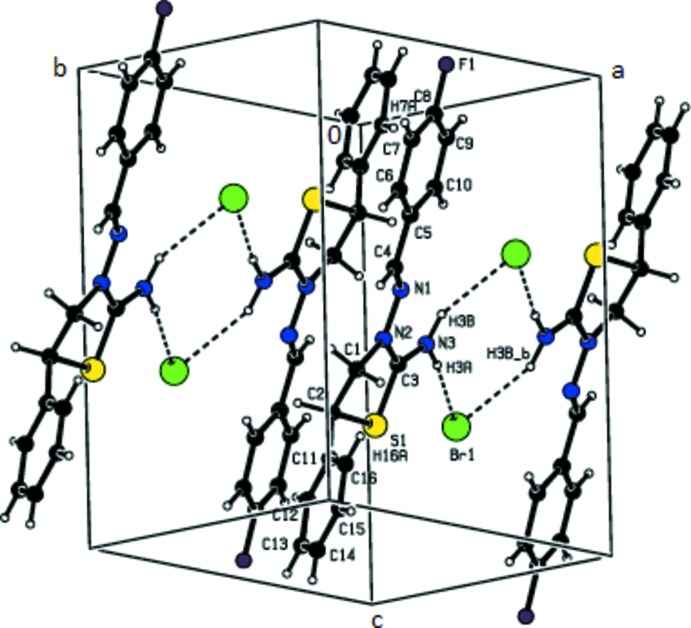
In the crystal of the title salt, cations and anions are linked by N–H⋯Br hydrogen bonds forming inversion-related dimers. The dimers are connected by weak C–H⋯Br hydrogen bonds into chains.
Keywords: crystal structure, charge assisted hydrogen bonding, thiazolidine ring, disorder, Hirshfeld surface analysis.
Abstract
In the cation of the title salt, C16H15FN3S+·Br−, the phenyl ring is disordered over two sets of sites with a refined occupancy ratio of 0.503 (4):0.497 (4). The mean plane of the thiazolidine ring makes dihedral angles of 13.51 (14), 48.6 (3) and 76.5 (3)° with the fluorophenyl ring and the major- and minor-disorder components of the phenyl ring, respectively. The central thiazolidine ring adopts an envelope conformation. In the crystal, centrosymmetrically related cations and anions are linked into dimeric units via N—H⋯Br hydrogen bonds, which are further connected by weak C—H⋯Br hydrogen bonds into chains parallel to [110]. Hirshfeld surface analysis and two-dimensional fingerprint plots indicate that the most important contributions to the crystal packing are from H⋯H (44.3%), Br⋯H/H⋯Br (16.8%), C⋯H/H⋯C (13.9%), F⋯H/H⋯F (10.3%) and S⋯H/H⋯S (3.8%) interactions.
Chemical context
Noncovalent interactions, both intermolecular and intramolecular, occur in virtually every substance and play an important role in the synthesis, catalysis, design of materials and biological processes (Akbari et al., 2017 ▸; Gurbanov et al., 2018 ▸; Kopylovich et al., 2011 ▸; Maharramov et al., 2010 ▸; Mahmoudi et al., 2018a
▸,b
▸,c
▸; Mahmudov et al., 2011 ▸, 2013 ▸, 2014a
▸,b
▸, 2015 ▸, 2017a
▸,b
▸, 2019 ▸; Shixaliyev et al., 2013 ▸, 2018 ▸). On the other hand, Schiff bases and related hydrazone ligands and their complexes have attracted attention over the past decades due to their potential biological, pharmacological and analytical applications (Kopylovich et al., 2011 ▸; Mahmoudi et al., 2018a
▸,b
▸,c
▸; Mahmudov et al., 2013 ▸). Hetercyclic amines are also widely used in the synthesis of Schiff bases, which provide different kinds of noncovalent interactions. As a further study in this field, we report herein the crystal structure and Hirshfeld surface analysis of the title compound.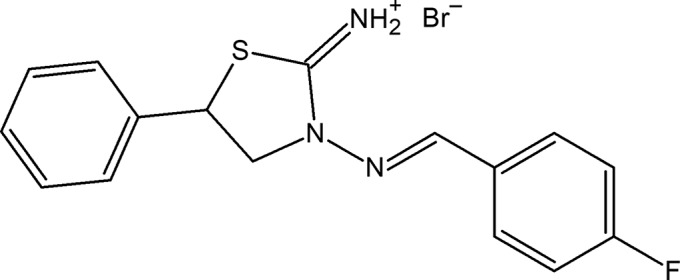
Structural commentary
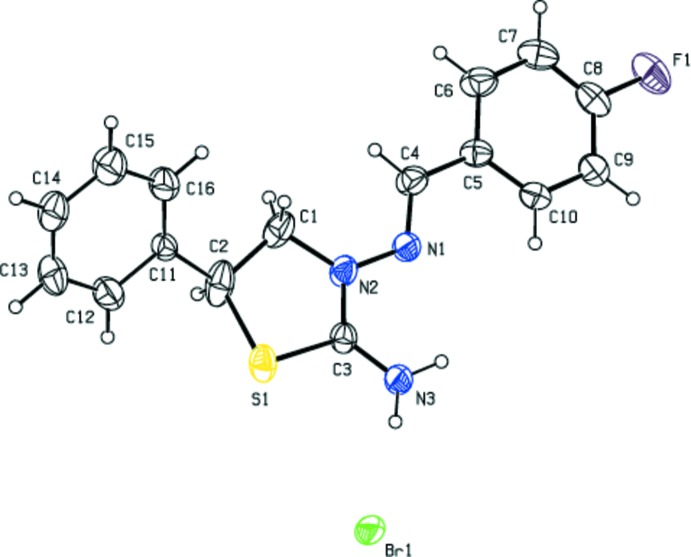
The thiazolidine ring (S1/N2/C1–C3) in the cation of the title salt (Fig. 1 ▸) adopts an envelope conformation, with puckering parameters of Q(2) = 0.321 (3) Å and φ(2) = 43.3 (5)°. The mean plane of the thiazolidine ring makes dihedral angles of 13.51 (14), 48.6 (3) and 76.5 (3)° with the fluorophenyl ring (C5–C10) and the major- and minor-disorder components (C11–C16 and C11′–C16′) of the phenyl ring, respectively. The N2—N1—C4—C5 bridge that links the thiazolidine and 4-fluorophenyl rings has a torsion angle of −177.36 (19)°.
