Abstract
Protein lysine crotonylation has emerged as an important post-translational modification (PTM) in the regulation of gene transcription through epigenetic mechanisms. Here we introduce a chemical probe, based on a water-soluble phosphine warhead, which specifically reacts with the crotonyl modification. We show that this reagent is complementary to antibody-based tools allowing detection of endogenous cellular proteins such as histones carrying the crotonylation PTM. The tool is also used to show that the histone acylation activity of the transcriptional co-activator, p300, can be activated by pre-existing lysine crotonylation through a positive feedback mechanism. This reagent provides a versatile and sensitive probe for the analysis of this PTM.
Graphical Abstract
Eukaryotic chromosomes are decorated with a diverse set of histone PTMs that function to regulate access to genomic DNA and that, as a consequence, are critical for the establishment and maintenance of cellular identity.1,2 An abundant PTM found on histones involves acetylation of the ε-amino group of lysine side-chains (Kac).3 This archetypal ‘epigenetic mark’ acts a positive regulator of gene transcription and functions either by directly altering chromatin structure, or through the recruitment of a myriad of trans-acting nuclear factors.4 Recent advances in mass spectrometry have revealed several other non-acetyl histone acylations, classified as short-chain Lys acylations.5–10 These include propionylation (Kpr), butyrylation (Kbu), ß-hydroxybutyrylation (Kbhb) and crotonylation (Kcr). Among this group, crotonylation has received the most attention since its initial discovery; it is found on all histones (H2A, H2B, H3, H4 and H1) and functions as a strong transcriptional activator.5 Intriguingly, the levels of histone crotonylation appear to mediate a crosstalk between the metabolic status of the cell, i.e. crotonyl-CoA levels, and gene expression.11,12 Moreover, recent proteomics studies indicate that treatment of cells with sodium crotonate, which elevates crotonyl-CoA levels, leads to the crotonylation of numerous non-histone nuclear proteins.13,14 Many questions remain regarding the functional implications of these protein crotonylation events.
The crotonyl group is unique among known short-chain lysine acylations, in that it contains an α,β unstaturated functionality. In the context of histones, this feature has been shown to mediate the interaction with protein ‘reader’ domains through either a π-π-π stacking mode, or a distinct hydrophobic encapsulation and hydrogen bonding mechanism.15–17 The unique chemical structure of the Kcr mark, in principle, endows it with the ability to undergo conjugate addition reactions with suitable nucleophiles. However, the very fact that the PTM exists argues that is largely unreactive to the common nucleophiles found in the cell, for example abundant thiol-containing molecules such as glutathione. Here we show that the crotonyl mark is highly reactive towards phosphine nucleophiles containing a pendent carboxylic acid group. Based on this observation, we go on to develop a covalent chemo-proteomic probe for the detection and functional analysis of protein crotonylation (Figure 1A). This simple reagent provides a versatile and cost-effective complement to antibody-based tools for the analysis of this PTM.
Figure 1.
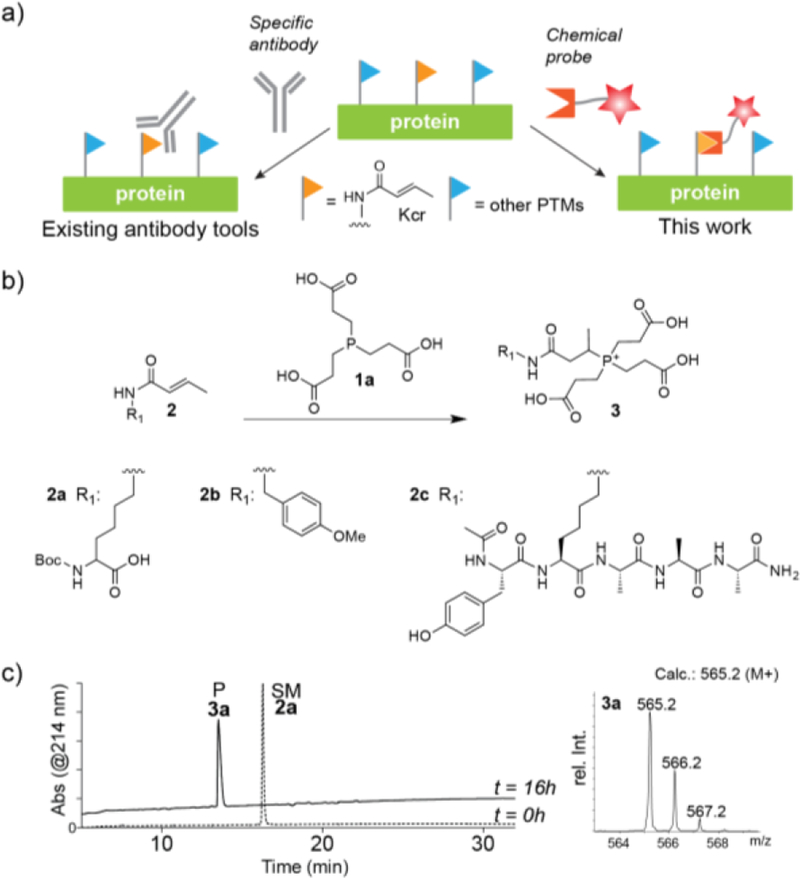
Detection of protein lysine crotonylation. (a) Left: the PTM is currently detected using specific antibodies. Right: in this study, we introduce a specific chemical probe based on a reactive phosphine warhead. (b) Scheme depicting the reaction between crotonylated model compounds, 2, and TCEP. (c) RP-HPLC analysis of the reaction between TCEP (1a) and model compound 2a. Inset: ESI-MS analysis of the product peak.
