatant was then collected and filtered through 0.22 µm, and 2% FBS (v/v) was added. To generate the P3 virus (used for protein production), 300 μL P2 virus was added to 30 mL of Sf9 cells in a sterile flask at 1.5 × 106 cells per mL. Cells were grown at 27 °C in suspension culture in the dark until they reached 50% viability as monitored by trypan blue staining. The culture supernatant was then collected and cleared by centrifugation, and 2% FBS (v/v) was added. During virus amplification Sf9 cell density was kept at around 1.5 × 106 cells per mL, diluting if needed, until growth arrested and viability dropped. Production of protein was carried out by adding a 1:100 dilution of each P3 virus to Sf9 suspension cultures at 1.5 × 106 cells per mL. Cells were harvested by centrifugation after 48 h at 27 °C in the dark. All recombinant PARP proteins were purified by Ni-affinity and gel filtration. Briefly, cells were lysed via sonication in a lysis buffer (25 mL per liter of cells; 40 mM Tris, pH 7.7, 1 M NaCl, 1.5 mM MgCl2, 10% glycerol, 5 mM β-ME, 1.0 mg of DNase per liter of cells, and 1 mM PMSF) and soluble lysates were isolated by centrifugation at 20,000 RCF for 30 minutes. The soluble lysate was then applied to pre-equilibrated Ni-affinity resin (2 mL of resin per liter of cells; Ni-NTA; Thermo Scientific) to capture the desired PARP protein. The column was washed with 300 mL of lysis buffer supplemented with 20 mM imidazole and subsequently eluted using 30 mL of Elution buffer (40 mM Tris, pH 7.7, 500 mM NaCl, 300 mM imidazole, 10% glycerol, and 5 mM β-ME). This elution was concentrated to 2 mL using an Amicon Ultra Centrifugal filter (Millipore; 30-kDa MWCO) and injected onto a gel filtration column (HiLoad 16/60 Superdex 200; GE Healthcare) that had been pre-equilibrated with a buffer containing 50 mM Tris, pH 7.5, 100 mM NaCl, 10% glycerol, 2 mM DTT. Pure fractions (as judged by SDS-PAGE gel electrophoresis) were concentrated to 100 μM as quantified by BCA assay, flash frozen in single-use aliquots and stored at −80 °C.
HPF1
The expression plasmid was transformed into Rosetta(DE3) cells, and 8 L were grown at 37 °C to OD600 = 0.6. The temperature was then changed to 18 °C, and expression was induced by addition of 0.5 mM IPTG for 16 hours. Cells were lysed via sonication in a lysis buffer containing 40 mM Tris, pH 7.5, 0.5 M NaCl, 5 mM β-ME, and 1 mM PMSF, and soluble lysate was isolated by centrifugation at 20,000 RCF for 30 minutes. The soluble lysate was then applied to pre-equilibrated Ni-affinity resin (2 mL of resin per liter of cells) to capture the 6xHis-Sumo-Flag-HPF1 protein. The column was washed with 300 mL of lysis buffer supplemented with 20 mM imidazole and subsequently eluted using 40 mL of lysis buffer supplemented with 300 mM imidazole. The eluted protein was dialyzed for 16 hours at 4 °C into dialysis buffer (40 mM Tris, pH 7.5, 0.3 M NaCl, 5 mM β-ME) in the presence of Ulp1 to remove the Sumo tag. A reverse nickel column was then run to remove the cleaved tag, and the flow-through containing Flag-HPF1 was collected. Next, the protein was concentrated to 2 mL using an Amicon Ultra Centrifugal filter (Millipore; 10-kDa MWCO) and injected onto a gel filtration column (HiLoad 16/60 Superdex 200; GE Healthcare) that had been pre-equilibrated with a buffer containing 50 mM Tris, pH 7.5, 100 mM NaCl, 10% glycerol, 2 mM DTT. Pure fractions (as judged by SDS-PAGE gel electrophoresis) were concentrated to 350 μM as quantified by BCA assay, flash frozen in single-use aliquots and stored at −80 °C.
ARH3
The expression plasmid was transformed into Rosetta(DE3) cells, and 8 L were grown at 37 °C to OD600 = 0.6. The temperature was then changed to 18 °C and, expression was induced by addition of 0.5 mM IPTG for 16 hours. Cells were lysed via sonication in a lysis buffer containing 40 mM Tris, pH 7.5, 0.5 M NaCl, 10 mM MgCl2, 5 mM β-ME, and 1 mM PMSF, and soluble lysate was isolated by centrifugation at 20,000 RCF for 30 minutes. The soluble lysate was then applied to pre-equilibrated Ni-affinity resin (2 mL of resin per liter of cells) to capture the 6xHis-Sumo-ARH3-HA protein. The column was washed with 300 mL of lysis buffer supplemented with 20 mM imidazole and subsequently eluted using 40 mL of lysis buffer supplemented with 300 mM imidazole. The eluted protein was dialyzed for 16 hours at 4 °C into dialysis buffer (40 mM Tris, pH 7.5, 0.3 M NaCl, 10 mM MgCl2, 5 mM β-ME) in the presence of Ulp1 to remove the Sumo tag. A reverse nickel column was then run to remove the cleaved tag, and the flow-through containing ARH3-HA was collected. Next, the protein was concentrated to 2 mL using an Amicon Ultra Centrifugal filter (Millipore; 10-kDa MWCO) and injected onto a gel filtration column (HiLoad 16/60 Superdex 200; GE Healthcare) that had been pre-equilibrated with a buffer containing 40 mM Tris, pH 7.5, 300 mM NaCl, 10 mM MgCl2, 10% glycerol, 2 mM DTT. Pure fractions (as judged by SDS-PAGE gel electrophoresis) were concentrated to 450 μM as quantified by BCA assay, flash frozen in single-use aliquots and stored at −80 °C.