Figure 1.
The molecular structure of the title salt. Displacement ellipsoids are drawn at the 30% probability level. H atoms are shown as spheres of arbitrary radius. The minor-disorder component has been omitted for clarity
Supramolecular features and Hirshfeld surface analysis
In the crystal, centrosymmetrically related cations and anions are linked via pairs of N—H⋯Br hydrogen bonds (Table 1 ▸) into dimeric units forming rings of 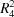 (8) graph-set motif (Fig. 2 ▸). The dimers are further connected by weak C—H⋯Br interactions to form chains running parallel to [110].
(8) graph-set motif (Fig. 2 ▸). The dimers are further connected by weak C—H⋯Br interactions to form chains running parallel to [110].
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N3—H3A⋯Br1 | 0.90 | 2.36 | 3.2557 (18) | 172 |
| N3—H3B⋯Br1i | 0.90 | 2.55 | 3.3552 (18) | 150 |
| C4—H4A⋯Br1ii | 0.93 | 2.99 | 3.726 (2) | 137 |
Symmetry codes: (i) 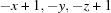 ; (ii)
; (ii) 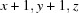 .
.
Figure 2.
A view of the intermolecular N—H⋯Br hydrogen bonds of the title salt in the unit cell. The minor-disorder component has been omitted for clarity
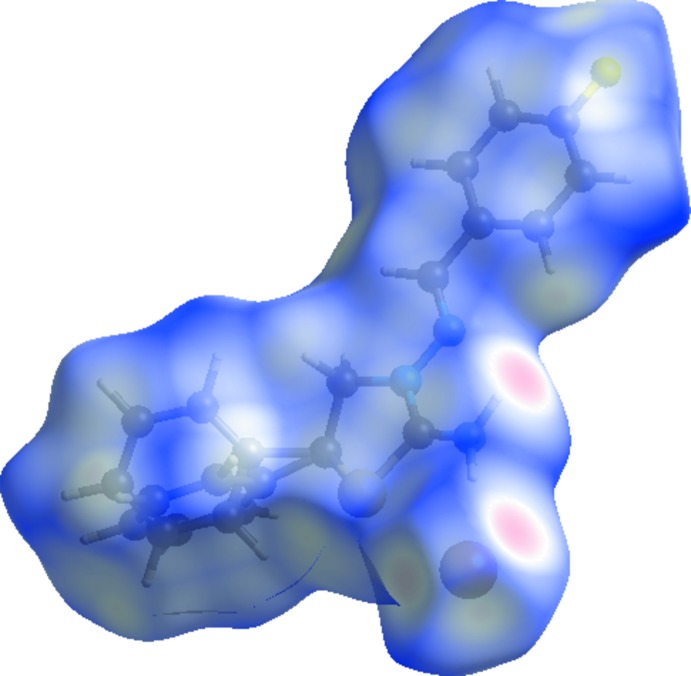
Hirshfeld surface analysis was used to investigate the presence of hydrogen bonds and intermolecular interactions in the crystal structure. The Hirshfeld surface analysis (Spackman & Jayatilaka, 2009 ▸) of the title salt was generated by CrystalExplorer3.1 (Wolff et al., 2012 ▸), and comprised d norm surface plots and 2D fingerprint plots (Spackman & McKinnon, 2002 ▸). The plots of the Hirshfeld surface mapped over d norm using a standard surface resolution with a fixed colour scale of −1.4747 (red) to 1.2166 a.u. (blue) is shown in Fig. 3 ▸. This plot was generated to quantify and visualize the intermolecular interactions and to explain the observed crystal packing.
Figure 3.
Hirshfeld surface of the title salt mapped with d norm.
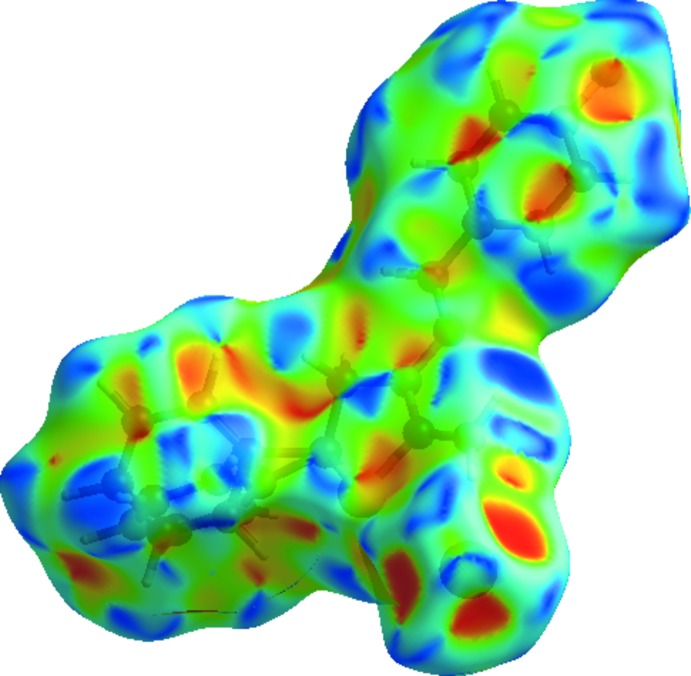
The shape index of the Hirshfeld surface is a tool to visualize π–π stacking interactions by the presence of adjacent red and blue triangles; if there are no adjacent red and/or blue triangles, then there are no π–π interactions. Fig. 4 ▸ clearly suggest that there are no π–π interactions present in the title salt.