In the course of our investigations into the chemical synthesis of crotonylated proteins, we observed a major side reaction between crotonylated lysine residues and tris(2-carboxyethyl)phosphine (TCEP; 1a), a commonly used water soluble reducing agent. An adduct of ~251Da was observed that was attributed to the covalent addition of TCEP to crotonylated lysine residues. We note that a similar addition has also been observed between TCEP and alkyne containing biomolecules.18 To further investigate the ability of TCEP to covalently add to crotonylated lysine residues in aqueous solution, a series of model compounds, 2a-d, were prepared and used in a set of exploratory reactions (Figure 1B). Treatment of 2a with TCEP at pH 8 led to clean conversion into a single product (3a) whose mass was consistent with conjugate addition of TCEP to the crotonyl group (Figure 1C). This assignment was confirmed by a series NMR studies using model compound 2b, which indicated the generation of an apparent β-phosphonium species (3b) during the course of the reaction with concomitant loss of the olefin signals from the crotonyl group (Figure S1). Other studies using model peptide 2c revealed that the reaction proceeded smoothly over the pH range 6–10 (Figure S2) and was tolerant to a range of additives commonly used in protein biochemistry, for example chaotropes and detergents (Figure S3). Kinetic studies indicated that the reaction at pH 8 proceeded with an associated second order rate constant of 6 (± 0.3) x 10−4 M−1s−1 (Figure S4). In keeping with a conjugate addition type process, we observed no reaction between TCEP and control compound 2d, containing the 3-butenoyl acyl group. Moreover, the oxidized version of TCEP, 1b, was unreactive toward the crotonyl group (Figure S5).
We were curious as to the driving force for this phosphine addition reaction. Phosphines are commonly used in organic chemistry as catalysts in conjugate addition reactions where they generate a β-phosphonium enolate species that acts as a strong base to deprotonate pronucleophiles.19 Indeed, trimethylphosphine has been shown to catalyze the reaction of water with methyl vinyl ketones.20 It is thus somewhat surprising that a β-phosphonium cation would be generated as the final product in the aqueous reaction between TCEP and the acyl crotonyl moiety. Consequently, we wondered whether there was something special about the TCEP structure that might account for this. With this in mind, we treated model compound 2a with several other water-soluble phosphines. In no case did we observe any reaction (Figure S6). Note that prolonged exposure of 2a to high concentrations of reduced glutathione (200 mM) did lead to trace amounts of an adduct (Figure S6). A unique feature of TCEP compared to the other unreactive phosphines tested is the presence of the three pendent carboxylate groups - these render the molecule