SIRT6
The expression plasmid (Addgene plasmid #41565, a kind gift from Cheryl Arrowsmith) was transformed into Rosetta(DE3) cells, and 8 L were grown at 37 °C to OD600 = 0.6. The temperature was then changed to 18 °C, and expression was induced by addition of 0.5 mM IPTG for 16 hours. Cells were lysed via sonication in a lysis buffer containing 40 mM Tris, pH 7.5, 0.5 M NaCl, 10 mM MgCl2, 5 mM β-ME, and 1 mM PMSF, and soluble lysate was isolated by centrifugation at 20,000 RCF for 30 minutes. The soluble lysate was then applied to pre-equilibrated Ni-affinity resin (2 mL of resin per liter of cells) to capture the 6xHis-SIRT6 protein. The column was washed with 300 mL of lysis buffer supplemented with 20 mM imidazole and subsequently eluted using 40 mL of lysis buffer supplemented with 300 mM imidazole. The eluted protein was concentrated to 2 mL using an Amicon Ultra Centrifugal filter (Millipore; 10-kDa MWCO) and injected onto a gel filtration column (HiLoad 16/60 Superdex 200; GE Healthcare) that had been pre-equilibrated with a buffer containing 50 mM Tris, pH 7.5, 100 mM NaCl, 1 mM MgCl2, 10% glycerol, and 2 mM DTT. Pure fractions (as judged by SDS-PAGE gel electrophoresis) were concentrated to 400 μM as quantified by BCA assay, flash frozen in single-use aliquots and stored at −80 °C.
Linker histone
Histone H1.0 was purchased from NEB (product #: M2501S).
Core histones
All purified histones were characterized by RP-HPLC and ESI-MS analysis (Supplementary Figs. 20-22). Wild type or mutant expression plasmids were transformed into Rosetta(DE3) cells, and 1 L was grown at 37 °C to OD600 = 0.6. Protein expression was then induced by addition of 0.5 mM IPTG for 3 hours. Cells were lysed via sonication in a lysis buffer containing 40 mM Tris, pH 7.5, 0.3 M NaCl, 1 mM EDTA, 5 mM β-ME, and 1 mM PMSF, and insoluble lysate was isolated by centrifugation at 20,000 RCF for 30 minutes. The insoluble fraction was then washed with lysis buffer supplemented with 1% Triton X-100, centrifuged at 20,000 RCF for 15 minutes. This wash was repeated two more times, with the final wash being performed in the absence of 1% Triton X-100. Next, recombinant histone protein was extracted from the insoluble pellet in a buffer containing 50 mM Tris, 7.5, 300 mM NaCl, 6 M guanidine hydrochloride, and 5 mM β-ME for 1–3 hours at room temperature and centrifuged at 20,000 RCF for 30 minutes. The soluble extract was then passed through a 0.45 μM syringe-top filter (Millipore), injected onto a preparative RP-HPLC column, and eluted over a 20–80% solvent B gradient in 30 minutes. Pure fractions (as determined by LC-MS) were lyophilized and stored at −80 °C prior to use in histone octamer assembly. The truncated 6xHis-Sumo-H3 (both 15–135 (A15C) and 29–135 (A29C)) histones were expressed and extracted as described above. Following extraction, the histone was immobilized on Ni-affinity resin in extraction buffer, washed with 50 mM Tris, 7.5, 300 mM NaCl, 6 M urea, 20 mM imidazole, and 5 mM β-ME, and eluted in wash buffer supplemented with 300 mM imidazole. The eluted protein was dialyzed for 16 hours at 4 °C into dialysis buffer (50 mM Tris, pH 7.5, 300 mM NaCl, 1.5 M urea, 5 mM β-ME) in the presence of Ulp1 to remove the Sumo tag. A reverse nickel column was then run to remove the cleaved tag, and the flow-through containing truncated H3 was collected. This construct was RP-HPLC purified as described above for other histones.
DNA and protein sequence information
601 DNA sequence with 15 base pair overhangs*
CTACTGGTACGGCAGACAGGATGTATATATCTGACACGTGCCTGGAGACTAGGGAGTAATCCCCTTGGCGGTTAAAACGCGGGGGACAGCGCGTACGTGCGTTTAAGCGGTGCTAGAGCTGTCTACGACCAATTGAGCGGCCTCGGCACCGGGATTCTCCAGTATTCGAGGCCGTTC
*601 sequence is underlined; 15 base pair overhangs are necessary for full PARP stimulation by acting as double strand breaks