Figure 4.
Hirshfeld surface of the title salt mapped with shape index.
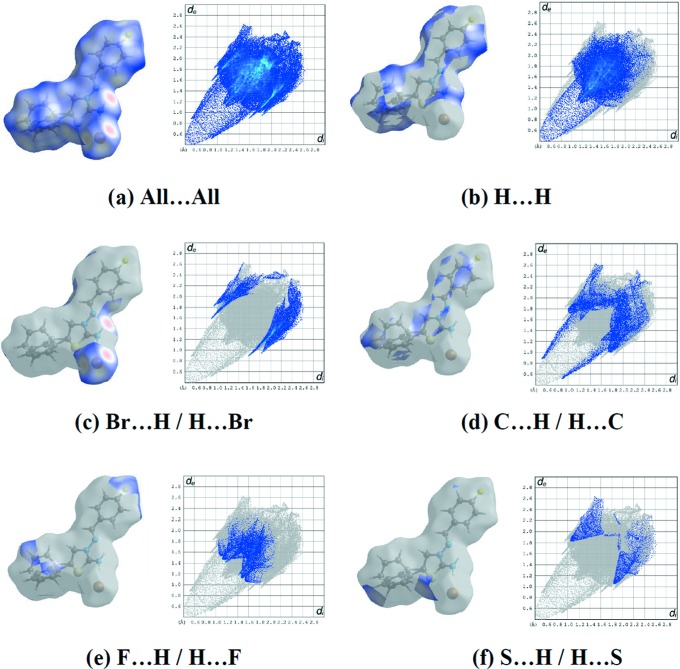
Fig. 5 ▸(a) shows the 2D fingerprint of the sum of the contacts contributing to the Hirshfeld surface represented in normal mode. These represent both the overall 2D fingerprint plots and those that represent H⋯H (44.3%), Br⋯H/H⋯Br (16.8%), C⋯H/H⋯C (13.9%), F⋯H/H⋯F (10.3%) and S⋯H/H⋯S (3.8%) contacts, respectively (Figs. 5 ▸ b–f). The most significant intermolecular interactions are the H⋯H interactions (44.3%) (Fig. 6b). All the contributions to the Hirshfeld surface are given in Table 2 ▸.
Figure 5.
The 2D fingerprint plots of the title salt, showing (a) all interactions, and delineated into (b) H⋯H, (c) Br⋯H/H⋯Br, (d) C⋯H/H⋯C, (e) F⋯H/H⋯F and (f) S⋯H/H⋯S interactions [d e and d i represent the distances from a point on the Hirshfeld surface to the nearest atoms outside (external) and inside (internal) the surface, respectively].
Table 2. Percentage contributions of interatomic contacts to the Hirshfeld surface for the title salt.
| Contact | Percentage contribution |
|---|---|
| H⋯H | 44.3 |
| Br⋯H/H⋯Br | 16.8 |
| C⋯H/H⋯C | 13.9 |
| F⋯H/H⋯F | 10.3 |
| S⋯H/H⋯S | 3.8 |
| N⋯C/C⋯N | 3.6 |
| S⋯C/C⋯S | 2.7 |
| N⋯H/H⋯N | 1.8 |
| C⋯C | 1.5 |
| N⋯N | 0.7 |
| Br⋯C/C⋯Br | 0.3 |
| S⋯N/N⋯S | 0.3 |
| F⋯C/C⋯F | 0.2 |
Database survey
A search of the Cambridge Structural Database (CSD, Version 5.40, update of November 2018; Groom et al., 2016 ▸) for 2-thiazolidiniminium compounds gave seven hits, viz. UDELUN (Akkurt et al., 2018 ▸), WILBIC (Marthi et al., 1994 ▸), WILBOI (Marthi et al., 1994 ▸), WILBOI01 (Marthi et al., 1994 ▸), YITCEJ (Martem’yanova et al., 1993a ▸), YITCAF (Martem’yanova et al., 1993b ▸) and YOPLUK (Marthi et al., 1995 ▸).
In the crystal of UDELUN (Akkurt et al., 2018 ▸), C—H⋯Br and N—H⋯Br hydrogen bonds link the components into a three-dimensional network with the cations and anions stacked along the b-axis direction. Weak C—H⋯π interactions, which only involve the minor-disorder component of the ring, also contribute to the molecular packing. In addition, there are also inversion-related Cl⋯Cl halogen bonds and C—Cl⋯π(ring) contacts.
In the remaining structures, the 3-N atom carries a C-substituent instead of an N-substituent, as found in the title compound. The first three crystal structures were determined for racemic (WILBIC; Marthi et al., 1994 ▸) and two optically active samples (WILBOI and WILBOI01; Marthi et al., 1994 ▸) of 3-(2′-chloro-2′-phenylethyl)-2-thiazolidiniminium p-toluenesulfonate. In all three structures, the most disordered fragment of these molecules is the asymmetric C atom and the Cl atom attached to it. The disorder of the cation in the racemate corresponds to the presence of both enantiomers at each site in the ratio 0.821 (3):0.179 (3). The system of hydrogen bonds connecting two cations and two anions into 12-membered rings is identical in the racemic and in the optically active crystals. YITCEJ (Martem’yanova et al., 1993a ▸) is a product of the interaction of 2-amino-5-methylthiazoline with methyl iodide, with alkylation at the endocylic N atom, while YITCAF (Martem’yanova et al., 1993b ▸) is a product of the reaction of 3-nitro-5-methoxy-, 3-nitro-5-chloro- and 3-bromo-5-nitrosalicylaldehyde with the heterocyclic base to form the salt-like complexes.