water soluble, but also influence the pKa of the phosphorus (pKa = 7.6)(Figure S7).21,22 In a key experiment, we found that the previously described tris-methyl ester analog of TCEP, 1c (phosphorus pKa= 4.7),22 was completely unreactive with compound 2a (Figure S8). This was unexpected since, 1c, acts a nucleophilic reducing agent over an extended pH range, and is in fact more reactive than TCEP itself at acidic pH.22 Notably, during the course of the incubation at pH 8 we observed slow hydrolysis of 1c to give the mono- and bis-methyl ester species (Figure S8). Unlike the parent tri-ester compound, these hydrolysis products reacted with 2a to give the corresponding phosphonium adducts. Collectively, these observations indicate that the presence of a free carboxylate group within the nucleophilic phosphine reagent facilitates the addition reaction. Conceivably, the negatively charged carboxylate group might stabilize the developing phosphonium cation through purely electrostatic means. However, other explanations are certainly possible, for example H-bonding interactions or even the involvement of a transient (i.e. rapidly exchanging) cyclic phosphorane species.23
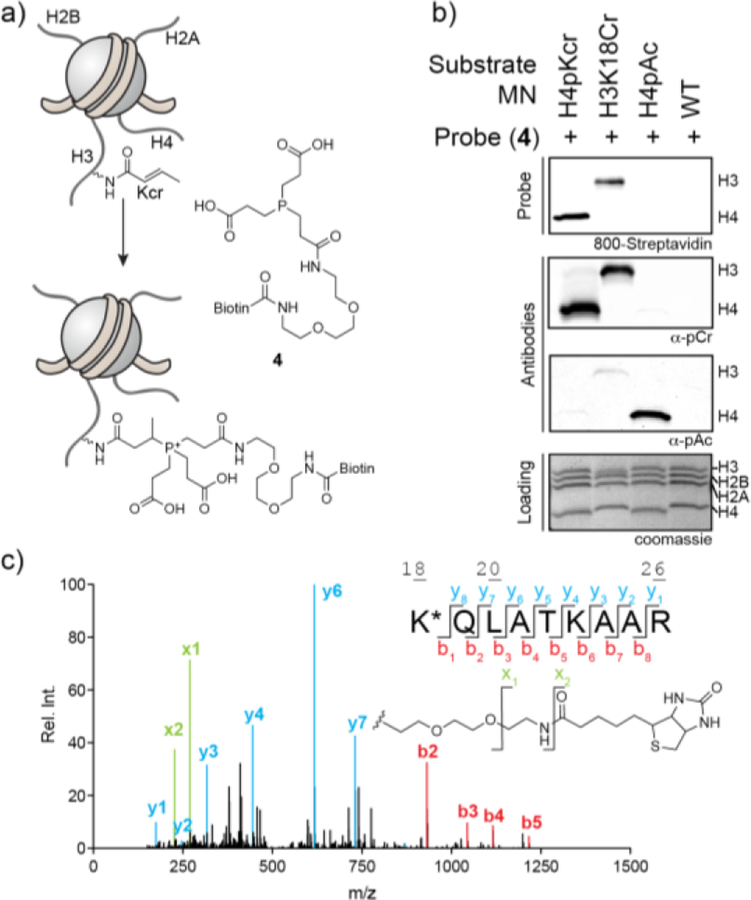
The observation that mono- and bis-ester derivatives of TCEP react with the crotonyl group suggested a strategy for the development of a chemical probe for detecting protein crotonylation (Figure 2a). Specifically, we introduced a biotin moiety onto one of the arms of TCEP to give probe compound 4. Model studies indicated that this modification did not significantly impact reactivity with the crotonyl group (Figure S4). We imagined that this molecule would allow for both the enrichment and detection of crotonylated proteins using standard avidin/streptavidin based affinity reagents. As an initial test of this idea, we generated semi-synthetic mononucleosomes containing either a single Kcr mark at position 18 on histone H3 (H3K18cr) or several Kcr marks on the tail of H4 (H4pCr). Treatment of these mononucleosomes with probe 4 led to robust incorporation of biotin into the crotonylated histone, as read out by streptavidin blotting (Figure 2b). Detection of the biotin signal was observed after as little as 1 hr reaction at 37°C and was dependent upon the presence of the reactive phosphine warhead (Figure S9). Importantly, no biotin was incorporated into unmodified nucleosomes, or nucleosomes with pre-installed Kac marks, even after overnight incubation. To further validate that the chemical probe indeed reacts with the crotonylated lysine residue, H3K18Cr mononucleosomes were treated with the probe and subjected to ArgC digestion followed by LC-MS/MS analysis. This confirmed that the probe reacted with the pre-installed crotonyl unit within the H3K18Cr protein (Figure 2c). Thus, probe 4 is able to detect crotonylation on purified histones.
Figure 2.