PARP1-Flag-6xHis (WT; Sf9 cell expression vector = pACEBac1; mammalian cell expression vector = pCMV)
MAESSDKLYRVEYAKSGRASCKKCSESIPKDSLRMAIMVQSPMFDGKVPHWYHFSCFWKVGHSIRHPDVEVDGFSELRWDDQQKVKKTAEAGGVTGKGQDGIGSKAEKTLGDFAAEYAKSNRSTCKGCMEKIEKGQVRLSKKMVDPEKPQLGMIDRWYHPGCFVKNREELGFRPEYSASQLKGFSLLATEDKEALKKQLPGVKSEGKRKGDEVDGVDEVAKKKSKKEKDKDSKLEKALKAQNDLIWNIKDELKKVCSTNDLKELLIFNKQQVPSGESAILDRVADGMVFGALLPCEECSGQLVFKSDAYYCTGDVTAWTKCMVKTQTPNRKEWVTPKEFREISYLKKLKVKKQDRIFPPETSASVAATPPPSTASAPAAVNSSASADKPLSNMKILTLGKLSRNKDEVKAMIEKLGGKLTGTANKASLCISTKKEVEKMNKKMEEVKEANIRVVSEDFLQDVSASTKSLQELFLAHILSPWGAEVKAEPVEVVAPRGKSGAALSKKSKGQVKEEGINKSEKRMKLTLKGGAAVDPDSGLEHSAHVLEKGGKVFSATLGLVDIVKGTNSYYKLQLLEDDKENRYWIFRSWGRVGTVIGSNKLEQMPSKEDAIEHFMKLYEEKTGNAWHSKNFTKYPKKFYPLEIDYGQDEEAVKKLTVNPGTKSKLPKPVQDLIKMIFDVESMKKAMVEYEIDLQKMPLGKLSKRQIQAAYSILSEVQQAVSQGSSDSQILDLSNRFYTLIPHDFGMKKPPLLNNADSVQAKAEMLDNLLDIEVAYSLLRGGSDDSSKDPIDVNYEKLKTDIKVVDRDSEEAEIIRKYVKNTHATTHNAYDLEVIDIFKIEREGECQRYKPFKQLHNRRLLWHGSRTTNFAGILSQGLRIAPPEAPVTGYMFGKGIYFADMVSKSANYCHTSQGDPIGLILLGEVALGNMYELKHASHISKLPKGKHSVKGLGKTTPDPSANISLDGVDVPLGTGISSGVNDTSLLYNEYIVYDIAQVNLKYLLKLKFNFKTSLWGGSGGDYKDDDDKGGSGVDELTGGSGHHHHHH
*Underlined residues represent PARP1 sequence
PARP1-Flag-6xHis (7xKQ; Sf9 cell expression vector = pACEBac1; mammalian cell expression vector = pCMV)
MAESSDKLYRVEYAKSGRASCKKCSESIPKDSLRMAIMVQSPMFDGKVPHWYHFSCFWKVGHSIRHPDVEVDGFSELRWDDQQKVKKTAEAGGVTGKGQDGIGSKAEKTLGDFAAEYAKSNRSTCKGCMEKIEKGQVRLSKKMVDPEKPQLGMIDRWYHPGCFVKNREELGFRPEYSASQLKGFSLLATEDKEALKKQLPGVKSEGKRKGDEVDGVDEVAKKKSKKEKDKDSKLEKALKAQNDLIWNIKDELKKVCSTNDLKELLIFNKQQVPSGESAILDRVADGMVFGALLPCEECSGQLVFKSDAYYCTGDVTAWTKCMVKTQTPNRKEWVTPKEFREISYLKKLKVKKQDRIFPPETSASVAATPPPSTASAPAAVNSSASADKPLSNMKILTLGKLSRNKDEVKAMIEKLGGKLTGTANKASLCISTKKEVEKMNKKMEEVKEANIRVVSEDFLQDVSASTKSLQELFLAHILSPWGAEVKAEPVEVVAPRGQSGAALSQQSQGQVKEEGINQSEQRMQLTLKGGAAVDPDSGLEHSAHVLEKGGKVFSATLGLVDIVKGTNSYYKLQLLEDDKENRYWIFRSWGRVGTVIGSNKLEQMPSKEDAIEHFMKLYEEKTGNAWHSKNFTKYPKKFYPLEIDYGQDEEAVKKLTVNPGTKSKLPKPVQDLIKMIFDVESMKKAMVEYEIDLQKMPLGKLSKRQIQAAYSILSEVQQAVSQGSSDSQILDLSNRFYTLIPHDFGMKKPPLLNNADSVQAKAEMLDNLLDIEVAYSLLRGGSDDSSKDPIDVNYEKLKTDIKVVDRDSEEAEIIRKYVKNTHATTHNAYDLEVIDIFKIEREGECQRYKPFKQLHNRRLLWHGSRTTNFAGILSQGLRIAPPEAPVTGYMFGKGIYFADMVSKSANYCHTSQGDPIGLILLGEVALGNMYELKHASHISKLPKGKHSVKGLGKTTPDPSANISLDGVDVPLGTGISSGVNDTSLLYNEYIVYDIAQVNLKYLLKLKFNFKTSLWGGSGGDYKDDDDKGGSGVDELTGGSGHHHHHH
*Underlined residues represent PARP1 sequence; highlighted residues represent K→Q mutation sites
PARP2-Flag-6xHis (WT; Sf9 cell expression vector = pACEBac1)
MAARRRRSTGGGRARALNESKRVNNGNTAPEDSSPAKKTRRCQRQESKKMPVAGGKANKDRTEDKQDGMPGRSWASKRVSESVKALLLKGKAPVDPECTAKVGKAHVYCEGNDVYDVMLNQTNLQFNNNKYYLIQLLEDDAQRNFSVWMRWGRVGKMGQHSLVACSGNLNKAKEIFQKKFLDKTKNNWEDREKFEKVPGKYDMLQMDYATNTQDEEETKKEESLKSPLKPESQLDLRVQELIKLICNVQAMEEMMMEMKYNTKKAPLGKLTVAQIKAGYQSLKKIEDCIRAGQHGRALMEACNEFYTRIPHDFGLRTPPLIRTQKELSEKIQLLEALGDIEIAIKLVKTELQSPEHPLDQHYRNLHCALRPLDHESYEFKVISQYLQSTHAPTHSDYTMTLLDLFEVEKDGEKEAFREDLHNRMLLWHGSRMSNWVGILSHGLRIAPPEAPITGYMFGKGIYFADMSSKSANYCFASRLKNTGLLLLSEVALGQCNELLEANPKAEGLLQGKHSTKGLGKMAPSSAHFVTLNGSTVPLGPASDTGILNPDGYTLNYNEYIVYNPNQVRMRYLLKVQFNFLQLWGGSGGDYKDDDDKGGSGVDELTGGSGHHHHHH
*Underlined residues represent PARP2 sequence
Flag-HPF1 (WT; E. coli expression vector = pET30)
SGSDYKDDDDKGGSGMVGGGGKRRPGGEGPQCEKTTDVKKSKFCEADVSSDLRKEVENHYKLSLPEDFYHFWKFCEELDPEKPSDSLSASLGLQLVGPYDILAGKHKTKKKSTGLNFNLHWRFYYDPPEFQTIIIGDNKTQYHMGYFRDSPDEFPVYVGINEAKKNCIIVPNGDNVFAAVKLFLTKKLKEITDKKKINLLKNIDEKLTEAARELGYSLEQRTVKMKQRDKKVVTKTFHGAGLVVPVDKNDVGYRELPETDADLKRICKTIVEAASDEERLKAFAPIQEMMTFVQFANDECDYGMGLELGMDLFCYGSHYFHKVAGQLLPLAYNLLKRNLFAEIIEEHLANRSQENIDQLAA
*Underlined residues represent HPF1 sequence
**An HPF1 construct lacking the Flag sequence (ΔDYKDDDDK) was used in the PARP/HPF1 library pull down assays to ensure only PARP constructs would be immobilized on Flag-agarose.