Synthesis and crystallization
To a solution of 3-amino-5-phenylthiazolidin-2-iminium bromide (1 mmol) in ethanol (20 ml) was added 4-fluorobenzaldehyde (1 mmol). The mixture was refluxed for 2 h and then cooled. The reaction product precipitated from the reaction mixture as colourless single crystals, was collected by filtration and washed with cold acetone (yield 64%; m.p. 544–545 K). Analysis calculated (%) for C16H15BrFN3S: C 50.53, H 3.98, N 11.05; found: C 50.47, H 3.93, N 11.00. 1H NMR (300 MHz, DMSO-d 6) : 4.56 (k, 1H, CH2, 3 J H-H = 6.6 Hz), 4,87 (t, 1H, CH2, 3 J H-H = 7.8 Hz), 5.60 (t, 1H, CH-Ar, 3J H-H = 7.8 Hz), 7.32–8.16 (m, 9H, 9Ar-H), 8.45 (s, 1H, CH=), 10.37 (s, 2H, NH2). 13C NMR (75 MHz, DMSO-d 6): 45.39, 55.97, 116.05, 127.81, 128.91, 129.13, 129.60, 131.05, 131.17, 137.55, 150.00, 167.89. MS (ESI), m/z: 300.36 [C16H15FN3S]+ and 79.88 Br−.
Refinement details
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. All H atoms were positioned geometrically and refined using a riding model, with N—H = 0.90 Å and C—H = 0.93–0.98 Å, and with U
iso(H) = 1.2U
eq(C,N). The phenyl ring in the cation is disordered over two sets of sites with an occupancy ratio of 0.503 (4):0.497 (4). Seven outliers (001; 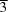 05;
05; 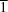 43; 010;
43; 010; 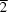 75;
75; 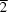 ,
,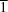 ,12; and 7
,12; and 7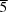 3) were omitted in the final cycles of refinement.
3) were omitted in the final cycles of refinement.
Table 3. Experimental details.
| Crystal data | |
| Chemical formula | C16H15FN3S+·Br− |
| M r | 380.28 |
| Crystal system, space group | Triclinic, P
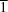
|
| Temperature (K) | 296 |
| a, b, c (Å) | 8.0599 (3), 8.6086 (4), 12.7608 (5) |
| α, β, γ (°) | 96.548 (2), 92.518 (2), 111.065 (2) |
| V (Å3) | 817.39 (6) |
| Z | 2 |
| Radiation type | Mo Kα |
| μ (mm−1) | 2.65 |
| Crystal size (mm) | 0.16 × 0.12 × 0.09 |
| Data collection | |
| Diffractometer | Bruker APEXII CCD |
| Absorption correction | Multi-scan (SADABS; Bruker, 2003 ▸) |
| T min, T max | 0.664, 0.782 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 12009, 3321, 2672 |
| R int | 0.025 |
| (sin θ/λ)max (Å−1) | 0.627 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.029, 0.077, 1.04 |
| No. of reflections | 3321 |
| No. of parameters | 254 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.37, −0.48 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989019004973/rz5254sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989019004973/rz5254Isup2.hkl
CCDC reference: 1909594
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
Ali Khalilov is grateful to Baku State University for the ‘50+50’ individual grant in support of this work.
supplementary crystallographic information
Crystal data
| C16H15FN3S+·Br− | Z = 2 |
| Mr = 380.28 | F(000) = 384 |
| Triclinic, P1 | Dx = 1.545 Mg m−3 |
| a = 8.0599 (3) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 8.6086 (4) Å | Cell parameters from 5655 reflections |
| c = 12.7608 (5) Å | θ = 2.6–26.3° |
| α = 96.548 (2)° | µ = 2.65 mm−1 |
| β = 92.518 (2)° | T = 296 K |
| γ = 111.065 (2)° | Plate, colorless |
| V = 817.39 (6) Å3 | 0.16 × 0.12 × 0.09 mm |
Data collection
| Bruker APEXII CCD diffractometer | 2672 reflections with I > 2σ(I) |
| φ and ω scans | Rint = 0.025 |
| Absorption correction: multi-scan (SADABS; Bruker, 2003) | θmax = 26.5°, θmin = 2.7° |
| Tmin = 0.664, Tmax = 0.782 | h = −10→7 |