A chemo-proteomic probe for protein lysine crotonylation. (a) Scheme depicting the application of TCEP analog 4 in the detection of protein crotonylation. (b) Semisynthetic mononucleosomes bearing indicated modifications were reacted with probe 4 (4 mM) before being analyzed by western-blot employing a biotin detection system (dye-conjugated streptavidin). For comparison, the same nucleosomes were analyzed by western blotting employing commonly used pan anti-lysine crotonyl (α-pCr) and pan anti-lysine acetyl (α-pAc) antibodies. A coomassie stained SDS-PAGE gel of the samples was used as a loading control. (c) Mass analysis of H3K18Cr conjugated with probe 4. Following the reaction, the protein was digested with ArgC and analyzed by LC-MS/MS. In addition to the expected y- and b-ions, fragmentation of the probe itself (lower right panel) is observed and denoted as x-ions.
Next we asked whether probe 4 could be used to analyze crotonylated proteins within a complex proteome. As an initial step, we showed that our probe could detect semisynthetic H3K18cr spiked into an E. coli lysates at nanomolar concentrations (Figure S10). Moreover, we were able to affinity capture the crotonylated protein, but not an unmodified control, from this lysate using streptavidin beads (Figure S10). Encouraged by these results we moved on to the detection of crotonylated proteins in mammalian cells. Indeed, we were able to detect endogenous crotonylated histone proteins in HEK293 cells using our probe (Figure 3a). Moreover, treatment of these cells with sodium crotonate, which as noted earlier up-regulates crotonyl-CoA levels,11,13 led to a substantial increase in the number of proteins reactive toward our probe (Figure 3a and Figure S11).
Figure 3.
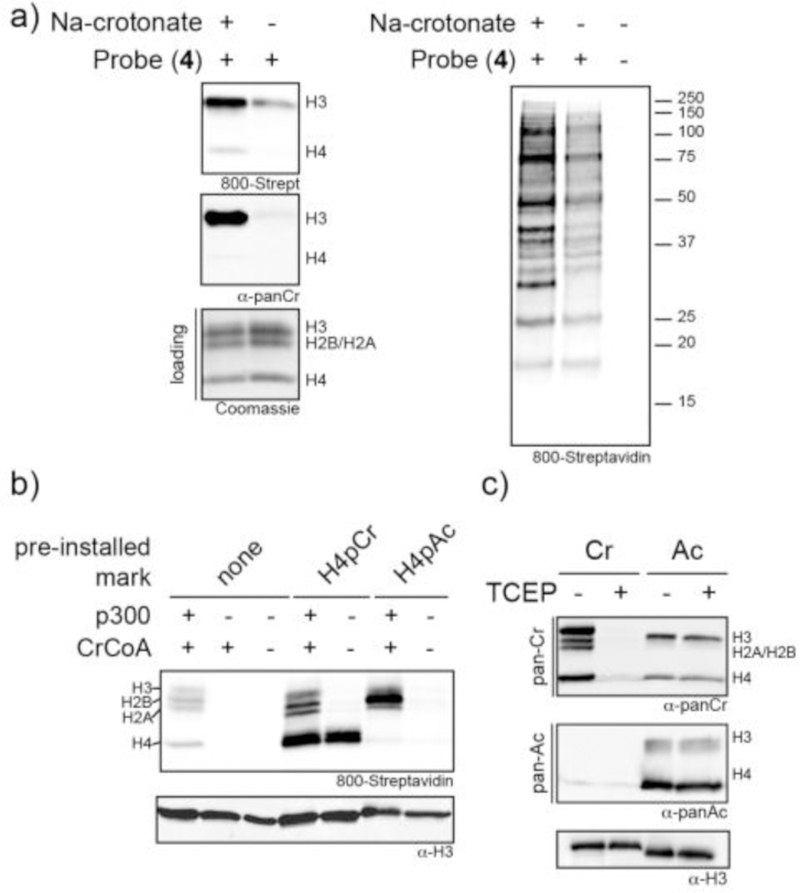
Analysis of protein crotonylation. (a) Western blot analysis of acid extracted histones (left panel) or nuclear extract (right panel) from HEK293 cells, either grown in the absence or presence of sodium crotonate. Protein crotonylation levels were detected using either probe 4 or α-pCr as in Figure 2. A coomassie stained SDS-PAGE gel served as a loading control. (b) In vitro p300 mediated crotonylation reactions. Wild-type 12-mer nucleosomal arrays as well as those pre-modified with H4pCr or H4pAc were treated with p300 in the presence of crotonyl-CoA. Protein crotonylation levels were detected using probe 4 prior to western blotting. (c) Effect of TCEP treatment on antibody recognition. 12-mer nucleosome arrays containing either crotonylation of acetylation marks (installed by p300 treatment) were treated with TCEP prior to western blotting using α-pCr or α-pAc antibodies. An α-H3 antibody was used as a loading control.