ARH3-HA (residues 14–363; E. coli expression vector = pET30)
AGAARSLSRFRGCLAGALLGDCVGSFYEAHDTVDLTSVLRHVQSLEPDPGTPGSERTEALYYTDDTAMARALVQSLLAKEAFDEVDMAHRFAQEYKKDPDRGYGAGVVTVFKKLLNPKCRDVFEPARAQFNGKGSYGNGGAMRVAGISLAYSSVQDVQKFARLSAQLTHASSLGYNGAILQALAVHLALQGESSSEHFLKQLLGHMEDLEGDAQSVLDARELGMEERPYSSRLKKIGELLDQASVTREEVVSELGNGIAAFESVPTAIYCFLRCMEPDPEIPSAFNSLQRTLIYSISLGGDTDTIATMAGAIAGAYYGMDQVPESWQQSCEGYEETDILAQSLHRVFQKSAQSLHRVFQKSYPYDVPDYARS
*Underlined residues represent ARH3 sequence
6xHis-SIRT6 (residues 3-355; E. coli expression vector = pET28a)
MGSSHHHHHHSSGLVPRGSVNYAAGLSPYADKGKCGLPEIFDPPEELERKVWELARLVWQSSSVVFHTGAGISTASGIPDFRGPHGVWTMEERGLAPKFDTTFESARPTQTHMALVQLERVGLLRFLVSQNVDGLHVRSGFPRDKLAELHGNMFVEECAKCKTQYVRDTVVGTMGLKATGRLCTVAKARGLRACRGELRDTILDWEDSLPDRDLALADEASRNADLSITLGTSLQIRPSGNLPLATKRRGGRLVIVNLQPTKHDRHADLRIHGYVDEVMTRLMKHLGLEIPAWDGPRVLERALPPLPRPPTPKLEPKEESPTRINGSIPAGPKQEPCAQHNGSEPASPKRERPTSPAPHRPPKRVKAKAVPS
*Underlined residues represent SIRT6 sequence
Linker histone H1.0 (residues 1–193; produced in E. coli by NEB)
TENSTSAPAAKPKRAKASKKSTDHPKYSDMIVAAIQAEKNRAGSSRQSIQKYIKSHYKVGENADSQIKLSIKRLVTTGVLKQTKGVGASGSFRLAKSDEPKKSVAFKKTKKEIKKVATPKKASKPKKAASKAPTKKPKATPVKKAKKKLAATPKKAKKPKTVKAKPVKASKPKKAKPVKPKAKSSAKRAGKKK
H3.1–3xFlag (mammalian cell expression vector = pCMV)
ARTKQTARKSTGGKAPRKQLATKAARKSAPATGGVKKPHRYRPGTVALREIRRYQKSTELLIRKLPFQRLVREIAQDFKTDLRFQSSAVMALQEAAEAYLVGLFEDTNLAAIHAKRVTIMPKDIQLARRIRGERADYKDDDDKADYKDDDDKADYKDDDDKA
*Underlined residues were sites of mutagenesis as described
Peptide synthesis and protein semi-synthesis
All peptides were synthesized via solid phase peptide synthesis on a CEM Discover Microwave Peptide Synthesizer (Matthews, NC) using FMOC chemistry. The histone and PARP1 automodification (AM) domain peptides were synthesized on a ChemMatrix Rink amide resin (0.5 mmol/g). The histone peptide employed in the semisynthesis of H3K9ac/K27ac and H3K9me3/K27me3 histones were synthesized on a ChemMatrix Trityl-OH PEG resin (0.49 mmol/g)*. General procedures are given first, followed by any specific methods for individual peptides. The resin was swollen in DMF and then deprotected with 20% piperidine in DMF (twice). In between all steps, the resin was washed with DMF and DCM. Each amino acid was dissolved in DMF (200 mM) and introduced to the resin after the deprotection step. DIPEA (2 M) in NMP and HOBt (500 mM)/HBTU (500 mM) in DMF were added to perform each coupling (twice). The final residue was deprotected, and the resin was then removed from the synthesizer. Cleavage was then performed with 92.5% TFA, 2.5% TIS, 2.5% EDT, and 2.5% H2O for two hours at room temperature. The cleavage reaction was precipitated with diethyl ether, dissolved in water with 0.1% TFA, and analyzed via RP-HPLC. The crude material was purified via prep scale RP-HPLC, and the desired fractions were analyzed, pooled and lyophilized. The AM domain peptides were N-terminally acetylated with acetic anhydride (20 eq) and DIPEA (40 eq) in DMF for 30 min at room temperature (twice). Then the resin was washed thoroughly with 20% piperidine, DMF, and then DCM to remove any remaining anhydride prior to cleavage. The H3(1–28) peptides were synthesized to contain a C-terminal hydrazide according to the following procedure. The Trityl-OH resin was washed with DCM and reacted with an excess of thionyl chloride (250 eq) in DCM for 16 h at room temperature. The resin was washed with DCM, and the chlorination reaction was repeated for 2 h at RT. The resin was washed with DCM, DMF, and 5% (v/v) DIPEA in DMF. On ice, the resin was reacted with 5% (v/v) hydrazine in DMF (30 eq) for 30 min. The resin was washed with DMF, and the reaction with hydrazine was repeated for 30 min. The resin was washed with DMF and then with 5% (v/v) anhydrous MeOH in DMF. See Supplementary Figures 20-24 for the RP-HPLC and ESI-MS characterization data of peptides and histone proteins.
*Ensuing native chemical ligation reactions with the H3 (A29C) construct to achieve modified, full-length histones were carried out as previously described26.
Peptide sequences
H3(1–21)WT: ARTKQTARKSTGGKAPRKQLA-amide
H3(1–21)S10A: ARTKQTARKATGGKAPRKQLA-amide
H3(1–21)K9A: ARTKQTARASTGGKAPRKQLA-amide
AM domain WT: acetyl-APRGKSGAALSKKSKGQVKEEGINKSEKRMKGGA-amide
AM domain 3xSA: acetyl-APRGKAGAALSKKAKGQVKEEGINKAEKRMKGGA-amide
AM domain 3xAc:acetyl-APRG(Kac)SGAALSK(Kac)SKGQVKEEGIN(Kac)SEKRMKGGA-amide
AM domain 3xAc-Random: acetyl-APRGKSGAALSKKSKGQV(Kac)EEGINKSE(Kac)RM(Kac)GGA-amide
H3(1–28)K9ac/K27ac: ARTKQTAR(Kac)STGGKAPRKQLATKAAR(Kac)S-NHNH2
H3(1–28)K9me3/K27me3: ARTKQTAR(Kme3)STGGKAPRKQLATKAAR(Kme3)S-NHNH2
H3 (1–28)K18ac: acetyl-ARTKQTARKSTGGKAPR(Kac)QLATKAARKS-NHNH2
H3(1–14)K9ac: ARTKQTAR(Kac)STGGK-NHNH2
Histone octamer preparation
Histone octamer and nucleosomes were prepared following a previously described workflow27. Purified histones were dissolved in histone unfolding buffer (6 M guanidine hydrochloride, 20 mM Tris, pH 7.5, and 5 mM DTT at 4 °C) and combined in equimolar ratios (50 nmol each of histones H2A, H2B, H3 (WT or mutant), and H4 (WT or mutant)). The total histone concentration was adjusted to 1 mg/mL, and the mixtures were placed in Slide-A-Lyzer MINI dialysis devices (3.5 kDa MW cutoff, ThermoFisher Scientific) and dialysed at 4 °C against 3 × 400 mL of octamer refolding buffer (2 M NaCl, 10 mM Tris, pH 7.6, 1.0 mM EDTA, 1 mM DTT at 4 °C) for at least 4 h for each step, with one dialysis step overnight. The mixtures were then concentrated to 500 μL and injected on a gel filtration column (Superdex 200 Increase 10/300 GL) that had been pre-equilibrated with a buffer containing 10 mM Tris, pH 7.6, 2 M NaCl, 1 mM EDTA, and 1 mM DTT. Fractions containing the octamer complex (as judged by FPLC elution chromatogram and SDS-PAGE gel electrophoresis) were concentrated to 30 μM as quantified by A280 (extinction coefficient = 44,700), diluted 2-fold with glycerol, and stored at a final concentration of 15 μM at −20 °C prior to use in nucleosome assembly.