| 12009 measured reflections | k = −10→10 |
| 3321 independent reflections | l = −14→15 |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.029 | H-atom parameters constrained |
| wR(F2) = 0.077 | w = 1/[σ2(Fo2) + (0.0376P)2 + 0.2228P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max = 0.001 |
| 3321 reflections | Δρmax = 0.37 e Å−3 |
| 254 parameters | Δρmin = −0.48 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| Br1 | 0.28837 (4) | −0.06846 (3) | 0.65093 (2) | 0.07274 (12) | |
| S1 | 0.50535 (9) | 0.40641 (8) | 0.72511 (4) | 0.06317 (17) | |
| F1 | 1.1652 (2) | 0.6889 (3) | 0.07197 (13) | 0.0994 (6) | |
| N1 | 0.7677 (2) | 0.5288 (2) | 0.48291 (13) | 0.0510 (4) | |
| C1 | 0.7491 (4) | 0.6768 (3) | 0.66453 (19) | 0.0676 (7) | |
| H1A | 0.765957 | 0.780305 | 0.635640 | 0.081* | |
| H1B | 0.859205 | 0.688530 | 0.704392 | 0.081* | |
| N2 | 0.6981 (3) | 0.5342 (2) | 0.57951 (13) | 0.0540 (4) | |
| C2 | 0.5974 (5) | 0.6381 (4) | 0.73431 (19) | 0.0803 (9) | |
| H2A | 0.506784 | 0.680819 | 0.710523 | 0.096* | |
| N3 | 0.5389 (3) | 0.2468 (2) | 0.54077 (14) | 0.0564 (5) | |
| H3A | 0.472588 | 0.153018 | 0.566419 | 0.068* | |
| H3B | 0.598218 | 0.240158 | 0.483319 | 0.068* | |
| C3 | 0.5851 (3) | 0.3881 (3) | 0.60334 (16) | 0.0489 (5) | |
| C4 | 0.8962 (3) | 0.6588 (3) | 0.46468 (18) | 0.0583 (6) | |
| H4A | 0.943400 | 0.751707 | 0.516854 | 0.070* | |
| C5 | 0.9692 (3) | 0.6619 (3) | 0.36204 (18) | 0.0532 (5) | |
| C6 | 1.0980 (3) | 0.8092 (3) | 0.3405 (2) | 0.0691 (7) | |
| H6A | 1.139096 | 0.902914 | 0.392330 | 0.083* | |
| C7 | 1.1660 (4) | 0.8188 (4) | 0.2431 (2) | 0.0767 (8) | |
| H7A | 1.254083 | 0.916970 | 0.229104 | 0.092* | |
| C8 | 1.1011 (3) | 0.6810 (4) | 0.1683 (2) | 0.0665 (7) | |
| C9 | 0.9756 (3) | 0.5314 (3) | 0.1862 (2) | 0.0659 (6) | |
| H9A | 0.935350 | 0.438646 | 0.133610 | 0.079* | |
| C10 | 0.9115 (3) | 0.5229 (3) | 0.28380 (19) | 0.0592 (6) | |
| H10A | 0.827792 | 0.422230 | 0.298028 | 0.071* | |
| C11 | 0.6893 (9) | 0.7289 (7) | 0.8505 (5) | 0.0447 (13) | 0.503 (4) |
| C12 | 0.5575 (7) | 0.6983 (7) | 0.9197 (5) | 0.0691 (15) | 0.503 (4) |
| H12A | 0.439229 | 0.635805 | 0.894732 | 0.083* | 0.503 (4) |
| C13 | 0.6019 (12) | 0.7611 (13) | 1.0269 (6) | 0.074 (2) | 0.503 (4) |
| H13A | 0.513251 | 0.739287 | 1.073481 | 0.089* | 0.503 (4) |
| C14 | 0.7747 (12) | 0.8546 (10) | 1.0639 (5) | 0.0740 (16) | 0.503 (4) |
| H14A | 0.804349 | 0.898834 | 1.135130 | 0.089* | 0.503 (4) |
| C15 | 0.9041 (8) | 0.8822 (8) | 0.9946 (4) | 0.0811 (17) | 0.503 (4) |
| H15A | 1.022734 | 0.943608 | 1.019215 | 0.097* | 0.503 (4) |
| C16 | 0.8600 (7) | 0.8201 (7) | 0.8893 (4) | 0.0660 (14) | 0.503 (4) |
| H16A | 0.949586 | 0.841027 | 0.843322 | 0.079* | 0.503 (4) |
| C11' | 0.5982 (9) | 0.7008 (8) | 0.8494 (5) | 0.0489 (14) | 0.497 (4) |
| C12' | 0.5168 (7) | 0.8137 (6) | 0.8805 (4) | 0.0631 (13) | 0.497 (4) |
| H12B | 0.447047 | 0.841350 | 0.831371 | 0.076* | 0.497 (4) |
| C13' | 0.5408 (8) | 0.8852 (7) | 0.9865 (5) | 0.0759 (18) | 0.497 (4) |
| H13B | 0.488252 | 0.962201 | 1.007970 | 0.091* | 0.497 (4) |
| C14' | 0.6390 (15) | 0.8437 (12) | 1.0570 (7) | 0.079 (3) | 0.497 (4) |
| H14B | 0.655646 | 0.893516 | 1.127240 | 0.094* | 0.497 (4) |
| C15' | 0.7133 (14) | 0.7321 (12) | 1.0286 (5) | 0.0843 (19) | 0.497 (4) |
| H15B | 0.779040 | 0.702274 | 1.078940 | 0.101* | 0.497 (4) |
| C16' | 0.6923 (8) | 0.6598 (8) | 0.9226 (5) | 0.0793 (17) | 0.497 (4) |
| H16B | 0.744737 | 0.582013 | 0.902974 | 0.095* | 0.497 (4) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1 | 0.0842 (2) | 0.05494 (16) | 0.05450 (16) | −0.00131 (12) | 0.02403 (12) | −0.00748 (11) |