We were also keen to explore the utility of our probe for studying the biochemistry surrounding histone crotonylation. Specifically, we asked whether the histone acyltransferase activity of the transcriptional co-activator, p300, which is known to crotonylate histones,11 is sensitive to the presence of pre-existing crotonylation marks on nucleosomes. This was motivated by previous studies that revealed a positive feedback mechanism for histone acetylation by p300.24 Accordingly, we generated 12-mer nucleosome arrays containing either unmodified histones, histones crotonylated on H4 (i.e. semisynthetic H4pCr) or histones acetylated on H4 (i.e. semisynthetic H4pAc). These chromatin substrates were then treated with recombinant full-length p300 supplemented with crotonyl-CoA. Analysis of the reaction mixtures using our probe revealed that the crotonylated and acetylated arrays were superior substrates compared to unmodified chromatin, as indicated by the levels of de novo crotonylation of all histones (Figure 3b). Analogous experiments were performed using acetyl-CoA as the cofactor, revealing that p300-mediated acetylation of the histones is stimulated by the presence of both Kcr and Kac marks on H4 (Figure S12). Thus, there appears to be a cross-stimulation of p300 acyltransferase activity by pre-existing Kcr or Kac marks on the chromatin substrate. Additional studies will be needed to ascertain whether this stimulation operates through a simple enzyme recruitment mechanism, or involves a more complex allosteric mode of activation.
Finally, in the course of our investigations we noticed that the pan anti-Kcr antibody cross-reacted with acetylated nucleosomes (Figure 3c). In principle, this cross-reactivity could complicate biochemical or proteomics studies relying on this reagent. We reasoned that treatment of crotonylated histones, but not their acetylated counterparts, with TCEP should selectively mask the ‘mark’ from antibody detection, due to the drastic changes in steric bulk and charge that accompanies the reaction. Indeed, treatment of crotonylated nucleosomes with TCEP blocked the recognition of the crotonyl mark by the anti-Kcr antibody, whereas the ability of the pan anti-Kac antibody to recognize acetylated nucleosomes was unaffected by TCEP treatment (Figure 3c, Figure S13). Thus, TCEP can be used in simple control reactions to verify that signals obtained from immunoblotting originate from the crotonylation mark. Based on this idea, it should be possible to selectively elute crotonylated proteins following immuno-precipitation of cellular extracts using anti-Kcr antibodies (or indeed other Kcr binding factors). Thus, we imagine that the TCEP-crotonyl reaction will prove useful in the context of existing antibody-based proteomics workflows. 25
The crotonyl group bears a unique structure among known protein PTMs in that it contains an α,β unsaturated functionality in conjugation with a carbonyl group. Here we exploit the serendipitous discovery that this PTM undergoes a conjugate addition reaction with the reducing agent TCEP to give a stable adduct. This unexpected reactivity of TCEP was traced to the presence of its pendent carboxylate groups, which we speculate stabilize the phosphonium species in the product. Based on this insight, we developed a sensitive and selective chemical probe of protein crotonylation and demonstrate its utility in the context of proteomics efforts and mechanistic biochemical studies. The new probe complements existing antibody-based tools for studying crotonylation.5 For example, the ability to covalently attach a biotin moiety to the modified protein would allow for the adoption of rigorous enrichment protocols within proteomics workflows employing readily available streptavidin reagents. Moreover, the chemical probe is unlikely to be sensitive to the local sequence or PTM environment around the crotonyl mark, and is thus expected to be less susceptible to epitope occlusion effects compared to antibodies.26 Thus, we imagine that our chemical probe will find utility in the study of this important PTM.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Zachary Brown, Robert Thompson and Felix Wojcik for valuable discussions. The work was supported by the U.S. National Institutes of Health (NIH grants GM107047 and CA196539).
Footnotes
ASSOCIATED CONTENT
The Supporting Information is available free of charge on the ACS Publications website. Full methods and experimental data, including Figures S1–S18 (PDF).