Nucleosome assembly
In a typical nucleosome assembly, octamer (150 pmol) was combined with 601 DNA (180 pmol, see Supplementary Information for DNA sequence) in 75 μL buffer (2 M NaCl, 10 mM Tris, 0.1 mM EDTA, 1 mM DTT, pH 7 at 4 °C). The mixtures were placed in Slide-A-Lyzer MINI dialysis devices (3.5 kDa MW cutoff, ThermoFisher Scientific) and dialysed at 4 °C against 200 mL nucleosome assembly start buffer (10 mM Tris, 1.4 M KCl, 0.1 mM EDTA, 1 mM DTT, pH 7 at 4 °C) for 1 h at 4 °C. Subsequently, 330 mL nucleosome assembly end buffer (10 mM Tris, 10 mM KCl, 0.1 mM EDTA, 1 mM DTT, pH 7 at 4 °C) was added at a rate of 1 mL/min using a peristaltic pump, followed by two final dialysis steps against nucleosome assembly end buffer (1 h and overnight). The dialysis mixture was transferred to a microcentrifuge tube, and any precipitate was removed by centrifugation. Final nucleosome preparations were quantified by UV spectroscopy at 260 nm. The quality of individual nucleosomes was assessed by native polyacrylamide gel electrophoresis (5% acrylamide gel, 0.5x TBE, 180 V, 40 min), followed by ethidium bromide staining. This resulted in a main band migrating around 500 bp (Supplementary Fig. 25).
Recombinant ADP-ribosylation assays
ADP-ribosylation assay conditions were adapted from a previously described protocol6.
Nucleosome substrates
For all nucleosome ADP-ribosylation assays, PARP/HPF1 complexes were pre-formed in PBS (supplemented with 1 mM DTT) at concentrations of 1 μM PARP1 or 2 μM PARP2 and 20 μM HPF1. These mixtures were treated as a 10x enzyme mix to achieve final assay concentrations of 100 nM PARP1 or 200 nM PARP2 and 2 μM HPF1. In assays devoid of either of these components, a similar 10x mixture was prepared without PARP½ or HPF1 as desired. In each assay, final NAD+ cofactor concentrations were either 100 μM (biotin-NAD+) or 500 μM (unlabeled NAD+), and final substrate nucleosome concentration was 500 nM. Reactions were performed in a final buffer of 50 mM Tris, pH 7.5, 20 mM NaCl, 2 mM MgCl2, and 2 mM DTT. Reactions were carried out for 20 minutes at 30 °C, quenched by the addition of 10 μM olaparib and standard 4X SDS sample loading buffer, and boiled at 95 °C for 4 minutes. This mixture (15 μL) was then loaded onto an SDS-PAGE gel (12% Bis-Tris unless otherwise noted) and the resolved gel was transferred to a PVDF membrane for western blot analysis of products (see ‘Western blot protocol’). An antibody for Histone H4 was used as a nucleosome substrate loading control for all assays. Three independent assays were performed for all densitometry-quantified blots (n = 3, replicate western blots shown in Supplementary Fig. 26). For PARP1-WT vs. PARP1–7xKQ autoribosylation assays, α-FLAG detection was used as a PARP loading control, and all components were identical except nucleosomes were replaced with 0.25 mg/mL sheared salmon sperm DNA to stimulate PARP1 activity.*
*In the case of nucleosomes, the blunt end DNA overhangs are sufficient to mimic a double strand break and stimulate ADPr by PARP½. For assays that lack nucleosome substrates, sheared salmon sperm DNA is added to assays to stimulate activity.
Peptide and linker histone substrates
For all peptide ADP-ribosylation assays, PARP1/HPF1 complexes were pre-formed in PBS (supplemented with 1 mM DTT) at concentrations of 10 μM PARP1 and 20 μM HPF1. These mixtures were treated as a 10x enzyme mix to achieve final assay concentrations of 1 μM PARP1 and 2 μM HPF1. In assays devoid of any of these components, a similar 10x mixture was prepared without PARP1 or HPF1 as desired. In each assay, final NAD+ cofactor concentration was 100 μM (biotin-NAD+) and final substrate peptide concentration was 300 μM and H1 substrate concentration was 5 μM. Reactions were performed in a final buffer of 50 mM Tris, pH 7.5, 20 mM NaCl, 2 mM MgCl2, 2 mM DTT, and 0.25 mg/mL sheared salmon sperm DNA. Reactions were carried out for 60 minutes at 30 °C, quenched by the addition of 10 μM olaparib and standard 4X SDS sample loading buffer, and boiled at 95 °C for 4 minutes. This mixture (15 μL) was then loaded onto an SDS-PAGE gel (4–12% Bis-Tris unless otherwise noted) and the resolved gel was transferred to a PVDF membrane for western blot analysis of products (see ‘Western blot protocol’). Three independent assays were performed for all densitometry-quantified blots (n = 3, replicate western blots shown in Supplementary Fig. 26).
Recombinant ARH3 hydrolysis assays
ARH3 assays were adapted from a previously described protocol13. Following ADP-ribosylation assays, reactions were quenched by the addition of 10 μM olaparib, and ARH3 was introduced to the reaction to a final concentration of 1 μM. Reactions were then incubated for an additional 30 minutes at 30 °C, quenched by the addition of standard 4X SDS sample loading buffer, and boiled at 95 °C for 4 minutes. This mixture (15 μL) was then loaded onto an SDS-PAGE gel (4–12% Bis-Tris unless otherwise noted) and the resolved gel was transferred to a PVDF membrane for western blot analysis of products (see ‘Western blot protocol’).