| S1 | 0.0911 (4) | 0.0681 (4) | 0.0419 (3) | 0.0401 (3) | 0.0203 (3) | 0.0126 (3) |
| F1 | 0.1003 (12) | 0.1287 (15) | 0.0746 (10) | 0.0375 (11) | 0.0439 (9) | 0.0375 (10) |
| N1 | 0.0588 (10) | 0.0484 (10) | 0.0428 (9) | 0.0154 (9) | 0.0093 (8) | 0.0067 (8) |
| C1 | 0.1015 (19) | 0.0518 (13) | 0.0448 (12) | 0.0257 (13) | −0.0005 (12) | −0.0010 (10) |
| N2 | 0.0706 (12) | 0.0494 (10) | 0.0385 (9) | 0.0183 (9) | 0.0088 (8) | 0.0032 (8) |
| C2 | 0.145 (3) | 0.0759 (18) | 0.0422 (13) | 0.0652 (19) | 0.0157 (15) | 0.0117 (12) |
| N3 | 0.0696 (11) | 0.0478 (10) | 0.0492 (10) | 0.0162 (9) | 0.0199 (9) | 0.0087 (8) |
| C3 | 0.0593 (12) | 0.0513 (12) | 0.0400 (10) | 0.0240 (10) | 0.0079 (9) | 0.0079 (9) |
| C4 | 0.0643 (14) | 0.0516 (13) | 0.0492 (12) | 0.0108 (11) | 0.0016 (10) | 0.0044 (10) |
| C5 | 0.0504 (12) | 0.0514 (12) | 0.0535 (12) | 0.0119 (10) | 0.0028 (10) | 0.0139 (10) |
| C6 | 0.0733 (16) | 0.0558 (14) | 0.0663 (15) | 0.0074 (12) | 0.0053 (13) | 0.0159 (12) |
| C7 | 0.0708 (16) | 0.0692 (17) | 0.0845 (19) | 0.0101 (13) | 0.0177 (14) | 0.0362 (16) |
| C8 | 0.0585 (14) | 0.0891 (19) | 0.0617 (15) | 0.0311 (14) | 0.0196 (12) | 0.0303 (14) |
| C9 | 0.0575 (14) | 0.0732 (16) | 0.0636 (15) | 0.0204 (12) | 0.0141 (11) | 0.0049 (12) |
| C10 | 0.0515 (12) | 0.0560 (13) | 0.0627 (14) | 0.0092 (10) | 0.0148 (11) | 0.0103 (11) |
| C11 | 0.052 (3) | 0.043 (3) | 0.043 (3) | 0.018 (3) | 0.015 (3) | 0.012 (2) |
| C12 | 0.060 (3) | 0.084 (4) | 0.059 (4) | 0.021 (3) | 0.014 (3) | 0.005 (3) |
| C13 | 0.084 (6) | 0.101 (6) | 0.045 (4) | 0.042 (5) | 0.018 (4) | 0.009 (4) |
| C14 | 0.100 (5) | 0.083 (5) | 0.042 (3) | 0.038 (4) | 0.006 (3) | 0.001 (3) |
| C15 | 0.085 (4) | 0.096 (4) | 0.055 (3) | 0.029 (3) | 0.003 (3) | 0.001 (3) |
| C16 | 0.066 (3) | 0.077 (3) | 0.049 (3) | 0.020 (3) | 0.011 (2) | 0.003 (2) |
| C11' | 0.051 (4) | 0.053 (3) | 0.036 (3) | 0.009 (3) | 0.016 (3) | 0.007 (2) |
| C12' | 0.066 (3) | 0.058 (3) | 0.065 (3) | 0.019 (2) | 0.009 (2) | 0.020 (2) |
| C13' | 0.092 (4) | 0.057 (3) | 0.083 (4) | 0.031 (3) | 0.041 (3) | 0.006 (3) |
| C14' | 0.095 (7) | 0.073 (5) | 0.042 (4) | 0.002 (5) | 0.013 (4) | −0.004 (3) |
| C15' | 0.087 (5) | 0.111 (7) | 0.055 (4) | 0.037 (5) | −0.011 (3) | 0.012 (4) |
| C16' | 0.084 (4) | 0.096 (4) | 0.072 (4) | 0.053 (4) | 0.008 (3) | 0.002 (3) |
Geometric parameters (Å, º)
| S1—C3 | 1.720 (2) | C9—C10 | 1.369 (3) |
| S1—C2 | 1.848 (3) | C9—H9A | 0.9300 |
| F1—C8 | 1.353 (3) | C10—H10A | 0.9300 |
| N1—C4 | 1.276 (3) | C11—C16 | 1.353 (8) |
| N1—N2 | 1.380 (2) | C11—C12 | 1.383 (7) |
| C1—N2 | 1.465 (3) | C12—C13 | 1.393 (10) |
| C1—C2 | 1.506 (4) | C12—H12A | 0.9300 |
| C1—H1A | 0.9700 | C13—C14 | 1.364 (14) |
| C1—H1B | 0.9700 | C13—H13A | 0.9300 |
| N2—C3 | 1.339 (3) | C14—C15 | 1.370 (9) |
| C2—C11' | 1.505 (6) | C14—H14A | 0.9300 |
| C2—C11 | 1.607 (7) | C15—C16 | 1.370 (7) |
| C2—H2A | 0.9800 | C15—H15A | 0.9300 |
| N3—C3 | 1.297 (3) | C16—H16A | 0.9300 |
| N3—H3A | 0.9001 | C11'—C16' | 1.333 (9) |
| N3—H3B | 0.9000 | C11'—C12' | 1.389 (8) |
| C4—C5 | 1.458 (3) | C12'—C13' | 1.394 (8) |
| C4—H4A | 0.9300 | C12'—H12B | 0.9300 |
| C5—C6 | 1.386 (3) | C13'—C14' | 1.334 (12) |
| C5—C10 | 1.390 (3) | C13'—H13B | 0.9300 |
| C6—C7 | 1.379 (4) | C14'—C15' | 1.329 (15) |
| C6—H6A | 0.9300 | C14'—H14B | 0.9300 |
| C7—C8 | 1.358 (4) | C15'—C16' | 1.399 (9) |
| C7—H7A | 0.9300 | C15'—H15B | 0.9300 |
| C8—C9 | 1.373 (4) | C16'—H16B | 0.9300 |
| C3—S1—C2 | 90.65 (11) | C10—C9—H9A | 121.0 |
| C4—N1—N2 | 117.98 (19) | C8—C9—H9A | 121.0 |
| N2—C1—C2 | 105.8 (2) | C9—C10—C5 | 121.2 (2) |
| N2—C1—H1A | 110.6 | C9—C10—H10A | 119.4 |