REFERENCES
- (1).Chen T; Dent SY R. Nat. Rev. Genet 2014, 15, 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Jenuwein T; Allis CD Science 2001, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- (3).Verdin E; Ott M Nat. Rev. Mol. Cell Biol 2015, 16, 258–264. [DOI] [PubMed] [Google Scholar]
- (4).Shahbazian MD; Grunstein M Annu. Rev. Biochem 2007, 76, 75–100. [DOI] [PubMed] [Google Scholar]
- (5).Tan M; Luo H; Lee S; Jin F; Yang JS; Montellier E; Buchou T; Cheng Z; Rousseaux S; Rajagopal N; et al. ; Cell 2011, 146, 1016–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Chen Y; Sprung R; Tang Y; Ball H; Sangras B; Kim SC; Falck JR; Peng J; Gu W; Zhao Y Mol. Cell. Proteomics 2007, 6, 812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Dai L; Peng C; Montellier E; Lu Z; Chen Y; Ishii H; Debernardi A; Buchou T; Rousseaux S; Jin F; et al. ; Nat. Chem. Biol 2014, 10, 365–370 [DOI] [PubMed] [Google Scholar]
- (8).Xie Z; Dai J; Dai L; Tan M; Cheng Z; Wu Y; Boeke JD; Zhao Y Mol. Cell. Proteomics 2012, 11, 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Tan M; Peng C; Anderson KA; Chhoy P; Xie Z; Dai L; Park J; Chen Y; Huang H; Zhang Y; et al. ; Cell Metab 2014, 19, 605–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Xie Z; Zhang D; Chung D; Tang Z; Huang H; Dai L; Qi S; Li J; Colak G; Chen Y; et al. ; Mol. Cell 2016, 62, 194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Sabari BR; Tang Z; Huang H; Yong-Gonzalez V; Molina H; Kong HE; Dai L; Shimada M; Cross JR; Zhao Y; et al. Mol. Cell 2015, 58, 203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Sabari BR; Zhang D; Allis CD; Zhao Y Nat. Rev. Mol. Cell Biol 2016, 18, 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Wei W; Mao A; Tang B; Zeng Q; Gao S; Liu X; Lu L; Li W; Du JX; Li J; et al. ;. J. Proteome Res 2017, 16, 1743–1752. [DOI] [PubMed] [Google Scholar]
- (14).Wu Q; Li W; Wang C; Fan P; Cao L; Wu Z; Wang FJ Proteome Res, 2017. 16, 3664–3671. [DOI] [PubMed]
- (15).Li Y; Sabari BR; Panchenko T; Wen H; Zhao D; Guan H; Wan L; Huang H; Tang Z; Zhao Y; et al. ; Mol. Cell 2016, 62, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Xiong X; Panchenko T; Yang S; Zhao S; Yan P; Zhang W; Xie W; Li Y; Zhao Y; Allis CD; Li H Nat. Chem. Biol 2016, 12, 1111–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Andrews FH; Shinsky SA; Shanle EK; Bridgers JB; Gest A; Tsun IK; Krajewski K; Shi X; Strahl BD; Kutateladze TG Nat. Chem. Biol 2016, 12, 396–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Qi YK; Chang HN; Pan KM; Tian CL; Zheng JS Chem. Commun 2015, 51, 14632–14635. [DOI] [PubMed] [Google Scholar]
- (19).Fan YC; Kwon O Chem. Commun 2013, 49, 11588–11619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Stewart IC; Bergman RG; Toste FD J. Am. Chem. Soc, 2003, 125, 8696–8697. [DOI] [PubMed] [Google Scholar]
- (21).Krezel A; Latajka R; Bujacz GD; Bal W Inorg. Chem 2003, 42, 1994–2003. [DOI] [PubMed] [Google Scholar]
- (22).Cline DJ, Redding SE, Brohawn SG, Psathas JN, Schneider JP, Thorpe C Biochem 2004, 43, 15195–15203. [DOI] [PubMed] [Google Scholar]
- (23).Enders D; Saint‐Dizier A; Lannou MI; Lenzen A Eur. J. Org. Chem 2006, 29–49.
- (24).Nguyen UTT; Bittova L; Müller MM; Fierz B; David Y; Houck-Loomis B; Feng V; Dann GP; Muir TW Nat. Methods 2014, 11, 834–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Wingren C Adv. Exp. Med. Biol 2016, 926, 163–179. [DOI] [PubMed] [Google Scholar]
- (26).Clayton AL; Hazzalin CA; Mahadevan LC, Mol. Cell 2006, 23, 289–296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.