Recombinant SIRT6 deacetylation assays
SIRT6 was added to the ADP-ribosylation assays (described above) to a final concentration of 1 μM. Assays were performed in the presence of 1 mM unlabeled NAD+ and analyzed as described in “Recombinant ADP-ribosylation assays; Nucleosome substrates” with the only exception being that reactions were quenched after 10 minutes. We note that previous reports detailing recombinant SIRT6 nucleosome deacetylation assays use a range of NAD+ concentrations from 1–5 mM28,29. This is not compatible with the commercially available biotin-NAD+ cofactor, which is prepared at a stock concentration of 250 μM and limited to a maximum working concentration of 100 μM in our assays. Therefore, nucleosome deacetylation/ADP-ribosylation assays were only performed with the unlabeled NAD+ cofactor to ensure previously published working NAD+ concentrations were utilized.
Western blot protocol
SDS-PAGE gels (12% Bis-Tris or 4–12% Bis-Tris; Biorad) were transferred to PVDF membranes and blocked in TBST (50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% tween-20) containing 3% milk for 1 hour. Membranes were then incubated in primary antibody in TBST overnight at 4 °C. The membranes were washed 3 × 5 minutes with TBST, and incubated in secondary antibody for 1 hour. Membranes were again washed 3 × 5 minutes with TBST and imaged. For detection of the biotin NAD+, the blocked membrane was directly incubated in IRDye streptavidin (Li-Cor) in TBST for 1 hour. The membrane was then washed 3 × 5 minutes with TBST and imaged. See ‘Supplementary Table 1‘ for antibodies and dilutions.
PARP/HPF1 nucleosome library assay
ADP-ribosylation activity assays
Reactions were performed as described above in ‘Recombinant ADP-ribosylation assays: nucleosome substrates’ in the presence of 300 nM DNA-barcoded mononucleosome library at a final reaction volume of 10 μL. Following the 20 minute incubation at 30 °C, reactions were quenched by the addition of 90 μL of wash buffer (50 mM Tris, pH 7.5, 100 nM NaCl, 2 mM MgCl2, 0.1% Triton X-100, 2 mM DTT, and 10 μM olaparib). The reaction was then added to streptavidin magnetic beads (300 μL of slurry suspended in 300 μL of wash buffer; resin source: New England Biolabs) and incubated on an end-over-end rotator at room temperature for 1 hour to capture biotinylated nucleosome products. Resin was then washed 10x with wash buffer (1 mL per wash) to ensure complete removal of unlabeled nucleosomes. Following the final wash, the resin was resuspended in DNA elution buffer (100 mM Tris, pH 7.5, 10 mM EDTA, 1% SDS, 200 μg/mL proteinase K) and incubated at 50 °C for 90 minutes. The mixture was then spun down to remove resin and the supernatant was added to 1 mL of Buffer PB (Qiagen) to enable purification of DNA by the Qiagen miniprep spin column protocol. Eluted DNA concentrations were calculated using a Qubit high-sensitivity dsDNA quantification kit. Next, DNA corresponding to each reaction was PCR amplified to incorporate multiplex barcodes and Illumina adaptor sequences, and Illumina sequencing/barcode sorting were performed as previously described15. For data processing, all nucleosome barcodes were first normalized to respective input values (barcode counts when high-throughput sequencing is performed on the library prior to assays) to control for differences in relative abundances. Subsequently all modified nucleosomes barcode counts were compared to that of a representative wild type nucleosome (Supplementary Dataset 1).
Nucleosome interaction assays
PARP/HPF1 complexes (2 nM PARP1-Flag or PARP2-Flag and 2 μM HPF1) were incubated with 20 nM mononucleosome library in 100 μL of binding buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.1% Triton X-100, and 1 mM DTT) for 60 minutes on an end-over-end rotator at 4 °C. Next, 20 μL of pre-equilibrated (in binding buffer) Flag-agarose affinity resin (G1 affinity resin; Genscript) was added to the binding reaction and incubated on an end-over-end rotator at 4 °C for an additional 90 minutes. Resin was then washed 5x with binding buffer (100 μL per wash). Following the final wash, the resin was resuspended in DNA elution buffer (100 mM Tris, pH 7.5, 10 mM EDTA, 1% SDS, 200 μg/mL proteinase K) and incubated at 50 °C for 90 minutes. The mixture was then spun down to remove resin, and the supernatant was added to 1 mL of Buffer PB (Qiagen) to enable purification of DNA by the Qiagen miniprep spin column protocol. Eluted DNA concentrations were calculated using a Qubit high-sensitivity dsDNA quantification kit. Next, DNA corresponding to each reaction was PCR amplified to incorporate multiplex barcodes and Illumina adaptor sequences, and Illumina sequencing/barcode sorting were performed as previously described15. For data processing, all nucleosome barcodes were first normalized to respective input values (barcode counts when high-throughput sequencing is performed on the library prior to assays) to control for differences in relative abundances. Subsequently all modified nucleosomes barcode counts were compared to that of a representative wild type nucleosome (Supplementary Dataset 1).
Circular dichroism
Circular dichroism spectra were collected on an Applied Photophysics ChiraScan CD spectrometer. Samples were prepared in 20 mM Tris, pH 7.5, 20 mM NaCl at 1 uM PARP1, and scans were recorded at 20 °C in a 1 mm path-length quartz cuvette. Data were recorded at a scan rate of 60 nm min−1 between 190 and 260 nm. Baselines recorded using the same buffer, cuvette, and instrument parameters were subtracted from each sample dataset. The spectra were converted from ellipticity (mdeg) to molar residue ellipticity (MRE; deg●cm2●dmol−1●res−1) by normalizing for the concentration of amide bonds and the cuvette path length.
Mammalian cell culturing and plasmid transfection
General procedures
U2OS cells were cultured as a monolayer in DMEM (ThermoFisher Scientific) supplemented with 10% FBS (Sigma-Aldrich), 100 units/mL of penicillin, and 100 µg/mL of streptomycin (ThermoFisher Scientific). All cells were maintained in an incubator at 37 °C and 5% CO2. Plasmid DNA was transfected into U2OS cells using Lipofectamine 2000 (Thermo-Fisher Scientific) according to the manufacturer’s instructions. These transfections were performed in OptiMEM (ThermoFisher Scientific).
Plasmid transfection
For a 6-well plate, the plasmid DNA (1 µg) was diluted in 125 uL of Optimem. Lipofectamine 2000 (3 µL) was diluted in 125 µL of Optimem. The solutions were mixed and allowed to incubate for 15 min at RT before being dripped over the cells. The cells were incubated at 37 °C. After 6 h, the cells were recovered in DMEM with 10% FBS without antibiotics. The cells were allowed to grow for 48 h before treating with hydrogen peroxide, as described below. For 10-cm plates, 5 µg of DNA and 15 µL of Lipofectamine 2000 were used per plate.