| C2—C1—H1A | 110.6 | C5—C10—H10A | 119.4 |
| N2—C1—H1B | 110.6 | C16—C11—C12 | 118.7 (5) |
| C2—C1—H1B | 110.6 | C16—C11—C2 | 133.2 (5) |
| H1A—C1—H1B | 108.7 | C12—C11—C2 | 108.2 (5) |
| C3—N2—N1 | 116.37 (17) | C11—C12—C13 | 120.0 (6) |
| C3—N2—C1 | 115.67 (18) | C11—C12—H12A | 120.0 |
| N1—N2—C1 | 127.52 (19) | C13—C12—H12A | 120.0 |
| C11'—C2—C1 | 129.4 (4) | C14—C13—C12 | 120.3 (7) |
| C1—C2—C11 | 104.7 (3) | C14—C13—H13A | 119.8 |
| C11'—C2—S1 | 104.9 (3) | C12—C13—H13A | 119.8 |
| C1—C2—S1 | 105.04 (17) | C13—C14—C15 | 119.0 (6) |
| C11—C2—S1 | 112.7 (2) | C13—C14—H14A | 120.5 |
| C1—C2—H2A | 111.4 | C15—C14—H14A | 120.5 |
| C11—C2—H2A | 111.4 | C16—C15—C14 | 120.5 (6) |
| S1—C2—H2A | 111.4 | C16—C15—H15A | 119.7 |
| C3—N3—H3A | 117.3 | C14—C15—H15A | 119.7 |
| C3—N3—H3B | 119.6 | C11—C16—C15 | 121.5 (5) |
| H3A—N3—H3B | 120.5 | C11—C16—H16A | 119.2 |
| N3—C3—N2 | 123.59 (19) | C15—C16—H16A | 119.2 |
| N3—C3—S1 | 123.18 (17) | C16'—C11'—C12' | 118.8 (6) |
| N2—C3—S1 | 113.23 (16) | C16'—C11'—C2 | 119.7 (5) |
| N1—C4—C5 | 120.0 (2) | C12'—C11'—C2 | 121.2 (5) |
| N1—C4—H4A | 120.0 | C11'—C12'—C13' | 119.1 (5) |
| C5—C4—H4A | 120.0 | C11'—C12'—H12B | 120.4 |
| C6—C5—C10 | 118.6 (2) | C13'—C12'—H12B | 120.4 |
| C6—C5—C4 | 119.1 (2) | C14'—C13'—C12' | 120.3 (6) |
| C10—C5—C4 | 122.3 (2) | C14'—C13'—H13B | 119.9 |
| C7—C6—C5 | 120.8 (3) | C12'—C13'—H13B | 119.9 |
| C7—C6—H6A | 119.6 | C15'—C14'—C13' | 121.0 (8) |
| C5—C6—H6A | 119.6 | C15'—C14'—H14B | 119.5 |
| C8—C7—C6 | 118.3 (2) | C13'—C14'—H14B | 119.5 |
| C8—C7—H7A | 120.9 | C14'—C15'—C16' | 119.7 (8) |
| C6—C7—H7A | 120.9 | C14'—C15'—H15B | 120.1 |
| F1—C8—C7 | 119.0 (2) | C16'—C15'—H15B | 120.1 |
| F1—C8—C9 | 117.9 (3) | C11'—C16'—C15' | 121.0 (6) |
| C7—C8—C9 | 123.1 (2) | C11'—C16'—H16B | 119.5 |
| C10—C9—C8 | 118.0 (3) | C15'—C16'—H16B | 119.5 |
| C4—N1—N2—C3 | −169.1 (2) | C6—C5—C10—C9 | −2.0 (4) |
| C4—N1—N2—C1 | 3.0 (3) | C4—C5—C10—C9 | 176.7 (2) |
| C2—C1—N2—C3 | −26.1 (3) | C1—C2—C11—C16 | −1.1 (7) |
| C2—C1—N2—N1 | 161.8 (2) | S1—C2—C11—C16 | 112.5 (6) |
| N2—C1—C2—C11' | 155.8 (4) | C1—C2—C11—C12 | −179.8 (4) |
| N2—C1—C2—C11 | 150.2 (3) | S1—C2—C11—C12 | −66.2 (5) |
| N2—C1—C2—S1 | 31.4 (2) | C16—C11—C12—C13 | 0.1 (10) |
| C3—S1—C2—C11' | −163.6 (3) | C2—C11—C12—C13 | 178.9 (6) |
| C3—S1—C2—C1 | −24.92 (19) | C11—C12—C13—C14 | 0.8 (14) |
| C3—S1—C2—C11 | −138.3 (3) | C12—C13—C14—C15 | −1.5 (15) |
| N1—N2—C3—N3 | −0.7 (3) | C13—C14—C15—C16 | 1.4 (12) |
| C1—N2—C3—N3 | −173.7 (2) | C12—C11—C16—C15 | −0.2 (9) |
| N1—N2—C3—S1 | 179.87 (14) | C2—C11—C16—C15 | −178.7 (5) |
| C1—N2—C3—S1 | 6.8 (3) | C14—C15—C16—C11 | −0.6 (10) |
| C2—S1—C3—N3 | −168.1 (2) | C1—C2—C11'—C16' | −64.5 (7) |
| C2—S1—C3—N2 | 11.35 (19) | S1—C2—C11'—C16' | 59.9 (7) |
| N2—N1—C4—C5 | −177.36 (19) | C1—C2—C11'—C12' | 109.3 (6) |
| N1—C4—C5—C6 | 174.4 (2) | S1—C2—C11'—C12' | −126.3 (5) |
| N1—C4—C5—C10 | −4.2 (4) | C16'—C11'—C12'—C13' | 2.2 (9) |
| C10—C5—C6—C7 | 0.8 (4) | C2—C11'—C12'—C13' | −171.6 (5) |
| C4—C5—C6—C7 | −177.9 (2) | C11'—C12'—C13'—C14' | −0.9 (9) |
| C5—C6—C7—C8 | 1.2 (4) | C12'—C13'—C14'—C15' | −1.0 (13) |
| C6—C7—C8—F1 | 179.3 (2) | C13'—C14'—C15'—C16' | 1.5 (16) |
| C6—C7—C8—C9 | −2.3 (4) | C12'—C11'—C16'—C15' | −1.7 (11) |
| F1—C8—C9—C10 | 179.6 (2) | C2—C11'—C16'—C15' | 172.2 (6) |
| C7—C8—C9—C10 | 1.1 (4) | C14'—C15'—C16'—C11' | −0.2 (14) |
| C8—C9—C10—C5 | 1.0 (4) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N3—H3A···Br1 | 0.90 | 2.36 | 3.2557 (18) | 172 |
| N3—H3B···Br1i | 0.90 | 2.55 | 3.3552 (18) | 150 |
| C4—H4A···Br1ii | 0.93 | 2.99 | 3.726 (2) | 137 |
Symmetry codes: (i) −x+1, −y, −z+1; (ii) x+1, y+1, z.