Olaparib treatment
Olaparib (10 mM in DMSO) was diluted in DMEM with 1% DMSO to 10 µM. This solution was diluted 1:10 in DMEM without DMSO to give a final concentration of 1 µM (0.1% DMSO), and this solution was added to the cells. The cells were incubated at 37 °C for 1 h.
Peroxide treatment
Hydrogen peroxide-induced DNA damage assays were adapted from a previously described protocol6. Hydrogen peroxide (10 M in water) was diluted in PBS to 1 mM. U2OS cells were washed with PBS and then placed in the 1 mM H2O2 for 20 min at 37 °C.
Generation of PARP1 knockout cell lines
U2OS cells were transfected with a plasmid containing Cas9, the appropriate sgRNA, and a puromycin resistance gene (pSpCas9(BB)-2A-Puro (PX459) V2.0) and were allowed to recover for 48 hours. The cells were treated with 1 µg/mL of puromycin for 48 h. The puromycin was removed, and the cells were allowed to grow for 7 days. The cells were plated by serial dilution to yield single colonies. Single colonies were screened for the knockout by treating with H2O2 and analyzing the lysates and histone extracts for ADP-ribosylation by western blot. Knockout was also confirmed by probing with a PARP1 antibody and by DNA sequencing*. Positive clones were counter-screened by treatment with 1 µg/mL puromycin to eliminate clones that stably integrated the transfected DNA. sgRNA sequences below were validated in a previous study6.
PARP1 sgRNA1: 5’-CCACCTCAACGTCAGGGTGC-3’ (exon 2)
PARP1 sgRNA2: 5’TGGGTTCTCTGAGCTTCGGT-3’ (exon 2)
*PARP1 knockout cell line genomic DNA was isolated using the Blood & Cell Culture DNA Mini Kit (Qiagen) following the manufacturer’s protocol. Target sequences were amplified (primers targeting 150 base pairs upstream and downstream of the target mutation site) and cloned into pET30 vectors for DNA sequencing. Target sites and allele mutations are shown below:
gRNA target 1
WT PARP1 sequence: CCATCCACCTCAACGTCAGGGTGCCGGATGGAGTGGCCCA
Mutant allele 1: CCATCCACCTCAACGTCAG*GTGCCGGATGGAGTGGCCCA
Mutant allele 2: CCATCCACCTCAACGTCAGG*TGCCGGATGGAGTGGCCCA
*represents site of base pair deletion
gRNA target 2
WT PARP1 sequence: GGGCCACTCCATCCGGCACCCTGACGTTGAGGTGGATGGGT
Mutant allele 1: GGGCCACTCCATCCGGCAC*CTGACGTTGAGGTGGATGGGT
Mutant allele 2: GGGCCACTCCATCCGGCACC*TGACGTTGAGGTGGATGGGT
*represents site of base pair deletion
Histone acid extraction
Cells were harvested in cold PBS. The histones were extracted using the Epiquik Histone Acid Extraction Kit (Epigentek, Farmingdale, NY) as indicated in the manufacturer’s instructions. The lysates were diluted in SDS loading buffer and boiled for 5 minutes to obtain samples for analysis. After the neutralization step, the extracted histones were diluted with SDS loading buffer and boiled for 5 min.
Immunoprecipitation of H3-Flag constructs from mammalian cells
U2OS cells were transfected as described above with plasmid DNA encoding a FLAG-tagged histone (i.e. H3.1WT, H3.1S10A, H3.1S28A, H3.1S10A,S28A, or H3.1K9QK27Q). After 48 hours, half of the plates were treated with 1 mM H2O2 for 20 min, and the histones were acid extracted. After the neutralization step in the histone acid extraction protocol (see ‘Histone acid extraction’) extracted histones (300 μL) were diluted 10x with immunoprecipitation buffer (25 mM Tris, pH 7.5, 500 mM NaCl, 1% Triton X-100, 10% glycerol, 1 mM DTT, 5 μM ADP-HPD, and 5 μM of olaparib) and added to 100 μL of pre-equilibrated Flag-agarose affinity resin (G1 affinity resin; Genscript). Incubation with the resin was performed on an end-over-end rotator for 1 hour at room temperature. Resin was then washed 4x with immunoprecipitation buffer (1 mL per wash and 1 minute per wash) and eluted with 100 mM glycine, pH 3.5 (3 × 100 μL treatments). We note that when elution buffer was incubated with the resin for extended periods of time, antibody leaching could be observed. Therefore, we tried to keep total time of elution to under 10 minutes (~3 minutes per 100 μL incubation). Elutions were then pooled and neutralized by the addition of 33 μL of 1 M Tris, pH 7.5 (final concentration 100 mM), and a standard TCA precipitation was performed (this was necessary for concentration of the eluted histones). The precipitated histones were resuspended in 40 μL of standard 4X SDS sample loading buffer and boiled at 95 °C for 4 minutes. Next, 20 μL of sample was then loaded onto an SDS-PAGE gel (12% Bis-Tris), and the resolved gel was transferred to a PVDF membrane for western blot analysis (see ‘Western blot protocol’). Relative efficiency of transfection (input; α-FLAG), DNA damage-induced ADP-ribosylation (input; α-ADPR), and immunoprecipitation (IP; α-FLAG) are also shown.
PARP1 rescue
Knockout U2OS cells were transfected as described above with plasmid DNA encoding a FLAG-tagged PARP1 (i.e. WT or 7xKQ). After 48 h, half of the plates were treated with 1 mM H2O2 for 20 min. The cells were treated with the Epiquik Histone Acid Extraction Kit, reserving the lysate for PARP1 analysis. The lysates and histones were analyzed by western blot (soluble fraction loading control = α-actin, histone fraction loading control = α-H4, transfection efficiency = α-FLAG, and PARP1 knockout validation = α-PARP1).
Statistics and reproducibility
All significance measurements that are reported for the mononucleosome library experiments represent P-values (two-tailed P-value ≤ 0.01, n = 3, three independent experiments). All other mononucleosomes and peptide assay replicates are from independent experiments (minimum n = 3, see figure captions). Reported errors represent standard error of the mean. All unlabeled NAD+ cofactor-based recombinant assay western blots were repeated once to ensure reproducibility.
Software and data analysis
The Princeton University installation of GALAXY and a custom script written in R (for simple barcode counting; available upon request) were used to process all sequencing data. GraphPad Prism was used to calculate errors/p-values and to generate heatmaps and bar graphs/dot-plots. ImageJ was used for densitometry measurements.
Reagent and data availability
The PARP½ plasmids (for Sf9 cell and mammalian cell expression) as well as the HPF1 plasmid (for E. coli expression) and the ARH3 plasmid (for E. coli expression) have been deposited at Addgene. All data generated and analyzed are reported in this published article, including uncropped gels/blots (Supplementary Fig. 26), complete mononucleosome library assay results (Supplementary Dataset 1), and replicate blots used for densitometry measurements (Supplementary Fig. 26). Barcode sorting scripts are available upon request.
Supplementary Dataset 1
Recombinant PARP1/HPF1 and PARP2/HPF1 binding and activity measurements for each member of the mononucleosome library from three independent experiments. Error measurements represent standard error of the mean from the three replicates. All measurements represent barcode counts relative to wild type. See ‘PARP/HPF1 nucleosome library assay’ for more details about data processing.
Supplementary Material
Acknowledgements.
We thank current members of the Muir laboratory as well as C.D. Allis for discussions and comments. We also thank the Princeton University Sequencing Core Facility. This work was supported by National Institutes of Health (NIH) Grants R37 GM086868, R01 GM107047 and P01 CA196539. G.P.L. and K.L.D. were supported by NIH Research Service Awards (1F32GM110880 and 5F32CA206418, respectively).
Footnotes
Competing financial interests. The authors declare no competing financial interests.
References
- 1.Chaudhuri AR & Nussenzweig A The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 18, 610–621, doi: 10.1038/nrm.2017.53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daniels CM, Ong SE & Leung AK The Promise of Proteomics for the Study of ADP-Ribosylation. Mol. Cell 58, 911–924, doi: 10.1016/j.molcel.2015.06.012 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogata N, Ueda K, Kawaichi M & Hayaishi O Poly(ADP-ribose) synthetase, a main acceptor of poly(ADP-ribose) in isolated nuclei. J. Biol. Chem. 256, 4135–4137 (1981). [PubMed] [Google Scholar]
- 4.Messner S & Hottiger MO Histone ADP-ribosylation in DNA repair, replication and transcription. Trends Cell Biol. 21, 534–542, doi: 10.1016/j.tcb.2011.06.001 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Bonfiglio JJ et al. Serine ADP-Ribosylation Depends on HPF1. Mol. Cell 65, 932–940 e936, doi: 10.1016/j.molcel.2017.01.003 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibbs-Seymour I, Fontana P, Rack JGM & Ahel I HPF1/C4orf27 Is a PARP-1-Interacting Protein that Regulates PARP-1 ADP-Ribosylation Activity. Mol. Cell 62, 432–442, doi: 10.1016/j.molcel.2016.03.008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krishnakumar R & Kraus WL The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol. Cell 39, 8–24, doi: 10.1016/j.molcel.2010.06.017 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ame JC et al. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 274, 17860–17868 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Pommier Y, O’Connor MJ & de Bono J Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci. Transl. Med. 8, 362ps317, doi: 10.1126/scitranslmed.aaf9246 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Jenuwein T & Allis CD Translating the histone code. Science 293, 1074–1080, doi: 10.1126/science.1063127 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Palazzo L et al. Serine is the major residue for ADP-ribosylation upon DNA damage. eLife 7, doi: 10.7554/eLife.34334 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leidecker O et al. Serine is a new target residue for endogenous ADP-ribosylation on histones. Nat. Chem. Biol. 12, 998–1000, doi: 10.1038/nchembio.2180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontana P et al. Serine ADP-ribosylation reversal by the hydrolase ARH3. eLife 6, doi: 10.7554/eLife.28533 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polo SE & Jackson SP Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 25, 409–433, doi: 10.1101/gad.2021311 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dann GP et al. ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference. Nature 548, 607–611, doi: 10.1038/nature23671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim MY, Mauro S, Gevry N, Lis JT & Kraus WL NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 119, 803–814, doi: 10.1016/j.cell.2004.11.002 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Mao Z et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science 332, 1443–1446, doi: 10.1126/science.1202723 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller KM et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 17, 1144–1151, doi: 10.1038/nsmb.1899 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tjeertes JV, Miller KM & Jackson SP Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 28, 1878–1889, doi: 10.1038/emboj.2009.119 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michishita E et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452, 492–496, doi: 10.1038/nature06736 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martello R et al. Proteome-wide identification of the endogenous ADP-ribosylome of mammalian cells and tissue. Nat. Commun. 7, 12917, doi: 10.1038/ncomms12917 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassa PO et al. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-kappaB-dependent transcription. J. Biol. Chem. 280, 40450–40464, doi: 10.1074/jbc.M507553200 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Muthurajan UM et al. Automodification switches PARP-1 function from chromatin architectural protein to histone chaperone. Proc. Natl. Acad. Sci. U.S.A 111, 12752–12757, doi: 10.1073/pnas.1405005111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Min A et al. Histone deacetylase inhibitor, suberoylanilide hydroxamic acid (SAHA), enhances anti-tumor effects of the poly (ADP-ribose) polymerase (PARP) inhibitor olaparib in triple-negative breast cancer cells. Breast Cancer Res. 17, 33, doi: 10.1186/s13058-015-0534-y (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ha K et al. Histone deacetylase inhibitor treatment induces ‘BRCAness’ and synergistic lethality with PARP inhibitor and cisplatin against human triple negative breast cancer cells. Oncotarget 5, 5637–5650, doi: 10.18632/oncotarget.2154 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Online methods references
- 26.Nguyen UT et al. Accelerated chromatin biochemistry using DNA-barcoded nucleosome libraries. Nat. Methods 11, 834–840, doi: 10.1038/nmeth.3022 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luger K, Mader AW, Richmond RK, Sargent DF & Richmond TJ Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260, doi: 10.1038/38444 (1997). [DOI] [PubMed] [Google Scholar]
- 28.Gil R, Barth S, Kanfi Y & Cohen HY SIRT6 exhibits nucleosome-dependent deacetylase activity. Nucleic Acids Res. 41, 8537–8545, doi: 10.1093/nar/gkt642 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang WW, Zeng Y, Wu B, Deiters A & Liu WR A Chemical Biology Approach to Reveal Sirt6-targeted Histone H3 Sites in Nucleosomes. ACS Chem. Biol. 11, 1973–1981, doi: 10.1021/acschembio.6b00243 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.