References
- Akbari Afkhami, F., Mahmoudi, G., Gurbanov, A. V., Zubkov, F. I., Qu, F., Gupta, A. & Safin, D. A. (2017). Dalton Trans. 46, 14888–14896. [DOI] [PubMed]
- Akkurt, M., Duruskari, G. S., Toze, F. A. A., Khalilov, A. N. & Huseynova, A. T. (2018). Acta Cryst. E74, 1168–1172. [DOI] [PMC free article] [PubMed]
- Bruker (2003). SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2007). APEX2 and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Gurbanov, A. V., Maharramov, A. M., Zubkov, F. I., Saifutdinov, A. M. & Guseinov, F. I. (2018). Aust. J. Chem. 71, 190–194.
- Kopylovich, M. N., Mahmudov, K. T., Haukka, M., Luzyanin, K. V. & Pombeiro, A. J. L. (2011). Inorg. Chim. Acta, 374, 175–180.
- Maharramov, A. M., Aliyeva, R. A., Aliyev, I. A., Pashaev, F. G., Gasanov, A. G., Azimova, S. I., Askerov, R. K., Kurbanov, A. V. & Mahmudov, K. T. (2010). Dyes Pigments, 85, 1–6.
- Mahmoudi, G., Seth, S. K., Bauzá, A., Zubkov, F. I., Gurbanov, A. V., White, J., Stilinović, V., Doert, Th. & Frontera, A. (2018a). CrystEngComm, 20, 2812–2821.
- Mahmoudi, G., Zangrando, E., Mitoraj, M. P., Gurbanov, A. V., Zubkov, F. I., Moosavifar, M., Konyaeva, I. A., Kirillov, A. M. & Safin, D. A. (2018b). New J. Chem. 42, 4959–4971.
- Mahmoudi, G., Zaręba, J. K., Gurbanov, A. V., Bauzá, A., Zubkov, F. I., Kubicki, M., Stilinović, V., Kinzhybalo, V. & Frontera, A. (2018c). Eur. J. Inorg. Chem. pp. 4763–4772.
- Mahmudov, K. T., Guedes da Silva, M. F. C., Kopylovich, M. N., Fernandes, A. R., Silva, A., Mizar, A. & Pombeiro, A. J. L. (2014a). J. Organomet. Chem. 760, 67–73.
- Mahmudov, K. T., Guedes da Silva, M. F. C., Sutradhar, M., Kopylovich, M. N., Huseynov, F. E., Shamilov, N. T., Voronina, A. A., Buslaeva, T. M. & Pombeiro, A. J. L. (2015). Dalton Trans. 44, 5602–5610. [DOI] [PubMed]
- Mahmudov, K. T., Gurbanov, A. V., Guseinov, F. I. & Guedes da Silva, M. F. C. (2019). Coord. Chem. Rev. 387, 32–46.
- Mahmudov, K. T., Kopylovich, M. N., Guedes da Silva, M. F. C. & Pombeiro, A. J. L. (2017a). Coord. Chem. Rev. 345, 54–72.
- Mahmudov, K. T., Kopylovich, M. N., Guedes da Silva, M. F. C. & Pombeiro, A. J. L. (2017b). Dalton Trans. 46, 10121–10138. [DOI] [PubMed]
- Mahmudov, K. T., Kopylovich, M. N., Maharramov, A. M., Kurbanova, M. M., Gurbanov, A. V. & Pombeiro, A. J. L. (2014b). Coord. Chem. Rev. 265, 1–37.
- Mahmudov, K. T., Kopylovich, M. N. & Pombeiro, A. J. L. (2013). Coord. Chem. Rev. 257, 1244–1281.
- Mahmudov, K. T., Maharramov, A. M., Aliyeva, R. A., Aliyev, I. A., Askerov, R. K., Batmaz, R., Kopylovich, M. N. & Pombeiro, A. J. L. (2011). J. Photochem. Photobiol. Chem. 219, 159–165.
- Martem’yanova, N. A., Chunaev, Y. M., Przhiyalgovskaya, N. M., Kurkovskaya, L. N., Filipenko, O. S. & Aldoshin, S. M. (1993a). Khim. Geterotsikl. Soedin. pp. 415–419.
- Martem’yanova, N. A., Chunaev, Y. M., Przhiyalgovskaya, N. M., Kurkovskaya, L. N., Filipenko, O. S. & Aldoshin, S. M. (1993b). Khim. Geterotsikl. Soedin. pp. 420–425.
- Marthi, K., Larsen, M., Ács, M., Bálint, J. & Fogassy, E. (1995). Acta Chem. Scand. 49, 20–27.
- Marthi, K., Larsen, S., Ács, M., Bálint, J. & Fogassy, E. (1994). Acta Cryst. B50, 762–771.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Shixaliyev, N. Q., Ahmadova, N. E., Gurbanov, A. V., Maharramov, A. M., Mammadova, G. Z., Nenajdenko, V. G., Zubkov, F. I., Mahmudov, K. T. & Pombeiro, A. J. L. (2018). Dyes Pigments, 150, 377–381.
- Shixaliyev, N. Q., Maharramov, A. M., Gurbanov, A. V., Nenajdenko, V. G., Muzalevskiy, V. M., Mahmudov, K. T. & Kopylovich, M. N. (2013). Catal. Today, 217, 76–79.
- Spackman, M. A. & Jayatilaka, D. (2009). CrystEngComm, 11, 19–32.
- Spackman, M. A. & McKinnon, J. J. (2002). CrystEngComm, 4, 378–392.
- Spek, A. L. (2003). J. Appl. Cryst. 36, 7–13.
- Wolff, S. K., Grimwood, D. J., McKinnon, J. J., Turner, M. J., Jayatilaka, D. & Spackman, M. A. (2012). CrystalExplorer. University of Western Australia.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989019004973/rz5254sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989019004973/rz5254Isup2.hkl
CCDC reference: 1909594
Additional supporting information: crystallographic information; 3D view; checkCIF report